Abstract
This study evaluated the inhibitory effects of calceolarioside B, extracted from the traditional Chinese herb Mutong (Akebia quinata Thumb), on the SARS-CoV-2 Omicron BA.2 variant. Molecular docking and molecular dynamics simulations predicted the binding sites and interactions between calceolarioside B and the Omicron BA.2 spike (S) protein. Biolayer interferometry (BLI) and immunofluorescence assays validated its high-affinity binding. Pseudovirus entry assays assessed the inhibitory effects of calceolarioside B on viral entry into host cells, while enzyme-linked immunosorbent assay (ELISA) measured inflammatory cytokine levels. Flow cytometry was used to analyze its effects on macrophage phenotype switching. Results demonstrated that calceolarioside B could bind to the Omicron BA.2 S protein with high affinity, and significantly inhibited viral entry into host cells by interfering with the binding of angiotensin-converting enzyme 2 (ACE2) receptor and S protein. Additionally, calceolarioside B reduced IL(interleukin)-6 expression levels and promoted the switch of macrophages from the pro-inflammatory M1 phenotype to the anti-inflammatory M2 phenotype. These findings suggest that calceolarioside B possesses antiviral and immunomodulatory effects, making it a potential dual-function inhibitor for the treatment of COVID-19.
Keywords: Calceolarioside B, SARS-CoV-2 Omicron BA.2, Molecular docking, Viral inhibition, Immunomodulation
Introduction
The global outbreak of COVID-19 began in Wuhan at the end of 2019. By July 2022, the cumulative number of infections worldwide had exceeded 500 million, with a death toll exceeding 6 million, posing a serious threat to global public health and economy (WHO).
COVID-19 is caused by the SARS-CoV-2, a single-strand RNA virus belonging to β-coronavirus [1]. The first two-thirds of its genome encode non-structural proteins, primarily involved in viral replication, while the remaining third encodes four structural proteins: spike (S), envelope (E), membrane (M), and nucleocapsid (N) [2]. The S protein contains the region responsible for mediating viral adsorption, which facilitates the entry of the virus into host cells by binding to the angiotensin-converting enzyme 2 (ACE2) receptor on the cell membrane [3, 4]. Furthermore, mutations of the S protein are a defining characteristic of SARS-CoV-2 variants. Presently, the Omicron BA.2 variant has spread rapidly across various regions of the world, becoming the predominant strain globally [5].
Vaccines remain the most crucial strategy for preventing SARS-CoV-2 infections. However, several COVID-19 vaccines have been developed specifically to inhibit the interaction between S protein and ACE2 receptor, which suggests that mutations in the S protein could increase the risk of vaccine failure [6, 7]. Compared with the reference strain Wuhan/Hu-1/2019, the BA.2 variant exhibits 31 amino acid substitutions in the S protein, with fewer than 10% of these mutations occurring on the internal side of the protein, the majority are on the surface. This extensive surface mutation confers a strong immune evasion ability, raising concerns that many neutralizing antibodies may become ineffective [5, 8, 9]. Certain monoclonal antibodies, such as REGEN-COV, Xevudy, and Ronapreve, have been reported to lose efficacy against this variant [5, 6]. Given the increasing frequency of mutations, the search for novel drug candidates for the prevention and treatment of COVID-19 has become an urgent priority.
Traditional Chinese medicine and its active ingredients exhibit antiviral effects. Calceolarioside B, extracted from the Chinese herb Mutong (Akebia quinata Thunb.), has demonstrated anti-HIV-gp41 activity with an IC50 of 0.1 mg/mL [10]. In addition, it has been predicted to possess potential anti-SARS-CoV-2 effects. In silico screening has shown that calceolarioside B can inhibit the interaction between the S protein of SARS-CoV-2 and ACE2 receptor [11]. The study also found that calceolarioside B has a binding affinity of 19.87 kcal/mol with the key enzyme 3CLpro involved in SARS-CoV-2 replication, suggesting its potential to disrupt important hydrogen bonds and alter the receptor-binding site, thereby exerting anti-virus effects [12]. While most studies have focused on structural analysis and predictions, there has been limited in-depth investigation on the anti-SARS-CoV-2 effects of calceolarioside B.
Cytokine storm is a pathological condition resulting from an overactive immune response, characterized by abnormally high levels of cytokine release. It is one of the severe complications of COVID-19 and a leading cause of mortality. Patients with severe COVID‐19 exhibit higher levels of IL (interleukin) ‐2, IL‐6, IL‐10, TNF (tumor necrosis factor) ‐α, and IFN-γ than those with mild infections [13, 14]. This heightened cytokine response can result in self-destructive inflammation at infection sites, causing collateral damage to the host cells, accompanied by increased vascular permeability, and circulatory disorders, potentially leading to multiple organ failure (MOF) [15]. Extracts from Akebia trifoliata pericarps contain calceolarioside B, which can modulate inflammation via the NF-κB/MAPK signaling pathway, and regulating the levels of inflammatory factors such as TNF-α, IL-6 and IL-1β [16, 17]. Moreover, calceolarioside B was identified as an IL-6 inhibitor, reducing IL-6 expression by 43.18% [18]. Although calceolarioside B has demonstrated anti-inflammatory properties, its effectiveness against inflammation induced by SARS-CoV-2 remains unclear. This study aims to evaluate the inhibitory effects of calceolarioside B on the SARS-CoV-2 Omicron BA.2 variant and to explore its potential in antiviral and anti-inflammatory activities. This research not only addresses the current knowledge gap regarding the anti-SARS-CoV-2 effects of calceolarioside B, but also has the potential to provide valuable scientific evidence for the development of new COVID-19 treatment strategies.
Materials and methods
Preparation of the target protein and ligand
The ligand calceolarioside B was downloaded from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/) [19]. The protein 7UB5, representing the SARS-CoV-2 Omicron BA.2 S protein trimer with the three-receptor-binding domain (RBD) configuration and lacking the P986-P987 stabilizing mutations (designated as S-GSAS-Omicron BA.2), was retrieved from the RCSB Protein Data Bank (https://www.rcsb.org/) [20]. The Schrödinger LigPrep tool (https://www.schrodinger.com/) was used to prepare a high-quality ligand for further molecular docking. Protein preparation, including preprocessing, optimization, water removal, and minimization, were undertaken using the Schrödinger Protein Preparation Wizard (https://www.schrodinger.com/) [21] SiteMap tools were used to evaluate the potential protein binding sites. A receptor grid was generated by applying the SiteMap results to Receptor grid generation in the Grid based Ligand Docking with Energetics (Glide) application, (version 9.1, Schrödinger, https://www.schrodinger.com/) of Maestro version 12.8 (Schrödinger). After screening, the receptor grid located in the RBD was used for molecular docking.
Molecular docking
The ligand is docked to the protein (7UB5) by employing the docking protocol of Glide version 9.1 with the receptor grid screened from the preparation process. After docking, each of the binding poses was then used in the Calculate Energy module of the MacroModel application (BatchMin V13.2; Schrödinger) to calculate the total energy.
Molecular dynamics simulation
The molecular dynamics of the interaction between calceolarioside B and SARS-CoV-2 Omicron BA.2 RBD were estimated using the GROMACS software (version 2022.2, https://manual.gromacs.org/2022.2/download.html) [22]. The topologies of RBD and calceolarioside B were generated using the OPSL-AA force field and LigParGen server (https://traken.chem.yale.edu/ligpargen/) [23], respectively. RBD and calceolarioside B complexes were constructed and then subjected to a dodecahedral water box SPC water model (https://www.gromacs.org/) [24]. The system was neutralized after adding suitable counter ions. Energy minimization was performed with a force constant of 1000 kJ mol−1 nm−1 and a maximum of 50,000 steps. During equilibration, NVT equilibration was set at 300 K and NPT at 1 bar. The molecular dynamics simulation was set to 1 ns. The root-mean-squared deviation (RMSD) and interaction energy were also ascertained by the GROMACS software.
Biolayer interferometry (BLI) assay
SARS-CoV-2 Omicron BA.2 Spike-RBD protein (His Tag) (Cat: 40592-V08H123; Sino Biological, Inc. Beijing, China) was diluted to 40 μg/mL and then immobilized on Ni–NTA biosensors (18–5114; Sartorius, NY, USA) for 300 s and an additional 60 s to obtain the baseline for verifying the protein-biosensor affinity. Calceolarioside B was serially diluted from 100 μM to 3.13 μM with PBS in a black plate (655997; Greiner Bio-One GmbH, Frickenhausen, Germany). After establishing the baseline for 120 s, the biosensors containing the RBD protein were immersed in wells with serial dilutions of calceolarioside B for 180 s to allow for association, followed by a dissociation step for another 180 s. The dissociation constant (KD) value was calculated by using a 1:1 binding model utilizing the Data Analysis Software 9.0 (Sartorius, USA).
Construction of the ACE2 receptor cell lines
Human embryonic kidney 293 T cell lines stably expressing ACE2-EGFP and ACE2-mcherry were constructed via lentiviral transfection. For this, ACE2-EGFP or ACE2-mcherry plasmids (Vectorbuilder Inc., Guangzhou, China) were used to co-transfect 293 T cells with the lentivirus packaging plasmids VSVG, GAG, and REV (#8454, #12251, #12253, Addgene Inc., MA, USA). After 12 h of incubation, the cells were supplied with a fresh medium. Following an additional 12 h, the supernatant containing the lentivirus was harvested and filtered through a 0.45 μm filter (SLHA033SB; Millipore, USA). Next, 5 × 104 293 T cells were seeded in 6-well plates and co-incubated with the lentivirus along with 5 μg/mL polybrene (Vectorbuilder Inc., China) for 48 h. After observing cell fluorescence, the cells were purified by treatment with 4 μg/mL puromycin (Cat: A1113803; Gibco, MT, USA) for 2 weeks.
Immunofluorescence of Omicron BA.2 protein binding to ACE2 293 T cells
1 × 104/well 293 T cells transfected with ACE2-EGFP plasmid were seeded on a coverslip placed inside a 24-well plate and incubated overnight. Spike-RBD protein (His Tag) (Cat: 40592-V08H123, Sino Biological, Inc.) and calceolarioside B (diluted from 200 to 25 μM) were premixed for 1 h and then incubated with the cells for another 1 h. The protein without premixing was the control. Subsequently, cells were washed and fixed with 1% paraformaldehyde (P804537-500 g; Shanghai Macklin Biochemical Co. Ltd., Shanghai, China) for 15 min, and blocked with 2% BSA (A801320-100 g; Shanghai Macklin) for 15 min. The cells were incubated with His Tag antibody (Cat: 12698S; Cell Signaling Technology, MA, USA) at room temperature for 2 h, followed by incubation with Alexa Fluor® 555 Phalloidin secondary antibody (Cat: 8953S; Cell Signaling Technology) for 2 h. After drying, the coverslips were mounted with the FluorSave™ reagent (345789; Calbiochem, Darmstadt, Germany). The binding of the S protein with the ACE2 receptors was visualized under a SP8 confocal microscope (Leica, Wetzlar, Germany). The semi-quantitative analysis was carried out using the ImageJ software (https://imagej.net/ij/download.html). The reduction in the density of secondary antibody-labeled Spike-RBD protein mediated by calceolarioside B was compared to the control group to assess its neutralizing potency.
Omicron BA.2 pseudovirus based virus neutralization assay
5 × 104/well 293T cells transfected with ACE2-mcherry plasmid were seeded overnight on a coverslip placed inside a 24-well plate. Calceolarioside B (diluted from 200 to 25 μM) was premixed with SARS-CoV-2 Omicron BA.2 spike pseudovirus and 5 μg/mL polybrene (Vectorbuilder Inc.) for 1 h to neutralize the virus. The mixtures were then added to the cells and incubated for 12 h. The medium was replaced and further incubated for 24 h. Next, the cells were washed and fixed with 1% PFA for 15 min and stained with DAPI (62248; Thermo-Fisher Scientific Inc., MA, USA) for 10 min. After the coverslips were dried, the slices were mounted using FluorSave reagent. The virus neutralization effect was visualized and analyzed employing the ImageXpress® Pico automated cell imaging system (Molecular Devices, CA, USA). The neutralizing potency was evaluated by calculating the percentage reduction of infected cells (marked in green).
MTT assay
Experiments were conducted using RLE-6TN rat type II alveolar epithelial cells (CRL-2300; ATCC, Manassas, VA, USA). To determine the half-maximal cytotoxic concentration (CC50) of calceolarioside B, cells were seeded into 96-well plates at a density of 5 × 10^3 cells per well. The cells were treated with calceolarioside B at concentrations of 0, 100, 200, 400, 800, 1600, and 3200 μM for 24 h, and cell viability was assessed using the MTT assay.
To determine the half-maximal effective concentration (EC50) of calceolarioside B, different concentrations of calceolarioside B (0, 12.5, 25, 50, 100, 200, and 400 μM) were mixed with pseudovirus and applied to RLE-6TN cells for 24 h. The EC50 was calculated based on the inhibition rate of viral infection. A selectivity index was calculated as the ratio CC50/IC50.
ELISA
To evaluate the effects of calceolarioside B on inflammation, RLE-6TN rat type II alveolar epithelial cells were induced with LPS (5 mg/L) to stimulate an inflammatory response. Subsequently, the cells were treated with calceolarioside B (200 μM) or left untreated. The Omicron BA.2 pseudovirus was added for stimulation. After 24 h, the cell culture supernatant was collected, and the IL-6 levels were measured using the Rat IL-6 ELISA Kit (E-EL-R0015c; Elabscience, Hubei, China) to determine the expression of the inflammation-related factors.
Flow cytometry
To observe the effect of calceolarioside B on the polarized state of macrophages, peripheral blood mononuclear cells (PBMCs) were purchased from Guangzhou Jennio Biotech Co. Ltd., Guangzhou, China. A transwell chamber composed of a 0.4 μm membrane was used to culture the monocytes in the lower chamber, and RLE-6TN rat type II alveolar epithelial cells in the upper chamber were then added with the medium (containing 200 μM calceolarioside) and Omicron BA.2 pseudovirus. After 72 h, the M1/M2 macrophages in the lower chambers of each group were stained with CD11b PE-Cy7 (60–0112; Tonbo Biosciences, CA, USA), CD163 (326510; BioLegend, CA, USA), CD206 (321104; BioLegend), and CD68 (333808; BioLegend) antibodies for 1 h. After incubation, the cells were washed with 2 mL PBS, then resuspended in 100 μL PBS, and detected by FACS Aria III flow cytometry (BD Inc., NJ, USA). Each experiment was repeated independently thrice.
Statistical analysis
Statistical analysis was conducted using the SPSS 22.0 software (IBM Corp., NY, USA), and the results were expressed as the mean ± standard deviation. The Shapiro–Wilk test was first used to judge whether the data conforms to normal distribution. All the data conformed to the normal distribution, one-way ANOVA was utilized to analyze the variations between multiple groups. If the results of the one-way ANOVA were significant, Tukey's method was utilized to perform the pairwise comparisons between the groups. A p value of < 0.05 was considered statistically significant.
Results
Calceolarioside B binds to the SARS-CoV-2 Omicron BA.2 RBD by molecular docking
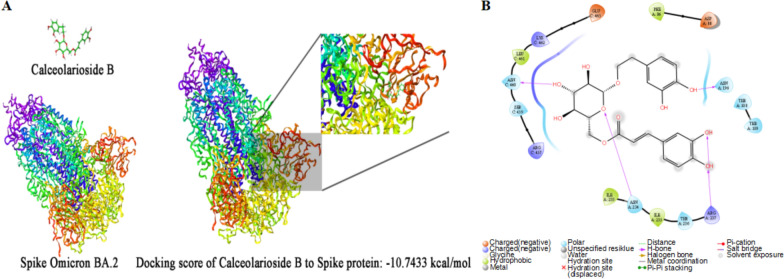
During the docking process, a total of 20 binding sites were generated from SARS-CoV-2 Omicron BA.2 protein using sitemap tools. However, only two of these were located in the RBD and showed rotational symmetry characteristics. One of the binding poses exhibited a lower total energy of − 10.74 kcal/mol after docking with calceolarioside B (Fig. 1A, B) and was subsequently included in the molecular dynamics analysis. In this analysis, RMSD was calculated as atomic distances over time and changes within 100 ps. The Lennard–Jones interaction energy was − 19.553 ± 6.9 kJ/mol, and the average interaction energy was − 96.6844 ± 17 kJ/mol. Thus, the results of molecular docking and molecular dynamics simulations together suggested that calceolarioside B was able to bind to and interact with the SARS-CoV-2 Omicron BA.2 RBD protein.
Fig. 1.
Molecular docking prediction of calceolarioside B with Omicron BA.2 spike protein. A Docking of calceolarioside B with the Omicron BA.2 spike protein using Schrödinger Maestro. B Molecular interactions were analyzed using the Schrödinger Ligand interaction diagram
Calceolarioside B exhibits high-affinity binding to Omicron BA.2 spike-RBD via BLI analysis
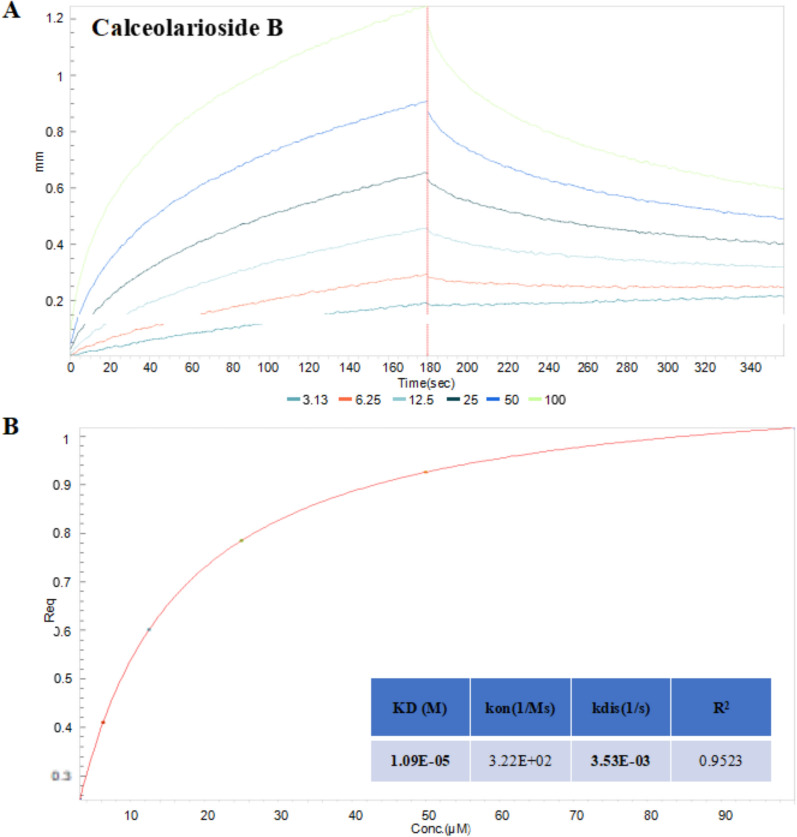
The interaction between calceolarioside B and Omicron BA.2 spike-RBD was further examined by using BLI assays to investigate the binding affinity. The spike-RBD protein was immobilized on Ni–NTA biosensors and exposed to wells containing calceolarioside B. The association and dissociation between the drug and protein were recorded and analyzed. As shown in Fig. 2A, B, calceolarioside B exhibited dose-dependent binding to the Omicron BA.2 spike-RBD in a 1:1 binding model, with a KD of 11 μM, and a steady-state R2 value of 0.9523. Consistent with the molecular docking results, calceolarioside B demonstrated high binding affinity for Omicron BA.2 spike-RBD, suggesting that it may inhibit virus entry by interacting with the S protein.
Fig. 2.
Interaction of calceolarioside B with the Omicron BA.2 spike-RBD protein. A Association and disassociation of calceolarioside B binding to the spike-RBD protein were monitored by employing the BLI assay. B Steady-state analysis and binding kinetics were calculated utilizing the Octet ForteBio data analysis software
Calceolarioside B inhibits the binding of spike protein to the ACE2-positive host cells
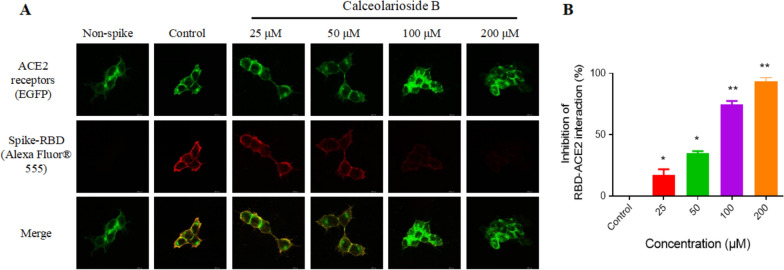
The viral inhibition of calceolarioside B was visualized by fluorescence staining. As shown in Fig. 3A, the EGFP labeled spike-RBD (red) colocalized with the ACE2 receptor (green). Moreover, calceolarioside B effectively inhibited this interaction in a concentration-dependent manner. Specifically, at a concentration of 100 μM, spike-RBD binding to ACE2 was reduced by over 70%, and at 200 μM, binding was suppressed by 90% (Fig. 3B).
Fig. 3.
Calceolarioside B suppressed the binding of the spike-RBD protein to the ACE2-positive host cells. A Spike-RBD was incubated with calceolarioside B for 1 h and then added to the ACE2-positive cells (green) and incubated for another hour. The protein not bound to ACE2 was washed, and the remaining was labeled with Alexa Fluor® 555 Phalloidin. Fluorescence staining showed reduced binding of spike-RBD (red) to ACE2-positive cells in the presence of calceolarioside B. B The images indicating the spike-RBD–ACE2 binding intensity were quantified by ImageJ. Data were presented as mean ± SD; n = 3; *p < 0.05, **p < 0.01 vs. Control, one-way ANOVA analysis
Calceolarioside B suppresses the entry of the Omicron BA.2 pseudovirus into the host cells
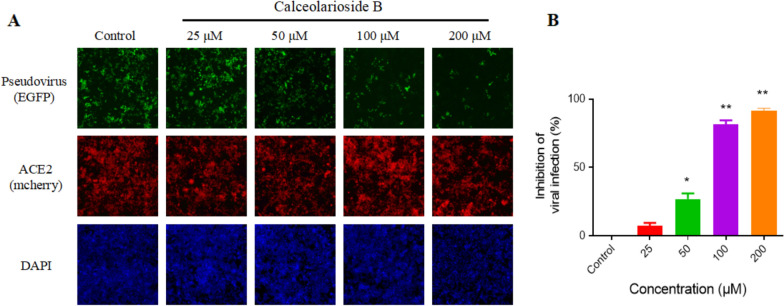
By adding the fluorescent reporter EGFP gene to the pseudovirus, the infection efficiency of the virus can be evaluated by counting the number of green fluorescent cells. The results showed that the number of virus-infected cells was inversely proportional to the concentration of calceolarioside B, whereas the number of recipient cells did not change significantly (Fig. 4A). The infection inhibition rates reached up to 80% and 90% with calceolarioside B concentrations of 100 μM and 200 μM, respectively (Fig. 4B).
Fig. 4.
Calceolarioside B blocked Omicron BA.2 pseudovirus infection. A Pseudovirus, calceolarioside B, and polybrene were premixed for 1 h and then co-incubated with the ACE2-positive cells for 12 h. The cells were replaced with fresh medium and incubated for two days for the fluorescence proteins to be expressed. ACE2 receptors were labeled with mCherry fluorescence (red). Pseudovirus-infected cells expressed the EGFP reporter protein (green). The cells were stained with DAPI (blue) for counting. The images were captured by ImageXpress Pico imaging system (20 × objective lens). B The infected and total number of cells in the images was quantified by the ImageXpress Pico imaging system. Data were presented as mean ± SD, n = 3; *p < 0.05, **p < 0.01 vs. Control, one-way ANOVA analysis
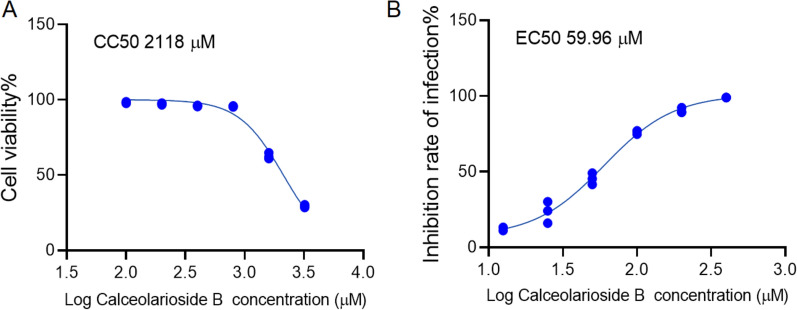
Determination of CC50 and EC50 for calceolarioside B in Rat Alveolar Cells
The CC50 and EC50 of calceolarioside B in RLE-6TN rat type II alveolar epithelial cells were determined using the MTT assay. Results indicated that increasing concentrations of calceolarioside B led to a progressive decrease in cell viability, with a CC50 value of approximately 2118 μM after 24 h of treatment (Fig. 5A). Furthermore, different concentrations of calceolarioside B were mixed with the pseudovirus, the inhibition rate of infection was found to increase as the concentration of calceolarioside B increased, with an EC50 value of approximately 60 μM (Fig. 5B). And selectivity index CC50/EC50 about 35.3.
Fig. 5.
MTT assay for measuring cell viability and inhibition rate of infection by calceolarioside B. A CC50 of calceolarioside B. B EC50 of calceolarioside B. Data were presented as mean ± SD
Calceolarioside B attenuates Omicron BA.2 pseudovirus-induced inflammation in lung cells
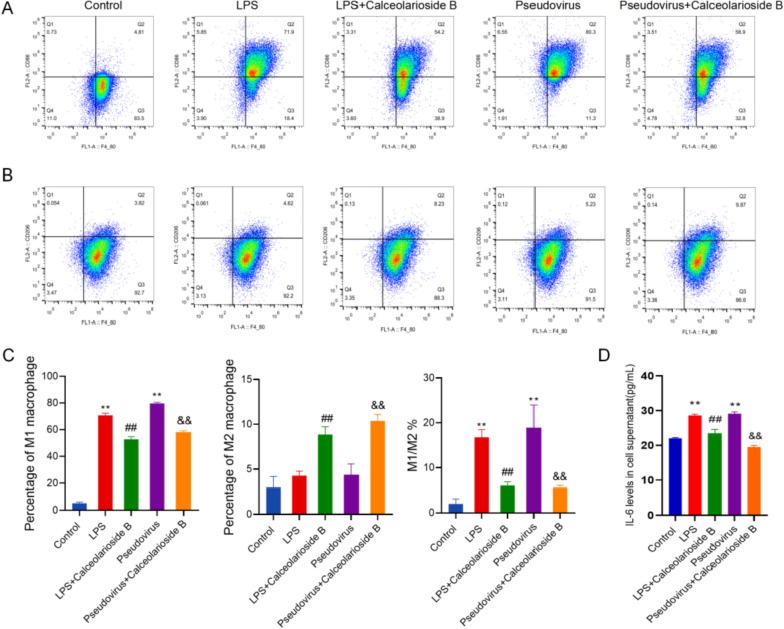
We assessed PBMC differentiation using flow cytometry. The results indicated that under LPS stimulation, the proportion of PBMCs differentiating into M1 type macrophages was significantly increased. A similar trend was observed in the pseudovirus group. However, compared to the LPS group, the proportion of PBMCs differentiating into M1 type macrophages was significantly reduced in the LPS + calceolarioside B group, while the proportion of M2 type macrophages was significantly increased. Similarly, in the pseudovirus + calceolarioside B group, the differentiation of PBMCs into M1 type macrophages was significantly decreased, and the proportion of M2 type macrophages was significantly increased compared to the pseudovirus group (Fig. 6A–C).
Fig. 6.
Calceolarioside B suppressed the inflammation induced by the Omicron BA.2 pseudovirus. A Flow cytometry analysis of the proportion of PBMCs differentiated into M1 macrophages. B Flow cytometry analysis of the proportion of PBMCs differentiated into M2 macrophages. C Quantitative analysis and M1/M2 macrophage ratio. D IL-6 levels in the cell supernatant. Data were presented as mean ± SD, **p < 0.01 vs. Control group, ##p < 0.01 vs LPS group, &&p < 0.01 vs pseudovirus group
We also measured the levels of inflammation-related factors using ELISA. As shown in Fig. 6D, both LPS-induced inflammation and pseudovirus infection significantly increased the levels of IL-6 in the cell supernatants compared to the control group. However, calceolarioside B treatment significantly reduced IL-6 levels compared to both the LPS and pseudovirus groups. Additionally, IL-6 levels were slightly higher in the pseudovirus group compared to the LPS group, though the difference was not statistically significant.
Discussion
Since the outbreak of the SARS-CoV-2 virus in December 2019, it has posed a severe global health threat [25]. The absence of specific drugs targeting SARS-CoV-2 has led to rising morbidity and mortality rates from COVID-19. Although some effective treatments have been identified to alleviate symptoms, a cure for COVID-19 remains elusive. However, China has managed to effectively control the COVID-19 outbreak, benefiting from the combined use of traditional Chinese medicine and synthetic drugs. For example, Huoxiangzhengqi capsules, Jinhua Qinggan granules, Lianhua Qingwen granules, and Shufeng Jiedu capsules are all traditional Chinese medicine preparations recommended by the “Diagnosis and Treatment Protocol for New Coronavirus Pneumonia” [26]. Therefore, identifying natural plant compounds that can prevent or treat infections caused by mutated SARS-CoV-2 strains is of considerable significance. Many such compounds, such as baicalein, pentaerythritol, sitosterol, termite, lupinol, amygdalin, epicatechin, and orange piedin, have been identified to possess potential therapeutic or preventive significance for SARS-CoV-2 [27–29]. This study evaluates the therapeutic efficacy of calceolarioside B against SARS-CoV-2 through a series of biological and pharmacological experiments.
Polyphenols exhibit various beneficial effects against viral diseases, including inhibiting viral replication and regulating the immune system [30, 31]. Research has investigated the potential of quercetin in preventing and treating SARS-CoV-2. Quercetin can inhibit several key SARS-CoV-2 enzymes, such as 3CLpro and PLpro, thereby blocking viral entry into host cells and replication. Additionally, quercetin impedes the binding of the virus to the ACE2 receptor, preventing viral entry [32]. Similarly, hesperidin inhibits the binding of the SARS-CoV-2 S protein to the ACE2 receptor [33]. These findings are consistent with our study, where we employed molecular docking and molecular dynamics simulations to predict the interaction between calceolarioside B and the S protein of the Omicron BA.2 variant. Our results demonstrated that calceolarioside B binds with high affinity to both RBDs of the Omicron BA.2 S protein, and one of these binding sites has lower total energy, which meets the stability requirements for molecular dynamics. Furthermore, calceolarioside B significantly inhibits viral entry into host cells by interfering with the binding of the S protein to the ACE2 receptor.
Due to the high pathogenicity of SARS-CoV-2, direct research is highly risky, leading to the widespread adoption of in vitro virus infection simulation systems. This system uses hACE2-expressing 293T cells to model infected cells and employs fluorescently labeled pseudoviruses expressing SARS-CoV-2 S protein or RBD protein to mimic the actual virus [34]. In this study, we constructed SARS-CoV-2 pseudovirus labeled with a fluorescent protein and assessed the inhibitory effect of calceolarioside B at various concentrations. The virus neutralization assay revealed an inverse relationship between the number of infected cells and calceolarioside B concentration, indicating that calceolarioside B effectively inhibits viral infection. Furthermore, we evaluated the toxicity of calceolarioside B on RLE-6TN rat type II alveolar epithelial cells, and determined the EC50 of calceolarioside B following pseudovirus infection. The selectivity index was approximately 35.3, indicating good selectivity and high safety.
In addition, the immunoregulatory properties of polyphenolic compounds offer additional advantages in combating COVID-19. Inflammation plays a dual role in the pathogenesis of SARS-CoV-2 infection: it is both a necessary defense mechanism for virus clearance and a potential cause of severe organ damage due to excessive response [35]. Therefore, effective management and regulation of inflammation are crucial for treating COVID-19. For example, curcumin alleviates severe inflammatory responses triggered by viral infection by inhibiting the NF-κB and MAPK signaling pathways, thus reducing the release of inflammatory factors [36]. To elucidate the specific mechanism of action of calceolarioside B in immune modulation, we compared the effects of calceolarioside B using both LPS-induced cell inflammation and pseudovirus infection models. The results demonstrated that calceolarioside B significantly reduces IL-6 levels in both models. Additionally, calceolarioside B decreased the proportion of M1 type macrophages while increasing the proportion of M2 type macrophages in PBMCs. This indicates that calceolarioside B effectively modulates the immune response by controlling inflammation.
Despite our study demonstrating the potential of calceolarioside B in inhibiting the SARS-CoV-2 Omicron BA.2 variant and modulating immune responses, there are still some limitations. Firstly, the absence of a positive control in our experimental design may affect the robustness of our conclusions, limiting our ability to compare our results with established standards. Additionally, the mechanism by which the pseudovirus induces an inflammatory response remains unclear. It is uncertain whether this response is mediated by the interaction between the S protein and ACE2 or by components of the retroviral vector used in the pseudovirus. Future research should explore the mechanisms underlying pseudovirus-induced inflammation to better understand the mode of action of calceolarioside B.
Conclusion
This study demonstrates that calceolarioside B has the potential to inhibit the entry of the Omicron BA.2 variant into host cells by interfering with the interaction between the S protein and the ACE2 receptor. Additionally, we explored the potential of calceolarioside B in modulating the inflammatory response, although the underlying mechanisms require further investigation. Future research should focus on more complex mechanistic studies and validate these findings through clinical trials, thereby supporting the development of novel antiviral therapies.
Acknowledgements
We appreciate each participant providing assistance in this study.
Abbreviations
- ACE2
Angiotensin-converting enzyme 2
- MOF
Multiple organ failure
- BLI
Biolayer interferometry
- RMSD
Root-mean-squared deviation
- IL-6
Interleukin 6
- TNF-α
Tumor necrosis factor α
- PBMCs
Peripheral blood mononuclear cells
- KD
Dissociation constant
Author contributions
All authors contributed to the study conception and design. Xiaobin Lin, Rui-hong Chen and Jin Han performed the data curation and formal analysis. Jin Han provided the funding acquisition and project administration. Xiaobin Lin, Yuzhi Yao, Qirong Wen, Fu-bin Liu and Yuanxuan Cai performed the methodology and software. All authors read and approved the final manuscript.
Funding
This work was supported by the doctoral start-up fund of Guangzhou Women and Children’s Medical Center (1600042–04), National Natural Science Foundation of China (82103420), Guangzhou Municipal Science and Technology Project (202102020182) and Innovation Team Project of Universities in Guangdong Province (2022KCXTD010).
Availability of data and materials
The data presented in this study are available upon request from the corresponding author.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Xiao-bin Lin and Yu-zhi Yao have contributed equally to this work as Co-first author.
Contributor Information
Rui-hong Chen, Email: guisitong2@hotmail.com.
Jin Han, Email: hanjin1123@163.com.
References
- 1.Mistry P, Barmania F, Mellet J, et al. SARS-CoV-2 variants, vaccines, and host immunity. Front Immunol. 2021;12: 809244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yadav R, Chaudhary JK, Jain N, et al. Role of structural and non-structural proteins and therapeutic targets of SARS-CoV-2 for COVID-19. Cells. 2021;10(4):821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Q, Zhang Y, Wu L, et al. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181(4):894-904.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Warner FJ, Rajapaksha H, Shackel N, et al. ACE2 from protection of liver disease to propagation of COVID-19. Clin Sci. 2020;134(23):3137–58. [DOI] [PubMed] [Google Scholar]
- 5.Uraki R, Kiso M, Iida S, et al. Characterization and antiviral susceptibility of SARS-CoV-2 Omicron BA.2. Nature. 2022;607(7917):119–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bruel T, Hadjadj J, Maes P, et al. Serum neutralization of SARS-CoV-2 Omicron sublineages BA.1 and BA.2 in patients receiving monoclonal antibodies. Nat Med. 2022;28(6):1297–302. [DOI] [PubMed] [Google Scholar]
- 7.Wilhelm A, Widera M, Grikscheit K, et al. Limited neutralisation of the SARS-CoV-2 Omicron subvariants BA.1 and BA.2 by convalescent and vaccine serum and monoclonal antibodies. EBioMedicine. 2022;82:104158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamasoba D, Kimura I, Nasser H, et al. Virological characteristics of the SARS-CoV-2 Omicron BA.2 spike. Cell. 2022;185(12):2103–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Willett BJ, Grove J, MacLean OA, et al. SARS-CoV-2 Omicron is an immune escape variant with an altered cell entry pathway. Nat Microbiol. 2022;7(8):1161–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim HJ, Yu YG, Park H, et al. HIV gp41 binding phenolic components from Fraxinus sieboldiana var. angustata. Planta Medica. 2002;68(11):1034–6. [DOI] [PubMed] [Google Scholar]
- 11.Sharma T, Zaman M, Rashid S et al. In Silico analysis of plant-derived medicinal compounds against spike protein of SARS-CoV-2 and Ace2. 2022. p. 299–313.
- 12.Tahir Ul Qamar M, Alqahtani SM, Alamri MA, et al. Structural basis of SARS-CoV-2 3CL and anti-COVID-19 drug discovery from medicinal plants. J Pharm Anal. 2020;10(4):313–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen G, Wu D, Guo W, et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J Clin Investig. 2020;130(5):2620–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu J, Li S, Liu J, et al. Longitudinal characteristics of lymphocyte responses and cytokine profiles in the peripheral blood of SARS-CoV-2 infected patients. EBioMedicine. 2020;55: 102763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao YM, Xu G, Wang B, et al. Cytokine storm syndrome in coronavirus disease 2019: a narrative review. J Intern Med. 2021;289(2):147–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang X, Yu N, Wang Z, et al. Akebia trifoliata pericarp extract ameliorates inflammation through NF-κB/MAPK signaling pathways and modifies gut microbiota. Food Funct. 2020;11(5):4682–96. [DOI] [PubMed] [Google Scholar]
- 17.Wang X, Yu N, Peng H, et al. The profiling of bioactives in Akebia trifoliata pericarp and metabolites, bioavailability and in vivo anti-inflammatory activities in DSS-induced colitis mice. Food Funct. 2019;10(7):3977–91. [DOI] [PubMed] [Google Scholar]
- 18.Jin H-G, Kim AR, Ko HJ, et al. Three new lignan glycosides with IL-6 inhibitory activity from Akebia quinata. Chem Pharm Bull. 2014;62(3):288–93. [DOI] [PubMed] [Google Scholar]
- 19.Li Q, Cheng T, Wang Y, et al. PubChem as a public resource for drug discovery. Drug Discov Today. 2010;15(23–24):1052–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burley SK, Bhikadiya C, Bi C, et al. RCSB protein data bank: tools for visualizing and understanding biological macromolecules in 3D. Protein Sci. 2022;31(12): e4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dey D, Kumar A. Structural-based Study to Identify the repurposed candidates against bacterial infections. Curr Med Chem 2024. [DOI] [PubMed]
- 22.Chavez Thielemann H, Cardellini A, Fasano M, et al. From GROMACS to LAMMPS: GRO2LAM : a converter for molecular dynamics software. J Mol Model. 2019;25(6):147. [DOI] [PubMed] [Google Scholar]
- 23.Dodda LS, Cabeza de Vaca I, Tirado-Rives J, et al. LigParGen web server: an automatic OPLS-AA parameter generator for organic ligands. Nucleic Acids Res. 2017;45(W1):W331–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu M, Sebastianelli F, Bacic Z. Quantum dynamics of H2, D2, and HD in the small dodecahedral cage of clathrate hydrate: evaluating H2-water nanocage interaction potentials by comparison of theory with inelastic neutron scattering experiments. J Chem Phys. 2008;128(24): 244715. [DOI] [PubMed] [Google Scholar]
- 25.The species Severe acute respiratory syndrome-related coronavirus. classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020;5(4):536–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Diagnosis and Treatment Protocol for Novel Coronavirus Pneumonia (Trial Version 7). Chin Med J (Engl) 2020, 133(9): 1087–1095. [DOI] [PMC free article] [PubMed]
- 27.Pei T, Yan M, Huang Y, et al. Specific flavonoids and their biosynthetic pathway in Scutellaria baicalensis. Front Plant Sci. 2022;13: 866282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He T, Qu R, Qin C, et al. Potential mechanisms of Chinese herbal medicine that implicated in the treatment of COVID-19 related renal injury. Saudi Pharm J. 2020;28(9):1138–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roshdy WH, Rashed HA, Kandeil A, et al. EGYVIR: an immunomodulatory herbal extract with potent antiviral activity against SARS-CoV-2. PLoS ONE. 2020;15(11): e0241739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jasso-Miranda C, Herrera-Camacho I, Flores-Mendoza LK, et al. Antiviral and immunomodulatory effects of polyphenols on macrophages infected with dengue virus serotypes 2 and 3 enhanced or not with antibodies. Infect Drug Resist. 2019;12:1833–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lalani S, Poh CL. Correction: Lalani, S. and Poh, C.L flavonoids as antiviral agents for enterovirus A71 (EV-A71) Viruses 2020, 12, 184. Viruses. 2020;12(7):712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gasmi A, Mujawdiya PK, Lysiuk R, et al. Quercetin in the prevention and treatment of coronavirus infections: a focus on SARS-CoV-2. Pharmaceuticals (Basel). 2022;15(9):1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheng FJ, Huynh TK, Yang CS, et al. Hesperidin is a potential inhibitor against SARS-CoV-2 infection. Nutrients. 2021;13(8):2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu T, Meng JR, Cheng W, et al. Discovery of honokiol thioethers containing 1,3,4-oxadiazole moieties as potential alpha-glucosidase and SARS-CoV-2 entry inhibitors. Bioorg Med Chem. 2022;67: 116838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gusev E, Sarapultsev A, Solomatina L, et al. SARS-CoV-2-specific immune response and the pathogenesis of COVID-19. Int J Mol Sci. 2022;23(3):1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thimmulappa RK, Mudnakudu-Nagaraju KK, Shivamallu C, et al. Antiviral and immunomodulatory activity of curcumin: a case for prophylactic therapy for COVID-19. Heliyon. 2021;7(2): e06350. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data presented in this study are available upon request from the corresponding author.