Abstract
Purpose
Pneumocystis jirovecii pneumonia (PJP) shows a high fatality rate in non-HIV patients. However, there are limited data on P. jirovecii drug resistance-related gene mutations in these patients. This study aimed to describe the prevalence of mutations in the dihydrofolate reductase (DHFR) and dihydropteroate synthase (DHPS) genes of P. jirovecii in non-HIV patients in China, providing a reference for drug usage.
Methods
We analyzed the polymorphisms of DHPS and DHFR genes from 45 non-HIV patients in China, including P. jirovecii infection (n = 14) and P. jirovecii colonization (n = 31). This analysis also considered clinical characteristics, P. jirovecii burden, treatment response, and prognosis.
Results
Compared to the P. jirovecii colonization, P. jirovecii infection had significantly altered blood indicators (GR%, LY%, HGB, TP, ALB, CRP, P<0.05) with higher P. jirovecii burden (P<0.05) and worse prognosis (P<0.05). Additionally, patients with P. jirovecii infection were more susceptible to infections, such as the Epstein-Barr virus, Cytomegalovirus, Mycoplasma and Klebsiella pneumoniae. Although no known drug-resistance mutations were detected in the DHPS gene in this study, 10 nonsynonymous mutations were identified. Furthermore, 10 nonsynonymous and 2 synonymous mutations were found in the DHFR gene. However, these mutations were not associated with a worse prognosis.
Conclusion
Our results implied that TMP-SMX prophylaxis is still recommended for PJP in high-risk non-HIV patients in China.
Keywords: Pneumocystis jirovecii pneumonia, mutation, drug resistance, prognosis, non-HIV
Introduction
Pneumocystis jirovecii pneumonia (PJP) is a severe fungal infection of the lungs caused by Pneumocystis jirovecii. It primarily affects individuals with weakened immune systems, and its mortality rate ranges from 21 to 50%.1–5 Over the years, preventive measures and advancements in antiretroviral therapy have significantly reduced the infection and mortality rates of PJP in HIV patients. However, the incidence of PJP has been increasing in non-HIV patients with organ transplants, tumors, rheumatic diseases, autoimmune diseases, as well as those using CD20 antibodies and Bruton’s tyrosine kinase inhibitors.6–8
Prophylactic medication is an effective measure in preventing PJP for individuals with weakened immune systems. Currently, the combination of trimethoprim-sulfamethoxazole (TMP-SMX) is considered as the first-line prophylactic treatment for P. jirovecii infections. TMP-SMX targets two folic acid biosynthesis enzymes, dihydrofolate reductase (DHFR) and dihydropteroate synthase (DHPS) of P. jirovecii. While mutations in DHFR have been associated with TMP resistance in various microbial pathogens, their effect on P. jirovecii is less clear.9,10 Two mutations in DHPS (Thr55Ala and Pro57Ser) are associated with the failure of sulfa prophylaxis and poor treatment outcomes and are therefore considered as TMP-SMX resistance loci.11–13 P. jirovecii can be transmitted through the air, and individuals with normal immune systems can be asymptomatic carriers, contributing to transmission.14,15 As patients treated with TMP-SMX might accumulate mutated strains, they could serve as hosts for the transmission of resistance strains. Therefore, environment factors such as this may also contribute to the selection of mutant P. jirovecii strains.16–18 Thus, by analyzing the drug resistance genes, including DHPS and DHFR, it can effectively guide PJP patient management and treatment decisions.
With the increasing number of immunosuppressed patients, P. jirovecii DHPS and DHFR mutations should be taken into account in order to utilize anti-P. jirovecii medication effectively. However, there is limited data available on the prevalence of these mutations in non-HIV patients. In this study, we analyzed the mutations of DHPS and DHFR in non-HIV patients in China, along with their clinical characteristics, treatment response and outcomes. Our findings indicated that no known drug resistance mutations were detected in these 45 patients (2018–2023), which may offer guidance for the appropriate utilization of TMP-SMX in patients at high risk of P. jirovecii infection.
Methods
Ethics Approval and Consent to Participate
Our study complies with the Declaration of Helsinki and has been approved by the Ethics Committee of Beijing Friendship Hospital, China (approval number 2023-P2-182-01); all clinical samples investigated were anonymized and obtained from an existing sample collection.
Patients and Samples
A total of 45 clinical samples were collected from patients at Beijing Friendship Hospital, Capital Medical University, between August 2018 and June 2023. These included 41 sputum samples and 4 bronchoalveolar lavage fluid (BALF) samples, obtained from the lungs during bronchoscopy procedures. Among these 45 patients, 14 were diagnosed with P. jirovecii infection and were treated with TMP-SMX, 31 patients were diagnosed with P. jirovecii colonization and did not receive any treatment with TMP-SMX. Sputum and BALF specimens were used to detect DHPS and DHFR gene mutations by sequence and assess the P. jirovecii load using qPCR testing.
DNA Extraction
DNA was extracted from fresh sputum or BALF specimens collected from patients. The specimens were pre-treated using 1N NaOH (1:1(v/v) for sputum, and 2:1(v/v) for BALF) 60°C for 1h to ensure complete liquefying before centrifugation at 8000 rpm for 5 min. The pellet was washed twice with saline, and DNA was extracted from the pellet using a kit (TIANGEN, DP705, Beijing, CHN) according to the manufacturer’s instruction and stored at −20°C for further assay.
Amplification of the P. jirovecii DHFR and DHPS Genes from Clinical Samples
The DHPS and DHFR genes were amplified in a 20μL reaction containing 10μL of GoTaq® G2 DNA Master Mix (Promega, M7823, Madison, WI, USA), 500nm primers, and 5–50 ng template DNA. The reaction was carried out in the Bio-Rad ALS1296 PCR system using touchdown procedure. Primers and amplification reaction conditions are listed in Table S1. A positive control and a negative control (distilled water) were used in the experiments.
Detection of Mutations by Sequencing
DNA sequencing was performed by Sangon Biotech Co., Ltd. in both forward and reverse directions to identify the DHPS and DHFR genes. The resulting sequences were assembled using Contig-Express software. To analyze the sequences, they were aligned with reference sequences using the CLUSTAL W Multiple alignment method in Bio-Edit software. Statistical support was assessed through 1000 bootstrap replications. The reference sequences’ GenBank accession numbers are as follows: for the nucleotide sequences, DHPS wild type (AF139132), DHPS Thr55Ala mutation type (U66278), DHPS Pro57Ser mutation type (U66281), and DHFR wild type (AF030968); and for the protein sequences, DHPS wild type (AAF14263), DHPS Thr55Ala and Pro57Ser mutation type (AAD05579), and DHFR wild type (AAF14071).
Quantitative the P. jirovecii Burden Using Real-Time PCR Assay
The fungal burden of 45 patients were analyzed using real-time PCR (Table S2). It was conducted in a 20μL reaction containing 300nm primers (PJ-F and PJ-R) and 200nm PJ-probe (5ʹFAM/3ʹBHQ1) with 10μL GoTaq Probe qPCR Master Mix (Promega, A6101, Madison, WI, USA) plus 1μL template DNA (5–50 ng). The reaction was performed in the Applied Biosystems 7500 Fast Real-Time PCR System (ABI) with 95°C for 2 min followed by 40 cycles of 95°C for 15 sec, 58°C for 50 sec.15 Each sample was tested with replicates, the plasmid mtSSU/pUC19 was used as positive control and reaction without template DNA (distilled water) was used as negative control in experiments.
Statistical Analysis
Statistical analysis was performed using the SPSS software version 22.0 and visualized in GraphPad Prism version 9.0. Continuous variables were presented as P50 (P25, P75) or average (±SD). Categorical variable differences between the groups were compared using chi-square test. The Mann–Whitney U-test or T-test was used to statistically compare between groups. P <0.05 was considered as statistical significance.
Results
Characteristics and Risk Factors of Patients
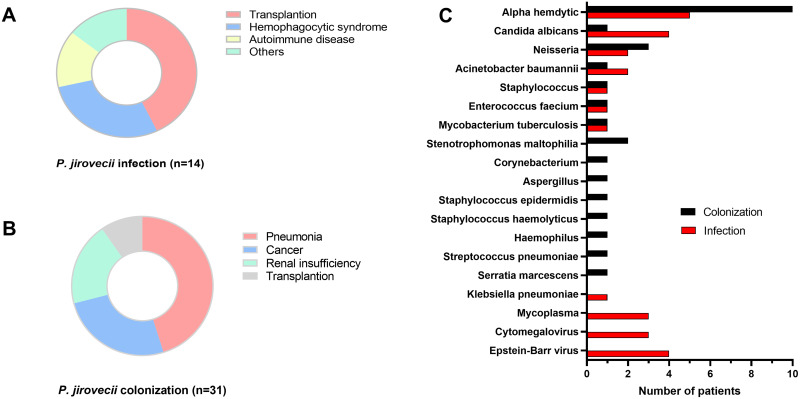
Among the 45 patients in this study, 14 (31.3%) were diagnosed with P. jirovecii infection, while 31(69.7%) exhibited P. jirovecii colonization, as per diagnostic criteria laid out in Table 1. There was no considerable difference in terms of age and gender between these two groups (P > 0.05) (Table 1). Observed symptoms in these patients consisted of respiratory failure (17.8%, 8/45), dyspnea (31.1%, 14/45), cough and/or hemoptysis (28.9%, 13/45), fever, defined as a body temperature exceeding 37.5°C (11.1%, 5/45), and others (11.1%, 5/45). Various underlying conditions were present in the P. jirovecii infection group (n = 14), including transplantation (n = 6), hemophagocytic syndrome (n = 4), autoimmune disease (n = 2) and other conditions (n = 2) (Figure 1A). The P. jirovecii colonization group (n = 31) contained conditions such as pneumonia (n = 14, including 7 newborns), cancer (n = 8), renal insufficiency (n = 6) and transplantation (n = 3) (Figure 1B).
Table 1.
Characteristics of 45 Patients with P. Jirovecii Infection and Colonization
| Groups | Age | Gender Number (%) | GR% | LY% | HGB | PLT | TP | ALB | CRP | PCT | LDH | Quantification of P. jirovecii/Copies | Treatment | Prognosis | Diagnostic Criteria | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Inpatient Days |
Death toll |
|||||||||||||||
| P. jirovecii infection (n=14, all immunosuppressed) | 59.5 (45.8, 68) | Female:1(7.1%) Male:13(92.9%) |
81.8±12.7 | 9.2 (5.7, 18.5) | 86 (73.5, 112.3) | 189 (80.3, 263.5) | 52.7±6.8 | 27.8±5.2 | 49.7 (21.3, 74.5) | 0.3 (0.2, 3.6) | 464 (148.3, 560.8) | 5.6×105 (2.5×104, 1.5×106) | TMP-SMX | 22 (15.8, 24.5) | 2 | ① clinical manifestation, ② imaging characteristics ③ conventional PCR which amplifies P. jirovecii mtLSU gene from sputum/BALF specimens. |
| P. jirovecii colonization (n=31, including 7 immunosuppressed) | 67 (15, 81) | Female:9 (29.0%) Male:22 (71.0%) |
72.1±13.9 | 16.3 (8.8, 27.8) | 122 (84.5, 146) | 220 (137, 270) | 59.4±8.3 | 31.7±5.1 | 3.5(0.6, 25.2) | 0.6 (0.4, 1.6) | 231.5 (193.8, 275.3) | 5.1×104 (1.6×104, 1.6×105) | Not treated according to PJP and treated according the underlying diseases. | 12 (9, 22.5) | 2 | These patients with clinical manifestation and P. jirovecii DNA positive using qPCR methods (Wu et al, 2021), while, the imaging characteristics did not support the diagnosis of PJP. |
| P value | 0.606 | 0.102 | 0.034 | 0.031 | 0.024 | 0.351 | 0.011 | 0.023 | 0.004 | 0.192 | 0.481 | 0.005 | – | 0.005 | 0.578 | – |
Notes: –: no data; Continuous variables were presented as P50 (P25, P75) or average (±SD). We compared between P. jirovecii infection and colonization groups and Bold indicates statistical significance (P<0.05).
Abbreviations: GR%, granulocyte percentage; LY%, lymphocyte percentage; HGB, hemoglobin; PLT, platelet; TP, total protein; ALB, albumin; CRP, C-reactive protein; PCT, procalcitonin; LDH, lactate dehydrogenase.
Figure 1.
The disease spectrum of 45 patients with P. jirovecii infection (A) and colonization (B) along with their co-infections (C).
We then analyzed the clinical characteristics, P. jirovecii burden and prognosis of patients with P. jirovecii infection versus colonization. Some of laboratory data (GR%, LY%, HGB, TP, ALB, CRP) were notably different between P. jirovecii infection and colonization groups (P<0.05). Also, patients in P. jirovecii infection group had a higher P. jirovecii burden and a worse prognosis than colonization group (P < 0.05) (Table 1).
Co-infections among these 45 patients were further analyzed. Except one case of Aspergillus infection in P. jirovecii colonization patients, patients with P. jirovecii infection were more susceptible to microbial infections, such as Epstein–Barr virus (EBV), Cytomegalovirus (CMV), Mycoplasma and Klebsiella pneumoniae (Figure 1C).
Prevalence of DHPS and DHFR Mutations
As the targets of TMP-SMX, DHPS and DHFR are associated with the effectiveness of the treatment.19 We analyzed the amino acid sequences of DHPS from the 45 patients discussed above by comparing them to the wild type DHPS and known mutant sequences from NCBI. No known drug resistance mutations (positions 55 and 57) were detected in the DHPS gene of these 45 patients. However, two mutations (P83L and D90N) were identified in one patient in the P. jirovecii infection group with TMP-SMX treatment. We also found several mutations, such as G51S, G52R, G58S, K76T, P83L, L86S, D90N, D90P, T91I, and V96I, in 8 patients that had P. jirovecii colonization without TMP-SMX treatment (Table 2). A similar analysis was carried out on the DHFR gene. The mutation S106L was found in one patient taking TMP/SMX, and mutations G18W, V45I, M57I, H68Y, A92G, S111P, F121S, A143D, and A143P were found in 5 patients not taking TMP-SMX (Table 2).
Table 2.
Dihydropteroate Synthase (DHPS) and Dihydropteridine Reductase (DHFR) Mutations of P. Jirovecii from 45 Patients
| Groups | Dihydropteroate Synthase (DHPS) | Dihydropteridine Reductase (DHFR) | ||||
|---|---|---|---|---|---|---|
| Nonsynonymous Mutations | Nonsynonymous Mutations | Synonymous Mutations | ||||
| Sites | Number (%) | Sites | Number (%) | Sites | Number (%) | |
| P. jirovecii infection and treatment with TMP-SMX (n=14) | P83L+D90N | 1/14 (7.14%) | S106L* | 1/14 (0%) | T312C* | 5/14(35.71%) |
| C352T | 1/14(7.14%) | |||||
| P. jirovecii colonization and without TMP-SMX prophylaxis (n=31) | G51S+G58S | 1/31 (7.14%) | G18W | 1/31 (7.14%) | T312C* | 12/31(38.71%) |
| G52R | 1/31 (7.14%) | V45I+N57I+A143D | 1/31 (7.14%) | C352T | 3/31(9.68%) | |
| K76T | 1/31 (7.14%) | H68Y | 1/31 (7.14%) | |||
| P83L+D90N | 1/31 (7.14%) | A92G+S111P+A143P | 1/31 (7.14%) | |||
| L86S | 1/31 (7.14%) | F121S | 1/31 (7.14%) | |||
| D90N | 1/31 (7.14%) | |||||
| D90P+T91I | 1/31 (7.14%) | |||||
| V96I* | 1/31 (7.14%) | |||||
Note: *Previously reported site.
We then examined the clinical characteristics, laboratory data and prognosis of these 14 patients that had P. jirovecii with DHPS and DHFR amino acid mutations. These patients (n = 14) exhibited severe underlying conditions, including hemophagocytic syndrome (n = 3), lung cancer (n = 4), acute kidney injury (n = 1), multiple infections (n = 2) and newborns with a lung infection, wet lungs or tetralogy of Fallot (n = 4). The main symptoms observed in these 14 patients were shortness of breath (5/14, 35.7%), bloody sputum (3/14, 21.4%), cough (3/14, 21.4%), fever (2/14, 14.3%) and jaundice (1/14, 7.1%). Additionally, the P. jirovecii burden of mutations and wild types were 7.4 × 104 (4.2 × 104, 2.7 × 105) and 4.1 × 104 (1.4 × 104, 3.4 × 105) copies, respectively, without significant difference (P=0.524). The number of inpatient days for patients with mutant and wild-type strains was 13(8, 22.3) and 19(11.5, 24) days, respectively, which showed no significant difference (P=0.116). These results indicated that the mutations in the P. jirovecii DHPS and DHFR genes showed only a limited relationship to prognosis.
The SNPs Analysis of P. jirovecii DHPS and DHFR Genes
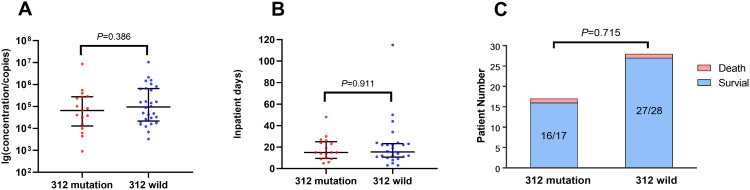
In addition, we analyzed the individual DHPS and DHFR gene sequences of P. jirovecii found in the 45 patients to define sequence profiles for these two genes. Our analysis indicated that no common SNPs were present in the DHPS genes of the 45 patient samples (Figure S1). It is interesting to note that two silent mutations (at positions 312 and 352) were identified in the DHFR genes (Table 2 and Figure S2). The mutation at position 312 was found in 35.7% (5/14) of patients with a P. jirovecii infection and 38.7% (12/31) of those with P. jirovecii colonization, which were independent of TMP-SMX medication. These patients had various underlying diseases, including hemophagocytic syndrome (n = 4), chronic renal allograft dysfunction and nephrotic syndrome (n = 4), cancer (n = 4), neonatal infection (n = 3), and pneumonia (n = 2). The quantitative data obtained from patients with the 312 mutation or wild-type revealed 1.3 × 105(9.1 × 102–5.3 × 107) and 1.7 × 105(3.3 × 103–3.9 × 108) copies, respectively. However, no significant difference was observed between mutation and wild-type (P=0.386) (Figure 2A). The prognosis of patients with isolates carrying either the wild-type gene or the 312-site mutation, including the number of inpatient days and mortality, showed no significant difference (Figure 2B and C). The mutation at position 352 was found in 7.1% (1/14) of patients with P. jirovecii infection and 9.7% (3/31) of those with P. jirovecii colonization (Table 2). Similarly, patients with the mutation 352 did not exhibit either a higher pathogen burden or a poorer prognosis, compared to those infected with the wild-type genotype strain.
Figure 2.
The comparison of P. Jirovecii burden (A), inpatient days (B) and mortality (C) between patients with the 312-site mutation and wild type of dihydrofolate reductase (DHFR) gene.
Discussion
P. jirovecii is a severe fungal pathogen in immunocompromised individuals. Although prophylactic medication is a crucial preventative measure, it poses challenges to the effectiveness of the drugs. In this study, we evaluated the prevalence of mutations of the P. jirovecii DHPS and DHFR genes from 45 non-HIV patients in China. We also explored these mutations related to clinical characteristics, drug prescription and treatment outcomes.
Previous studies have shown geographical diversity in the frequency of DHPS and DHFR mutations in patient samples globally. The prevalence of DHPS mutations is higher in developed countries compared to developing countries, and these mutations are associated with exposure to sulfa drugs and transmission via person-to-person.12,13,17,20 In our study, the DHPS sequences from 45 patients revealed no mutations at positions 55 and 57 (known drug resistance loci in DHPS). This result aligns with previous research on HIV patients with PJP in China, suggesting that the absence of TMP-SMX prophylaxis may be the reason that no drug resistance mutations arise.21 Another mutation of DHPS (V96I) has been reported as a drug resistance site and associated with increased mortality.22 In this study, only one patient with lung cancer in P. jirovecii colonization group carried this mutation and experienced severe episodes of PJP, requiring admission to the intensive care unit (ICU) and mechanical ventilation. This patient was hospitalized for 48 days but ultimately died. Other DHPS mutations were not found to be associated with a poor prognosis. Importantly, the detected mutations P83L and D90N of DHPS were observed in patients regardless of drug prescription. It implied that these mutations may be acquired through person-to-person transmission rather than drug associated selective pressure.
Among the DHFR mutation sites identified in this study (Table 2), the S106L coincides with changes involved in drug resistance of P. falciparum and S. pneumoniae.23 DHFR drug resistance mutations are often linked with specific features located on two adjacent helices (residues K30-S41 and Q172-V181), which impact the interaction with TMP by altering the function of these two alpha helices.24 In our research, these DHFR mutations were not situated on these two alpha helices. At the nucleic acid level, we observed that two silent mutations (positions 312 and 352) were highly prevalent in the DHFR gene sequences of both the drug-treated and untreated group samples. The DHFR silent mutation (312T) has been shown to be associated with PJP cases with moderate-to-high fungal burdens and poor prognosis in HIV patients.21,25,26 However, in our study, these two mutations are not associated with a higher P. jirovecii burden or a more unfavorable prognosis (Table 2 and Figure 2). It is possible that, as for the DHFR 312 mutation of P. jirovecii, HIV-positive individuals may exhibit different clinical manifestations from HIV–negative patients. Our data implied that these two sites maybe be applied as markers for source tracing and conducting evolutionary analysis of P. jirovecii in the future investigations.
Commonly found in patients undergoing immunosuppressive drug treatment, the typical PJP clinical symptoms include fever, dry cough, and progressive shortness of breath. The mortality rate among non-HIV PJP patients remains notably high. Factors implicated in a poor prognosis include age, white blood cell count, existing pulmonary diseases at the time of PJP diagnosis, solid tumors, CMV co-infection, elevated LDH (Lactate dehydrogenase) levels, low lymphocyte count, need for invasive ventilation during hospital stay, pneumomediastinum, and pneumothorax.27,28 Our data showed that patients with P. jirovecii infection exhibited a higher P. jirovecii burden and longer hospital stays than patients with P. jirovecii colonization (Table 1). Additionally, P. jirovecii infection patients were more susceptible to microbial infections such as EBV, CMV, Mycoplasma and Klebsiella pneumoniae (Figure 1C).
Although immunosuppressive drug usage can increase infection risk, it is noteworthy that adjunctive treatment with corticosteroids may possibly lead to more favorable clinical outcomes in non-HIV PJP patients suffering from respiratory failure. However, corticosteroids adjunctive treatment should not be administered to non-HIV PJP patients unless they are dealing with hypoxemia.29 The monitoring of CD4 cell counts (<200/μL) can assist clinicians in evaluating patients who could potentially benefit from TMP-SMX prophylaxis for PJP.30 Although β-(1-3)-D-glucan acts as a critical marker for pneumocystis infection, its specificity is limited.31 Some studies have pointed that β-(1-3)-D-glucan values (>200pg/mL) typically indicate clinically significant P. jirovecii infections in PCR-positive oncology patients, and the median BDG level was 500pg/mL in renal transplantation patients.32 Our results revealed significant differences in GR% (granulocyte ratio), LY% (lymphocyte ratio), HGB (hemoglobin), TP (total protein), ALB (albumin) and CRP (C-reactive protein) between patients with P. jirovecii infection versus those with P. jirovecii colonization. Future investigations would be performed to analyze these indicators, along with the CD4 cell count and β-(1-3)-D-glucan level of more patients to build a prognostic model for PJP patients.
Conclusion
In conclusion, our study shows that the previously reported drug resistance mutations (T55A and P57S) of P. jirovecii DHPS were not detected in any of the 45 clinical samples collected from non- HIV patients in China between 2018 and 2023. Although several mutations and SNPs in DHPS and DHFR were identified here, they did not appear to have a strong association with drug susceptibility and selection pressure. Moreover, these mutations showed only a limited correlation with poor prognosis. Our data implied that prophylactic TMP-SMX is still suitable for individuals at high risk of P. jirovecii infection in China.
Funding Statement
This work was supported by the [National Natural Science Foundation of China] under Grant [No. 82072234] to GY. The founders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Disclosure
The authors declare that there are no conflicts of interest in this article.
References
- 1.Bienvenu AL, Traore K, Plekhanova I, Bouchrik M, Bossard C, Picot S. Pneumocystis pneumonia suspected cases in 604 non-HIV and HIV patients. Int J Infect Dis. 2016;46:11–17. doi: 10.1016/j.ijid.2016.03.018 [DOI] [PubMed] [Google Scholar]
- 2.Cillóniz C, Dominedò C, Álvarez-Martínez MJ, et al. Pneumocystis pneumonia in the twenty-first century: HIV-infected versus HIV-uninfected patients. Expert Rev Anti Infect Ther. 2019;17(10):787–801. doi: 10.1080/14787210.2019.1671823 [DOI] [PubMed] [Google Scholar]
- 3.Su YS, Lu JJ, Perng CL, Chang FY. Pneumocystis jirovecii pneumonia in patients with and without human immunodeficiency virus infection. J Microbiol Immunol Infect. 2017;41(6):478–482. doi: 10.1086/593140 [DOI] [PubMed] [Google Scholar]
- 4.Wickramasekaran RN, Jewell MP, Sorvillo F, Kuo T. The changing trends and profile of pneumocystosis mortality in the United States, 1999–2014. Mycoses. 2017;60(9):607–615. doi: 10.1111/myc.12636 [DOI] [PubMed] [Google Scholar]
- 5.Grønseth S, Rogne T, Hannula R, Åsvold BO, Afset JE, Damås JK. Epidemiological and clinical characteristics of immunocompromised patients infected with Pneumocystis jirovecii in a twelve-year retrospective study from Norway. BMC Infect Dis. 2021;21(1):659. doi: 10.1186/s12879-021-06144-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang L, Fu Q, Ye Y, Lin Y, Yan Q, Chen S. High incidence and mortality of Pneumocystis jirovecii infection in anti-MDA5-antibody-positive dermatomyositis: experience from a single center. Arthritis Res Therapy. 2021;23(1):232. doi: 10.1186/s13075-021-02606-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Monnet X, Vidal-Petiot E, Osman D, et al. Critical care management and outcome of severe Pneumocystis pneumonia in patients with and without HIV infection. Crit Care. 2008;12(1):R28. doi: 10.1186/cc6806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xue T, Kong X, Ma L. Trends in the epidemiology of pneumocystis pneumonia in immunocompromised patients without HIV infection. J Fungi. 2023;9(8):812. doi: 10.3390/jof9080812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dale GE, Broger C, D’Arcy A, et al. A single amino acid substitution in staphylococcus aureus dihydrofolate reductase determines trimethoprim resistance. J Mol Biol. 1997;266(1):23–30. doi: 10.1006/jmbi.1996.0770 [DOI] [PubMed] [Google Scholar]
- 10.da Silva C, Boene S, Datta D, et al. Targeted and whole-genome sequencing reveal a north-south divide in P. falciparum drug resistance markers and genetic structure in Mozambique. Commun Biol. 2023;6(1):619. doi: 10.1038/s42003-023-04997-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang L, Crothers K, Atzori C, et al. Dihydropteroate synthase gene mutations in Pneumocystis and sulfa resistance. Emerg Infect Dis. 2004;10(10):1721–1728. doi: 10.3201/eid1010.030994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kazanjian P, Armstrong W, Hossler PA, et al. Pneumocystis carinii mutations are associated with duration of sulfa or sulfone prophylaxis exposure in AIDS patients. J Infect Dis. 2000;182(2):551–557. doi: 10.1086/315719 [DOI] [PubMed] [Google Scholar]
- 13.Ponce CA, Chabé M, George C, et al. High prevalence of Pneumocystis jirovecii Dihydropteroate synthase gene mutations in patients with a first episode of pneumocystis pneumonia in Santiago, Chile, and clinical response to trimethoprim-sulfamethoxazole therapy. Antimicrob Agents Chemother. 2017;61(2):e01290–01216. doi: 10.1128/aac.01290-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cissé OH, Ma L, Jiang C, Snyder M, Kovacs JA. Humans are selectively exposed to Pneumocystis jirovecii. mBio. 2020;11(2):e03138–03119. doi: 10.1128/mBio.03138-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu Y, Wang F, Wang C, et al. Detection of Pneumocystis jirovecii and Toxoplasma gondii in patients with lung infections by a duplex qPCR assay. PLoS Negl Trop Dis. 2021;15(12):e0010025. doi: 10.1371/journal.pntd.0010025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rabodonirina M, Vanhems P, Couray-Targe S, et al. Molecular evidence of interhuman transmission of Pneumocystis pneumonia among renal transplant recipients hospitalized with HIV-infected patients. Emerg Infect Dis. 2004;10(10):1766–1773. doi: 10.3201/eid1010.040453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takahashi T, Hosoya N, Endo T, et al. Relationship between mutations in dihydropteroate synthase of Pneumocystis carinii f. sp. hominis isolates in Japan and resistance to sulfonamide therapy. J Clin Microbiol. 2000;38(9):3161–3164. doi: 10.1128/JCM.38.9.3161-3164.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hauser PM, Nahimana A, Taffe P, et al. Interhuman transmission as a potential key parameter for geographical variation in the prevalence of Pneumocystis jirovecii dihydropteroate synthase mutations. Clinl Infect Dis. 2010;51(4):e28–e33. doi: 10.1086/655145 [DOI] [PubMed] [Google Scholar]
- 19.Baker FN, Cushion MT, Porollo A. A quantitative model to estimate drug resistance in pathogens. J Fungi. 2016;2(4). doi: 10.3390/jof2040030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de la Horra C, Friaza V, Morilla R, et al. Update on Dihydropteroate Synthase (DHPS) mutations in pneumocystis jirovecii. J Fungi. 2021;7(10):856. doi: 10.3390/jof7100856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang M, Xu X, Guo Y, et al. Polymorphisms involving the Pneumocystis jirovecii-related genes in AIDS patients in eastern China. Infect Genet Evol. 2019;75:103955. doi: 10.1016/j.meegid.2019.103955 [DOI] [PubMed] [Google Scholar]
- 22.Singh Y, Mirdha BR, Guleria R, et al. Novel dihydropteroate synthase gene mutation in Pneumocystis jirovecii among HIV-infected patients in India: putative association with drug resistance and mortality. J Global Antimicrob Resist. 2019;17:236–239. doi: 10.1016/j.jgar.2019.01.007 [DOI] [PubMed] [Google Scholar]
- 23.Nahimana A, Rabodonirina M, Bille J, Francioli P, Hauser PM. Mutations of Pneumocystis jirovecii Dihydrofolate Reductase Associated with Failure of Prophylaxis. Antimicrob Agents Chemother. 2004;48(11):4301–4305. doi: 10.1128/aac.48.11.4301-4305.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leidner F, Kurt Yilmaz N, Schiffer CA. Deciphering antifungal drug resistance in Pneumocystis jirovecii DHFR with molecular dynamics and machine learning. J Chem Inf Model. 2021;61(6):2537–2541. doi: 10.1021/acs.jcim.1c00403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Esteves F, de Sousa B, Calderon EJ, et al. Multicentre study highlighting clinical relevance of new high-throughput methodologies in molecular epidemiology of Pneumocystis jirovecii pneumonia. Clin Microbiol Infect. 2016;22(6):566e569–566e519. doi: 10.1016/j.cmi.2016.03.013 [DOI] [PubMed] [Google Scholar]
- 26.Esteves F, Gaspar J, Marques T, et al. Identification of relevant single-nucleotide polymorphisms in Pneumocystis jirovecii: relationship with clinical data. Clin Microbiol Infect. 2010;16(7):878–884. doi: 10.1111/j.1469-0691.2009.03030.x [DOI] [PubMed] [Google Scholar]
- 27.Wang Y, Zhou X, Saimi M, et al. Risk factors of mortality from Pneumocystis Pneumonia in Non-HIV patients: a meta-analysis. Front Public Health. 2021;9:680108. doi: 10.3389/fpubh.2021.680108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iriart X, Challan Belval T, Fillaux J, et al. Risk factors of Pneumocystis pneumonia in solid organ recipients in the era of the common use of posttransplantation prophylaxis. Am J Transplant. 2015;15(1):190–199. doi: 10.1111/ajt.12947 [DOI] [PubMed] [Google Scholar]
- 29.Ding L, Huang H, Wang H, He H. Adjunctive corticosteroids may be associated with better outcome for non-HIV Pneumocystis pneumonia with respiratory failure: a systemic review and meta-analysis of observational studies. Ann Intens Care. 2020;10(1):34. doi: 10.1186/s13613-020-00649-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Messiaen PE, Cuyx S, Dejagere T, van der Hilst JC. The role of CD4 cell count as discriminatory measure to guide chemoprophylaxis against Pneumocystis jirovecii pneumonia in human immunodeficiency virus-negative immunocompromised patients: a systematic review. Transpl Infect Dis. 2017;19(2). doi: 10.1111/tid.12651 [DOI] [PubMed] [Google Scholar]
- 31.Mercier T, Aissaoui N, Gits-Muselli M, et al. Variable correlation between bronchoalveolar lavage fluid fungal load and serum-(1,3)-beta-d-glucan in patients with pneumocystosis-A multicenter ECMM excellence center study. J Fungi. 2020;6(4):327. doi: 10.3390/jof6040327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morjaria S, Frame J, Franco-Garcia A, Geyer A, Kamboj M, Babady NE. Clinical performance of (1,3) Beta-D glucan for the diagnosis of pneumocystis pneumonia (PCP) in cancer patients tested with PCP polymerase chain reaction. Clinl Infect Dis. 2019;69(8):1303–1309. doi: 10.1093/cid/ciy1072 [DOI] [PMC free article] [PubMed] [Google Scholar]