Abstract
Background
Yes-associated protein 1 (YAP1) regulates the survival, proliferation, and stemness of cells, and contributes to the development of metabolic dysfunction associated fatty liver disease (MAFLD). However, the regulatory role of intestinal YAP1 in MAFLD still remains unclear.
Methods
Terminal ileal specimens were used to compare intestinal YAP1 activation in patients with and without MAFLD. Mice targeted for knocking out YAP1 in the intestinal epithelium were fed a high-fat diet (HFD) for 8 consecutive weeks. In a separate group, the mice were fed an HFD supplemented with the bile acid binder cholestyramine (CHO) or a low-fat diet with deoxycholic acid (DCA). Immunofluorescence, Immunohistochemistry, Western blot, RT-qPCR, ELISA, 16S rDNA sequencing, tissue and enteroid culture techniques were used to evaluate the effects of an HFD or DCA on the gut‒liver axis in mice or humans.
Results
Intestinal YAP1 was activated in both humans with MAFLD and mice fed an HFD. In in vivo studies, YAP1 knockout in intestinal epithelial cells of mice alleviated the hepatic steatosis induced by an HFD, and mitigated the adverse effects of HFD on the gut‒liver axis, including the upregulation of lipopolysaccharide (LPS) and inflammation levels, enrichment of intestinal Gram-negative bacteria, and inhibition of intestinal stem cell (ISC) differentiation into the goblet and Paneth cells. High-fat feeding (HFF) produced high concentrations of DCA. The consumption of DCA mimics these HFF-induced changes, and is accompanied by the activation of Abelson tyrosine-protein kinase 1 (ABL1) and its direct substrate, YAP1, in the terminal ileum. In vitro studies further confirmed that DCA upregulated the tyrosine phosphorylation of YAP1Y357 in ISC by activating ABL1, which inhibited the differentiation of ISCs into secretory cells.
Conclusions
Our findings reveal that the activation of the ABL1‒YAP1 axis in ISCs by DCA contributes to hepatic steatosis through the gut‒liver axis, which may provide a potential intestinal therapeutic target for MAFLD.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12967-024-05865-6.
Keywords: Yes-associated protein 1, Deoxycholic acid, High-fat diet, Intestinal stem cell, Hepatic steatosis
Introduction
Nonalcoholic fatty liver disease (NAFLD) is a metabolic-related disorder that has been renamed metabolic dysfunction-associated fatty liver disease (MAFLD). The pathogenesis of MAFLD involves factors such as lipotoxicity, endoplasmic reticulum stress, oxidative stress, impaired intestinal mucosal barrier function, disturbance of gut microbiota, and epigenetic inheritance [1]. Exposure to a high-fat diet (HFD) and diet-induced obesity are major causes of MAFLD.
The chronic low-grade inflammation observed in patients with MAFLD may result from the invasion of intestinal pathogens into the gut‒liver axis [1, 2]. The intestinal mucosal barrier is an important host defence system against external antigens [3] and includes the physical barrier composed of mucin secreted by intestinal epithelial goblet cells and tight junctions, and the functional barrier composed of antimicrobial peptides secreted by Paneth cells, as well as immunoglobulin and complement proteins produced by mucosal immune cells [4]. The integrity of the intestinal mucosal barrier depends on the function of LGR5+ intestinal stem cells (ISCs) located in epithelial crypts [5]. Impaired differentiation of ISCs into secretory cell lines, such as goblet and Paneth cells, is associated with the occurrence and progression of inflammatory and metabolic diseases [6, 7]. Chronic exposure to HFD induces increased permeability of the intestinal mucosal barrier [8, 9] that leads to the translocation of intestinal bacteria and their metabolites such as endotoxins into the blood and the liver, thereby activating the liver immune system and leading to hepatic inflammation and steatosis [10]. Therefore, abnormal regulation of gut‒liver axis plays an important role in the development of MAFLD.
Exposure to HFD can induce changes in the bile acid pool, especially the elevation in deoxycholic acid (DCA) levels [11, 12]. Our previous study had confirmed that 2 weeks of high-fat feeding increased fecal DCA levels in mice [6]. Significantly elevated circulating levels of DCA have been reported in patients with NAFLD and nonalcoholic steatohepatitis (NASH) [13, 14], and disease activity and degree of hepatic fibrosis are positively correlated with increasing levels of DCA and its derivatives [15]. The enterohepatic circulation of bile acids is well known, with about 95% of the primary bile acids being re-absorbed in the terminal ileum, and a small portion of the unabsorbed primary bile acids being converted by bacteria colonizing the caecum into the secondary bile acids DCA or lithocholic acid (LCA) [16]. Different bile acid components and their concentrations have been shown to have different effects on intestinal inflammation and mucosal permeability [17]. DCA that is more hydrophobic has a protective effect on the gut at physiological concentrations (e.g. < 50 μM), displaying anti-inflammatory and mucosal renewal attributes [18, 19]. Conversely, high concentrations of DCA have cytotoxic effects [19], inducing DNA damage in esophageal, intestinal epithelial, or stem cells that leads to carcinogenesis [20, 21], or DNA damage in intestinal epithelial cells that results in disruption the intestinal mucosal barrier [22]. According to previous studies, DNA damage can be sensed by ABL1 [23], a member of the nonreceptor tyrosine kinase SRC family [24]. DNA damage activates Yes-associated protein 1 (YAP1), a direct substrate of ABL1 [23, 25], which is one of the mechanisms of apoptosis in stressed cells [26, 27]. Abnormal bile acid levels have also been shown to promote YAP1 activation [18].
YAP1 is a transcriptional coactivator present in cells and acts as a nuclear transducer of the Hippo pathway [28]. After activation, YAP1 promotes the transcription of target genes by binding to transcriptional enhanced associate domain (TEAD) proteins [28]. It regulates cell survival, proliferation and stemness, and participates in the maintenance of metabolic homeostasis and the development of diseases [28]. YAP1 plays an important role in the regulation of liver and intestinal pathophysiology. In the liver, YAP1 promotes the development of MAFLD, cirrhosis, and carcinogenesis [29, 30]. In the intestine, in addition to its proven carcinogenic effects [31], YAP1 also affects intestinal mucosal epithelial barrier function [32]. ISCs have been reported to be regulated by both WNT and YAP1 signals [33]. The nuclear input of YAP1 plays a key role in the proliferation and differentiation of ISCs after intestinal injury [34].
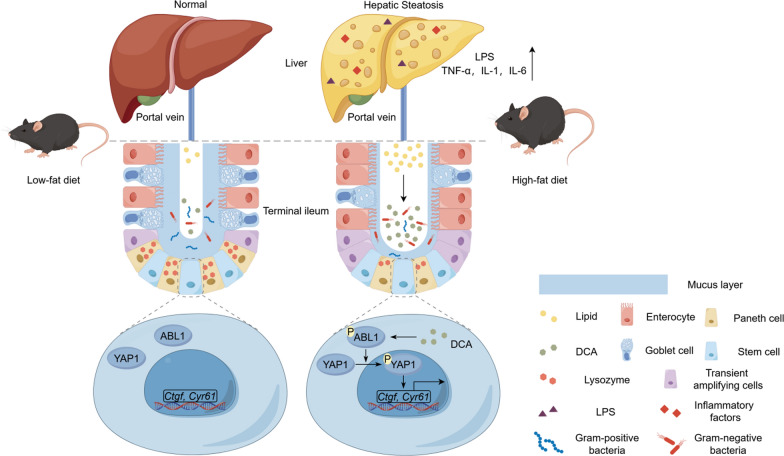
Taken together, this raises the possibility that in chronic exposure to HFD, high concentrations of DCA in the intestinal cavity persistently activate YAP1 to abnormally regulate ISCs, thereby compromising the terminal ileal mucosal barrier and triggering hepatic steatosis via the gut‒liver axis. The terminal ileal mucosa, adjacent to the caecal region, displays a marked increase in intestinal bacteria and may be more susceptible to the initial high concentrations of DCA, and thus may be the site of an important intestinal origin of HFD-induced hepatic steatosis.
Materials and methods
Mice and diets
Male C57BL/6 J mice, aged 6 weeks and weighing 18–22 g, were obtained from the Animal Experimentation Centre of the Shanghai General Hospital. After a week of acclimatisation with a normal diet, the mice were randomly divided into four groups and fed for 8 consecutive weeks with the following: (i) a control low-fat diet (LFD) containing 10 kcal% fat, (ii) a high-fat diet (HFD) containing 60 kcal% fat, (iii) HFD + cholestyramine (CHO, a bile acid binder, C4650, MilliporeSigma, Saint Louis, MO, USA), an HFD mixed with 6% (w/w) CHO, (iv) an LFD + deoxycholic acid (DCA), LFD + 0.2% DCA (D2510, Sigma‒Aldrich, Saint Louis, MO, USA). Yap1fl/fl mice were provided by Zhang Laboratory (Center for Excellence in Molecular Cell Science, Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences, Shanghai, China).The preparation of the mice is based on previous literature reports [35]. And the Yap1fl/fl Vil1-Cre mice were purchased from Cyagen Biosciences. The mice were raised in an SPF environment. After a week of acclimatisation to a normal diet, they were fed either an LFD (10 kcal% fat) or HFD (60 kcal% fat) for 8 consecutive weeks. At the end of the 8-week period, the mice in each group were fasted for 12 h and then euthanised through cervical dislocation. Fresh faeces, plasma, liver tissues, and terminal ileum tissues were collected using aseptic equipment and were stored at − 80 °C for subsequent experiments. Lgr5-EGFP-IRES-CreERT2 mice were abtained from The Jackson Laboratory, USA (https://www.jax.org/strain/008875#). All the diets were purchased from Trophic Animal Feed High-Tech Co., Ltd., Nantong, China.
Measurement of faecal bile acid concentration
Methanol (400 µL) was added to mouse faeces and thoroughly mixed. The samples were ground for 60 s at a frequency of 55 Hz in a high-throughput tissue grinder; this process was repeated three times. The samples were subjected to ultrasonic treatment at room temperature for 30 min. After centrifugation, the supernatant was extracted, methanol was added, and the mixture was vortexed and centrifuged again. The obtained supernatant was injected into a liquid chromatograph for separation, and the separated compounds were analysed using a mass spectrometer. After adjusting the ion modes and analytical parameters of the mass spectrometer, a standard curve was established by measuring the known concentrations of bile acids, and the concentrations of different bile acids in the samples were calculated according to the regression equation [36, 37].
Histological analysis
Fresh liver and intestinal tissues obtained from mice were fixed in 4% paraformaldehyde, followed by paraffin embedding and sectioning into 4-µm thickness. Sections were de-paraffinised, rehydrated, and stained with haematoxylin and eosin (HE) or Picrosirius red solutions. Frozen liver tissues stored at − 80 °C were used for cryosectioning and stained with Oil Red O staining solution (Sigma‒Aldrich, USA).
Periodic acid-Schiff staining
Fresh terminal ileum tissues from mice were fixed in 4% paraformaldehyde, dehydrated, embedded in paraffin, followed by sectioning into 4-µm slices. Periodic acid-Schiff staining was used to quantitatively analyse goblet cells in ileal villi. The average number of goblet cells per villus crypt unit was recorded for each field of view, with five fields of view selected per sample.
Immunohistochemistry
Human terminal ileal tissues were obtained from patients who underwent surgery for right colon cancer (6 NonMAFLD patients and 3 MAFLD patients). The paraffin-embedded mouse or human ileum tissue samples were separated into slices (4 µm), followed by antigen retrieval and incubation with primary antibodies against pYAP1 at a dilution of 1:200. A secondary antibody was then used, and the slices were subsequently stained with a 3, 3’-diaminobenzidine solution. Simultaneously, counterstaining of the nuclei was performed with haematoxylin (Beyotime). Finally, the representational areas were photographed via a light microscope at the appropriate magnification.
Immunofluorescence
Fresh mouse or human ileal and mouse liver tissues were fixed in 4% paraformaldehyde for 24 h, embedded in paraffin, and sectioned. Antigen repair was performed after deparaffinisation and rehydration. After being subjected to a blocking buffer (Beyotime, Nanjing, China) for 1 h, tissue sections were incubated with primary antibodies (YAP1, phosphorylated YAP1 [pYAP1], MUC2, lysozyme [LYZ], KI67, F4/80, TGR5) at 4 °C for 16 h. The primary antibodies used are listed in Supplementary Table 1. The sections were allowed to reach room temperature (20–25℃) for 1 h and then incubated with a secondary antibody (1:200, Yeasen, China) at 37 °C for 1 h. After washing the slides, they were incubated with DAPI (Yeasen, Shanghai, China) for 5 min, coverslipped, and imaged under a fluorescence microscope (Leica DMI8, Germany) or Leica TCS SP8 confocal laser scanning microscope (Leica TCS NT, Wetzlar, Germany).
ELISA
Mouse liver tissues were retrieved from the − 80 °C freezer, and 100 mg was excised and added to PBS containing 1% PMSF. The tissues were homogenised through ultrasonication (30 Hz, 2 min), placed on ice for 1 h, and centrifuged at 4 °C, 12,000 rpm for 10 min. The concentration of lipopolysaccharide (LPS) in the liver tissue was determined using an ELISA kit (Abclonal, Shanghai, China). The protein concentration was measured using a BCA kit (Yeason, Shanghai, China).
D-lactate was used as an indicator of intestinal barrier permeability [38]. D-lactate in mouse serum was measured according to the instructions of a mouse ELISA kit (BYabscience, Nanjing, China).
In vitro culture of ileal tissue
Mouse or human ileal tissues were immersed in PBS, longitudinally opened, and cut into 5 mm fragments, which were then homogenised and placed in six-well plates with DMEM containing 2% penicillin–streptomycin and amphotericin (Beyotime, Shanghai, China). Intestine tissues from Yap1fl/fl and Yap1fl/fl Vil1-Cre mice were treated with vehicle (DMSO) or 100 μM DCA (D2510, Sigma‒Aldrich, USA). After 24 h, the total protein was extracted from the cultured tissue pieces for further analysis. Tissue samples from the human terminal ileum were cocultured with vehicle (DMSO), 100 μM DCA (D2510, Sigma‒Aldrich, USA), 100 μM DCA combined with the YAP1 antibody (1:1000, 14074, CST, USA), or 100 μM DCA combined with the ABL1 antagonist, Imatinib (100 μM, MCE, HY-15463), in a cell incubator for 24 h. The culture medium was replaced every 12 h. Total RNA was extracted from the cultured tissue samples for further analysis [6].
In vitro enteroid culture
The extraction of crypts and the cultivation of enteroids were carried out as described in the article [39]. Fresh terminal ileal tissues (4–5 cm) taken from mice or humans were cut lengthwise into 0.5 cm segments after soaking in PBS. Pieces of small intestinal tissue were placed in a centrifuge tube containing cold PBS and gently shaken to remove mucus from the mucosal surface of the intestine. After discarding the PBS, 5 mmol/L ethylenediaminetetraacetic acid (EDTA) solution for mice or 40 mmol/L EDTA solution for humans was added to the tube, and it was then incubated at 4 ℃ for 1 h. After incubation, the EDTA in the centrifuge tube was discarded. Cold PBS was then added to the centrifuge tube and vigorously shaken. The suspension was filtered through a 70 µm filter and centrifuged at 4 ℃, 1200 rpm for 5 min (mice) or 3000 rpm for 5 min (human) to obtain the ileal crypt precipitation. The crypt precipitate was diluted to a concentration of 200 per 10 µl with PBS buffer. The 20 µl diluted crypts were mixed with 20 µl matrix gel (356231, Corning, NY, USA), and the mixture was seeded into a 48-well plate. After fixation in an incubator at 37 °C for 15 min, 200 µl of culture medium (IntestiCult™ Organoid Growth Medium, STEMCELL Technologies, Vancouver, BC, Canada) was added to each well. The 48-well plates were then placed in an incubator. After 24 h, enteroid formation was observed under a bright-field microscope. For mice fed different diets for 8 weeks, we recorded enteroid budding rates at different time points at 24, 48, 72, and 96 h. In another separate experiment, we cultured the extracted normal mice or human small intestinal crypts for 24 h and then added 100 μM DCA for co-incubation. Finally, the budding rates of enteroids were recorded at 24, 48 and/or 72-h time points after the intervention, or immunofluorescence staining of enteroids was performed.
Immunofluorescence staining of enteroids
The culture-medium was discarded and then washed with 200 µl PBS buffer. After discarding the PBS, 100 µl of 4% paraformaldehyde was added to each well and fixed for 1 h. The fixation solution was discarded, and the implantation wells were rinsed with PBS. One hour after adding the blocking solution (Beyotime, Shanghai, China) to the wells, primary antibodies (YAP1, pYAP1, MUC2, and LYZ) were introduced to the wells and co-incubated with enteroids at 4 °C overnight. The primary antibodies used are listed in Supplementary Table 1. The following day, the enteroids were allowed to equilibrate to room temperature for 1 h. Subsequently, each well was rinsed with PBS buffer and then incubated with secondary antibodies (34206ES60, 1:200, Yeasen, China) at 37 °C for 1 h. Following the removal of the secondary antibodies and a wash with PBS buffer, the enteroids were incubated with DAPI (Yeasen, Shanghai, China). Finally, images were captured using a fluorescence microscope.
Intervention of HT-29 cell line
HT-29 cell lines (CL0118, Pricella, China) were cocultured with vehicle (DMSO), DCA (100 μM, D2510, Sigma‒Aldrich), or DCA combined with ABL1 antagonist, Imatinib (100 μM, MCE, HY-15463) in a cell incubator. A pre-experiment was conducted to determine the optimal time point for DCA treatment, and 15 min was identified as a suitable time point for intervention. Imatinib was added 2 h before DCA intervention. Protein and RNA were collected from the cultured HT-29 cells for further study.
RNA Extraction and RT‒qPCR
Total RNA was extracted from the ileum and liver tissues of mice and humans using the TRIzol reagent (Takara, Shiga, Japan), with 1 ml added per 50 mg of tissue. The tissues were ground in a high-throughput tissue grinder at 70 Hz for 30 s, which was repeated twice. Chloroform was added to the mixture, and the aqueous phase was collected after centrifugation. Isopropanol was added to precipitate the RNA, and the supernatant was removed after centrifugation. RNA was washed with 75% ethanol, centrifuged again, and the precipitate was dissolved in distilled water for concentration measurements. A total of 1000 ng RNA was reverse-transcribed into cDNA using the HyperScript III RT SuperMix kit (EnzyArtisan, Shanghai, China). The target primers were then quantified through real-time qPCR analysis using Universal SYBR qPCR Mix kit (EnzyArtisan, Shanghai, China) in a 10 µL reaction system. The primer sequences are listed in Supplementary Table 2. The relative expression levels of the target genes were normalised against GAPDH and quantified using 2−ΔΔCt method.
Western Blot analysis
Proteins from mouse ileal tissues were lysed using RIPA lysis buffer (EpiZyme, Shanghai, China) containing protease and phosphatase inhibitors (Beyotime, Nanjing, China). The tissues were ground in a high-throughput tissue grinder at 70 Hz for 60 s, which was repeated twice. The resulting solution was placed on ice for 1 h and then centrifuged at 4 °C at 12,000 rpm for 15 min to collect the supernatant. The protein supernatant was mixed with 5 × SDS sample buffer (EpiZyme, Shanghai, China) at a 4:1 ratio, heated at 100 °C for 10 min, and stored at − 80 °C. Equal amounts of protein were electrophoresed in a 7.5% SDS-PAGE gel and transferred to PVDF membranes (Millipore, Tullagreen, Ireland). Five percent non-fat milk or quick blocking solution (EpiZyme, Shanghai, China) was used to block nonspecific epitopes on the membrane. After incubation overnight with primary antibodies (YAP1, pYAP1, ABL1, phosphorylated ABL1 [pABL1], Leucine-rich repeat-containing G protein-coupled receptor 5 [LGR5], Cytokeratin 18 [KRT18], ACTIN) at 4 °C, the protein was incubated with enzyme-labelled goat anti-rabbit antibody (1:100,000, Jackson ImmunoResearch, Baltimore, MD, USA) at room temperature for 1 h the following day. The target bands were observed using ECL chemiluminescence and quantitatively analysed using ImageJ software. The primary antibodies used in this study are listed in Supplementary Table 1.
16S rDNA sequencing of faecal microbiota
DNA was isolated and extracted from faecal specimens of mice. Amplification and sequencing of the V3-V4 region of the 16S rDNA gene were conducted by the Genedenovo company (International Biological Island, Huangpu District, Guangzhou, China).
Statistical analysis
Data are expressed as mean ± SEM. Significant differences in the data were analysed using GraphPad Prism 9.0. One-way ANOVA was used to compare the differences between multiple groups, and the unpaired t-test was used to compare the differences between two groups that conformed to a normal distribution. The Mann–Whitney U test was used to compare two groups that did not conform to a normal distribution. Metabolomic data were analysed using the R software (version 3.5) and SIMCA-P 14.1 (Umetrics, Umeå, Sweden). Differences were considered statistically significant at p < 0.05.
Results
Intestinal YAP1 is elevated in HFD-fed mice and MAFLD patients
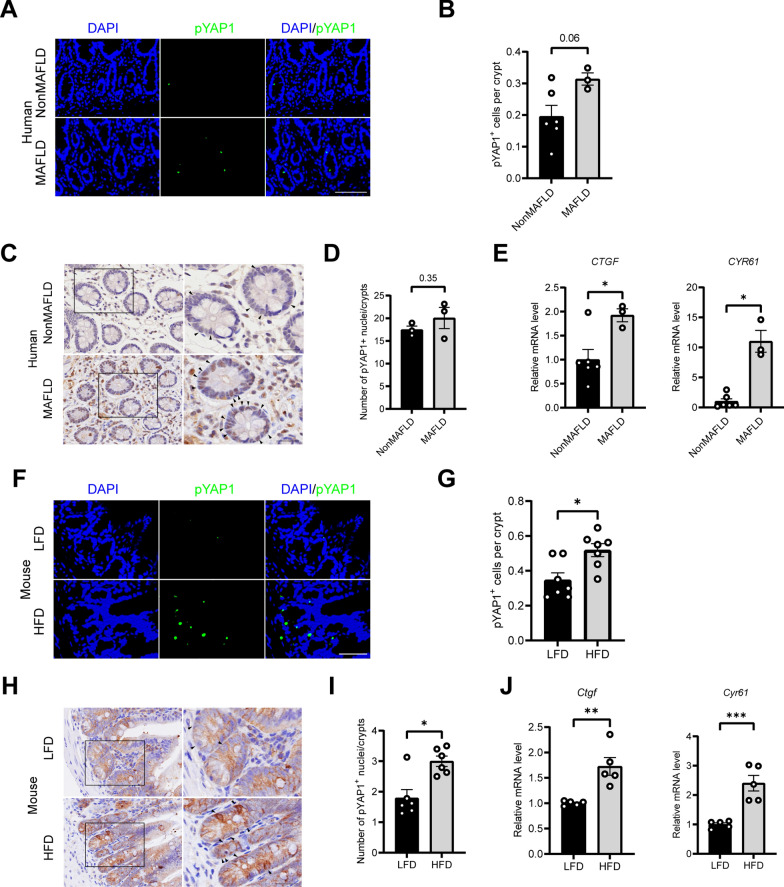
To understand the role of intestinal YAP1 in MAFLD, immunofluorescence and immunohistochemistry were used to detect the expression of phosphorylated YAP1Y357 (pYAP1) in the terminal ileum of MAFLD patients. Similar to the previous morphological observations of YAP1 [32, 33], the result of this study revealed that pYAP1 is mainly located in the crypts where ISCs reside, and compared to NonMAFLD patients, the expression of pYAP1 was upregulated in MAFLD patients (Fig. 1A–D). At the gene level, downstream target genes of YAP1, including Ctgf and Cyr61, were upregulated in MAFLD patients compared with those in NonMAFLD patients (Fig. 1E). To further validate these findings, we examined pYAP1 and its target genes in terminal ileum specimens from mice. Similarly, compared with LFD group, immunofluorescence showed that pYAP1 positive cells at the bottom of small intestine crypt of HFD mice were increased (Fig. 1F, G); Immunohistochemical study further confirmed that the number of pYAP1 positive nucleus at the bottom of small intestine crypt of HFD mice were significantly increased (Fig. 1H, I). The expressions of Ctgf and Cyr61, downstream target genes of YAP1, were also significantly increased in HFD mice (Fig. 1J). These results suggest that intestinal YAP1 is activated in HFD mice and MAFLD patients.
Fig. 1.
YAP1 is activated in HFD-fed mice and MAFLD patients. A Representative micrographs of immunofluorescence staining for pYAP1 in the crypt region of the ileum tissue from humans with and without MAFLD. Scale bars: 100 μm. B Quantitative analysis of A (n = 6 or 3). C, D Representative micrographs of immunohistochemistry results for nuclear pYAP1+ cells in the ileal crypts of the NonMAFLD and MAFLD patients and the quantitative analysis (n = 3). The arrows point to nuclear pYAP1+ cells. Scale bars: 25 μm. E mRNA expression of CTGF and CYR61 in the ileum tissues of humans with and without MAFLD (n = 6 or 3). F Representative micrographs of immunofluorescence staining for pYAP1 in the crypt region of the ileum tissue in mice. Scale bars: 50 μm. G Quantitative analysis of the data in F (n = 7). H, I Representative micrographs of immunohistochemistry results for nuclear pYAP1+ cells in the ileal crypts of the LFD- and HFD-fed mice and the quantitative analysis (n = 6). The arrows point to nuclei of pYAP1+ cells. Scale bars: 25 μm. J mRNA expression of Ctgf and Cyr61 in mouse ileum tissue (n = 5). LFD, low-fat diet; HFD, high-fat diet; pYAP1, phosphorylated YAP1; NonMAFLD, without MAFLD. *P < 0.05, **P < 0.01, ***P < 0.001
An intestinal YAP1 knockout improves hepatic inflammation and steatosis in the HFD-fed mice
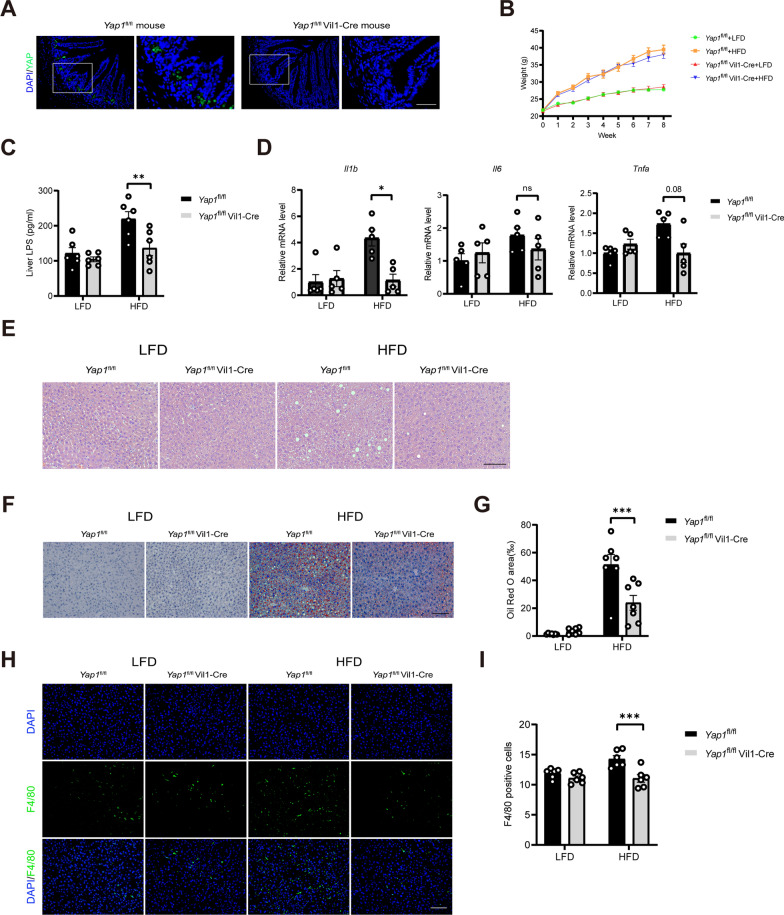
To investigate the role of intestinal YAP1 in hepatic inflammation and hepatic steatosis, YAP1 was knocked out in the murine intestine. The results revealed that YAP1 was expressed at the base of the crypt where ISCs were located in the Yap1fl/fl mice, which is consistent with the findings of previous studies (Fig. 2A). YAP1 expression was not detected in the intestinal epithelium of the Yap1fl/fl Vil1-Cre mice. The YAP1 knockout did not affect the body weight of the HFD-fed mice (Fig. 2B). In mice fed an HFD, the YAP1 knockout decreased the LPS content as well as the mRNA expression levels of the inflammation-related genes (IL-1β and TNF-α) in the liver (Fig. 2C and D). Hematoxylin eosin staining and Oil Red O staining of the liver tissue revealed that the YAP1 knockout reduced the lipid droplet content (Fig. 2E–G), and immunostaining revealed that the YAP1 knockout downregulated liver F4/80 expression in the livers of mice fed an HFD (Fig. 2H and I). These results suggest that knocking out YAP1 in the intestinal epithelial cells reduces LPS accumulation, inflammation, and lipid droplet deposition in the livers of mice.
Fig. 2.
An intestinal YAP1 knockout improves hepatic inflammation and steatosis in the HFD-fed mice. A Representative micrographs of immunofluorescence staining for YAP1 in the intestines of the Yap1fl/fl and Yap1fl/fl Vil1-Cre mice. Scale bars: 50 μm. B Body weight changes of the Yap1fl/fl and Yap1fl/fl Vil1-Cre mice in 8 weeks (n = 6). C LPS concentration in the liver tissue of the Yap1fl/fl and Yap1fl/fl Vil1-Cre mice (n = 6). D The mRNA expression of IL-1β, IL-6 and TNF-α in liver tissue of Yap1fl/fl and Yap1fl/fl Vil1-Cre mice (n = 5). E Representative micrographs of H&E staining in liver tissues from the Yap1fl/fl and Yap1fl/fl Vil1-Cre mice. Scale bars: 100 μm. F Representative micrographs of Oil Red O-stained liver tissue from Yap1fl/fl and Yap1fl/fl Vil1-Cre mice. Scale bars: 100 μm. G Quantitative analysis of F (n = 7). H Representative micrographs of immunofluorescence staining for F4/80 in the liver tissues of the Yap1fl/fl and Yap1fl/fl Vil1-Cre mice. Scale bars: 100 μm. I Quantitative analysis of the data in H (n = 6). LFD, low-fat diet; HFD, high-fat diet. *P < 0.05, **P < 0.01, ***P < 0.001, ns: not significant
An intestinal YAP1 knockout improves intestinal inflammation and dysbiosis in the HFD-fed mice
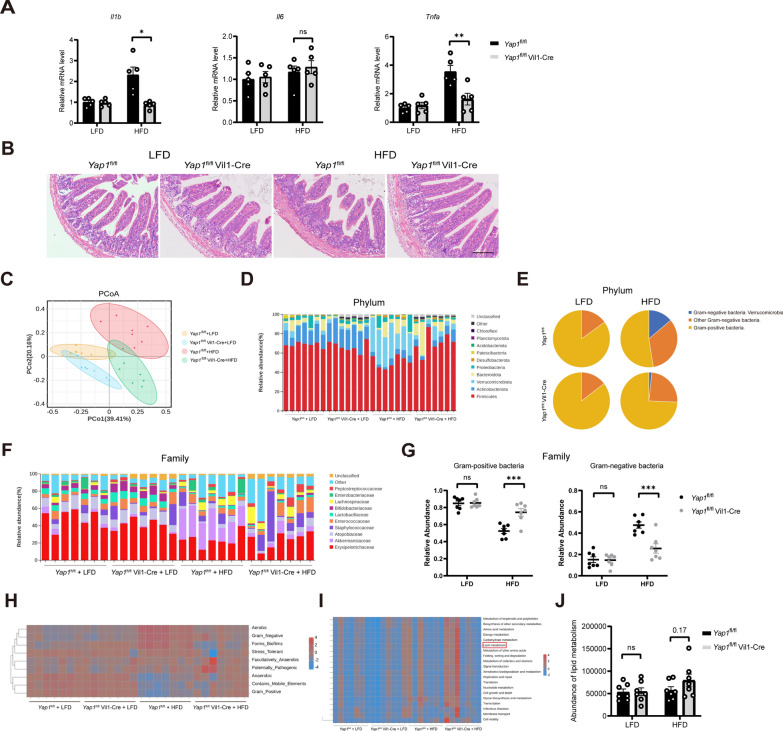
We evaluated the effects of YAP1 on intestinal inflammation and the microbiota in the HFD-fed mice. First, the results of quantitative reverse transcriptase‒polymerase chain reaction assay revealed that an intestinal YAP1 knockout downregulated the expression of genes associated with intestinal inflammation (IL-1β and TNF-α) in HFD-fed mice (Fig. 3A). The damage to the ileum mucosa and the shedding of intestinal villi were significantly improved in the Yap1fl/fl Vil1-Cre mice compared to Yap1fl/fl mice fed an HFD (Fig. 3B). Sequencing analysis of the gut microbiota revealed that the faecal bacterial community composition of the HFD-fed mice was altered by the YAP1 knockout. In the HFD-fed mice, the relative abundance of Firmicutes was significantly greater, and that of Verrucomicrobia was significantly lower, in the Yap1fl/fl Vil1-Cre mice than in the Yap1fl/fl mice fed an HFD. (Fig. 3C and D). Interestingly, we found that the knocking out YAP1 increased the relative abundance of Gram-positive bacteria, and decreased the abundance of Gram-negative bacteria (specifically Verrucomicrobia) in mice fed HFD (Fig. 3E). Similar findings were observed at the family level in the Yap1fl/fl mice and Yap1fl/fl Vil1-Cre mice fed an HFD (Fig. 3F and G). Further functional analysis via Bugbase revealed a significant increase in aerobic and Gram-negative bacteria in the Yap1fl/fl mice fed an HFD, which could be reversed by a Yap1 gene knockout in the intestine (Fig. 3H). Predictive analysis of functional pathways based on PICRUSt2 revealed significant changes in functional pathways by a Yap1 gene knockout, such as lipid metabolism (Fig. 3I, J). These results suggest that a YAP1 knockout in intestinal epithelial cells improves intestinal inflammation and the faecal bacterial community composition in HFD-fed mice.
Fig. 3.
An intestinal YAP1 knockout improves intestinal inflammation and dysbiosis in the HFD-fed mice. A The mRNA expression of IL-1β, IL-6 and TNF-α in Yap1fl/fl and Yap1fl/fl Vil1-Cre mice ileum tissue (n = 5). B Representative micrographs of H&E staining of ileal tissues from the Yap1fl/fl and Yap1fl/fl Vil1-Cre mice. Scale bars: 100 μm. C Principal coordinate analysis illustrates the characteristics of the changes in the gut microbiota among the Yap1fl/fl and Yap1fl/fl Vil1-Cre mice subjected to LFD and HFD respectively (n = 7). D Relative abundance of intestinal bacteria at the phylum level in Yap1fl/fl and Yap1fl/fl Vil1-Cre mice (n = 7). E Proportion of Gram-negative and Gram-positive bacteria in the gut microbiota of the intestinal microflora of the Yap1fl/fl and Yap1fl/fl Vil1-Cre mice at the phylum level (n = 7). F Relative abundance of intestinal bacteria at the family level in Yap1fl/fl and Yap1fl/fl Vil1-Cre mice (n = 7). G Relative abundance of Gram-positive bacteria and Gram-negative bacteria in the Yap1fl/fl and Yap1fl/fl Vil1-cre mice (n = 7). H Bacterial phenotypic analysis based on BugBase (n = 7). I Functional analysis based on PICRUSt2 (n = 7). J Abundance of lipid metabolism pathway in the Yap1fl/fl and the Yap1fl/fl Vil1-Cre mice (n = 7). LFD, low-fat diet; HFD, high-fat diet. *P < 0.05, **P < 0.01, ***P < 0.001, ns: no significant
An intestinal YAP1 knockout improves ISC differentiation dysfunction induced by an HFD
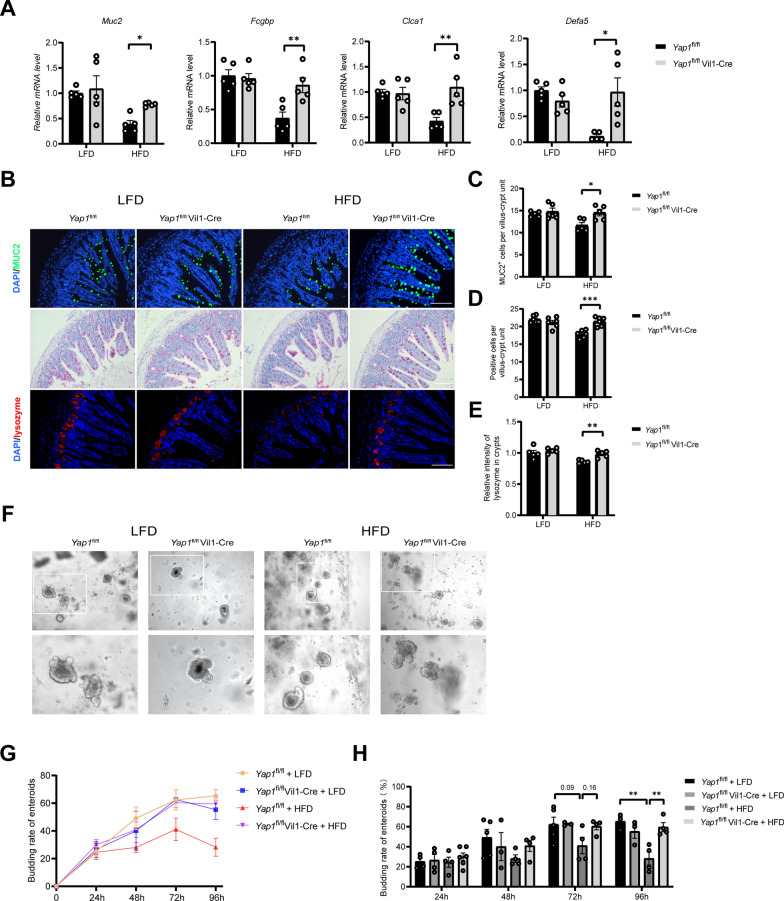
We evaluated the effects of YAP1 on ISC differentiation in HFD-fed mice. The results revealed that knocking out YAP1 significantly increased the mRNA levels of the loci involved in goblet cell functions (Muc2, Fcgbp and Clca1) and those related to Paneth cell functions (Defa5) in the intestines of HFD-fed mice (Fig. 4A). Immunofluorescence staining or periodic acid–Schiff (PAS) staining revealed that knocking out YAP1 in HFD-fed mice increased the number of goblet and Paneth cells (Fig. 4B–E). At 96 h of culture, the ileal enteroid budding rate of the Yap1fl/fl mice was significantly lower in the HFD-fed group than in the LFD-fed group; however, this reduction in the budding rate induced by the HFD could be reversed by a YAP1 knockout, suggesting that YAP1 regulates the differentiation of ISCs during high-fat feeding (Fig. 4F–H). Moreover, detection of the intestinal tight junction barrier indicators Tjp1 and Occludin revealed that the YAP1 knockout disrupted the intestinal tight junction barrier (Supplementary Fig. 1A). These results suggest that YAP1 downregulates the differentiation of ISCs into goblet and Paneth cells in HFD-fed mice.
Fig. 4.
An intestinal YAP1 knockout improves ISC differentiation in HFD-fed mice. A The mRNA levels of Muc2, Fcgbp, Clca1 and Defa5 in ileum tissue of Yap1fl/fl and Yap1fl/fl Vil1-Cre mice (n = 5). B Representative micrographs depicting immunofluorescence staining for MUC2 and lysozyme in mouse ileum tissue, as well as Periodic acid–Schiff (PAS) staining in mouse ileum tissue; Scale bars: 100 μm. C–E Quantitative analysis of B (n = 5 or 6). F Representative micrographs of enteroids in Yap1fl/fl and Yap1fl/fl Vil1-Cre mice fed LFD or HFD after 96-h culture. Scale bars: 100 μm. G Line graph of changes in enteroid budding rates at 24, 48, 72, and 96 h (n = 3–6). H Comparison of enteroid budding rates at 24, 48, 72, and 96 h (n = 3–6). LFD, low-fat diet; HFD, high-fat diet. *P < 0.05, **P < 0.01, ***P < 0.001
High concentrations of DCA induced by an HFD activate intestinal YAP1 signalling
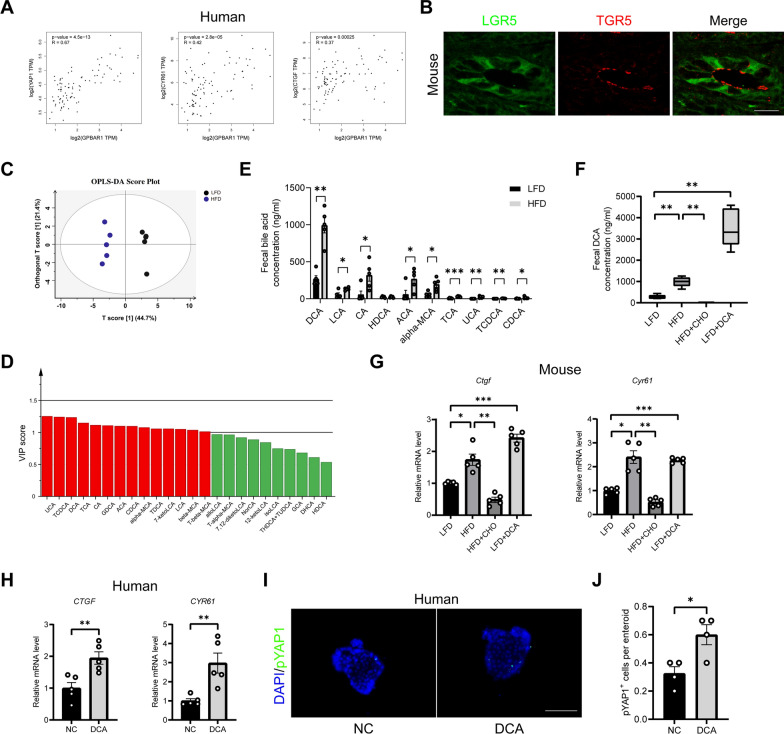
The GTEx database revealed a positive correlation between the G Protein-coupled bile acid receptor, GPBAR1 (TGR5) and YAP1, as well as its target genes CTGF, and CYR61 (Fig. 5A). The immunofluorescence images captured by confocal microscopy revealed that the membrane receptor TGR5 of bile acids was expressed in the LGR5-positive cells at the base of the ileal crypts (Fig. 5B).
Fig. 5.
High concentrations of DCA activate intestinal YAP1 signaling. A Correlation analysis of TGR5 with YAP1 and its target genes in the human intestine using data from the GTEx database. B Representative micrographs of immunofluorescence staining for LGR5 and TGR5 showed colocalization. Scale bars: 20 μm. C Orthogonal partial least squares discriminant analysis (OPLS-DA) of faecal bile acid in LFD and HFD groups. D Variable importance in the projection (VIP) value (red: VIP > 1). E Faecal individual bile acid concentration in LFD and HFD groups (n = 5). F Faecal DCA concentrations in LFD, HFD, HFD + CHO, and LFD + DCA groups (n = 5). G The mRNA expression levels of Ctgf and Cyr61 in mouse ileum tissue (n = 5). H The mRNA expression levels of CTGF, and CYR61 in human ileum tissue treated with 100 μM DCA for 24 h (n = 5). I Representative micrographs of immunofluorescence staining for pYAP1 in human ileal enteroids treated with or without 100 μM DCA. Scale bars: 100 μm. J Quantitative analysis of I (n = 4). DCA, deoxycholic acid; LFD, low-fat diet; HFD, high-fat diet; CHO, cholestyramine. *P < 0.05, **P < 0.01, ***P < 0.001
We quantified bile acid fractions in HFD-fed mice for 8 weeks. Bile acid-targeted metabolomics was used to analyse the bile acid composition of mouse faeces. Orthogonal partial least squares discriminant analysis (OPLS-DA) revealed a good separation between the LFD and the HFD groups, which was caused by the variable importance of projection (VIP, value > 1) of more than a dozen bile acids such as UCA, TCDCA, and DCA (Fig. 5C and D). Faecal total bile acids, especially DCA, alpha-MCA, and TCA, were significantly greater in the HFD group than in the LFD group (Fig. 5E). DCA is a potent TGR5 agonist. These data suggest that 8 weeks of high-fat feeding rapidly altered the composition of bile acids and significantly elevated the DCA concentrations.
To further investigate the role of DCA in HFD-induced hepatic steatosis, we orally administered either cholestylamine (CHO, a bile acid binder) to mice fed an HFD, or administered DCA orally to LFD-fed mice. Compared with LFD, the DCA supplementation increased the faecal DCA concentration (Fig. 5F). Compared with the HFD group, the concurrent oral feeding with CHO significantly reduced the concentration of DCA in the faeces (Fig. 5F). Abnormal bile acid levels have been shown to promote YAP1 activation [18]. CHO downregulated the mRNA levels of genes downstream of YAP1 (Ctgf and Cyr61) in the intestines of the HFD-fed mice, whereas DCA supplementation upregulated the expression of Ctgf and Cyr61 in the LFD-fed mice (Fig. 5G). In vitro studies revealed that the expression of the YAP1 downstream genes (CTGF and CYR61) was significantly increased in human ileal tissues cocultured with 100 μM DCA for 24 h (Fig. 5H). A significant increase in the number of pYAP1+ cells was further confirmed in human ileal enteroids cocultured with DCA for 72 h (Fig. 5I and J). However, DCA had no effect on YAP1 expression in the mouse ileal enteroids (Supplementary Fig. 2A and B). These results suggest that high concentrations of DCA induced by an HFD are key factors in activating the YAP1 pathway in ISCs.
High concentrations of DCA impair the differentiation functions of ISCs
To probe the mechanism by which an HFD affects the intestinal barrier via DCA, we examined the relevant indicators of ISC differentiation and proliferation.
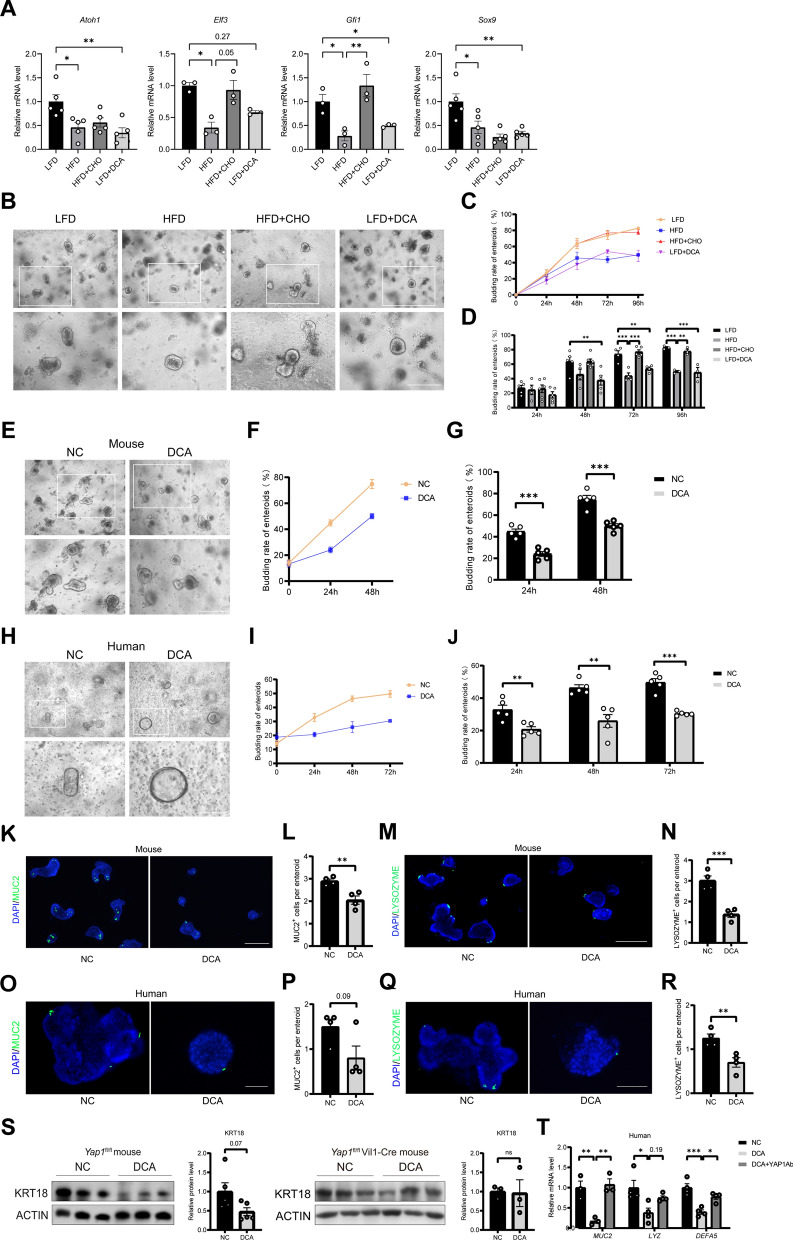
Compared with those in the LFD group, the mRNA levels of Atoh1, Elf3, and Gfi1, the key factors involved in ISC differentiation into goblet cells, and Sox9, the key factor involved in ISC differentiation into Paneth cells, were significantly downregulated by high-fat feeding, and concurrent oral feeding with the bile acid binder CHO partially reversed these changes. Compared with LFD supplementation, DCA supplementation downregulated the mRNA levels of Atoh1, Gfi1 and Sox9 in the ISCs of the LFD- and DCA-fed mice (Fig. 6A). Representative micrographs of intestinal enteroids in the four groups (LFD, HFD, HFD + CHO, and LFD + DCA) after 96 h of culture are shown in Fig. 6B. At both the 72-h and 96-h culture time points, the rate of ileal enteroid budding was significantly lower in the HFD-fed group than in the LFD group, and supplementation of the HFD with CHO prevented this decrease (Fig. 6C and D). Compared with LFD alone, LFD with DCA significantly reduced the budding rate of ileal enteroids in these mice at 48, 72, and 96 h (Fig. 6C and D). Immunofluorescence staining and periodic acid–Schiff (PAS) staining revealed that high-fat feeding or DCA supplementation downregulated the number of goblet and lysozyme+ Paneth cells compared with those in the LFD group, while CHO administration reversed these changes (Supplementary Fig. 3A–F). The mRNA levels of the Muc2, Fcgbp and Clca1 loci related to goblet cell function were significantly lower in the intestines of the HFD group than in those of the LFD group, and concurrent feeding with CHO partially reversed these changes. The addition of DCA to the LFD decreased the mRNA levels of Muc2, Fcgbp and Clca1 in the intestines of the LFD mice (Supplementary Fig. 3G). The gene expression levels of Defa5 and Lyz, which are related to Paneth cell function, were significantly lower in the intestines of the HFD-fed mice than in those of the LFD-fed mice, and cofeeding with CHO partially reversed these changes. Compared with the LFD supplementation, DCA supplementation of LFD downregulated the mRNA levels of Defa5 and Lyz in the intestines of these mice (Supplementary Fig. 3H). These results suggest that DCA induced by high-fat feeding reduces the ability of ISCs to differentiate into goblet and Paneth cells.
Fig. 6.
High concentrations of DCA inhibit the ISC differentiation into goblet and Paneth cells, which is prevented by YAP1 antibody. A The mRNA expression levels of Atoh1, Elf3, Gfi1 and Sox9 in mouse ileum tissue (n = 3 or 5). B Representative micrographs of intestinal enteroids in four groups after 96 h of culture in mice. Scale bars: 200 μm. C Line graph of changes in enteroids budding rates at 24, 48, 72 and 96 h (n = 3–6). D Comparison of enteroids budding rates at 24, 48, 72 and 96 h (n = 3–6). E Representative micrographs of mouse intestinal enteroids treated with or without DCA. Scale bars: 200 μm. F Line graph of changes in enteroids budding rates after DCA intervention with 24, 48 h (n = 5). G Comparison of enteroids budding rates after DCA intervention with 24, 48 h (n = 5). H Representative micrographs of human intestinal enteroids treated with or without DCA. Scale bars: 100 μm. I Line graph of changes in human enteroids budding rates after DCA intervention with 24, 48 and 72 h (n = 5). J Comparison of human enteroids budding rates after DCA intervention with 24, 48 and 72 h (n = 5). K Representative micrographs of immunofluorescence staining for MUC2 in mouse intestinal enteroids. Scale bars: 200 μm. L Quantitative analysis of K (n = 4). M Representative micrographs of immunofluorescence staining for lysozyme in mouse intestinal enteroids. Scale bars: 200 μm. N Quantitative analysis of M (n = 4). O Representative micrographs of immunofluorescence staining for MUC2 in human intestinal enteroids. Scale bars: 100 μm. P Quantitative analysis of O (n = 4). Q Representative micrographs of immunofluorescence staining for lysozyme in human intestinal enteroids. Scale bars: 100 μm. R Quantitative analysis of Q (n = 4). S Protein expression of goblet cell marker KRT18 in the terminal ileum of Yap1fl/fl mice and Yap1fl/fl Vil1-Cre mice after 24 h coculture with 100 μM DCA or DMSO (n = 5 or 3). T In vitro culture of human ileal tissue, the mRNA levels of MUC2, LYZ, and DEFA5 were assessed in human ileum tissue with various treatments (n = 3 or 4). DCA, deoxycholic acid; LFD, low-fat diet; HFD, high-fat diet; CHO, cholestyramine. *P < 0.05, **P < 0.01, ***P < 0.001
To provide direct evidence that DCA induces damage in ISCs, we incubated intestinal enteroids from mice or humans with DCA (100 μM), and the results revealed a decrease in the budding rate of these enteroids at different time points (Fig. 6E–J). Immunofluorescence staining revealed that the expression of MUC2 and lysozyme in the terminal ileal enteroids of both mice and humans was significantly reduced by coculture with DCA (Fig. 6K–R). Cytokeratin 18 (KRT18) is a marker of goblet cells and the loss of KRT18 can cause a breakdown of the keratin cytoskeleton [40]. In vitro tissue culture experiments revealed that DCA downregulated KRT18 protein expression in the terminal ileum of the Yap1fl/fl mice; however, no such decrease was detected in the Yap1fl/fl Vil1-Cre mice (Fig. 6S). These results suggest that DCA impairs the differentiation of ISCs. In vitro studies revealed that the mRNA levels of MUC2, LYZ, and DEFA5 in human ileum tissues were significantly reduced after 24 h of coculture with DCA, and these changes were partially reversed by preadministration of an anti-YAP1 antibody (Fig. 6T). These findings provide direct evidence that YAP1 is involved in the regulatory effect of DCA on ISCs.
Additional studies have confirmed the effect of DCA on ISC proliferation. Compared with the LFD group, the HFD group presented upregulated expression of LGR5 and KI67 in the ileum; the addition of oral bile acid binder CHO reversed this changes. Compared with the LFD group, the addition of DCA increased the expression of LGR5 and KI67 in the ileum of the mice (Supplementary Fig. 4A–D). Interestingly, DCA increased the diameter of the human intestinal enteroids (Supplementary Fig. 4E and F). These results suggest that DCA is involved in the abnormal proliferation of ISCs.
High concentrations of DCA cause hepatic inflammation and steatosis through the gut‒liver axis
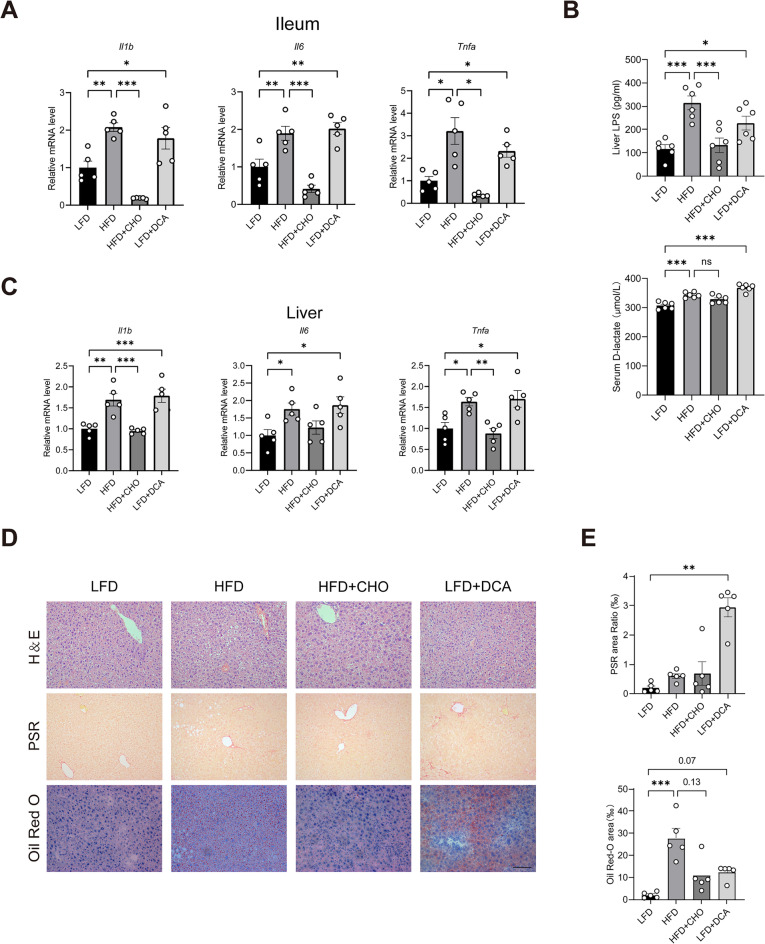
We studied the role of DCA in HFD-induced hepatic steatosis by varying the concentration of DCA in the intestine via supplementation with DCA and CHO. Mouse body weight was greater in the HFD group than in the LFD group; oral CHO reversed this change. Compared with LFD, the addition of DCA reduced the weight of the mice (Supplementary Fig. 5A and B). CHO downregulated the mRNA levels of inflammation-related genes, such as IL-1β, IL-6, and TNF-α, in the terminal ileum of the HFD-fed mice. Compared with LFD alone, DCA supplementation upregulated the expression of inflammation-related genes such as IL-1β, IL-6, and TNF-α in the terminal ileum of the LFD-fed mice (Fig. 7A). These results suggest that bile acids, especially DCA, play important roles in HFD-induced hepatic steatosis. Enzyme-linked immunosorbent assays revealed significant increases in the D-lactate content in the serum and the lipopolysaccharide (LPS) content in the livers of HFD-fed mice, and these increases were prevented by cofeeding with CHO. Compared with a LFD alone, DCA supplementation increased the content of D-lactate in the serum and LPS in the livers of the LFD-fed mice (Fig. 7B). It is well known that LPS is derived from intestinal Gram-negative bacteria, suggesting that LPS may be an important substance associated with the gut‒liver axis. Changes in the levels of IL-1β, IL-6, and TNF-α in the terminal ileum of four groups of mice (LFD, HFD, HFD + CHO, LFD + DCA) were also observed in the liver, suggesting a possible link to the gut-liver axis (Fig. 7C). In addition, haematoxylin‒eosin (HE) staining revealed significant inflammatory infiltrates in the livers of the HFD-fed mice, and these infiltrates were markedly improved by concurrent CHO feeding. LFD supplementation with DCA resulted in a significant inflammatory infiltrate in the livers of these mice compared with those in the LFD group (Fig. 7D). Picrosirius red (PSR) histochemical staining revealed that DCA significantly increased collagen deposition in the mouse liver. Oil red histochemical staining revealed that CHO reduced the content of lipid droplets in the livers of the HFD-induced mice (p = 0.13). LFD supplementation with DCA increased the content of the lipid droplets in the livers of these mice compared with those in the LFD group (p = 0.07) (Fig. 7D and E). Notably, a dietary DCA treatment decreased the levels of hepatic Yap1 or its target genes (Supplementary Fig. 5C). These results indicate that an HFD may impair intestinal barrier function and increase the abundance of Gram-negative bacteria through DCA, thereby increasing LPS accumulation in the liver, leading to inflammation and steatosis.
Fig. 7.
High concentrations of DCA cause liver inflammation and steatosis by disrupting the intestinal barrier. A mRNA expression of IL-1β, IL-6 and TNF-α in mouse ileum tissue (n = 5). B Serum D-lactate concentrations and LPS concentrations in mice liver tissues (n = 6). C mRNA expression of IL-1β, IL-6 and TNF-α in mouse liver tissue (n = 5). D Representative micrographs of haematoxylin‒eosin, picrosirius red and Oil Red histochemical staining. Scale bars: 100 μm. E Quantitative analysis of D (n = 5). Lipopolysaccharide, LPS; LFD, low-fat diet; HFD, high-fat diet; HFD + CHO, HFD + cholestyramine; DCA, deoxycholic acid. *P < 0.05, **P < 0.01, ***P < 0.001
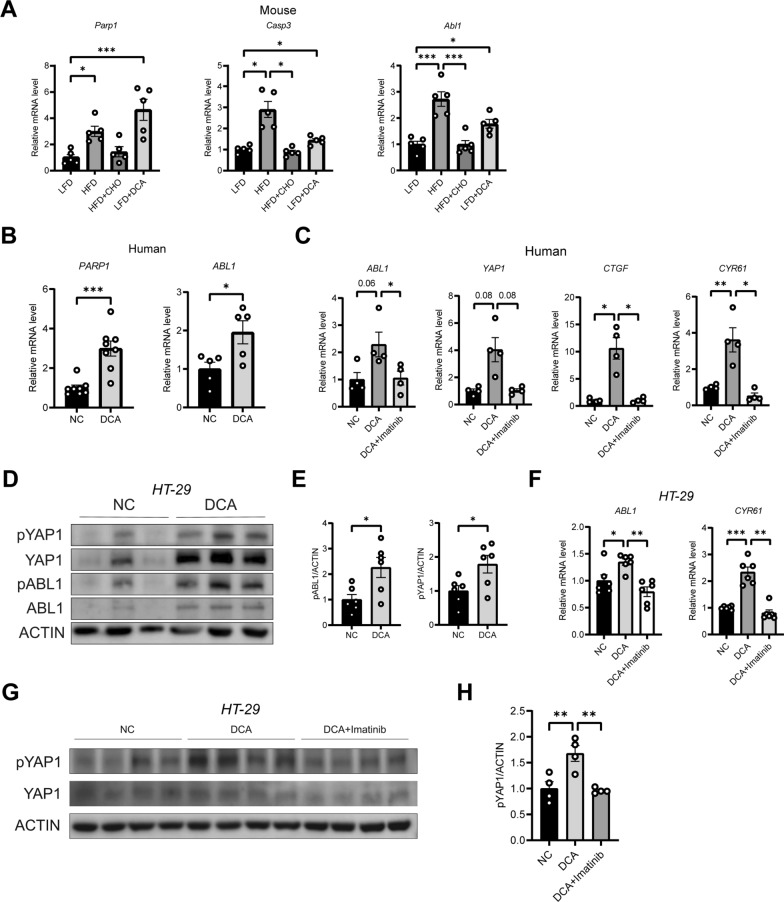
DCA regulates YAP1 signalling through ABL1
Furthermore, we investigated how DCA activates YAP1 and impairs the differentiation function of ISCs. As mentioned earlier, high concentrations of DCA can cause DNA damage [19]. According to previous studies, DNA damage can be sensed by ABL1 [23], and YAP1 is one of the substrates of ABL1 [23, 25]. Therefore, we hypothesized that DCA promotes YAP1 phosphorylation by activating ABL1. We found that CHO decreased the mRNA levels of DNA damage-related genes (Parp1 and Caspase3) and a YAP1 upstream-related gene (Abl1) in the ileums of the HFD-fed mice (Fig. 8A). Compared with a LFD alone, DCA supplementation increased the mRNA levels of Parp1, Caspase3, and Abl1 in the ileums of LFD mice compared to those in the LFD-fed group (Fig. 8A). In addition, the expression of PARP1 and ABL1 was significantly increased in human ileal tissues cocultured with DCA for 24 h (Fig. 8B). In addition, the increased expression of ABL1, YAP1, and its downstream CTGF, and CYR61 genes induced by the administration of DCA was reversed by the ABL1 antagonist Imatinib in the in vitro coculture with human distal ileal tissue (Fig. 8C). In HT-29 cells, coculture with DCA changed the phosphorylation of YAP1 and ABL1 in a time-dependent manner (Supplementary Fig. 2C and D), and the phosphorylation levels of both were significantly increased after just 15 min (Fig. 8D and E). Moreover, DCA upregulated the expression of CYR61, the target gene of YAP1, and the phosphorylation of the YAP1 protein, which was reversed by treatment with an ABL1 antagonist (Fig. 8F–H). These results suggest that DCA regulates YAP1 phosphorylation through ABL1.
Fig. 8.
High concentrations of DCA activate YAP1 via ABL1. A mRNA expression levels of Parp1, Caspase3, and Abl1 in the ileum tissue of the mice (n = 5). B mRNA expression levels of PARP1 and ABL1 in human ileum tissue treated with 100 μM DCA for 24 h (n = 8 or 5). C Relative mRNA expression of ABL1, YAP1, CTGF, and CYR61 in human ex vivo tissue culture with DCA, or DCA combined with the ABL1 inhibitor Imatinib (n = 4). D Relative protein expressions of pYAP1, YAP1, pABL1, and ABL1 in HT-29 cells treated with or without DCA for 15 min. E Quantitative analysis of D (n = 6). F Relative mRNA expression of ABL1 and CYR61 in HT-29 cells treated with DCA alone, or with DCA combined with the ABL1 inhibitor Imatinib (n = 6). G The relative protein expressions of pYAP1 and YAP1 in HT-29 cells treated with DCA alone, or with DCA combined with the ABL1 inhibitor, Imatinib. H Quantitative analysis of G (n = 4). LFD, low-fat diet; HFD, high-fat diet; HFD + CHO, HFD + cholestyramine; DCA, deoxycholic acid. *P < 0.05, **P < 0.01, ***P < 0.001
Discussion
This study confirmed that the induction of high intestinal DCA levels by chronic HFD consumption activated the ABL1‒YAP1 signalling pathway, impaired the differentiation of ISCs into goblet and Paneth cells, resulting in intestinal barrier function dysfunction, enrichment of intestinal Gram-negative bacteria and excessive accumulation of bacterial metabolite LPS in the livers, which may be an important enterogenic mechanism of hepatic steatosis.
The chronic low-grade inflammatory state prevailing in patients with MAFLD may be due to an invasion by intestinal pathogens. There are significant differences in the composition and abundance of gut microbiota between MAFLD patients and healthy people [41–43]. Patients with MAFLD presented reduced gut microbiota diversity, accompanied by an decreased proportion of beneficial bacteria, and an increased abundance of harmful bacteria such as the enrichment of some Gram-negative bacteria [44, 45]. Overgrowth of intestinal bacteria has been reported in 50% of patients with MAFLD, with significantly elevated levels of endotoxins, LPS, and inflammatory markers associated with the histological progression of liver injury, along with increased intestinal permeability [46]. Exposure to an HFD is an important cause of MAFLD. Similar to the results of Shen et al. [47], we found in the present study that high-fat feeding for 8 weeks resulted in an increase in hepatic steatosis in mice, accompanied by the upregulation of liver inflammatory factors and the accumulation of LPS. The increased LPS load in the liver is caused by an increase in the number of LPS-producing bacteria in the gut. This study demonstrated the enrichment of Gram-negative bacteria in the terminal ileum of HFD-fed mice, accompanied by a decrease in the number and function of goblet cells that maintain the intestinal epithelial mucosal barrier and Paneth cells that maintain the immune barrier. Moreover, we found that the intestinal YAP1 signalling pathway was significantly activated into the nucleus in the HFD-fed mice compared with the LFD-fed mice, and the upregulation of YAP1 signalling was also found in the terminal ileum specimens from patients with MAFLD.
To validate the regulatory role of YAP1 in intestinal mucosal barrier function, we constructed Yap1 gene knockout mice, in which the gene deletion was targeted to the intestinal epithelium. Our in vivo experiments revealed that YAP1 knockout significantly reduced HFD-induced liver inflammation and LPS accumulation, also alleviated hepatic steatosis. This exciting new finding suggests that intestinal YAP1 could be a potential target for intervening in the development of hepatic steatosis through the gut-liver axis, which appears to play a critical regulatory role. At the intestinal level, knocking out YAP1 not only alleviated HFD-induced gut inflammation, but also altered the composition and function of the gut microbiota: At the phylum level, the HFD-induced decrease in Firmicutes and Bacteroidetes, along with the increases in Verrucobacteria and Proteobacteria, was reversed. At the family level, the elevated abundance of Akkmansiaceae and Enterobacteriacea caused by an HFD was also reversed, indicating improved enrichment of Gram-negative bacteria and a subsequent reduction in hepatic LPS accumulation. Additionally, the analysis of bacterial metabolic functions revealed a trend toward reversing the decline in lipid metabolism caused by an HFD. The above evidence from gut-liver axis supports the notion that targeting intestinal epithelial YAP1 knockout can mitigate hepatic inflammation and steatosis.
Similar to the previous finding [32, 33], we also confirmed that intestinal epithelial YAP1 is predominantly expressed in ISCs located in the crypts. In this study, a YAP1 knockout improved HFD-induced damage to goblet and Paneth cells, as well as the proliferation and differentiation function of ISCs [48]. The YAP1-TEAD transcription factor complex has been reported to be a key downstream effector of WNT signalling in the gut [32]. In physiological homeostasis, the regulation of LGR5+ ISC function is driven by WNT signalling [33], and the YAP1 protein expressed in LGR5+ ISCs is inhibited by upstream Hippo signalling under high WNT activity, preventing the uncontrolled overgrowth of intestinal crypt cells [49]. In the case of intestinal injury, YAP1 expressed in ISCs is activated and translocated into the nucleus to increase ISC proliferation and reduce ISC differentiation into the goblet and Paneth cells by inhibiting the nuclear translocation of β-CATENIN, downstream of the WNT signalling pathway [49]. A previous study revealed that the upregulation of YAP1, through the elimination of the Lipolysis-stimulated lipoprotein receptor in ISCs, can inhibit Paneth cell differentiation, thereby altering the composition of the intestinal flora [50]. We found that a YAP1 knockout significantly modified the HFD-induced intestinal flora structure, which may be attributed to the effect of YAP1 on the differentiation of ISCs into Paneth cells. In animal models of dextran sulfate sodium-induced enteritis and radiation-induced acute, short-term intestinal injury, YAP1 was found to play an active role in promoting intestinal mucosal repair and regeneration [33, 49]. In vitro studies by Giovanni's team [18] revealed that an agonist of TGR5 promoted the regeneration of LGR5+ cells and enteroid growth by activating YAP1 and its upstream regulator, the proto-oncogene tyrosine-protein kinase SRC. The GTEx database revealed a positive correlation between TGR5 and YAP1 in normal human ileums, suggesting that bile acids in the intestinal cavity may regulate intestinal YAP1. However, abnormal acute activation and long-term chronic activation of bile acid signalling have been shown to be harmful [19]. For gastrointestinal mucosa exposed to high concentrations of DCA derived from chronic HFD consumption, previous studies have shown an impairment of the intestinal barrier function, chronic low-grade inflammation, and the consequent expansion of cancer cells with epithelial stem cell properties [20, 21]. What role does bile acid play in the regulation of YAP1 in ISCs during this pathological process?
Alteration in the bile acid pool and its metabolism have been reported in patients with MAFLD [13–15]. Similar to the results of previous studies [6, 12, 51], our data also revealed that high-fat or DCA feeding for 8 weeks changed the composition of the intestinal bile acid pool and induced the production of more hydrophobic secondary bile acid, namely DCA. Bernstein et al. [11] have reported that high-fat intake led to colonic mucosal exposure to DCA concentrations ranging from 0.07–0.73 mM. High concentrations of DCA have been reported to induce oxidative stress in ISCs, causing DNA damage and inducing apoptosis, thereby disrupting the intestinal mucosal barrier [19, 22]. In this study, we found that a DCA diet mimicked HFD-induced hepatic steatosis, which was accompanied by increased hepatic LPS levels and intestinal inflammation. Moreover, we observed that 8 weeks of DCA feeding, significantly increased the expression of YAP1 downstream functional genes Ctgf and Cyr61 in the intestinal tissue of mice. But surprisingly, DCA feeding reduced the expression of Ctgf and Cyr61 at the gene level in liver tissue of mice. Previous studies have indicated that the activation of YAP1 signalling in hepatocytes is often associated with the development of steatosis and steatohepatitis [52, 53]. Therefore, we speculated that the direct effect of DCA on liver tissue may not play a decisive role in the onset of hepatic steatosis. In in vitro studies, significant upregulation of CTGF and CYR61 was observed in human terminal ileal tissues cultured with 100 μM DCA for 24 h. The results revealed that high concentrations of DCA were involved in the adverse regulation of the gut‒liver axis during high-fat feeding through YAP1 signalling.
The normal function of ISCs is based on maintaining intestinal barrier homeostasis as well as regeneration and repair after intestinal mucosal injury [5]. We observed that exposure to an HFD or DCA supplementation for 8 weeks abnormally enhanced the proliferative function of ISCs, while reducing the differentiation of ISCs into goblet and Paneth cells that make up the intestinal epithelial barrier. Notably, supplementation of an HFD with CHO reversed the abovementioned changes induced by high-fat feeding. We also found that the budding rates of ileal enteroids in the HFD-fed and DCA-fed mice were significantly lower than that in the LFD-fed mice, and supplementation of an HFD with CHO reversed the decline in the budding rate induced by the HFD. Knocking out the Yap1 gene increased the germination rate of ileal enteroids in the HFD-fed mice, and reversed the inhibitory effect of an HFD on ISC differentiation. In vitro coculture experiments with mouse or human terminal ileal enteroids demonstrated that DCA decreased the budding rate of ileal enteroids and the numbers of goblet and Paneth cells. However, a YAP1 knockout targeting the intestinal epithelium or the use of YAP1 inhibitors eliminated inhibition effects of DCA on intestinal goblet and Paneth cells. Our data revealed that DCA induced by high-fat feeding was involved in dysregulating the differentiation of intestinal LGR5+ ISCs into goblet and Paneth cells, whereas YAP1 may be involved in the regulation of ISCs by DCA.
In a preliminary study of the regulatory mechanisms of YAP1 induced by high concentrations of DCA, we observed that after 8 weeks of exposure to an HFD or DCA, the activation of intestinal YAP1 signalling was accompanied by increased expression of Parp1 (a marker of DNA damage) and Abl1 (a sensor that responds to DNA damage). High DCA concentrations can cause DNA damage by inducing the generation of reactive oxygen species [19, 20, 22]. ABL1 is a member of the SRC family of nonreceptor tyrosine kinases. The YAP1 protein is a direct substrate of ABL1, and endogenous or recombinant ABL1 can increase the tyrosine phosphorylation of YAP1 at Y357, and increase its stability and activity [23, 25]. In vitro studies using human ileal tissue and cell line cultures directly confirmed that 100 μM DCA induced DNA damage and enhanced the expression of PARP1 and ABL1. Human cell line culture also revealed that DCA promoted the phosphorylation of YAP1Y357 in a time-dependent manner, accompanied by increased ABL1 phosphorylation. To determine the regulatory role of ABL1 on YAP1, we used the selective ABL1 kinase inhibitor Imatinib in our in vitro studies. Beyond its well-documented antitumor effects [54, 55], Imatinib has been reported to slow the progression of hepatic steatosis and fibrosis [56, 57]. In vivo and in vitro studies by AlAsfoor et al. demonstrated that Imatinib reduced pro-inflammatory markers in mouse macrophages and human monocytes and downregulated the gene expression of the SREBP transcription factor, known for activating the lipogenic transcription program, thereby improving liver steatosis in obese mice [56]. Similarly, the results of Kim et al. 's study revealed that the stimulation of overnutrition can promote the phosphorylation of PPARγ2 through ABL1 kinase, enhancing its interaction with MLL4, contributing to hepatic steatosis in mice, which can be improved by Imatinib [58]. In this study, using in vitro models of human distal ileum tissue and the HT-29 cell line, we observed that Imatinib reversed DCA-induced upregulation of the YAP1 signalling pathway. Our results suggest that ABL1 is involved in the upstream regulation of YAP1 by DCA in the gut. Future studies are needed to knockout Abl1 gene in the intestinal epithelium of mice or administering Imatinib in vivo to confirm the involvement of ABL1 in the regulation of YAP1Y357 phosphorylation by DCA, as well as to clarify the selectivity and specificity of ABL1 inhibition sites.
Our study has several limitations. The current study revealed that high concentrations of DCA induced by high-fat feeding led to ISC dysfunction by inducing DNA damage and activating YAP1 translocation into the nucleus. CHO is an anion exchange resin that sequesters a variety of bile acid subclasses including DCA. Hence, further studies are needed to confirm the direct effects of DCA by feeding it to Yap1 gene knockout mice. DNA damage markers and Abl1 levels were found to be increased in mice fed HFD or DCA. Future studies using germ-free mice are needed to elucidate the effects of YAP1 knockout targeted to the intestinal epithelium on the gut microbiota. High-fat feeding for 8 weeks mimics hepatic inflammation and steatosis in the initial stage of MAFLD development, and the role of YAP1 in the subsequent progression of this disease needs to be validated.
Conclusions
We found that HFD-mediated elevation of DCA levels dysregulates the ISC function. DCA downregulates ISC differentiation into goblet and Paneth cells through upregulation of the ABL1–YAP1 signalling pathway, thereby disrupting the ileal mucosal barrier function, which may be one of the initiating mechanisms of hepatic steatosis induced by an HFD via the gut‒liver axis (Fig. 9). Targeting the ABL1‒YAP1 pathway, or reducing DCA is expected to block the above enterogenic mechanisms of MAFLD.
Fig. 9.
A schematic depicting the mechanism involving HFD-mediated increase in DCA levels leading to dysregulation of ISC function and hepatic steatosis via ABL1‒YAP pathway
Supplementary Information
Acknowledgements
We thank Dr. Chen Huang (Department of General Surgery of Shanghai General Hospital) for his support in the collection of human specimens. We also thank Prof. Lei Zhang (Excellence in Molecular Cell Science, Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences, Shanghai, China) for the generous provision of Yap1fl/fl mice.
Abbreviations
- YAP1
Yes-associated protein 1
- ABL1
Abelson tyrosine-protein kinase 1
- NAFLD
Nonalcoholic fatty liver disease
- MAFLD
Metabolic dysfunction-associated fatty liver disease
- HFD\HFF
High-fat diet\high-fat feeding
- LFD
Low-fat diet
- CHO
Cholestyramine
- DCA
Deoxycholic acid
- LPS
Lipopolysaccharide
- ISC
Intestinal stem cell
- EDTA
Ethylenediaminetetraacetic acid
- pYAP1
Phosphorylated Yes-associated protein 1
- pABL1
Phosphorylated abelson tyrosine-protein kinase 1
- KRT18
Cytokeratin 18
Author contributions
Tiancheng Mao: Writing-original draft, Formal analysis, Data curation, Investigation, Methodology, Visualization. Xianjun Xu: Writing-original draft, Data curation, Investigation, Visualization. Leheng Liu: Writing-original draft, Conceptualization, Investigation. Methodology; Yulun Wu: Investigation, Visualization, Formal analysis. Xiaowan Wu: Formal analysis, Data curation. Wenlu Niu: Investigation, Formal analysis. Dandan You: Investigation, Formal analysis. Xiaobo Cai: Investigation, Project administration, Supervision, Writing-Review & Editing. Lungen Lu: Project administration, Supervision, Writing-Review & Editing. Hui Zhou: Conceptualization, Funding acquisition, Project administration, Supervision, Writing-Review & Editing.
Funding
This work was financially supported by the National Natural Science Foundation of China (82370558).
Data availability
All data from this article are available from the corresponding authors upon request.
Declarations
Ethics approval and consent to participate
All animal procedures were approved by the Institutional Animal Care and Use Committee of the Shanghai General Hospital (protocol code 2021SQ205, 1 Match 2021). The human study was approved by the Ethics Committee of the Shanghai General Hospital (approval ID 20240226025226875). The study protocol adhered to the ethical principles of medical research involving human subjects as outlined in the 2013 Helsinki Declaration and the 2018 Istanbul Declaration.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no conflict of interest or financial conflicts to disclose.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Tiancheng Mao, Xianjun Xu and Leheng Liu are contributed equally as the first authors.
Contributor Information
Xiaobo Cai, Email: caixiaobo19790719@126.com.
Lungen Lu, Email: lungenlu1965@163.com.
Hui Zhou, Email: mdzhouhui@163.com.
References
- 1.Ji J, Wu L, Wei J, Wu J, Guo C. The gut microbiome and ferroptosis in MAFLD. J Clin Transl Hepatol. 2023;11(1):174–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoyles L, Fernández-Real J-M, Federici M, Serino M, Abbott J, Charpentier J, et al. Molecular phenomics and metagenomics of hepatic steatosis in non-diabetic obese women. Nat Med. 2018;24(7):1070–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bischoff SC, Barbara G, Buurman W, Ockhuizen T, Schulzke J-D, Serino M, et al. Intestinal permeability—a new target for disease prevention and therapy. BMC Gastroenterol. 2014;14:189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Turner JR. Intestinal mucosal barrier function in health and disease. Nat Rev Immunol. 2009;9(11):799–809. [DOI] [PubMed] [Google Scholar]
- 5.Gehart H, Clevers H. Tales from the crypt: new insights into intestinal stem cells. Nat Rev Gastroenterol Hepatol. 2019;16(1):19–34. [DOI] [PubMed] [Google Scholar]
- 6.Liu L, Xu J, Xu X, Mao T, Niu W, Wu X, et al. Intestinal stem cells damaged by deoxycholic acid via AHR pathway contributes to mucosal barrier dysfunction in high-fat feeding mice. Int J Mol Sci. 2022. 10.3390/ijms232415578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martel J, Chang S-H, Ko Y-F, Hwang T-L, Young JD, Ojcius DM. Gut barrier disruption and chronic disease. Trends Endocrinol Metab. 2022;33(4):247–65. [DOI] [PubMed] [Google Scholar]
- 8.Loomba R, Friedman SL, Shulman GI. Mechanisms and disease consequences of nonalcoholic fatty liver disease. Cell. 2021;184(10):2537–64. [DOI] [PubMed] [Google Scholar]
- 9.Serino M, Luche E, Gres S, Baylac A, Bergé M, Cenac C, et al. Metabolic adaptation to a high-fat diet is associated with a change in the gut microbiota. Gut. 2012;61(4):543–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rohr MW, Narasimhulu CA, Rudeski-Rohr TA, Parthasarathy S. Negative effects of a high-fat diet on intestinal permeability: a review. Adv Nutr. 2020;11(1):77–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bernstein H, Bernstein C, Payne CM, Dvorakova K, Garewal H. Bile acids as carcinogens in human gastrointestinal cancers. Mutat Res. 2005;589(1):47–65. [DOI] [PubMed] [Google Scholar]
- 12.Yoshimoto S, Loo TM, Atarashi K, Kanda H, Sato S, Oyadomari S, et al. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature. 2013. 10.1038/nature12347. [DOI] [PubMed] [Google Scholar]
- 13.Jiao N, Baker SS, Chapa-Rodriguez A, Liu W, Nugent CA, Tsompana M, et al. Suppressed hepatic bile acid signalling despite elevated production of primary and secondary bile acids in NAFLD. Gut. 2018;67(10):1881–91. [DOI] [PubMed] [Google Scholar]
- 14.Puri P, Daita K, Joyce A, Mirshahi F, Santhekadur PK, Cazanave S, et al. The presence and severity of nonalcoholic steatohepatitis is associated with specific changes in circulating bile acids. Hepatology. 2018;67(2):534–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smirnova E, Muthiah MD, Narayan N, Siddiqui MS, Puri P, Luketic VA, et al. Metabolic reprogramming of the intestinal microbiome with functional bile acid changes underlie the development of NAFLD. Hepatology. 2022;76(6):1811–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ridlon JM, Harris SC, Bhowmik S, Kang D-J, Hylemon PB. Consequences of bile salt biotransformations by intestinal bacteria. Gut Microbes. 2016;7(1):22–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Calzadilla N, Comiskey SM, Dudeja PK, Saksena S, Gill RK, Alrefai WA. Bile acids as inflammatory mediators and modulators of intestinal permeability. Front Immunol. 2022;13:1021924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sorrentino G, Perino A, Yildiz E, El Alam G, Bou Sleiman M, Gioiello A, et al. Bile acids signal via TGR5 to activate intestinal stem cells and epithelial regeneration. Gastroenterology. 2020;159(3):956. [DOI] [PubMed] [Google Scholar]
- 19.Zeng H, Umar S, Rust B, Lazarova D, Bordonaro M. Secondary bile acids and short chain fatty acids in the colon: a focus on colonic microbiome, cell proliferation, inflammation, and cancer. Int J Mol Sci. 2019. 10.3390/ijms20051214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li D, Cao W. Bile acid receptor TGR5, NADPH oxidase NOX5-S and CREB mediate bile acid-induced DNA damage in Barrett’s esophageal adenocarcinoma cells. Sci Rep. 2016;6:31538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Glinghammar B, Inoue H, Rafter JJ. Deoxycholic acid causes DNA damage in colonic cells with subsequent induction of caspases, COX-2 promoter activity and the transcription factors NF-kB and AP-1. Carcinogenesis. 2002;23(5):839–45. [DOI] [PubMed] [Google Scholar]
- 22.Liu L, Dong W, Wang S, Zhang Y, Liu T, Xie R, et al. Deoxycholic acid disrupts the intestinal mucosal barrier and promotes intestinal tumorigenesis. Food Funct. 2018;9(11):5588–97. [DOI] [PubMed] [Google Scholar]
- 23.Levy D, Adamovich Y, Reuven N, Shaul Y. Yap1 phosphorylation by c-Abl is a critical step in selective activation of proapoptotic genes in response to DNA damage. Mol Cell. 2008;29(3):350–61. [DOI] [PubMed] [Google Scholar]
- 24.Plattner R, Kadlec L, DeMali KA, Kazlauskas A, Pendergast AM. c-Abl is activated by growth factors and Src family kinases and has a role in the cellular response to PDGF. Genes Dev. 1999;13(18):2400–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li B, He J, Lv H, Liu Y, Lv X, Zhang C, et al. c-Abl regulates YAPY357 phosphorylation to activate endothelial atherogenic responses to disturbed flow. J Clin Invest. 2019;129(3):1167–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ren F, Ning H, Ge Y, Yin Z, Chen L, Hu D, et al. Bisphenol A induces apoptosis in response to DNA damage through c-Abl/YAPY357/ p73 pathway in P19 embryonal carcinoma stem cells. Toxicology. 2022;470: 153138. [DOI] [PubMed] [Google Scholar]
- 27.Strano S, Monti O, Pediconi N, Baccarini A, Fontemaggi G, Lapi E, et al. The transcriptional coactivator Yes-associated protein drives p73 gene-target specificity in response to DNA Damage. Mol Cell. 2005;18(4):447–59. [DOI] [PubMed] [Google Scholar]
- 28.Ibar C, Irvine KD. Integration of hippo-YAP signaling with metabolism. Dev Cell. 2020;54(2):256–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qing J, Ren Y, Zhang Y, Yan M, Zhang H, Wu D, et al. Dopamine receptor D2 antagonism normalizes profibrotic macrophage-endothelial crosstalk in non-alcoholic steatohepatitis. J Hepatol. 2022;76(2):394–406. [DOI] [PubMed] [Google Scholar]
- 30.Zheng K, Zhou W, Ji J, Xue Y, Liu Y, Li C, et al. Si-Ni-San reduces lipid droplet deposition associated with decreased YAP1 in metabolic dysfunction-associated fatty liver disease. J Ethnopharmacol. 2023;305: 116081. [DOI] [PubMed] [Google Scholar]
- 31.Johnson R, Halder G. The two faces of Hippo: targeting the Hippo pathway for regenerative medicine and cancer treatment. Nat Rev Drug Discov. 2014;13(1):63–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guillermin O, Angelis N, Sidor CM, Ridgway R, Baulies A, Kucharska A, et al. Wnt and Src signals converge on YAP-TEAD to drive intestinal regeneration. EMBO J. 2021;40(13): e105770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deng F, Peng L, Li Z, Tan G, Liang E, Chen S, et al. YAP triggers the Wnt/β-catenin signalling pathway and promotes enterocyte self-renewal, regeneration and tumorigenesis after DSS-induced injury. Cell Death Dis. 2018;9(2):153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seo Y, Park S-Y, Kim H-S, Nam J-S. The Hippo-YAP signaling as guardian in the pool of intestinal stem cells. Biomedicines. 2020. 10.3390/biomedicines8120560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang N, Bai H, David KK, Dong J, Zheng Y, Cai J, et al. The Merlin/NF2 tumor suppressor functions through the YAP oncoprotein to regulate tissue homeostasis in mammals. Dev Cell. 2010;19(1):27–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu T, An Z, Shi C, Li P, Liu L. A sensitive and efficient method for simultaneous profiling of bile acids and fatty acids by UPLC-MS/MS. J Pharm Biomed Anal. 2020;178: 112815. [DOI] [PubMed] [Google Scholar]
- 37.Zhang J, Gupte J, Gong Y, Weiszmann J, Zhang Y, Lee KJ, et al. Chronic over-expression of fibroblast growth factor 21 increases bile acid biosynthesis by opposing FGF15/19 action. EBioMedicine. 2017;15:173–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wen ZS, Du M, Tang Z, Zhou TY, Zhang ZS, Song HH, et al. Low molecular seleno-aminopolysaccharides protect the intestinal mucosal barrier of rats under weaning stress. Int J Mol Sci. 2019. 10.3390/ijms20225727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beyaz S, Mana MD, Roper J, Kedrin D, Saadatpour A, Hong S-J, et al. High-fat diet enhances stemness and tumorigenicity of intestinal progenitors. Nature. 2016;531(7592):53–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hesse M, Grund C, Herrmann H, Bröhl D, Franz T, Omary MB, et al. A mutation of keratin 18 within the coil 1A consensus motif causes widespread keratin aggregation but cell type-restricted lethality in mice. Exp Cell Res. 2007;313(14):3127–40. [DOI] [PubMed] [Google Scholar]
- 41.Del Chierico F, Nobili V, Vernocchi P, Russo A, De Stefanis C, Gnani D, et al. Gut microbiota profiling of pediatric nonalcoholic fatty liver disease and obese patients unveiled by an integrated meta-omics-based approach. Hepatology. 2017;65(2):451–64. [DOI] [PubMed] [Google Scholar]
- 42.Hernández-Ceballos W, Cordova-Gallardo J, Mendez-Sanchez N. Gut microbiota in metabolic-associated fatty liver disease and in other chronic metabolic diseases. J Clin Transl Hepatol. 2021;9(2):227–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shen F, Zheng RD, Sun XQ, Ding WJ, Wang XY, Fan JG. Gut microbiota dysbiosis in patients with non-alcoholic fatty liver disease. Hepatobiliary Pancreat Dis Int. 2017;16(4):375–81. [DOI] [PubMed] [Google Scholar]
- 44.Albillos A, de Gottardi A, Rescigno M. The gut-liver axis in liver disease: pathophysiological basis for therapy. J Hepatol. 2020;72(3):558–77. [DOI] [PubMed] [Google Scholar]
- 45.Zhong HJ, Zhuang YP, Xie X, Song JY, Wang SQ, Wu L, et al. Washed microbiota transplantation promotes homing of group 3 innate lymphoid cells to the liver via the CXCL16/CXCR6 axis: a potential treatment for metabolic-associated fatty liver disease. Gut Microbes. 2024;16(1):2372881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Soppert J, Brandt EF, Heussen NM, Barzakova E, Blank LM, Kuepfer L, et al. Blood endotoxin levels as biomarker of nonalcoholic fatty liver disease: a systematic review and meta-analysis. Clin Gastroenterol Hepatol. 2023;21(11):2746–58. [DOI] [PubMed] [Google Scholar]
- 47.Shen Z, Shen B, Dai W, Zhou C, Luo X, Guo Y, et al. Expansion of macrophage and liver sinusoidal endothelial cell subpopulations during non-alcoholic steatohepatitis progression. iScience. 2023;26(5): 106572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sato T, van Es JH, Snippert HJ, Stange DE, Vries RG, van den Born M, et al. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature. 2011;469(7330):415–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gregorieff A, Liu Y, Inanlou MR, Khomchuk Y, Wrana JL. Yap-dependent reprogramming of Lgr5(+) stem cells drives intestinal regeneration and cancer. Nature. 2015;526(7575):715–8. [DOI] [PubMed] [Google Scholar]
- 50.An Y, Wang C, Fan B, Wang Z, Li Y, Kong F, et al. LSR targets YAP to modulate intestinal Paneth cell differentiation. Cell Rep. 2023;42(9): 113118. [DOI] [PubMed] [Google Scholar]
- 51.Zhou H, Zhou S-Y, Gillilland M, Li J-Y, Lee A, Gao J, et al. Bile acid toxicity in Paneth cells contributes to gut dysbiosis induced by high-fat feeding. JCI Insight. 2020. 10.1172/jci.insight.138881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee NY, Choi MG, Lee EJ, Koo JH. Interplay between YAP/TAZ and metabolic dysfunction-associated steatotic liver disease progression. Arch Pharm Res. 2024;47(6):558–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhu C, Tabas I, Schwabe RF, Pajvani UB. Maladaptive regeneration—the reawakening of developmental pathways in NASH and fibrosis. Nat Rev Gastroenterol Hepatol. 2021;18(2):131–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cohen MH, Williams G, Johnson JR, Duan J, Gobburu J, Rahman A, et al. Approval summary for imatinib mesylate capsules in the treatment of chronic myelogenous leukemia. Clin Cancer Res. 2002;8(5):935–42. [PubMed] [Google Scholar]
- 55.Lam TJR, Udonwa SA, Masuda Y, Yeo MHX, Farid Bin Harunal Ras M, Goh BKP. A systematic review and meta-analysis of neoadjuvant imatinib use in locally advanced and metastatic gastrointestinal stromal tumors. World J Surg. 2024;48(7):1681–91. [DOI] [PubMed] [Google Scholar]
- 56.AlAsfoor S, Rohm TV, Bosch AJT, Dervos T, Calabrese D, Matter MS, et al. Imatinib reduces non-alcoholic fatty liver disease in obese mice by targeting inflammatory and lipogenic pathways in macrophages and liver. Sci Rep. 2018;8(1):15331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kuo WL, Yu MC, Lee JF, Tsai CN, Chen TC, Chen MF. Imatinib mesylate improves liver regeneration and attenuates liver fibrogenesis in CCL4-treated mice. J Gastrointest Surg. 2012;16(2):361–9. [DOI] [PubMed] [Google Scholar]
- 58.Kim DH, Kim J, Kwon JS, Sandhu J, Tontonoz P, Lee SK, et al. Critical Roles of the Histone Methyltransferase MLL4/KMT2D in Murine Hepatic Steatosis Directed by ABL1 and PPARγ2. Cell Rep. 2016;17(6):1671–82. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data from this article are available from the corresponding authors upon request.