ABSTRACT
With global protein prices on the rise, lowering protein levels in animal feed, together with balancing diet composition and reducing nitrogen emissions, can both reduce the environmental impact of agriculture and save on feed costs. However, the formulation of an ideal amino acid (AA) composition is crucial for better protein utilization by livestock. This study aimed to investigate the effects of different lysine to methionine ratios on the antioxidant capacity and immune function of the rumen in Tibetan sheep. Ninety male Tibetan sheep, weaned at 2 months of age, were randomly divided into three groups (1:1, 2:1 and 3:1 lysine ratios) and subjected to a 100‐day feeding trial. RNA sequencing (RNA‐seq) was utilized to analyse the impact of different AA ratios on gene expression in rumen tissue, whereas the levels of antioxidant enzymes (total antioxidant capacity [T‐AOC], superoxide dismutase [SOD], glutathione peroxidase [GSH‐Px] and catalase [CAT]) and immunoglobulins (immunoglobulin A [IgA], immunoglobulin G [IgG] and immunoglobulin M [IgM]) were evaluated. The results indicated that the 1:1 group significantly upregulated the expression of PTGS2, PLA2G12A and PLA2G4 genes, enhancing antioxidant enzyme activity, reducing free radical production and modulating systemic immune responses. COL16A1 and KCNK5 were highly expressed in the protein digestion and absorption pathway, maintaining the structural integrity and function of the rumen epithelium. BMP4 and TGFBR2 were significantly enriched in the cytokine–cytokine receptor interaction pathway and positively correlated with CAT and T‐AOC. ITGA8 was upregulated in the 1:1 group, participating in the regulation of various cellular signalling pathways. ATP2B1 was enriched in the cyclic guanosine monophosphate (cGMP)– protein kinase G (PKG) signalling and mineral absorption pathways, primarily influencing oxidative stress and immune responses by regulating intracellular calcium ion concentration. This study demonstrates that a 1:1 lysine to methionine ratio is most beneficial for enhancing the antioxidant capacity and immune function of the rumen in Tibetan sheep.
Keywords: amino acids, anti‐oxidation, immunity, low protein, Tibetan sheep, transcriptomics
This study investigates the impact of different lysine to methionine ratios on the antioxidant capacity and immune function in the rumen of Tibetan sheep using RNA‐Seq analysis. A 1:1 ratio significantly enhances the expression of key genes and antioxidant enzymes, reducing oxidative stress and boosting immune responses. These findings provide a scientific basis for optimizing amino acid supplementation in sheep diets to improve health and reduce environmental impact.
1. Introduction
Amino acids (AAs) are critical factors influencing animal growth (Wu 2009). Their supplementation in animal diets enhances feed utilization and promotes the development of protein feed resources, among other benefits (Stein and Shurson 2009). However, both deficiency and excess of AA can cause an imbalance, negatively impacting protein utilization efficiency and increasing nitrogen emissions (Higgs et al. 2023). The balance of AA is crucial for maximizing their utilization efficiency (Stein and Shurson 2009). In feed production, the amounts of added AA are strictly controlled, ensuring balanced relationships between AA, both common and rare, and thus determination of the appropriate proportions of the added AA is necessary to maintain the optimal AA balance (Maqsood et al. 2022).
The rumen is the largest compartment of the ruminant stomach. It functions as both a large fermenter and a storage tank, containing vast numbers of bacteria, fungi and protozoa (Ushida and Jouany 1985). These microbial communities are always in a state of dynamic equilibrium, and through anaerobic fermentation, the food in the rumen will be degraded into nutrients to meet the growth and developmental needs of the host (Fliegerova et al. 2021). Because of this efficient digestion in the rumen, ruminants can maintain their body and production requirements by consuming low‐quality crude high‐fibre forage for long periods of time (Blümmel, Karsli, and Russell 2003). Therefore, the rumen plays a significant role in the health, digestion and absorption of the animal. Several studies have shown that after reducing the level of crude protein in the diet of dairy cows, there was a significant reduction in milk production which could be improved by supplementation with lysine, methionine and histidine that enhanced nitrogen utilization and reduced nitrogen emissions (Lee et al. 2012). Therefore, determining the appropriate AA ratio in low‐protein diets is critical for growth, nutrient digestibility and reduction in feeding costs in ruminants, including the Tibetan sheep.
Oxidative stress caused by excess ROS disrupts cellular redox homeostasis, induces cellular autophagy, triggers apoptosis and causes irreversible tissue damage, and the body's antioxidant defence system effectively scavenges these robot operating system (ROS) (Snider et al. 2013). Moreover, as the immune response is boosted, the compounds begin to multiply until they reach a critical mass capable of fighting infectious pathogens (Li, Hou et al. 2007). This process results in a feedback ‘suppression’ of the immune response in the animal, ensuring that the host fights inflammation without overstimulating or suppressing immune activity (Cui et al. 2023). However, the antioxidant and immunological capacity is more dependent on the nutrients consumed, and the mechanism of action of consuming different ratios of AAs in Tibetan sheep is not clear.
Tibetan sheep (Ovis aries) are found on the Tibetan Plateau, China. They are a valuable germplasm resource (Gui et al. 2021) and the development of the Tibetan sheep industry is necessary for improving the quality of life of local herders as well as the competitiveness of the society (Li et al. 2021). In recent years, transcriptome sequencing (RNA sequencing [RNA‐Seq]) methods have been widely used to explore the molecular mechanisms underlying animal phenotypes from the level of gene expression, including the biological functions of AA in animals. Therefore, in this experiment, RNA‐Seq was used to analyse the effects of different AA proportions on the antioxidant capacity and immune function of the Tibetan sheep rumen while consuming low‐protein diets.
2. Materials and Methods
2.1. Animals and Diets
The experiment was conducted from April to July 2022 at Jinzang Ranch, Haiyan County, Haibei Tibetan Autonomous Prefecture, Qinghai Province, China. Ninety male plateau lambs with similar body weight (body weight = 15.37 ± 0.92 kg), weaned at 2 months of age, were randomly selected and divided into three groups. Each group contained 30 lambs, with 5 replicates per group and 6 sheep per replicate. The three groups were the LP‐L group (1:1 lysine: methionine ratio), LP‐M group (2:1 lysine: methionine ratio) and LP‐H group (3:1 lysine: methionine ratio). The experimental diet consisted of 70% concentrate and 30% roughage (consisting of oat silage and oat hay, with a dry‐matter ratio of 1:1); additionally, soybean meal, rapeseed meal, cottonseed meal, maize germ meal and palm meal were used as the protein source to provide crude protein in the diet. Table 1 shows the composition and nutritional content of the diet. The experimental animals were housed in a shed with natural sunlight and access to a large field. They were fed daily at 08:00 and 17:00 and could eat and drink freely. The experiment lasted for 100 days, including 10 days of pre‐feeding and 90 days of formal feeding. At the end of the experiment, one sheep was randomly selected from each replicate in each group, and a total of 15 sheep were slaughtered. Rumen tissues were collected from the sheep. Part of the tissue sample was snap‐frozen in liquid nitrogen and stored in a freezer at −80°C for RNA separation, whereas the rest was fixed in 4% paraformaldehyde for tissue sectioning.
TABLE 1.
Dietary concentrate composition and nutrient levels (dry matter basis).
| Items | LP‐L | LP‐M | LP‐H |
|---|---|---|---|
| Ingredient (%) | |||
| Oat hay | 15.000 | 15.000 | 15.000 |
| Oat silage | 15.000 | 15.000 | 15.000 |
| Corn | 36.533 | 37.100 | 37.100 |
| Wheat | 7.700 | 7.700 | 7.700 |
| Soybean meal | 0.700 | 0.700 | 0.700 |
| Rapeseed meal | 7.000 | 7.000 | 7.000 |
| Cottonseed meal | 0.700 | 0.700 | 0.700 |
| Maize germ meal | 0.700 | 0.700 | 0.700 |
| Palm meal | 11.200 | 11.200 | 11.200 |
| NaCl | 0.350 | 0.350 | 0.602 |
| Limestone | 0.350 | 0.441 | 0.700 |
| Baking soda | 0.070 | 0 | 0.070 |
| Premix a | 2.940 | 2.940 | 2.940 |
| Lys | 1.386 | 0.931 | 0.483 |
| Met | 0.371 | 0.238 | 0.105 |
| Total | 100.000 | 100.000 | 100.000 |
| Nutrient levels | |||
| DE (MJ/kg) b | 10.760 | 10.840 | 10.840 |
| Crude protein | 9.940 | 9.980 | 9.980 |
| Ether extract | 2.850 | 2.870 | 2.870 |
| Crude fibre | 22.470 | 22.61 | 22.610 |
| Neutral detergent fibre | 33.720 | 33.77 | 33.770 |
| Acid detergent fibre | 23.370 | 23.39 | 23.390 |
| Ca | 0.421 | 0.424 | 0.424 |
| P | 0.171 | 0.172 | 0.172 |
Provided per kilogram of diets: Cu 15 mg, Fe 55 mg, Zn 25 mg, Mn40 mg, Se 0.30 mg, I 0.5 mg, Co 0.20 mg, VA 20 000 IU, VD 4 000 IU, VE 40 IU.
Digestive energy is the calculated value, whereas the rest are measured values.
2.2. Histological Analysis of the Rumen
After slaughter, the rumen tissues were removed. The contents of the rumen were cleaned, and a tissue sample of approximately 1 cm2 of tissue was cut from the same position in each rumen with scissors, rinsed with normal saline and immediately immersed in 10 mL of 4% paraformaldehyde in a centrifuge tube for the preparation of tissue sections. All sections were prepared, stained, photographed and measured by Tianjin Keritai Biological Co. Ltd.
2.3. Antioxidant and Immune Response Indices
The antioxidant and immune response indices were determined by double‐antibody one‐step sandwich enzyme‐linked immunosorbent assays (ELISAs). The antioxidant indicators included the total antioxidant capacity (T‐AOC), superoxide dismutase (SOD) activity, glutathione peroxidase (GSH‐Px) activity, catalase (CAT) activity and malondialdehyde (MDA) contents. Indicators of immune response included immunoglobulin A (IgA), immunoglobulin M (IgM) and immunoglobulin G (IgG). The ELISA kits used in this experiment were purchased from Jiangsu Enzyme Label Biotechnology Co. Ltd. (China).
2.4. RNA Extraction, Library Preparation and Sequencing
Total RNA was extracted from the rumen tissue using TRIzol reagent according to the manufacturer's instructions (Magen). Paired‐end libraries were prepared using an ABclonal mRNA‐seq Lib Prep Kit (ABclonal, China), following the manufacturer's instructions. The mRNA was purified from 1 µg of total RNA using Oligo (dT) magnetic beads followed by fragmentation using divalent cations at elevated temperatures in ABclonal first strand synthesis reaction buffer. The mRNA fragments were then reverse‐transcribed into cDNA and amplified by PCR. The PCR products were purified (AMPure XP system; Beckman Coulter, Brea, CA, USA) and library quality was assessed on an Agilent Bioanalyzer 4150 system (Santa Clara, CA, USA). Finally, the library preparations were sequenced on an Illumina Novaseq 6000 (or MGISEQ‐T7) (San Diego, CA, USA), and 150 bp paired‐end reads were generated.
2.5. Data Analysis
The data generated from the Illumina (or BGI) platform were used for bioinformatics analysis. All the analyses were performed using an in‐house pipeline from Shanghai Applied Protein Technology (Shanghai, China). The major software and parameters were as follows.
2.5.1. Quality Control
Raw data (or raw reads) in the FASTA format were first processed using in‐house Perl scripts. In this step, the adapter sequences were removed, and low‐quality reads, as well as those where the N ratio was greater than 5%, were filtered to obtain clean reads that could be used for subsequent analysis.
2.5.2. Mapping to the Reference Genome
The clean reads were then individually aligned to the reference genome using the orientation mode in HISAT2 software (http://daehwankimlab.github.io/hisat2/) to obtain the mapped reads. These mapped reads were then mapped to the O. aries reference genome.
2.5.3. Quantification of Gene Expression
FeatureCounts (http://subread.sourceforge.net/) was used to determine the number of reads mapped to each gene. The fragments per kilobase of transcript per million mapped reads (FPKM) of each gene were calculated on the basis of the length of the gene and the number of mapped reads to the gene.
2.5.4. Differential Gene Expression, Trend Review and KEGG Enrichment Analysis
Differential expression of genes was analysed using the DESeq2 package in R (http://bioconductor.org/packages/release/bioc/html/DESeq2.html). Genes with | log2FC | > and Padj < 0.05 were considered differentially expressed genes (DEGs). The trend analysis uses linear regression model and LOESS regression to fit the change trend of gene expression with time and identifies the key gene regulation network. The ggplot2 package was used to map gene expression trends in R language environment to show the dynamic changes of different genes during development. Using the clusterProfiler R package for KEGG pathway enrichment analysis, a KEGG pathway is considered significantly enriched when p < 0.05.
2.5.5. Real‐Time Fluorescence Quantitative PCR Verification
Six DEGs were randomly selected, with GAPDH as the internal control. Primers were designed and synthesized by Shanghai Bioengineering Co. Ltd (Table 2). RNA was extracted from tissue samples using TransZol Up Plus RNA Kit and then reverse‐transcribed into cDNA using Universal SYBR Green qPCR Mix. Real‐time fluorescence quantitative PCR was performed, with relative gene expression calculated by the 2−ΔΔCt method.
TABLE 2.
Primers used in RT‐qPCR.
| Name | Primer sequence (5′–3′) | Tm (°C) | Product length ( bp) |
|---|---|---|---|
| GAPDH | F‐GACCTGCCGCCTGGAGAAAC | 60 | 120 |
| R‐AGAGTGAGTGTCGCTGTTGAAGTC | |||
| MYH11 | F‐AGCCAGAGACGAGAGGACCTTC | 60 | 120 |
| R‐AAGCCGTTGGAGAGGAATGTGTAG | |||
| TAGLN | F‐TCTTCAAGCAGATGGAGCAGGTG | 60 | 93 |
| R‐AGAGGTCAACGGTCTGGAACATATC | |||
| ACTA1 | F‐GCGTGGCTACTCCTTCGTGAC | 60 | 171 |
| R‐TTGCCGATGGTGATGACCTGAC | |||
| ACTG2 | F‐GCCACAGCAGCCTCCTCTTC | 60 | 191 |
| R‐TTGCGGATGTCAATGTCACACTTC | |||
| FLNC | F‐GGTGGAGGTGCTGTACGATGAC | 60 | 141 |
| R‐GTGAAGCGGTTAGCCTTGTTGAC | |||
| FLNA | F‐CGAGAAGCCACCACCGAGTTC | 60 | 196 |
| R‐CGTCGTAGGTCACATCCACAGAG |
Note: The melting temperature (Tm ) of each primer pair is 60°C, ensuring optimal annealing during PCR. Product length is specified for accurate amplification and subsequent analysis.
2.6. Statistical Analysis
The phenotypic data were sorted using Microsoft Excel 2019, and Duncan's method was used to perform multiple comparison analyses using SPSS 26.0 (IBM Corp., Armonk, NY, USA). Data are expressed as mean ± standard deviation, with p < 0.05 considered statistically significant.
3. Results
3.1. Histomorphological Observations of the Rumen
The images from the morphological analysis of rumen tissue present histological sections and measured parameters from three groups: LP‐L, LP‐M and LP‐H. The measured parameters include muscularis thickness, papillae length, papillae width, corneal thickness and nipple density. Histological sections (Figure 1a) reveal distinct differences in rumen structure among the three groups. The LP‐L group exhibits well‐developed and prominent papillae, indicating robust rumen morphology. The LP‐M group shows moderately developed papillae and a slightly reduced overall structure. The LP‐H group exhibits less developed papillae, suggesting possible atrophy or reduced growth. Measured parameters (Figure 1b) indicate that papillae length and width in the LP‐L group are significantly greater than in the LP‐M and LP‐H groups (p < 0.05). The muscularis thickness in the LP‐L group is significantly greater than in the LP‐M group (p < 0.05). The corneal thickness in the LP‐M group is significantly greater than in the LP‐L group (p < 0.05).
FIGURE 1.
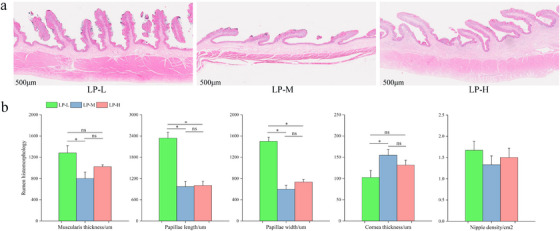
Histological analysis of the rumen tissue. (a) Representative histological sections of the rumen from the three experimental groups (LP‐L, LP‐M and LP‐H). Scale bars represent 500 µm. (b) Quantitative measurements of muscularis thickness, papillae length, papillae width, corneal thickness and nipple density. Data are presented as mean ± SD. Statistical significance is indicated as follows: *p < 0.05.
3.2. Determination of Antioxidant Capacity and Immune Level
As shown in Table 3, the activity of the antioxidant enzyme GSH‐Px was significantly higher in the LP‐L group compared with the LP‐M and LP‐H groups (p < 0.05). The MDA content in the LP‐L group was significantly lower than that in the LP‐M and LP‐H groups (p < 0.05). However, there were no significant differences in T‐AOC, CAT and SOD among the three groups (p > 0.05). In terms of immune levels, the levels of IgG were markedly higher in the LP‐L group relative to the LP‐M and LP‐H groups (p < 0.05), whereas IgM levels were significantly greater in the LP‐L and LP‐M groups compared with the LP‐H group (p < 0.05), with no significant difference between LP‐L and LP‐M (p > 0.05). However, IgA levels were no significant difference among the three groups (p > 0.05).
TABLE 3.
Antioxidant capacity and immune level measurement.
| Items | LP‐L | LP‐M | LP‐H | p value |
|---|---|---|---|---|
| Oxidation index | ||||
| T‐AOC | 53.88 ± 0.92 | 54.71 ± 1.28 | 51.35 ± 1.89 | 0.252 |
| SOD | 171.52 ± 4.69 | 179.10 ± 11.43 | 184.68 ± 3.48 | 0.410 |
| GSH‐Px | 1001.74 ± 78.84a | 930.43 ± 57.52b | 923.06 ± 33.44b | 0.048 |
| CAT | 222.26 ± 8.37 | 207.63 ± 14.93 | 211.89 ± 5.50 | 0.602 |
| MDA | 12.98 ± 0.31b | 15.32 ± 0.39a | 14.78 ± 0.43a | 0.001 |
| Immune indices | ||||
| IgA | 258.19 ± 8.08 | 258.89 ± 19.57 | 251.25 ± 10.51 | 0.910 |
| IgG | 684.31 ± 57.02a | 610.00 ± 44.83b | 630.83 ± 42.60b | 0.036 |
| IgM | 1507.05 ± 45.13a | 1321.82 ± 33.45b | 1499.09 ± 52.39a | 0.027 |
Note: Values are expressed as mean ± standard deviation. Different superscript letters (a, b) indicate significant differences between groups (p < 0.05).
Abbreviations: CAT, catalase; GSH‐Px, glutathione peroxidase; IgA, immunoglobulin A; IgG, immunoglobulin G; IgM, immunoglobulin M; MDA, malondialdehyde; SOD, superoxide dismutase; T‐AOC, total antioxidant capacity.
3.3. Analysis of Sequencing Data
3.3.1. Quality Control
Transcriptome sequencing was performed on rumen tissues from the LP‐L, LP‐M and LP‐H groups (three samples per group). As shown in Table 4, the number of raw reads from all samples ranged between 40,403,834 and 66,760,508. The number of clean reads ranged between 40,265,522 and 66,709,028, the error was below 0.05% for all samples, the Q20 content was above 97.5%, and the GC content was in the range of 43.19%–47.47%. This indicated that the quality of the library construction was good, and the base content was close enough to meet the requirements of the subsequent analysis.
TABLE 4.
Sample sequencing data evaluation.
| Sample name | Raw_reads | Clean_reads | Error (%) | Q20 (%) | GC (%) |
|---|---|---|---|---|---|
| LP‐L‐1 | 58,802,530 | 58,667,696 | 0.02 | 98.75 | 44.71 |
| LP‐L‐2 | 57,625,582 | 57,581,386 | 0.04 | 99.08 | 45.63 |
| LP‐L‐3 | 66,760,508 | 66,709,028 | 0.04 | 99.09 | 46.96 |
| LP‐M‐1 | 40,403,834 | 40,265,522 | 0.05 | 97.68 | 43.74 |
| LP‐M‐2 | 58,486,784 | 58,352,622 | 0.02 | 98.62 | 47.47 |
| LP‐M‐3 | 66,226,352 | 66,137,412 | 0.04 | 98.84 | 44.81 |
| LP‐H‐1 | 59,862,156 | 59,716,872 | 0.02 | 98.83 | 43.19 |
| LP‐H‐2 | 60,540,092 | 60,395,842 | 0.02 | 98.67 | 46.05 |
| LP‐H‐3 | 77,868,552 | 77,807,648 | 0.04 | 98.11 | 44.93 |
Note: Error rate indicates the proportion of bases that were called incorrectly. Q20 represents the percentage of bases with a Phred score of 20 or higher, indicating 99% accuracy. GC content is the percentage of guanine and cytosine bases in the sample.
3.3.2. Sequencing Levels of Gene Expression
From Table 5, it can be seen that the total read content of all samples ranged between 47,737,716 and 70,781,462, with the number of sequences localized to the genome in the range of 45,698,446–69,071,913, which accounted for more than 94.80% of the total reads. Of the total mapped reads, the segmentation sequenced sequences on the two exons were in the range of 1477,135–3338,288. As shown in Figure 2a, the average correlation coefficients (R 2) among individuals in the LP‐L, LP‐M and LP‐H groups were 0.9556, 0.9733 and 0.9733, respectively. Figure 2b shows that each group exhibited consistent and concentrated expression, with no entanglement or overlap observed between groups. Figure 2c illustrates the differences among the three groups by reducing data complexity through transformation into principal components (PCs) that capture the maximum variance. In conclusion, these analyses indicate that the genes in the three sequencing groups exhibit high expression levels.
TABLE 5.
Sample sequencing for gene expression levels.
| Sample name | Total reads | Total mapped | Mapping rates (%) | Splice reads |
|---|---|---|---|---|
| LP‐L‐1 | 47,737,716 | 45,698,446 | 95.73 | 3068,892 |
| LP‐L‐2 | 62,588,774 | 61,285,357 | 97.92 | 3338,288 |
| LP‐L‐3 | 70,781,462 | 69,071,913 | 97.58 | 2246,050 |
| LP‐M‐1 | 65,491,848 | 64,253,592 | 98.11 | 2688,837 |
| LP‐M‐2 | 48,129,642 | 46,474,944 | 96.56 | 1477,135 |
| LP‐M‐3 | 62,858,234 | 61,820,109 | 98.35 | 1731,206 |
| LP‐H‐1 | 56,946,024 | 55,785,896 | 97.96 | 2565,278 |
| LP‐H‐2 | 52,258,500 | 50,307,052 | 96.27 | 2468,710 |
| LP‐H‐3 | 53,929,202 | 51,122,488 | 94.80 | 3261,286 |
Note: Total mapped reads refer to the number of reads that aligned to the reference genome. Mapping rates indicate the percentage of total reads that successfully mapped. Splice reads represent the reads that span exon‐exon junctions.
FIGURE 2.

RNA sequencing (RNA‐seq) data quality and expression levels. (a) Pearson correlation heatmap showing the correlation between samples in the LP‐L, LP‐M and LP‐H groups. (b) Density distribution of FPKM (Fragments per Kilobase of transcript per million mapped reads) values across the samples. (c) Principal component analysis (PCA) plot illustrating the variance in gene expression among the LP‐L, LP‐M and LP‐H groups.
3.3.3. UpRegulation and DownRegulation of DEGs
As illustrated in the volcano plot in Figure 3, 157 genes are upregulated and 455 genes are downregulated between the LP‐M and LP‐L groups. The comparison between the LP‐H and LP‐L groups reveals 39 upregulated genes and 174 downregulated genes. Compared to the LP‐M group, the LP‐H group exhibits six upregulated genes and five downregulated genes.
FIGURE 3.
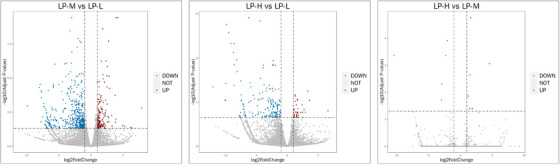
Differential gene expression analysis. Volcano plots displaying differentially expressed genes between LP‐M versus LP‐L, LP‐H versus LP‐L and LP‐H versus LP‐M groups. Genes with significant upregulation are shown in red, and those with significant downregulation are shown in blue.
3.3.4. Trend Analysis
Figure 4a illustrates the gene expression profiles at various time points or conditions. Each line represents a distinct gene, with the y‐axis denoting gene expression levels. Each line represents a distinct gene, with the y‐axis denoting gene expression levels. Figure 4b presents the clustering of gene expression profiles. Genes are clustered according to their expression trends, and the representative trend for each cluster is depicted along with the corresponding gene count. Notably, profile0, profile1 and profile3 exhibit significantly enriched trends.
FIGURE 4.
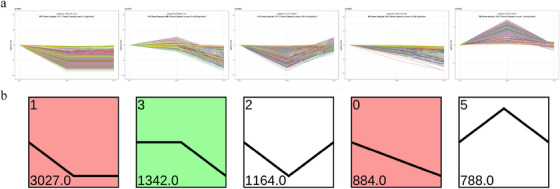
Trend analysis of gene expression. (a) Gene expression profiles over various conditions or time points. Each line represents a distinct gene, with the y‐axis denoting gene expression levels. (b) Clustering of gene expression profiles. Clusters are grouped on the basis of their expression trends, with the representative trend for each cluster and corresponding gene count shown.
3.3.5. KEGG Enrichment Analysis
KEGG enrichment analysis was conducted for all DEGs (Figure 5). The analysis identified extracellular matrix (ECM)‐receptor interaction and arachidonic acid metabolism as commonly enriched pathways across the three groups. In the LP‐H versus LP‐L group, significant enrichment was found in ECM‐receptor interaction, cyclic guanosine monophosphate (cGMP)– protein kinase G (PKG) signaling pathway, arachidonic acid metabolism, mineral absorption and complement and coagulation cascades pathways. In the LP‐M versus LP‐L group, significant enrichment was observed in ECM‐receptor interaction, protein digestion and absorption, arachidonic acid metabolism and cytokine–cytokine receptor interaction pathways. In the LP‐H versus LP‐M group, significant enrichment was observed in intestinal immune network for IgA production, arginine and proline metabolism, antigen processing and presentation and Th1 and Th2 cell differentiation pathways. These pathways regulate rumen phenotypic traits, including tissue development, antioxidant activity and immunity. Additionally, core genes regulating rumen phenotypic traits, such as tissue development, antioxidant activity and immunity, were identified from significantly enriched pathways. These genes include CFB, ATP2B1, PTGS2, PLA2G12A, ITGA8, PLA2G4A, COL16A1, BMP4, KCNK5 and TGFBR2.
FIGURE 5.
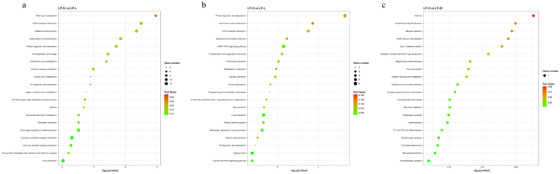
KEGG pathway enrichment analysis. KEGG pathway enrichment of differentially expressed genes in (a) LP‐H versus LP‐L, (b) LP‐M versus LP‐L and (c) LP‐H versus LP‐M groups. Pathways significantly enriched are indicated with varying colours and sizes.
3.3.6. RT‐qPCR Verification
The RT‐qPCR verification of the transcriptome sequencing data (Figure 6) showed that the expression of ACTA1, FLNA, FLNC, ACTG2, TAGLN, and MYH11 was consistent with the expression patterns of the transcriptome sequencing data, confirming the accuracy and credibility of the data.
FIGURE 6.
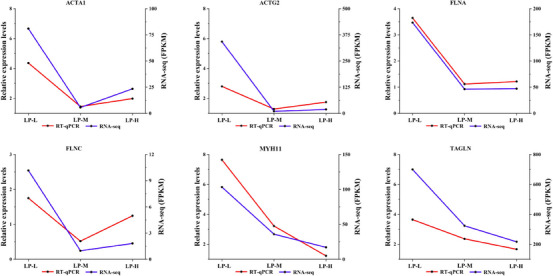
Validation of RNA sequencing (RNA‐seq) data by RT‐qPCR. Relative expression levels of ACTA1, ACTG2, FLNA, FLNC, MYH11 and TAGLN measured by RT‐qPCR and RNA‐seq. Data are presented as mean ± SD from three biological replicates.
3.4. Correlation Analysis of Phenotypic Traits and Core Genes
Figure 7a illustrates a significant correlation between antioxidant enzymes (CAT, T‐AOC, SOD and GSH‐Px) and multiple genes. It is noteworthy that CAT and T‐AOC exhibit a highly significant positive correlation with CFB, ATP2B1, PTGS2, PLA2G12A, ITGA8, PLA2G4A, COL16A1 and BMP4. These findings suggest that these antioxidant enzymes may play a critical role in modulating oxidative stress responses via these genes. Figure 7b underscores the relationship between immunoglobulins (IgA, IgG and IgM) and gene expression: IgA, IgG and IgM exhibit significant positive correlations with ATP2B1, PTGS2, PLA2G12A, ITGA8 and COL16A1. These associations suggest that these genes may play a role in the immune response of Tibetan sheep. Figure 7c shows that muscle layer thickness, teat length and teat width were all highly significantly and positively correlated with CFB, ATP2B1, PTGS2, PLA2G12A, ITGA8 and PLA2G4A. These results suggest that these genes may influence tissue development and structural characteristics of Tibetan sheep.
FIGURE 7.
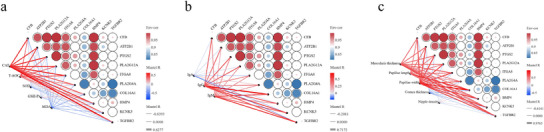
Correlation analysis of phenotypic traits and core genes. (a) Significant correlations between antioxidant enzymes (catalase [CAT], total antioxidant capacity [T‐AOC], superoxide dismutase [SOD], glutathione peroxidase [GSH‐Px]) and core genes. (b) Significant correlations between immunoglobulins (immunoglobulin A [IgA], immunoglobulin G [IgG], immunoglobulin M [IgM]) and gene expression. (c) Correlation between histomorphological parameters of the rumen and core gene expression. Red lines indicate positive correlations, and blue lines indicate negative correlations.
4. Discussion
The efficiency of nutrient absorption and utilization in animal feed can be assessed through the morphological observation of rumen tissue (Beiranvand et al. 2014). In the morphology of rumen tissue, the length, width and density of papillae influence ion transport and nutrient absorption in the rumen epithelium (Montoro et al. 2013). This study found that the Lys/Met ratio of 1:1 was significantly more effective than the 2:1 and 3:1 groups. It indicates that a Lys/Met ratio of 1:1 can provide a balanced AA profile, ensuring sufficient substrates for protein synthesis. This balance can promote cell division and growth, especially in the epithelial cells of the papillae, thereby enhancing the development of the rumen epithelium (Guoyao 2010). Moreover, lysine is essential for collagen synthesis, a major component of connective tissue, which enhances tissue strength and elasticity, thereby promoting papillae growth (Fuller et al. 2007). A thicker muscle layer enhances feed retention and facilitates the optimal mixing of feed with rumen microorganisms, which is crucial for the animal's growth and development (Lv et al. 2020). Research has shown that lysine is a critical AA for the synthesis of muscle proteins, including actin and myosin (Brosnan and Brosnan 2006). Methionine is a precursor in the methionine pathway that initiates protein synthesis and plays a crucial role in the mTOR (mammalian target of rapamycin) signalling pathway, regulating cell growth and metabolism (Wu and Morris 1998). Therefore, a 1:1 ratio of lysine and methionine can optimize muscle protein synthesis, enhancing the volume and strength of rumen tissue and leading to increased muscle layer thickness. A balanced intake of lysine and methionine can regulate the synthesis and degradation of keratin, preventing excessive thickening of the stratum corneum (Kimball and Jefferson 2006). The stratum corneum may provide some protective function for the rumen, but excessive thickness does not significantly impact nutrient absorption (Greenwood et al. 1997). In this experiment, the LP‐M group showed higher values compared to the LP‐L and LP‐H groups, consistent with the aforementioned results.
When the body is under oxidative stress, the rumen function is impaired, which will affect the normal growth and development of the animal (Engler et al. 2022). Changes in the T‐AOC reflect both the body's metabolism and its ability to compensate for free radical generation when it is subjected to external stimuli (Gu, Hao, and Wang 2012). GSH‐Px and SOD cooperate to eliminate harmful free radicals and thus protect cell membranes from damage. GSH‐Px shows widespread expression in the body where it catalyses the decomposition of peroxides (Battin and Brumaghim 2009). CAT is an antioxidant enzyme that acts downstream of SOD to eliminate reactive oxygen radicals and hydrogen peroxide, and its activity level is a measure of the free radical‐scavenging ability of the body (Abdollahi, Rezaei, and Fazaeli 2020). MDA is an end‐product of lipid peroxidation, and its levels are an indication of both oxidative stress and antioxidant capacity, with higher MDA levels indicating weaker antioxidant capacity (Akhavan‐Salamat and Ghasemi 2016). It has been found that methionine and lysine supplementation affects the antioxidant capacity in sheep, whereas methionine supplementation can reduce MDA contents and increase the levels of SOD, CAT, phosphoglycolate phosphatase and glutathione (GSH) S‐transferase (Mavrommatis et al. 2021). GSH is synthesized from glutamate, cysteine and glycine (Bauchart‐Thevret et al. 2009). The availability of cysteine is the rate‐limiting factor in GSH synthesis, and methionine can be converted into cysteine via its metabolic pathway (Bertolo et al. 2013). When the Lys/Met ratio is 1:1, methionine is adequately supplied and can be effectively converted into cysteine, thereby promoting GSH synthesis. Adequate levels of methionine support the function of antioxidant enzymes and GSH, thereby reducing oxidative stress and subsequently decreasing MDA production.
Immunoglobulin levels represent a measure of immunity, and changes in immunoglobulin concentrations can also reflect the level of resistance to infectious diseases and epidemics (Wu et al. 2012). AAs are necessary for the regulation of key metabolic pathways associated with the immune response and thus play extremely important roles in the resistance to stress and disease mediated by protein synthesis and cell signalling mechanisms (Cuca and Jensen 1990). IgM is the first antibody type produced during the humoral immune response and has a potent bactericidal effect (Liu et al. 2020). IgA is mainly involved in mucosal immune processes and local resistance to infection (Terayama, Terada, and Nakanuma 1996). IgG, on the other hand, is the most important antibody in the humoral immunity process and can act against bacteria, viruses and toxins (Bhaskara et al. 2021). It has been shown that methionine affects the immune system through the modulation of intracellular GSH and cysteine levels, influencing the proliferation of immune cells and enhancing immunity (Deng, Wong, and Nolan 2007; Rubin et al. 2007). Swain and Johri (2000) found that the addition of methionine to the diets of laying hens at 3.0 and 6.5 g/kg increased the humoral and cellular immune responses, respectively. The results of the present study were consistent with these findings, and a Lys/Met ratio of 1:1 significantly enhances immune activity in rumen tissue and reduces immune stress. This suggests that a 1:1 Lys/Met ratio can modulate immune cell metabolism and signal transduction pathways, promote the expression of anti‐inflammatory cytokines and inhibit the release of pro‐inflammatory cytokines, thereby mitigating inflammatory responses and immune stress. Heat shock protein 70 (HSP70) may also play a protective role in cellular stress responses. Research indicates that an optimal AA balance can increase HSP70 expression, enhance cellular stress tolerance and reduce immune stress (Kumaraguru et al. 2003).
This study utilized RNA‐seq to examine the transcriptome of rumen tissues in Tibetan sheep subjected to three distinct AA ratios, aiming to understand the impact of these varying proportions. KEGG analysis revealed significant enrichment in pathways that regulate phenotypic traits, including the cGMP–PKG signalling pathway, ECM‐receptor interaction, mineral absorption, arachidonic acid metabolism, protein digestion and absorption and cytokine–cytokine receptor interaction. The ECM‐receptor interaction and protein digestion and absorption pathways play crucial roles in the growth and repair of rumen tissue. In the ECM‐receptor interaction pathway, signals are transmitted by binding ECM to cell membrane receptors, regulating cell growth, proliferation and differentiation (Novoseletskaya, Evdokimov, and Efimenko 2023). Within the protein digestion and absorption pathway, the rumen efficiently breaks down ingested proteins into AAs and small peptides, which are absorbed and utilized. These AAs are essential for the growth and repair of rumen tissue (Zhang, Liu, and Zhang 2021). The cGMP–PKG signalling pathway influences various downstream targets such as ion channels, phosphodiesterases and transcription factors, collectively impacting the metabolism of rumen microorganisms (Amado et al. 2011). Activation of PKG regulates enzymes involved in nitrogen metabolism, potentially enhancing the assimilation of supplemental AAs into microbial proteins (Stenvang et al. 2018). Furthermore, the cGMP–PKG pathway improves blood flow and nutrient delivery to the rumen by regulating smooth muscle relaxation and contraction, promoting rumen development, modulating cellular antioxidant responses and reducing oxidative stress damage (Brady et al. 2007). Notably, the cGMP–PKG pathway interconnects with the mineral absorption, ECM‐receptor interaction and cytokine–cytokine receptor interaction pathways. The cGMP–PKG pathway produces cGMP under the influence of nitric oxide (NO) or natriuretic peptides. cGMP activates PKG, which phosphorylates target proteins to regulate various cellular functions (Scholten et al. 2006). In the mineral absorption pathway, calcium and magnesium are vital for ECM protein function and integrin‐mediated cell adhesion mechanisms (Powell, Jugdaohsingh, and Thompson 1999). The cGMP–PKG pathway regulates mineral absorption and ECM‐receptor interactions, suggesting a coordinated mechanism for maintaining cellular integrity and signal transduction (Shen, Johnson, and Gobe 2016). This pathway influences mineral absorption by regulating ion channels and transporters. PKG phosphorylates and regulates transient receptor potential melastatin (TRPM) channels involved in magnesium and calcium transport (Hofmann et al. 2009). The pathway also affects ECM‐receptor interactions by regulating integrin and ECM receptor expression. PKG‐mediated phosphorylation modulates receptor affinity for ECM proteins, influencing cell‐matrix interactions and signalling pathways that control cell proliferation and differentiation (Hiura and Nakagawa 2012). In the cytokine–cytokine receptor interaction pathway, ECM‐receptor interactions influence cytokine signal transduction by regulating the cellular environment (Hussain et al. 2014). Integrin signalling can enhance or inhibit cytokine receptor signalling, thus impacting immune responses and inflammation (Mcvie and Ringborg 2012). Additionally, the cGMP–PKG pathway modulates cytokine and receptor expression. PKG influences the production of inflammatory cytokines like TNF‐α and IL‐6 and their receptors, affecting immune cell signalling and inflammatory responses (Manchope et al. 2016).
The core genes involved in regulating rumen tissue development, antioxidation and immunity (CFB, ATP2B1, PTGS2, PLA2G12A, ITGA8, PLA2G4A, COL16A1, BMP4, KCNK5 and TGFBR2) were identified on the basis of the above pathways. Notably, PTGS2, PLA2G12A and PLA2G4 are enriched in the arachidonic acid metabolism pathway. PLA2G12A and PLA2G4 are subtypes of phospholipase A2, capable of catalysing phospholipids in the cell membrane and releasing arachidonic acid, thereby providing substrates for downstream PTGS2 (Lee et al. 2011). Chen et al. (2024) demonstrated that PTGS2 might be a potential regulator of immune and inflammatory responses in CSU patients, suggesting that targeting PTGS2 could offer a novel therapeutic perspective for CSU (Chen et al. 2024). PTGS2 catalyses the conversion of arachidonic acid to prostaglandin G2 (PGG2), which is subsequently converted to prostaglandin H2 (PGH2), a precursor of various prostaglandins and thromboxanes. The synthesis and release of prostaglandins modulate oxidative stress and inflammatory responses (Smith, Garavito, and DeWitt 1996). In this study, a Lys/Met ratio of 1:1 resulted in the upregulation of PTGS2, PLA2G12A and PLA2G4 gene expression. This upregulation enhances the expression and activity of antioxidant enzymes by regulating arachidonic acid metabolism and prostaglandins, reducing free radical production, modulating systemic immune responses and maintaining rumen immune homeostasis. COL16A1 and KCNK5 are enriched in the protein digestion and absorption pathway and exhibit high expression levels in the 1:1 group. Research has indicated that during cold‐season nutritional stress, COL16A1 expression in the rumen epithelium of Tibetan sheep increases. This elevation helps maintain the structural integrity and function of the rumen epithelium, thereby enhancing antioxidative capacity and immune defence mechanisms (Sha et al. 2024). KCNK5 regulates cellular redox state, promoting the production of antioxidant enzymes like SOD and CAT, thereby enhancing cellular antioxidative capacity (Liu et al. 2022). This suggests that a Lys/Met ratio of 1:1 enhances the synthesis of ECM protein (COL16A1) and potassium ion channel protein (KCNK5). BMP4 and TGFBR2 can both bind to receptors, initiating downstream SMAD protein pathways, regulating cyclins and inhibiting the NF‐κB signalling pathway to reduce oxidative stress and inflammatory responses (Zhang et al. 2023). They are enriched in the cytokine–cytokine receptor interaction pathway, with BMP4 exhibiting a highly significant positive correlation with CAT and T‐AOC. ITGA8 is upregulated in the Lys/Met 1:1 group, indicating that an AA ratio of 1:1 participates more in regulating various cellular signalling pathways. Lysine regulates protein synthesis through the mTOR pathway, whereas methionine serves as a precursor of S‐adenosylmethionine (SAM), a major methyl donor in methylation reactions, thereby affecting gene expression (Li, Yin et al. 2007). This gene binds to ECM proteins (such as fibronectin and laminin), facilitating cell adhesion, migration and signal transduction, which in turn influences tissue development and repair (Zhang et al. 2020). Interestingly, the ITGA8 gene enhances cellular antioxidative capacity through activation of cell signalling pathways, such as the PI3K/Akt pathway. It also plays a critical role in the adhesion and activation of immune cells, including macrophages and T cells, thereby promoting immune responses (Hynes 2002; Marek et al. 2016). In this study, ATP2B1 was found to be enriched in the cGMP–PKG signalling pathway and the mineral absorption pathway. ATP2B1 primarily regulates tissue development and antioxidative capacity within the cGMP–PKG signalling pathway. As a plasma membrane calcium pump, it influences the proliferation and differentiation of rumen epithelial cells via phosphorylation, thereby promoting tissue development and modulating calcium ion concentration, which affects cellular oxidative stress (Singh and Webster 1976). Within the mineral absorption pathway, ATP2B1 primarily regulates immune responses by modulating intracellular calcium ion concentration, influencing the activation and function of immune cells, including macrophages and T cells. Proper calcium ion levels facilitate the activation of immune cells and cytokine secretion, thereby enhancing the body's immune defence mechanisms (Berridge, Lipp, and Bootman 2000).
5. Conclusions
Through RNA‐seq and biochemical analysis, this study systematically evaluated the impact of different lysine to methionine ratios on the antioxidant and immune functions of the rumen in Tibetan sheep. The results demonstrated that a 1:1 ratio significantly enhances the expression and activity of antioxidant enzymes and immunoglobulins, reducing oxidative stress and inflammatory responses. Key genes, such as CFB, ATP2B1, PTGS2, PLA2G12A, ITGA8, PLA2G4A, COL16A1, BMP4, KCNK5 and TGFBR2, play crucial roles in maintaining rumen epithelial health and function across different signalling pathways. The study highlights the critical influence of an appropriate lysine to methionine ratio on rumen health, providing a scientific basis for optimizing the feeding strategies for Tibetan sheep.
Author Contributions
Fengshuo Zhang: conceptualization, data curation, formal analysis, methodology, validation, writing–original draft, writing–review and editing. Quyangangmao Su: formal analysis, investigation, methodology. Zhanhong Gao: software, supervision. Zhenling Wu: investigation, methodology, validation. Qiurong Ji: project administration, software. Tingli He: software, visualization. Kaina Zhu: formal analysis, visualization. Xuan Chen: formal analysis, investigation. Yu Zhang: methodology, software. Shengzhen Hou: resources, supervision. Linsheng Gui: resources, supervision.
Disclosure
I would like to declare on behalf of my co‐authors that the work described was original research that has not been published previously, and not under consideration for publication elsewhere, in whole or in part. All the authors listed have approved the manuscript that is enclosed.
Conflicts of Interest
The authors declare no conflicts of interest.
Ethics Statement
Ethics approval and consent to participate our study was carried out in compliance with the ARRIVE guidelines (AVMA Guidelines for the Euthanasia of Animals: 2020 Edition). All animal procedures for experiments were approved by the Committee of Experimental Animal care and handling techniques were approved (QUA‐2020‐0710) by the Qinghai University of Animal Care Commit‐tee. Moreover, all applicable rules and regulation of the organization and government were followed regarding the ethical use of experimental animal.
Peer Review
The peer review history for this article is available at https://publons.com/publon/10.1002/vms3.70173.
Funding: This research was supported by Qinghai Science and Technology Department Project (2022NK169).
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
References
- Abdollahi, M. , Rezaei J., and Fazaeli H.. 2020. “Performance, Rumen Fermentation, Blood Minerals, Leukocyte and Antioxidant Capacity of Young Holstein Calves Receiving High‐Surface ZnO Instead of Common ZnO.” Archives of Animal Nutrition 74: 189–205. [DOI] [PubMed] [Google Scholar]
- Akhavan‐Salamat, H. , and Ghasemi H. A.. 2016. “Alleviation of Chronic Heat Stress in Broilers by Dietary Supplementation of Betaine and Turmeric Rhizome Powder: Dynamics of Performance, Leukocyte Profile, Humoral Immunity, and Antioxidant Status.” Tropical Animal Health and Production 48: 181–188. [DOI] [PubMed] [Google Scholar]
- Amado, L. L. , Garcia M. L., Pereira T. C., Yunes J. S., Bogo M. R., and Monserrat J. M.. 2011. “Chemoprotection of Lipoic Acid Against Microcystin‐Induced Toxicosis in Common Carp (Cyprinus carpio, Cyprinidae).” Comparative Biochemistry and Physiology. Toxicology & Pharmacology 154: 146–153. [DOI] [PubMed] [Google Scholar]
- Battin, E. E. , and Brumaghim J. L.. 2009. “Antioxidant Activity of Sulfur and Selenium: A Review of Reactive Oxygen Species Scavenging, Glutathione Peroxidase, and Metal‐Binding Antioxidant Mechanisms.” Cell Biochemistry and Biophysics 55: 1–23. [DOI] [PubMed] [Google Scholar]
- Bauchart‐Thevret, C. , Stoll B., Chacko S., and Burrin D. G.. 2009. “Sulfur Amino Acid Deficiency Upregulates Intestinal Methionine Cycle Activity and Suppresses Epithelial Growth in Neonatal Pigs.” American Journal of Physiology. Endocrinology and Metabolism 296: E1239–E1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beiranvand, H. , Ghorbani G. R., Khorvash M., et al. 2014. “Interactions of Alfalfa Hay and Sodium Propionate on Dairy Calf Performance and Rumen Development.” Journal of Dairy Science 97: 2270–2280. [DOI] [PubMed] [Google Scholar]
- Berridge, M. J. , Lipp P., and Bootman M. D.. 2000. “The Versatility and Universality of Calcium Signalling.” Nature Reviews Molecular Cell Biology 1: 11–21. [DOI] [PubMed] [Google Scholar]
- Bertolo, R. F. , and Mcbreairty L. E.. 2013. “The Nutritional Burden of Methylation Reactions.” Current Opinion in Clinical Nutrition and Metabolic Care 16: 102–108. [DOI] [PubMed] [Google Scholar]
- Bhaskara, V. , Leal M. T., Seigner J., et al. 2021. “Efficient Production of Recombinant Secretory IgA Against Clostridium Difficile Toxins in CHO‐K1 Cells.” Journal of Biotechnology 331: 1–13. [DOI] [PubMed] [Google Scholar]
- Blümmel, M. , Karsli A., and Russell J. R.. 2003. “Influence of Diet on Growth Yields of Rumen Micro‐Organisms In Vitro and In Vivo: Influence on Growth Yield of Variable Carbon Fluxes to Fermentation Products.” British Journal of Nutrition 90: 625–634. [DOI] [PubMed] [Google Scholar]
- Brady, C. A. , Dover T. J., Massoura A. N., Princivalle A. P., Hope A. G., and Barnes N. M.. 2007. “Identification of 5‐HT3A and 5‐HT3B Receptor Subunits in Human Hippocampus.” Neuropharmacology 52: 1284–1290. [DOI] [PubMed] [Google Scholar]
- Brosnan, J. T. , and Brosnan M. E.. 2006. “The Sulfur‐Containing Amino Acids: An Overview.” Journal of Nutrition 136: 1636S–1640S. [DOI] [PubMed] [Google Scholar]
- Chen, Y. , Jian X., Zhu L., et al. 2024. “PTGS2: A Potential Immune Regulator and Therapeutic Target for Chronic Spontaneous Urticaria.” Life Sciences 344: 122582. [DOI] [PubMed] [Google Scholar]
- Cuca, M. , and Jensen L. S.. 1990. “Arginine Requirement of Starting Broiler Chicks.” Poultry Science 69: 1377–1382. [DOI] [PubMed] [Google Scholar]
- Cui, X. , Yang Y., Zhang M., et al. 2023. “Transcriptomics and Metabolomics Analysis Reveal the Anti‐Oxidation and Immune Boosting Effects of Mulberry Leaves in Growing Mutton Sheep.” Frontiers in Immunology 13: 1088850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, K. , Wong C. W., and Nolan J. V.. 2007. “Carry‐Over Effects of Early‐Life Supplementary Methionine on Lymphoid Organs and Immune Responses in Egg‐Laying Strain Chickens.” Animal Feed Science and Technology 134: 66–76. [Google Scholar]
- Engler, P. , Desguerets C., Benarbia M. E. A., and Mallem Y.. 2022. “Supplementing Young Cattle With a Rumen‐Protected Grape Extract Around Vaccination Increases Humoral Response and Antioxidant Defenses.” Veterinary and Animal Science 15: 100232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fliegerova, K. O. , Podmirseg S. M., Vinzelj J., et al. 2021. “The Effect of a High‐Grain Diet on the Rumen Microbiome of Goats With a Special Focus on Anaerobic Fungi.” Microorganisms 9: 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller, B. , Davis T. A., Jaeger L. A., et al. 2007. “Important Roles for the Arginine Family of Amino Acids in Swine Nutrition and Production.” Livestock Science 112, no. 1–2: 8–22. [Google Scholar]
- Greenwood, R. H. , Morrill J. L., Titgemeyer E. C., and Kennedy G. A.. 1997. “A New Method of Measuring Diet Abrasion and Its Effect on the Development of the Forestomach.” Journal of Dairy Science 80: 2534–2541. [DOI] [PubMed] [Google Scholar]
- Gu, X. H. , Hao Y., and Wang X. L.. 2012. “Overexpression of Heat Shock Protein 70 and Its Relationship to Intestine Under Acute Heat Stress in Broilers: 2. Intestinal Oxidative Stress.” Poultry Science 91: 790–799. [DOI] [PubMed] [Google Scholar]
- Gui, L.‐S. , Raza S. H. A., Ahmed Allam F. A. E., et al. 2021. “Altered Milk Yield and Rumen Microbial Abundance in Response to Concentrate Supplementation During the Cold Season in Tibetan Sheep.” Electronic Journal of Biotechnology 53: 80–86. [Google Scholar]
- Guoyao, W. 2010. “Functional Amino Acids in Growth, Reproduction, and Health.” Advances in Nutrition 1: 31–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs, R. J. , Chase L. E., Schwab C. G., et al. 2023. “Balancing Dairy Cattle Diets for Rumen Nitrogen and Methionine or All Essential Amino Acids Relative to Metabolizable Energy.” Journal of Dairy Science 106: 1826–1836. [DOI] [PubMed] [Google Scholar]
- Hiura, A. , and Nakagawa H.. 2012. “Changes in Response Behaviors to Noxious Heat and Mechanical Stimuli After Carrageenan‐Induced Inflammation in Mice Treated With Capsaicin 2 or 15 Days After Birth.” WebmedCentral Neurosciences 1: 331–347. [Google Scholar]
- Hofmann, F. , Bernhard D., Lukowski R., and Weinmeister P.. 2009. “cGMP Regulated Protein Kinases (cGK).” Handbook of Experimental Pharmacology 191: 137–162. [DOI] [PubMed] [Google Scholar]
- Hussain, F. , Martinez‐Zaguilan R., Sennoune S. R., Arutunyan A., Castro‐Marin R., and del Rosario C.. 2014. “V‐ATPase Regulates Communication Between Microvascular Endothelial Cells and Metastatic Cells.” Cellular and Molecular Biology 60, no. 1: 19–25. [PubMed] [Google Scholar]
- Hynes, R. O. 2002. “Integrins: Bidirectional, Allosteric Signaling Machines.” Cell 110: 673–687. [DOI] [PubMed] [Google Scholar]
- Kimball, S. R. , and Jefferson L. S.. 2006. “Signaling Pathways and Molecular Mechanisms Through Which Branched‐Chain Amino Acids Mediate Translational Control of Protein Synthesis.” Journal of Nutrition 136, no. 1: 227S–231S. [DOI] [PubMed] [Google Scholar]
- Kumaraguru, U. , Gouffon C. A., Ivey R. A., Rouse B. T., and Bruce B. D.. 2003. “Antigenic Peptides Complexed to Phylogenically Diverse Hsp70s Induce Differential Immune Responses.” Cell Stress & Chaperones 8: 134–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, C. , Hristov A. N., Cassidy T. W., et al. 2012. “Rumen‐Protected Lysine, Methionine, and Histidine Increase Milk Protein Yield in Dairy Cows Fed a Metabolizable Protein‐Deficient Diet.” Journal of Dairy Science 95: 6042–6056. [DOI] [PubMed] [Google Scholar]
- Lee, J. C. , Simonyi A., Sun A. Y., and Sun G. Y.. 2011. “Phospholipases A2 and Neural Membrane Dynamics: Implications for Alzheimer's Disease.” Journal of Neurochemistry 116: 813–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, L. L. , Hou Z. P., Yin Y. L., Liu Y. H., and Wang S. X.. 2007. “Intramuscular Administration of Zinc Metallothionein to Preslaughter Stressed Pigs Improves Anti‐Oxidative Status and Pork Quality.” Asian‐Australasian Journal of Animal Sciences 20: 761–767. [Google Scholar]
- Li, L. L. , Ma S. K., Peng W., et al. 2021. “Genetic Diversity and Population Structure of Tibetan Sheep Breeds Determined by Whole Genome Resequencing.” Tropical Animal Health and Production 53: 174. [DOI] [PubMed] [Google Scholar]
- Li, P. , Yin Y. L., Li D., Kim S. W., and Wu G.. 2007. “Amino Acids and Immune Function.” British Journal of Nutrition 98: 237–252. [DOI] [PubMed] [Google Scholar]
- Liu, J. , Wang Y., Min Q., Xiong E., Heyman B., and Wang J. Y.. 2020. “Regulation of Humoral Immune Responses and B Cell Tolerance by the IgM Fc Receptor (FcmuR).” Advances in Experimental Medicine and Biology 1254: 75–86. [DOI] [PubMed] [Google Scholar]
- Liu, X. , Sha Y., Lv W., et al. 2022. “Multi‐Omics Reveals That the Rumen Transcriptome, Microbiome, and Its Metabolome Co‐Regulate Cold Season Adaptability of Tibetan Sheep.” Frontiers in Microbiology 13: 859601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv, F. , Wang X., Pang X., and Liu G.. 2020. “Effects of Supplementary Feeding on the Rumen Morphology and Bacterial Diversity in Lambs.” PeerJ 8: e9353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manchope, M. F. , Calixto‐Campos C., Coelho‐Silva L., et al. 2016. “Naringenin Inhibits Superoxide Anion‐Induced Inflammatory Pain: Role of Oxidative Stress, Cytokines, Nrf‐2 and the NO−cGMP−PKG−KATP Channel Signaling Pathway.” PLoS ONE 11: e0153015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maqsood, M. A. , Khan E. U., Qaisrani S. N., et al. 2022. “Interactive Effect of Amino Acids Balanced at Ideal Lysine Ratio and Exogenous Protease Supplemented to Low CP Diet on Growth Performance, Carcass Traits, Gut Morphology, and Serum Metabolites in Broiler Chicken.” Tropical Animal Health and Production 54: 186. [DOI] [PubMed] [Google Scholar]
- Marek, I. , Lichtneger T., Cordasic N., et al. 2016. “Alpha8 Integrin (Itga8) Signalling Attenuates Chronic Renal Interstitial Fibrosis by Reducing Fibroblast Activation, Not by Interfering With Regulation of Cell Turnover.” PLoS ONE 11: e0150471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrommatis, A. , Mitsiopoulou C., Christodoulou C., et al. 2021. “Effects of Supplementing Rumen‐Protected Methionine and Lysine on Milk Performance and Oxidative Status of Dairy Ewes.” Antioxidants (Basel) 10: 654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcvie, G. , and Ringborg U.. 2012. “EurocanPlatform, an FP7 Project of the European Commission—First Year Commentary.” Ecancermedicalscience 6: ed13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montoro, C. , Miller‐Cushon E. K., DeVries T. J., and Bach A.. 2013. “Effect of Physical Form of Forage on Performance, Feeding Behavior, and Digestibility of Holstein Calves.” Journal of Dairy Science 96: 1117–1124. [DOI] [PubMed] [Google Scholar]
- Novoseletskaya, E. S. , Evdokimov P. V., and Efimenko A. Y.. 2023. “Extracellular Matrix‐Induced Signaling Pathways in Mesenchymal Stem/Stromal Cells.” Cell Communication and Signaling 21: 244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell, J. J. , Jugdaohsingh R., and Thompson R. P. H.. 1999. “The Regulation of Mineral Absorption in the Gastrointestinal Tract.” Proceedings of the Nutrition Society 58: 147–153. [DOI] [PubMed] [Google Scholar]
- Rubin, L. L. , Ribeiro A. M. L., Canal C. W., et al. 2007. “Influence of Sulfur Amino Acid Levels in Diets of Broiler Chickens Submitted to Immune Stress.” Brazilian Journal of Poultry Science 9: 53–59. [Google Scholar]
- Scholten, A. , Poh M. K., van Veen T. A., van Breukelen B., Vos M. A., and Heck A. J. R.. 2006. “Analysis of the cGMP/cAMP Interactome Using a Chemical Proteomics Approach in Mammalian Heart Tissue.” Journal of Proteome Research 5, no. 6: 1435–1447. [DOI] [PubMed] [Google Scholar]
- Sha, Y. , Liu X., He Y., et al. 2024. “Multi‐Omics Revealed Rumen Microbiota Metabolism and Host Immune Regulation in Tibetan Sheep of Different Ages.” Frontiers in Microbiology 15: 1339889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, K. , Johnson D. W., and Gobe G. C.. 2016. “The Role of cGMP and Its Signaling Pathways in Kidney Disease.” American Journal of Physiology. Renal Physiology 311: F671–F681. [DOI] [PubMed] [Google Scholar]
- Singh, M. , and Webster P. J.. 1976. “A Review of Macromolecular Transport and Secretion at the Cellular Level.” American Journal of Digestive Diseases 21: 346–355. [DOI] [PubMed] [Google Scholar]
- Smith, W. L. , Garavito R. M., and DeWitt D. L.. 1996. “Prostaglandin Endoperoxide H Synthases (Cyclooxygenases)‐1 and ‐2.” Journal of Biological Chemistry 271: 33157–33160. [DOI] [PubMed] [Google Scholar]
- Snider, G. W. , Ruggles E., Khan N., and Hondal R. J.. 2013. “Selenocysteine Confers Resistance to Inactivation by Oxidation in Thioredoxin Reductase: Comparison of Selenium and Sulfur Enzymes.” Biochemistry 52: 5472–5481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein, H. H. , and Shurson G. C.. 2009. “Board‐Invited Review: The Use and Application of Distillers Dried Grains With Solubles in Swine Diets.” Journal of Animal Science 87: 1292–1303. [DOI] [PubMed] [Google Scholar]
- Stenvang, M. , Schafer N. P., Malmos K. G., et al. 2018. “Corneal Dystrophy Mutations Drive Pathogenesis by Targeting TGFBIp Stability and Solubility in a Latent Amyloid‐Forming Domain.” Journal of Molecular Biology 430: 1116–1140. [DOI] [PubMed] [Google Scholar]
- Swain, B. K. , and Johri T. S.. 2000. “Effect of Supplemental Methionine, Choline and Their Combinations on the Performance and Immune Response of Broilers.” British Poultry Science 41: 83–88. [DOI] [PubMed] [Google Scholar]
- Terayama, N. , Terada T., and Nakanuma Y.. 1996. “An Immunohistochemical Study of Tumour Vessels in Metastatic Liver Cancers and the Surrounding Liver Tissue.” Histopathology 29: 37–43. [DOI] [PubMed] [Google Scholar]
- Ushida, K. , and Jouany J. P.. 1985. “Effect of Protozoa on Rumen Protein Degradation in Sheep.” Reproduction Nutrition Development (1980) 25: 1075–1081. [DOI] [PubMed] [Google Scholar]
- Wu, B.‐Y. , Cui H.‐M., Peng X., Fang J., Cui W., and Liu X.‐D.. 2012. “Effect of Methionine Deficiency on the Thymus and the Subsets and Proliferation of Peripheral Blood T‐Cell, and Serum IL‐2 Contents in Broilers.” Journal of Integrative Agriculture 11: 1009–1019. [Google Scholar]
- Wu, G. 2009. “Amino Acids: Metabolism, Functions, and Nutrition.” Amino Acids 37: 1–17. [DOI] [PubMed] [Google Scholar]
- Wu, G. , and Morris S. M.. 1998. “Arginine Metabolism: Nitric Oxide and Beyond.” Biochemical Journal 336: 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L. , Duan Y., Guo Q., Wang W., and Li F.. 2020. “A Selectively Suppressing Amino Acid Transporter: Sodium‐Coupled Neutral Amino Acid Transporter 2 Inhibits Cell Growth and Mammalian Target of Rapamycin Complex 1 Pathway in Skeletal Muscle Cells.” Animal Nutrition 6: 513–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, T. , Ren Y., Yang C., et al. 2023. “An Integrated Transcriptome and Microbial Community Analysis Reveals Potential Mechanisms for Increased Immune Responses When Replacing Silybum marianum Meal With Soybean Meal in Growing Lambs.” Frontiers in Microbiology 14: 1093129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, W. , Liu Y., and Zhang H.. 2021. “Extracellular Matrix: An Important Regulator of Cell Functions and Skeletal Muscle Development.” Cell & Bioscience 11: 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.


