Abstract
Background
Deep sowing has emerged as a vital agricultural strategy, particularly in arid and semi-arid regions, as it allows seeds to access water stored in deeper soil layers. This approach facilitates successful germination and establishment of crops, even in challenging environmental conditions. Previous studies have shown that the length of the maize mesocotyl is an important trait influencing deep-sowing tolerance. Several factors play a crucial role in regulating mesocotyl elongation, primarily including light, hormones, metabolites, and reactive oxygen species (ROS). Therefore, further understanding the regulatory mechanisms of mesocotyl elongation is essential for enhancing maize germination and growth under deep sowing conditions.
Results
In this study, we identified a deep sowing-tolerant inbred line, DH65232, which showed significantly increased mesocotyl length compared to B73 under deep sowing conditions. Transcriptome analysis revealed that differentially expressed genes in the mesocotyl of the two inbred lines were mainly enriched in three pathways: hormone regulation, intermediate metabolites, and redox enzymes. Measurements of hormone content and phenotypic analysis following GA3 treatment indicated that GA3 plays a positive role in promoting mesocotyl elongation under deep-sowing stress in the inbred line DH65232. Additionally, untargeted metabolomics revealed that DH65232 exhibited a higher number of differential metabolites related to antioxidant pathway under deep-sowing stress compared to normal sowing. In deep sowing conditions, the determination of POD, CAT, SOD activities, and MDA content in the mesocotyl of B73 and DH65232 shows that DH65232 has a stronger ability to scavenge ROS.
Conclusions
Above all, the inbred line DH65232 exhibits a greater tolerance to deep sowing due to its stronger antioxidant activity. Our study has contributed to a deeper understanding of the complex tolerance mechanisms in maize and provided new insights for the development of new maize varieties under deep sowing conditions.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12870-024-05994-6.
Keywords: Maize, Deep sowing, Mesocotyl, Hormone, Metabolism, ROS, Antioxidant activity
Background
Maize is one of the three most important food crops globally, along with rice and wheat [1, 2]. It plays a crucial role in food security and is cultivated across a vast range of environments due to its adaptability and high yield potential [2]. Maize is not only a staple food for human consumption but also serves as a critical source of livestock feed, industrial raw materials, and biofuel. Given its significance, ensuring its productivity is vital, especially in regions facing challenging growing conditions. A substantial portion of the global maize-growing regions are located in arid and semi-arid climates [3, 4], where drought conditions frequently occur. These regions are characterized by low and erratic rainfall, which imposes severe stress on maize during both germination and subsequent growth stages [5–7]. Drought stress, especially during early growth, can lead to poor seedling establishment, stunted growth, and ultimately reduced yields [8, 9].
In most common maize varieties, the optimal planting depth is between 3 and 5 cm, where moisture is usually more readily available [1, 10, 11]. However, in arid and semi-arid regions, the topsoil often lacks sufficient water, especially after long dry spells or during seasonal droughts [12]. This moisture deficit in the surface soil creates a major barrier to successful maize germination [13]. Seeds may fail to absorb enough water to initiate the germination process, leading to poor crop establishment and subsequent yield losses [12]. As is known to all, deep sowing (≥ 20 cm) is beneficial for maize germination and growth in water-scarce regions [10, 14, 15], however, this planting method imposes great pressure on maize seedlings to break through the soil and grow normally during germination. Therefore, understanding and improving maize tolerance to deep-sowing stress is of great importance, particularly for regions where water scarcity is a persistent issue. Advances in this research area could lead to the development of maize varieties with enhanced deep-sowing tolerance, ensuring better seedling emergence, and higher yields in drought-prone environments.
Different species have evolved distinct mechanisms to push the seedlings through the soil when crops experience deep-sowing stress. In maize, sorghum, and rice, the mesocotyl elongates to adapt to deep sowing conditions [14, 16–19], while in wheat and barley, both the mesocotyl and the first internode contribute to this process [20–23]. In maize, the mesocotyl is a critical organ that connects the base of the radicle to the coleoptile node [2, 24, 25].
In recent years, QTLs associated with mesocotyl length have been discovered in various crops, including rice and maize, providing valuable references for studying mesocotyl development [26–31]. Additionally, several functional genes have been identified as being closely related to mesocotyl elongation. For example, in maize, ZmSRO1e promotes mesocotyl elongation and enhances tolerance to deep sowing by inhibiting the activity of ZmbZIP61 [2]. In rice, overexpression of the OsEXP4 has been shown to promote mesocotyl elongation by regulating cell wall loosening [32]. OsPAO5, has also been identified in rice, where it regulates mesocotyl length by oxidizing polyamines to release hydrogen peroxide, which leads to reduced ethylene synthesis and subsequently shortens the mesocotyl [33]. These discoveries highlight the complexity of mesocotyl elongation and its regulation by multiple genetic pathways across different crop species. Understanding these mechanisms can provide new insights for breeding crop varieties with enhanced tolerance to deep sowing conditions, which is especially valuable in drought-prone regions where deep sowing is required for successful germination and growth.
The elongation of the mesocotyl is regulated by both light and hormones [34, 35]. Previous studies have found that red light [36] and phytochromes [37] are key factors influencing mesocotyl elongation. Hormones play an important role in helping plants resist abiotic stress [38, 39]. Among the various hormones involved in regulating mesocotyl elongation are auxin [1, 40–42], gibberellin (GA) [19, 43, 44], abscisic acid (ABA) [45], ethylene [33, 46], cytokinin (CK) [47], jasmonic acid (JA) [46], strigolactones [48], and brassinosteroids [41, 48]. In sweet corn, when sown at a depth of 10 cm, the endogenous GA content in the mesocotyl significantly increases, and exogenous GA application promotes mesocotyl elongation [43]. In maize, the ZmMYB59 negatively regulates mesocotyl elongation through the gibberellin signaling pathway [19]. Auxin can promote mesocotyl elongation, while cycloheximide in maize inhibits mesocotyl elongation by reducing auxin levels [41]. Under low water potential stress, the accumulation of ABA in maize helps maintain root elongation but inhibits mesocotyl elongation [45].
Drought stress triggers a range of physiological responses in plants, one of the most significant being the increased production of ROS [49–51]. The production of ROS is tightly linked to the activity of key antioxidant enzymes, including peroxidase (POD), catalase (CAT), and superoxide dismutase (SOD), which help mitigate oxidative damage by neutralizing excess ROS [52]. In maize, the ZmNAC17 positively regulates the elongation of the mesocotyl by mediating the auxin and ROS synthesis pathways [53]. In rice, the polyamine oxidase encoded by the OsPAO5 can oxidize polyamines to release hydrogen peroxide, leading to reduced ethylene synthesis and shorter mesocotyls [33]. In maize, POD also plays a crucial role in lignin biosynthesis, a process that significantly impacts cell wall composition and rigidity, which may ultimately influence mesocotyl elongation [54]. Despite this understanding above, the precise interaction between hormonal regulation and oxidative stress tolerance for deep-sowing tolerance in maize remains poorly understood.
In this study, we report the identification and characterization of a high-quality deep-sowing tolerant maize inbred line, DH65232. This line demonstrates a unique combination of hormonal balance and enhanced ROS scavenging capacity, which together contribute to its superior mesocotyl elongation. Specifically, DH65232 exhibits hormone levels that are more conducive to promoting elongation, possibly through optimized GA signaling, along with a more efficient antioxidant system that reduces the damaging effects of ROS. This dual advantage allows seedlings to emerge from deeper soil layers, overcoming the mechanical resistance imposed by the surrounding soil. Our findings provide valuable insights into the mechanisms underlying deep-sowing tolerance in maize and offer novel perspectives for the improvement of drought-resistant varieties.
Results
The maize inbred line DH65232 exhibits Tolerance to Deep Sowing conditions
We identified a maize inbred line, DH65232, exhibiting strong deep-sowing tolerance through screening a maize Genome-Wide Association Study (GWAS) population. To further validate the deep-sowing tolerance phenotype of DH65232, we conducted a comparative experiment using DH65232 alongside the well-known inbred line B73, which was used as a control. Both inbred lines were sown under two distinct planting conditions: a normal sowing depth of 5 cm and a challenging deep-sowing depth of 30 cm. These depths were chosen to simulate standard and extreme planting environments, respectively. After 10 days of growth, we collected and measured various seedling phenotypic traits, including mesocotyl length, total seedling length, coleoptile length, plumule length, root length, and fresh weight. These traits are critical indicators of the ability to emerge and establish itself under deep sowing conditions, where the seedlings must elongate their mesocotyl to push through the deeper soil layers to reach the surface.
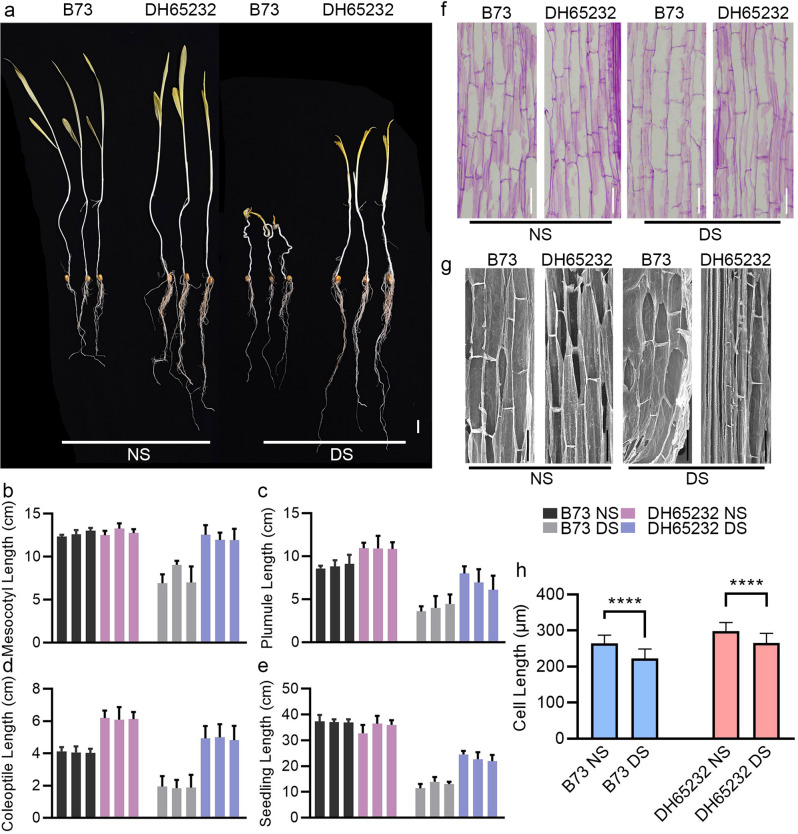
The statistical analysis of the collected data revealed significant differences between the two inbred lines under deep sowing conditions. Specifically, DH65232 showed superior mesocotyl elongation and overall seedling vigor compared to B73, which struggled to emerge from the 30 cm depth. These results confirm that DH65232 possesses an enhanced capacity for deep-sowing tolerance, making it a promising candidate for further breeding programs aimed at improving maize performance in challenging agricultural environments (Fig. 1A). When the planting depth shifted from normal sowing to deep sowing, the mesocotyl length of DH65232 remained stable, while B73 decreased by approximately 28.3% (Fig. 1B). Under deep sowing conditions, the plumule length of DH65232 was reduced by 36%, whereas the plumule length of B73 decreased by 58% (Fig. 1C). In line with these observations, we also found that the coleoptile length and seedling length of DH65232 under deep sowing conditions were reduced by 20% and 25%, respectively. In contrast, B73 showed more pronounced reductions, with a 55% decrease in coleoptile length and a 70% reduction in seedling length (Fig. 1D, E). In addition, we examined the root length and fresh weight of both DH65232 and B73 under normal and deep sowing conditions. The results showed no significant changes in root length between the normal and deep sowing conditions for either DH65232 or B73 (Additional file 1: Fig. S1A). However, under deep-sowing, the fresh weight of B73 decreased by approximately 11.9%, while DH65232 showed no significant difference (Additional file 1: Fig. S1B).
Fig. 1.
Seedling morphology and cell length of inbred lines B73 and DH65232. A Morphology of B73 and DH65232 under both treatments, Bar = 2 cm. B-E Mesocotyl length, plumule length, coleoptile length, and seedling length under different depth conditions. F and G are longitudinal structures of mesocotyl cells of B73 and DH65232 under NS and DS conditions, respectively, with observed by paraffin section and by scanning electron microscopy. Bar = 100 μm. H Cell length of mesocotyl. Asterisks indicate significant differences (****, P < 0.0001; t-Test)
Mesocotyl elongation is considered one of the primary factors that enable maize seedlings to adapt to deep sowing conditions [14, 16, 17]. To gain further insight into the cellular basis underlying the deep-sowing tolerance of DH65232, we conducted histological sectioning and scanning electron microscopy (SEM) to observe the cell length of the mesocotyl in both DH65232 and B73 (Fig. 1F, G). The histological analysis revealed that under deep sowing conditions, the mesocotyl cell length in B73 decreased by approximately 15.9%, while the reduction in DH65232 was only 10.6% (Fig. 1H). This suggests that DH65232 maintains better cellular integrity in response to deep-sowing stress compared to B73. Additionally, our statistical analysis showed that under normal sowing conditions, the mesocotyl cell length of DH65232 was 12.9% longer than that of B73. Notably, this difference was even more pronounced under deep sowing conditions, where the mesocotyl cell length of DH65232 was 19.3% longer than that of B73 (Fig. 1H).
These findings provide compelling evidence that the deep-sowing tolerance of DH65232 is associated with its ability to maintain greater mesocotyl elongation and less reduction in cell length under deep-sowing stress, contributing to its enhanced adaptability.
Transcriptomic analysis of DH65232 and B73 under normal and deep sowing conditions
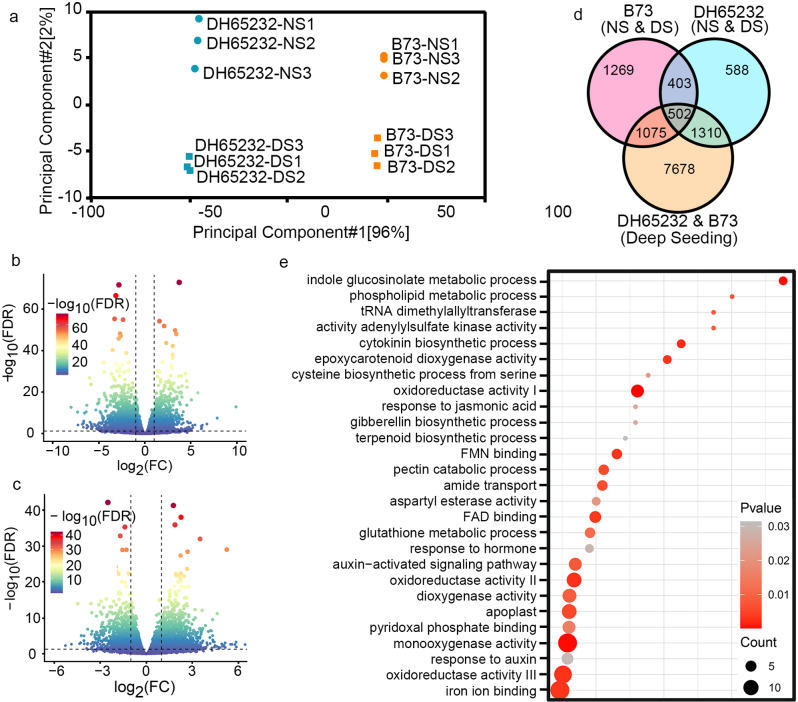
In order to investigate the differentially expressed genes that contribute to the longer mesocotyl in DH65232 under deep sowing conditions, we performed transcriptome sequencing on the mesocotyl organs of DH65232 and B73 under normal and deep sowing conditions. The transcriptome sequencing included four sets of samples: normal sowing B73 (B73-NS), normal sowing DH65232 (DH65232-NS), deep sowing B73 (B73-DS), and deep sowing DH65232 (DH65232-DS), with each set containing three biological replicates. Principal component analysis of the transcriptome data showed that the four sets of samples were in different quadrants, and there was good aggregation among the data within each group (Fig. 2A). A total of 46,083 genes were detected in the transcriptome analysis. Between normal and deep sowing conditions for B73, a total of 32,262 differentially expressed genes (DEGs) were identified, of which 3,249 exhibited significant differences. Among these, 1,303 genes were significantly upregulated, while 1,946 genes were downregulated (Fig. 2B). Between normal and deep sowing DH65232, there were 34,348 DEGs, with 2,803 genes showing significant differences. Among these, 1,813 genes were significantly upregulated, and 990 genes were downregulated (Fig. 2C). There were 905 shared genes among the significantly differentially expressed genes between different sowing depths in B73 and DH65232 (Fig. 2D), of which 502 genes exhibited significant differential expression between deep sowing B73 and DH65232.
Fig. 2.
Transcriptomic analysis under different treatment conditions. A Principal Component Analysis (PCA) of B73 and DH65232 under NS and DS treatments. 1, 2, and 3 represent three biological replicates. B Volcano plot of upregulated and downregulated DEGs in the B73 inbred line under different sowing conditions. C Volcano plot of up-regulated and down-regulated DEGs in DH65232 under different sowing conditions. D Venn diagram of DEGs in B73 under different sowing depth treatments, DEGs in DH65232 under different depth treatments, and DEGs between B73 and DH65232 under deep sowing conditions. E GO enrichment analysis of the 502 overlapping DEGs in the Venn diagram
Subsequently, we conducted Gene Ontology (GO) analysis on these 502 DEGs (Fig. 2E; Additional file 1: Fig. S2). The results revealed that they were primarily enriched in six categories: redox enzymes, hormone regulation, intermediates of metabolism, protein modifications, glycosyltransferases, and ion transmembrane transport. Among these, hormone regulation exhibited the most extensive enrichment, involving the synthesis, metabolism, and signal transduction of hormones, such as auxins, cytokinins, gibberellins, abscisic acid, jasmonic acid, and ethylene. In “redox enzymes”, it included peroxidases, monooxygenases, dioxygenases, coenzymes involved in oxidative defense, and the synthesis of terpenoid compounds that can participate in the antioxidant processes. Additionally, there was significant enrichment in the category of intermediates of metabolism, which encompassed the synthesis and metabolism of carbohydrates, peptides, amino acids, and lipids. These results suggest that DH65232 may adapt to deep sowing through three pathways: hormone regulation, redox enzymes, and the accumulation of intermediates of metabolism.
The qRT-PCR results revealed notable changes in gene expression when comparing the DH65232 inbred line to the B73 inbred line. Specifically, genes associated with hormone regulation displayed significant differences in expression. For example, Zm00001d007255 was downregulated in DH65232, while Zm00001d014617, Zm00001d038695, and Zm00001d045026 were upregulated (Additional file 1: Fig. S3A). These findings are consistent with the RNA-seq data and further confirm the involvement of these genes in the regulation of plant hormones.In terms of genes involved in intermediate metabolite accumulation, we observed upregulation of Zm00001d03446 and Zm00001d022460 in DH65232, while Zm00001d045340 was downregulated (Additional file 1: Fig. S3B). These expression patterns align closely with the RNA-seq results and suggest that these genes are involved in the regulation of metabolic processes in maize, potentially influencing key pathways such as nutrient utilization and energy production.Additionally, genes related to redox regulation showed distinct expression changes. For instance, Zm00001d03584 was downregulated in DH65232, whereas Zm00001d016842, Zm00001d024234, and Zm00001d039533 were upregulated (Additional file 1: Fig. S3C). This further supports the RNA-seq findings and highlights the role of redox enzymes in regulating stress responses and cellular homeostasis in maize. Taken together, the qRT-PCR results are in full agreement with the trends observed in the RNA-seq data, further confirming the accuracy and reliability of our RNA-seq findings.
Alterations in multiple hormones in DH65232 under deep sowing conditions
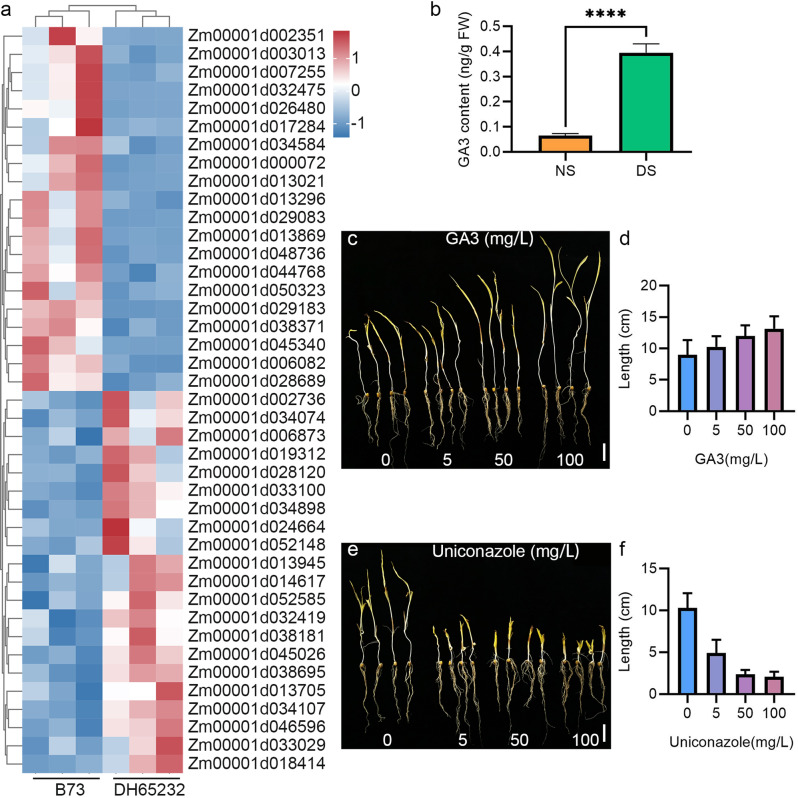
Previous studies have demonstrated that various hormones, including GA, ABA, and IAA, facilitate the development of the mesocotyl by promoting the elongation of cells under deep sowing conditions [1, 19, 33, 40, 41, 43–48]. In GO analysis, we identified pathways related to hormone synthesis, metabolism, and response (Figs. 2E and 3A). Based on this evidence, we hypothesize that alterations in hormone levels may effectively enable DH65232 to adapt deep sowing conditions (Additional file 2: Table S1). To investigate this further, we measured the hormone levels in the mesocotyl of DH65232 under different sowing conditions, including auxin, cytokinin, abscisic acid, gibberellins (GA1, GA3, GA4), and the ethylene precursor (ACC). The results indicated that, compared to normal sowing, the levels of GA3, GA4, and ACC (Fig. 3B; Additional file 1: Fig. S4A, B) were significantly elevated in the mesocotyl of DH65232 under deep sowing conditions. In contrast, the levels of GA1, ABA, and IAA (Additional file 1: Fig. S4C, D, E) were significantly reduced, while no significant difference was observed in cytokinin (Additional file 1: Fig. S4F).
Fig. 3.
Results of DEGs involved in hormone regulatory pathways, hormone content detection, and exogenous hormone treatments. A Expression profiles of genes involved in hormone regulatory pathways. Three independent biological samples were used. B Content of GA3 in the mesocotyls of the inbred line DH65232 under different depth treatments. Asterisks indicate significant differences (****, P < 0.0001; t-Test). C and D Morphology of DH65232 treated with different concentrations of GA3 and uniconazole at a sowing depth of 30 cm. Bar = 2 cm. E and F Mesocotyl length of seedlings grown under the treatments of two exogenous hormones
To validate the effects of these hormones, we selected GA3, which exhibited the most significant changes, for exogenous hormone application experiments. Seeds of DH65232 harvested from the same batch were soaked in different concentrations of hormone solutions and then planted in the same container. After 10 days, we measured the lengths of the mesocotyl and seedlings. The results indicated that with increasing concentrations of GA3 treatment, the length of the mesocotyl in DH65232 under deep sowing conditions increased (Fig. 3C, D; Additional file 1: Fig. S4G). We also conducted similar treatments using different concentrations of a GA3 inhibitor (uniconazole). As we initially hypothesized, the results aligned with our expectations. With the application of increasing concentrations of uniconazole, we observed a consistent and gradual decrease in the length of the mesocotyl in DH65232. This finding supports our assumption that higher doses of uniconazole would lead to a reduction in mesocotyl elongation. (Fig. 3E, F; Additional file 1: Fig. S4H).
Based on these results, we propose that the alterations in endogenous hormone levels within the mesocotyl are a critical factor enabling DH65232 to successfully adapt to deep sowing conditions. In particular, GA appears to play a pivotal role in this process, likely by promoting mesocotyl elongation, which helps the seedling overcome the mechanical resistance of deeper soil layers.
Changes in Intermediate metabolites of DH65232 under different sowing conditions
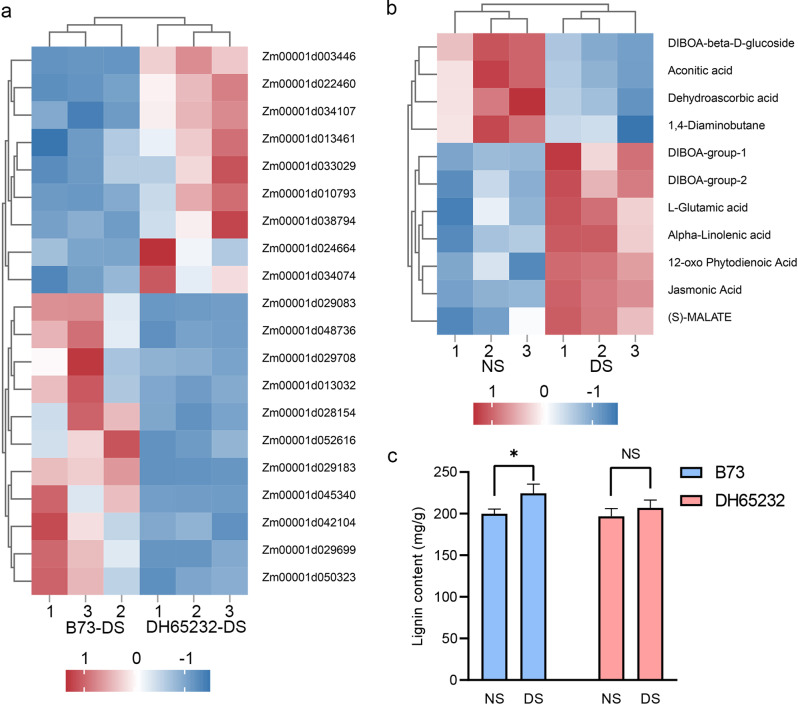
The GO analysis also revealed a significant enrichment of differentially expressed genes associated with intermediate metabolites (Figs. 2E and 4A). To explore this further, we conducted untargeted metabolomics analysis on mesocotyl organ samples from DH65232 under normal and deep sowing conditions. Principal Component Analysis (PCA) of the untargeted metabolomics data showed that the two sample groups were in distinct quadrants, with good clustering within each group (Additional file 1: Fig. S5A). The analysis detected a total of 522 differential metabolites, of which 133 were found to be significantly different (P-value < 0.05, VIP > 1). Among these, 95 metabolites showed decreased levels, while 38 metabolites exhibited increased levels (Additional file 1: Fig. S5B). We performed KEGG pathway analysis on the 133 differential metabolites, identifying 31 enriched biological pathways (Additional file 1: Fig. S5C). The most significantly enriched pathway was the benzoxazines biosynthesis pathway, which included two key differential metabolites, “DIBOA” and “DIBOA-glucoside”. These compounds have been demonstrated to be involved in scavenging reactive oxygen species, facilitating cell wall loosening to promote cell elongation, and modulating hormone signaling pathways [55].
Fig. 4.
DEGs involved in the regulation of intermediate metabolite pathways, metabolite content, and lignin content. A Expression of genes involved in the regulation of intermediate metabolite pathways. Three biologically independent samples were used. B Content of selected differential metabolites in the metabolomics analysis. Three biologically independent samples were used. C Lignin content in the mesocotyls of inbred lines B73 and DH65232 under different depth treatments. Asterisks indicate significant differences (NS, no significant difference; *, P < 0.05; t-Test)
Another noteworthy pathway is the glutathione metabolism pathway, which includes three distinct intermediate metabolites, “L-glutamate”, “putrescine”, and “dehydroascorbic acid” (Fig. 4B, Additional file 4: Table S3). All three of these metabolites exhibit antioxidant functions [56–59]. We also identified another biological pathway, α-linolenic acid metabolism, which also comprises three intermediate metabolites: “linoleic acid”, “12-OPDA”, and “jasmonic acid”. These three metabolites activate antioxidant enzyme activity through jasmonic acid, thereby enhancing the antioxidant capacity [60–62]. Interestingly, among the various regulatory pathways, we identified the metabolism pathway of acetylsalicylate and dicarboxylate, which also contains three differential metabolites: “aconitic acid”, “malic acid”, and “L-glutamate”. These metabolites are involved directly or indirectly in the tricarboxylic acid (TCA) cycle. Furthermore, all three metabolites possess the ability to scavenge reactive oxygen species, increase plant antioxidant capacity, and maintain cell wall homeostasis [63–68].
Among these pathways associated with differential metabolites, one important function, aside from antioxidant capacity, relates to cell wall loosening. The degree of cell wall loosening corresponds directly to the lignin content within the cells. In general, lower lignin content leads to a more relaxed cell wall, facilitating easier cell elongation [69–71]. There are significant differences in cell length between B73 and DH65232 under different sowing conditions (Fig. 1H). To verify whether the changes in cell length of B73 and DH65232 are indeed related to lignin content, we measured the lignin content in the mesocotyls of different inbred lines under normal and deep sowing conditions (Fig. 4C). The results showed that, compared to normal sowing, the lignin content in the mesocotyls of B73 significantly changed under deep sowing conditions, increasing by approximately 11%. Unlike B73, DH65232 showed no significant differences in lignin content across various sowing conditions, while the lignin content in DH65232 significantly decreased compared to B73 under deep sowing conditions.
The above results suggest that under deep sowing conditions, there are significant changes in the intermediate metabolites in DH65232. Notably, the reduction in lignin content may be one of the key factors contributing to the tolerance of DH65232 to deep sowing conditions.
Improved antioxidant ability of DH65232 under deep sowing conditions
The GO analysis indicates that the most significantly altered regulatory pathway is about redox enzymes (Figs. 2E and 5A). Additionally, research conducted by Wang Yang et al. has shown that maize mesocotyls accumulate excessive peroxides under deep sowing conditions, negatively affecting their elongation [72]. Based on this evidence, we hypothesize that the oxidative defense capacity of DH65232 is altered under deep sowing conditions.
Fig. 5.
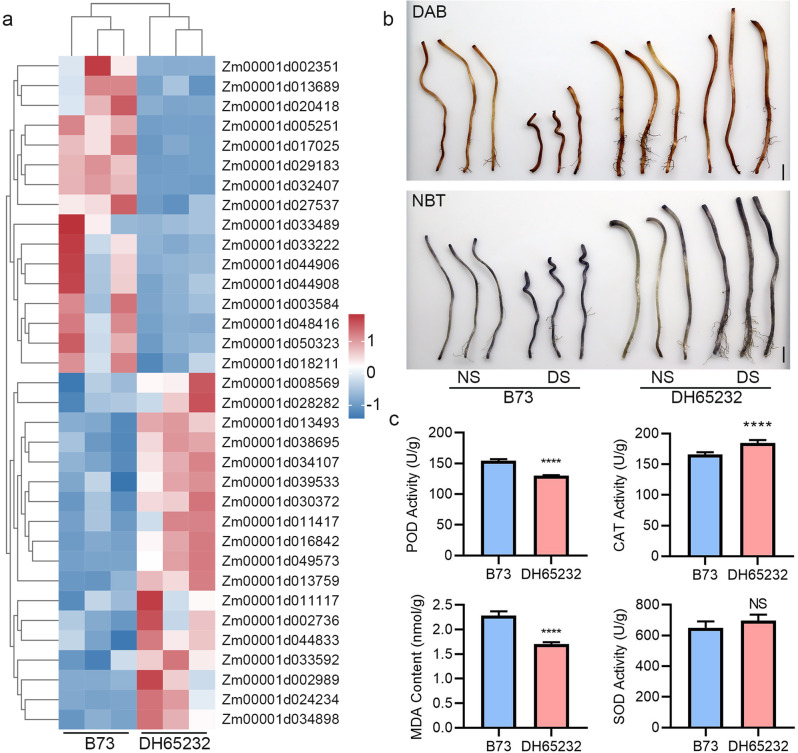
DEGs involved in the redox enzyme pathway and physiological indicators related to oxidative defense. A Expression profiles of genes involved in the redox enzyme pathway. Three independent biological samples were used. B DAB and NBT staining of inbred lines B73 and DH65232 under different depth treatments. Bar = 1 cm. C Detection of POD, CAT, SOD activities, and MDA content in the mesocotyls of B73 and DH65232 under different sowing conditions. Asterisks indicate significant differences (NS, no significant difference; ****, P < 0.0001; t-Test)
To explore this hypothesis, we performed diaminobenzidine (DAB) and nitrotetrazolium blue chloride (NBT) staining on the mesocotyls of B73 and DH65232 under both normal and deep sowing treatments (Fig. 5B). The results reveal that at a depth of 30 cm, the mesocotyls of DH65232 exhibit a lighter DAB coloration compared to B73, suggesting a greater accumulation of peroxides in B73. However, the differences observed in NBT staining were less pronounced. This indicates that, during deep sowing, B73 has higher peroxide levels compared to DH65232, while the levels of superoxide remain relatively stable. Additionally, we investigated the activities and contents of key antioxidase in the mesocotyls of B73 and DH65232 under deep sowing conditions. The enzymes of interest included POD, SOD, and CAT, alongside the assessment of MDA content, which serves as an indicator of oxidative stress (Fig. 5C). The results of our measurements revealed a notable difference in enzyme activities between B73 and DH65232. Specifically, the POD activity in the mesocotyls of B73 was significantly higher than that in DH65232, exhibiting an increase of approximately 18.5%. Conversely, we observed that the CAT content in B73 was lower compared to DH65232, showing a decrease of about 10.8%. This reduction in CAT levels indicates that B73 may have a diminished capacity to decompose hydrogen peroxide into water and oxygen, which could lead to increased oxidative damage if the accumulation of ROS is not adequately managed. The MDA content in B73 was markedly higher than that in DH65232, with an increase of 33.7%. The elevated MDA levels suggest a greater degree of lipid peroxidation occurring in B73, further implicating it in higher oxidative stress compared to DH65232. Elevated MDA levels are often associated with membrane damage and may adversely affect cellular integrity and function. However, despite these differences in POD, CAT, and MDA levels, our analysis revealed no significant difference in SOD activity between B73 and DH65232. This lack of variation in SOD activity indicates that both lines may be similarly effective in converting superoxide radicals into hydrogen peroxide, a crucial step in the antioxidant defense pathway.
Overall, these findings highlight the contrasting oxidative defense strategies employed by B73 and DH65232 under deep sowing conditions, with B73 exhibiting heightened POD activity alongside increased oxidative stress, as evidenced by elevated MDA levels, while DH65232 appears to maintain a more balanced oxidative status.
Discussion
Maize, recognized as one of the three primary staple crops worldwide, plays a crucial role in global food security and agricultural economies [1, 2]. A large proportion of maize cultivation is arid and semi-arid regions [3, 4], where water scarcity presents significant challenges to crop establishment and yield. Expanding the planting area and increasing the yield of maize in these regions is considered one of the most effective strategies for boosting global maize production, which is essential to meet the growing demand for food, feed, and biofuel. One promising approach to enhance maize productivity in water-limited regions is the deep sowing of seeds [12–15]. However, this also introduces another challenge: the increased depth creates a greater barrier for seedlings as they attempt to break through the soil surface, which can hinder emergence and reduce plant survival rates if the crop lacks adequate resilience [73–75]. Consequently, understanding the physiological and genetic mechanisms that enable maize tolerance to deep sowing has emerged as a key research priority. Uncovering these mechanisms could also guide the development of maize varieties with enhanced tolerance to drought and hold great potential to support agricultural sustainability by expanding maize planting areas and boosting yields in arid and semi-arid regions, thereby contributing to global food security and resilience against climate variability.
In our study, we identified a deep-sowing-tolerant maize inbred line, DH65232, by screening a maize GWAS population under deep sowing conditions (30 cm) (Fig. 1A). Compared to normal sowing, the commonly used inbred line B73 showed a reduction in mesocotyl length under deep sowing, whereas DH65232 displayed no significant change (Fig. 1B). Mesocotyl elongation is considered closely related to deep-sowing tolerance [16–19], as it provides the primary force for pushing the seedling through the soil to the surface, thereby enhancing emergence rates under deep sowing conditions [76, 77]. Our statistical analysis further revealed that, under deep sowing conditions, both inbred lines showed significant reductions in coleoptile and seedling lengths compared to normal sowing (Fig. 1B, C, D, E). This reduction may be a general response across all inbred lines. The deep-sowing tolerance observed in DH65232 may be attributed to its ability to sustain mesocotyl growth with minimal impact under deep sowing conditions. This resilience in mesocotyl elongation likely provides the seedling with sufficient upward growth force to push through deeper soil layers, ensuring successful emergence.
The genetic network underlying mesocotyl elongation in maize remains largely unknown [77]. Hormones are recognized as key regulators in this process [78], and research has shown that multiple plant hormones, including GA, JA, ABA, CKs, auxins, and brassinosteroids, play essential roles in regulating maize mesocotyl elongation [77]. Among these hormones, GA is known to significantly promote mesocotyl elongation [3, 79]. This effect may be linked to GA enhancement of starch-metabolizing enzyme activity, which provides the energy and building blocks necessary for cell expansion [19]. Conversely, ABA inhibits mesocotyl elongation by activating the wall-loosening enzyme XET [80]. This enzyme modifies the plant cell wall structure, limiting the expansion of mesocotyl cells and thereby reducing elongation [80]. The deep-sowing tolerance of maize inbred line 3681-4 is attributed to rapid mesocotyl elongation regulated by auxins [81]. In rice, the ethylene core signaling factor OsEIL1 regulates Semi-Dwarf1 (SD1), which encodes GA20-oxidase, to jointly promote mesocotyl elongation [78].
In the RNA-seq data, many hormone-related genes exhibited significantly differential expression, including those related to auxins and gibberellins (Figs. 2E and 3A). Hormone content analysis revealed that, compared to normal sowing, the levels of GA3, GA4, and ACC were significantly elevated in DH65232 under deep sowing conditions (Fig. 3B; Additional file 1: Fig. S4A, B), while the levels of GA1, ABA and IAA decreased significantly (Additional file 1: Fig. S4C, D, E). These changes in hormone levels align with previous findings that hormones can regulate mesocotyl elongation [77]. Previous studies have identified CK as factors influencing mesocotyl elongation, however, our experimental results showed no significant changes in CK levels. This lack of variation could imply that, despite their known role in growth regulation, cytokinins may not be a primary influencing factor in the mesocotyl elongation of the DH65232 inbred line under deep sowing conditions. Additionally, the absence of change in CK levels might be related to the specific types of CKs we measured or the timing of our measurements in relation to the developmental stages of the seedlings.
Abiotic stress induces the production of a substantial amount of ROS within plant cells [82]. There is a notable connection between hormone signaling pathways and ROS, which helps coordinate plant development and stress responses [83–86]. For instance, hormones such as GA and auxin have been shown to influence the activity of ROS, modulating their levels and effects on the plant [87–91]. GO analysis of DEGs revealed that genes involved in responding to deep-sowing stress are primarily enriched in three pathways: hormone regulation, intermediary metabolism, and redox enzymes (Fig. 2E). Furthermore, our untargeted metabolomic analysis identified several significantly different intermediary metabolites, many of which are involved in the ROS scavenging pathways (Fig. 4B). However, it remains unclear which specific genes in DH65232 are transcriptionally or translationally altered to directly enhance its deep-sowing tolerance. Additionally, our untargeted metabolomic analysis revealed various intermediary metabolites whose roles in pathways beyond hormone regulation and oxidative defense need further investigation.
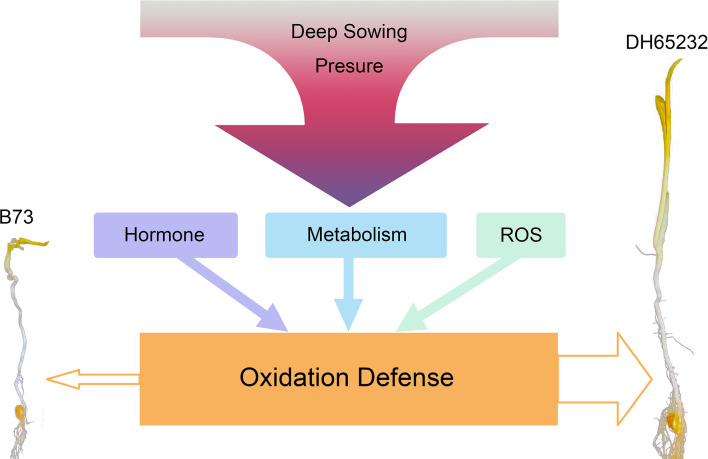
In light of our findings, we propose a hypothesis that during the deep sowing of the maize inbred line DH65232, alterations in hormone levels, intermediate metabolites, and antioxidase activity play a significant role in activating the oxidative defense pathway. This enhancement in oxidative defense could be the fundamental reason for the superior tolerance to deep sowing of DH65232 (Fig. 6).
Fig. 6.
Increasing the seeding depth exerts stress on the emergence of maize seedlings, with the mesocotyl being the primary organ for responding to this stress. This stress stimulates changes in hormone content, metabolite content, and enzyme activity related to ROS within the mesocotyl. Both hormones and metabolites, as well as antioxidant enzymes, are closely related to the oxidative defense of plants. The results of this study reveal that the enhancement of oxidative defense capacity is the fundamental reason why the mesocotyl is longer and the tolerance to deep sowing is stronger in the inbred line DH65232
Conclusions
In this study, we screened a maize inbred line, DH65232, exhibiting notable tolerance to deep sowing from a maize GWAS population. Through transcriptome analysis combined with measurements of hormone levels, untargeted metabolite analysis, and antioxidant enzyme activity assay, we uncovered potential mechanisms regulating maize tolerance to deep sowing. Based on these findings, we propose a hypothesis: during deep sowing of DH65232, changes in hormone and intermediate metabolite levels, along with related enzyme activities, ultimately enhance its antioxidant ability, thereby increasing the tolerance to deep sowing. Our findings provide novel insights into the basis of deep sowing tolerance, identifying potential targets for breeding maize varieties better adapted to challenging conditions.
Methods
Plant materials, growth conditions, and phenotypic measurements
The plant materials used in this study are maize inbred lines B73 and DH65232. B73, released in 1972, was the product of breeding efforts led by Wilbert Russell, an emeritus professor of agronomy at Iowa State University (ISU). The development of the inbred line DH65232 involved crossing the introduced Mozambique maize variety with the local variety “Dahongpao” from southern China, resulting in the creation of “6237”. DH65232 line was then further developed using the pedigree method, with continuous self-pollination over eight generations to stabilize and refine its genetic traits of the line 6237. The two inbred lines were sown in sand, with sowing depths of 5 cm and 30 cm, respectively. Under dark conditions at 28 °C, the plants were grown for ten days. Sowing in sand was chosen to avoid the influence of different nutrient compositions around the roots of the seedlings on growth, and dark treatment was used to prevent the effects of light on mesocotyl elongation.
Ten days after sowing, the inbred seedlings were carefully removed from sand, rinsed with clean water, and wiped dry. We measured six phenotypic data, include mesocotyl length, seedling length, coleoptile length, plumule length, root length, and fresh weight.
Cytological analysis
The mesocotyls of B73 and DH65232 from normal sowing and deep sowing at 10 DAS were used for paraffin sectioning and scanning electron microscopy (SEM) observation.
For paraffin section observation, mesocotyls need to be sectioned into approximately 3 mm cylindrical samples, fixed in FAA buffer (formaldehyde: acetic acid: ethanol: water = 10:5:50:35, v/v/v/v), and vacuum infiltrated twice for 30 min. After dehydration with graded ethanol concentrations, the samples were embedded in paraffin for sectioning. Sections were stained with Schiff’s reagent and photographed under bright field using an Olympus BX51 microscope. Cell length of the mesocotyls at 10 DAS was measured using Image J.
For SEM observation, samples were fixed in glutaraldehyde solution. After dehydration with graded ethanol concentrations, samples were critical point dried with isoamyl acetate, adhered to metal sample stages with conductive adhesive, sputter coated with metal, and photographed in a scanning electron microscope (JEOL). Cell length of the mesocotyls was measured using Image J.
RNA extraction and transcriptome sequencing
Mesocotyl samples from B73 and DH65232 were collected under normal sowing and deep sowing conditions at 10 DAS. These samples were preserved in RNase-free 50 mL centrifuge tubes and stored in a -80 °C freezer. The mesocotyls were ground into powder under liquid nitrogen environment, and total RNA was extracted using the Ultrapure RNA Kit (CWBIO).
RNA samples meeting the requirements were then submitted to Wuhan Huada Company for database construction and transcriptome sequencing. The raw sequencing data were trimmed using Fastp with default parameters. Clean reads were then aligned to the Zea mays AGPv4 reference genome. Gene read counts were obtained using FeatureCounts2. Differentially expressed genes were identified using DESeq2 with criteria of FDR < 0.05 and TPM > 2.
Quantitative real-time PCR
Total RNA reverses transcription by a HiScript II Q RT SuperMix (Vazyme). qRT-PCR was performed with ChamQ Blue Universal SYBR qPCR Master Mix (Vazyme) and a CFX Connect Real-Time System (Bio-Rad). The primers of DEGs and the maize Actin gene were showed in Table S4 (Additional file 5: Table S4). Data were generated from three biological replicates of each sample.
Hormone measurement and hormone exogenous treatment
Mesocotyls of DH65232 preserved in a -80 °C freezer were submitted to Nanjing Ruiyuan Biotechnology Company for hormone content measurement. The measured indicators included IAA, ABA, Zeatin, GA1, GA3, GA4, and ACC (ethylene synthesis precursor).
GA3 and Uniconazole (GA synthesis inhibitor) were selected for hormone exogenous application experiments. First, the seeds of DH65232 material were disinfected. The seeds were immersed in 75% ethanol for 1 min, followed by immersion in 7% sodium hypochlorite solution for 10 min, with shaking every 2 min to ensure thorough soaking. The seeds were then rinsed with water until no residual sodium hypochlorite odor was detected.
After thorough disinfection, the seeds were soaked in GA3 solutions at concentrations of 5, 50, and 100 mg/L [92, 93], Uniconazole solutions at concentrations of 5, 50, and 100 mg/L [94], and a control solution without any hormones for 12 h. The soaked seeds were then planted in sand at a depth of 30 cm, and subjected to dark treatment for 10 days to observe phenotypic changes.
Untargeted metabolomics measurement
The mesocotyl of DH65232, stored in a -80℃ freezer, was subjected to untargeted metabolite detection by Zhengzhou Zhongpu Biotechnology. After grinding the frozen sample into powder, it was extracted overnight at a ratio of fresh weight to extraction solution of 0.1 g/ml using 80% HPLC-grade methanol containing 1µM chrysanthemin as an internal standard. Subsequently, UHPLC-MS/MS analysis was conducted, ultimately obtaining the content of differential metabolites under different treatment conditions, which were then used for subsequent pathway analysis.
Lignin content detection
The mesocotyls of inbred lines B73 and DH65232 at 10 DAS under normal and deep sowing conditions were dried to a constant weight. The dried mesocotyls were ground into a fine powder using a sample grinder and sieved through a 50-mesh screen. Approximately 5 mg of the powder was weighed into 2 ml centrifuge tubes, prepared for three biological replicates. The Lignin Content Assay Kit (Solarbio) was used to extract lignin. The spectrophotometer was adjusted to a wavelength of 280 nm, and the absorbance was measured with glacial acetic acid as the zero reference. The lignin content was calculated using the absorbance values according to the provided formula.
DAB and NBT staining
The mesocotyls at different sowing depths at 10 DAS were placed in DAB and NBT staining solutions. Under a vacuum pressure of 0.6 atmospheres for 30 min, the mesocotyls were then incubated on a shaker at 28 °C overnight for staining. Afterward, they were decolorized with anhydrous ethanol for 30 min. Finally, a camera was used to record the color changes in the mesocotyls of different materials.
Extraction and determination of the activities of POD, CAT, and SOD, and MDA content
The kits used for extraction and detection were sourced from Suzhou Grace Biotech, with catalog numbers G0107W, G0105W, G0109W, and G0101W, respectively. The mesocotyl samples from different sowing depths at 10 DAS were collected using liquid nitrogen and temporarily stored in a -80 °C freezer. The frozen samples were ground into powder in a liquid nitrogen environment, and then extraction solution was added. The extraction was performed according to the instructions provided in the manual. The extracted sample solutions were processed in different ways. Using a pre-heated microplate reader, the absorbance of the processed samples was measured under appropriate wavelength conditions. Enzyme activities or contents were calculated based on the provided formulas.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
Not applicable.
Abbreviations
- NS
Normal sowing
- DS
Deep sowing
- DAS
Days after sowing
- DEGs
Differentially expressed genes
- GO
Gene ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- DAB
Diaminobenzidine
- NBT
Nitrotetrazolium blue chloride
- POD
Peroxidase
- CAT
Catalase
- SOD
Superoxide dismutase
- MDA
Malondialdehyde
Author contributions
Z.C., L.H.J., Z.X.Y., Z.X.S., and Y.X.R. conceived the project. S.M.F., P.M.L., and Z.C. wrote the paper. P.M.L. prepared the figures. Z.G.M. analyzed the data. S.M.F. and T.Z.A. performed the experiments. Z.M.Y., H.X.F., and Z.Y.J. counted the phenotypes. All authors reviewed the manuscript.
Funding
This work was funded by Shandong Natural Science Foundation (ZR2023MC107); National Key Research and Development Program of China (2022YFD1201700).
Data availability
The dataset supporting the conclusions of this article is available in the NCBI Sequence Read Archive (SRA) platform under the accession number PRJNA1180223 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1180223/).
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Mingfei Sun, Menglin Pu and Guangming Zheng contributed equally to this work.
Contributor Information
Hongjun Liu, Email: HongjunL@sdau.edu.cn.
Chao Zhou, Email: zhouc@sdau.edu.cn.
References
- 1.Zhao X, Li J, Niu Y, Hossain Z, Gao X, Bai X, Mao T, Qi G, He F. Exogenous serotonin (5-HT) promotes mesocotyl and coleoptile elongation in maize seedlings under deep-seeding stress through enhancing auxin accumulation and inhibiting lignin formation. Int J Mol Sci 2023; 24(23). [DOI] [PMC free article] [PubMed]
- 2.Qin L, Kong F, Wei L, Cui M, Li J, Zhu C, Liu Y, Xia G, Liu S. Maize ZmSRO1e promotes mesocotyl elongation and deep sowing tolerance by inhibiting the activity of ZmbZIP61. J Integr Plant Biol. 2024;66(8):1571–86. [DOI] [PubMed] [Google Scholar]
- 3.Pan B, Zhong T, Zhao G. Promoting deep-sowing germinability of corn (Zea mays) by seed soaking with gibberellic acid. Arch Agron Soil Sci. 2016;63(9):1314–23. [Google Scholar]
- 4.Troyer AF. The location of genes governing long first internode of corn. 1996(0016-6731). [DOI] [PMC free article] [PubMed]
- 5.Rasheed A, Jie H, Ali B, He P, Zhao L, Ma Y, Xing H, Qari SH, Hassan MU, Hamid MR et al. Breeding drought-tolerant maize (Zea mays) using molecular breeding tools: recent advancements and future prospective. Agronomy 2023; 13(6).
- 6.Peer LA, Bhat MY, Lone AA, Dar ZA, Mir BA. Genetic, molecular and physiological crosstalk during drought tolerance in maize (Zea mays): pathways to resilient agriculture. Planta 2024; 260(4). [DOI] [PubMed]
- 7.Hui D, Daryanto S, Wang L, Jacinthe P-A. Global synthesis of drought effects on maize and wheat production. PLoS ONE 2016; 11(5). [DOI] [PMC free article] [PubMed]
- 8.Leng G. Maize yield loss risk under droughts in observations and crop models in the United States. Environ Res Lett 2021; 16(2).
- 9.Agunbiade VF, Babalola OO. Endophytic and rhizobacteria functionalities in alleviating drought stress in maize plants. Plant Prot Sci. 2023;59(1):1–18. [Google Scholar]
- 10.Zhao X, Niu Y. The combination of conventional QTL analysis, bulked-segregant analysis, and RNA-sequencing provide new genetic insights into maize mesocotyl elongation under multiple deep-seeding environments. Int J Mol Sci 2022; 23(8). [DOI] [PMC free article] [PubMed]
- 11.Zhao X, Zhong Y, Zhou W. Molecular mechanisms of mesocotyl elongation induced by brassinosteroid in maize under deep-seeding stress by RNA-sequencing, microstructure observation, and physiological metabolism. Genomics. 2021;113(6):3565–81. [DOI] [PubMed] [Google Scholar]
- 12.Rida S, Maafi O, López-Malvar A, Revilla P, Riache M, Djemel A. Genetics of germination and seedling traits under drought stress in a MAGIC population of maize. Plants 2021; 10(9). [DOI] [PMC free article] [PubMed]
- 13.Sousa LIS, Brito AEA, Souza LC, Teixeira KBS, Nascimento VR, Albuquerque GDP, Oliveira Neto CF, Okumura RS, Nogueira GAS, Freitas JMN et al. Does silicon attenuate PEG 6000-induced water deficit in germination and growth initial the seedlings corn. Brazilian J Biology 2023; 83. [DOI] [PubMed]
- 14.Yang Y, Ma Y-t, Liu Y-y, Lyle D, Li D-d, Wang P-x, Xu J-l, Zhen S-h et al. Lu J-w, Peng Y-l: Dissecting the genetic basis of maize deep-sowing tolerance by combining association mapping and gene expression analysis. Journal of Integrative Agriculture 2022; 21(5):1266–1277.
- 15.Kutschera U, Khanna R. Auxin action in developing maize coleoptiles: challenges and open questions. Plant Signal Behav 2020; 15(6). [DOI] [PMC free article] [PubMed]
- 16.Dungan GH. Response of corn to extremely deep planting. Agron J. 1950;42(5):256–7. [Google Scholar]
- 17.Qi X, Zhuang Z, Ji X, Bian J, Peng Y. The mechanism of exogenous salicylic acid and 6-benzylaminopurine regulating the elongation of maize mesocotyl. Int J Mol Sci 2024; 25(11). [DOI] [PMC free article] [PubMed]
- 18.Ju L, Lv N, Yin F, Niu H, Yan H, Wang Y, Fan F, Lv X, Chu J, Ping J. Identification of key genes regulating sorghum mesocotyl elongation through transcriptome analysis. Genes 2023; 14(6). [DOI] [PMC free article] [PubMed]
- 19.Wang Y, Wang Y, Yang R, Wang F, Fu J, Yang W, Bai T, Wang S, Yin H. Effects of gibberellin priming on seedling emergence and transcripts involved in mesocotyl elongation in rice under deep direct-seeding conditions. J Zhejiang University-SCIENCE B. 2021;22(12):1002–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blackburn A, Sidhu G, Schillinger WF, Skinner D, Gill K. QTL mapping using GBS and SSR genotyping reveals genomic regions controlling wheat coleoptile length and seedling emergence. Euphytica 2021; 217(3).
- 21.Singh K, Shukla S, Kadam S, Semwal VK, Singh NK, Khanna-Chopra R. Genomic regions and underlying candidate genes associated with coleoptile length under deep sowing conditions in a wheat RIL population. J Plant Biochem Biotechnol. 2014;24(3):324–30. [Google Scholar]
- 22.Paynter BH, Clarke GPY. Coleoptile length of barley (Hordeum vulgare L.) cultivars. Genet Resour Crop Evol. 2009;57(3):395–403. [Google Scholar]
- 23.Hidekazu Takahashi KS, Kazuyoshi T. Maping genes for deep-seeding tolerance in barely. Euphytica. 2001;122:37–43. [Google Scholar]
- 24.Hochholdinger F, Tuberosa R. Genetic and genomic dissection of maize root development and architecture. Curr Opin Plant Biol. 2009;12(2):172–7. [DOI] [PubMed] [Google Scholar]
- 25.Hochholdinger F, Yu P, Marcon C. Genetic control of root system development in maize. Trends Plant Sci. 2018;23(1):79–88. [DOI] [PubMed] [Google Scholar]
- 26.Lee HS, Sasaki K, Higashitani A, Ahn SN, Sato T. Mapping and characterization of quantitative trait loci for mesocotyl elongation in rice (Oryza sativa L). Rice. 2012;5(1939–8425):13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang H, Ma P, Zhao Z, Zhao G, Tian B, Wang J, Wang G. Mapping QTL controlling maize deep-seeding tolerance-related traits and confirmation of a major QTL for mesocotyl length. Theor Appl Genet. 2011;124(1):223–32. [DOI] [PubMed] [Google Scholar]
- 28.Wu J, Feng F, Lian X, Teng X, Wei H, Yu H, Xie W, Yan M, Fan P, Li Y et al. Genome-wide Association Study (GWAS) of mesocotyl elongation based on re-sequencing approach in rice. BMC Plant Biol 2015;15(1). [DOI] [PMC free article] [PubMed]
- 29.Lu Q, Zhang M, Niu X, Wang C, Xu Q, Feng Y, Wang S, Yuan X, Yu H, Wang Y, et al. Uncovering novel loci for mesocotyl elongation and shoot length in indica rice through genome-wide association mapping. Planta. 2015;243(3):645–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Edzesi WM, Dang X, Liu E, Bandoh WKN, Gakpetor PM, Ofori DA, Hong D. Screening germplasm and detecting QTLs for mesocotyl elongation trait in rice (Oryza sativa L.) by association mapping. BMC Genomic Data 2023;24(1). [DOI] [PMC free article] [PubMed]
- 31.Zhao X, Niu Y, Hossain Z, Shi J, Mao T, Bai X. Integrated QTL mapping, meta-analysis, and RNA-sequencing reveal candidate genes for maize deep-sowing tolerance. Int J Mol Sci 2023, 24(7). [DOI] [PMC free article] [PubMed]
- 32.Choi D, Lee Y, Cho H-T, Kende H. Regulation of expansin gene expression affects growth and development in transgenic rice plants. Plant Cell. 2003;15(6):1386–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lv Y, Shao G, Jiao G, Sheng Z, Xie L, Hu S, Tang S, Wei X, Hu P. Targeted mutagenesis of POlyamine Oxidase 5 that negatively regulates mesocotyl elongation enables the generation of direct-seeding rice with improved grain yield. Mol Plant. 2021;14(2):344–51. [DOI] [PubMed] [Google Scholar]
- 34.Lyu Y, Wei X, Zhong M, Niu S, Ahmad S, Shao G, Jiao G, Sheng Z, Xie L, Hu S, et al. Integrated transcriptome, small RNA, and degradome analysis to elucidate the regulation of rice seedling mesocotyl development during the passage from darkness to light. Crop J. 2020;8(6):918–28. [Google Scholar]
- 35.Zhan J, Lu X, Liu H, Zhao Q, Ye G. Mesocotyl elongation, an essential trait for dry-seeded rice (Oryza sativa L.): a review of physiological and genetic basis. Planta 2019;251(1). [DOI] [PubMed]
- 36.Sawers RJH, Linley PJ, Farmer PR, Hanley NP, Costich DE, Terry MJ, Brutnell TP. elongated mesocotyl1, a phytochrome-deficient mutant of maize. Plant Physiol. 2002;130(1):155–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sheehan MJ, Kennedy LM, Costich DE, Brutnell TP. Subfunctionalization of PhyB1 and PhyB2 in the control of seedling and mature plant traits in maize. Plant J. 2006;49(2):338–53. [DOI] [PubMed] [Google Scholar]
- 38.Waadt R, Seller CA, Hsu P-K, Takahashi Y, Munemasa S, Schroeder JI. Plant hormone regulation of abiotic stress responses. Nat Rev Mol Cell Biol. 2022;23(10):680–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Verma V, Ravindran P, Kumar PP. Plant hormone-mediated regulation of stress responses. BMC Plant Biol 2016;16(1). [DOI] [PMC free article] [PubMed]
- 40.Chen L, Huang X-X, Zhao S-M, Xiao D-W, Xiao L-T, Tong J-H, Wang W-S, Li Y-J, Ding Z, Hou B-K. IPyA glucosylation mediates light and temperature signaling to regulate auxin-dependent hypocotyl elongation in Arabidopsis. Proc Natl Acad Sci. 2020;117(12):6910–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kutschera U, Wang Z-Y. Growth-limiting proteins in maize coleoptiles and the auxin-brassinosteroid hypothesis of mesocotyl elongation. Protoplasma. 2015;253(1):3–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu Z, Duan X, Luo L, Dai S, Ding Z, Xia G. How plant hormones mediate salt stress responses. Trends Plant Sci. 2020;25(11):1117–30. [DOI] [PubMed] [Google Scholar]
- 43.Leng B, Li M, Mu C, Yan Z, Yao G, Kong X, Ma C, Zhang F, Liu X. Molecular mechanism of gibberellins in mesocotyl elongation response to deep-sowing stress in sweet maize. Curr Issues Mol Biol. 2022;45(1):197–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mo W, Tang W, Du Y, Jing Y, Bu Q, Lin R. PHYTOCHROME-INTERACTING FACTOR-LIKE14 and SLENDER RICE1 interaction controls seedling growth under salt stress. Plant Physiol. 2020;184(1):506–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saab IN, Ho T, Fau - Sharp RE, Sharp RE. Translatable RNA populations associated with maintenance of primary root elongation and inhibition of mesocotyl elongation by abscisic acid in maize seedlings at low water potentials. Plant Physiol. 1995;109(2):593–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xiong Q, Ma B, Lu X, Huang Y-H, He S-J, Yang C, Yin C-C, Zhao H, Zhou Y, Zhang W-K, et al. Ethylene-inhibited jasmonic acid biosynthesis promotes mesocotyl/coleoptile elongation of etiolated rice seedlings. Plant Cell. 2017;29(5):1053–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hu Z, Yamauchi T, Yang J, Jikumaru Y, Tsuchida-Mayama T, Ichikawa H, Takamure I, Nagamura Y, Tsutsumi N, Yamaguchi S, et al. Strigolactone and cytokinin act antagonistically in regulating rice mesocotyl elongation in darkness. Plant Cell Physiol. 2014;55(1):30–41. [DOI] [PubMed] [Google Scholar]
- 48.Sun S, Wang T, Wang L, Li X, Jia Y, Liu C, Huang X, Xie W, Wang X. Natural selection of a GSK3 determines rice mesocotyl domestication by coordinating strigolactone and brassinosteroid signaling. Nat Commun 2018;9(1). [DOI] [PMC free article] [PubMed]
- 49.Gill SS, Tuteja N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol Biochem. 2010;48(12):909–30. [DOI] [PubMed] [Google Scholar]
- 50.Sato H, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K. Complex plant responses to drought and heat stress under climate change. Plant J. 2024;117(6):1873–92. [DOI] [PubMed] [Google Scholar]
- 51.Gao H, Cui J, Liu S, Wang S, Lian Y, Bai Y, Zhu T, Wu H, Wang Y, Yang S, et al. Natural variations of ZmSRO1d modulate the trade-off between drought resistance and yield by affecting ZmRBOHC-mediated stomatal ROS production in maize. Mol Plant. 2022;15(10):1558–74. [DOI] [PubMed] [Google Scholar]
- 52.Tarkowski ŁP, Signorelli S, Considine MJ, Montrichard F. Integration of reactive oxygen species and nutrient signalling to shape root system architecture. Plant Cell Environ. 2022;46(2):379–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang R, Li K, Wang M, Sun M, Li Q, Chen L, Xiao F, Zhang Z, Zhang H, Jiao F et al. ZmNAC17 regulates mesocotyl elongation by mediating auxin and ROS biosynthetic pathways in maize. Int J Mol Sci 2024;25(9). [DOI] [PMC free article] [PubMed]
- 54.Zhao X, Niu Y, Bai X, Mao T. Transcriptomic and metabolic profiling reveals a lignin metabolism network involved in mesocotyl elongation during maize seed germination. Plants 2022, 11(8). [DOI] [PMC free article] [PubMed]
- 55.Wang J, Tao M, Xu L, Fan N, Zhao C, Xiao Z, Wang Z. Chitosan nanocarriers loaded with salicylic acid for controlling fall armyworm (Spodoptera frugiperda) and alleviating oxidative stress in maize plants. Environ Science: Nano. 2023;10(12):3295–306. [Google Scholar]
- 56.Fawole OA, Atukuri J, Arendse E, Opara UO. Postharvest physiological responses of pomegranate fruit (cv. Wonderful) to exogenous putrescine treatment and effects on physico-chemical and phytochemical properties. Food Sci Hum Wellness. 2020;9(2):146–61. [Google Scholar]
- 57.Asgher M, Sehar Z, Rehaman A, Rashid S, Ahmed S, Per TS, Alyemeni MN, Khan NA. Exogenously-applied L-glutamic acid protects photosynthetic functions and enhances arsenic tolerance through increased nitrogen assimilation and antioxidant capacity in rice (Oryza sativa L). Environ Pollut 2022;301. [DOI] [PubMed]
- 58.Chavan SN, De Kesel J, Desmedt W, Degroote E, Singh RR, Nguyen GT, Demeestere K, De Meyer T, Kyndt T. Dehydroascorbate induces plant resistance in rice against root-knot nematode Meloidogyne graminicola. Mol Plant Pathol. 2022;23(9):1303–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang X, Zhu L, Qian M, Jiang L, Gu P, Jia L, Qian C, Luo W, Ma M, Wu Z et al. PbrWRKY62-PbrADC1 module involves in superficial scald development of Pyrus Bretschneideri Rehd. Fruit via regulating putrescine biosynthesis. Mol Hortic 2024;4(1). [DOI] [PMC free article] [PubMed]
- 60.Yang B, Pan F, Yasmeen F, Shan L, Pan J, Zhang M, Weng X, Wang M, Li M, Wang Q et al. Integrated multi-omic analysis reveals the cytokinin and sucrose metabolism-mediated regulation of flavone glycoside biosynthesis by MeJA exposure in Ficus Pandurata Hance. Food Res Int 2023;174. [DOI] [PubMed]
- 61.Deng J, Liu Q, Zhang Q, Zhang C, Liu D, Fan D, Yang H. Comparative study on composition, physicochemical and antioxidant characteristics of different varieties of kiwifruit seed oil in China. Food Chem. 2018;264:411–8. [DOI] [PubMed] [Google Scholar]
- 62.Wang K-D, Borrego EJ, Kenerley CM, Kolomiets MV. Oxylipins other than jasmonic acid are xylem-resident signals regulating systemic resistance induced by Trichoderma virens in maize. Plant Cell. 2020;32(1):166–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fardus J, Hossain MS, Fujita M. Modulation of the antioxidant defense system by exogenous L-glutamic acid application enhances salt tolerance in Lentil (Lens culinaris Medik). Biomolecules 2021;11(4). [DOI] [PMC free article] [PubMed]
- 64.La VH, Lee B-R, Islam MT, Mamun MA, Park S-H, Bae D-W, Kim T-H. Characterization of glutamate-mediated hormonal regulatory pathway of the drought responses in relation to proline metabolism in Brassica napus L. Plants 2020, 9(4). [DOI] [PMC free article] [PubMed]
- 65.Liu H, Zhao X, Bi J, Dong X, Zhang C. A natural mutation in the promoter of the aconitase gene ZjACO3 influences fruit citric acid content in jujube. Hortic Res 2024;11(3). [DOI] [PMC free article] [PubMed]
- 66.Yu M, Wang S, Gu G, Shi TL, Zhang J, Jia Y, Ma Q, Porth I, Mao JF, Wang R. Integration of mitoflash and time-series transcriptomics facilitates energy dynamics tracking and substrate supply analysis of floral thermogenesis in lotus. Plant, Cell & Environment; 2024. [DOI] [PubMed]
- 67.Zhang Y, Giese J, Kerbler SM, Siemiatkowska B, Perez de Souza L, Alpers J, Medeiros DB, Hincha DK, Daloso DM, Stitt M, et al. Two mitochondrial phosphatases, PP2c63 and Sal2, are required for posttranslational regulation of the TCA cycle in Arabidopsis. Mol Plant. 2021;14(7):1104–18. [DOI] [PubMed] [Google Scholar]
- 68.Zhang H, Du W, Peralta-Videa JR, Gardea-Torresdey JL, White JC, Keller A, Guo H, Ji R, Zhao L. Metabolomics reveals how cucumber (Cucumis sativus) reprograms metabolites to cope with silver ions and silver nanoparticle-induced oxidative stress. Environ Sci Technol. 2018;52(14):8016–26. [DOI] [PubMed] [Google Scholar]
- 69.Yuan L, Dang J, Zhang J, Wang L, Zheng H, Li G, Li J, Zhou F, Khan A, Zhang Z et al. A glutathione S-transferase regulates lignin biosynthesis and enhances salt tolerance in tomato. Plant Physiol 2024. [DOI] [PubMed]
- 70.Zhao X, Zhao Y, Zeng Q-y, Liu C-J. Cytochrome b5 diversity in green lineages preceded the evolution of syringyl lignin biosynthesis. Plant Cell. 2024;36(7):2709–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu C, Yu H, Rao X, Li L, Dixon RA. Abscisic acid regulates secondary cell-wall formation and lignin deposition in Arabidopsis thaliana through phosphorylation of NST1. Proceedings of the National Academy of Sciences 2021;118(5). [DOI] [PMC free article] [PubMed]
- 72.Wang Y, He J, Ye H, Ding M, Xu F, Wu R, Zhao F, Zhao G. Transcriptome analysis revealed the key genes and pathways involved in seed germination of maize tolerant to deep-sowing. Plants 2022; 11(3). [DOI] [PMC free article] [PubMed]
- 73.Jie Y, Wang W, Wu Z, Ren Z, Li L, Zhou Y, Zhang M, Li Z, Yi F, Duan L. Deciphering physiological and transcriptional mechanisms of maize seed germination. Plant Mol Biol 2024;114(5). [DOI] [PubMed]
- 74.Ma L, Wang C, Hu Y, Dai W, Liang Z, Zou C, Pan G, Lübberstedt T, Shen Y. GWAS and transcriptome analysis reveal MADS26 involved in seed germination ability in maize. Theor Appl Genet. 2022;135(5):1717–30. [DOI] [PubMed] [Google Scholar]
- 75.Han C, Yang P. Studies on the molecular mechanisms of seed germination. Proteomics. 2015;15(10):1671–9. [DOI] [PubMed] [Google Scholar]
- 76.Zhao J, Liu S, Zhao X, Huang Z, Sun S, Zeng Z, He Y, Wang Z. Rice gene OsUGT75A regulates seedling emergence under deep-sowing conditions. Crop J. 2024;12(1):133–41. [Google Scholar]
- 77.Niu L, Hao R, Wu X, Wang W, Tuberosa R. Maize mesocotyl: role in response to stress and deep-sowing tolerance. Plant Breeding. 2020;139(3):466–73. [Google Scholar]
- 78.Lyu Y, Dong X, Niu S, Cao R, Shao G, Sheng Z, Jiao G, Xie L, Hu S, Tang S, et al. An orchestrated ethylene–gibberellin signaling cascade contributes to mesocotyl elongation and emergence of rice direct seeding. J Integr Plant Biol. 2024;66(7):1427–39. [DOI] [PubMed] [Google Scholar]
- 79.Hu S, Sanchez DL, Wang C, Lipka AE, Yin Y, Gardner CAC, Lübberstedt T. Brassinosteroid and gibberellin control of seedling traits in maize (Zea mays L). Plant Sci. 2017;263:132–41. [DOI] [PubMed] [Google Scholar]
- 80.Fry SC. Smith Rc Fau - Renwick KF, Renwick Kf Fau - Martin DJ, Martin Dj Fau - Hodge SK, Hodge Sk Fau - Matthews KJ, Matthews KJ: Xyloglucan endotransglycosylase, a new wall-loosening enzyme activity from plants. 1992;282 (3)(0264–6021):821–828. [DOI] [PMC free article] [PubMed]
- 81.Zhao G-W, Wang J-H. Effect of auxin on mesocotyl elongation of dark-grown maize under different seeding depths. Russ J Plant Physiol. 2010;57(1):79–86. [Google Scholar]
- 82.Fichman Y, Mittler R. Rapid systemic signaling during abiotic and biotic stresses: is the ROS wave master of all trades? Plant J. 2020;102(5):887–96. [DOI] [PubMed] [Google Scholar]
- 83.Choudhury FK, Rivero RM, Blumwald E, Mittler R. Reactive oxygen species, abiotic stress and stress combination. Plant J. 2016;90(5):856–67. [DOI] [PubMed] [Google Scholar]
- 84.Ding Y, Shi Y, Yang S. Advances and challenges in uncovering cold tolerance regulatory mechanisms in plants. New Phytol. 2019;222(4):1690–704. [DOI] [PubMed] [Google Scholar]
- 85.Gupta AA-O, Rico-Medina AA-OX, Caño-Delgado AA-O. The physiology of plant responses to drought. Science. 2020;368(Electronic):1095–9203. [DOI] [PubMed] [Google Scholar]
- 86.Lamers J, van der Meer T, Testerink C. How plants sense and respond to stressful environments. Plant Physiol. 2020;182(4):1624–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tognetti VB, MÜHlenbock PER, Van Breusegem F. Stress homeostasis – the redox and auxin perspective. Plant Cell Environ. 2011;35(2):321–33. [DOI] [PubMed] [Google Scholar]
- 88.Wang C, Yang A, Yin H, Zhang J. Influence of water stress on endogenous hormone contents and cell damage of maize seedlings. J Integr Plant Biol. 2008;50(4):427–34. [DOI] [PubMed] [Google Scholar]
- 89.Colebrook EH, Thomas SG, Phillips AL, Hedden P, Davies SA, Dow JAT, Lukowiak K. The role of gibberellin signalling in plant responses to abiotic stress. J Exp Biol. 2014;217(1):67–75. [DOI] [PubMed] [Google Scholar]
- 90.Achard P, Renou J-P, Berthomé R, Harberd NP, Genschik P. Plant DELLAs restrain growth and promote survival of adversity by reducing the levels of reactive oxygen species. Curr Biol. 2008;18(9):656–60. [DOI] [PubMed] [Google Scholar]
- 91.Achard P, Gusti A, Cheminant S, Alioua M, Dhondt S, Coppens F, Beemster GTS, Genschik P. Gibberellin signaling controls cell proliferation rate in Arabidopsis. Curr Biol. 2009;19(14):1188–93. [DOI] [PubMed] [Google Scholar]
- 92.Xie Z, Jin L, Sun Y, Zhan C, Tang S, Qin T, Liu N, Huang J. OsNAC120 balances plant growth and drought tolerance by integrating GA and ABA signaling in rice. Plant Commun 2024;5(3). [DOI] [PMC free article] [PubMed]
- 93.Ahmad I, Kamran M, Ali S, Bilegjargal B, Cai T, Ahmad S, Meng X, Su W, Liu T, Han Q. Uniconazole application strategies to improve lignin biosynthesis, lodging resistance and production of maize in semiarid regions. Field Crops Res. 2018;222:66–77. [Google Scholar]
- 94.Lee B-D, Yim Y, Cañibano E, Kim S-H, García-León M, Rubio V, Fonseca S. Paek N-C: CONSTITUTIVE PHOTOMORPHOGENIC 1 promotes seed germination by destabilizing RGA-LIKE 2 in Arabidopsis. Plant Physiol. 2022;189(3):1662–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The dataset supporting the conclusions of this article is available in the NCBI Sequence Read Archive (SRA) platform under the accession number PRJNA1180223 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1180223/).