Abstract
FUS-mediated alternative splicing and METTL3-regulated RNA methylation play crucial roles in RNA processing. The purpose of this study was to investigate the interactive roles of FUS and METTL3 in gastric cancer (GC) progression. RNA sequencing data were obtained from the TCGA-STAD dataset. Differentially expressed genes (DEGs) were analyzed across groups stratified by the medians of FUS, METTL3, and NEAT1, respectively. Endoplasmic reticulum (ER) stress markers PERK, IRE1, pIRE1, Bip, and CHOP, as well as related apoptosis stress markers PARP, cleaved-PARP, (Cleaved) Caspase 7, and (Cleaved) Caspase 3, were assessed through western blotting. Alternative splicing and N6-methyladenosine (m(6)A) methylation of specific genes were detected with MeRIP-PCR. Finally, in vivo experiments were conducted using nude mice bearing sh-FUS-transfected HGC27 xenograft tumors. FUS and METTL3 expression levels were elevated in GC tissues. A significant overlap of DEGs was observed between the FUS- and METTL3-stratified groups. These overlapping DEGs were predominantly enriched in mRNA processing and protein processing in the ER. ER stress and apoptosis were induced by sh-FUS or sh-METTL3, which was further enhanced by ER stress inducer tunicamycin in both MKN45 and HGC27 cells. Similarly, DEGs for NEAT1 high- and low-expressed groups were enriched in protein processing in the ER and spliceosome. To a lesser extent, ER stress was also induced by sh-NEAT1 and enhanced by tunicamycin in HGC27 cells. Furthermore, sh-FUS or sh-METTL3 influenced alternative splicing and methylation of specific mRNAs, including FUS, NEAT1, PCNA, MCM2, and BIRC5. Tumor progression was inhibited by sh-FUS in mice, and ER stress and apoptosis were induced, which were further enhanced by tunicamycin. FUS and METTL3 collaborate to facilitate RNA maturation. Inhibiting FUS or METTL3 promoted ER stress and apoptosis and inhibited progression in GC.
Graphical abstract
Aberrant levels of FUS and METTL3 can evoke endoplasmic reticulum (ER) stress and apoptosis by generating splicing and methylation variants of mRNAs in gastric cancer, supporting the therapeutic potential of inducing ER stress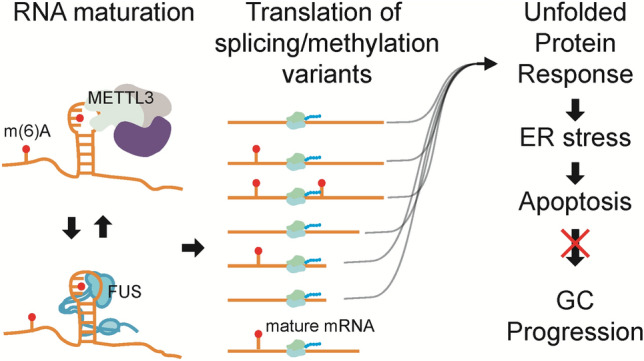
Supplementary Information
The online version contains supplementary material available at 10.1007/s10238-024-01525-7.
Keywords: Alternative splicing, Unfolded protein response, FUS, METTL3, NEAT1, RNA processing
Introduction
Gastric cancer (GC) causes the fifth and third leading cancer incidences and deaths worldwide [1]. Characterized by significant intertumor and intratumor heterogeneity, GC advances rapidly, metastasizes extensively, and often displays resistance to chemotherapy [2]. The five-year overall survival (OS) rate for GC patients spans from 70% to a mere 4%, largely contingent upon the disease stage at diagnosis [3]. The survival rate for locally advanced GC can be markedly improved to about 74% by open distal gastrectomy (ODG) with D2 lymph node dissection when performed by skilled surgeons [4]. This underscores the limited efficacy of current (neoadjuvant) chemotherapies for late-stage GC patients.
Fused in sarcoma (FUS), also known as TSL or hnRNP P, represents a nuclear RNA-binding protein exhibiting broad nucleic acid-binding activity. FUS orchestrates various cellular processes such as alternative splicing (AS) of RNA, mRNA transport, microRNA processing, and DNA repair by selectively binding to specific motifs [5,6]. Its low complexity domain (LCD), or prion-like domain in the C-terminal region, facilitates liquid–liquid phase separation (LLPS) and self-assembly [7], forming paraspeckles in collaboration with other components through direct interaction with the long non-coding RNA NEAT1 as a scaffold [8, 9]. LLPS is pivotal for FUS functionality, as its suppression or mutations lead to compromised AS, autoregulation, and paraspeckle dysfunction [10, 11]. Recent findings have implicated FUS in promoting GC progression, particularly through interactions with a cascade of regulatory RNAs. For instance, the long non-coding RNA DLX6-AS1 stabilizes MAP4K1 by upregulating FUS expression, consequently fostering proliferation, migration, and epithelial–mesenchymal transition (EMT) of GC cells [12]. However, the involvement of FUS-mediated AS in GC progression remains elusive.
Methyltransferase-like 3 (METTL3) serves as a core component of the N6-methyladenosine (m(6)A) methyltransferase complex (MTC), functioning as an m(6)A "writer" to modulate RNA degradation, translation, splicing, stability, and export [13]. Extensive evidence has established the role of METTL3 in promoting GC by augmenting proliferation, invasion, migration, EMT, stemness, and drug resistance [14–16]. Notably, alterations in m(6)A modifications induced by METTL3 can trigger genome-wide AS, including cell cycle-related genes [17].
The maturation of mRNA can be concurrently regulated by both FUS and METTL3. For instance, non-SMC condensin I complex subunit H (NCAPH) is upregulated in clear cell renal cell carcinomas, which is dependent on FUS-associated splicing and METTL3-mediated m(6)A modification [18]. Direct interaction between METTL3 and FUS has been observed [19], as evidenced by the alleviation of FUS-associated granules in amyotrophic lateral sclerosis (ALS) neurons upon treatment with STM2457 (a METTL3 inhibitor) [20], suggesting that METTL3 regulates FUS levels. Moreover, extensive crosstalk between AS and m(6)A modification has been identified in RNA maturation [21], potentially triggering endoplasmic reticulum (ER) stress, unfolded protein response (UPR), and apoptosis [22]. In this study, we hypothesize that such collaboration between FUS and METTL3 is prevalent in promoting mRNA maturation and GC progression. This hypothesis was predicted through bioinformatics analysis and validated in GC cells treated with sh-FUS and sh-METTL3, as well as in nude mice bearing HGC27 xenograft tumors transfected with sh-FUS.
Methods
Bioinformatics analysis
Transcriptome sequencing data from 370 GC patients and 34 paired adjacent normal tissues were acquired from TCGA-STAD (https://portal.gdc.cancer.gov/projects/) on 06/25/2023. The log2FC for FUS, METTL3, and NEAT1 in the patients were compared between adjacent tissues, stages I & II, and stages III & IV. Subsequently, Pearson correlations of FUS, METTL3, and NEAT1 were analyzed in the GC tissues. FUS, METTL3, and NEAT1 high- and low-expressed groups were stratified by their respective medians. Differentially expressed genes (DEGs) were defined as Log2FC > 2 and adjusted P value < 0.05 (Table S1), with volcano plots illustrating these findings. The ggvenn package was employed to identify overlapping DEGs. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genome (KEGG) enrichments were applied to the interested DEGs, particularly the intersection genes, using the Limma package in R (v3.2.5).
Cell culture and transfection
MKN45 (FH0267) and HGC27 (FH0271) cells were procured from FuHeng Biology (Shanghai, China) and cultured in RPMI1640 (C3010-0500, Viva Cell, Shanghai, China) or DMEM (C3113-0500, Viva Cell), respectively. The culture medium was supplemented with 10% FBS (04–001-1ACS, Viva Cell) and 1 × penicillin–streptomycin solution. Plasmids of FUS were designed based on its siRNAs. The sequences for sh-METTL3 and sh-NEAT1 were as reported [23, 24]. These shRNAs were packaged into the pLV-THM lentiviral vector (Addgene, USA), which underwent amplification in Escherichia coli (TOP10; Invitrogen, USA) and cultured for 24 h before recombinant plasmid extraction. Subsequently, the vector was packaged with 293T cells. For transfection, MKN45 or HGC27 cells were infected with the vector-loading lentivirus, followed by screening with Puromycin (2 μg/mL, P8833, Sigma-Aldrich, USA) and identification via qPCR and western blotting. The shRNAs and si-RNAs used are listed in Table S2.
Cell viability
Cells in the logarithmic growth phase were isolated, and approximately 5,000 cells were seeded in each well of a 96-well plate. ER stress was enhanced with tunicamycin. The IC50 of tunicamycin (HY-A0098, 200–1,200 ng/mL, MCE, China) at 48 h was evaluated by adding 10 μL of CCK-8 (C0037, Beyotime, China) 1 h before measuring the optical density at 450 nm. Tunicamycin concentrations used for co-treatment assays were 1,552 ng/mL for HGC27 cells and 1,191 ng/mL for MKN45 cells, corresponding to their calculated IC50 values, respectively.
qPCR
RNA was extracted from cultured cells by directly adding 1 mL of Trizol (15,596,026, Invitrogen) or from xenograft tumor tissues by grinding in liquid nitrogen with Trizol (50 mg tissue per 1 mL). After extraction with chloroform and isopropanol, the RNAs were reverse transcribed into cDNA using HiScript III All-in-one RT SuperMix Perfect for qPCR (R333-01, Vazyme, China). Finally, real-time PCR was performed with ChamQ Universal SYBR qPCR Master Mix (Q711-02, Vazyme), following a protocol of 95 °C for 30 s, followed by 40 cycles of 95 °C for 5 s and 60 °C for 30 s. GAPDH was utilized as an internal control. Relative RNA levels were determined using the 2−∆∆Ct method. Primers used are listed in Table S2.
Western blotting
Total protein was extracted with lysis buffer (P0013, Beyotime) directly from cell samples on ice, or from xenograft tumor tissues by grinding in liquid nitrogen. The lysis buffer was added with PMSF (G2008, Servicebio, China) and a protease and phosphatase inhibitor cocktail (P1045, Beyotime). After quantification with a BCA kit (P0010, Beyotime) and addition of loading buffer, the proteins were loaded onto SDS-PAGE gel and electrophoresed at 90 V for 20 min, followed by 120 V for 1 h. The proteins were then transferred onto an NC membrane (66,485, Pall, USA). The membrane was incubated with 5% skim milk blocking solution for 2 h, followed by overnight incubation with primary antibodies at 37 °C, and subsequent incubation with secondary antibodies for 1 h. Finally, the blots were visualized with ECL reagent (G2014, Servicebio) and semi-quantified using ImageJ software (1.53t, NIH, USA). Antibodies used were: HRP-labeled Goat Anti-Mouse IgG (A0216, 1:1000, Beyotime), HRP-labeled Goat Anti-Rabbit IgG (A0208, 1:1000, Beyotime), METTL3 (CY7240, 1:1000, Abways, China), FUS/TLS (68,262–1-Ig, 1:30,000, Proteintech, USA), GAPDH (AB0036, AB0037, 1:5000, Abways), PERK (24,390–1-AP, 1:3000, Proteintech), BIP (ab108615, 1:5000, Abcam, UK), IRE1 (27,528–1-AP, 1:2000, Proteintech), p-IRE1 (ab124945, 1:2000, Abcam), Caspase 7 (AF5118, 1:1000, Affinity, China), Cleaved-Caspase 7 (AF4023, 1:2000, affinity), CHOP (66,741–1-Ig, 1:500, Proteintech), PARP (13,371–1-AP, 1:4000, Proteintech), Cleaved-PARP (AF7023, 1:1000, Affinity), Caspase 3 (DF6879, 1:500, Affinity), Cleaved Caspase 3 (AF7022, 1:1000, Affinity).
Methylated RNA immunoprecipitation (MeRIP)-PCR
Total RNA extraction followed the above-described method, wherein it was treated with Fragmentation Buffer via PCR at 70 °C for 6 min. The resulting RNA fragments were collected and incubated with 30 µL of PC Buffer, 1 µL of PC Enhancer, and 750 µL of ethanol overnight at − 80 °C. The centrifuged RNA was then incubated with m(6)A antibody-conjugated PGM beads at 4 °C for 1 h. After washing with IP buffer, elution, and ethanol precipitation, the target RNAs were subjected to qPCR analysis, as described above. The primer sequences are also listed in Table S2. Finally, the qPCR products were separated with northern blotting.
Animal experiments
Animal experiments were conducted under the approval of the Ethics Committee of General Hospital of Ningxia Medical University (approval no.: KYLL-2021-766). All procedures were performed in accordance with the guidelines provided by the Institutional Animal Care and Use Committee (IACUC). Each five nude mice (aged 4–6 weeks, sourced from Shanghai Lab. Animal Research Center) were randomly assigned to the NC, NC + tunicamycin, shFUS, and shFUS + Tunicamycin groups. Normal HGC27 cells or sh-FUS-transfected HGC27 cells were inoculated into the armpits of the mice, which received weekly injections of tunicamycin (0.25 mg/kg, intraperitoneal injection, on days 0, 7, 14, and 21) or equivalent saline. Weekly observations were made regarding the condition of the mice, including measurements of tumor volume and body weight. On the 30th day, the mice were euthanized by cervical dislocation, and tumor tissues were collected. Tumor volume and weight were measured.
The tumor tissues were divided into three parts for RNA and protein extraction, as described earlier, or fixed in 4% PFA (G1101, Servicebio) for histological staining. The fixed tissues were routinely embedded in paraffin, cut into 4 μm sections, and mounted on slides. Sections were rehydrated sequentially: xylene I for 15 min, xylene II for 15 min, ethanol I for 5 min, ethanol II for 5 min, 85% ethanol for 5 min, 75% ethanol for 5 min, and distilled water for 10 min.
For TUNEL staining, sections were treated with 20 μg/mL DNase-free protease K (ST533, Beyotime), incubated with TUNEL working solution (C1088, Beyotime) for 1 h, and observed under a fluorescence microscope (XSP-C204, CIC, China).
For Ki67 staining (AF0198, Affinity), sections were sequentially incubated with citric acid (pH 6.0) antigen repair solution, 3% H2O2, 3% BSA (A8020, Solarbio, China), and the Ki67 primary and secondary antibodies. After counterstaining with Harris hematoxylin for approximately 3 min, the sections were observed. Finally, the positive staining area for Ki67 and TUNEL was determined using ImageJ software.
Statistics
Each experiment was repeated at least three times. Quantitative data are presented as mean ± standard deviation. Group differences were analyzed using Student's t test or one-way analysis of variance (one-way ANOVA) followed by Tukey's multiple comparisons test. Statistical analyses were performed using GraphPad Prism (v8.0, La Jolla, USA), with P < 0.05 considered statistically significant.
Results
FUS and METTL3 are related to RNA maturation, ER stress and apoptosis in GC
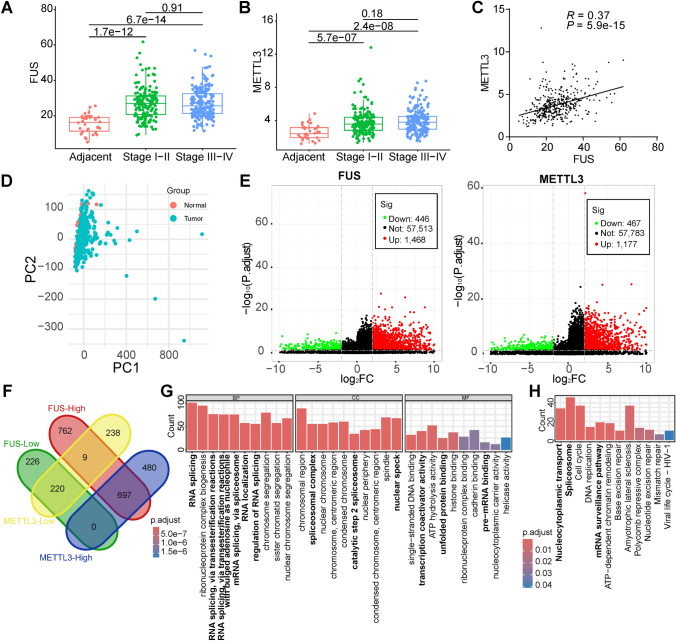
The potential interaction between FUS and METTL3 was predicted through bioinformatics analysis. FUS expression was elevated in the 370 GC patients compared to the 34 adjacent normal tissues (Fig. 1A), across both stages I-II and stages III-IV. Similar trends were observed for METTL3 (Fig. 1B). However, neither FUS nor METTL3 showed significant changes between group I-II and group III-IV patients. Despite a relatively modest correlation coefficient, a clear correlation between FUS and METTL3 was evident in these GC samples (Fig. 1C). Acceptable sample variation was confirmed by a PCA plot, generated after batch effects were removed using the ComBat method (Fig. 1D). The differentially expressed genes (DEGs) for FUS and METTL3 groups were characterized (Fig. 1E, Table S1), revealing a significant proportion of intersection genes (Fig. 1F). Specifically, 917 genes were identified as FUS-Low/METTL3-Low or FUS-High/METTL3-High, while only 9 genes were FUS-Low/METTL3-High or FUS-High/METTL3-Low, indicating a high correlation or consistency in the functions of FUS and METTL3. The intersection DEGs were enriched in GO and KEGG pathways (Fig. 1G, H, Table S1), notably related to RNA splicing, spliceosomal complex, nuclear speckle, cell cycle, mRNA surveillance pathway, and unfolded protein binding. These results suggest that FUS and METTL3 work in concert to promote RNA maturation, potentially contributing to ER stress and UPR in GC.
Fig. 1.
Potential interaction of FUS and METTL3 predicted by bioinformatics. RNA sequencing data were obtained from the TCGA-STAD dataset. A FUS mRNA expression in adjacent non-tumor tissues and GC tissues of the patients; B METTL3 mRNA expression in adjacent non-tumor tissues and GC tissues of the patients; C the correlation of FUS and METTL3 in GC tissues; D PCA plot of the involved samples; E volcano plot of the DEGs for FUS and METTL3, characterized as Log2FC > 2 and adjusted P value < 0.05; F intersection of the DEGs; G enrichment of the intersectional DEGs with GO; H Enrichment of the intersectional DEGs with KEGG
ER stress and apoptosis were induced with sh-FUS and tunicamycin to inhibit GC
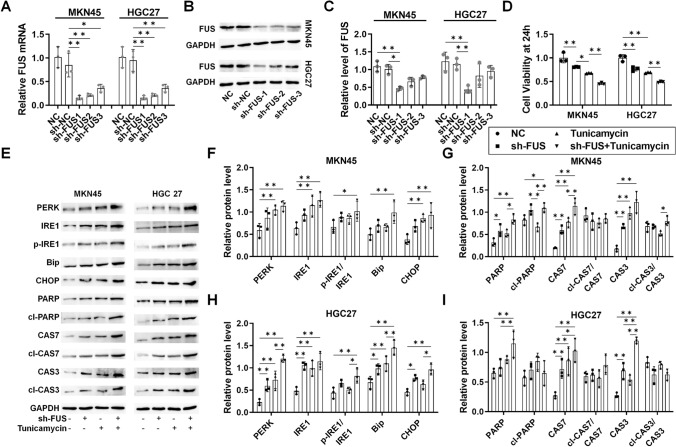
To assess the impact of FUS on RNA maturation, UPR, and apoptosis, FUS was silenced using shRNA. Both mRNA and protein levels of FUS were reduced by sh-FUS in MKN45 and HGC27 cells (Fig. 2A–C). sh-FUS led to a modest reduction in the viability of GC cells, which was significantly enhanced with co-treatment of tunicamycin (Fig. 2D). Western blotting analysis revealed induction of ER stress (PERK, IRE1, pIRE1/IRE1, Bip, CHOP) and apoptosis markers (PARP, cleaved-PARP/PARP, Caspase 7, Cleaved Caspase 7/Caspase7; Caspase 3, and Cleaved Caspase 3/Caspase3) by sh-FUS, further augmented with tunicamycin in both MKN45 and HGC27 cells (Fig. 2E–I). These findings support the role of sh-FUS in inducing ER stress and apoptosis in GC cells.
Fig. 2.
The role of FUS-regulated alternative splicing (AS) in ER stress and apoptosis in GC. For both MKN45 and HGC27 cells, samples were collected after stable transfection was established for 72 h, followed by 48 h of tunicamycin treatment. A mRNA level of FUS in sh-FUS-transfected cells; B protein level of FUS in sh-FUS-transfected cells; C protein level of FUS in sh-FUS-transfected cells analyzed by ImageJ; D viability of the sh-FUS-transfected cells within 48 h; E western blotting analysis of UPR and apoptosis protein markers; F Analysis of UPR markers in MKN45 cells with ImageJ; G analysis of apoptosis markers in MKN45 cells with ImageJ; H analysis of UPR markers in HGC27 cells with ImageJ; I analysis of apoptosis markers in HGC27 cells with ImageJ. N = 3. *P < 0.05 and **P < 0.01 by one-way ANOVA followed by Tukey’s multiple comparisons
ER stress and apoptosis were induced with sh-METTL3 and tunicamycin to inhibit GC
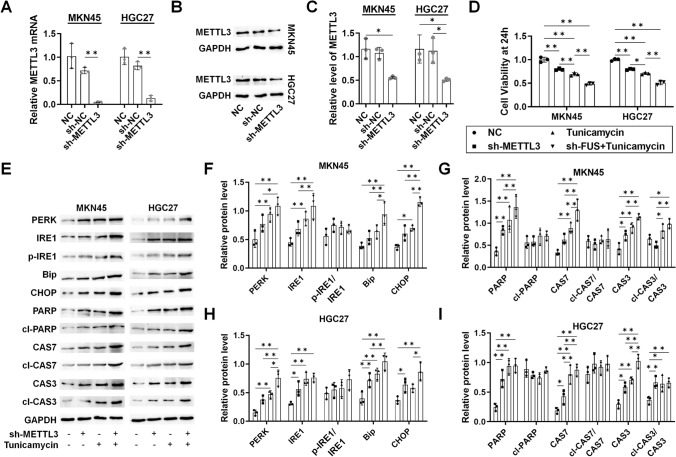
Similarly, the impact of METTL3 on RNA maturation, UPR, and ER stress was assessed by silencing METTL3 with shRNA. Both mRNA and protein levels of METTL3 were reduced by sh-METTL3 in MKN45 and HGC27 cells (Fig. 3A–C). sh-METTL3 led to reduced viability of GC cells, which was further decreased with tunicamycin co-treatment (Fig. 3D). Western blotting analysis revealed induction of ER stress and apoptosis markers by sh-METTL3, which were further enhanced with tunicamycin in both cell lines (Fig. 3E–I), supporting the induction of ER stress and apoptosis in GC cells.
Fig. 3.
The role of METTL3-regulated m(6)A methylation in ER stress and apoptosis in GC. For both MKN45 and HGC27 cells, samples were collected after stable transfection was established for 72 h, followed by 48 h of tunicamycin treatment. A mRNA level of METTL3 in sh-METTL3-transfected cells; B protein level of METTL3 in sh-METTL3-transfected cells; C protein level of METTL3 in sh-METTL3-transfected cells analyzed by ImageJ; D viability of the sh-METTL3-transfected cells within 48 h; E Western blotting analysis of UPR and apoptosis protein markers; F analysis of UPR markers in MKN45 cells with ImageJ; G analysis of apoptosis markers in MKN45 cells with ImageJ; H analysis of UPR markers in HGC27 cells with ImageJ; I Analysis of apoptosis markers in HGC27 cells with ImageJ. N = 3. *P < 0.05 and **P < 0.01 by one-way ANOVA followed by Tukey’s multiple comparisons
ER stress and apoptosis were induced with sh-NEAT1 and tunicamycin
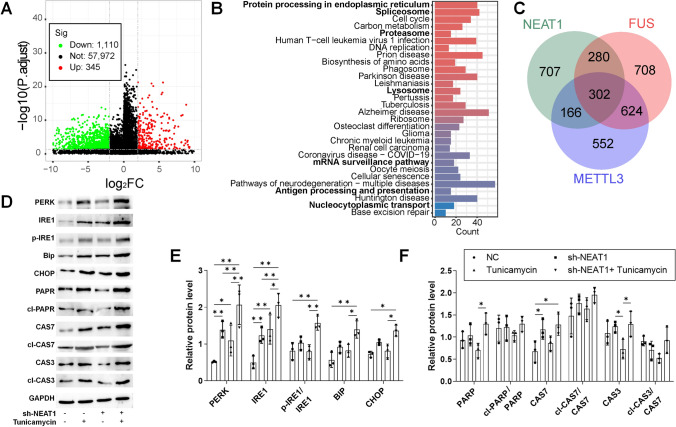
Given the reported association of NEAT1 with FUS and METTL3 in RNA maturation (25, 26), DEGs for NEAT1 were analyzed in the TCGA-STAD dataset (Fig. 4A). Similar to FUS and METTL3, DEGs for NEAT1 were enriched in pathways related to protein processing in the ER, cell cycle, spliceosome, mRNA surveillance pathway, and nucleocytoplasmic transport (Fig. 4B), though with a reduced intersection of DEGs relative to FUS and METTL3 (Fig. 4C). Treatment of HGC27 cells with sh-NEAT1 induced UPR and increased expression of ER stress markers (Fig. 4D–F), indicating NEAT1's potential contribution to FUS/METTL3-regulated RNA maturation in GC cells, albeit with less significant effects on ER stress.
Fig. 4.
The role of NEAT1 in ER stress and apoptosis in GC. HGC27 cells were stably transfected with sh-NEAT1 for 72 h, and samples were collected after 48 h of tunicamycin treatment. A Volcano plot of the DEGs for NEAT1 differently expressed groups in TCGA-STAD, characterized as Log2FC > 2 and adjusted P value < 0.05; B KEGG enrichment of the DEGs; C intersection of the DEGs for FUS, METTL3, and NEAT1 differently expressed groups; D western blotting analysis of UPR and apoptosis protein markers; E analysis of UPR markers with ImageJ; F analysis of apoptosis markers with ImageJ. N = 3 for western blotting. *P < 0.05 and **P < 0.01 by one-way ANOVA followed by Tukey’s multiple comparisons
FUS-mediated AS coordinates METTL3-dependent m(6)A modification in RNA maturation
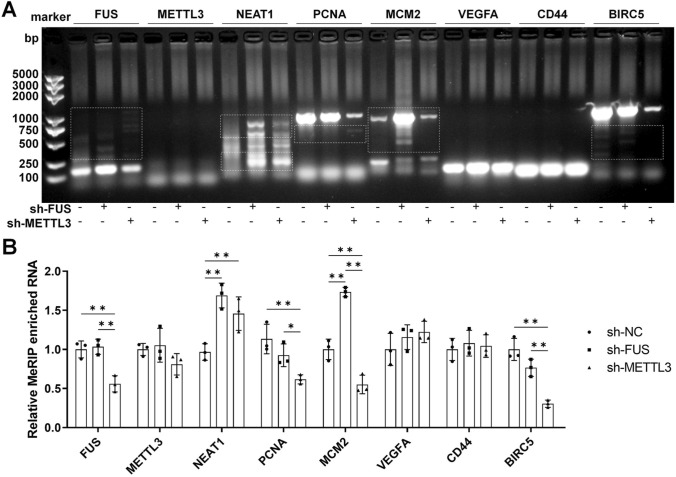
The predicted coordination of FUS-mediated AS and METTL3-dependent m(6)A modification can be widespread and interactive [19, 21]. So, we intended to observe the typical alternatively spliced variants in MeRIP enriched RNAs to confirm this coordination. FUS, METTL3, NEAT1, and several reported key regulators that potentially experience AS (PCNA, MCM2, VEGFA, CD44, and BIRC5) in GC was tested with MeRIP-qPCR followed by northern blotting [27], in HGC27 cells transfected with sh-FUS or sh-METTL3 (Fig. 5A). Alterations in splice variants were observed for FUS, NEAT1, PCNA, MCM2, and BIRC5, accompanied by significant changes in m(6)A modification, including reduced methylated FUS, MCM2, and BIRC5 with ch-METTL3; increased methylated NEAT1 and MCM2 with sh-FUS; and increased methylated NEAT1 with sh-METTL3 (Fig. 5B). The results confirmed the presence of coordinated regulation of RNA maturation by FUS and METTL3 in GC cells.
Fig. 5.
The levels of typical transcripts were tested with MeRIP-PCR. HGC27 cells were transfected with an empty vector, sh-FUS, or sh-METTL3 for 72 h. A Alternative splicing (AS) and m(6)A methylation of specific mRNAs were tested by MeRIP-PCR, with AS directly observed (marked with a white dotted frame); B analysis of the relative RNA levels of the main blot lanes with ImageJ, which indicates different methylation level of the target genes. N = 3. *P < 0.05 and **P < 0.01 by one-way ANOVA followed by Tukey’s multiple comparisons
Enhanced ER stress and apoptosis by sh-FUS and tunicamycin inhibited GC in vivo
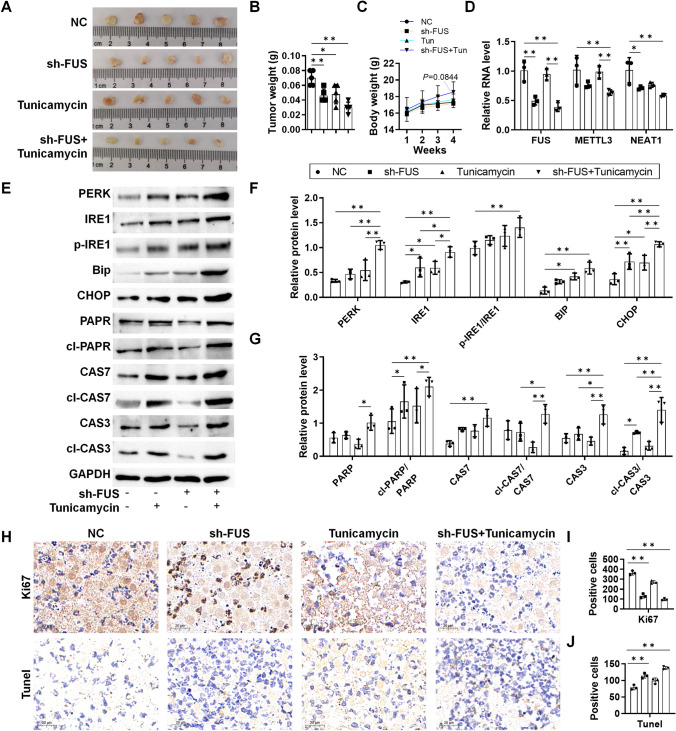
Finally, the induction of ER stress and apoptosis by sh-FUS was validated in xenograft mice bearing sh-FUS-transfected HGC27 cells. Tumor tissues were smaller in sh-FUS xenografts, and their size was further reduced with tunicamycin co-treatment (Fig. 6A, B), with no significant changes observed in body weight (Fig. 6C). Reduced expression of FUS, METTL3, and NEAT1 was observed during the 30-day period (Fig. 6D). Western blotting analysis confirmed the activation of ER stress and apoptosis with sh-FUS and tunicamycin treatment in vivo (Fig. 6E–G). Histological staining revealed reduced Ki67 and increased TUNEL with sh-FUS, which is enhanced with tunicamycin treatment (Fig. 6H–J), supporting the inhibition of proliferation and enhancement of ER stress and apoptosis by sh-FUS and tunicamycin in GC.
Fig. 6.
In vivo validation of ER stress and apoptosis in GC xenograft mice. sh-NC or sh-FUS-transfected HGC27 cells were inoculated into the armpits of the mice, which were dosed with tunicamycin on days 0, 7, 14, and 21. Tumor tissues were collected on the 30th day. A Tumor tissues harvested on day 30; B tumor volume of the mice; C body weight of the mice measured every week; D relative RNA level of FUS, METTL3, and NEAT1 in the tumor tissues; E western blotting analysis of UPR and apoptosis protein markers; F analysis of UPR markers with ImageJ; G analysis of apoptosis markers with ImageJ; H Ki67, and TUNEL staining of the tumor tissues; I Ki67 positive cells per field; J TUNEL positive cells per field. The cells were counted with ImageJ. N = 3–5 as indicated by each scattered dot. *P < 0.05 and **P < 0.01 by one-way ANOVA followed by Tukey’s multiple comparisons
Discussion
Large-scale genomic studies widely support the occurrence of aberrant RNA processing in promoting cancer progression and unveiling novel therapeutic vulnerabilities [28]. Simple alterations in RNA processing genes or proteins are anticipated to generate isoforms within specific segments of the transcriptome and proteome, potentially triggering ER stress and apoptosis, sensitizing chemotherapy, and eliciting immunogenic responses [29–31]. In GC, various modes of RNA processing are significantly linked to tumor prognosis, stemness, and chemoresistance [32]. Here, we investigated the therapeutic potential of inducing ER stress in GC by evoking UPR through the silencing of RNA processors FUS and METTL3. A significant proportion of intersecting DEGs between the FUS and METTL3 differentially expressed samples was identified. These intersecting genes were enriched in mRNA processing and maturation-related GO pathways, including RNA splicing, RNA localization, spliceosomal complex, nuclear speck, transcription coactivator activity, unfolded protein binding, and pre-mRNA binding. Similarly, mRNA processing and maturation-associated KEGG pathways, such as nucleocytoplasmic transport, spliceosome, and mRNA surveillance pathway, were also enriched. Other significantly enriched pathways, including cell cycle, chromosome segregation, base excision repair, and mismatch repair, are potentially related to the cellular DNA damage response induced by UPR, as regulated by p53 [33]. This enrichment strongly suggests that FUS and METTL3 interact extensively to promote mRNA maturation in GC, and that abnormal levels of FUS and METTL3 can trigger ER stress and apoptosis by generating splicing and methylation transcript variants. We further validated this hypothesis by showing that silencing FUS or METTL3 significantly induced ER stress and apoptosis, as indicated by the increased protein levels of PERK, IRE1, p-IRE1/IRE1, Bip, CHOP, PARP, cleaved PARP, Caspase 7, cleaved Caspase 7/Caspase 7, Caspase 3, and cleaved Caspase 3/Caspase 3 in the sh-FUS and sh-METTL3 groups. These effects were validated in nude mice bearing sh-FUS-transfected HGC27 cells, where decreased RNA levels of FUS, METTL3, and NEAT1 were observed in vivo. Additionally, mice in the sh-FUS group exhibited smaller tumor sizes, increased expression of UPR and ER stress-related apoptosis marker proteins, lower tissue Ki67 expression, and higher TUNEL levels. These effects were all augmented with tunicamycin co-treatment, suggesting that dysregulated RNA maturation regulators such as FUS and METTL3 can serve as therapeutic vulnerabilities.
More than 95% of all human genes undergo AS [28]. FUS has primarily been characterized as an alternative RNA splicer extensively involved in the maturation of nascent RNA within the central nervous system [34]. By forming condensates with other splicing factors and noncoding RNAs such as EWS, TAF15, and NEAT1, FUS undergoes phase separation to form paraspeckles [35, 36]. Rearrangements of the FUS gene with CREB1 or ATF1 are frequently observed in malignant gastrointestinal neuroectodermal tumors [37]. AS induced by FUS contributes to heterogeneity in GC, resulting in different epithelial–mesenchymal transition subtypes and survival outcomes [38]. FUS has been shown to significantly promote GC progression by forming a LBX2-AS1/miR-219a-2-3p/FUS/LBX2 positive feedback loop [26]. We further showed that silencing FUS inhibited GC progression by inducing ER stress. The MeRIP-PCR experiments confirmed the induction of isoforms of NEAT1, MCM2, and BIRC5 with sh-FUS. These effects were mediated by dysregulated RNA splicing and maturation, potentially generating numerous RNA and protein isoforms with altered RNA splicing, translation, and protein folding and processing.
The total m(6)A content significantly increases in patients infected with H. pylori, a key inducer of GC in humans [39]. METTL3 is responsible for catalyzing over 95% of all m(6)A modifications in mRNA [40]; high METTL3 expression is associated with poor clinical outcomes [41]. However, patients from the STAD dataset with lower METTL3 expression exhibited shorter OS (Table S1). This paradox may be explained by the fact that both increased and decreased m6A levels can contribute to GC progression and drug resistance [42]. The activity of METTL3 depends on its localization and co-localized proteins. Cytoplasmic METTL3 tends to directly bind to mRNA, initiating translation and promoting tumor progression, independent of RNA methylation [43]. In the nucleus, METTL3 can suppress GC when METTL14 is highly expressed [44], but tends to promote GC when METTL3 predominates over METTL14, crucially influencing EMT and metastasis in GC [45]. Additionally, METTL3 plays critical roles in proliferation, angiogenesis, glycolysis, and drug resistance in GC [14, 16, 46]. We also showed that silencing METTL3 inhibited GC progression both in vitro and in vivo. The tumor-suppressive effect of sh-METTL3 was associated with induced ER stress and apoptosis. Reduced m(6)A levels of FUS, PCNA, MCM2, and BIRC5 were achieved with sh-METTL3, while the m(6)A level of NEAT1 increased. Moreover, AS changes of FUS, NEAT1, PCNA, MCM2, and BIRC5 were induced with sh-METTL3. As these tested RNAs were chosen for their potential effects on GC and certain DEGs (PCNA, MCM2, BIRC5) of them exhibited significant changes in m(6)A modification and AS levels, it is reasonable to conclude that both FUS and METTL3 are necessary for the RNA processing of a subset of RNAs in GC. Silencing FUS or METTL3 resulted in dysregulated RNA maturation, UPR, and apoptosis. METTL3 potentially acts upstream of FUS, as evidenced by the induction of splicing variants of FUS with sh-METTL3, while methylated METTL3 mRNA was less significantly influenced by sh-FUS.
NEAT1 serves as the scaffold for RNA processing-related paraspeckles, frequently observed at active transcription sites [47]. In addition to forming phase separations and directly interacting with FUS in the cell nucleus, mutations in the FUS gene can disrupt paraspeckles, leading to excessive free NEAT1 [48]. Conversely, METTL3 can regulate m(6)A modification of NEAT1, altering its secondary structure [49]. The m(6)A "reader" protein hnRNPA2B1 stabilizes NEAT1, promoting metastasis and chemoresistance in GC [25, 50], Considering these potential interactions among FUS, METTL3, and NEAT1, along with their functional overlaps in RNA maturation, it is reasonable to suspect that these three regulators form a cooperative complex within paraspeckles. However, we found that although sh-NEAT1 induced UPR, it was not significantly affected. Previous research in GC has indicated the promotion of ER stress with NEAT1, consistent with our observations [51]. This was also supported by our bioinformatics analysis (Table S1), where fewer intersection genes were identified in the FUS-high/METTL3-high/NEAT1-high subset (32 DEGs) compared to the FUS-high/METTL3-high/NEAT1-low subset (240 DEGs). These results suggest that although the interaction between FUS and NEAT1 is well-characterized, the interaction between FUS and METTL3 may be more significant, at least in GC.
There are limitations to this work. Downregulation of target genes was achieved with shRNAs, which may have been incomplete. FUS and METTL3 may exhibit redundancy in their effects [52, 53], allowing regular RNA processing to continue even upon silencing. Recently, a novel inhibitor of METTL3 has been developed [54], which warrants further investigation. Ideally, MeRIP followed bottom-up transcriptome sequencing could be employed to explore the targeting RNAs and interactive mechanisms for FUS and METTL3.
Conclusion
Collaborations between FUS and METTL3 appear to be widespread, facilitating RNA maturation, which in turn mitigates ER stress and apoptosis, thereby promoting progression in GC. Inhibiting FUS/METTL3 to induce ER stress and apoptosis could be a potential strategy to hinder GC progression.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
None.
Abbreviations
- GC
Gastric cancer
- OS
Overall survival
- ODG
Open distal gastrectomy
- FUS
Fused in sarcoma
- AS
Alternative splicing
- LCD
Low complexity domain
- LLPS
Liquid–liquid phase separation
- EMT
Epithelial–mesenchymal transition
- m(6)A
N6-methyladenosine
- MTC
Methyltransferase complex
- NCAPH
Non-SMC condensin I complex subunit H
- ALS
Amyotrophic lateral sclerosis
- UPR
Unfolded protein response
- ER
Endoplasmic reticulum
- DEGs
Differentially expressed genes
- GO
Gene ontology
- KEGG
Kyoto Encyclopedia of Genes and Genome
Author contributions
DL, BD, and GL performed the experiments, data collection and data analysis. DL was a major contributor in writing the manuscript. BD and GL were responsible for data visualization and literature search. ZY contributed to the study conception and design and was a major contributor in critically revising the manuscript. All authors read and approved the final manuscript.
Funding
This study was funded by the Key Research and Development Project of Ningxia Science and Technology Department (Grant No.: 2022BEG03154).
Data availability
Data will be made available from the corresponding author on reasonable request.
Declarations
Conflict of interest
The authors declare no competing interests.
Ethical approval
Animal experiments were conducted under the approval of the Ethics Committee of General Hospital of Ningxia Medical University (approval no.: KYLL-2021–766).
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Mukkamalla SKR, Recio-Boiles A, Babiker HM. Gastric Cancer. StatPearls. Treasure Island (FL): StatPearls Publishing Copyright© 2024, StatPearls Publishing LLC. 2024
- 2.Gao JP, Xu W, Liu WT, Yan M, Zhu ZG. Tumor heterogeneity of gastric cancer: from the perspective of tumor-initiating cell. World J Gastroenterol. 2018;24(24):2567–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Afrash MR, Mirbagheri E, Mashoufi M, Kazemi-Arpanahi H. Optimizing prognostic factors of five-year survival in gastric cancer patients using feature selection techniques with machine learning algorithms: a comparative study. BMC Med Inform Decis Mak. 2023;23(1):54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Etoh T, Ohyama T, Sakuramoto S, Tsuji T, Lee S-W, Yoshida K, et al. Five-year survival outcomes of laparoscopy-assisted vs open distal gastrectomy for advanced gastric cancer: the JLSSG0901 randomized clinical trial. JAMA Surg. 2023;158(5):445–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fujino Y, Ueyama M, Ishiguro T, Ozawa D, Ito H, Sugiki T, et al. FUS regulates RAN translation through modulating the G-quadruplex structure of GGGGCC repeat RNA in C9orf72-linked ALS/FTD. Elife. 2023;12:RP84338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takeda JI, Masuda A, Ohno K. Six GU-rich (6GU(R)) FUS-binding motifs detected by normalization of CLIP-seq by Nascent-seq. Gene. 2017;618:57–64. [DOI] [PubMed] [Google Scholar]
- 7.Portz B, Lee BL, Shorter J. FUS and TDP-43 phases in health and disease. Trends Biochem Sci. 2021;46(7):550–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamada A, Toya H, Tanahashi M, Kurihara M, Mito M, Iwasaki S, et al. Species-specific formation of paraspeckles in intestinal epithelium revealed by characterization of NEAT1 in naked mole-rat. RNA (New York, NY). 2022;28(8):1128–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Riva P, Ratti A, Venturin M. The long non-coding RNAs in neurodegenerative diseases: novel mechanisms of pathogenesis. Curr Alzheimer Res. 2016;13(11):1219–31. [DOI] [PubMed] [Google Scholar]
- 10.Wang Y, Chen LL. Organization and function of paraspeckles. Essays Biochem. 2020;64(6):875–82. [DOI] [PubMed] [Google Scholar]
- 11.Zhou Y, Liu S, Liu G, Oztürk A, Hicks GG. ALS-associated FUS mutations result in compromised FUS alternative splicing and autoregulation. PLoS Genet. 2013;9(10):e1003895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu Q, Ma J, Meng W, Hui P. DLX6-AS1 promotes cell proliferation, migration and EMT of gastric cancer through FUS-regulated MAP4K1. Cancer Biol Ther. 2020;21(1):17–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu X, Ye W, Gong Y. The role of RNA methyltransferase METTL3 in normal and malignant hematopoiesis. Front Oncol. 2022;12:873903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin S, Liu J, Jiang W, Wang P, Sun C, Wang X, et al. METTL3 promotes the proliferation and mobility of gastric cancer cells. Open Med (Warsaw, Poland). 2019;14:25–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu P, Liu K, Huang S, Lv J, Yan Z, Ge H, et al. N(6)-methyladenosine-modified MIB1 promotes stemness properties and peritoneal metastasis of gastric cancer cells by ubiquitinating DDX3X. Gastric Cancer: Off J Int Gastric Cancer Assoc Jpn Gastric Cancer Assoc. 2024;27(2):275–91. [DOI] [PubMed] [Google Scholar]
- 16.Okugawa Y, Toiyama Y, Yin C, Ruiya M, Goel A, Ichikawa T, et al. Prognostic potential of METTL3 expression in patients with gastric cancer. Oncol Lett. 2023;25(2):64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim Y, Shin S, Kwon S, Moon K, Baek SV, Jo A, et al. METTL3 regulates alternative splicing of cell cycle-related genes via crosstalk between mRNA m(6)A modifications and splicing factors. Am J Cancer Res. 2023;13(4):1443–56. [PMC free article] [PubMed] [Google Scholar]
- 18.Chen Z, Ruan W, Guo C, Chen K, Li L, Tian J, et al. Non-SMC condensin I complex subunit H participates in anti-programmed cell death-1 resistance of clear cell renal cell carcinomas. Cell Prolif. 2023;56:e13400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Di Timoteo G, Giuliani A, Setti A, Biagi MC, Lisi M, Santini T, et al. M6A reduction relieves FUS-associated ALS granules. Nat Commun. 2024;15(1):5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Timoteo GD, Giuliani A, Setti A, Biagi MC, Lisi M, Grandioso A, et al. M(6)A reduction relieves FUS-associated ALS granules. bioRxiv. 2023:2023.10.25.563954.
- 21.Zhu ZM, Huo FC, Zhang J, Shan HJ, Pei DS. Crosstalk between m6A modification and alternative splicing during cancer progression. Clin Transl Med. 2023;13(10):e1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jaud M, Philippe C, Di Bella D, Tang W, Pyronnet S, Laurell H, et al. Translational regulations in response to endoplasmic reticulum stress in cancers. Cells. 2020;9(3):540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu T, Feng Y, Yang S, Ge Y, Zhang T, Li J, et al. Depicting the profile of METTL3-mediated lncRNA m6A modification variants and identified SNHG7 as a prognostic indicator of MNNG-induced gastric cancer. Toxics. 2023;11(11):944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma P, Pan Y, Yang F, Fang Y, Liu W, Zhao C, et al. KLF5-modulated lncRNA NEAT1 contributes to tumorigenesis by acting as a scaffold for BRG1 to silence GADD45A in gastric cancer. Mol Therapy—Nucleic Acids. 2020;22:382–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jia Y, Yan Q, Zheng Y, Li L, Zhang B, Chang Z, et al. Long non-coding RNA NEAT1 mediated RPRD1B stability facilitates fatty acid metabolism and lymph node metastasis via c-Jun/c-Fos/SREBP1 axis in gastric cancer. J Exp Clin Cancer Res: CR. 2022;41(1):287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang Z, Dong X, Pu M, Yang H, Chang W, Ji F, et al. LBX2-AS1/miR-219a-2-3p/FUS/LBX2 positive feedback loop contributes to the proliferation of gastric cancer. Gastric Cancer: Official J Int Gastric Cancer Assoc Jpn Gastric Cancer Assoc. 2020;23(3):449–63. [DOI] [PubMed] [Google Scholar]
- 27.Li Y, Yuan Y. Alternative RNA splicing and gastric cancer. Mutat Res Rev Mutat Res. 2017;773:263–73. [DOI] [PubMed] [Google Scholar]
- 28.Obeng EA, Stewart C, Abdel-Wahab O. Altered RNA processing in cancer pathogenesis and therapy. Cancer Discov. 2019;9(11):1493–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hung C, Patani R. Elevated 4R tau contributes to endolysosomal dysfunction and neurodegeneration in VCP-related frontotemporal dementia. Brain. 2024;147(3):970–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wong TL, Loh JJ, Lu S, Yan HHN, Siu HC, Xi R, et al. ADAR1-mediated RNA editing of SCD1 drives drug resistance and self-renewal in gastric cancer. Nat Commun. 2023;14(1):2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fang M, Yao J, Zhang H, Sun J, Yin Y, Shi H, et al. Specific deletion of Mettl3 in IECs triggers the development of spontaneous colitis and dysbiosis of T lymphocytes in mice. Clin Exp Immunol. 2024;217:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang X, Wu L, Jia L, Hu X, Yao Y, Liu H, et al. The implication of integrative multiple RNA modification-based subtypes in gastric cancer immunotherapy and prognosis. iScience. 2024;27(2):108897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.López I, Tournillon AS, Nylander K, Fåhraeus R. p53-mediated control of gene expression via mRNA translation during endoplasmic reticulum stress. Cell Cycle. 2015;14(21):3373–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rogelj B, Easton LE, Bogu GK, Stanton LW, Rot G, Curk T, et al. Widespread binding of FUS along nascent RNA regulates alternative splicing in the brain. Sci Rep. 2012;2(1):603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ernst EH, Nielsen J, Ipsen MB, Villesen P, Lykke-Hartmann K. Transcriptome analysis of long non-coding RNAs and genes encoding paraspeckle proteins during human ovarian follicle development. Front Cell Dev Biol. 2018;6:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnson CN, Sojitra KA, Sohn EJ, Moreno-Romero AK, Baudin A, Xu X, et al. Insights into molecular diversity within the FUS/EWS/TAF15 protein family: unraveling phase separation of the N-terminal low-complexity domain from RNA-binding protein EWS. J Am Chem Soc. 2024;146(12):8071–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boland JM, Folpe AL. Oncocytic variant of malignant gastrointestinal neuroectodermal tumor: a potential diagnostic pitfall. Hum Pathol. 2016;57:13–6. [DOI] [PubMed] [Google Scholar]
- 38.Jun Y, Suh Y-S, Park S, Lee J, Kim J-I, Lee S, et al. Comprehensive analysis of alternative splicing in gastric cancer identifies epithelial-mesenchymal transition subtypes associated with survival. Can Res. 2022;82(4):543–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li H, Lin J, Cheng S, Chi J, Luo J, Tang Y, et al. Comprehensive analysis of differences in N6-methyladenosine RNA methylomes in helicobacter pylori infection. Front Cell Dev Biol. 2023;11:1136096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poh HX, Mirza AH, Pickering BF, Jaffrey SR. Alternative splicing of METTL3 explains apparently METTL3-independent m6A modifications in mRNA. PLoS Biol. 2022;20(7):e3001683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Su Z, Xu L, Dai X, Zhu M, Chen X, Li Y, et al. Prognostic and clinicopathological value of m6A regulators in human cancers: a meta-analysis. Aging. 2022;14(21):8818–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen D, Gu X, Nurzat Y, Xu L, Li X, Wu L, et al. Writers, readers, and erasers RNA modifications and drug resistance in cancer. Mol Cancer. 2024;23(1):178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wei X, Huo Y, Pi J, Gao Y, Rao S, He M, et al. METTL3 preferentially enhances non-m(6)A translation of epigenetic factors and promotes tumourigenesis. Nat Cell Biol. 2022;24(8):1278–90. [DOI] [PubMed] [Google Scholar]
- 44.Fan HN, Chen ZY, Chen XY, Chen M, Yi YC, Zhu JS, et al. METTL14-mediated m(6)A modification of circORC5 suppresses gastric cancer progression by regulating miR-30c-2-3p/AKT1S1 axis. Mol Cancer. 2022;21(1):51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yue B, Song C, Yang L, Cui R, Cheng X, Zhang Z, et al. METTL3-mediated N6-methyladenosine modification is critical for epithelial-mesenchymal transition and metastasis of gastric cancer. Mol Cancer. 2019;18(1):142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Q, Chen C, Ding Q, Zhao Y, Wang Z, Chen J, et al. METTL3-mediated m(6)A modification of HDGF mRNA promotes gastric cancer progression and has prognostic significance. Gut. 2020;69(7):1193–205. [DOI] [PubMed] [Google Scholar]
- 47.Lamond AI, Spector DL. Nuclear speckles: a model for nuclear organelles. Nat Rev Mol Cell Biol. 2003;4(8):605–12. [DOI] [PubMed] [Google Scholar]
- 48.An H, Skelt L, Notaro A, Highley JR, Fox AH, La Bella V, et al. ALS-linked FUS mutations confer loss and gain of function in the nucleus by promoting excessive formation of dysfunctional paraspeckles. Acta Neuropathol Commun. 2019;7(1):7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wen S, Wei Y, Zen C, Xiong W, Niu Y, Zhao Y. Long non-coding RNA NEAT1 promotes bone metastasis of prostate cancer through N6-methyladenosine. Mol Cancer. 2020;19(1):171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang J, Zhang J, Liu H, Meng L, Gao X, Zhao Y, et al. N6-methyladenosine reader hnRNPA2B1 recognizes and stabilizes NEAT1 to confer chemoresistance in gastric cancer. Cancer Commun (London, England). 2024;44(4):469–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou Y, Sha Z, Yang Y, Wu S, Chen H. lncRNA NEAT1 regulates gastric carcinoma cell proliferation, invasion and apoptosis via the miR‑500a‑3p/XBP‑1 axis. Molecular medicine reports. 24(1), 2021 [DOI] [PMC free article] [PubMed]
- 52.Kapeli K, Pratt GA, Vu AQ, Hutt KR, Martinez FJ, Sundararaman B, et al. Distinct and shared functions of ALS-associated proteins TDP-43, FUS and TAF15 revealed by multisystem analyses. Nat Commun. 2016;7(1):12143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang P, Wei L, Du B, Qin K, Zheng J, Wang J. Unraveling the independent role of METTL3 in m6A modification and tumor progression in esophageal squamous cell carcinoma 2024 [DOI] [PMC free article] [PubMed]
- 54.Li Z, Feng Y, Han H, Jiang X, Chen W, Ma X, et al. A stapled peptide inhibitor targeting the binding interface of N6-adenosine-methyltransferase subunits METTL3 and METTL14 for cancer therapy. Weinheim: Angewandte Chemie International Edition; 2024. p. e202402611. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available from the corresponding author on reasonable request.