Abstract
Background
Previous clinical studies have suggested an increased risk of tumor development in patients with pulmonary arterial hypertension (PAH). However, it remains unclear whether there is a causal relationship between PAH and tumor occurrence. This study investigates the causal link between PAH and cancer from a genetic perspective using Mendelian randomization (MR).
Method
Genome-wide association study (GWAS) summary data for PAH and various common cancer types were obtained from the GWAS Catalog. Single nucleotide polymorphisms (SNPs) significantly associated with PAH at the genome-wide significance threshold (P < 1 × 10−6) were selected as instrumental variables (IVs). Inverse-variance weighted (IVW) was used as the primary method for MR analysis, with sensitivity analyses including tests for heterogeneity and horizontal pleiotropy.
Results
The results from the IVW analysis indicate that genetically proxied PAH is associated with an increased risk of liver cancer [odd ratio (OR) 1.11, 95% confidence interval (CI) 1.01–1.22, P = 0.025), while showing no significant causal relationship with other common types of tumors (thyroid cancer: OR 0.95, 95% CI 0.86–1.06, P = 0.360; lung cancer: OR 0.95, 95% CI 0.90–1.01, P = 0.129; gastric cancer: OR 0.97, 95% CI 0.93–1.02, P = 0.243; colorectal cancer: OR 1.01, 95% CI 0.98–1.05, P = 0.412). Except for the MR analysis examining the causal effect of PAH on lung cancer (P = 0.049), the remaining MR analyses displayed no significant heterogeneity (P > 0.05). Additionally, the MR-Egger intercept test did not find evidence of horizontal pleiotropy (P > 0.05).
Conclusion
This study highlights that PAH may serve as a potential risk factor for this liver cancer. Future research should aim to elucidate the biological mechanisms at play and explore the potential for early interventions that could mitigate cancer risk in this vulnerable population.
Supplementary Information
The online version contains supplementary material available at 10.1007/s12672-024-01727-1.
Keywords: Pulmonary arterial hypertension, Cancer, GWAS, Mendelian randomization
Introduction
Pulmonary arterial hypertension (PAH) is a progressive vascular disease characterized by elevated pulmonary artery pressure and vascular remodeling, leading to right heart failure and decreased exercise capacity [1–3]. The etiology of PAH is complex and involves a combination of genetic [4, 5], environmental [6], and health-related factors [7]. Genetic predispositions, such as mutations in the BMPR2 gene [8, 9], are strongly associated with familial forms of PAH. Environmental influences, including exposure to toxins, certain medications, and infections, can also act as triggers for the development of the disease [10, 11]. Additionally, PAH may arise secondary to various underlying health conditions [10, 11]. While much research has focused on the cardiopulmonary implications of PAH, its potential role as a risk factor for various malignancies has garnered increasing interest [12]. Cancer remains a leading cause of morbidity and mortality globally [13], and understanding the relationships between chronic diseases, such as PAH, and cancer risk is crucial for developing effective prevention and management strategies.
Emerging evidence suggests a complex interplay between pulmonary hypertension and cancer biology. Patients with chronic lung diseases, including PAH, often exhibit systemic inflammation [14, 15], hypoxia [16], and altered metabolism [17, 18]—factors known to influence cancer development [18–21]. Recent epidemiological studies have indicated that PAH significantly increases the risk of developing cancer, highlighting the need for further investigation into this association. However, the causal mechanisms underlying these associations remain poorly understood, necessitating rigorous investigation.
To address this gap, Mendelian randomization (MR) offers a powerful tool for assessing causal relationships between exposures and outcomes by utilizing genetic variants as instrumental variables (IVs) [22, 23]. This approach mitigates confounding and reverse causation, providing more reliable insights into the causal pathways linking PAH and cancer. In this study, we aim to evaluate the causal effects of genetically proxied PAH on various cancer types including thyroid cancer, lung cancer, gastric cancer, liver cancer, and colorectal cancer.
This study aims to investigate the causal relationship between PAH and the risk of developing various common cancers by integrating genetic data and employing MR methodologies. Specifically, we seek to determine whether PAH acts as an independent risk factor for cancers such as thyroid, lung, gastric, liver, and colorectal cancers. By addressing this question, our research aims to provide insights that could inform clinical practices, guide targeted screening strategies for at-risk populations, and ultimately improve patient outcomes in both PAH and cancer management. Furthermore, this study contributes to the growing understanding of the connections between chronic diseases and cancer, laying the groundwork for future research and potential therapeutic innovations.
Materials and methods
The assumptions of MR
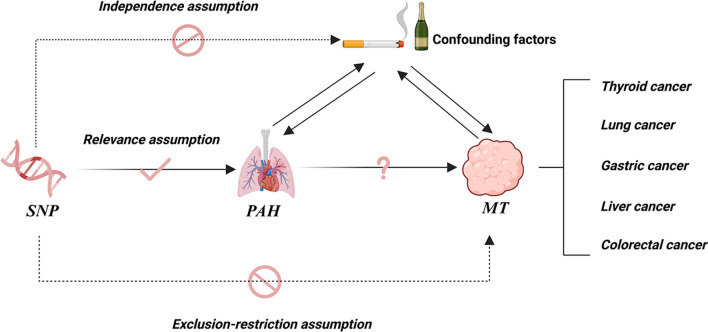
In this study, we explored the causal relationship between PAH and various common cancers based on the three main assumptions of MR study (Fig. 1) [24]. First, we identified genetic variants associated with PAH that showed significant correlation in large-scale genome-wide association study (GWAS), serving as effective IVs to reflect the exposure status of PAH. Subsequently, we assumed that IVs are independent of cancer risk, meaning that the influence of these single nucleotide polymorphisms (SNPs) on cancer risk is not confounded by other factors. Additionally, we ensured that the selected IVs are not directly related to cancer risk; specifically, these genetic variants influence cancer risk solely through their effect on PAH, without direct association with the cancers themselves. This approach allows us to minimize the potential impact of reverse causation, thereby enhancing the credibility of our findings.
Fig. 1.
Diagram of MR study design. SNP single nucleotide polymorphisms, PAH pulmonary arterial hypertension, MT malignant tumor
Data sources
The large-scale GWAS summary data related to PAH and cancer were obtained online from the GWAS Catalog (https://www.ebi.ac.uk/gwas/) on December 5, 2023. The GWAS Catalog is a comprehensive and well-curated resource for genetic association studies, offering detailed information on genetic variants associated with a broad spectrum of traits and diseases [25]. This platform enables researchers to access high-quality, curated data on SNPs and their phenotypic associations, providing a robust foundation for the analysis conducted in this study. The IDs are respectively “GCST007228” (PAH, N = 11,744), “GCST90018929” (thyroid cancer, N = 491,974), “GCST90018875” (lung cancer, N = 492,803), “GCST90018849” (N = 476,116), and “GCST90018858” (N = 475,638), “GCST90018808” (N = 470,002). The GWAS population for PAH and the GWAS population for cancer have no sample overlap. To minimize the genetic background differences among diverse populations, all GWAS samples included in this analysis were from European cohorts. Furthermore, since the GWAS data utilized in this study are publicly accessible, no additional ethical approval was required. Detailed information regarding the GWAS used in this MR analysis is summarized in Table 1.
Table 1.
Data sources of summary GWAS datasets
| Exposure/Outcome | Traits | ID | Sample size | Year | Population |
|---|---|---|---|---|---|
| Exposure | PAH | GCST007228 | 11,744 | 2018 | European |
| Outcome | Thyroid cancer | GCST90018929 | 491,974 | 2021 | European |
| Outcome | Lung cancer | GCST90018875 | 492,803 | 2021 | European |
| Outcome | Gastric cancer | GCST90018849 | 476,116 | 2021 | European |
| Outcome | Hepatic cancer | GCST90018858 | 475,638 | 2021 | European |
| Outcome | Colorectal cancer | GCST90018808 | 470,002 | 2021 | European |
GWAS genome-wide association study, PAH pulmonary arterial hypertension
SNP selection
First, PAH was used as the exposure factor, and various common cancer types were designated as the outcome factors. Data on PAH were extracted from the GWAS database, including SNPs that met the significance threshold of P < 1 × 10−6 [26]. The thresholds for analysis were set with r2 = 0.001 and a genetic distance of 10,000 kb to ensure that the IVs satisfied the assumption of independence, thereby eliminating the influence of linkage disequilibrium on the analysis results. Subsequently, to minimize the interference of confounding factors on the outcome data, the PhenoScanner [27], an online database designed to facilitate the exploration of genetic associations with various phenotypes and diseases, was used to filter and exclude SNPs associated with confounding factors. SNPs selected for use as IVs were uploaded to the PhenoScanner database, with parameters set to P = 1 × 10−5 and r2 = 0.8, to assess whether the selected SNPs were associated with smoking and alcohol consumption. These rigorously selected SNPs served as IVs for subsequent MR analysis, enhancing the robustness and credibility of the results. Additionally, the F-statistic was calculated to assess the strength of the included SNPs, with only IVs having F > 10 retained to avoid potential weak instrument in this MR analysis. The formula for calculating F is as follows: F = R2 (N-2)/(1-R2), R2 = 2 × (1-MAF) × MAF × β2 [28], where R2 is the proportion of variance explained by the instruments, N is the sample size, β represents the effect size of SNP, and MAF refers to the minor allele frequency. Finally, reverse causality was assessed through the MR Steiger test [29]. The remaining SNPs were then retained for following MR analysis.
MR estimates
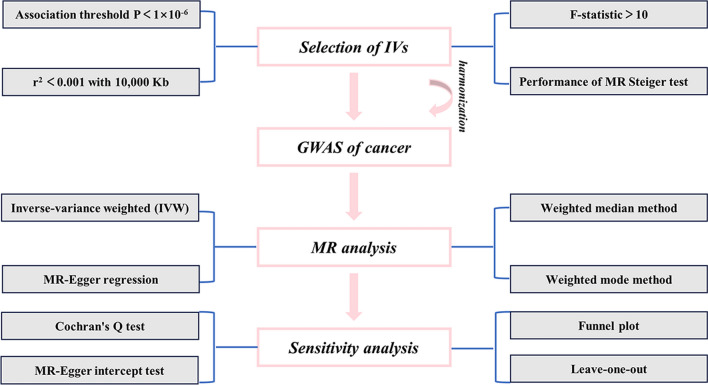
Using R (Version 4.3.2) software, MR analyses were conducted on the selected SNPs employing inverse variance weighted (IVW) [18], MR-Egger regression [30], weighted median [31], and weighted mode methods to infer the causal relationships between PAH and cancers. Among these, the IVW method is the primary approach in MR analysis. When horizontal pleiotropy is absent, the IVW method provides a relatively stable and accurate assessment of the causal effect of PAH on the cancers by combining the Wald estimates of each IV through meta-analysis. When heterogeneity is present, random-effect models are chosen; otherwise, fixed-effect models are applied. The MR-Egger regression, weighted median, and weighted mode methods are generally used as supplementary analyses to validate the main results. Additionally, the intercept of the MR-Egger regression could be utilized to detect horizontal pleiotropy, while the slope of the line could assess causal relationships. P-value > 0.05 in the heterogeneity analysis by Cochran's Q test indicates the absence of heterogeneity. Leave-one-out analysis was employed for sensitivity analysis to determine whether the overall estimate is influenced by specific SNPs. The symmetry of the funnel plot could help assess the presence of bias in the results (Fig. 2). P-value < 0.05 is considered statistically significant.
Fig. 2.
Flowchart of this MR study process. IVs instrumental variables, MR Mendelian randomization, GWAS genome-wide association study, IVW inverse-variance weighted
Result
IVs validity
A total of 9, 9, 9, 8, and 9 SNPs were selected as genetic proxies for PAH to analyze the causal effects of PAH on thyroid cancer, lung cancer, gastric cancer, hepatic cancer, and colorectal cancer, respectively. All SNPs passed the MR Steiger test, and their F-statistics were > 10. Detailed information regarding the SNPs used as instrumental variables can be found in Supplementary Table 1.
Effect of genetically predicated PAH on cancers
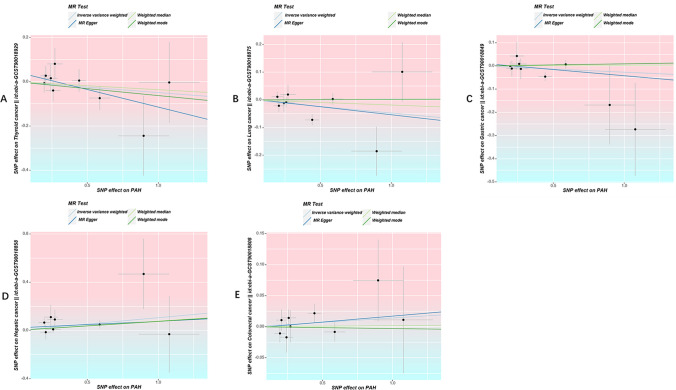
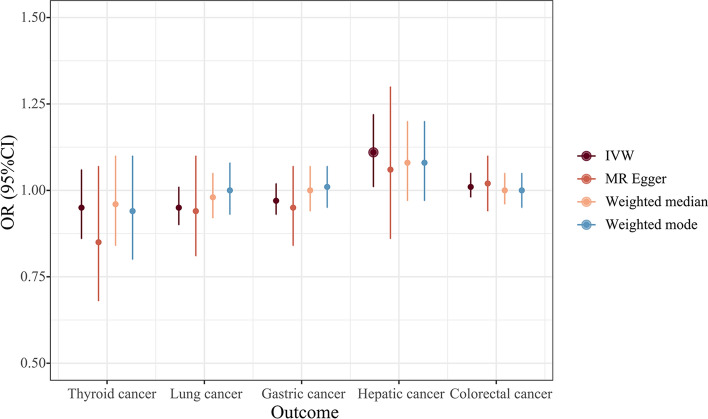
The IVW results indicated that PAH is associated with approximately an 11% increase in the risk of liver cancer [odd ratio (OR) 1.11, 95% confidence interval (CI) 1.01–1.22, P = 0.025); however, there is no evidence to suggest that PAH is related to the other four common cancer types. (Thyroid cancer: OR 0.95, 95% CI 0.86–1.06, P = 0.360; lung cancer: OR 0.95, 95% CI 0.90–1.01, P = 0.129; gastric cancer: OR 0.97, 95% CI 0.93–1.02, P = 0.243; colorectal cancer: OR 1.01, 95% CI 0.98–1.05, P = 0.412) (Figs. 3–4).
Fig. 3.
Scatter plots of SNPs associated with PAH and the risk of cancer. A Scatter plots of SNPs associated with PAH and the risk of thyroid cancer. B Scatter plots of SNPs associated with PAH and the risk of lung cancer. C Scatter plots of SNPs associated with PAH and the risk of gastric cancer. D Scatter plots of SNPs associated with PAH and the risk of hepatic cancer. E Scatter plots of SNPs associated with PAH and the risk of colorectal cancer. MR Mendelian randomization, PAH pulmonary arterial hypertension, IVW inverse-variance weighted
Fig. 4.
Forest plot of SNPs associated with PAH and the risk of cancer. IVW inverse-variance weighted, OR odds ratio, CI confidence interval
Sensitivity analysis
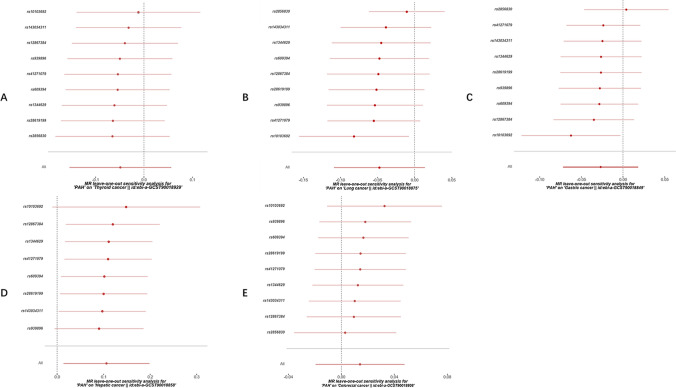
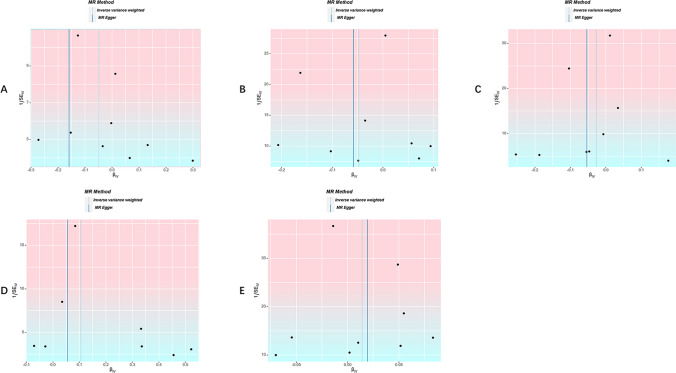
Cochran’s Q test indicated that there is mild heterogeneity in the MR analysis exploring the casual effect of PAH on lung cancer (P = 0.049), while all other MR analyses showed no evidence of heterogeneity (P > 0.05). Additionally, the MR-Egger intercept test suggested that there is no evidence of significant heterogeneity in this MR analysis (Table 2). Leave-one-out analysis results revealed no SNPs with a significant impact on the causal association (Fig. 5). The funnel plot displayed symmetry, indicating that there is no evidence of substantial bias (Fig. 6).
Table 2.
Sensitivity analysis in this MR study
| Exposure | Outcome | Heterogeneity test (IVW) | Pleiotropy test | |||
|---|---|---|---|---|---|---|
| Q | P-value | Intercept | P-value | |||
| PAH | Thyroid cancer | 5.30 | 0.725 | 0.04 | 0.317 | |
| PAH | Lung cancer | 15.54 | 0.049 | 0.00 | 0.882 | |
| PAH | Gastric cancer | 8.89 | 0.352 | 0.01 | 0.651 | |
| PAH | Hepatic cancer | 5.55 | 0.594 | 0.02 | 0.607 | |
| PAH | Colorectal cancer | 5.34 | 0.720 | 0.00 | 0.889 | |
PAH pulmonary arterial hypertension, IVW inverse-variance weighted, Q heterogeneity statistic Q
Fig. 5.
The leave-one-out sensitivity analysis for the effect of PAH on cancer risk. A the leave-one-out sensitivity analysis for the effect of PAH on thyroid cancer risk. B the leave-one-out sensitivity analysis for the effect of PAH on lung cancer risk. C the leave-one-out sensitivity analysis for the effect of PAH on gastric cancer risk. D the leave-one-out sensitivity analysis for the effect of PAH on hepatic cancer risk. E the leave-one-out sensitivity analysis for the effect of PAH on colorectal cancer risk
Fig. 6.
Funnel plots of the causal effect of PAH on cancer risk. A funnel plot of the causal effect of PAH on thyroid cancer risk. B funnel plot of the causal effect of PAH on lung cancer risk. C funnel plot of the causal effect of PAH on gastric risk. D funnel plot of the causal effect of PAH on hepatic risk. E funnel plot of the causal effect of PAH on colorectal risk
Discussion
This study aimed to investigate the causal relationship between PAH and various common cancers using (MR methods. Our findings suggest that genetically proxied PAH is significantly associated with an increased risk of liver cancer, while no substantial causal links were found with other common cancer types, including thyroid, lung, gastric, and colorectal cancers. These results have important implications for understanding how chronic conditions like PAH may influence cancer risk.
Previous epidemiological studies have also indicated a potential association between PAH and tumors. A cohort study using Danish nationwide registries that included over 5,000 PAH patients found that the risk of developing cancer in PAH patients was significantly increased in the first year, nearly doubling compared to the general population [32]. Coincidentally, another study has explored the potential association between Pulmonary hypertension and cancer within a German outpatient cohort. Utilizing the Disease Analyzer database (IQVIA), this research identified over 11,000 treated PH patients, revealing a significantly higher cancer incidence (23.2%) compared to non-PH patients (8.5%) over a 10-year period, with notable associations observed in both male and female patients, as well as specific cancer types such as respiratory and skin cancers [33]. However, our study did not find a significant causal relationship between PAH and lung cancer. This discrepancy could be explained by several factors. First, MR analysis relies on genetic variants as IVs, which may not fully capture the complexities of the relationship observed in clinical studies, where confounding factors such as smoking or environmental exposures could play a significant role. Second, clinical studies often reflect direct observational associations, which are more susceptible to bias from confounding and reverse causality. In contrast, MR analysis seeks to infer causality based on genetic variation, which may not be directly comparable to the findings of observational studies. Additionally, the absence of a significant result in our MR analysis could also be due to insufficient power or the lack of relevant genetic variants strongly associated with both PAH and lung cancer. Future studies with larger datasets and more refined genetic instruments could help clarify this relationship.
The observed association between PAH and cancer raises questions about the underlying mechanisms. One potential explanation is that chronic hypoxia, a hallmark of PAH, may promote tumorigenesis in the liver. Hypoxia could lead to the activation of hypoxia-inducible factors (HIFs), which are transcription factors that regulate various genes involved in angiogenesis, metabolism, and cell survival [34]. This process could enhance the growth and spread of liver tumors. Additionally, the hemodynamic changes associated with PAH may lead to liver dysfunction, contributing to an environment conducive to cancer development. Inflammation and fibrosis, often seen in both PAH and liver disease, may also play a role in this relationship by promoting cellular changes that facilitate cancer progression. Understanding the potential pathways linking PAH and cancer is critical for developing preventive strategies and therapeutic interventions. For instance, targeted therapies that address the underlying pathophysiology of PAH may also mitigate cancer risk. Furthermore, monitoring liver function in PAH patients could provide early indicators of increased cancer risk, enabling timely interventions. It is also important to consider the implications of genetic and environmental heterogeneity in different populations, as genetic variants and environmental exposures influencing PAH and cancer risk may vary significantly across diverse cohorts. The findings of this study, based on a European population, may not fully capture these variations. Replicating these analyses in non-European populations is essential to validate the observed associations and ensure their generalizability. Such efforts could provide a deeper understanding of the interplay between genetic predisposition, environmental factors, and disease outcomes, ultimately leading to more personalized and inclusive preventive strategies.
The use of MR in this study offers several advantages. MR is powerful for establishing causal relationships as it leverages genetic variants as IVs, which are less prone to confounding and reverse causation. This method provides more accurate causal effect estimates compared to traditional observational studies and allows exploration of the effects of long-term exposure, such as PAH, since genetic variants are fixed at conception and remain constant throughout life. However, MR is not without its limitations [35]. One significant concern is the assumption of independence among the IVs. If genetic variants are associated with confounding factors, the results may be biased. Additionally, while we did not observe evidence of horizontal pleiotropy in our analysis, this could still be a concern in MR studies. Horizontal pleiotropy occurs when genetic variants affect the outcome through pathways other than the exposure, potentially leading to misleading conclusions. The use of sensitivity analyses, such as MR-Egger regression and leave-one-out analyses, helps to address these issues, but they cannot completely eliminate the risk of bias. Another significant limitation of this study is the lack of exploration into the underlying mechanisms linking pulmonary arterial PAH to cancer. While our findings suggest a potential association between PAH and certain cancer types, the biological pathways that may mediate this relationship remain unclear.
In conclusion, this study highlights a significant association between genetically proxied PAH and an increased risk of liver cancer, suggesting that chronic conditions like PAH may have important implications for cancer risk. Understanding the underlying mechanisms linking PAH to cancer can inform preventive strategies and therapeutic approaches. While MR provides a robust framework for establishing causal relationships, researchers must remain vigilant about the potential limitations and biases inherent in this method. Further research is needed to explore the mechanisms linking PAH to cancer and to validate these findings in diverse populations. This knowledge will be critical for improving patient outcomes and informing public health strategies aimed at reducing cancer risk in individuals with chronic conditions.
Conclusion
This study establishes a significant causal relationship between genetically proxied PAH and an increased risk of liver cancer, while finding no notable associations with other common cancer types. By leveraging Mendelian randomization, our findings provide robust evidence supporting the role of PAH in cancer risk, particularly liver cancer, and highlight the potential for targeted screening and preventive strategies in PAH patients. These insights contribute to the growing understanding of the interplay between chronic diseases and cancer, laying a foundation for future research into the underlying mechanisms and therapeutic interventions.
Supplementary Information
Acknowledgements
No.
Author contributions
WZ designed the study. YF performed data analysis and drafted the manuscript. WZ and XD revised the manuscript. All authors read and approved the final manuscript.
Funding
None.
Data availability
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Naeije R, Richter MJ, Rubin LJ. The physiological basis of pulmonary arterial hypertension[J]. Eur Respir J. 2022. 10.1183/13993003.02334-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hassoun PM. Pulmonary arterial hypertension[J]. N Engl J Med. 2021;385(25):2361–76. [DOI] [PubMed] [Google Scholar]
- 3.Montani D, Günther S, Dorfmüller P, et al. Pulmonary arterial hypertension[J]. Orphanet J Rare Dis. 2013;8:1–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Soubrier F, Chung WK, Machado R, et al. Genetics and genomics of pulmonary arterial hypertension[J]. J Am Coll Cardiol. 2013;62(25S):D13–21. [DOI] [PubMed] [Google Scholar]
- 5.Ma L, Chung WK. The role of genetics in pulmonary arterial hypertension[J]. J Pathol. 2017;241(2):273–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lichtblau M, Reimann L, Piccari L. Pulmonary vascular disease, environmental pollution, and climate change[J]. Pulm Circ. 2024;14(2):e12394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Humbert M, Nunes H, Sitbon O, et al. Risk factors for pulmonary arterial hypertension[J]. Clin Chest Med. 2001;22(3):459–75. [DOI] [PubMed] [Google Scholar]
- 8.Cuthbertson I, Morrell NW, Caruso P. BMPR2 mutation and metabolic reprogramming in pulmonary arterial hypertension[J]. Circ Res. 2023;132(1):109–26. [DOI] [PubMed] [Google Scholar]
- 9.Hong KH, Lee YJ, Lee E, et al. Genetic ablation of the BMPR2 gene in pulmonary endothelium is sufficient to predispose to pulmonary arterial hypertension[J]. Circulation. 2008;118(7):722–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schermuly RT, Ghofrani HA, Wilkins MR, et al. Mechanisms of disease: pulmonary arterial hypertension[J]. Nat Rev Cardiol. 2011;8(8):443–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lai YC, Potoka KC, Champion HC, et al. Pulmonary arterial hypertension: the clinical syndrome[J]. Circ Res. 2014;115(1):115–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bravos E, Cottin V, Dauphin C, et al. Cancer incidence in patients with pre-capillary pulmonary hypertension[J]. J Heart Lung Transplant. 2019;38(7):778–80. [DOI] [PubMed] [Google Scholar]
- 13.Global Burden of Disease Cancer Collaboration. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 29 cancer groups, 1990 to 2017: a systematic analysis for the global burden of disease study[J]. JAMA oncology, 2019. [DOI] [PMC free article] [PubMed]
- 14.Kumar R, Graham B. How does inflammation contribute to pulmonary hypertension?[J]. Eur Respir J. 2018. 10.1183/13993003.02403-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Price LC, Wort SJ, Perros F, et al. Inflammation in pulmonary arterial hypertension[J]. Chest. 2012;141(1):210–21. [DOI] [PubMed] [Google Scholar]
- 16.Nathan SD, Barbera JA, Gaine SP, et al. Pulmonary hypertension in chronic lung disease and hypoxia[J]. Eur Respir J. 2019. 10.1183/13993003.01914-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu W, Janocha AJ, Erzurum SC. Metabolism in pulmonary hypertension[J]. Annu Rev Physiol. 2021;83(1):551–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dang CV. Links between metabolism and cancer[J]. Genes Dev. 2012;26(9):877–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Michels N, van Aart C, Morisse J, et al. Chronic inflammation towards cancer incidence: a systematic review and meta-analysis of epidemiological studies[J]. Crit Rev Oncol Hematol. 2021;157:103177. [DOI] [PubMed] [Google Scholar]
- 20.Chen Z, Han F, Du Y, et al. Hypoxic microenvironment in cancer: molecular mechanisms and therapeutic interventions[J]. Signal Transduct Target Ther. 2023;8(1):70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stine ZE, Schug ZT, Salvino JM, et al. Targeting cancer metabolism in the era of precision oncology[J]. Nat Rev Drug Discovery. 2022;21(2):141–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Birney E. Mendelian randomization[J]. Cold Spring Harbor Perspect Med. 2021. 10.1101/cshperspect.a041302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Richmond RC, Smith GD. Mendelian randomization: concepts and scope[J]. Cold Spring Harb Perspect Med. 2022;12(1):a040501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanderson E, Glymour MM, Holmes MV, et al. Mendelian randomization[J]. Nat Rev Methods Primers. 2022;2(1):6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sollis E, Mosaku A, Abid A, et al. The NHGRI-EBI GWAS catalog: knowledgebase and deposition resource[J]. Nucleic Acids Res. 2023;51(D1):D977–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Choi KW, Chen CY, Stein MB, et al. Assessment of bidirectional relationships between physical activity and depression among adults: a 2-sample Mendelian randomization study[J]. JAMA Psychiat. 2019;76(4):399–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kamat MA, Blackshaw JA, Young R, et al. PhenoScanner V2: an expanded tool for searching human genotype–phenotype associations[J]. Bioinformatics. 2019;35(22):4851–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou P, Hao Z, Chen Y, et al. Association between gut microbiota and diabetic microvascular complications: a two-sample Mendelian randomization study[J]. Front Endocrinol. 2024;15:1364280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hemani G, Tilling K, Davey SG. Orienting the causal relationship between imprecisely measured traits using GWAS summary data[J]. PLoS Genet. 2017;13(11):e1007081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burgess S, Thompson SG. Interpreting findings from Mendelian randomization using the MR-Egger method[J]. Eur J Epidemiol. 2017;32:377–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bowden J, Davey Smith G, Haycock PC, et al. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator[J]. Genet Epidemiol. 2016;40(4):304–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sørensen HT, Skajaa N, Klok FA, et al. Cancer risk in pulmonary hypertension patients[J]. Clin Epidemiol. 2022. 10.2147/CLEP.S345054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roderburg C, Loosen SH, Hippe HJ, et al. Pulmonary hypertension is associated with an increased incidence of cancer diagnoses[J]. Pulm Circ. 2022;12(2):e12000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Amelio I, Melino G. The p53 family and the hypoxia-inducible factors (HIFs): determinants of cancer progression[J]. Trends Biochem Sci. 2015;40(8):425–34. [DOI] [PubMed] [Google Scholar]
- 35.Smith GD, Ebrahim S. Mendelian randomization: prospects, potentials, and limitations[J]. Int J Epidemiol. 2004;33(1):30–42. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.