Abstract
Background
Existing biomarkers and models for predicting response to programmed cell death protein 1 monoclonal antibody in advanced squamous-cell non-small cell lung cancer (sqNSCLC) did not have enough accuracy. We used data from the ORIENT-3 study to construct artificial neural network (ANN) systems to predict the response to sintilimab for sqNSCLC.
Methods
Four ANN systems based on bulk RNA data to predict disease control (DC), immune DC (iDC), objective response (OR) and immune OR (iOR) were constructed and tested for patients with sqNSCLC treated with sintilimab. The mechanism exploration on the bulk and the spatial level were performed in patients from the ORIENT-3 study and the real world, respectively.
Findings
sqNSCLC patients with different responses to sintilimab showed each unique transcriptomic spectrum. Four ANN systems showed high accuracy in the test cohort (AUC of DC, iDC, OR and iOR were 0.83, 0.89, 0.93 and 0.94, respectively). The performance of ANN systems was better than that of linear model systems and showed high stability. The mechanism exploration on the bulk level suggested that patients with lower ANN system scores (worse response) had a higher ratio of immune-related pathways enrichment. The mechanism exploration on the spatial level indicated that patients with better response to immunotherapy had fewer clusters of both tumor and cytotoxicity T cell spots.
Interpretation
The four ANN systems showed high accuracy, robustness and stability in predicting the response to sintilimab for patients with sqNSCLC.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00262-024-03886-0.
Keywords: Squamous-cell non-small-cell lung cancer, Artificial neural network, Sintilimab, Response
Introduction
Lung cancer, one of the most frequently diagnosed cancer types, is the leading cause of cancer-related deaths around the world [1, 2], and non-small cell lung cancer (NSCLC) accounts for 85% of patients with lung cancer [3]. Immunotherapy, especially programmed cell death protein 1 (PD-1)/programmed death ligand 1 (PD-L1) monoclonal antibody (mAb), brought benefits to patients with advanced NSCLC. The efficacy of PD-1/PD-L1 mAb monotherapy or combined with chemotherapy in the first- and posterior-line treatment was limited, with objective response rate (ORR) of 43.3–64.8% [4–6] and 13.6–25.5% [7, 8], respectively. To identify patients could benefit from PD-1/PD-L1 mAb, many studies focus on developing biomarkers, and the most common used in patients with NSCLC were PD-L1 [5], tumor mutation burden (TMB) [9] and fraction of copy number-altered genome (FGA) [10]. However, the existing biomarkers did not show enough accuracy [11], and some of them might play different roles in squamous-cell non-small cell lung cancer (sqNSCLC) and non-sqNSCLC [12].
Recently, deep learning had been used in the diagnosis of malignant tumor and show high accuracy [13]. Therefore, predictive model constructed by deep learning might help to identify eligible patients who will benefit most from PD-1/PD-L1 mAb. Some studies had provided signatures or models to predict the response to immunotherapy for pan-cancer [14, 15], but the robustness of these were limited. Perhaps, due to the heterogeneity between different types of malignant tumors, models established based on a specific tumor type only have better predictive ability for that type of tumor, and generalized use of models might result in incorrect results. In NSCLC, there are many difference between sqNSCLC and non-sqNSCLC, including cell of origin, genomic alteration, pathway alteration, etc. [16]. Besides, because of lack of driver gene alteration, PD-1/PD-L1 mAb plays more important roles in sqNSCLC than non-sqNSCLC. For this reason, it is necessary to construct specialized predictive models for sqNSCLC.
ORIENT-3 was an open-label, multicenter, randomized controlled phase 3 study that recruited patients with stage IIIB/IIIC/IV sqNSCLC after failure with first-line platinum-based chemotherapy, which had response records and RNA data [8]. This study used the RNA data of ORIENT-3 study to construct artificial neural network (ANN) systems to predict the response to sintilimab for patients with sqNSCLC, and tested the accuracy, robustness and stability of these ANN systems. Besides, both bulk and spatial transcriptomic data were used to explore the potential mechanism of these ANN systems and provide possible therapy targets.
Methods
Study design
There were three cohorts designed in this study. The cohort 1 and the cohort 2 were 61 and 49 patients with RNA data from sintilimab arm and docetaxel arm of the ORIENT-3 study [8], respectively. The cohort 3 was three patients treated with PD-1 mAb combined with chemotherapy from the real world. The efficacy and RNA data of the cohort 1 and 2 were obtained from the ORIENT-3 study [8]. The clinicopathological characteristics of the cohort 3 were obtained from the clinical records, and the tumor specimens were formalin-fixed paraffin-embedded (FFPE) samples from the tissue bank of Department of Pathology, Cancer Hospital, Chinese Academy of Medical Science (CAMS). In the cohort 3, the tumor responses were determined according to the Response Evaluation Criteria in Solid Tumors version 1.1, and the progression-free survival (PFS) was defined as the duration from the initial of first time of immunotherapy in the advanced tumor therapy to the date of disease progression or the last follow-up.
Transcriptomic spectrum between different responses
The 59 patients with response data in the cohort 1 were used to performed transcriptomic spectrum. The confirmed best of response (BORC) results were classed into response (R, including complete response [CR] and partial response [PR]), stable disease (StD, S) and progressive disease (PD, P). For immune BOR (iBOR), the results were classed into immune response (iR, including immune CR [iCR] and immune PR [iPR]), immune StD (iStD, iS) and immune unconfirmed PD (iUPD, iP). Uni-variable logistic regression was used to calculate the odds ratio (OR) of different BORC (R versus S + P, S versus R + P and P versus R + S) and iBOR (iR versus iS + iP, iS versus iR + iP and iP versus iR + iS) results to the residue other patients for each gene.
Gene set enrichment analysis (GSEA) was performed by “clusterProfiler” package [17], information of gene sets in “HALLMARK”, “Kyoto Encyclopedia of Genes and Genomes (KEGG)” and “Gene Ontology (GO)” was obtained from www.gsea-msigdb.org/gsea/msigdb/index.jsp. A total of 1350 gene sets associated tumor and its microenvironment were analyzed as a point of importance. The 1,350 gene sets were summarized into six classes by us according to the information of these gene sets provided on the molecular signatures database (MSigDB), including 421 “Immunity,” 127 “Cell cycle,” 315 “Metabolism & energy,” 286 “Genetic and epigenetic information,” 129 “extracellular matrix (ECM) & metastasis” and 72 “Cell death” gene sets. The gene sets with raw P < 0.05 & FDR < 0.25 were considered as statistically significant.
ANN systems construction
RNA data were normalized based on the expression of the sum of two house keeping genes (ACTB and GAPDH), then were converted logarithmically. The ANN systems construction flowchart was shown in Figure S1 and performed independently for disease control (DC, R + S), immune DC (iDC, iR + iS), objective response (OR, R) and immune OR (iOR, iR). The 59 patients from the cohort 1 with both response and RNA data were randomly divided into the training cohort (70%) and the test cohort (30%). Receiver operating characteristic (ROC) curve were used in the training cohort to select genes, and the top 30 significant (P < 0.05, area under the curve [AUC] of ROC curve > 0.75 or < 0.25, ranked by random forest method [“randomForest” package, https://cran.r-project.org/doc/Rnews/Rnews_2002-3.pdf]) genes were used to construct ANN models. If the significant genes were fewer than 30, all significant genes would be used to construct ANN models. The “neuralnet” package (https://CRAN.R-project.org/package=neuralnet) was used to construed ANN models. The number of hidden neurons was based on Nh = (4n2 + 3)/(n2 − 8) (Nh, the number of hidden neurons; n, the number of input neurons) [13]. Three hundred times of threefold cross validation were performed to obtain 900 ANN models and test the accuracy of them. The final predicted scores of the ANN system (900 ANN models) were equal to the weighted average of output values of all ANN models. Finally, the accuracy of the ANN system was tested in the test cohort.
ANN systems evaluation
First, according linear models’ system was also built and was compared with the ANN system to evaluate the effectiveness of the ANN system. Second, to evaluate the clinical practicality, the best cut-off of the final predicted scores of the ANN system and according linear models’ system were determined by the Youden indexes of both the training and the test cohort. Third, the correlation between the expression of each gene constructing the ANN system and the final predicted scores were explored by Pearson correlation coefficient to evaluated the internal correlation of the ANN system. Finally, the AUC and width of according 95% confidence interval [CI] of each ANN system with all combination of gene expression missing were tested to evaluate the stability of the ANN system. In the stability evaluation, the number of missing genes were ranged from one to seven, and the expression of these missing genes were replaced by the mean normalized expression of them in 59 patients from the cohort 1.
Mechanism exploration on bulk level
The final predicted scores of the four ANN systems were calculated for patients from the cohort 1 and cohort 2, and the correlation between the final predicted scores and the activation of pathways as well as tumor immune microenvironment (TIME) were explored. In pathway enrichment analysis, GSEA and gene set variation analysis (GSVA) were performed by “clusterProfiler” [17] and “GSVA” [18] package, respectively. The information and classification of reference gene sets were the same as the part of “Transcriptomic spectrum between different responses.” In TIME analysis, “ESTIMATE” algorithm was used to calculate the stromal and immune scores by “estimate” package [19]. Besides, “CIBERSORT” [20], “quanTIseq” [21] and single sample GSEA (ssGSEA) [18] were performed to calculate the infiltration fractions or scores of immune cells. The reference gene sets of ssGSEA contained the marker genes of 28 kinds of immune cells [14].
Mechanism exploration on spatial level
5μm FFPE slides of patients from the cohort 3 were prepared and incubated followed by hematoxylin–eosin (HE) staining. Tissue imaging was conducted following the application of approximately 100 µL of 85% Glycerol (Thermofisher, Catalog number 15514011). The Visium slide was inserted into a cassette and incubated. Pre-hybridization, library preparation, encompassing probe ligation, probe release and extension, probe elution, and FFPE library assembly were processed according to the Visium Spatial Gene Expression User Guide (10× Genomics, User Guide CG000407 Rev C, human transcriptome Product number 1000338) and all reagents were from the Visium Spatial Gene Expression for FFPE Reagent Kit (10× Genomics). The completed libraries were subjected to sequencing on the Novaseq6000 platform (Illumina). The raw spatial sequencing RNA data were normalized by the “Seurat” [22] package for the following analysis.
The final predicted scores of the four ANN systems were calculated for patients from the cohort 3, and the correlation between the final predicted scores and the activation of pathways (calculated by “GSVA” [18]) were explored. The “ESTIMATE” [19], “CIBERSORT” [20] and “quanTIseq” [21] were used to evaluated tumor purity and the infiltration of cytotoxic T lymphocyte (CTL), then the tumor cells dominant spots (tumor spots) and spots with relative high infiltration level of CTL (CTL spots) were identified. RNA data of tumor and CTL spots from each patients were scaled and used to performed principal component analysis (PCA). Then, the principal components were used to performed uniform manifold approximation and projection (UMAP) to further dimension reduction. K-means method was used to group tumor and CTL spots into different clusters, the optimal number of clusters was determined by the “NbClust” [23] package.
Cell–cell communication analysis was performed by the “CellChat” [24] package, and the “CellChatDB.human” was used as the reference. Pseudotime analysis was performed by “monocle” [25–27] package, and the expression of S100A2 and some of keratin-related genes was used to determine the direction of trajectory.
Statistical analysis
R software (version 4.2.1, https://www.r-project.org) was employed for all the statistical analysis. Loupe Browser (version 6.5.0, https://support.10xgenomics.com) was used to assist tumor spots identification. Spearman correlation analysis was performed for the final predicted score, to calculate the relationship between the final predicted scores and gene expression, GSVA score as well as infiltration fraction or score of immune cells. In this study, raw P < 0.05 & FDR < 0.25 were considered statistically significant.
Role of the funding source
This work was supported by the National Science and Technology Major Project for Key New Drug Development (2017ZX09304015). Professor Yuankai Shi had roles in data collection, data analysis, data interpretation, and writing of the manuscript. Professor Yuankai Shi had full access to all the dataset of this study and the decision to submit for publication.
Findings
Patient baseline characteristics
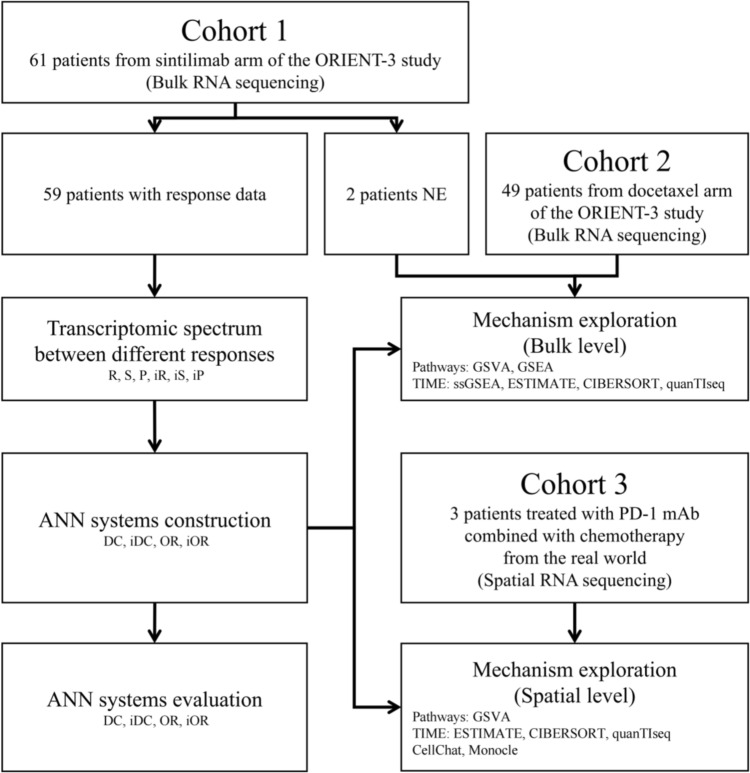
Study flowchart was shown in Fig. 1. Patients’ clinicopathologic characteristics of the cohort 1 (n = 61), the cohort 2 (n = 49) and the cohort 3 (n = 3) were summarized in Table 1. Most of the patients in the cohort 1 and 2 as well as all patients in the cohort 3 were male and smoker. The age distribution of the three cohorts were similar. All patients suffered locally advanced or metastatic sqNSCLC. In the cohort 1 and 3, patients received PD-1 mAb, and all patients in the cohort 1 received sintilimab alone, whereas patients in the cohort 3 received PD-1 mAb (two received tislelizumab and one received pembrolizumab) combined with chemotherapy. The ORR and disease control rate (DCR) of the cohort 1 were 23.7% and 74.6%, respectively. There were two patients with PR and one patient with StD in the cohort 3.
Fig. 1.
Study flowchart. NE: not evaluable; R: response; S: stable; P: progressive; iR: immune response; iS: immune stable; iP: immune progressive; ANN: artificial neural network; DC: disease control; iDC: immune disease control; OR: objective response; iOR: immune objective response; GSVA: gene set variation analysis; GSEA: gene set enrichment analysis; TIME: tumor immune microenvironment; ssGSEA: single sample gene set enrichment analysis; PD-1: programmed cell death protein 1; mAb: monoclonal antibody
Table 1.
Clinicopathologic characteristics of all patients in this study
| Characteristics | Cohort 1 (n = 61) | Cohort 2 (n = 49) | Cohort 3 (n = 3) |
|---|---|---|---|
| Sex, n (%) | |||
| Male | 57 (93.4) | 42 (85.7) | 3 (100.0) |
| Female | 4 (6.6) | 7 (14.3) | 0 |
| Age (years) | |||
| Mean ± SD | 59.5 ± 8.8 | 59.4 ± 8.3 | 62.7 ± 3.5 |
| Median (min, max) | 61.0 (43.0, 74.0) | 59.0 (34.0, 72.0) | 63.0 (59.0, 66.0) |
| > 60, n (%) | 32 (52.5) | 21 (42.9) | 2 (66.7) |
| ≤ 60, n (%) | 29 (47.5) | 28 (57.1) | 1 (33.3) |
| Smoking status, n (%) | |||
| Current smoker/quitted | 55 (90.2) | 38 (77.6) | 3 (100.0) |
| Non-smoker | 6 (9.8) | 11 (22.4) | 0 |
| Staging, n (%) | |||
| Stage IIIB | 8 (13.1) | 3 (6.1) | 1 (33.3) |
| Stage IIIC | 1 (1.6) | 2 (4.1) | 0 |
| Stage IV A | 29 (47.5) | 27 (55.1) | 1 (33.3) |
| Stage IVB | 23 (37.7) | 17 (34.7) | 1 (33.3) |
| Best of response to PD-1 mAb | |||
| CR | 1 (1.6) | NA | 0 |
| PR | 13 (21.3) | NA | 2 (66.7) |
| StD | 30 (49.2) | NA | 1 (33.3) |
| PD | 15 (24.6) | NA | 0 |
| NE | 2 (3.3) | NA | 0 |
SD standard deviation, PD-1 programmed cell death protein 1, mAb monoclonal antibody, CR complete response, PR partial response, StD stable disease, PD progressive disease, NE not evaluable according to Response Evaluation Criteria in Solid Tumors version 1.1 criteria, NA not available
Transcriptomic spectrum between different responses
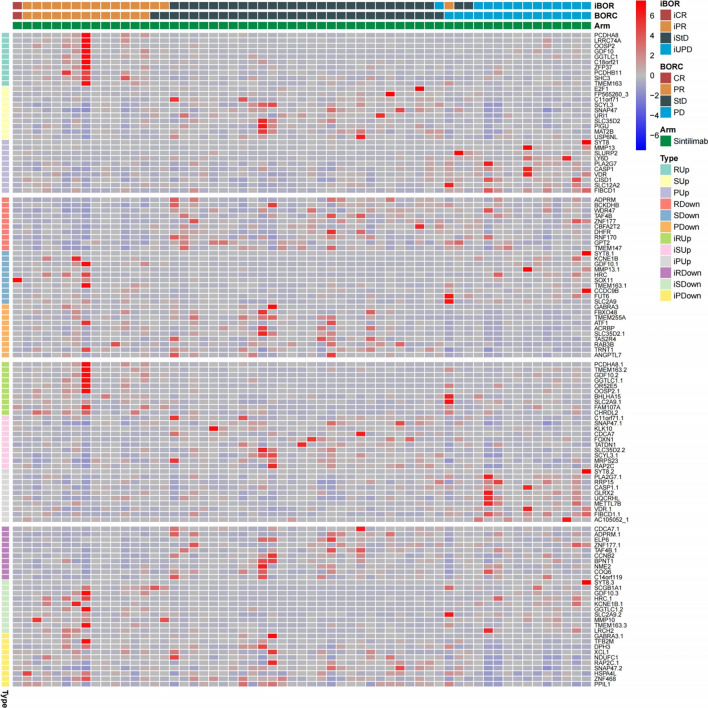
The top 10 up-regulated and down-regulated genes between different responses were shown in Fig. 2. To further understand the transcriptomic characteristics between different responses, the GSEA were performed. There were 11.4% (36/315) “Metabolism & energy”, 11.2% (47/420, one gene set was not available [NA]) “Immunity” and 10.5% (30/286) “Genetic and epigenetic information” gene sets were enriched in the R group, 96.9% (123/127) “Cell cycle”, 84.6% (242/286) “Genetic and epigenetic information” and 77.8% (56/72) “Cell death” gene sets were enriched in the S group, 59.7% (77/129) “ECM & metastasis” gene sets were enriched in the P group (Fig. S2A). In terms of iBOR, the results were similar to BORC (Fig. S2B). When the statistically significant gene sets were selected, there were 79.5% (101/127) “Cell cycle”, 56.3% (161/286) “Genetic and epigenetic information” and 23.6% (17/72) “Cell death” gene sets were enriched in the S group, whereas almost no gene set was enriched in the R and P group, and this result was also observed in the iBOR (Fig. S2C&D). Normalized enrichment score (NES) of all 1350 gene set between different responses were shown in Fig. S2E, and the S and iS group had the most enrichment gene sets.
Fig. 2.
The top 10 up-regulated and down-regulated genes between different responses in the cohort 1. iBOR: immune best of response; BORC: confirmed best of response; iCR: immune complete response; iPR: immune partial response; iStD: immune stable disease; iUPD: immune unconfirmed progressive disease; CR: complete response; PR: partial response; StD: stable disease; PD: progressive disease; R: response; S: stable; P: progressive; iR: immune response; iS: immune stable; iP: immune progressive
Construction and evaluation of ANN systems
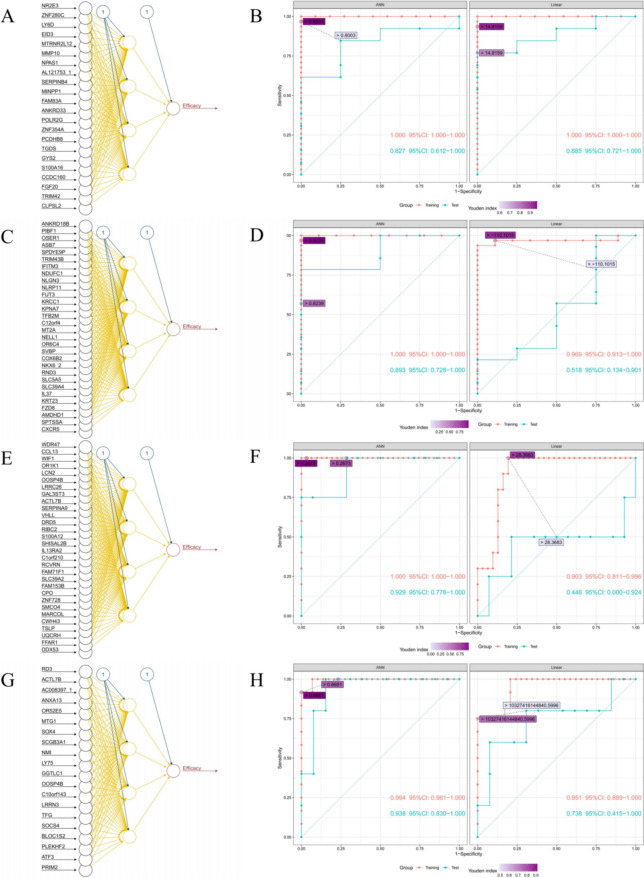
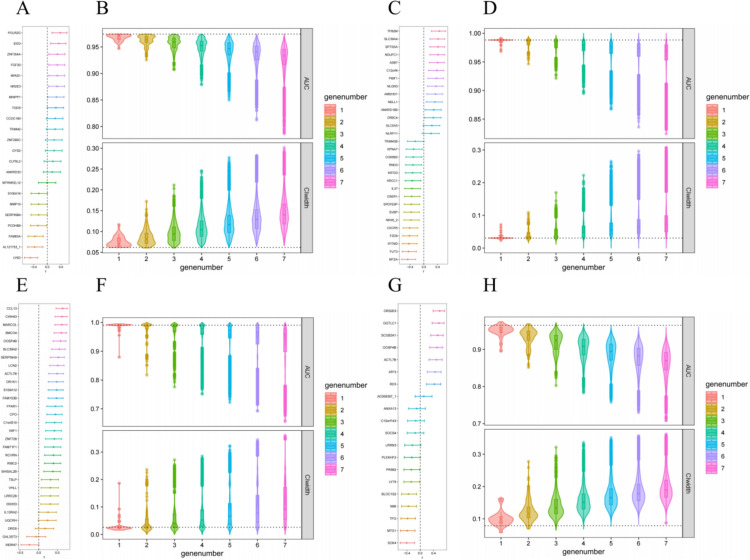
In the construction of ANN system for DC, 22 genes were screened by ROC (Fig. S3A), and the importance of these 22 genes were ranked by random forest method (Fig. S3B). Normalized expression was used as input neurons to constructed ANN system for DC. Each ANN model in this system contained three layers with 22 input neurons, four hidden neurons and one output neuron, respectively (Fig. 3A). The ANN system showed high accuracy in both the training cohort and the test cohort, with the AUC of 1.000 (95% CI 1.000–1.000) and 0.827 (95% CI 0.612–1.000), respectively, whereas the according linear models’ system also showed high accuracy in both the training cohort and the test cohort, with the AUC of 1.000 (95% CI 1.000–1.000) and 0.885 (95% CI 0.721–1.000), respectively (Fig. 3B). The best cut-off of the final predicted scores of the ANN system and according linear models’ system were 0.8003 and 14.8159, respectively (Fig. S3C). The correlation between the expression of each gene constructing the ANN system and the final predicted scores were weak, with Pearson correlation coefficient ranged from − 0.50 to 0.38 (Fig. 4A). The AUC of the ANN system with all combination of one to seven missing genes were 0.947–0.974, 0.926–0.974, 0.908–0.974, 0.879–0.974, 0.850–0.976, 0.812–0.974 and 0.786–0.974, respectively (Fig. 4B, the AUC without missing gene was 0.974); the width of 95% CI of the ANN system with all combination of one to seven missing genes were 0.062–0.117, 0.060–0.173, 0.060–0.212, 0.062–0.246, 0.062–0.278, 0.064–0.283 and 0.065–0.302, respectively (Fig. 4B, the width of 95% CI without missing gene was 0.062). The AUC of the ANN system and the width of according 95% CI with all combination of one to two missing genes were showed in Fig. S4A.
Fig. 3.
Structure of ANN systems and ROC curve of ANN systems and linear model systems for DC A and B, iDC (C and D), OR (E and F) and iOR (G and H). ANN: artificial neural network; ROC: receiver operating characteristic; DC: disease control; iDC: immune disease control; OR: objective response; iOR: immune objective response
Fig. 4.
Pearson correlation coefficients between expression of input genes and predicted score, and stability of ANN systems for DC (A and B), iDC (C and D), OR (E and F) and iOR (G and H). ANN: artificial neural network; DC: disease control; iDC: immune disease control; OR: objective response; iOR: immune objective response; AUC: area under the curve; CI: confidence interval
In the construction of ANN system for iDC, 47 genes were screened by ROC (Fig. S3D), and the importance of these 47 genes were ranked by random forest method (Fig. S3E). Normalized expression of the top 30 genes were used as input neurons to constructed ANN system for iDC. Each ANN model in this system contained three layers with 30 input neurons, four hidden neurons and one output neuron, respectively (Fig. 3C). The ANN system showed high accuracy in both the training cohort and the test cohort, with the AUC of 1.000 (95% CI 1.000–1.000) and 0.893 (95% CI 0.728–1.000), respectively, whereas the according linear models’ system did not show high accuracy in the test cohort, the AUC were 0.969 (95% CI 0.913–1.000) and 0.518 (95% CI 0.134–0.901) in the training cohort and the test cohort, respectively (Fig. 3D). The best cut-off of the final predicted scores of the ANN system and according linear models’ system were 0.8239 and -110.1015, respectively (Fig. S3F). The correlation between the expression of each gene constructing the ANN system and the final predicted scores were weak, with Pearson correlation coefficient ranged from − 0.47 to 0.45 (Fig. 4C). The AUC of the ANN system with all combination of one to seven missing genes were 0.968–0.992, 0.946–0.995, 0.921–0.997, 0.895–1.000, 0.868–1.000, 0.836–1.000 and 0.824–1.000, respectively (Fig. 4D, the AUC without missing gene was 0.988); the width of 95% CI of the ANN system with all combination of one to seven missing genes were 0.023–0.072, 0.015–0.109, 0.011–0.170, 0.000–0.223, 0.000–0.265, 0.000–0.296 and 0.000–0.307, respectively (Fig. 4D, the width of 95% CI without missing gene was 0.030). The AUC of the ANN system and the width of according 95% CI with all combination of one to two missing genes were showed in Fig. S4B.
In the construction of ANN system for OR, 48 genes were screened by ROC (Fig. S3G), and the importance of these 48 genes were ranked by random forest method (Fig. S3H). Normalized expression of the top 30 genes were used as input neurons to constructed ANN system for OR. Each ANN model in this system contained three layers with 30 input neurons, four hidden neurons and one output neuron, respectively (Fig. 3E). The ANN system showed high accuracy in both the training cohort and the test cohort, with the AUC of 1.000 (95% CI 1.000–1.000) and 0.929 (95% CI 0.776–1.000), respectively; whereas the according linear models’ system didn’t show high accuracy in the test cohort, the AUC were 0.903 (95% CI 0.811–0.996) and 0.446 (95% CI 0.000–0.924) in the training cohort and the test cohort, respectively (Fig. 3F). The best cut-off of the final predicted scores of the ANN system and according linear models’ system were 0.2673 and 28.3683, respectively (Figure S3I). The correlation between the expression of each gene constructing the ANN system and the final predicted scores was weak, with Pearson correlation coefficient ranged from − 0.25 to 0.64 (Fig. 4E). The AUC of the ANN system with all combination of one to seven missing genes were 0.879–0.995, 0.817–1.000, 0.776–1.000, 0.751–1.000, 0.722–1.000, 0.692–1.000 and 0.656–1.000, respectively (Fig. 4F, the AUC without missing gene was 0.990); the width of 95% CI of the ANN system with all combination of one to seven missing genes were 0.015–0.187, 0.000–0.236, 0.000–0.272, 0.000–0.285, 0.000–0.322, 0.000–0.345 and 0.000–0.361, respectively (Fig. 4F, the width of 95% CI without missing gene was 0.026). The AUC of the ANN system and the width of according 95% CI with all combination of one to two missing genes were showed in Fig. S4C.
In the construction of ANN system for iOR, 20 genes were screened by ROC (Figure S3J), and the importance of these 20 genes were ranked by random forest method (Figure S3K). Normalized expression was used as input neurons to constructed ANN system for iOR. Each ANN model in this system contained three layers with 20 input neurons, four hidden neurons and one output neuron, respectively (Fig. 3G). The ANN system showed high accuracy in both the training cohort and the test cohort, with the AUC of 0.994 (95% CI 0.981–1.000) and 0.938 (95% CI 0.830–1.000), respectively, whereas the according linear models’ system did not show high accuracy in the test cohort, the AUC were 0.951 (95% CI 0.889–1.000) and 0.738 (95% CI 0.415–1.000) in the training cohort and the test cohort, respectively (Fig. 3H). The best cut-off of the final predicted scores of the ANN system and according linear models’ system were 0.6681 and 10,327,416,144,840.5996, respectively (Figure S3L). The correlation between the expression of each gene constructing the ANN system and the final predicted scores were weak, with Pearson correlation coefficient ranged from − 0.41 to 0.59 (Fig. 4G). The AUC of the ANN system with all combination of one to seven missing genes were 0.895–0.975, 0.852–0.969, 0.803–0.972, 0.783–0.969, 0.758–0.965, 0.730–0.961 and 0.709–0.959, respectively (Fig. 4H, the AUC without missing gene was 0.965); the width of 95% CI of the ANN system with all combination of one to seven missing genes were 0.057–0.164, 0.068–0.278, 0.065–0.322, 0.068–0.337, 0.077–0.351, 0.083–0.357 and 0.087–0.363, respectively (Fig. 4H, the width of 95% CI without missing gene was 0.079). The AUC of the ANN system and the width of according 95% CI with all combination of one or two missing genes were showed in Fig. S4D.
Mechanism exploration on bulk level
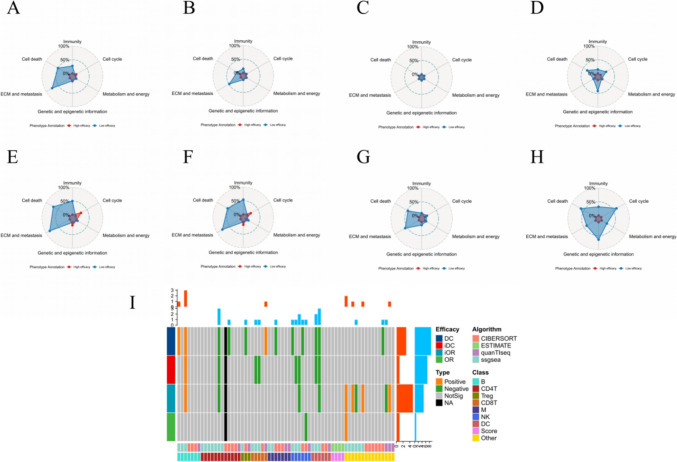
In GSVA results of ANN system predicted scores, almost no gene set was enriched in the high efficacy group, more “Immunity” (27.4% [115/420] vs. 0 [0/420]), “ECM & metastasis” (74.4% [96/129] vs. 0 [0/129]) and “Cell death” (50.0% [36/72] vs. 2.8% [2/72]) gene sets were enriched in patients without DC (Fig. 5A); more “Immunity” (15.2% [64/420] vs. 0 [0/420]), “ECM & metastasis” (48.1% [62/129] vs. 0 [0/129]) and “Cell death” (15.3% [11/72] vs. 1.4% [1/72]) gene sets were enriched in patients without iDC (Fig. 5B); no gene set was enriched in patients without OR (Fig. 5C); more “Immunity” (14.8% [62/420] vs. 0.5% [2/420]), “Cell cycle” (23.6% [30/127] vs. 0.8% [1/127]), “Genetic and epigenetic information” (41.6% [119/286] vs. 1.7% [5/286]) and “Cell death” (33.3% [24/72] vs. 0 [0/72]) gene sets were enriched in patients without iOR (Fig. 5D). The GSEA results (Fig. 5E–H) of ANN system prediction scores were similar with the GSVA results. The pathway enrichment results without consideration of statistical significance were showed in Fig. S5.
Fig. 5.
Mechanism exploration on bulk level. GSVA results of ANN system for DC (A), iDC (B), OR (C) and iOR (D). GSEA results of ANN system for DC (E), iDC (F), OR (G) and iOR (H). (I) TIME results. GSVA: gene set variation analysis; ANN: artificial neural network; DC (Efficacy): disease control; iDC: immune disease control; OR: objective response; iOR: immune objective response; GSEA: gene set enrichment analysis; TIME: tumor immune microenvironment; ECM: extracellular matrix; NA: not available; ssGSEA: single sample gene set enrichment analysis; B: B cell; CD4T: CD4+ T cell; Treg: regulatory T cell; CD8T: CD8+ T cell; M: macrophage; NK: natural killer cell; DC (Class): dendritic cell
The TIME results showed that most immune cells and scores were not related with efficacy of sintilimab, some negative relationships were observed, while positive relationships were rare (Fig. 5I).
Mechanism exploration on spatial level
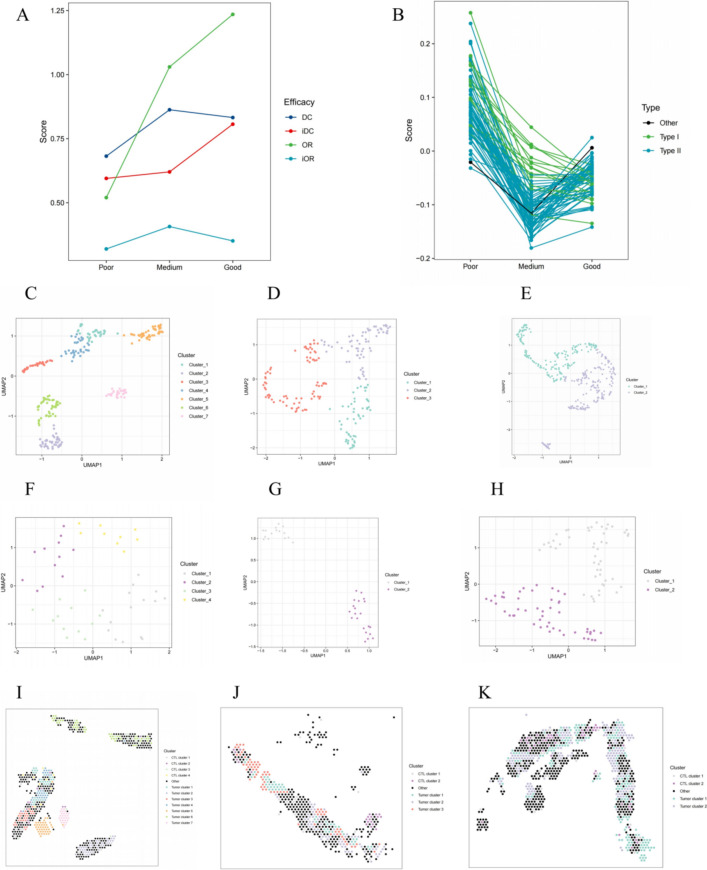
In the cohort 3, the patient with BOR of StD and PFS of 3.4 months (progressed) was defined as P1 (poor response); the patient with BOR of PR and PFS of 6.1 months (progressed) was defined as P2 (medium response); the patient with BOR of PR and PFS of 9.2 months (not progressed) was defined as P3 (good response). The four ANN system prediction scores of three patients were in Fig. 6A. The GSVA scores of most T cell related gene sets (98.6% [70/71]) in the two patients with PR were lower than that with StD, and 21.1% (15/71) gene sets (defined as type I gene sets) showed lowest scores in the patient with longer PFS (9.2 months), whereas 77.5% (55/71) gene sets (defined as type II gene sets) showed lowest scores in the patient with shorter PFS (6.1 months, Fig. 6B).
Fig. 6.
Mechanism exploration on spatial level. Predicted scores of four ANN systems (A) and GSVA results of 71 T cell related gene sets (B) in three patients from the cohort 3. Clustering result of tumor spots for P1 (C), P2 (D) and P3 (E). Clustering result of CTL spots for P1 (F), P2 (G) and P3 (H). Spatial distribution of tumor spots and CTL spots for P1 (I), P2 (J) and P3 (K). ANN: artificial neural network; GSVA: gene set variation analysis; CTL: cytotoxic T lymphocyte; DC: disease control; iDC: immune disease control; OR: objective response; iOR: immune objective response; UMAP: uniform manifold approximation and projection
There were 586, 487 and 865 spots detected from the P1, P2 and P3, respectively. Moreover, 249, 196 and 362 spots from the P1, P2 and P3, respectively, were identified as tumor spots by results of HE staining and “ESTIMATE” [19], 48, 36 and 98 spots from the P1, P2 and P3, respectively, were identified as CTL spots by results of “CIBERSORT” [20] and “quanTIseq” [21]. The best clustering number of tumor spots was seven (Fig. S6A and Fig. 6C), three (Fig. S6B and Fig. 6D) and two (Fig. S6C and Fig. 6E) in the P1, P2 and P3, respectively; the best clustering number of CTL spots was four (Fig. S6D and Fig. 6F), two (Fig. S6E and Fig. 6G) and two (Fig. S6F and Fig. 6H) in the P1, P2 and P3, respectively. Spots in the same cluster were also close in the spatial distribution (Fig. S6G–I and Fig. 6I–K).
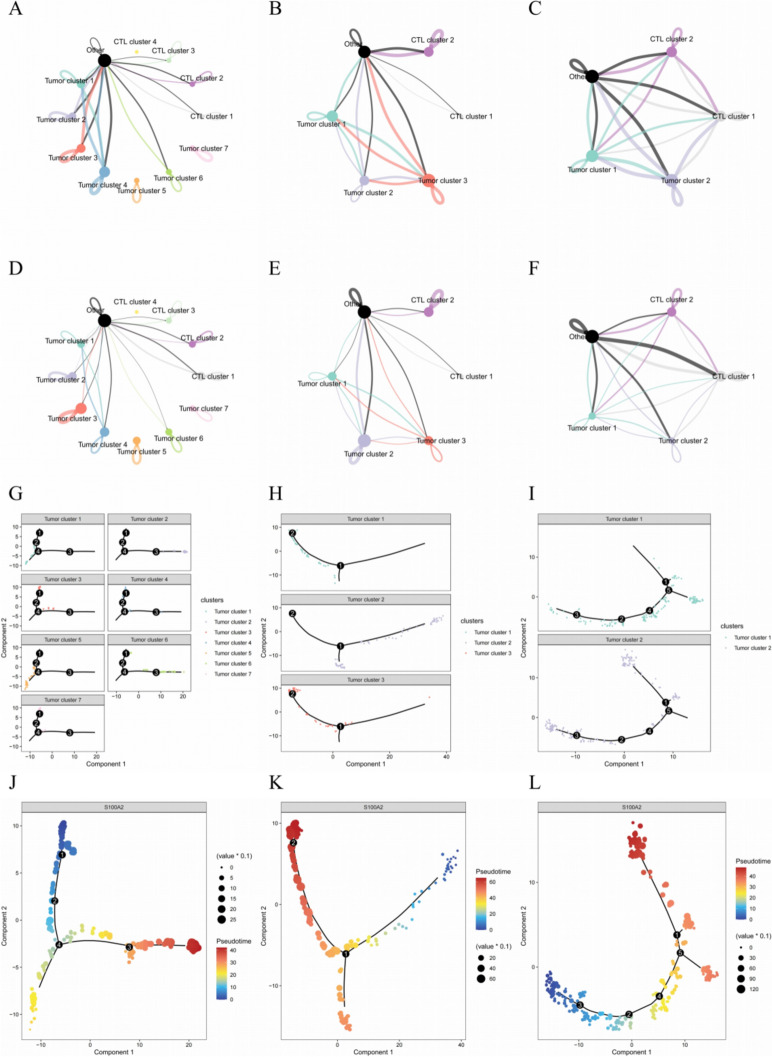
Results of cell–cell communication analysis showed that P1 with poor response to immunotherapy had the minimum communication count (Fig. 7A and Fig. S7A); P2 with medium response to immunotherapy had the medium communication count (Fig. 7B and Fig. S7B); P3 with good response to immunotherapy had the maximum communication count (Fig. 7C and Fig. S7C). The communication strength showed similar results (Fig. 7D–F and Fig. S7D–F). In terms of the complex of communications, the P1 were mainly had internal signals of each clusters, whereas the P2 had signals between tumor clusters at the same time; as for the P3, the communications linked different tumor and CTL clusters together. Therefore, P1 with the maximum number of clusters showed minimum communication, which represented the highest heterogeneity, while the P3 had the opposite.
Fig. 7.
Results of cell–cell communication analysis and pseudotime analysis. Net plot of interaction counts for P1 (A), P2 (B) and P3 (C). Net plot of interaction strength for P1 (D), P2 (E) and P3 (F). Distribution of different tumor spot clusters in trajectory for P1 (G), P2 (H) and P3 (I). Identification of the direction of trajectory by the expression of S100A2 for P1 (J), P2 (K) and P3 (L). S100A2: S100 calcium binding protein A2; CTL: cytotoxic T lymphocyte
Results of pseudotime analysis showed that different tumor clusters were located in different sites of trajectory (Fig. 7G–I). The expression of S100A2 and some of keratin-related genes was used to determine the direction of trajectory (Fig. 7J–L and Fig. S7G–L), and the distribution of pseudotime values were different among tumor clusters (Fig. 7M–O).
Discussion
Immunotherapy had brought clinical benefits to patients with lung cancer, but tools on predicting the efficacy of immunotherapy were still limited both in quantity and accuracy. The most commonly used predictive tool was PD-L1 [5], which was an index determined by the immunohistochemistry (IHC) staining of PD-L1. Some predictive tools were calculated by multiple indexes and complex methods, like (TMB) [9], FGA [10], immunophenoscore (IPS) [14], Tumor Immune Dysfunction and Exclusion (TIDE) score [15] and IFN-γ-related mRNA profile [28]. These predictive tools were built in one or some types of cancer, then were generically used in other types of cancer, which might brought inaccuracy and misleading results. For example, TMB played different roles in different types of cancer, in the type I cancers (including lung adenocarcinoma, bladder cancer, melanoma and colorectal cancer), TMB was related to better response to immunotherapy, while TMB was unrelated or related to worse response to immunotherapy in the type II cancers, including esophageal cancer, sqNSCLC, breast cancer, glioma, clear cell renal cell carcinoma, head and neck squamous cell carcinoma and gastric cancer [12]. Therefore, the same predictive tool had different predictive values among types of cancer. Although, the TIDE score had distinguished input types as NSCLC, melanoma and other, sqNSCLC and non-sqNSCLC were still mixed together [15]. Our previous study showed that better response to immunotherapy in lung adenocarcinoma was related to more antitumor immune cell (including CD8+ T cell and type I macrophage) and less protumor immune cell (regulatory T cell and type II macrophage) infiltration, moreover better response was also related to more active immune-related gene sets [29]. In this study, better response to immunotherapy in sqNSCLC almost not related to immune cell infiltration, and better response was related to less active immune-related gene sets, which suggested that sqNSCLC and non-sqNSCLC were different on transcriptomic level and the mechanism of response to immunotherapy might be different. As a result, it is necessary to build predictive tools for sqNSCLC excluding other histologic types of lung cancer.
This study had built four ANN systems to predict response to sintilimab in sqNSCLC, which showed higher accuracy than according linear models’ systems except the DC model. The correlation between the expression of each gene constructing the ANN system and the final predicted scores were weak because of the nonlinear fitting of ANN model. Moreover, stability of the four ANN systems was evaluated to simulate the real world usage scenarios, for example expression of some genes were NA because of different panel of test. We used the mean normalized expression of missing genes in 59 patients from the cohort 1 to replace the missing values, and evaluated influences of missing genes with the number from one to seven. Results of stability evaluation showed that the AUC of most combinations of gene expression missing were over than 0.80, and the width of according 95% CI were less than 0.20, which meant the four ANN systems had high stability. This study had explored the relationship between final predicted scores of the four ANN systems and pathways activities. Interestingly, more “Immunity” gene sets were enriched in the low efficacy group with lower predicted scores. Moreover, results of TIME didn’t provide any valuable explanation.
To further validate this seemingly contradictory phenomenon, spatial sequencing was performed in the independent cohort 3. Similar to results of the bulk level, the GSVA scores of most T cell related gene sets in the two patients with PR were lower than that with StD on the spatial level. Further analysis based on the spatial information was performed, and results showed that patient with worse response to immunotherapy accompanied with more clusters of tumor and CTL spots and weaker cell–cell communications. In other words, worse response might be related to higher heterogeneity. A recent study suggested that although higher heterogeneity resulted by more subclones of tumor provided more mutation and neoantigen burden, but response to these subclones might not prime an efficient antitumor immunity [30]. Therefore, higher heterogeneity of tumor might lead to higher heterogeneity of CTL in TIME, the latter would result in higher diversity of immune response and might performance as more “Immunity” gene sets enrichment, which represented lower join force of cytotoxic and was not conducive to the effectiveness of immunotherapy. Many genes used as input neurons to constructed ANN systems were related to intratumoral heterogeneity and TIME.
This study constructed four ANN systems to predict the response to immunotherapy specially in sqNSCLC and explored potential mechanism on the bulk and the spatial level. Compared with constructing gene signatures in other studies, ANN system with high fitting efficiency could avoid the instability caused by different reference background genes. This study used 300 times of threefold cross validation to reduce the randomness in model construction. Removing input genes and replacing them with mean values was used to evaluate the stability of ANN systems, providing experience for the use in the future when some input genes are missing. For patients with sqNSCLC who will receive sintilimab, the baseline tissue samples can be sequenced and the RNA data are used as input to calculate the prediction scores according to our ANN systems. The binomial prediction results can also acquire base on our recommended thresholds. For those with better efficacy prediction results, sintilimab could bring benefits, whereas other treatments would be better for those with worse efficacy prediction results. There were still some limits in this study. The sample size used in model construction and mechanism exploration needed to be improved. Moreover, due to the nonlinear fitting of ANN model, it was difficult to explain the relationship between input genes and final predicted scores.
The four ANN systems showed high accuracy, robustness and stability in predicting the response to sintilimab for patients with sqNSCLC, and lower intratumoral heterogeneity and higher join force of cytotoxic might be the potential mechanism of patients with better response.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors thank the patients and their families who made this study possible and the clinical study teams who participated in the study. The authors would also like to thank Innovent Biologics, Inc.provide the research data.
Abbreviations
- ANN
Artificial neural network
- DC
Disease control
- FFPE
Formalin-fixed paraffin-embedded
- FGA
Fraction of copy number-altered genome
- GO
Gene ontology
- GSEA
Gene set enrichment analysis
- GSVA
Gene set variation analysis
- IHC
Immunohistochemistry
- KEGG
Kyoto encyclopedia of genes and genomes
- mAb
Monoclonal antibody
- NES
Normalized enrichment score
- PD-1
Programmed cell death protein 1
- PD-L1
Programmed death ligand 1
- sqNSCLC
Squamous-cell non-small cell lung cancer
- TIDE
Tumor immune dysfunction and exclusion
- TIME
Tumor immune microenvironment
- TMB
Tumor mutation burden
Author contributions
Study design, Data acquisition and manuscript review: Yuankai Shi, Puyuan Xing. Data analysis, methodology, software and visualization: Tongji Xie and Guangyu Fan. Manuscript writing:Tongji Xie, Guangyu Fan and Le Tang. Funding acquisition, investigation, project administration and supervision: Yuankai Shi.
Funding
This work was supported by the National Science and Technology Major Project for Key New Drug Development (2017ZX09304015). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed. No writing assistance was used in the production of this manuscript.
Data availability
No datasets were generated or analyzed during the current study.
Declarations
Conflict of interest
The authors declare no competing interests.
Consent for publication
All authors agreed to submit for consideration for publication in this journal.
Ethics approval and consent to participate
The ORIENT-3 study was approved by the Ethics Committee of each study center. Written informed consent was obtained from all patients of the ORIENT-3 study. The designation of the cohort 3 was approved by the institutional review board of the Cancer Hospital, CAMS (No.23/262-4004). The study was performed in full accordance with the guidelines for Good Clinical Practice and the Declaration of Helsinki.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Tongji Xie, Guangyu Fan and Le Tang contributed equally to this study.
Contributor Information
Puyuan Xing, Email: xingpuyuan@cicams.ac.cn.
Yuankai Shi, Email: syuankai@cicams.ac.cn.
References
- 1.Bray F, Laversanne M, Sung H et al (2024) Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 74(3):229–263. 10.3322/caac.21834 [DOI] [PubMed] [Google Scholar]
- 2.Xia C, Dong X, Li H et al (2022) Cancer statistics in China and United States, 2022: profiles, trends, and determinants. Chin Med J 135(5):584–590. 10.1097/CM9.0000000000002108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thai AA, Solomon BJ, Sequist LV, Gainor JF, Heist RS (2021) Lung cancer. Lancet 398(10299):535–554. 10.1016/S0140-6736(21)00312-3 [DOI] [PubMed] [Google Scholar]
- 4.Gogishvili M, Melkadze T, Makharadze T et al (2022) Cemiplimab plus chemotherapy versus chemotherapy alone in non-small cell lung cancer: a randomized, controlled, double-blind phase 3 trial. Nat Med 28(11):2374–2380. 10.1038/s41591-022-01977-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reck M, Rodríguez-Abreu D, Robinson AG et al (2016) Pembrolizumab versus chemotherapy for PD-L1-positive non-small-cell lung cancer. N Engl J Med 375(19):1823–1833. 10.1056/NEJMoa1606774 [DOI] [PubMed] [Google Scholar]
- 6.Ren S, Chen J, Xu X et al (2022) Camrelizumab plus carboplatin and paclitaxel as first-line treatment for advanced squamous NSCLC (CameL-Sq): a phase 3 trial. J Thorac Oncol 17(4):544–557. 10.1016/j.jtho.2021.11.018 [DOI] [PubMed] [Google Scholar]
- 7.Rittmeyer A, Barlesi F, Waterkamp D, et al. (2017) Atezolizumab versus docetaxel in patients with previously treated non-small-cell lung cancer (OAK): a phase 3, open-label, multicentre randomised controlled trial [published correction appears in Lancet. 389(10077):e5]. Lancet. 389(10066):255–265. 10.1016/S0140-6736(16)32517-X [DOI] [PMC free article] [PubMed]
- 8.Shi Y, Wu L, Yu X et al (2022) Sintilimab versus docetaxel as second-line treatment in advanced or metastatic squamous non-small-cell lung cancer: an open-label, randomized controlled phase 3 trial (ORIENT-3). Cancer Commun 42(12):1314–1330. 10.1002/cac2.12385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rizvi NA, Hellmann MD, Snyder A et al (2015) Cancer immunology: mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 348(6230):124–128. 10.1126/science.aaa1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rizvi H, Sanchez-Vega F, La K, et al. (2018) Molecular Determinants of Response to Anti-Programmed Cell Death (PD)-1 and Anti-Programmed Death-Ligand 1 (PD-L1) Blockade in Patients With Non-Small-Cell Lung Cancer Profiled With Targeted Next-Generation Sequencing [published correction appears in J Clin Oncol. 2018 Jun 1;36(16):1645]. J Clin Oncol. 36(7):633–641. 10.1200/JCO.2017.75.3384. [DOI] [PMC free article] [PubMed]
- 11.Dempke WCM, Fenchel K, Dale SP (2018) Programmed cell death ligand-1 (PD-L1) as a biomarker for non-small cell lung cancer (NSCLC) treatment-are we barking up the wrong tree? Transl Lung Cancer Res 7(Suppl 3):S275–S279. 10.21037/tlcr.2018.04.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McGrail DJ, Pilié PG, Rashid NU et al (2021) High tumor mutation burden fails to predict immune checkpoint blockade response across all cancer types. Ann Oncol 32(5):661–672. 10.1016/j.annonc.2021.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang S, Liu Y, Chen J et al (2020) Autoantibody signature in hepatocellular carcinoma using seromics. J Hematol Oncol 13(1):85. 10.1186/s13045-020-00918-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Charoentong P, Finotello F, Angelova M et al (2017) Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep 18(1):248–262. 10.1016/j.celrep.2016.12.019 [DOI] [PubMed] [Google Scholar]
- 15.Jiang P, Gu S, Pan D et al (2018) Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat Med 24(10):1550–1558. 10.1038/s41591-018-0136-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Swanton C, Govindan R (2016) Clinical implications of genomic discoveries in lung cancer. N Engl J Med 374(19):1864–1873. 10.1056/NEJMra1504688 [DOI] [PubMed] [Google Scholar]
- 17.Wu T, Hu E, Xu S et al (2021) clusterProfiler 40: a universal enrichment tool for interpreting omics data. Innovation 2(3):100141. 10.1016/j.xinn.2021.100141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hänzelmann S, Castelo R, Guinney J (2013) GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinform 14:7. 10.1186/1471-2105-14-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoshihara K, Shahmoradgoli M, Martínez E et al (2013) Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun 4:2612. 10.1038/ncomms3612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Newman AM, Liu CL, Green MR et al (2015) Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 12(5):453–457. 10.1038/nmeth.3337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sturm G, Finotello F, List M (2020) Immunedeconv: an R package for unified access to computational methods for estimating immune cell fractions from bulk RNA-sequencing data. Methods Mol Biol 2120:223–232. 10.1007/978-1-0716-0327-7_16 [DOI] [PubMed] [Google Scholar]
- 22.Hao Y, Hao S, Andersen-Nissen E et al (2021) Integrated analysis of multimodal single-cell data. Cell 184(13):3573-3587.e29. 10.1016/j.cell.2021.04.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Charrad M, Ghazzali N, Boiteau V, Niknafs A (2014) NbClust: an R package for determining the relevant number of clusters in a data set. J Stat Softw 61(6):1–36. 10.18637/jss.v061.i06 [Google Scholar]
- 24.Jin S, Guerrero-Juarez CF, Zhang L et al (2021) Inference and analysis of cell-cell communication using Cell Chat. Nat Commun 12(1):1088. 10.1038/s41467-021-21246-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trapnell C, Cacchiarelli D, Grimsby J et al (2014) The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol 32(4):381–386. 10.1038/nbt.2859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qiu X, Hill A, Packer J, Lin D, Ma YA, Trapnell C (2017) Single-cell mRNA quantification and differential analysis with Census. Nat Methods 14(3):309–315. 10.1038/nmeth.4150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qiu X, Mao Q, Tang Y et al (2017) Reversed graph embedding resolves complex single-cell trajectories. Nat Methods 14(10):979–982. 10.1038/nmeth.4402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ayers M, Lunceford J, Nebozhyn M, Murphy E, Loboda A, Kaufman DR, Albright A, Cheng JD, Kang SP, Shankaran V, Piha-Paul SA, Yearley J, Seiwert TY, Ribas A, McClanahan TK (2017) IFN-γ-related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest 127(8):2930–2940. 10.1172/JCI91190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xie T, Fan G, Huang L, Lou N, Han X, Xing P, Shi Y (2022) Analysis on methylation and expression of PSMB8 and its correlation with immunity and immunotherapy in lung adenocarcinoma. Epigenomics 14(22):1427–1448. 10.2217/epi-2022-0282 [DOI] [PubMed] [Google Scholar]
- 30.Westcott PMK, Muyas F, Hauck H, Smith OC, Sacks NJ, Ely ZA, Jaeger AM, Rideout WM 3rd, Zhang D, Bhutkar A, Beytagh MC, Canner DA, Jaramillo GC, Bronson RT, Naranjo S, Jin A, Patten JJ, Cruz AM, Shanahan SL, Cortes-Ciriano I, Jacks T (2023) Mismatch repair deficiency is not sufficient to elicit tumor immunogenicity. Nat Genet 55(10):1686–1695. 10.1038/s41588-023-01499-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No datasets were generated or analyzed during the current study.