Abstract
Background
Spinal cord injury (SCI) inflicts a severe burden on patients and lacks effective treatments. Owing to the poor regenerative capabilities of endogenous oligodendrocyte precursor cells (OPCs) following SCI, there is a growing interest in alternative sources, such as human umbilical cord mesenchymal stem cells (HUCMSCs). TET3 is a key DNA demethylase that plays an important role in neural differentiation, but its role in OPC formation is not well understood. This study aimed to explore the TET3-mediated one-step induction of HUCMSCs into OPCs.
Methods
In vitro, HUCMSCs were induced into OPCs following TET3 overexpression. Changes of methylation and hydroxymethylation during differentiation were monitored, mechanisms involved in the TET3-driven HUCMSC differentiation into OPCs were identified by RNA sequencing. Methylation levels in NG2 and PDGFRA promoter region were detected using Bisulfite Polymerase Chain Reaction (BSP).In vivo, therapeutic effects of iOPCs were evaluated through a rat Allen’s SCI model.
Results
The in vitro analysis confirmed that TET3 enhances HUCMSC differentiation into OPCs, validitied by specific marker expression. The induced OPCs (iOPCs) exhibited methylation and hydroxymethylation patterns similar to native OPCs. BSP analysis demonstrated that TET3 overexpression significantly reduced CpG island methylation in the NG2 and PDGFRA promoter regions. RNA sequencing revealed that TET3 induces iOPCs to express a series of genes essential for OPC formation while inhibiting the signaling pathways that hinder OPC development. In a rat model of SCI, TET3-overexpressing HUCMSCs appear to have the potential to differentiate into iOPCs in vivo, suppressed secondary injury, and promoted functional recovery. The therapeutic effects of iOPCs on SCI were superior to those of standard mesenchymal stem cell treatments.
Conclusions
Our study demonstrated that TET3-mediated demethylation reshapes the methylation patterns of HUCMSCs, enabling their efficient one-step conversion into OPCs and significantly reducing the time required for cell preparation. This approach offers a potential strategy for early intervention in SCI. In an SCI model, TET3-induced OPCs contributed to spinal cord repair, providing novel insights into cell therapy strategies for SCI through the lens of methylation regulation.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12967-024-05929-7.
Keywords: Human umbilical cord mesenchymal stem cells, Oligodendrocyte precursor cells, Spinal cord injury, TET3, 5hmC, Induction
Background
Spinal cord injury (SCI) involves complex pathophysiological responses triggered by mechanical damage to the spinal column, causing severe sensory, motor, and autonomic nervous system dysfunctions commonly associated with vehicular accidents and workplace injuries [1]. The incidence of SCI has been progressively increasing over the years [2, 3], imposing a high economic strain on families and the society. Therefore, developing effective treatments for SCI to alleviate patient suffering and reduce the societal burden has emerged as a pressing issue.
In the pathological process of SCI, the loss of oligodendrocytes (OLs) is a major cause of axonal demyelination and worsened damage [4]. Myelin regeneration is widely recognized as a critical therapeutic goal for promoting recovery after SCI [5]. Oligodendrocyte precursor cells (OPCs) are critical to this process. Following SCI, OPCs migrate to the injury site and differentiate into OLs to promote remyelination [6]. However, the regenerative capacity of endogenous OPCs after SCI is limited, and the use of exogenous OPCs for treatment is hindered by ethical and sourcing issues, making it challenging to apply in clinical practice. As a result, the search for effective alternative cells to OPCs has become a focal point in neuroscience research [7].
Among the various promising seed cells, human umbilical cord mesenchymal stem cells (HUCMSCs) stand out due to their ease of access, strong immunomodulatory properties, low immunogenicity, and reduced tumorigenesis risk [8]. Despite these advantages, the neural differentiation potential of HUCMSCs is relatively limited, and the process of inducing neural differentiation is complex and time-consuming [9, 10]. This challenge is particularly evident when differentiating into OLs, as the prolonged induction period often causes SCI patients to miss the optimal therapeutic window [11–14]. Therefore, finding strategies to enhance the neural differentiation potential of HUCMSCs is crucial.
Advancements in epigenetics have opened new possibilities for the application of HUCMSCs. 5-methylcytosine (5mC) is the most common form of DNA methylation, which can be converted into 5-hydroxymethylcytosine (5hmC) through demethylation by the Ten-Eleven Translocation (TET) family. 5hmC can act as a stable product, regulating gene expression and contributing to cell differentiation [15]. Studies show that 5hmC is highly expressed in embryonic stem cells (ESCs) and helps maintain pluripotency [16]. Further research highlights 5hmC’s significant role in neural development, with high enrichment in the brain, suggesting a specific function related to the nervous system [17].
Additionally, 5hmC exhibits tissue-specific and age-dependent patterns during neural development, particularly in the cerebellum and hippocampus, where levels increase progressively with development, suggesting its regulatory role in neurogenesis and neural differentiation [18]. Moreover, studies have found that 5hmC levels are higher in the neuronal genome, suggesting that the mechanisms of 5hmC in the central nervous system (CNS) may differ from those in ESCs. In the nervous system, 5hmC is specifically enriched, with a marked reduction in 5mC in these regions. This suggests the presence of cell-specific epigenetic regulatory mechanisms in the nervous system, where 5hmC and 5mC may function like a switch to control gene expression [18].
TET3, a key enzyme in the TET family, catalyzes the conversion of 5mC to 5hmC, playing a crucial role in neural development. Although TET3 is nearly undetectable in ESCs, its expression rapidly increases during neural differentiation. While TET3 knockout ESCs can maintain normal self-renewal and pluripotency, their ability to undergo neural differentiation is significantly impaired [19]. This indicates that TET3 is essential for sustaining the neural differentiation process. Research by Li demonstrated that TET3 is significantly upregulated in the prefrontal cortex, leading to the redistribution of 5hmC and alterations in gene expression, which support rapid behavioral adaptation. Loss of TET3, however, results in marked deficits in memory retention [20]. Additionally, overexpression of TET3 in olfactory sensory neurons significantly alters genome-wide 5hmC levels and gene expression, affecting the expression of olfactory receptors and axonal targeting to the olfactory bulb [21] TET3-mediated demethylation has been shown to activate regeneration-related genes, promoting neural tissue repair [22]. TET3 is also crucial in the direct conversion of somatic cells into neurons, enabling fibroblasts to efficiently convert into mature neurons through DNA demethylation [23]. Additionally, TET3 exhibits unique spatial and temporal expression patterns during oligodendrocyte (OL) development. As OLs mature, 5hmC levels change dynamically, and TET3 knockout impairs OL development [24]. These findings underscore TET3’s role in neural system maturation, with DNA hydroxymethylation being a key mechanism in neural development.
However, a notable gap on the regulation of HUCMSC differentiation into OPCs through methylation modifications still exists. Given the critical role of TET3 in neural differentiation, we hypothesize that TET3 may promote the differentiation of HUCMSCs into OPCs by mediating DNA hydroxymethylation.
In this study, we established a TET3-overexpressing HUCMSC model (TET3-OE) and harnessed the neurogenic effects of TET3 to drive the differentiation of these cells into induced OPCs (iOPCs). By investigating the underlying mechanisms, we evaluated the therapeutic potential of TET3-OE-derived iOPCs for SCI treatment. Our findings offer novel insights into the application of HUCMSCs in SCI therapy, providing promising strategies for enhancing treatment outcomes. Importantly, this research underscores the clinical relevance of TET3-mediated epigenetic modifications, paving the way for more effective cell-based therapies that could significantly improve the quality of life for SCI patients.
Materials and methods
Identification of HUCMSCs
The HUCMSCs were procured from HeiLongJiang Alliancells North Bioscience Co., Ltd (Batch number: Gg2200002) and their differentiation potential was assessed.The cell morphology remained stable after multiple passages of culture. All the cells used in this study are between passage 3 and passage 5.
To verify the surface markers on HUCMSCs, we used passage 3 HUCMSCs and followed the method published by Mukai T [25] to identify their immunophenotype. Expression of surface markers CD73 (13160, Cell Signaling Technology), CD105 (4335, Cell Signaling Technology), immunoglobulin (Ig)G1 (5415, Cell Signaling Technology), IgG2a (61656, Cell Signaling Technology), CD45 (13917, Cell Signaling Technology), and HLA-DR (78397, Cell Signaling Technology) were identified by flow Cytometer (FACSA Canto II; BD Biosciences, USA).
To assess the multi-lineage differentiation potential of HUCMSCs, we induced osteogenic and adipogenic differentiation using osteogenic and adipogenic differentiation medium according to the manufacturer’s respective protocols, For osteogenic differentiation, Seed Passage 3 (P3) HUCMSCs at a density of 1 × 104/cm2 in a 6-well plate. Once the cells reach 100% confluence, replace the culture medium with the osteogenic induction differentiation medium for HUCMSCs (PD-017, Procell, Wuhan, China), refreshing the medium every 72 h. After four weeks, perform Alizarin Red staining to identify differentiation and capture images for observation. For adipogenic differentiation, HUCMSCs of P3 were seeded at a density of 1 × 104/cm2 in a 6-well plate. Once the HUCMSCs reach 100% confluence, replace the culture medium with the adipogenic differentiation medium for HUCMSCs (PD-019, Procell, Wuhan, China), refreshing the medium every 72 h. After four weeks, stain with Oil Red O, then observe and capture images under a microscope (BX51, Olympus).
Adenovirus infection
The TET3 overexpression (TET3-OE) adenovirus was acquired from GeneChem Co. (Shanghai, China). Before transfection, a pre-transfection experiment was conducted to determine the optimal multiplicity of infection (MOI) for the TET3-OE adenovirus. Subsequently, the formal transfection was initiated according to the transfection method provided by GeneChem. Both the TET3-OE adenovirus and control adenovirus were added at the optimal MOI value determined in the pre-experiment. Quantitative polymerase chain reaction (qPCR) and western blot analyses were conducted to assess changes in TET3 expression levels. Cells with high TET3 expression were designated as TET3-OE. Cells with the control vector (GV269) were designated as the Control.
Neuronal differentiation
TET3-OE was constructed as described above. After infection, the media of the TET3-OE and Control groups were replaced with a neural induction medium: DMEM/F12 medium (Corning) supplemented with 2% B27 (17504044, Gibco), 20 ng/ml bFGF (10014-HNAE, Sino Biological), 20 ng/ml EGF (10014-HNAE, Sino Biological), and 10 ng/ml PDGF-AA (100-13A, Peprotech) [26, 27], 24 h post-transfection to initiate induction culture. The neural induction medium was replaced every 48 h, and cellular morphological changes were periodically observed.
Quantitative PCR
RNA samples were extracted using the TRIzol (15596026, Invitrogen, USA) method according to the manufacturer’s protocol, and the purity and concentration of RNA were determined using a UV–Vis spectrophotometer (DU640 BECKMAN, USA). Then, cDNA synthesis was performed using the PrimeScript™ RT reagent kit with gDNA Eraser (Perfect Real Time) (TAKARA Code No. RRO47A, Japan) according to the manufacturer’s instructions. Next, qPCR was performed using TB Green Premix Ex Taq II (Tli RNase H Plus) (TAKARA Code No. RR820A, Japan). The reaction mixture, comprising reverse-transcribed cDNA, primers, and SYBR Green, was prepared and assayed in triplicate. Expression was normalized to that of GAPDH. Relative gene expression was calculated using the 2−ΔΔCt method. The primer sequences are described in Supplementary Table 1.
Bisulfite sequencing PCR
Genomic DNA was extracted using the Genomic DNA Extraction Kit (TAKARA, 9765). DNA samples were pretreated with the EpiTect Bisulfite Kit (QIAGEN, 59041) according to the manufacturer’s instructions. Primers were designed using Methprimer to amplify the promoter region of the target gene. The PCR products were purified using the Agarose Gel DNA Purification Kit (DP219). The purified PCR products were then cloned into the pUC18 cloning vector, and 10 clones were randomly selected for Sanger sequencing. All Methprimers are listed in Supplementary Table 2.
Western blotting
Western blot was conducted, as reported previously by Li et al. [28]. The primary antibodies NG2 (1:1000, Abcam ab275024), PDGFRA (1:1000, Abcam ab203491), TET3 (1:1000, GeneTex gtx121453), mouse anti-DNMT1 (1:1000, Abcam ab13537), DNMT3A (1:1000, Proteintech 20954-1-ap), GAPDH (1:1000, Abcam ab8245) were used to identify the target proteins.
Dot blotting
To investigate the changes in methylation levels during the differentiation of HUCMSCs into induced OPCs (iOPCs) under the influence of TET3, Dot blot analysis was conducted on TET3-OE collected on days 1, 3, 5, and 7 days post induction (dpi), as well as on the Control group before and after the 7-day induction period, following the method described by Zhang et al. [23]. Primary antibodies against 5hmC (1:10,000; active motif 39,769) and 5mC (1:1000; Epigentek A10141) were used for the assay.
Immunocytochemistry staining
The immunocytochemistry assay was conducted under the protocol described by Zhang et al. [23]. Primary antibodies NG2 (1:200, Abcam ab275024), PDGFRA (1:400, Abcam ab203491), A2B5 (1:200, Abcam ab53521), and anti-5hmC (1:500, active motif 39769) and secondary antibodies Alexa 488 (1:800, Abcam ab150113) and Alexa 594 (1:800, Abcam ab150080) were used for the assay. The prepared samples were observed under an immunofluorescence microscope (CKX41-A329H, Olympus) or a confocal microscope (LSM 510 Meta, Carl Zeiss, Germany) for detailed examination.
RNA sequencing
Cells from the TET3-OE and Control groups 7 days after induction (7 dpi) were sent to Biomarker Technologies (Beijing, China) for library construction and sequencing. Libraries were sequenced with 2 × 150 bp paired-end reads on the Illumina platform. Raw data were filtered to remove adapter and low-quality reads. High-quality reads were aligned to the reference genome using HISAT2 with default parameters. Transcript and gene expression levels were quantified and normalized by FPKM using StringTie. Differential expression was determined using fold change (FC) ≥ 1.5 and p values < 0.05.
Establishment of a rat SCI model
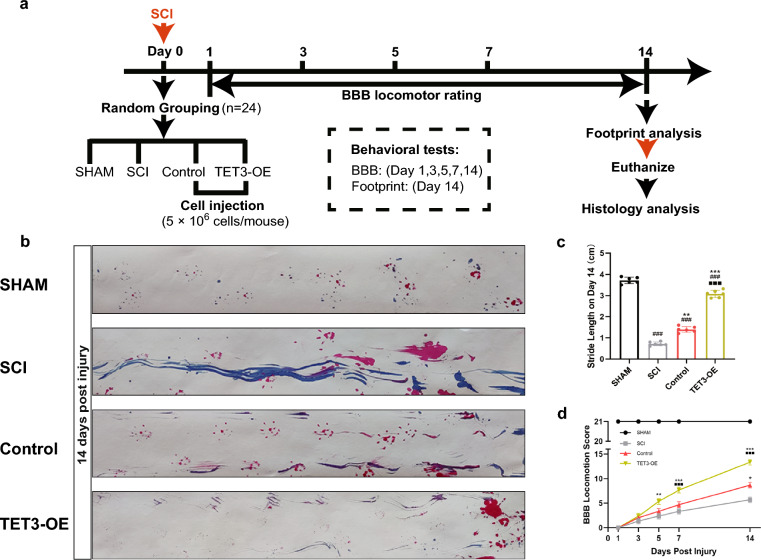
All animal experiments, including surgical interventions and postoperative care, were conducted following Animal Welfare Act regulations and approved by the Animal Care and Use Committee of The First Hospital Affiliated to Harbin Medical University (No. 2020052). Eight-week-old female Sprague–Dawley rats (n = 24) weighing 250–300 g were obtained from the Experimental Animal Technology Center of the Second Affiliated Hospital of Harbin Medical University, Harbin, China. Animals were randomly divided into four groups: SCI, Sham, Control, and TET3-OE. An SCI model was established using Allen’s method [29]. Cells for transplantation were immediately injected intrathecally into the site of the SCI using a microsyringe after the surgery, with each animal receiving an injection of 5 × 106 cells, in a total volume of 40 μl. The SCI group underwent the modeling procedure without cell injection. The TET3-OE group received HUCMSCs overexpressing TET3, which were transfected with TET3 adenoviruses 24 h prior to transplantation. The Control group were injected with HUCMSCs with control adenovirus, which were transfected with Control adenoviruses 24 h prior to transplantation. The Sham group underwent similar surgical procedures without a weight drop. The Basso, Beattie, and Bresnahan (BBB) locomotor rating scale was used to assess the recovery of locomotor function in rats on the 1st, 3rd, 5th, 7th, and 14th days post-surgery. On the 14th day post-surgery, following behavioral experiments (BBB locomotor rating scale and footprint analysis), the rats were euthanized by overdose of anesthesia, and spinal cord tissue samples were prepared for HE, Nissl, and immunofluorescence staining. The detailed protocols of the behavioral experiments and the histological and immunofluorescence staining are described in the Supplementary Material 1.
Statistical analysis
All experiments were replicated three or more times and analyzed using SPSS 19.0. One-way analysis of variance was employed for comparisons among three or more groups followed by Tukey’s test, while comparisons between two groups were assessed using unpaired t test. For BBB scores, two-way ANOVA with repeated measures was used followed by Sidak’s test. Significance was defined as p < 0.05.
Results
Characterization of HUCMSCs and establishment of the TET3-OE cell model
The HUCMSCs exhibited a typical spindle-shaped morphology and whirlpool-like growth patterns, stable across multiple passages (Figure S1a). Flow cytometry analysis confirmed the positive expression of CD73 and CD105, while IgG1, IgG2a, CD45, and HLA-DR were negative expressed, consistent with the characteristics of HUCMSCs (Figure S1b). Multilineage differentiation assays demonstrated the potential of HUCMSCs to differentiate into osteocytes and adipocytes, validated by Alizarin Red and Oil Red O staining, respectively (Figure S1c, d).
To establish the TET3 overexpressing model, adenoviral transfection was optimized, with an MOI of 100 selected for achieving high TET3 expression while maintaining cell viability (Figure S1e). Quantitative PCR (qPCR) and Western blot analyses confirmed significant upregulation of TET3 at both the transcriptional and protein levels (Fig. 1b–d). These results demonstrate the successful construction of the TET3-OE cell model. The HUCMSCs with confirmed high expression of TET3 were designated as the TET3-OE group, and those with control adenovirus (GV269) were designated as the Control group.
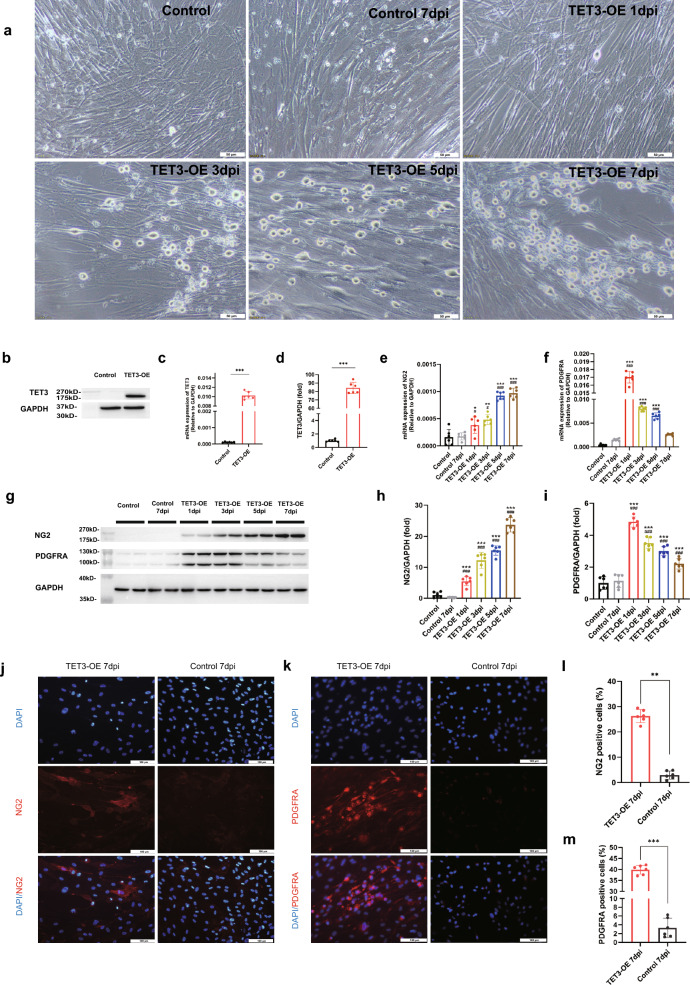
Fig. 1.
Neural induction transforming TET3 overexpression (TET3-OE) HUCMSCs into iOPCs. a TET3-OE exhibiting morphological transformation resembling an OPC-like morphology and the Control group showing no change (scale bar = 50 μm). b TET3 gene expression following transfection (n = 6 samples), as detected by qPCR. c, d TET3 expression following transfection detected by western blot (n = 6 samples). Data are shown as mean ± standard deviation. statistical significance was determined using unpaired t-test (***p < 0.001). e, f TET3-OE significantly expressing OPC-specific markers NG2 and PDGFRA, as detected by qPCR (n = 6 samples), Statistical analysis was performed using one-way ANOVA with Tukey’s HSD post-hoc test. *p < 0.05, **p < 0.01,***p < 0.001 vs Control group. #p < 0.05, ###p < 0.001 vs Control group 7 dpi. g–i TET3-OE significantly expressing OPC-specific markers NG2 and PDGFRA post-induction, as observed by western blot (n = 6 samples). j, l Immunofluorescence staining showing a significantly increased number of NG2-positive cells in TET3-OE 7 days post-induction (7 dpi) compared to that in the Control group 7 dpi (n = 6 samples). k, m Immunofluorescence staining showing a significantly increased number of PDGFRA-positive cells in TET3-OE 7 dpi- compared to that in the Control group 7 dpi (n = 6 samples, Scale bar = 100 μm). Data were shown as mean ± standard deviation. Statistical analysis was performed using one-way ANOVA with Tukey’s HSD post-hoc test. *p < 0.05, **p < 0.01,***p < 0.001 vs Control group. #p < 0.05, ##p < 0.01, ###p < 0.001 vs Control group 7 dpi
Transition of HUCMSCs overexpressing TET3 into OPCs under neural induction
By the third day of induction, TET3-OE cells exhibited a transition from spindle-shaped to OPC-like morphology, characterized by bipolar structures and retracted cytoplasm. By day 7, the cells gradually displayed a spherical body with multiple elongated projections, resembling OPCs. In contrast, the Control group maintained the retained their spindle-shaped morphology throughout the induction period (Fig. 1a).
To evaluate the ability of TET3 to induce HUCMSCs into OPCs, we assessed the expression of OPC-specific markers NG2, PDGFRA, and A2B5. NG2, a transmembrane protein abundantly expressed in OPCs, is critical for cell migration and proliferation. PDGFRA, a hallmark receptor of OPCs, regulates proliferation and sustains OPC activity and differentiation potential [30, 31]. A2B5, an antibody that specifically recognizes gangliosides on OPC surfaces, is commonly used to identify early oligodendrocyte differentiation [32].
The qPCR revealed that the expression of NG2 (Fig. 1e) and PDGFRA (Fig. 1f) was significantly upregulated following the initiation of induction. specifically, NG2 expression progressively increased over the course of induction, Reaching to its highest level on day 7. whereas PDGFRA maintained a high level at the onset of induction but gradually decreased during the process. These findings were consistent with the western blot results, suggesting a phenotypic shift toward OPC in TET3-OE (Fig. 1g–i, Figure S2a-c).
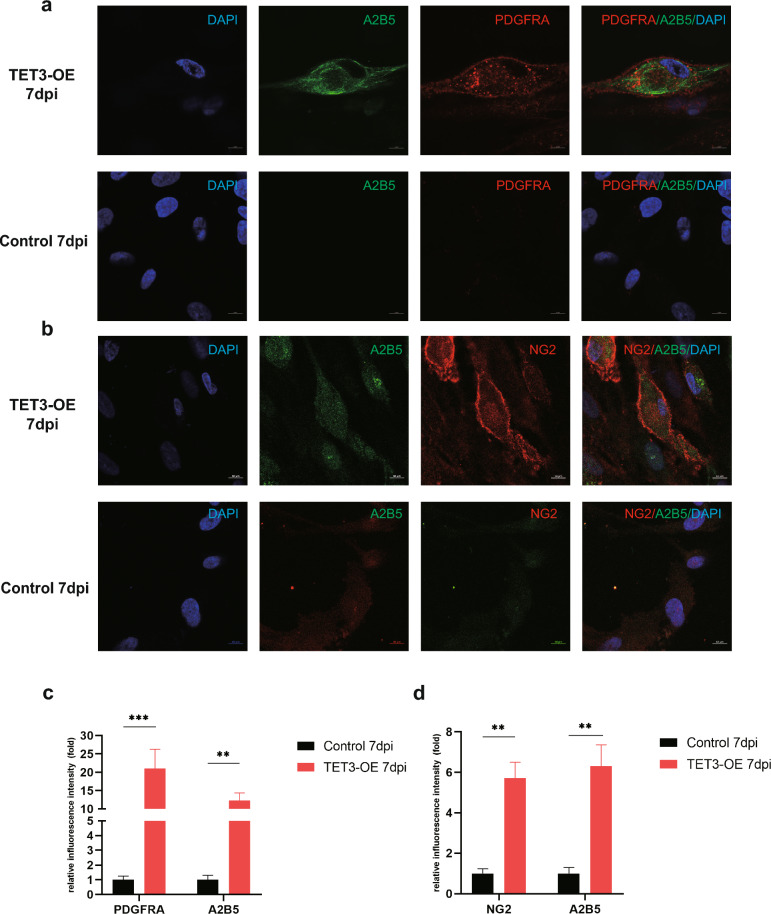
Immunofluorescence confirmed significantly higher positive rates for NG2 and PDGFRA in the TET3-OE 7 dpi group than in the Control 7 dpi group. validating TET3-induced iOPC differentiation (Fig. 1j–m). To further confirm the transformation of TET3-OE cells into iOPCs, we observed cells double-stained with NG2/A2B5 (Fig. 2b, d) and PDGFRA/A2B5 (Fig. 2a, c) using confocal microscopy. After 7 dpi, TET3-OE revealed bright NG2 signals distributed along the cell periphery and localized to the cell membrane. PDGFRA also showed bright signals along the cell periphery and localized to the cell membrane, along with scattered vesicle-like signals in the cytoplasm. A2B5 was observed as a bright signal on the cell membrane. These results indicated that the expression of NG2, PDGFRA, and A2B5 in iOPCs was significantly upregulated and was consistent with the localization characteristics of OPCs.
Fig. 2.
Confocal imaging of iOPCs. a, b At 7 dpi, the expression patterns of NG2, PDGFRA, and A2B5 in TET3-OE display localization resembling that of OPCs. c, d The expression levels of NG2, PDGFRA, and A2B5 in TET3-OE are significantly higher than those in Control (n = 6 samples, scale bar = 10 μm). Data were shown as mean ± standard deviation. Statistical analysis was performed unpaired t-test, **p < 0.01, ***p < 0.001
Changes in methylation patterns during differentiation
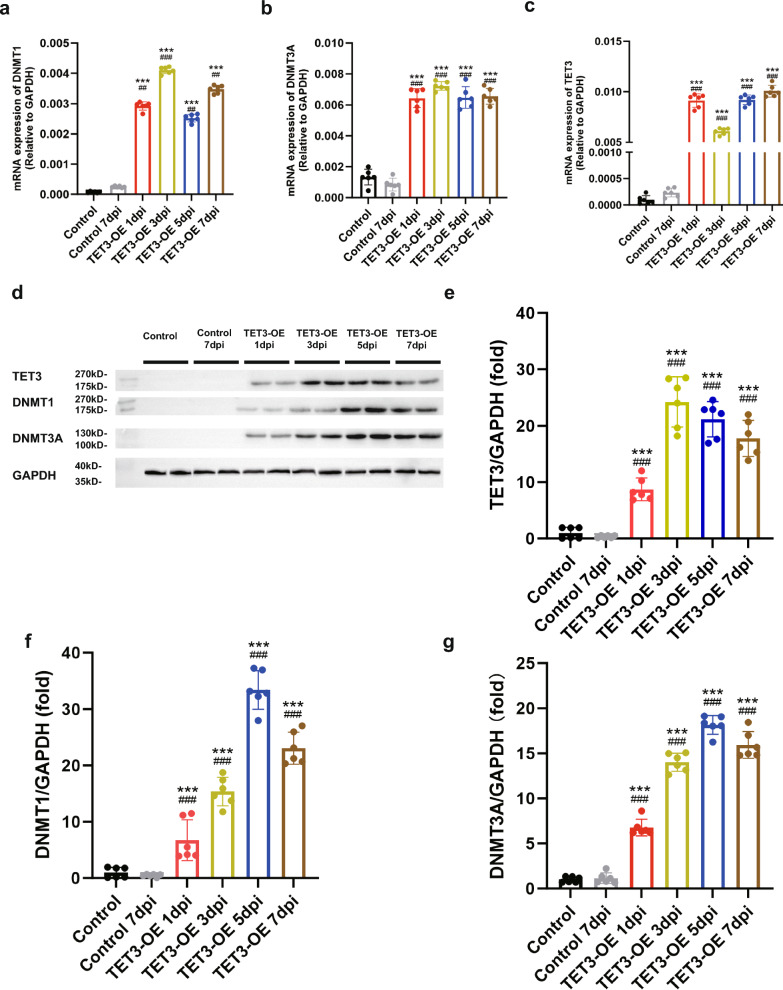
To elucidate the changes in methylation patterns during differentiation, we analyzed the expression of DNA methyltransferases (DNMT1, DNMT3A) and TET3. As the induction period progressed, the expression levels of DNMT1 and DNMT3A gradually increased (Fig. 3a, b, d, f, g). TET3 remained highly expressed throughout the induction period (Fig. 3c, d, e), suggesting a shift in methylation and hydroxymethylation patterns during the differentiation of TET3-OE cells.
Fig. 3.
Changes in methylation regulatory patterns during differentiation. a–c mRNA expression of methylation regulatory enzymes (a) DNMT1, (b) DNMT3A, and (c) TET3 during differentiation (n = 6 samples). d–g Expression of corresponding (d) methylation regulatory proteins and their (e–g) quantification (e) TET3, (f) DNMT1, (g) DNMT3A, (n = 6 samples). Data were shown as mean ± standard deviation. Statistical analysis was performed using one-way ANOVA with Tukey’s HSD post-hoc test. ***p < 0.001 vs Control group. ##p < 0.01, ###p < 0.001 vs Control 7dpi group
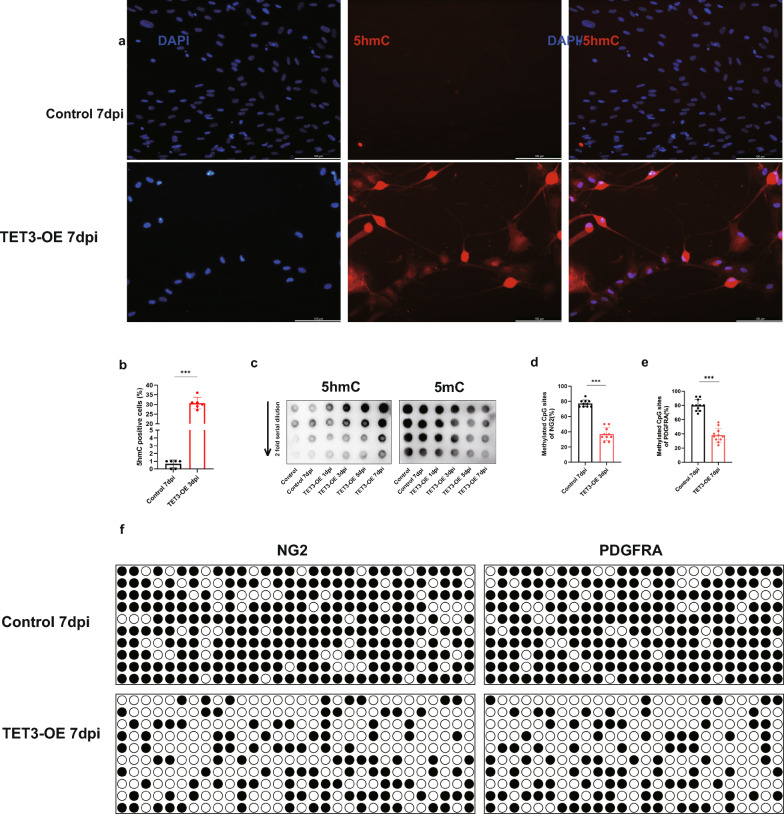
To directly observe the relationship between 5hmC and iOPCs differentiation, we performed immunofluorescence staining to detect 5hmC in TET3-OE 7 dpi. 5hmC levels significantly increased in the OPC-like cells of the TET3-OE group. This indicates that TET3 and its product 5hmC play crucial roles in the differentiation of HUCMSCs into iOPCs (Fig. 4a, b).
Fig. 4.
5hmC plays a crucial role in the differentiation of TET3-OE HUCMSCs to iOPCs. a, b Immunofluorescence staining for 5hmC in TET3-OE 7dpi and Control 7dpi (scale bar = 100 μm, n = 6 samples). Data were shown as mean ± standard deviation. Statistical significance was determined using unpaired t-test, ***p < 0.001.c Dynamic monitoring of 5hmC and 5mC changes during the differentiation process via dot blot. d, e Quantification of methylation levels in NG2 (d) and PDGFRA (e) promoters. Data were shown as mean ± standard deviation. Statistical significance was determined using unpaired t-test, ***p < 0.001. f Bisulfite Sequencing PCR analysis of NG2 and PDGFRA promoter regions
Dynamic changes in methylation levels during neural differentiation were further evaluated using dot blot analyses at different induction time points (1, 3, 5, and 7 dpi). A progressive increase in 5hmC levels, coupled with a gradual decrease in 5mC levels, was observed in the TET3-OE group, indicating that TET3 effectively facilitates demethylation by converting 5mC to 5hmC during differentiation (Fig. 4c).
To further investigate the role of TET3-regulated methylation changes during differentiation, we analyzed the methylation levels of CpG islands in the promoter regions of NG2 and PDGFRA using BSPCR. The results showed a significant reduction in CpG island methylation levels in the TET3-OE group compared to the Control group at 7 dpi (Fig. 4d, e, f). These findings further confirm the pivotal role of TET3 in promoting the differentiation of HUCMSCs into OPCs.
Transcriptomic insights into TET3-mediated differentiation of HUCMSCs into OPCs
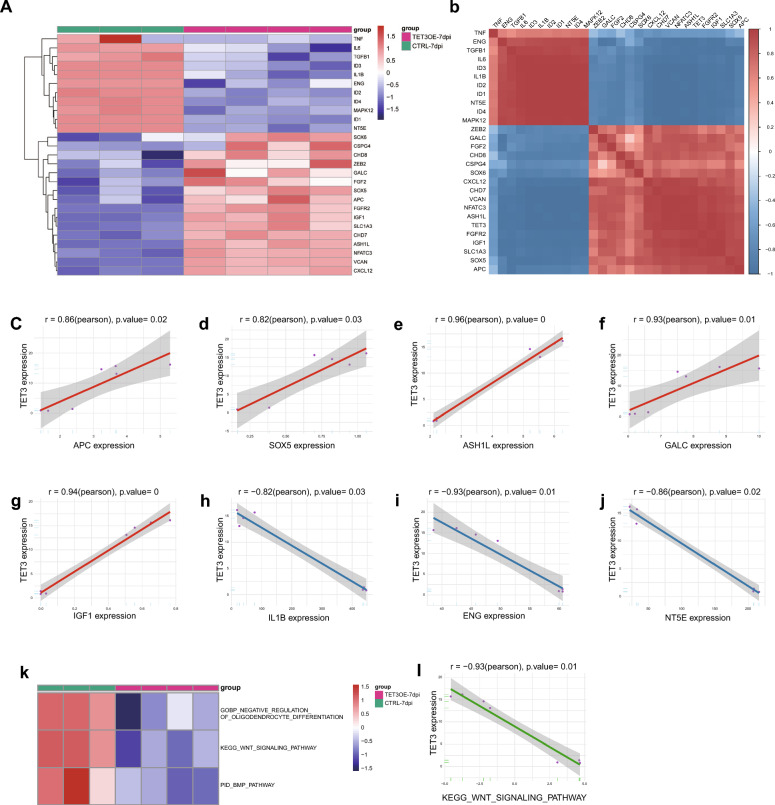
Based on a review of literatures, we have identified a series of genes relatied to the formation and differentiation of OPCs, as summarized in supplementary Table 3. RNA sequencing results suggest that TET3 overexpression significantly upregulates genes that are potentially critical for the development of the oligodendrocyte lineage, Concurrently, TET3 downregulates genes known to inhibit OPC formation and myelination, as well as certain MSC markers, reflecting a possible lineage transition from HUCMSCs to OPCs (Fig. 6a). To further investigate the potential relationship between TET3 and OPC formation, we selected a set of genes previously reported to play pivotal roles in OPC biology and analyzed their correlation with TET3 expression. The results revealed strong positive correlations with genes promoting OPC formation and differentiation (APC, SOX5, ASH1L, GALC, IGF1) (Fig. 5c–g), while showing a significant negative correlation with genes (IL1B), as well as MSC markers (ENG, NT5E) (Fig. 5h–j). These data suggest that HUCMSCs might undergo a lineage transition toward OPCs under the regulatory influence of TET3.
Fig. 6.
SCI model rat recovery improved when treated with TET3-OE. a Experimental timeline for the rat spinal cord injury (SCI) model. Eight-week-old female Sprague–Dawley rats (n = 24; 250–300 g) were randomly divided into four groups: SCI, Sham, Control, and TET3-OE. SCI was induced using Allen’s method on Day 0. Intrathecal injections (5 × 106 cells) were administered immediately after surgery. Behavioral assessments using the Basso, Beattie, and Bresnahan (BBB) locomotor rating scale were performed on Days 1, 3, 5, 7, and 14 post-surgery. On Day 14, footprint analysis was conducted, followed by euthanasia. Spinal cord tissues were collected for histology analysis. b, c Footprint analysis on Day 14 post injury showed that the TET3-OE group exhibited the most significant improvement (n = 6 rats). Representative images for each group were randomly chosen, avoiding selection of the most severe or mild cases. Data were shown as mean ± standard deviation, statistical analysis was performed using one-way ANOVA with Tukey’s HSD post-hoc test. **p < 0.01, ***p < 0.001 vs SCI group, ###p < 0.001 vs sham group, ■■■p < 0.001 vs TET3-OE group. d The Basso, Beattie, and Bresnahan score showed significant locomotion function recovery in the TET3-OE group (n = 6 rats). Data were shown as mean ± standard deviation. Statistical analysis was performed using two-way ANOVA with repeated measures, followed by Sidak’s test. *p < 0.05, **p < 0.01, ***p < 0.001 vs SCI group, ###p < 0.001 vs sham group, ■■■p < 0.001 vs Control group
Fig. 5.
Transcriptome analysis of the role of TET3 in HUCMSC differentiation towards oligodendrocyte progenitor cells (OPCs). a Heatmap showing upregulation of genes promoting oligodendrocyte lineage formation and differentiation and downregulation of inhibitory genes. b Correlation analysis between TET3 expression and multiple genes related to the oligodendrocyte lineage. c–g Positive correlation between TET3 and (c) APC, (d) SOX5, (e) ASH1L, (f) GALC, and (g) IGF1. h–j Negative correlation between TET3 and (h) IL1B, (i) ENG (CD105), and (j) NT5E (CD73). k Suppression of pathways inhibiting oligodendrocyte lineage formation during differentiation. l Negative correlation between TET3 and the Wnt signaling pathway
During differentiation, several pathways inhibiting OL lineage formation appeared to be suppressed (Fig. 5k). Notably, the Wnt signaling pathway was significantly downregulated, negatively correlated with TET3 expression, this observation suggests that TET3 may facilitate neural differentiation by mitigating Wnt-mediated inhibitory signals (Fig. 5l).
Improved recovery of SCI rats treated with TET3-OE
To assess hind limb motor recovery in rats after SCI, footprint analysis was performed on day 14 post-surgery. The Sham group exhibited normal gaits. Among the other groups, the TET3-OE treatment group showed the greatest improvement, with a significant increase in stride length and distinct hind limb separation. The Control group showed slight improvement compared to the SCI group (Fig. 6b, c).
We also evaluated BBB scores at various time points before and after the development of SCI in rat models. The time points assessed were 1 day before surgery and 1, 3, 5, 7, and 14 days post-surgery. Both treatment groups (Control and TET3-OE) showed significant motor function recovery compared to the SCI group, with the TET3-OE group displaying markedly better outcomes than the Control group. However, a significant difference remained between the treatment and Sham groups (Fig. 6d).
Pathological and morphological changes in SCI rats treated with TET3-OE HUCMSCs
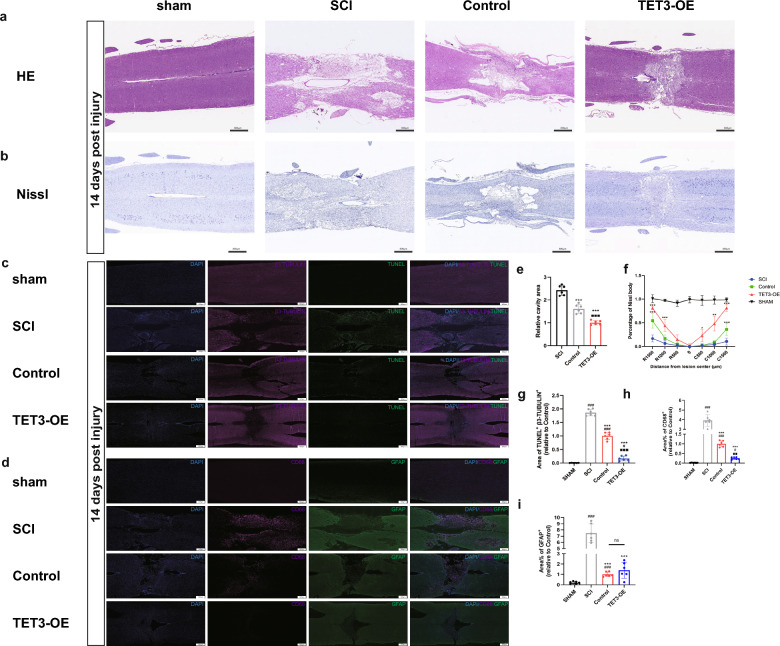
HE staining revealed the structure of spinal cord tissue. The Sham group showed a normal structure with intact cellular morphology. The SCI group exhibited substantial structural disruption and a large spinal cavity. Between the treatment groups (TET3-OE group, Control group), the TET3-OE group demonstrated the smallest spinal cavity area and the most well-preserved tissue architecture compared to the Control and SCI groups, suggesting reduced tissue damage and potential enhanced repair in the TET3-OE group (Fig. 7a, e).
Fig. 7.
Morphological and Inflammatory Changes in SCI Rats. a Morphological changes of Hematoxylin and eosin (HE) staining at 14 days post injury (Scale bar = 800 μm, n = 6 rats). Representative images for each group were randomly chosen, avoiding selection of the most severe or mild cases. b Distribution of neurons by Nissl staining at 14 days post injury (Scale bar = 800 μm, n = 6 rats). Representative images for each group were randomly chosen, avoiding selection of the most severe or mild cases. c Double immunostaining of TUNEL and β3-TUBULIN at 14 days post injury (Scale bar = 500 μm, n = 6 rats). d Double staining of GFAP and CD68 at 14 days post injury (Scale bar = 500 μm, n = 6 rats). e Quantitative analysis of spinal cavity area in HE staining. f Quantitative analysis of Nissl at different distances from the injury center. g Quantitative analysis of TUNEL and β3-TUBULIN double staining cells showed reduced neuronal apoptosis in the TET3-OE group. h Quantitative analysis of CD68 showed fewest macrophages infiltration in the TET3-OE group. i Quantitative analysis of GFAP demonstrates reduced astrocyte reactivity in the TET3-OE group and Control group. Data were presented as mean ± standard deviation. Statistical analysis was performed using one-way ANOVA with Tukey’s HSD post-hoc test. *p < 0.05, **p < 0.01, ***p < 0.001 vs SCI group, #p < 0.05, ###p < 0.001 vs sham group, ■■p < 0.01, ■■■p < 0.001 vs TET3-OE group.ns, p > 0.05 between TET3-OE group and Control group
To evaluate neuronal distribution and morphology, we performed Nissl staining. The Sham group exhibited typical neuronal structure with intact and densely packed Nissl bodies, while the SCI group showed a reduced and uneven distribution of Nissl bodies, indicative of neural damage. In the TET3-OE group, the distribution of Nissl bodies was more consistent, and a relatively higher density was observed compared to the SCI group, reflecting differences in neuronal morphology and distribution (Fig. 7b, f).
To assess neuronal survival, we conducted double staining of TUNEL and β3-TUBULIN to evaluate neuronal apoptosis at the injury site. The TET3-OE group exhibited fewer TUNEL/β3-TUBULIN double-positive cells cells compared to the Control group and SCI group, suggesting a reduction in neuronal apoptosis. This finding, when considered alongside the observed Nissl body distribution, indicates a potential preservation effect of TET3-OE on neuronal cells (Fig. 7c, g).
To investigate the inflammatory response and astrogliosis following spinal cord injury, double staining of GFAP and CD68 was performed. CD68 staining, marking macrophages, revealed that the TET3-OE group had the fewest macrophages infiltration compared to the SCI and Control groups, indicating a reduced inflammatory response in the TET3-OE group (Fig. 7d, h). Meanwhile, GFAP staining was used to assess astrocytic reactivity and astrogliosis. Compared to the SCI group, the TET3-OE group and Control group had fewer astrocytes in the injury area. This may indicate a controlled astrocytic response, which could contribute to minimizing spinal cavity formation. However, no significant differences were found between the TET3-OE group and Control group (Fig. 7d, i).
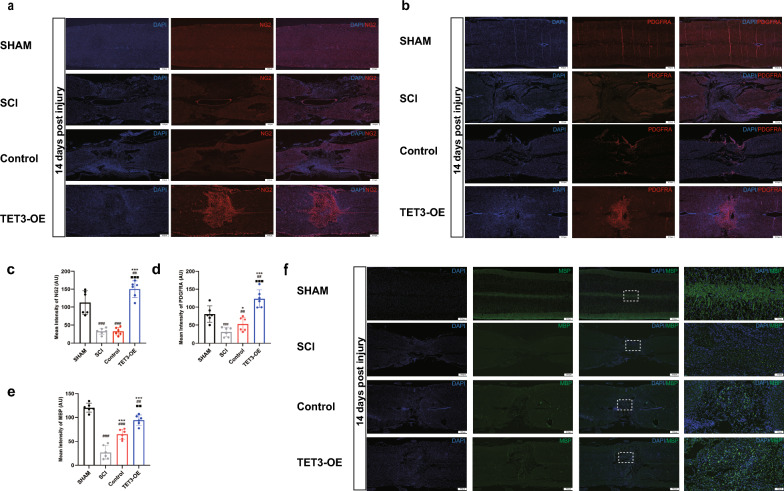
A high number of NG2 (+) (Fig. 8a, c) and PDGFRA (+) (Fig. 8b, d) cells were detected in the injured areas treated with TET3-OE, suggesting that TET3-OE cells can differentiate into OPCs in vivo. Moreover, Immunofluorescence staining for MBP demonstrated significantly elevated MBP expression in the TET3-OE group, indicating active myelination (Fig. 8e, f). These findings suggest that TET3-OE cells not only differentiate into OPCs but may also facilitate myelin regeneration in the injured spinal cord.
Fig. 8.
OPC formation and myelination in SCI rats. a–d Immunofluorescence assay indicated that TET3-OE can express the OPC-specific markers NG2 (a, c) and PDGFRA (b, d) in vivo at 14 days post injury. e, f MBP immunofluorescence staining reveals significantly higher MBP expression in the TET3-OE group. Data are shown as mean ± standard deviation. n = 6 rats, scale bar = 500 μm. Statistical analysis was performed using one-way ANOVA with Tukey’s HSD post-hoc test. *p < 0.05, ***p < 0.001 vs SCI group, ##p < 0.01, ###p < 0.001 vs sham group, ■■p < 0.01, ■■■p < 0.001 vs TET3-OE group
Discussion
Our study highlights the therapeutic potential of TET3-mediated reprogramming of HUCMSCs into OPCs, offering a novel pathway for enhancing the regenerative capacity of these cells for treating SCI.
Our findings reveal that the overexpression of TET3 not only facilitates the differentiation of HUCMSCs into iOPCs but also enhances motor function recovery post-SCI. This efficacy is underpinned by a pivotal shift in epigenetic regulation marked by the conversion of 5mC to 5hmC [33, 34], Previous research has established that TET3 plays a vital role in growth and development, particularly in the CNS, where it is essential for maintaining and advancing neural differentiation [19, 35]. TET3 is expressed in both neurons and oligodendrocytes [36], several studies have highlighted its critical role in regulating neuronal development and function [23, 35, 36], However, while TET3 has been implicated in oligodendrocyte formation, its specific role during the early stages of oligodendrocyte development remains poorly understood. Our research addresses this gap by exploring the role of TET3 in oligodendrocyte formation, building on prior findings. We highlight the potential of TET3 in regenerative medicine applications involving adult stem cells and propose a novel therapeutic strategy for spinal cord injury based on TET3-mediated epigenetic regulation.
Our study also highlights the advantages of the one-step TET3-mediated induction method over traditional MSC neural induction techniques. Through overexpressing TET3 in HUCMSCs, we induced considerable morphological changes during neural induction, triggering the transformation of these cells into iOPCs within 1 week, far surpassing the several weeks to months required by conventional methods [26, 37, 38]. Notably, during induction, PDGFRα and NG2 expression exhibited a specific temporal pattern, with PDGFRα significantly increasing upon induction but decreasing as NG2 expression gradually increased, reaching its peak on the 7th day (Fig. 3b–d). PDGFRα, a specific receptor on the surface of OPCs, binds with its ligand PDGF and supports OPC formation, promoting cell proliferation and survival, while its decreasing expression indicates a transition from proliferation to maturation. Subsequently, NG2 expression facilitates OPC migration and positionin[39, 40]. This sequential expression reflects the dynamic transition of cells from a proliferative state to a specific cellular fate, highlighting the role of TET3 in gene expression modulation during HUCMSC differentiation into OPCs.
Confocal microscopy observations of NG2 (+)/A2B5 (+) and PDGFRA (+)/A2B5 (+) double-stained OPC-like cells, with NG2, PDGFRA, and A2B5 localization consistent with OPCs [41–43], strongly supported the results that HUCMSCs transform into OPCs. Our findings, validated through morphological changes, nucleic acid and protein expression, and specific marker localization, further confirm successful cellular differentiation and the functional potential of the differentiated cells.
Our study reveals a dynamic methylation modification phenomenon during cellular differentiation in that 5hmC levels gradually increased while 5mC levels correspondingly decreased, indicating a continuous transformation facilitated by TET3. Immunofluorescence analysis after 7 days of induction revealed 5hmC predominantly expressed in cells resembling OPCs, suggesting that TET3-OE drives the elevation of 5hmC levels during OPC differentiation [44]. Surprisingly, DNMT1 and DNMT3A expression levels did not decrease but were co-upregulated with TET3, potentially reflecting the intricate nature of epigenetic modifications during neural differentiation. Previous studies confirmed the critical roles of DNMT1 and DNMT3A in the differentiation of OPCs to OLs, verifying that the epigenetic pattern of TET3-OE cells involves transitioning toward OLs [45, 46]. Moreover, the co-upregulation may reflect cell adjustment to maintain epigenetic homeostasis during differentiation, aiming for precise cell fate determination. Recent research reported that TET3 regulates DNMT1 expression by binding to its promoter, effectively suppressing mesodermal genes and ensuring precise control of neural differentiation. This regulation aligns with the observed HUCMSCs-to-OPCs transition upon TET3 overexpression in this study [47].
Additionally, our BSPCR results corroborate these findings, providing direct evidence of TET3’s role in modifying specific gene promoter regions, By reducing methylation in these regions, TET3 effectively activates NG2 and PDGFRA expression, which are pivotal for OPC differentiation. The upregulation of NG2 and PDGFRA underscores TET3’s ability to regulate the differentiation of HUCMSCs into OPCs by altering their methylation landscapes. Previous studies have shown that TET3-mediated demethylation is a critical mechanism for activating genes associated with cellular differentiation [35, 47, 48], Our findings build upon this understanding by suggesting that TET3 may play a pivotal role in guiding HUCMSCs toward OPC differentiation, potentially shedding light on its regulatory influence in oligodendrocyte development. This insight offers a deeper perspective on how TET3-driven epigenetic mechanisms could contribute to neural differentiation.
RNA sequencing revealed that high TET3 expression significantly upregulated APC, SOX5, ASH1L, and GALC, which are reported to be highly expressed in OPCs according to existing literature [49–52], while downregulating the HUCMSC-specific markers ENG (CD105) and NT5E (CD73) [52]. TET3 expression showed a positive correlation with APC, SOX5, ASH1L, and GALC and negative correlation with ENG and NT5E. These findings suggest that TET3 may play a role in accelerating the neural differentiation of HUCMSCs into OPCs and potentially promoting their further maturation.
Key genes such as APC, SOX5, ASH1L, and GALC were highly expressed in the oligodendroglial lineage, showing significant temporal regulation. APC and SOX5 are expressed early in OPCs, with APC modulating OPC differentiation and proliferation via the Wnt/β-catenin signaling pathway [49]. SOX5 potentially enhances proliferation, migration, and initial differentiation [50]. while the expression of ASH1L implicated in the transition to OLs [50]. while GALC, initially expressed at low levels, peaks during myelination to support myelin synthesis and function [51]. These findings suggest that the temporal dynamics of these genes, modulated by TET3, could reflect the progression of HUCMSCs toward an oligodendroglial fate. TET3-mediated gene regulation accelerates HUCMSCs to iOPC conversion by modulating OPC differentiation-related genes and relieving the Wnt pathway inhibitory effect on OPC differentiation. Additionally, the role of TET3 in environmental regulation was evident in the downregulation of pro-inflammatory cytokines, such as IL1B, TNF, and IL6 [53–55], along with the upregulation of factors that promote OPC formation and differentiation, such as IGF1, FGF2, and CXCL12 [56–58]. TET3 might create a more supportive milieu for OPC survival and maturation.
Consistent with the RNA sequencing results, the improved functional recovery and histological outcomes observed in our study suggest that TET3-OE HUCMSCs not only differentiate into OPCs but also but may also further mature into MBP (+) oligodendrocytes with myelination potential. These findings support the burgeoning evidence that cell transplantation, coupled with targeted methylation modification, can enhance repair mechanisms within the injured spinal cord, potentially offering a dual modality for SCI treatment that leverages both cellular replacement and epigenetic reconditioning.
Numerous studies have demonstrated that transplanting exogenous stem cells into the SCI site effectively mitigates secondary injury and promotes tissue repair. For instance, implanting MSCs has been shown to significantly reduce glial scar formation and cavity development [59–62]. Similarly, Lai et al. reported that transplanting spinal cord-like tissue into the injury site attenuated inflammation, reduced scar formation, and facilitated functional recovery of the spinal cord [63]. Our findings are consistent with these observations. In our in vivo experiments, we noted that compared to the SCI group, both the TET3-OE and control HUCMSC groups exhibited significantly reduced astrocyte proliferation at the injury site. However, there was no significant difference in astrocyte content between the TET3-OE and control groups, suggesting that the suppression of astrocyte proliferation may not be directly mediated by TET3 but rather stem from the inherent immunomodulatory properties of HUCMSCs.
Furthermore, TET3-OE HUCMSCs appeared to improve the microenvironment of the injury site by suppressing inflammation and reducing inflammatory infiltration. Compared to other groups, the TET3-OE group exhibited markedly lower levels of inflammation and neuronal apoptosis in the injury region. This indicates that TET3, through its role in regulating DNA methylation, contributes to creating an environment more conducive to spinal cord repair.
In our study, we successfully achieved one-step differentiation of HUCMSCs into OPCs through TET3 overexpression, significantly reducing the induction time required by traditional methods. By integrating molecular, epigenetic, and animal model analyses, it provides robust evidence for the role of TET3 in promoting OPC differentiation and its therapeutic potential in SCI. However, there are still some limitations. First, BSPCR analysis verified TET3’s regulatory effects on the methylation status of promoter regions of key OPC-related genes (NG2 and PDGFRA), However, this method focuses on specific gene promoter regions and does not comprehensively cover all potentially involved genes and genomic regions. Future studies could employ high-throughput techniques such as WGBS to further expand our understanding of TET3’s epigenetic regulatory mechanisms and its role in OPC differentiation. Second, this study primarily utilized a rat spinal cord contusion model, which, while informative, does not fully replicate the complex pathological characteristics of human SCI. To address this, Further validation in larger animal models and long-term follow-up studies will be essential to more comprehensively evaluate the therapeutic potential of TET3 and to ensure the safety and efficacy of cell transplantation for future clinical applications. Third, while this study focused on the therapeutic potential of HUCMSCs derived OPCs in the SCI model, other cell sources, such as ESC- or iPSC-derived OPCs, are also considered to have promising potential. Future studies could build upon this work by conducting comparative analyses across various cell sources to further delineate the unique advantages of TET3-OE HUCMSCs and better define their potential role in clinical applications.
Conclusions
In conclusion, our study demonstrated that TET3-mediated demethylation reshapes the methylation patterns of HUCMSCs, enabling their efficient one-step conversion into OPCs and significantly reducing the time required for cell preparation. This approach offers a potential strategy for early intervention in SCI. In an SCI model, TET3-induced OPCs contributed to spinal cord repair, providing novel insights into cell therapy strategies for SCI through the lens of methylation regulation.
Supplementary Information
Acknowledgements
We would like to extend our sincere gratitude to Yanbo Teng for her timely assistance with RNA sequencing analyses.
Abbreviations
- SCI
Spinal cord injury
- OPCs
Oligodendrocyte precursor cells
- HUCMSCs
Human umbilical cord mesenchymal stem cells
- iOPCs
Induced OPCs
- OPCs
OL precursor cells
- 5mC
5-Methylcytosine
- 5hmC
5-Hydroxymethylcytosine
- Ig
Immunoglobulin
- TET3-OE
TET3 overexpression
- MOI
Multiplicity of infection
- qPCR
Quantitative polymerase chain reaction
- BBB
Basso, Beattie, and Bresnahan
Author contributions
YSW provided guidance for study design. YZ performed experiments, analyzed data, and drafted the manuscript. MG helped with the culture and characterization of HUCMSCs. ZP, YYW, ML, RL, and PL established the rat SCI model. JL and YL offered support during the statistical analysis. TW and YWZ helped with the BBB scores. All authors read and approved the final paper.
Funding
This study was financially supported by a Grant from the Natural Science Foundation of China (Project No. 81871781) and the Key Project of the Natural Science Foundation of Heilongjiang Province of China (Project No. ZD2021H003).
Availability of data and materials
The data that support the findings of this study are available from the corresponding author upon request.
Declarations
Ethics approval and consent to participate
All animal experiments, including surgical interventions and postoperative care, were conducted following Animal Welfare Act regulations and approved by the Animal Care and Use Committee of The First Affiliated Hospital of Harbin Medical University (No. 2020052).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.NSCIS Center. Spinal cord injury (SCI) 2016 facts and figures at a glance. J Spinal Cord Med. 2016;39:493–4. [DOI] [PMC free article] [PubMed]
- 2.Cripps RA, Lee BB, Wing P, Weerts E, Mackay J, Brown D. A global map for traumatic spinal cord injury epidemiology: towards a living data repository for injury prevention. Spinal Cord. 2011;49:493–501. [DOI] [PubMed] [Google Scholar]
- 3.Jin MC, Medress ZA, Azad TD, Doulames VM, Veeravagu A. Stem cell therapies for acute spinal cord injury in humans: a review. Neurosurg Focus. 2019;46:E10. [DOI] [PubMed] [Google Scholar]
- 4.Rolls A, Shechter R, London A, Segev Y, Jacob-Hirsch J, Amariglio N, et al. Two faces of chondroitin sulfate proteoglycan in spinal cord repair: a role in microglia/macrophage activation. PLoS Med. 2008;5: e171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alizadeh A, Dyck SM, Karimi-Abdolrezaee S. Myelin damage and repair in pathologic CNS: challenges and prospects. Front Mol Neurosci. 2015;8:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baydyuk M, Morrison VE, Gross PS, Huang JK. Extrinsic factors driving oligodendrocyte lineage cell progression in CNS development and injury. Neurochem Res. 2020;45:630–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mackay-Sim A, St John JA. Olfactory ensheathing cells from the nose: clinical application in human spinal cord injuries. Exp Neurol. 2011;229:174–80. [DOI] [PubMed] [Google Scholar]
- 8.Liau LL, Looi QH, Chia WC, Subramaniam T, Ng MH, Law JX. Treatment of spinal cord injury with mesenchymal stem cells. Cell Biosci. 2020;10:112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang Y, Jahagirdar BN, Reinhardt RL, Schwartz RE, Keene CD, Ortiz-Gonzalez XR, et al. Pluripotency of mesenchymal stem cells derived from adult marrow. Nature. 2002;418:41–9. [DOI] [PubMed] [Google Scholar]
- 10.Barzilay R, Ben-Zur T, Bulvik S, Melamed E, Offen D. Lentiviral delivery of LMX1a enhances dopaminergic phenotype in differentiated human bone marrow mesenchymal stem cells. Stem Cells Dev. 2009;18:591–601. [DOI] [PubMed] [Google Scholar]
- 11.Hachem LD, Ahuja CS, Fehlings MG. Assessment and management of acute spinal cord injury: from point of injury to rehabilitation. J Spinal Cord Med. 2017;40:665–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Santos AK, Gomes KN, Parreira RC, Scalzo S, Pinto MCX, Santiago HC, et al. Mouse neural stem cell differentiation and human adipose mesenchymal stem cell transdifferentiation into neuron- and oligodendrocyte-like cells with myelination potential. Stem Cell Rev Rep. 2022;18:732–51. [DOI] [PubMed] [Google Scholar]
- 13.Zhang HT, Fan J, Cai YQ, Zhao SJ, Xue S, Lin JH, et al. Human Wharton’s jelly cells can be induced to differentiate into growth factor-secreting oligodendrocyte progenitor-like cells. Differentiation. 2010;79:15–20. [DOI] [PubMed] [Google Scholar]
- 14.Mikaeili Agah E, Parivar K, Nabiuni M, Hashemi M, Soleimani M. Induction of human umbilical Wharton’s jelly-derived stem cells toward oligodendrocyte phenotype. J Mol Neurosci. 2013;51:328–36. [DOI] [PubMed] [Google Scholar]
- 15.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Szulwach KE, Li X, Li Y, Song CX, Wu H, Dai Q, et al. 5-hmC-mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nat Neurosci. 2011;14:1607–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mellén M, Ayata P, Dewell S, Kriaucionis S, Heintz N. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell. 2012;151:1417–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li T, Yang D, Li J, Tang Y, Yang J, Le W. Critical role of Tet3 in neural progenitor cell maintenance and terminal differentiation. Mol Neurobiol. 2015;51:142–54. [DOI] [PubMed] [Google Scholar]
- 20.Li X, Wei W, Zhao QY, Widagdo J, Baker-Andresen D, Flavell CR, et al. Neocortical Tet3-mediated accumulation of 5-hydroxymethylcytosine promotes rapid behavioral adaptation. Proc Natl Acad Sci USA. 2014;111:7120–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Colquitt BM, Allen WE, Barnea G, Lomvardas S. Alteration of genic 5-hydroxymethylcytosine patterning in olfactory neurons correlates with changes in gene expression and cell identity. Proc Natl Acad Sci USA. 2013;110:14682–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weng YL, An R, Cassin J, Joseph J, Mi R, Wang C, et al. An intrinsic epigenetic barrier for functional axon regeneration. Neuron. 2017;94:337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang J, Chen S, Zhang D, Shi Z, Li H, Zhao T, et al. Tet3-mediated DNA demethylation contributes to the direct conversion of fibroblast to functional neuron. Cell Rep. 2016;17:2326–39. [DOI] [PubMed] [Google Scholar]
- 24.Zhao X, Dai J, Ma Y, Mi Y, Cui D, Ju G, et al. Dynamics of ten-eleven translocation hydroxylase family proteins and 5-hydroxymethylcytosine in oligodendrocyte differentiation. Glia. 2014;62:914–26. [DOI] [PubMed] [Google Scholar]
- 25.Mukai T, Nagamura-Inoue T, Shimazu T, Mori Y, Takahashi A, Tsunoda H, et al. Neurosphere formation enhances the neurogenic differentiation potential and migratory ability of umbilical cord-mesenchymal stromal cells. Cytotherapy. 2016;18:229–41. [DOI] [PubMed] [Google Scholar]
- 26.Kim DS, Jung SJ, Lee JS, Lim BY, Kim HA, Yoo JE, et al. Rapid generation of OPC-like cells from human pluripotent stem cells for treating spinal cord injury. Exp Mol Med. 2017;49: e361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang J, Cheng X, Qi J, Xie B, Zhao X, Zheng K, et al. EGF enhances oligodendrogenesis from glial progenitor cells. Front Mol Neurosci. 2017;10:106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Z, Li B, Yu H, Wang P, Wang W, Hou P, et al. DNMT1-mediated epigenetic silencing of TRAF6 promotes prostate cancer tumorigenesis and metastasis by enhancing EZH2 stability. Oncogene. 2022;41:3991–4002. [DOI] [PubMed] [Google Scholar]
- 29.Li S, Zhou J, Zhang J, Wang D, Ma J. Construction of rat spinal cord injury model based on Allen’s animal model. Saudi J Biol Sci. 2019;26:2122–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang H, He W, Tang T, Qiu M. Immunological markers for central nervous system glia. Neurosci Bull. 2023;39:379–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Antel JP, Lin YH, Cui QL, Pernin F, Kennedy TE, Ludwin SK, et al. Immunology of oligodendrocyte precursor cells in vivo and in vitro. J Neuroimmunol. 2019;331:28–35. [DOI] [PubMed] [Google Scholar]
- 32.Zhou H, He Y, Wang Z, Wang Q, Hu C, Wang X, et al. Identifying the functions of two biomarkers in human oligodendrocyte progenitor cell development. J Transl Med. 2021;19:188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu X, Zhang Y. TET-mediated active DNA demethylation: mechanism, function and beyond. Nat Rev Genet. 2017;18:517–34. [DOI] [PubMed] [Google Scholar]
- 34.Feng LL, Liu RY, An K, Tang S, Wu J, Yang Q. TET3 as a non-invasive screening tool for the detection of fibrosis in patients with chronic liver disease. Sci Rep. 2023;13:6382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsukada YI, Akiyama T, Nakayama KI. Maternal TET3 is dispensable for embryonic development but is required for neonatal growth. Sci Rep. 2015;5:15876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Antunes C, Da Silva JD, Guerra-Gomes S, Alves ND, Ferreira F, Loureiro-Campos E, et al. Tet3 ablation in adult brain neurons increases anxiety-like behavior and regulates cognitive function in mice. Mol Psychiatry. 2021;26:1445–57. [DOI] [PubMed] [Google Scholar]
- 37.Goldman SA, Kuypers NJ. How to make an oligodendrocyte. Development. 2015;142:3983–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim HS, Kim JY, Song CL, Jeong JE, Cho YS. Directly induced human Schwann cell precursors as a valuable source of Schwann cells. Stem Cell Res Ther. 2020;11:257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiang C, Qiu W, Yang Y, Huang H, Dai ZM, Yang A, et al. ADAMTS4 enhances oligodendrocyte differentiation and remyelination by cleaving NG2 proteoglycan and attenuating PDGFRα signaling. J Neurosci. 2023;43:4405–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fletcher JL, Makowiecki K, Cullen CL, Young KM. Oligodendrogenesis and myelination regulate cortical development, plasticity and circuit function. Semin Cell Dev Biol. 2021;118:14–23. [DOI] [PubMed] [Google Scholar]
- 41.Kucharova K, Stallcup WB. The NG2 proteoglycan promotes oligodendrocyte progenitor proliferation and developmental myelination. Neuroscience. 2010;166:185–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Đặng TC, Ishii Y, Nguyen VD, Yamamoto S, Hamashima T, Okuno N, et al. Powerful homeostatic control of oligodendroglial lineage by PDGFRα in adult brain. Cell Rep. 2019;27:1073. [DOI] [PubMed] [Google Scholar]
- 43.Windrem MS, Nunes MC, Rashbaum WK, Schwartz TH, Goodman RA, McKhann G, et al. Fetal and adult human oligodendrocyte progenitor cell isolates myelinate the congenitally dysmyelinated brain. Nat Med. 2004;10:93–7. [DOI] [PubMed] [Google Scholar]
- 44.Zhang M, Wang J, Zhang KX, Lu GZ, Liu YM, Ren KK, et al. Ten-eleven translocation 1-mediated DNA hydroxymethylation is required for myelination and remyelination in the mouse brain. Nat Commun. 2021;12. [DOI] [PMC free article] [PubMed]
- 45.Moyon S, Ma D, Huynh JL, Coutts DJC, Zhao C, Casaccia P, et al. Efficient remyelination requires DNA methylation. eNeuro. 2017;4. [DOI] [PMC free article] [PubMed]
- 46.Moyon S, Huynh JL, Dutta D, Zhang F, Ma D, Yoo S, et al. Functional characterization of DNA methylation in the oligodendrocyte lineage. Cell Rep. 2016;15:748–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ketchum HC, Suzuki M, Dawlaty MM. Catalytic-dependent and -independent roles of TET3 in the regulation of specific genetic programs during neuroectoderm specification. Commun Biol. 2024;7:415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Varzideh F, Gambardella J, Kansakar U, Jankauskas SS, Santulli G. Molecular mechanisms underlying pluripotency and self-renewal of embryonic stem cells. Int J Mol Sci. 2023;24:8386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lang J, Maeda Y, Bannerman P, Xu J, Horiuchi M, Pleasure D, et al. Adenomatous polyposis coli regulates oligodendroglial development. J Neurosci. 2013;33:3113–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gao Y, Duque-Wilckens N, Aljazi MB, Wu Y, Moeser AJ, Mias GI, et al. Loss of histone methyltransferase ASH1L in the developing mouse brain causes autistic-like behaviors. Commun Biol. 2021;4:756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pooyan P, Karamzadeh R, Mirzaei M, Meyfour A, Amirkhan A, Wu Y, et al. The dynamic proteome of oligodendrocyte lineage differentiation features planar cell polarity and macroautophagy pathways. Gigascience. 2020;9. [DOI] [PMC free article] [PubMed]
- 52.Shang Y, Guan H, Zhou F. Biological characteristics of umbilical cord mesenchymal stem cells and its therapeutic potential for hematological disorders. Front Cell Dev Biol. 2021;9: 570179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li Y, Liu L, Ding X, Liu Y, Yang Q, Ren B. Interleukin-1β attenuates the proliferation and differentiation of oligodendrocyte precursor cells through regulation of the microRNA-202-3p/β-catenin/Gli1 axis. Int J Mol Med. 2020;46:1217–24. [DOI] [PubMed] [Google Scholar]
- 54.Bai X, Zhao N, Koupourtidou C, Fang LP, Schwarz V, Caudal LC, et al. In the mouse cortex, oligodendrocytes regain a plastic capacity, transforming into astrocytes after acute injury. Dev Cell. 2023;58:1153. [DOI] [PubMed] [Google Scholar]
- 55.Bernardo A, Giammarco ML, De Nuccio C, Ajmone-Cat MA, Visentin S, De Simone R, et al. Docosahexaenoic acid promotes oligodendrocyte differentiation via PPAR-γ signalling and prevents tumor necrosis factor-α-dependent maturational arrest. Biochim Biophys Acta Mol Cell Biol Lipids. 2017;1862:1013–23. [DOI] [PubMed] [Google Scholar]
- 56.Hsieh J, Aimone JB, Kaspar BK, Kuwabara T, Nakashima K, Gage FH. IGF-I instructs multipotent adult neural progenitor cells to become oligodendrocytes. J Cell Biol. 2004;164:111–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Akiyama M, Hasegawa H, Hongu T, Frohman MA, Harada A, Sakagami H, et al. Trans-regulation of oligodendrocyte myelination by neurons through small GTPase Arf6-regulated secretion of fibroblast growth factor-2. Nat Commun. 2014;5:4744. [DOI] [PubMed] [Google Scholar]
- 58.Li Y, Tang G, Liu Y, He X, Huang J, Lin X, et al. CXCL12 gene therapy ameliorates ischemia-induced white matter injury in mouse brain. Stem Cells Transl Med. 2015;4:1122–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Boido M, Ghibaudi M, Gentile P, Favaro E, Fusaro R, Tonda-Turo C. Chitosan-based hydrogel to support the paracrine activity of mesenchymal stem cells in spinal cord injury treatment. Sci Rep. 2019;9. [DOI] [PMC free article] [PubMed]
- 60.Okuda A, Horii-Hayashi N, Sasagawa T, Shimizu T, Shigematsu H, Iwata E, et al. Bone marrow stromal cell sheets may promote axonal regeneration and functional recovery with suppression of glial scar formation after spinal cord transection injury in rats. J Neurosurg Spine. 2017;26:388–95. [DOI] [PubMed] [Google Scholar]
- 61.Shende P, Subedi M. Pathophysiology, mechanisms and applications of mesenchymal stem cells for the treatment of spinal cord injury. Biomed Pharmacother. 2017;91:693–706. [DOI] [PubMed] [Google Scholar]
- 62.Li X, Tan J, Xiao Z, Zhao Y, Han S, Liu D, et al. Transplantation of hUC-MSCs seeded collagen scaffolds reduces scar formation and promotes functional recovery in canines with chronic spinal cord injury. Sci Rep. 2017;7:43559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lai BQ, Bai YR, Han WT, Zhang B, Liu S, Sun JH, et al. Construction of a niche-specific spinal white matter-like tissue to promote directional axon regeneration and myelination for rat spinal cord injury repair. Bioact Mater. 2022;11:15–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon request.