Abstract
Background
Radiation-induced heart disease (RIHD) is one of the most serious complications of radiation therapy (RT) for thoracic tumors, and new interventions are needed for its prevention and treatment. Small extracellular vesicles (sEVs) from stem cells have attracted much attention due to their ability to repair injury. However, the role of umbilical cord mesenchymal stem cell (UCMSC)-derived sEVs in protecting cardiac organoids from radiation-induced injury and the underlying mechanisms are largely unknown.
Methods
A radiation-induced cardiac organoid injury model was established by using X-ray radiation, and the optimal radiation dose of 20 Gy was determined by live/dead staining. After radiation, the cardiac organoids were treated with sEVs derived from UCMSCs, and energy metabolism, calcium transient changes and the ultrastructure of the organoids were assessed through Seahorse analysis, optical mapping and transmission electron microscopy, respectively. Confocal microscopy was used to observe the changes in mitochondrial ROS and mitochondrial membrane potential (ΔΨm). Furthermore, real-time quantitative PCR was used to verify the RNA-seq results.
Results
After X-ray radiation, the mortality of cardiac organoids significantly increased, energy metabolism decreased, and calcium transients changed. We also observed that the mitochondrial structure of cardiac organoids was disrupted and that ΔΨm was decreased. These effects could be inhibited by sEVs treatment. sEVs may protect against radiation-induced cardiac organoid injury by regulating oxidative phosphorylation and the p53 signaling pathway.
Conclusion
sEVs derived from UCMSCs can be used as a potential therapeutic strategy for radiation-induced heart disease.
Graphical Abstract
Supplementary Information
The online version contains supplementary material available at 10.1186/s13287-024-04115-2.
Keywords: Radiation-induced heart disease, Mesenchymal stem cells, Small extracellular vesicles, Cardiac organoids, Mitochondrial function
Introduction
The heart is traditionally regarded as a radiation-tolerant organ [1]. However, the advent of radiotherapy for thoracic malignancies has led to increased attention being given to RIHD [2]. RIHD causes irreversible damage to the cardiac structure, the conduction system, and the circulatory system [3]. Presently, there is no effective intervention for RIHD [4, 5]. Despite the efficacy of statins (inhibitors of the hydroxymethylglutaryl-CoA reductase enzyme), angiotensin-converting enzyme inhibitors (ACEIs), and antioxidants in mitigating radiation-induced cardiac injury, surgical intervention is inevitable [6–8]. Consequently, the development of efficacious treatments for RIHD is urgently needed.
Small extracellular vesicles (sEVs), originally believed to be cellular remnants originating from platelets and red blood cells [9], have garnered increasing interest from researchers due to their potential for use in tissue repair, regeneration, signaling, and the induction of cell differentiation [10]. UCMSCs-sEVs can alleviate fibrosis [11], inhibit inflammatory responses [12], suppress oxidative stress damage [13], promote tissue repair [14], and reduce apoptosis [15]. These comprehensive advantages are incomparable to single drug therapy. sEVs derived from UCMSCs have demonstrated therapeutic efficacy and no adverse responses in various conditions, including membranous proliferative glomerulonephritis [16], liver ischemia‒reperfusion injury [17], and cartilage damage associated with osteoarthritis [18]. Notably, there is currently a dearth of research investigating the use of sEVs for the treatment of RIHD.
Organoids are 3D cell culture systems established in vitro that closely mimic the structural characteristics of the source tissue or organ in vivo [19]. Three-dimensional organoids can be cultivated from induced pluripotent stem cells (iPSCs) or adult stem cells (AdSCs) [20]. Specifically, cardiac organoids not only recapitulate the chamber structure of the heart but also encompass diverse cell types; these organoids exhibit the capacity to generate electrophysiological responses [21], among other characteristics.
In the present study, we subjected cardiac organoids to X-ray radiation to establish a model of RIHD. After radiation exposure, UCMSCs-sEVs were used to treat radiation-injured cardiac organoids, and the protective effect of sEVs on RIHD and its potential mechanism were investigated.
Methods and materials
Generation of cardioids containing cardiomyocytes (CMs), endocardial cells (ECs) and mesenchymal cells
The WTC iPS cell line (male, skin fibroblast-derived) [22] was obtained from the CORIELL Institute (GM25256, USA). Cardioids were generated as described by Hofbauer et al. [23]. Briefly, 6000 hiPSCs were seeded directly into ultralow attachment 96-well plates (Corning) in CDM medium [24] supplemented with bFGF (30 ng/mL, R&D Systems, 233-FB-025/CF), LY294002 (5 µM, Tocris, 1130), activin A (50 ng/mL, Sigma‒Aldrich, SRP3003), BMP4 (10 ng/mL, R&D Systems, RD-314-BP-050), CHIR99021 (5 µM, R&D Systems, RD-4423/50) and ROCKi (5 µM) for 36–40 h. On Day 2, the medium was exchanged for CDM medium supplemented with BMP4 (10 ng/mL), bFGF (8 ng/mL), insulin (10 µg/mL, Sigma‒Aldrich, I9278), IWR1 (1 µM, Tocris, 3532/10), retinoic acid (0.5 µM, Sigma‒Aldrich, R2625) and VEGF-A (200 ng/mL, PeproTech, AF-100-20) for 4 days, with medium change every day. Subsequently, the medium was changed to CDM containing BMP4 (10 ng/mL), bFGF (8 ng/mL), insulin (10 µg/mL) and VEGF-A (100 ng/mL) for 2 days, after which the medium was changed every day. For maintenance, CDM medium supplemented with 100 ng/mL VEGF-A was used and exchanged every other day.
Immunofluorescence analysis
For immunostaining, the samples were washed with Dulbecco’s phosphate-buffered saline (DPBS), fixed with 4% paraformaldehyde, and embedded in paraffin to prepare paraffin sections. The sections were blocked in 10% normal goat serum for 1 h at room temperature. Primary antibodies against MYL7 (Santa Cruz, sc-365255), cTnT (Abcam, ab45932), CDH5 (Invitrogen, 14-1449-82), PECAM1 (R&D, AF806), IRX4 (Thermo, PA5-97879), and VIM (Abcam, ab24525) were then added at the indicated dilutions to 3% normal goat serum and incubated overnight at 4 °C. After three washes with phosphate-buffered saline (PBS), appropriate secondary antibodies diluted 1:500 in 3% normal goat serum were added for 2 h at room temperature protected from light. After the previous washing steps were repeated, Hoechst 33,258 (Sigma‒Aldrich, 94403) was added to DPBS for 15 min at room temperature protected from light. The samples were mounted with fluorescence mounting medium (Dako, S3023) and stored at 4 °C until imaging with an inverted or confocal laser-scanning microscope (Nikon A1R).
Extraction and isolation of UCMSCs-sEVs
Clinical-grade UCMSCs was obtained from China Medical Management Consulting (Beijing) LTD. CO. Human umbilical cords were provided by women who gave birth at Yantai Laiyang Central Hospital (2023-002-01, Yantai, China). Written informed consent was obtained from the donors participating in the study.
UCMSCs culture media were collected and centrifuged at 300 × g for 10 min at 4 °C, followed by sequential centrifugation steps at 2,000 × g for 20 min and subsequently at 10,000 × g for 30 min. The supernatant was subsequently transferred to a sterile ultraspeed centrifuge tube (Beckman Coulter, 355654) and subjected to centrifugation at 100,000 × g for 70 min at 4 °C 2 times using an ultraspeed centrifuge (Beckman Coulter, XPN-90). Ultimately, the precipitate containing UCMSCs-sEVs was diluted to 200 µl in PBS.
Nanoparticle tracking analysis (NTA)
Two microliters of UCMSCs-sEVs was diluted in PBS to achieve a final volume of 8 ml. The authors subsequently adhered to the manufacturer’s instructions and chose the default software settings for sEVs or nanospheres. The NTA (Particle Metrix, ZetaView PMX 110) was configured with the specified parameters, and the acquired data were analyzed by using built-in ZetaView software (version 8.04.02 SP2).
Transmission electron microscopy (TEM) analysis
The UCMSCs-sEV sample was diluted with PBS and applied to a 300-mesh copper grid. Droplets were generated during the application, allowing the sample to adsorb for 5 min. Excess liquid was removed using filter paper, followed by staining by dropping 10 µl of uranyl acetate onto the copper grid for 2 min. Then, surplus liquid was eliminated with filter paper, followed by drying, and the samples were observed under a TEM (RuliTEM, HT7800). Images were captured for documentation.
For ultrastructural observation of cardiac organoids, 20 cardiac organoids per group were centrifuged, fixed in 2.5% glutaraldehyde for 2 h, immersed in 1% osmium acid for 1 h, dehydrated using a gradient of ethanol, and transitioned with acetone. The organoids were subsequently embedded in Epon 812 resin and cut into semithin sections. Adjacent Sect. (70 nm) were cut and stained with titanium lead, after which changes in the ultrastructure were observed, and TEM images were collected.
Mouse apical heart tissue was collected and observed with TEM according to the above-mentioned procedure.
Western blot (WB)
WB analysis was also conducted to quantify the relative protein expression of the sEVs. Briefly, UCMSCs-sEVs were subjected to sodium dodecyl sulfate‒polyacrylamide gel electrophoresis (SDS‒PAGE) to separate proteins, which were subsequently transferred onto a polyvinylidene fluoride (PVDF) membrane. Blocking was carried out using Tris-buffered saline with Tween 20 (TBST) containing 5% bovine serum albumin (BSA), and the membrane was incubated with antibodies against CD9 (Abcam, ab307085; dilution ratio: 1:1,000), CD63 (Abcam, ab216130; dilution ratio: 1:1,000), CD90 (Cell Signaling Technology, 13801 S; dilution ratio: 1:1,000), TSG101 (Abcam, ab133586; dilution ratio: 1:1,000), and β-tubulin (Abcam, ab59680; dilution ratio: 1:5,000) at 4 °C overnight. Subsequently, the membrane was incubated with horseradish peroxidase (HRP)-labeled secondary antibodies (Boster, BA1054; dilution ratio: 1:1,0000), and the protein bands were visualized using an enhanced chemiluminescence (ECL) kit (Zomanbio, ZD310) and an automatic chemiluminescence imaging system (Tanon, 5200).
Radiation-induced cardiac organoid injury model and treatment
Cardiac organoids were exposed to radiation using a small animal X-ray irradiator (Rad Source, RS 2000). The high-voltage generator had a maximum tube voltage of 160 kV, a maximum power of 25 mA, a dose rate of 1.175 Gy/min, and a radiation exposure time of 1022 s. After radiation of 20 Gy X-rays, the culture medium in the ultra-low-attachment 96-well plates was completely replaced with fresh medium. Then the cardiac organoids were treated with 0.2 µl (1 × 109 P/ml), 1 µl (5 × 109 P/ml), 2 µl (1 × 1010 P/ml), and 4 µl (2 × 1010 P/ml) UCMSCs-sEVs or 4 µl PBS for 48 h at 37 °C in a 5% CO2 incubator in optimization of the therapeutic concentration of UCMSCs-sEVs experiment. In other experiments, the cardiac organoids were treated with 4 µl (2 × 1010 P/ml) UCMSCs-sEVs or 4 µl PBS for either 24–48 h at 37 °C in a 5% CO2 incubator.
Radiation-induced heart injury model and treatment
6-8-week-old male C57BL/6J mice were purchased from Vital River Laboratory Animal Technology Co., Ltd. (Beijing, China). The mice were maintained under a 12-h light/dark cycle, with controlled humidity ranging from 50 to 60% and a stable temperature (21–23 °C). After 1-week acclimatization, 15 Mice were anesthetized with pentobarbital sodium (60 mg/kg) and fixed in a radiation box. 10 mice received single-dose radiation of 20 Gy X-ray locally to the heart and were then randomly divided into two groups, radiation and UCMSCs-sEVs group. PBS (100 µl) or UCMSCs-sEVs (1 × 1010 P/100µl) were injected through the tail vein respectively after the mice woke up, and then given four consecutive alternate-day injections. Another 5 mice were used as non-radiation sham control, and PBS were injected same as radiation group. At the end of the experiment, animals were euthanized by intraperitoneal injection of excessive pentobarbital sodium. Control group, radiation group, and UCMSCs-sEVs group were marked separately at the feeding cage positions. The group allocation of experiments is blind to the sampling and analysis personnel. If there is an average weight loss of 50% and a 50% decrease in feed intake in mice during the experiment, the experiment should be terminated in advance under the guidance of a veterinarian. All procedures that involved animals were approved by the Institutional Animal Care and Use Committee of the Laboratory Animal Center of Academy of Military Medical Sciences (IACUC-DWZX-2023-532). The work has been reported in line with the ARRIVE guidelines 2.0.
Live/dead staining
The viability of the cardiac organoids was assessed using the LIVE/DEAD® Viability/Cytotoxicity Kit for mammalian cells (Invitrogen, L3224) according to the manufacturer’s protocol. Briefly, LIVE/DEAD staining working solutions containing 2 µM calcein AM (4 mM, C-AM) and 4 µM calcein homodimer-1 (2 mM, EthD-1) were added to the organoids, which were subsequently incubated at 37 ℃ for 30 min with 5% CO2. After the incubation, the organoids were washed with preheated PBS. The fluorescence intensities of FITC (green) and TRITC (red) were examined at wavelengths of 488 nm and 561 nm, respectively, using a laser confocal microscope. Additionally, fluorescence intensity measurements were conducted using ImageJ (version 2.14.0/1.54f, Java 1.8.0_322), and the relative fluorescence intensities of C-AM and EthD-1 were calculated.
Mitochondrial membrane potential (ΔΨm) detection
The ΔΨm of cardiac organoids was measured using tetramethylrhodamine (TMRM, Invitrogen, I34361) staining. Cardiac organoids were stained with TMRM (dilution ratio of 1:1000) at 37 °C in the dark for 30 min and then observed under a laser confocal microscope (CrestOptics, Italy) at a wavelength of 561 nm. ImageJ software was used to analyze the mean fluorescence intensity (MFI).
Determination of mitochondrial ROS
MitoSOX (Invitrogen, M36008), a reactive oxygen superoxide ion (ROI) probe, was utilized to investigate reactive oxygen species (ROS) production in cardiac organoids. Specifically, cardiac organoids were stained with MitoSOX (dilution ratio of 1:1000) at 37 ℃ in the dark for 30 min and subsequently examined via laser confocal microscopy at a wavelength of 561 nm. ImageJ software was used to analyze the images.
Detection of energy metabolism
A Seahorse XFe24 Extracellular Flux Analyzer (Agilent Technologies, USA) was used for the glycolytic stress test and mitochondrial stress test assays following the manufacturer’s protocol. These assays were performed to quantitatively measure the extracellular acidification rate (ECAR) and oxygen consumption rate (OCR) of the cardiac organoids. In brief, cardiac organoids (20 per well) were cultured in a Seahorse XF 24-well microplate, and baseline measurements were performed. Subsequently, a Glycolysis Rate Assay Kit (Alicelligent, ALS22022) was used according to the manufacturer’s instructions, and 10 mM glucose, 10 µM oligomycin, and 50 mM 2-DG were sequentially added to the cardiac organoids at specified time points to measure the ECAR. Similarly, for the mitochondrial stress test, the OCR was determined with a CellMito Stress Test Kit (Alicelligent, ALS22012), and the cells were treated sequentially with 20 µM oligomycin, 10 µM FCCP, or 15 µM rotenone/antimycin A, which are inhibitors of mitochondrial complex I and mitochondrial complex III, respectively. The data obtained from these experiments were analyzed using Seahorse XF-24 Wave software (version 2.6.3), with OCR and ECAR values expressed in picomoles per minute (pmol/min) and milli-pH per minute (mpH/min), respectively.
Calcium transient test
Cardiac organoids were rinsed with Tyrode’s salt solution (T2397, Sigma, USA), followed by an incubation period in Tyrode’s salt solution containing 2 µM Fluo 4-AM (F14217, Thermo, USA) and Pluronic F127 (P3000MP, Invitrogen, USA) for 25 min at 37 °C. Afterward, the organoids were washed twice with Tyrode’s salt solution. Subsequently, the cardiac organoids were placed in a dish containing 200 µl of normal Tyrode’s salt solution and subjected to observation via optical mapping (Mapping Lab, OMS-PCIE-2002). Firstly, we continuously photographed cardiac organs for 18 s to collect the activation time of calcium signals. Through pixel merging, we collected the degree of dispersion in the direction and order of calcium signal transduction during this period. Starting from the first activation of calcium signals, the waveform changes of calcium signal conduction amplitude and rate within 10s are recorded uninterruptedly. At the same time, signal frequency spectrum analysis was conducted on the calcium signals of cardiac organoids beating. The times of 30%, 80% and 90% repolarization of calcium signals were recorded respectively.
RNA-seq analysis
RNA isolation was conducted using TRIzol reagent (T9424, Sigma, USA), and the samples were subsequently sent to Berry Genomics (Beijing, China) for sequencing. The RNA concentration was detected and preliminarily quantified using a Qubit 3.0. Then, qPCR was employed to accurately quantify the effective concentration of the library. Sequencing was carried out on the Illumina NovaSeq 6000 platform in PE150 mode. The complete sequencing data can be downloaded from NCBI (PRJNA1113873).
RT-qPCR assay
The extracted RNA was reverse transcribed into complementary DNA (cDNA) using the EVo M-NLV RT Mix Kit with gDNA Clean for qPCR Ver.2 (AG11728, Accurate Biotechnology, China). The mRNA expression of related genes was quantified using a SYBR Green Premix Pro Taq HS qPCR Kit III (AG11739, Accurate Biotechnology, China) on a CFX Connect real-time PCR system (Bio-Rad, USA). qPCR was conducted in quadruplicate, and the relative expression levels were analyzed by the 2−ΔΔCt method, with β-tubulin serving as the control. PCR primers (Supplementary Table 1) were designed using Primer 5.0 software.
Myocardial enzyme detection
14 and 28 days after radiation, blood were collected from the inner canthus of the eye and serum were isolated, then Creatine Kinase (CK), Creatine Kinase-MB (CK-MB), Lactate Dehydrogenase (LDH) and Lactate Dehydrogenase 1 (LDH1) were analyzed using an automatic biochemical analyzer (Shenzhen Leadman Life Science, Chemray 800).
Echocardiography
At day 42 after radiation, mice were shaved off the hair of chest and limb and were anesthetized with 1.5% isoflurane (RWD, R510-22-10), and then cardiac function was evaluated by two-dimensional transthoracic echocardiography using a Vevo2100 imaging system (VisualSonics, Canada).
Hematoxylin-eosin and Masson staining
Mice were sacrificed at day 42 after radiation, the heart were collected and fixed, embedded in paraffin, and then sectioned at a thickness of 5 μm and stained with hematoxylin-eosin (H&E) and Masson.
Statistical analysis
All the data were analyzed with one-way analysis of variance (ANOVA), and graphs were generated with GraphPad Prism 8.0.2. The data are expressed as the mean ± SD, and P < 0.05 was considered to indicate statistical significance. Normality of data were evaluated through parameter testing to assess whether the data met the assumptions of the statistical approach. If the data does not follow a normal distribution, then increase the sample size. *means P values less than 0.05; **means P values less than 0.01; ***means P values less than 0.001.
Results
Generation of cardioids from human iPSCs (hiPSCs)
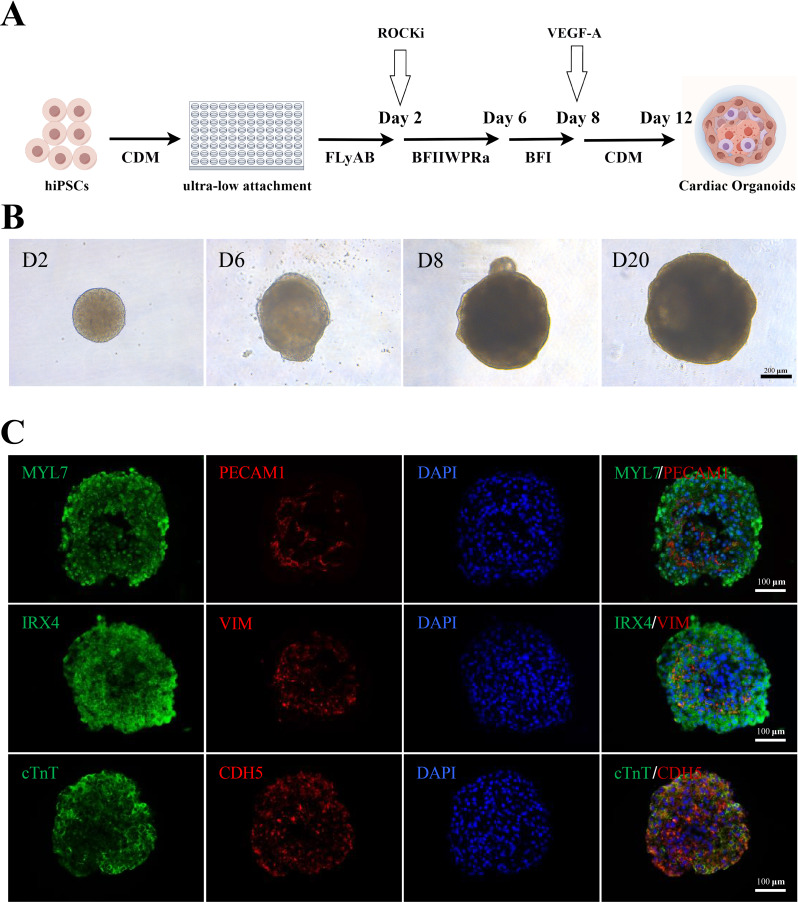
The formation of organoids relies on the intrinsic self-patterning of cells through minimal stimulation of signaling pathways that mimic organ development in vivo. Following the protocol of Hofbauer et al. [23], we established cardiac organoids (Fig. 1A) by generating hiPSC spheroids in 96-well U-bottom plates. These spheroids were then subjected to a differentiation protocol in which the midanterior primitive streak, cardiac mesoderm, and cardiovascular derivatives were sequentially induced. This effect was achieved by modulating WNT, activin A, bone morphogenic protein 4 (BMP4), and basic fibroblast growth factor (bFGF) signaling via the addition of 0.5 µM retinoic acid (RA) from Days 2 to 5. By Day 12, the spheroids initiated spontaneous beating (Fig. 1B and Supplementary Video 1).
Fig. 1.
Preparation of cardiac organoids. (A) Schematic of the preparation of cardiac organoids, which started beating on Day 12. By Figdraw. (B) Time course of cardioid formation. Bright-field images of spheroids showing the increase in size and degree of cardioid formation over time. Scale bars: 200 μm. (C) Immunostaining for the CM markers cTnT and MYL7, the endocardium markers PECAM1 and CDH5, the ventricular-specific marker IRX4, and the mesenchymal marker vimentin (VIM) in cardioids on Day 30. Scale bars: 100 μm
Immunofluorescence analysis on Day 30 of differentiation revealed the formation of an inner core composed of densely packed CMs, identified by cardiac troponin T (cTnT) and myosin light chain 7 (MYL7) expression. Furthermore, the CMs within the spheroids exhibited ventricular specificity, as indicated by the expression of the ventricular-specific marker iroquois homeobox 4 (IRX4). Additionally, the spheroids developed an envelope containing cells expressing the endocardium marker cadherin 5 (CDH5) and platelet and endothelial cell adhesion molecule 1 (PECAM1), as did vimentin-positive mesenchymal cells. This structural arrangement closely resembled the ventricular endocardium of early human embryos (Fig. 1C). These findings are consistent with the results reported by Hofbauer et al. [23], confirming the successful construction of cardiac organoids in our study.
Extraction and identification of UCMSCs-sEVs
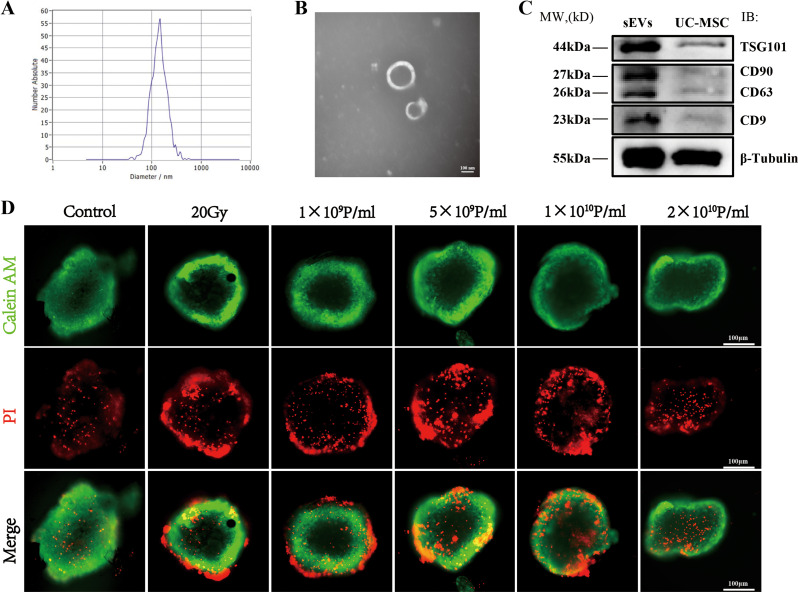
sEVs were extracted from the supernatant of UCMSCs through differential centrifugation. The particle diameter distribution and concentration of the UCMSCs-sEVs were subsequently analyzed using NTA, revealing an average diameter of approximately 138 nm and a concentration of 5.5 E 11 particles/mL (Fig. 2A). TEM images showed that UCMSCs-sEVs had a round or oval structure and a double-membrane configuration and had a diameter of approximately 100 nm (Fig. 2B). WB results (Fig. 2C) demonstrated that sEVs expressed the tetraspanin proteins CD63 and CD9, and tumor susceptibility gene 101 (TST101). sEVs also expressed CD90, which is a marker of their derived cells UCMSCs [25].
Fig. 2.
Identification of UCMSCs-sEVs and optimization of the therapeutic concentration. (A) NTA revealed the particle size distribution of UCMSCs-sEVs. (B) TEM image demonstrating the bilayer membrane structure of sEVs with a diameter of approximately 100 nm. Scale bar: 100 nm. (C) WB image showing the expression of UCMSCs-sEV markers, including TSG101, CD63, CD9, and CD90. (D) Optimization of the therapeutic concentration of UCMSCs-sEVs for radiation-induced cardiac organoid injury. A concentration of 2 × 1010 particles/ml resulted in a notable decrease in red fluorescence intensity. Scale bar: 100 μm
UCMSCs-sEVs improved the energy metabolic capability of cardiac organoids following radiation injury
The results of live/dead staining revealed that the most significant damage to cardiac organoids occurred 48 h after exposure to 20 Gy of X-ray radiation (Supplementary Fig. 1A-C). We subsequently optimized the therapeutic concentration of UCMSCs-sEVs for radiation-induced cardiac organoid injury using live/dead staining and found that the therapeutic effect of UCMSCs-sEVs was most pronounced at a concentration of 2 × 1010 particles/mL (Fig. 2D).
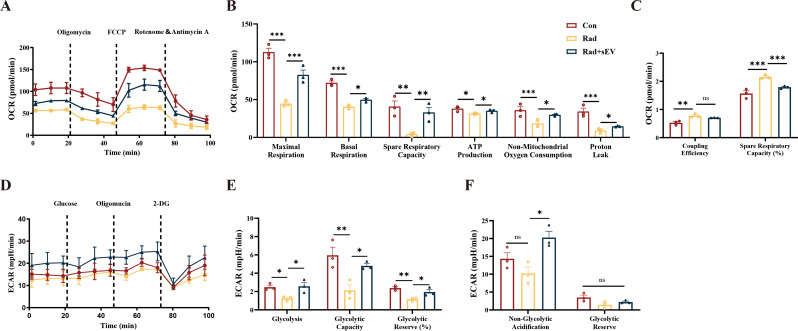
Further analysis of the therapeutic effects was performed using Seahorse analysis. Following 20 Gy of X-ray radiation, the mitochondrial function of cardiac organoids decreased and intervention of UCMSCs-sEVs could substantially improve the maximum respiratory capacity, basal respiration and spare respiratory capacity (Fig. 3A and B). UCMSCs-sEVs also enhanced ATP production, increased non-mitochondrial oxygen consumption values, and restored proton leak in cardiac organoids (Fig. 3B). Furthermore, it was found that UCMSCs-sEVs could reduce the coupling efficiency of radiation-damaged cardiac organoids, and decrease the proportion of spare respiratory capacity (Fig. 3C). These results indicated that UCMSCs-sEVs improved the ATP production capacity of cardiac organoids post-radiation damage.
Fig. 3.
UCMSCs-sEVs improved the energy metabolic capacity of radiation-injured cardiac organoids. (A) Changes in the OCR in cardiac organoids after radiation and UCMSCs-sEVs treatment. (B) Statistical analysis of maximal respiration, basal respiration, spare respiratory capacity, ATP production, non-mitochondrial oxygen consumption and proton leak. (C) Changes in coupling efficiency and percentage in spare respiratory capacity. (D) ECAR levels in cardiac organoids after radiation and UCMSCs-sEVs treatment. (E) Glycolysis levels, maximum glycolytic capacity and total glycolytic capacity reserve in cardiac organoids after radiation injury and UCMSCs-sEVs treatment. (F) Nonglycolytic acidification value and glycolytic capacity reserve after radiation and UCMSCs-sEVs treatment. n = 3
To explore whether UCMSCs-sEVs could exert a therapeutic effect on anaerobic respiration, we evaluated the ECAR (Fig. 3D). The glycolytic level of cardiac organoids decreased after X-ray radiation, and the maximum glycolytic capacity and the glycolytic capacity reserve ratio decreased (Fig. 3E). The nonglycolytic acidification value and glycolysis capacity reserve of cardiac organoids decreased after radiation, but these changes were not statistically significant compared with those in the control group (Fig. 3F). UCMSCs-sEVs treatment reversed the radiation-induced ECAR dysfunction in cardiac organoids (Fig. 3E and F).
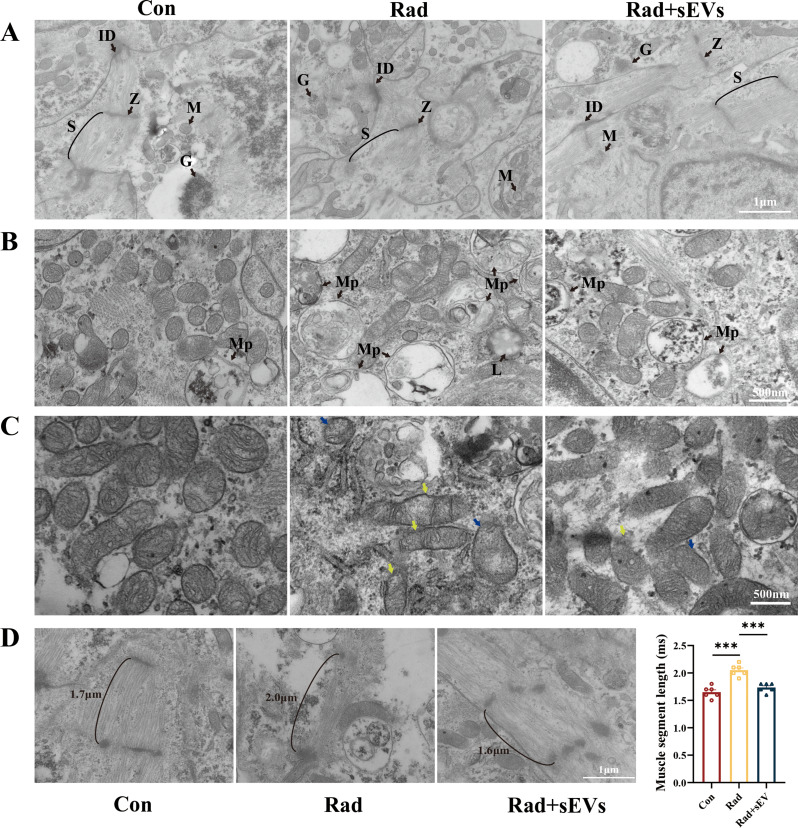
UCMSCs-sEVs treatment alleviated radiation-induced ultrastructural injury in cardiac organoids
Mitochondria provide energy for the heart, while myofibrils are the functional basis for the autonomous activity of the heart. Mitochondria and myofibrils are indispensable structures in cardiomyocytes. TEM was used to observe the ultrastructure of cardiac organoids after radiation and UCMSCs-sEVs treatment. TEM images revealed that the cardiac organoids possessed abundant glycogen and exhibited well-defined intercalated discs and sarcomeres. Compared with those in the radiation group, disordered myofiber alignment, marked autophagy of mitochondria, and heightened lysosomal activity were mitigated in the UCMSCs-sEVs treatment group (Fig. 4A and B). Furthermore, we observed vacuoles in the mitochondria of the cardiac organoids, accompanied by a reduction in the number of inner membrane folds after radiation. UCMSCs-sEVs treatment resulted in a notable decrease in abnormally vacuolated mitochondria and a significant increase in the number of inner mitochondrial cristae in radiation-injured cardiac organoids, albeit not as neatly arranged as in control organoids (Fig. 4C).
Fig. 4.
Ultrastructural features of cardiac organoids after radiation and UCMSCs-sEVs treatment. (A) TEM was used to observe the ultrastructure of cardiac organoids, including myofiber alignment and mitochondria. Z = Z line, ID = intercalated disc, S = sarcomere, M = mitochondria, G = glycogen. Scale bar: 1 μm. (B) Representative images of mitochondrial autophagy and lysosomal activity in cardiac organoids post-irradiation. Mp = Mitophagy, L = lysosomal. Scale bar: 500 nm. (C) Mitochondrial changes observed via TEM. Reductions in mitochondrial cristae (yellow arrow), mitochondrial vacuolation (blue arrow), and enhanced lysosomal activity were shown in radiation-injured cardiac organoids. Scale bar: 500 nm. (D) Myofibril length of cardiac organoids after radiation and UCMSCs-sEVs treatment. n = 6. Scale bar: 1 μm
Furthermore, after radiation, the myofibers in cardiac organoids appeared disordered and scarce, accompanied by a significant increase in the length of myofibrils. After treatment with UCMSCs-sEVs, the myofibers exhibited a neat arrangement, and both the quantity and length of the myofibrils were nearly restored to normal (Fig. 4D). Conversely, other cellular ultrastructure features, including the Golgi apparatus, endoplasmic reticulum, and nucleus, did not significantly differ (Supplementary Fig. 2).
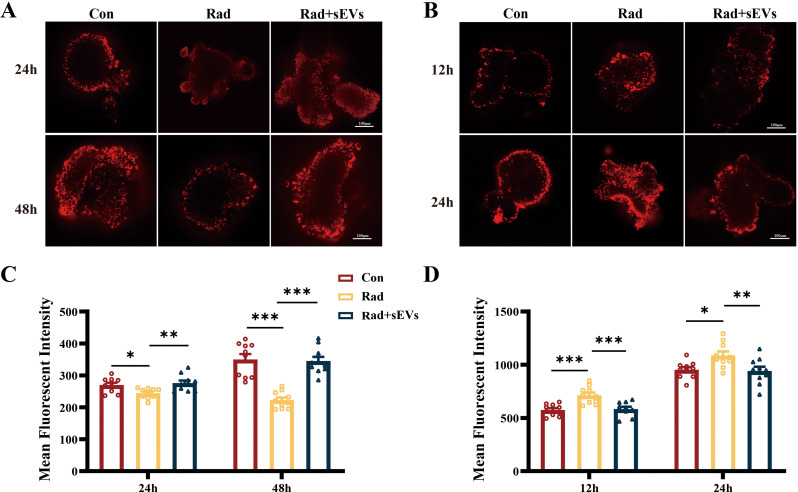
UCMSCs-sEVs protected cardiac organoids from oxidative stress and mitochondrial injury induced by radiation
TMRM staining was used to evaluate the mitochondrial membrane potential (ΔΨm) of cardiac organoids after radiation and treatment with UCMSCs-sEVs at different time points. The results revealed a decrease in the ΔΨm (red fluorescence intensity) at 24 h and 48 h following 20 Gy X-ray radiation, which was restored after UCMSCs-sEVs treatment (Fig. 5A and B). Mitochondrial ROS were assessed by MitoSOX staining and observed using a confocal microscope. At 12 h and 24 h after radiation, the MFI of red fluorescence, which represents the ROS level, increased significantly. Treatment with UCMSCs-sEVs resulted in an apparent decrease in the MFI of ROS (Fig. 5C and D). However, we did not observe the same trend of increase in ROS at 48 h after radiation, similar to that observed at 12 h and 24 h. We speculate that the effect of radiation on ROS may be rapid and transient and that this effect diminishes in the late stage of radiation, but that radiation sustains damage to the ΔΨm.
Fig. 5.
UCMSCs-sEVs treatment improved the mitochondrial membrane potential and decreased oxidative stress in radiation-injured cardiac organoids. At 24 h and 48 h after radiation and UCMSCs-sEVs treatment, the cardiac organoids were stained with TMRM. (A) Representative confocal microscopy images showing the ΔΨm, and (B) the histogram shows the quantitative results. At 12 h and 24 h after radiation and UCMSCs-sEVs treatment, (C) confocal microscopy was used to detect mitochondrial ROS production (red fluorescence), and (D) the histogram shows the quantitative results. n = 10. Scale bar: 100 μm
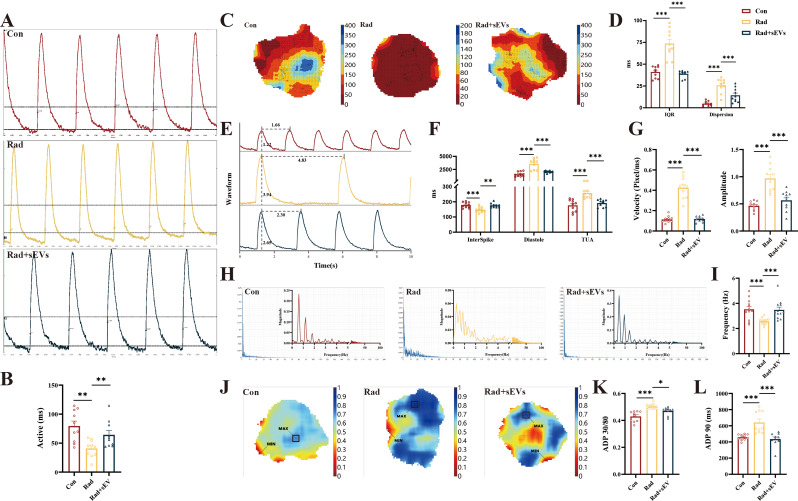
UCMSCs-sEVs restored normal calcium transients in radiation-injured cardiac organoids
The entry of calcium ions is a characteristic of excitation contraction coupling in the myocardium. Cardiac organoids have pacemaker sites that function similarly to the sinoatrial node, driving the contraction of the entire 3D spheroid. After X-ray radiation, changes in myofibrils were observed under an electron microscope, allowing us to examine calcium signals in cardiac organoids. Optical mapping was conducted to assess changes in calcium transients resulting from alterations in myotomes post-irradiation. We studied the speed, dispersion, amplitude, frequency, and repolarization time of calcium signal activity. The results revealed that the activation of calcium physiological activity in cardiac organoids was significantly accelerated after radiation, and the administration of UCMSCs-sEVs restored calcium activation (Fig. 6A and B, Supplementary Video 2). In radiation-injured cardiac organoids, the direction of calcium signal conduction was disrupted, exhibiting multiple excitation phenomena, and reentry occurred during conduction. Following treatment with UCMSCs-sEVs, the 90% calcium ion duration interquartile range discrete time (IQR) and time of Calcium Ion Dispersion during conduction (Dispersion) of cardiac organoids have both recovered (Fig. 6C and D). The rate of release of activated calcium ions (InterSpike: time taken for the calcium wave to reach its peak) was accelerated by X-rays, the diastole (Calcium Signal Release Interval) decreased with a longer rate, and the effective period of inactivity (TAU: Calcium reabsorption) increased in radiation-injured cardiac organoids (Fig. 6E and F). Moreover, the calcium signal transduction rate (Velocity) faster, and the amplitude of calcium signal propagation increased (Fig. 6E and G). Compared to that in the control cardiac organoids, the frequency of calcium activity and the magnitude of calcium signaling reaching threshold has become more chaotic (Fig. 6H and I). In contrast, treatment with UCMSCs-sEVs restored all these abnormal calcium physiological processes (Fig. 6E-I). The ratio of recovery time for 30% repolarization to recovery time for 80% repolarization (Fig. 6J and K) and the time for 90% repolarization of calcium ions (Fig. 6L) decreased after UCMSCs-sEVs treatment. All the experimental data were obtained under uniform signal-to-noise ratio conditions.
Fig. 6.
UCMSCs-sEVs restored the normal calcium transients of radiation-injured cardiac organoids. The calcium signal was detected by using optical mapping. (A) Representative images showing the calcium signaling activation time and (B) the histogram showing the quantitative results. (C) Representative images showing the direction of calcium signal conduction and the degree of dispersion with the vertical coordinate of conduction time. (D) Quantitative results of the discrete degree of calcium transients and the calcium signaling dispersion index. (E) Representative images showing calcium signal conduction waveforms and (F-I) the histogram shows the quantitative results of physiological parameters of the calcium signal. (J) Schematic of the change in ADP 30/80. The vertical coordinate is time in seconds. (K) Quantitative results for ADP 30/80. (L) Changes in ADP 90. n = 10
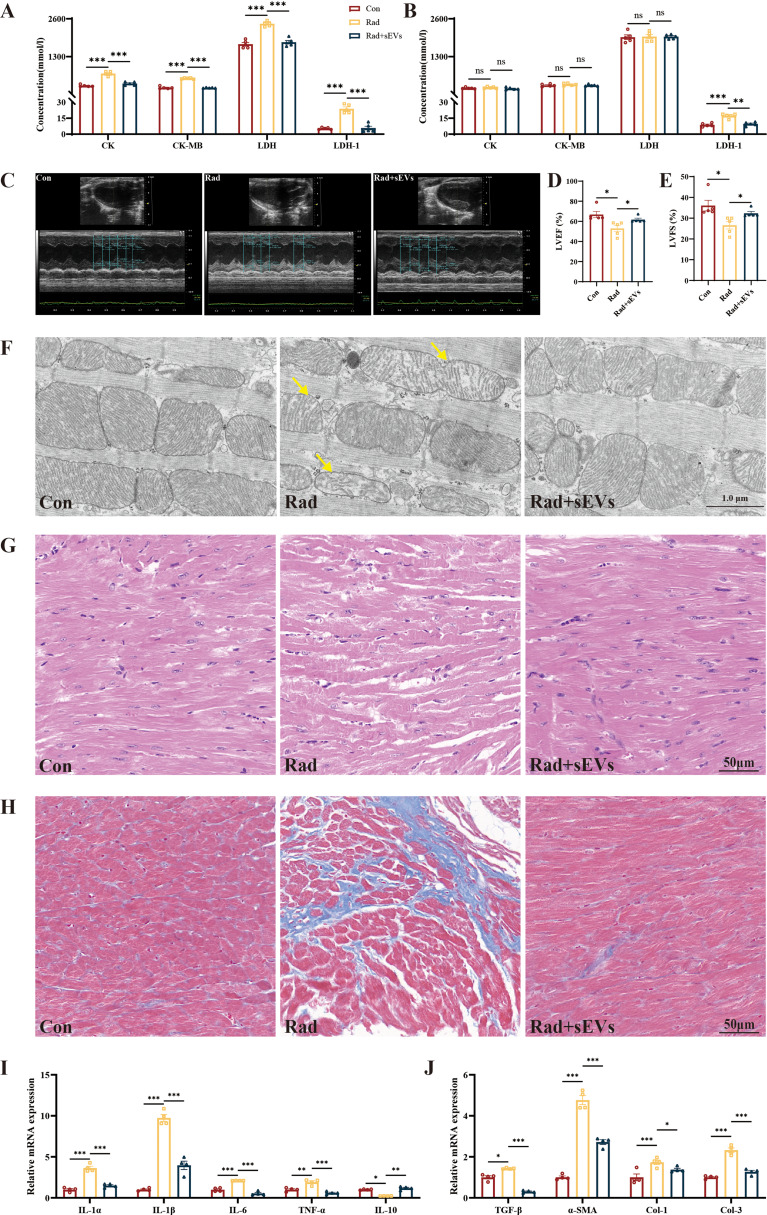
UCMSCs-sEVs alleviated the radiation-induced cardiac damage in mice
The concentration of myocardial enzymes CK, CK-MB, LDH, and LDH1 was elevated at day 14 after radiation, and returned to normal levels at day 28, except for LDH1 which remained elevated. Treatment with UCMSCs-sEVs significantly recovered the myocardial enzyme profiles (Fig. 7A and B). Cardiac function tests revealed that left ventricular ejection fraction (LVEF) and left ventricular fractional shortening (LVFS) reduced in radiation group, and improved after treated with UCMSCs-sEVs (Fig. 7C and E). UCMSCs-sEVs treatment could also ameliorate reduction of mitochondrial cristae (Fig. 7F), reduce disarray of myofibrillar (Fig. 7G) and decrease collagen deposition (Fig. 7H) caused by radiation damage.
Fig. 7.
The therapeutic effects of UCMSCs-sEVs on radiation-induced cardiac damage in mice. Changes in myocardial enzyme profile on day 14 (A) and day 28 (B) after radiation. n = 5. (C) Representative images of echocardiography at day 42 after radiation. Statistical results of LVEF (D) and LVFS (E). n = 5. (F) Electron microscopy images of mouse apical heart tissue. Yellow arrow: reduced mitochondrial cristae. Scale bar: 1 μm. Representative images of mouse heart HE staining (G) and Masson staining (H). Expression of inflammation related genes (I) and fibrosis related genes (J) in mouse heart. n = 4
We also detected the expression of inflammation- and fibrosis-related genes by using RT-qPCR, and the results showed that the levels of inflammatory cytokines (IL-1α, IL-1β, IL-6, and TNF-α) and fibrosis related factors (TGF-β, α-SMA, type I collagen, and type III collagen) increased, and the level of anti-inflammatory cytokine IL-10 decreased post-radiation but were significantly recovered after UCMSCs-sEVs treatment (Fig. 7I and J). The expression of inflammation and fibrosis related genes in cardiac organoids were consistent with results observed in mouse hearts (Supplementary Fig. 3).
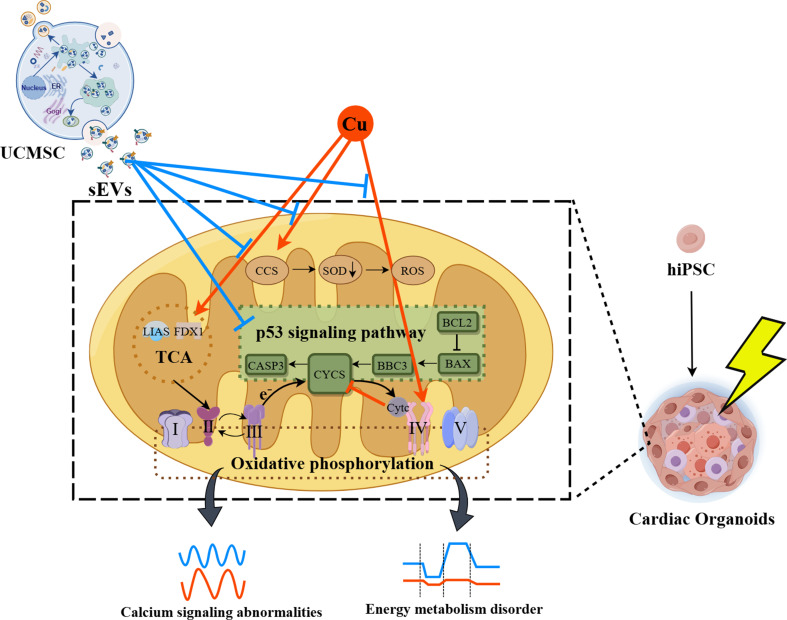
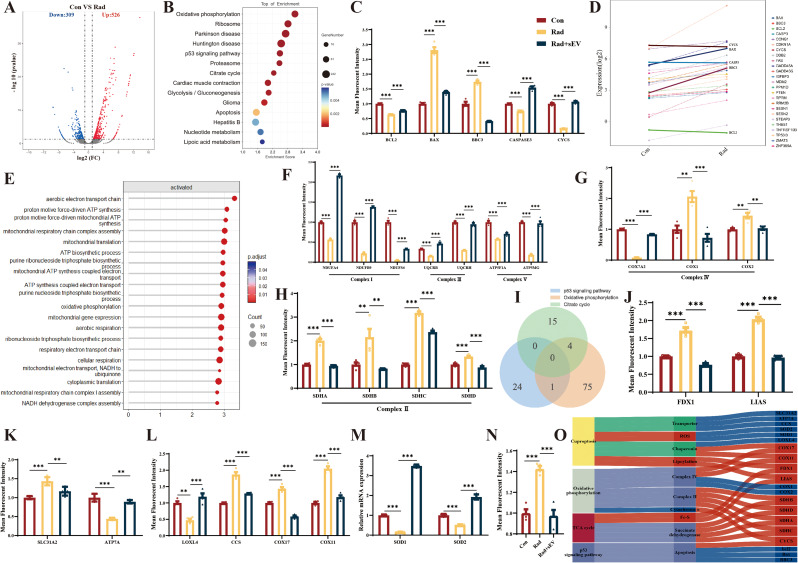
Copper metabolism interacts with the oxidative phosphorylation pathway and the p53 signaling pathway may be key to radiation-induced cardiac organoids injury
To investigate the potential mechanisms underlying radiation-induced injury and the therapeutic effect of UCMSCs-sEVs on cardiac organoids, we subjected control cardiac organoids and radiation-injured cardiac organoids to RNA-seq analysis. A total of 835 DEGs were identified in the radiation group compared to the control group by differential gene analysis, 526 genes exhibited increased expression, and 309 genes exhibited decreased expression (Fig. 8A). According to the KEGG enrichment analysis, we selected the p53 signaling pathway associated with X-ray-induced apoptosis (Fig. 8B). RT‒qPCR was used to verify the expression of p53 signaling pathway-associated apoptotic genes. The results showed that the expression of Bcl2 decreased and that of Bax and Bbc3 increased after radiation, and UCMSCs-sEVs treatment upregulated the expression of Bcl-2 and downregulated the expression of Bax and Bbc3, which was consistent with the results of live/dead staining of cardiac organoids (Fig. 8C). However, after radiation, the expression of caspase 3 and cytochrome C (CYCS) was decreased instead of increased (Fig. 8C), moreover, the same results were obtained by analyzing the differential expression of p53 signaling pathway genes (Fig. 8D). These findings suggested that there might be other death paradigms after X-ray exposure in addition to the caspase 9/caspase 3 pathway. We also detected the expression level of necroptosis (caspase-independent apoptosis) related genes, but have not found the expected changes (Supplementary Fig. 4).
Fig. 8.
The potential mechanism of action of UCMSCs-sEVs in radiation-induced cardiac organoid injury. (A) Volcano map of differentially expressed genes. There were a total of 835 DEGs, 526 of which were upregulated and 309 of which were downregulated. (B) KEGG enrichment analysis of RNA-seq data 48 h post-radiation. (C) Caspase 3-related apoptotic genes in the p53 signaling pathway. (D) Differential gene expression in the p53 signaling pathway. (E) GO enrichment was used to analyze gene function. (F) Complex I, complex III and complex V involved in oxidative phosphorylation. (G) Complex IV involved in oxidative phosphorylation. (H) Succinate dehydrogenase and complex II involved in the TCA cycle and oxidative phosphorylation. (I) Venn diagram showing the p53 signaling pathway with overlapping oxidative phosphorylation genes, such as CYCS; the TCA cycle with overlapping oxidative phosphorylation genes, such as SDHA, SDHB, SDHC and SDHD. (J) Lipoylated genes in the TCA cycle. (K) Expression of copper ion transport proteins (SLC31A and ATP7A). (L) The expression of copper-dependent monoamine oxidase (LOXL4), a copper chaperone for superoxide dismutase (CCS), the cytochrome c oxidase copper chaperone COX17 and the cytochrome c oxidase copper chaperone COX11. (M) The expression of superoxide dismutase. (N) The expression of the cytochrome C oxidase 1. (O) Sankey diagram describing the potential mechanisms of X-ray-induced radiation damage in cardiac organoids. n = 4
CYCS is the upstream gene of Caspase 3, therefore, we focused on the oxidative phosphorylation pathway associated with CYCS. According to the GO enrichment analysis, the functions of the genes related to CYCS and genes involved in oxidative respiratory chain signaling pathways were found to be related to the electron transport chain and ATP synthesis (Fig. 8E). Genes related to the oxidative respiratory chain were assessed via RT‒qPCR. The results showed that radiation downregulated the expression of complex I (NDUFA4, NDUFS4 and NDUFB9), complex III (UQCR8H and UQCRH) and complex V (ATP5MG and ATP5F1A)-associated genes, and UCMSCs-sEVs treatment reversed these changes (Fig. 8F). The expression of complex IV-related genes in the oxidative respiratory chain (COX7A2, COX1, and COX2) varied after radiation, whereas the expression of these genes was normalized after treatment with UCMSCs-sEVs (Fig. 8G). Similarly, the expression of complex II (SDHA, SDHB, SDHC, and SDHD)-related genes was normalized after treatment with UCMSCs-sEVs (Fig. 8H).
To investigate the paradoxical changes in complexes II and IV in cardiac organoids after X-ray radiation and in the other causes of cardiac organoid death, we performed a Venn plot analysis and revealed that succinate dehydrogenase changed with oxidative phosphorylation and the tricarboxylic acid (TCA) cycle (complex II), and that CYCS changed with oxidative phosphorylation in the p53 signaling pathway (Fig. 8I). We examined the genes involved in lipoylation at the initiation site of the TCA cycle (FDX1 and LIAS) and found that their expression was also increased (Fig. 8J). Changes in the expression of lipoylated genes in the TCA cycle are associated with cuproptosis, therefore, we examined the expression of copper ion transporters (SLC31A2 and ATP7A), and unexpectedly, the expression of both SLC31A2 and ATP7A increased (Fig. 8H). The expression of copper ion chaperones (LOXL4, CCS, COX11 and COX17) was also increased (Fig. 8L). The increased expression of CCS downregulated the expression of SOD1 and SOD2 (Fig. 8M) and caused the generation of reactive oxygen species (ROS). Copper ions are transported into mitochondria by COX17 and bind to SCO1 (Fig. 8N). A Sankey diagram describing the potential mechanisms of X-ray-induced radiation damage in cardiac organoids is shown in Fig. 8O. UCMSCs-sEVs treatment reversed the changes induced by X-ray radiation (Fig. 8I and N).
Discussion
With the increasing development of radiation therapy, the incidence of RIHD is rising [26]. While existing drugs have some therapeutic effects, they cannot fundamentally ameliorate myocardial oxygen consumption and prevent myocardial hypertrophy. It’s deserved deep study of new therapeutic strategy.
Conventional models for studying RIHD typically involve animals and cells, but these models have limitations, including species-derived differences and complex spatial structures. 3D organoids offer a solution to these limitations [27]. The formation of organoids from hiPSCs avoids the issue of inconsistent experimental results caused by different species [28]. Organoids derived from genetically knockout mouse strains are more capable of becoming important and visualizable tools for answering the unsolved mysteries in biological processes [29]. According to the experimental protocol of Hofbauer P et al. [23], cardiac organoids derived from hiPSCs possess cardiomyocytes, endothelial cells, and mesenchymal cells and can form chamber-like structures [30, 31]. These organoids can also spontaneously contract in a rhythmic pattern. In simulating RIHD, cardiac organoids have an advantage over cardiomyocyte cell lines because they better mimic the overall structure and conduction system of the heart. Therefore, cardiac organoids were used in this study to replace cardiomyocytes and establish a radiation injury model.
Our results verified that X-ray radiation could lead to a decrease in ATP production and glycolytic capacity. Additionally, we observed that X-ray radiation caused alterations in mitochondrial membrane structure, which in turn led to an increase in ROS production within the mitochondria and a decrease in mitochondrial membrane potential to increase proton permeability. TEM revealed the disappearance of mitochondrial inner cristae and substantial vacuolation of mitochondria after radiation. After treatment with UCMSCs-sEVs, mitochondrial function was restored, and energy metabolism in cardiac organoids was significantly improved.
The abnormal calcium signaling caused by X-ray radiation in cardiac organoids and changes in myofibrils have become potential inducers of ventricular arrhythmias [32]. Calcium ions affect muscle fiber contraction [33], and the contraction of cardiomyocytes affects calcium ion flow through excitation-contraction coupling [34]. A delay after depolarization, which occurs after complete repolarization, accelerates the rate of calcium ion activation and release [35]. This finding is consistent with the findings of Weiss JN [36] et al. that calcium signaling disorders lead to ventricular tachyarrhythmias and QT syndrome. Disordered calcium homeostasis promotes the occurrence of myocardial hypertrophy and plays an important role in cardiac remodeling [37–39]. The strength of cardiac systole depends on the amplitude of Ca2+ transients [40]. X-rays cause an increase in amplitude, activating excitation-contraction coupling, exacerbating contractile dysfunction and cardiac volume load, and inhibiting heart function [41]. Decreased cardiac pumping function activates the renin-angiotensin-aldosterone system and β-adrenergic system, increasing ventricular volume and promoting pumping function recovery [42]. Although the production of angiotensin II and aldosterone solves the problem of insufficient cardiac output [43], it also exacerbates cardiac preload and promotes myocardial fibrosis maturation. This change ultimately leads to late manifestations of RIHD, such as diastolic dysfunction and restrictive cardiomyopathy [44]. Maturation of myocardial fibrosis not only limits ventricular expansion but also hinders calcium ion flow [45]. Prolongation of the effective refractory period of calcium ions further promotes the occurrence of arrhythmia [46–48]. The changes in myofibrils and calcium transients in RIHD complement each other, jointly leading to heart dysfunction. UCMSCs-sEVs therapy can not only repair myofibrils, but also improve calcium signal. In other words, UCMSCs-sEVs may treat myocardial hypertrophy and dilation caused by RIHD and improve the occurrence of arrhythmia.
Through RNA sequencing, the p53 signaling pathway related to apoptosis was studied. Apoptosis is not the main cause of death in RIHD, as discovered through RT-qPCR. To further explore the true cause of death in cardiac organoids, GO enrichment analysis revealed that these genes are primarily involved in the electron transport chain and ATP synthesis. The increased expression of genes related to Complex II and IV in the oxidative phosphorylation signaling pathway, also associated with the TCA cycle. The study of Tsvetkov et al. [49] showed that the elevated expression of FDX1 and LIAS was associated with cuproptosis, this inferred us that cuproptosis may be related to RIHD. Copper ions enter mitochondria through COX17 and affect the core subunits COX1 and COX2 of cytochrome C oxidase (cCo) in complex IV [49, 50], leading to an increase in the expression of copper metabolism-related genes in complex IV. The increased expression of the core subunit of cCo may lead to abnormal assembly of cCo [51], affecting the electron transport chain and ATP synthesis. Abnormal cCo assembly leads to decreased CYCS expression, which inhibits the caspase 3-mediated apoptosis signaling pathway. Cuproptosis disorders could inhibit the production of caspases to some extent, which might be a possible characteristic death pattern of cardiac organoids after X-ray exposure. However, whether radiation-induced cuproptosis truly inhibits apoptosis and the underlying mechanism require further study.
Although the specific mechanism of RIHD injury has not been fully elucidated, this is the first report on the occurrence of cuproptosis in cardiac organoids during X-ray-induced injury. Moreover, the results suggested the presence of a multifaceted repair mechanism involving energy metabolism, electrophysiology, the p53 pathway, cuproptosis and oxidative phosphorylation in UCMSCs-sEVs during radiation-induced cardiac organoid injury. These results indicate that UCMSCs-sEVs have the potential to become a new therapeutic drug for RIHD.
This study has several limitations. The 3D cardiac organoids used to simulate RIHD, while advanced, cannot fully replace human hearts for research purposes. Additionally, these studies focused on gene expression levels without detecting changes in protein expression, and the mechanism of action of UCMSCs-sEVs was not explored in detail. Future research will address these limitations, further explore the therapeutic mechanisms of action of sEVs in RIHD and validate the specific mechanism by which cuproptosis occurs in radiologic heart disease to facilitate translational applications in clinical settings and provide new insights into the true mechanisms of RIHD injury.
Conclusions
UCMSCs-sEVs were used in this study to treat radiation-induced cardiac organoid injury (RICoI), and subsequent observations of mitochondrial structure and function and electrical signal transmission revealed promising therapeutic effects on RICoI. The observed therapeutic effects might be attributed to the combined effects of the p53 signaling pathway, the oxidative phosphorylation signaling pathway and copper ion accumulation. This is the first report that radiation induces cuproptosis in cardiac organoids and that UCMSCs-sEVs alleviate RICoI and RIHD. These findings not only contribute to the understanding of radiogenic cardiac injury but also provide a new therapeutic strategy for RIHD.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We are grateful to China Medical Management Consulting (Beijing) LTD. CO for gifting us Clinical-grade UCMSCs. The authors thank AJE for language polishing.
Abbreviations
- RIHD
Radiation-induced heart disease
- RT
Radiation therapy
- sEVs
Small extracellular vesicles
- UCMSCs
Umbilical cord mesenchymal stem cells
- ACEI
Angiotensin-converting enzyme inhibitors
- EVs
Extracellular vesicles
- iPSC
Induced pluripotent stem cells
- AdSC
Adult stem cells
- CMs
Cardiomyocytes
- ECs
Endocardial cells
- DPBS
Dulbecco’s phosphate-buffered saline
- PBS
Phosphate-buffered saline
- NTA
Nanoparticles Tracking Analysis
- TEM
Transmission electron microscopy
- WB
Western blot
- SDS
PAGE-Sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- PVDF
Polyvinylidene fluoride
- TBST
Tris-buffered saline with Tween 20
- BSA
Bovine serum albumin
- HRP
Horseradish peroxidase
- ECL
Enhanced chemiluminescence
- TMRM
Tetramethylrhodamine
- MFI
Mean fluorescence intensity
- ROI
Reactive oxygen superoxide ion
- ROS
Reactive oxygen species
- ECAR
Extracellular acidification rate
- OCR
Oxygen consumption rate
- Rote/AA
Rotenone/antimycin A
- cDNA
Complementary DNA
- hiPSCs
Human induced pluripotent stem cells
- BMP4
Bone morphogenic protein 4
- bFGF
Basic fibroblast growth factor
- RA
Retinoic acid
- cTnT
Cardiac troponin T
- MYL7
Myosin light chain 7
- IRX4
Iroquois homeobox 4
- CDH5
Cadherin 5
- PECAM1
Platelet and endothelial cell adhesion molecule 1
- VIM
Vimentin
- TST101
Tumor susceptibility gene 101
Author contributions
Hu Cao: Study design; execution, acquisition of data, analysis and interpretation; drafting and revising the article; Liang Yue and Jingyuan Shao: Execution, acquisition of data, analysis and interpretation; Fanxuan Kong, Shenghua Liu, Hongyu Huai, Zhichao He and Yuefeng Yang: Execution, acquisition of data; Yingxia Tan and Hua Wang; Conception, study design, revising and critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This research was supported by Zhejiang Provincial Natural Science Foundation of China under Grant No.LY19H160010.
Data availability
All data generated and analyzed during this study are included in this manuscript and supplementary information file. We have uploaded the complete sequencing data to the NCBI public database, BioProject: PRJNA1113873 (https://dataview.ncbi.nlm.nih.gov/object/PRJNA1113873?reviewer=mvm4itj9d4qgu3kj4uijohl3pn). After the publication of the article, we will make the complete sequencing data openly accessible.
Declarations
Ethics approval and consent to participate
The iPSCs used in our research was obtained from CORIELL Institute (GM25256, USA), which appropriate consent was obtained at the time of biospecimen collection. Clinical-grade UCMSCs was obtained from China Medical Management Consulting (Beijing) LTD. CO. Human umbilical cords were provided by women who gave birth at Yantai Laiyang Central Hospital (Yantai, China). The collection of umbilical cords and the development of mesenchymal stem cells were reviewed and approved by Laiyang Central Hospital of Yantai City on November 14, 2023, with approval number of 2023-002-001. Title of the approved project was “Umbilical cord collection and development of mesenchymal stem cells”. Written informed consent was obtained from the donors participating in the study. All procedures that involved human samples were approved by the ethics committee of Academy of Military Medical Sciences (AF/SC-08/02.425). Title of the approved project was “Experimental study on intervention effect of mesenchymal stem cells derived small extracellular vesicles on radiation-induced cardiac organoid injury”. All procedures that involved animals were approved by the Institutional Animal Care and Use Committee of the Laboratory Animal Center of Academy of Military Medical Sciences (IACUC-DWZX-2023-532). Title of the approved project was “Study on protective effect of stem cells derived small extracellular vesicles on radiation-induced heart damage in mice”.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing financial interest.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Hu Cao, Liang Yue and Jingyuan Shao contributed equally to this work.
Contributor Information
Yingxia Tan, Email: tanhu333@126.com.
Hua Wang, Email: 18511712135@163.com.
References
- 1.Wang H, Wei J, Zheng Q, et al. Radiation-induced heart disease: a review of classification, mechanism and prevention. Int J Biol Sci. 2019;15(10):2128–38. 10.7150/ijbs.35460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee PJ, Mallik R. Cardiovascular effects of radiation therapy: practical approach to radiation therapy-induced heart disease. Cardiol Rev. 2005;13(2):80–6. 10.1097/01.crd.0000131188.41589.c5. [DOI] [PubMed] [Google Scholar]
- 3.Donnellan E, Phelan D, McCarthy CP, et al. Radiation-induced heart disease: a practical guide to diagnosis and management. Cleve Clin J Med. 2016;83(12):914–22. 10.3949/ccjm.83a.15104. [DOI] [PubMed] [Google Scholar]
- 4.Boero IJ, Paravati AJ, Triplett DP, et al. Modern Radiation Therapy and Cardiac outcomes in breast Cancer. Int J Radiat Oncol Biol Phys. 2016;94(4):700–8. 10.1016/j.ijrobp.2015.12.018. Epub 2015 Dec 17. [DOI] [PubMed] [Google Scholar]
- 5.Boekel NB, Schaapveld M, Gietema JA, et al. Cardiovascular Disease Risk in a large, Population-based cohort of breast Cancer survivors. Int J Radiat Oncol Biol Phys. 2016;94(5):1061–72. Epub 2015 Dec 14. [DOI] [PubMed] [Google Scholar]
- 6.Zhang K, He X, Zhou Y, et al. Atorvastatin ameliorates Radiation-Induced Cardiac Fibrosis in rats. Radiat Res. 2015;184(6):611–20. 10.1667/RR14075.1. [DOI] [PubMed] [Google Scholar]
- 7.van der Veen SJ, Ghobadi G, de Boer RA, et al. ACE inhibition attenuates radiation-induced cardiopulmonary damage. Radiother Oncol. 2015;114(1):96–103. 10.1016/j.radonc.2014.11.017. [DOI] [PubMed] [Google Scholar]
- 8.O’Herron T, Lafferty J. Prophylactic use of colchicine in preventing radiation induced coronary artery disease. Med Hypotheses. 2018;111:58–60. 10.1016/j.mehy.2017.12.021. [DOI] [PubMed] [Google Scholar]
- 9.Johnstone RM, Adam M, Hammond JR, et al. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes). J Biol Chem. 1987;262(19):9412–20. [PubMed] [Google Scholar]
- 10.Li M, Fang F, Sun M, et al. Extracellular vesicles as bioactive nanotherapeutics: an emerging paradigm for regenerative medicine. Theranostics. 2022;12(11):4879–903. 10.7150/thno.72812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li H, Rong P, Ma X, et al. Mouse umbilical cord mesenchymal stem cell paracrine alleviates Renal Fibrosis in Diabetic Nephropathy by reducing myofibroblast transdifferentiation and cell proliferation and upregulating MMPs in Mesangial cells. J Diabetes Res. 2020;2020:3847171. 10.1155/2020/38471712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi Y, Wang Y, Li Q, et al. Immunoregulatory mechanisms of mesenchymal stem and stromal cells in inflammatory diseases. Nat Rev Nephrol. 2018;14(8):493–507. 10.1038/s41581-018-0023-5. [DOI] [PubMed] [Google Scholar]
- 13.Yao J, Zheng J, Cai J, et al. Extracellular vesicles derived from human umbilical cord mesenchymal stem cells alleviate rat hepatic ischemia-reperfusion injury by suppressing oxidative stress and neutrophil inflammatory response. FASEB J. 2019;33(2):1695–710. 10.1096/fj.. [DOI] [PubMed] [Google Scholar]
- 14.Zhao S, Xiu G, Wang J, et al. Engineering exosomes derived from subcutaneous fat MSCs specially promote cartilage repair as miR-199a-3p delivery vehicles in Osteoarthritis. J Nanobiotechnol. 2023;21(1):341. 10.1186/s12951-023-02086-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Z, Yan K, Ge G, et al. Exosomes derived from mir-155-5p-overexpressing synovial mesenchymal stem cells prevent osteoarthritis via enhancing proliferation and migration, attenuating apoptosis, and modulating extracellular matrix secretion in chondrocytes. Cell Biol Toxicol. 2021;37(1):85–96. 10.1007/s10565-020-09559-9. [DOI] [PubMed] [Google Scholar]
- 16.Chen W, Zhang F, Hou X, et al. Ameliorating role of microRNA-378 carried by umbilical cord mesenchymal stem cells-released extracellular vesicles in mesangial proliferative glomerulonephritis. Cell Commun Signal. 2022;20(1):28. 10.1186/s12964-022-00835-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng J, Lu T, Zhou C, et al. Extracellular vesicles derived from human umbilical cord mesenchymal stem cells protect Liver Ischemia/Reperfusion Injury by reducing CD154 expression on CD4 + T cells via CCT2. Adv Sci (Weinh). 2020;7(18):1903746. 10.1002/advs.201903746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cao H, Chen M, Cui X, et al. Cell-free Osteoarthritis treatment with sustained-release of chondrocyte-targeting exosomes from umbilical cord-derived mesenchymal stem cells to rejuvenate aging chondrocytes. ACS Nano. 2023;17(14):13358–76. 10.1021/acsnano.3c01612. [DOI] [PubMed] [Google Scholar]
- 19.Lehmann R, Lee CM, Shugart EC, et al. Human organoids: a new dimension in cell biology. Mol Biol Cell. 2019;30(10):1129–37. 10.1091/mbc.E19-03-0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shamir ER, Ewald AJ. Three-dimensional organotypic culture: experimental models of mammalian biology and disease. Nat Rev Mol Cell Biol. 2014;15(10):647–64. 10.1038/nrm3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim H, Kamm RD, Vunjak-Novakovic G. Progress in multicellular human cardiac organoids for clinical applications. Cell Stem Cell. 2022;29(4):503–14. 10.1016/j.stem.2022.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kreitzer FR, Salomonis N, Sheehan A, et al. A robust method to derive functional neural crest cells from human pluripotent stem cells. Am J Stem Cells. 2013;2(2):119–31. [PMC free article] [PubMed] [Google Scholar]
- 23.Hofbauer P, Jahnel SM, Papai N, et al. Cardioids reveal self-organizing principles of human cardiogenesis. Cell. 2021;184(12):3299–e331722. 10.1016/j.cell.2021.04.034. [DOI] [PubMed] [Google Scholar]
- 24.Mendjan S, Mascetti VL, Ortmann D, et al. NANOG and CDX2 pattern distinct subtypes of human mesoderm during exit from pluripotency. Cell Stem Cell. 2014;15(3):310–25. 10.1016/j.stem.2014.06.006. [DOI] [PubMed] [Google Scholar]
- 25.Meng Y, Li C, Liang Y, et al. Umbilical cord mesenchymal-stem-cell-derived exosomes exhibit anti-oxidant and antiviral effects as cell-free therapies. Viruses. 2023;15(10):2094. 10.3390/v15102094. PMID: 37896871; PMCID: PMC10612094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yi J, Yue L, Zhang Y, et al. PTPMT1 protects cardiomyocytes from necroptosis induced by γ-ray irradiation through alleviating mitochondria injury. Am J Physiol Cell Physiol. 2023;324(6):C1320–31. 10.1152/ajpcell.00466.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fatehullah A, Tan SH, Barker N. Organoids as an in vitro model of human development and disease. Nat Cell Biol. 2016;18(3):246–54. 10.1038/ncb3312. [DOI] [PubMed] [Google Scholar]
- 28.Sugimoto S, Sato T. Organoid vs in vivo mouse model: which is Better Research Tool to Understand the Biologic mechanisms of Intestinal Epithelium? Cell Mol Gastroenterol Hepatol. 2022;13(1):195–7. 10.1016/j.jcmgh.2021.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Farin HF, Jordens I, Mosa MH, et al. Visualization of a short-range wnt gradient in the intestinal stem-cell niche. Nature. 2016;530(7590):340–3. 10.1038/nature16937. [DOI] [PubMed] [Google Scholar]
- 30.Kim H, Kamm RD, Vunjak-Novakovic G, Wu JC. Progress in multicellular human cardiac organoids for clinical applications. Cell Stem Cell. 2022;29(4):503–14. 10.1016/j.stem.2022.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu L, Liu K, Feng Q, Liao Y, Cardiac Organoids. A 3D technology for modeling Heart Development and Disease. Stem Cell Rev Rep. 2022;18(8):2593–605. 10.1007/s12015-022-10385-1. [DOI] [PubMed] [Google Scholar]
- 32.Landstrom AP, Dobrev D, Wehrens XHT. Calcium Signaling and Cardiac Arrhythmias. Circ Res. 2017;120(12):1969–93. 10.1161/CIRCRESAHA.117.310083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vikhorev PG, Song W, Wilkinson R, et al. The dilated cardiomyopathy-causing mutation ACTC E361G in cardiac muscle myofibrils specifically abolishes modulation of ca(2+) regulation by phosphorylation of troponin I. Biophys J. 2014;107(10):2369–80. 10.1016/j.bpj.2014.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jin JP. Myofilament and cytoskeleton proteins: fine machineries of biological movements. Arch Biochem Biophys. 2013;535(1):1–2. 10.1016/j.abb.2013.02.011. [DOI] [PubMed] [Google Scholar]
- 35.Sato D, Shiferaw Y, Garfinkel A, et al. Spatially discordant alternans in cardiac tissue: role of calcium cycling. Circ Res. 2006;99(5):520–7. 10.1161/01.RES.0000240542.03986.e7. [DOI] [PubMed] [Google Scholar]
- 36.Weiss JN, Garfinkel A, Karagueuzian HS, et al. Perspective: a dynamics-based classification of ventricular arrhythmias. J Mol Cell Cardiol. 2015;82:136–52. 10.1016/j.yjmcc.2015.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jarcho JA, McKenna W, Pare JA, et al. Mapping a gene for familial hypertrophic cardiomyopathy to chromosome 14q1. N Engl J Med. 1989;321(20):1372–8. 10.1056/NEJM198911163212005. [DOI] [PubMed] [Google Scholar]
- 38.Geisterfer-Lowrance AA, Kass S, Tanigawa G, et al. A molecular basis for familial hypertrophic cardiomyopathy: a beta cardiac myosin heavy chain gene missense mutation. Cell. 1990;62(5):999–1006. 10.1016/0092-8674(90)90274-i. [DOI] [PubMed] [Google Scholar]
- 39.Kapplinger JD, Landstrom AP, Bos JM, et al. Distinguishing hypertrophic cardiomyopathy-associated mutations from background genetic noise. J Cardiovasc Transl Res. 2014;7(3):347–61. 10.1007/s12265-014-9542-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bers DM. Cardiac excitation-contraction coupling. Nature. 2002;415(6868):198–205. 10.1038/415198a. [DOI] [PubMed] [Google Scholar]
- 41.Hill JA, Olson EN. Cardiac plasticity. N Engl J Med. 2008;358(13):1370–80. 10.1056/NEJMra072139. [DOI] [PubMed] [Google Scholar]
- 42.Weber KT, Brilla CG. Pathological hypertrophy and cardiac interstitium. Fibrosis and renin-angiotensin-aldosterone system. Circulation. 1991;83(6):1849–65. 10.1161/01.cir.83.6.1849. [DOI] [PubMed] [Google Scholar]
- 43.Halliday BP, Prasad SK. The Interstitium in the Hypertrophied Heart. JACC Cardiovasc Imaging. 2019;12(11 Pt 2):2357–68. 10.1016/j.jcmg.2019.05.033. [DOI] [PubMed] [Google Scholar]
- 44.Lee Chuy K, Nahhas O, Dominic P, et al. Cardiovascular complications Associated with Mediastinal Radiation. Curr Treat Options Cardiovasc Med. 2019;21(7):31. 10.1007/s11936-019-0737-0. [DOI] [PubMed] [Google Scholar]
- 45.Swynghedauw B, Chevalier B, Charlemagne D, et al. Cardiac hypertrophy, arrhythmogenicity and the new myocardial phenotype. II. The cellular adaptational process. Cardiovasc Res. 1997;35(1):6–12. 10.1016/s0008-6363(97)00076-x. [DOI] [PubMed] [Google Scholar]
- 46.Dobrev D, Wehrens XHT. Calcium-mediated cellular triggered activity in atrial fibrillation. J Physiol. 2017;595(12):4001–8. 10.1113/JP273048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Splawski I, Timothy KW, Sharpe LM, et al. Ca(V)1.2 calcium channel dysfunction causes a multisystem disorder including arrhythmia and autism. Cell. 2004;119(1):19–31. 10.1016/j.cell.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 48.Vercellino I, Sazanov LA. The assembly, regulation and function of the mitochondrial respiratory chain. Nat Rev Mol Cell Biol. 2022;23(2):141–61. 10.1038/s41580-021-00415-0. [DOI] [PubMed] [Google Scholar]
- 49.Tsvetkov P, Coy S, Petrova B, et al. Copper induces cell death by targeting lipoylated TCA cycle proteins. Science. 2022;375(6586):1254–61. 10.1126/science.abf0529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Horng YC, Cobine PA, Maxfield AB, et al. Specific copper transfer from the Cox17 metallochaperone to both Sco1 and Cox11 in the assembly of yeast cytochrome C oxidase. J Biol Chem. 2004;279(34):35334–40. 10.1074/jbc.M404747200. [DOI] [PubMed] [Google Scholar]
- 51.Nývltová E, Dietz JV, Seravalli J, Khalimonchuk O, Barrientos A. Coordination of metal center biogenesis in human cytochrome c oxidase. Nat Commun. 2022;13(1):3615. 10.1038/s41467-022-31413-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated and analyzed during this study are included in this manuscript and supplementary information file. We have uploaded the complete sequencing data to the NCBI public database, BioProject: PRJNA1113873 (https://dataview.ncbi.nlm.nih.gov/object/PRJNA1113873?reviewer=mvm4itj9d4qgu3kj4uijohl3pn). After the publication of the article, we will make the complete sequencing data openly accessible.