Abstract
We explored the biological mechanisms by which curcumin (Cur) confronts osteosarcoma (OS) tumorigenesis and potential drug gene targets based on network pharmacology and in vitro cell experiments. Cur has been recognized for its significant role in combating various types of tumors. However, the intrinsic molecular mechanisms through which it affects OS remain uncharted. In this study, we performed network pharmacology methods including protein-protein interaction (PPI) and core target screening, Functional Enrichment Analysis and Network Construction, Molecular Docking, which obtained the potential target of Cur. Meanwhile, cell experiments (wound healing assay, Transwell assay, Western blots, immunofluorescence, et al.) in vitro were performed to verify the targets, and reveal the biological mechanisms. A total of 18 hub genes were identified through our network pharmacological analysis. In vitro studies show that Cur inhibits the proliferation, migration, invasion capabilities of MG63 and U2OS cells. Western blot reveals a down-regulation of p-PI3K, PI3K, p-Akt, Akt, EGFR, Src, p-Src (Tyr416) and STAT3 expression when treated with Cur. Additionally, Cur upregulated epithelial proteins (E-cadherin and Occludin) while decreasing the expression of the mesenchymal protein (N-cadherin). In addition, Cur treatment decreases the EGFR/Src signaling pathway in the presence of active Src overexpression. Cur inhibits the proliferation, migration, invasion, epithelial-mesenchymal transition (EMT) by down-regulating EGFR/Src signaling axis, also resulting in coordinated weakening of its downstream regulatory genes, including Akt, STAT3, Bcl2, ERK1/2, among others signal axis (PI3K/Akt signaling pathway).
Keywords: Curcumin, Osteosarcoma, EGFR, Src, PI3K/Akt
Highlights
Curcumin inhibits the proliferation, migration, invasion, and epithelial-mesenchymal transition capabilities of MG63 and U2OS cells.
The anti-osteosarcoma effect of curcumin depends on the regulation of EGFR and Src based on network pharmacological analysis.
Curcumin downregulates the EGFR/Src signaling axis, which leads to a coordinated weakening of its downstream regulatory genes and other signaling axes (PI3K/Akt signaling pathway) in osteosarcoma cells.
Introduction
Osteosarcoma (OS) is one of the most common primary bone cancers and tends to occur in children and adolescents. 1 Currently, surgical treatment is the primary therapeutic strategy for OS in clinical practice. 2 However, studies conducted at the Rizzoli Institute have shown that the mortality rate remains high for patients with extremity OS who have lung metastases, even when treated with multiple treatments such as chemotherapy and surgery. 3 While chemotherapy treatment is often associated with collateral toxic effects, which impair the health of organs, 4 radiotherapy can serve as a local treatment option for unresectable tumors, as well as palliative care for symptomatic metastases following intralesional resection.5–7 Even with the assistance of existing imaging techniques, the improvement in patients with metastatic, recurrent, or unresectable OS is still not optimistic. 8 Therefore, it is imperative to investigate novel anti-OS drugs and explore their mechanisms of action.
Curcumin (Cur), a yellow compound derived from the natural spice turmeric, has been found to have multifarious pharmacological effects, including antioxidant, anti-inflammatory, and radical-scavenging capabilities, which are beneficial to human health. A wealth of studies over the past few decades has underscored the health benefits of this ingredient.9–11 Although relevant studies have demonstrated the anti-cancer activity of Cur against OS,12–14 the intrinsic molecular mechanisms through which it affects OS remain uncharted. Mechanistically, the association among the roles of epidermal growth factor receptor (EGFR) and Src, as well as their involvement in the epithelial-mesenchymal transition (EMT), remain unclear.
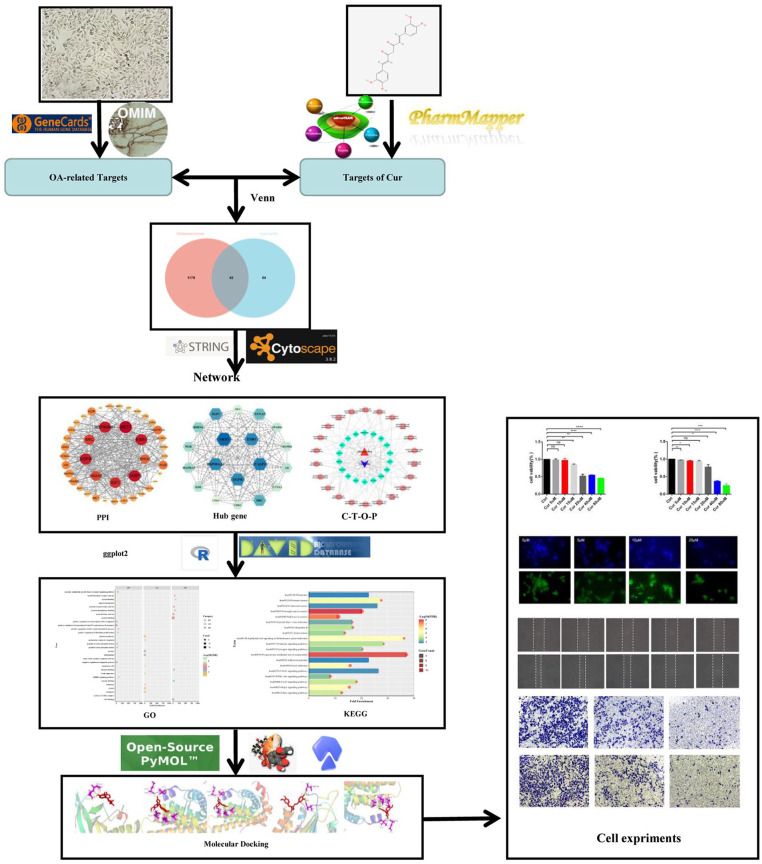
Network pharmacology is an interdisciplinary approach combining computer science and medicine. It differs from conventional drug discovery methods by focusing on drugs’ ability to target multiple proteins or networks involved in a disease.15,16 Therefore, network pharmacology may facilitate our analysis to explore the interactions between target proteins and small molecules from a biological perspective. 17 The procedures used in this study are depicted in Figure. 1. In detail, We utilized network pharmacology to explore the regulatory mechanisms of Cur on OS through molecular dynamics simulations. Our analysis identified specific targets of Cur for suppressing OS, including EGFR, Akt, Src, CDK2, among others. We also confirmed and validated the computational findings in wet lab. Our in vitro study shows that low doses of Cur exhibit significant inhibitory effects on the proliferation, migration, and invasion in MG63 and U2OS cells. The inhibitory mechanism is primarily attributed to the down-regulation of the EGFR/Src signaling axis. This, in turn, weakens the downstream regulatory proteins Akt and STAT3, and related signaling pathways such as PI3K/Akt, ultimately inhibiting migration, invasion, proliferation, and EMT of OS cells.
Figure 1.
Workflow chart of the research.
Materials and methods
Potential targets collection
Potential targets for Cur were retrieved from PharmMapper (http://www.lilab-ecust.cn/pharmmapper), 18 and we selected the reliable targets under the condition of norm fit > 0.5. The chemical composition structure was collected from PubChem (https://pubchem.ncbi.nlm.nih.gov/). The admetSAR (http://lmmd.ecust.edu.cn/admetsar2/) platform was used to predict the properties of Cur, such as absorbance. 19 OS-related targets were obtained via screening both GeneCards (https://www.genecards.org) and OMIM (https://omim.org) databases. In the Genecards database, closer relationship analysis to the disease through a higher score value. Targets with a relevance score >1 were considered as OS-related targets. The intersection of drug and disease targets was viewed as potential targets. Finally, we normalized the targets to their equivalent nomenclature and converted them into the corresponding gene names specific to Homo sapiens using the Uniprot database (https://sparql.uniprot.org/).
Protein-protein interaction and core targets screening
The shared gene targets were incorporated into the STRING database (https://cn.string-db.org/). The organism was specified as Homo sapiens, and a confidence score for the association network was set to medium confidence (0.4). The protein-protein interaction (PPI) network was then saved as a TSV file and loaded into CytoScape 3.8.2 for further analysis. CentiScape, a Cytoscape plug, was used to examine the topological properties of network, namely “degree”, “betweenness centrality”, and “closeness centrality”. Genes with degree values greater than the average were identified as hub genes.
Functional enrichment analysis and network construction
The shared targets and Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were analyzed online using the DAVID Database (https://david.ncifcrf.gov/). GO terms and KEGG pathways with a p-value less than or equal to 0.05 were recognized as significant. To directly analyze the association between Curcumin (Cur) and Overall Survival (OS), and the candidate targets and OS-related signaling pathways, a Cur-Target-OS-Pathway (C-T-O-P) network was constructed using Cytoscape 3.8.2 software.
Molecular docking
AutoDockTools-1.5.6 can perform molecular docking analysis according to the semi-flexible docking principle. PyMOL is software used for preparing small molecule components and proteins before docking. OpenBabelGui is software that converts chemical structure formats.
First, we selected core targets in the PPI as potential receptors based on the degree value. Cur’s 3D structures in SDF format were obtained from the PubChem database, and the hub targets were inputted into the RCSB PDB database (https://www.rcsb.org/) to create the protein’s PDB format. Second, both the water molecules and original residues were removed from the target protein using PyMOL, and OpenBabel was used to convert the SDF format of Cur into MOL2 format. Next, the target proteins were inputted into AutoDockTools-1.5.6 for hydrogenation, charge calculation, and determination of an atom’s rigid structure. Simultaneously, the ligand was processed and saved in PDBQT format. Lastly, AutoDock was executed for molecular docking, and PyMOL was used to visualize the results.
Reagents
Cur (SC8670) and CCK-8 (CA1210) were purchased from Solarbio (Beijing, China). For western blots, primary antibody against GAPDH (60004-1-Ig), PI3K (20584-1-AP), EGFR (66455-1-Ig), Src (11097-1-AP), p-Akt (66444-1-Ig), Akt (10176-2-AP), Bcl2 (68103-1-Ig), STAT3 (10253-2-AP), α-tubulin (66031-1-Ig) were obtained from Proteintech group (Wuhan, China). Rabbit polyclonal antibodies p-Src (Tyr416) (#6943) were obtained from Cell Signaling Technology (Shanghai, China). p-PI3K (ab182651) were purchased from Abcam (Cambridge, USA). The secondary antibody against Rabbit polyclonal antibodies (SA00001-2), the secondary antibody against monoclonal antibodies (SA00001-1) were obtained from Proteintech group (Wuhan, China). The construction of Src-WT, Src-CA mutants and Cherry-Rab7 was carried out as per previously reported methods. 20 EdU assay kit (C0071S) was obtained from Beyotime biotechnology co, ltd (shanghai, China). Phalloidin (HY-P0028) was gained from MedChemExpress (New Jersey, USA).
Cell culture
MG63 and U2OS cells were obtained from Chinese Academy of Sciences (Shanghai China) and were cultured in high-glucose DMEM containing 10% fetal bovine serum (FBS). The culture conditions were maintained in an incubator to ensure an optimal environment, which required a constant temperature of 37°C, a stable CO2 concentration of 5%, a pH range of 7.2-7.4, and a high relative humidity of 95%.
Cell counting kit-8 (CCK-8) assay
we estimated the viability of MG63 and U2OS cells. The cells (2000 cells / well) were seeded into 96-well plates for 12 h at 37℃ with 5% CO2 in a atmosphere. Following incubation, the cells were washed with Dulbecco’s Phosphate Buffer Saline (PBS) three times before being exposed to the following treatments for 24 hours: 0 (containing 0.1% DMSO), 5, 10, 15, 20, and 40 μM Cur. And the cell medium was replaced with fresh medium containing 10% CCK8 after treatment. Subsequently, the OD value was measured via performing using a microplate reader at 450 nm (Huisong, MB-580) after an additional 2 hours of incubation at 37°C.
Transwell assay for migration and invasion
The procedure for the migration and invasion assays is described as follows: The cells were placed into a serum-free upper chamber for migration evaluation. An appropriate number of cells were added to 200 μL of serum-free DMEM in the upper chamber. After incubation for 24 h at 37℃, we observed the OS cells that migrated to lower chamber (DMEM containing 10% FBS). the chambers were fixed with 4% paraformaldehyde (PFA) for 30 min and stained with 0.1% crystal violet for 15 min. Subsequently, the cells in the upper chamber were wiped away, and the cells on the bottom were used for observation, photography, and counting under an inverted microscope (OLYMPUS, CKX53). The invasion test was replaced with a pre-paved Matrigel chamber, following the same procedure as the migration assay.
5′-ethynyl-2′-deoxyuridine (EdU) assay
we assessed the proliferation ability of cells according to the manufacturer’s instructions. Briefly, U2OS and MG63 cells were seeded into 12-well plates at an appropriate density about 48 h. EdU buffer was added into the wells and were label about 2 h. Then, we fixed and permeated with 4% formaldehyde and 0.1% Triton X-100, respectively. The cellular nuclei were stained with Hoechst 33342 after the EdU reaction, and photographed and count the stained cells using a fluorescence microscope (OLYMPUS, IX83).
Wound healing assay
The experimental steps are described below: Cells were seeded at an optimal concentration (5 x105 cells / mL) into a 6-well plate. A “Wound” was scraped into the cell layer using a 200 μL pipette tip. Then, The cells were then washed with PBS. The OS cells were treated with varying concentrations (0, 5, 10, 15, 20 μM) of cur for 24 h under high-glucose DMEM (containing 2% FBS) after PBS cleaning. The distance of the cells was measured and recorded using a microscope (OLYMPUS, IX83).
Colony formation experiment
MG63 and U2OS cells (1000 cells/well) in the logarithmic growth phase were seeded into 6-well plates, and treated with varying concentrations of Cur (0, 5, 10, 20 μM) and cultured for 14 days at 37℃. Subsequently, they were fixed with 4% formaldehyde, and stained with 0.1% crystal violet solution after PBS cleaning. Finally, the plates were rinsed with PBS solution and the cells were observed using a microscope after drying.
Western blotting
The Western Blot procedure is as follows: Cells were lysed with RIPA lysis buffer (containing phosphatase inhibitor Cocktail II and phenylmethanesulfonyl fluoride) on ice. The concentration of the protein was quantified using a BCA kit. An equal amount of total protein was loaded onto the gel and separated via SDS-PAGE electrophoresis. Subsequently, the membranes were blocked with 5% skim milk or bovine serum albumin (BSA) at room temperature after the protein transfer process. The membranes were then incubated with primary antibodies overnight at 4°C, followed by incubation with secondary antibodies (1:5000) for 1 hour after washing. The blots were detected using an ECL kit and photographed using an imaging system (BIO-RAD, ChemiDoc MP). The gray values of the proteins were analyzed and quantified using ImageJ software.
Immunofluorescence
The samples were fixed and permeabilized with 4% formaldehyde and 0.1% Triton X-100, respectively. After sealing with 3% BSA, they were incubated with a primary antibody. Subsequently, a secondary antibody (1:100) was applied, followed by PBS washing. Finally, the cells were sealed with an antifade solution containing DAPI and observed using a fluorescence microscope (OLYMPUS, IX83).
Statistical analyses
We performed quantitative analysis of the results from at least three independent experiments using Prism 9.0. A two-tailed Student’s t-test was conducted to estimate statistical significance. The bar graph represents the mean ± SEM from three independent experiments. Differences between the mean values of two groups were considered statistically significant at *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001, while “ns” indicates no statistically significant difference between the two groups.
Results
Network pharmacology analysis
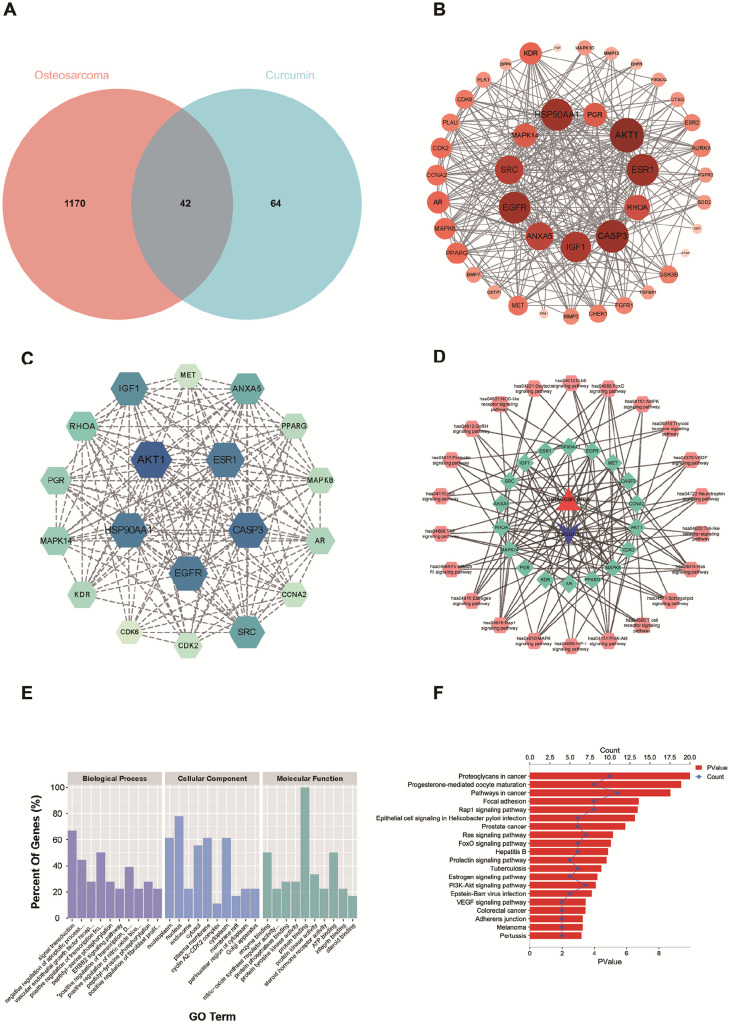
The chemical structure of Cur and its corresponding ADMET information are detailed in Table S1. As shown in Figure. 2A, 1212 disease targets were selected from GeneCards and OMIM. The predicted targets were compiled using the PharmMapper database, yielding 106 drug targets. The Venn diagram indicates that there are 42 overlapping targets for OS and Cur. These common targets were submitted to the STRING database for protein-protein interaction (PPI) network construction (Figure. 2B). Interestingly, we found no proteins that did not interact with others within this network. The resulting PPI network comprises 42 proteins and 309 edges, with an average node degree of 14.7. Degree, betweenness, and closeness are considered important parameters based on topological analysis. As shown in Figure. 2C, we selected genes with degree values above 14.7 as core genes. There are 18 core genes, and some of the parameters are presented in Table S2. It can be concluded that AKT1, CASP3, EGFR, HSP90A1, ESR1, IGF1, Src, ANXA5, RHOA, MAPK14, PGR, KDR, AR, PPARG, MAPK8, CDK2, CCNA2, and MET are significant targets for Cur intervention in OS.
Figure 2.
(A) Venn diagram depicting the number of commen targets of between Cur and OS; (B) Protein-protein interaction network analysis; (C) GO functional enrichment and KEGG pathway enrichment of commen targets; (D) Construction of C-T-O-P network; (E, F) GO functional enrichment and KEGG pathway enrichment of common gene targets.
We extracted all signaling pathways from the results to construct a C-T-O-P network, which is displayed in Figure. 2D. The diagram includes 22 signaling pathways and 18 targets. The top five significant signaling pathways are the Rap1 signaling pathway (hsa04015), Ras signaling pathway (hsa04014), FoxO signaling pathway (hsa04068), PI3K/Akt signaling pathway (hsa04151), and VEGF signaling pathway (hsa04370). In addition, results from GO enrichment analysis identified 118 biological processes (BP), 15 cellular components (CC), and 32 molecular functions (MF). The top 10 significantly enriched items in BP, CC, and MF are displayed in Figure. 2E. The anti-osteosarcoma (anti-OS) mechanisms of Cur primarily involve enzyme binding, signal transduction, nitric oxide synthase regulator activity, negative regulation of the apoptotic process, protein phosphatase binding, and vascular endothelial growth factor receptor (VEGFR) signaling. KEGG enrichment analysis yielded 60 pathways. The top 20 significant pathways are listed in Figure. 2F. These pathways include proteoglycans in cancer, progesterone-mediated oocyte maturation, other pathways in cancer, focal adhesion, the Rap1 signaling pathway, Ras signaling pathway, FoxO signaling pathway, PI3K/Akt signaling pathway, and others.
Molecular docking
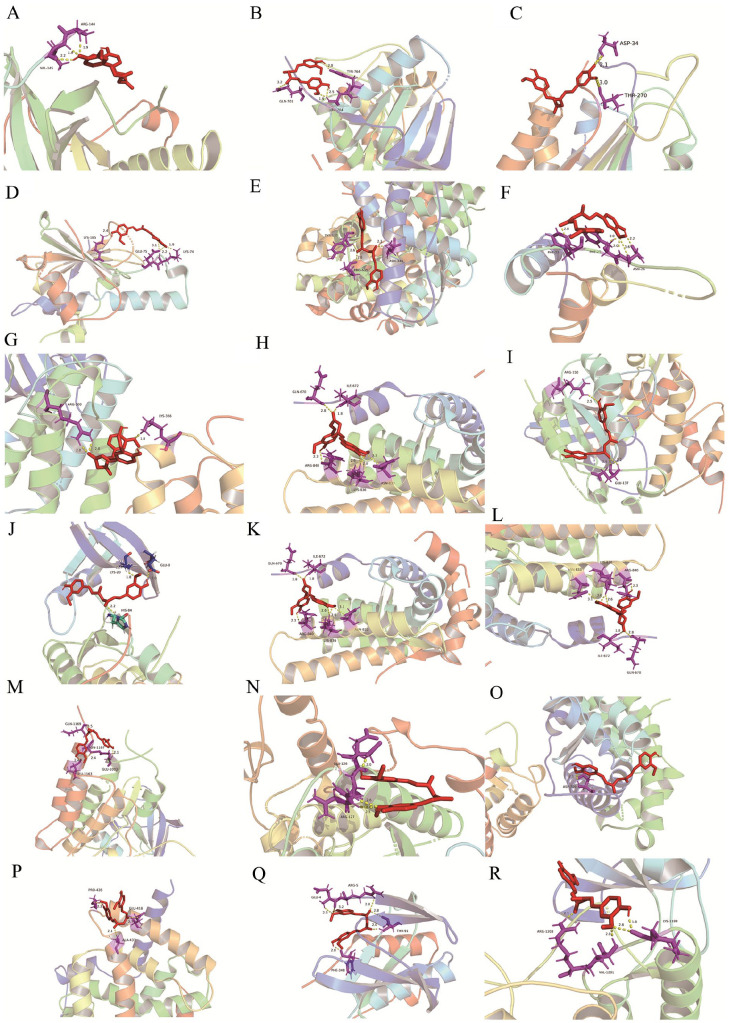
Affinity is commonly used to evaluate the strength of the interaction between a ligand and its receptor. In general, a binding energy of less than -1.2 kcal/mol (or -5 kJ/mol) suggests that the interaction is likely to be stable. Figure. 3 shows the results of molecular docking performed between Cur and the hub gene targets. As shown in Table S3, all binding energy values are less than zero, indicating a strong affinity between the compound and the hub target genes. This suggests that Cur may interfere with the bonding of the docking pocket to the target receptor, thus playing a pivotal role in the treatment of OS.
Figure 3.
Binding of Cur with (A) AKT (4ejn), (B) EGFR (5xdk), (C) CASP3 (3kjf), (D) HSP90AA1 (3bm9), (E) ESR1 (4pxm), (F) IGF1 (limx), (G) Src (4mxo), (H) ANA5 (lnvr), (I) RHOA (3lxr), (J) CDK2 (laq1), (K) AR (lt5z), (L) CCNA (2c5n), (M) KDR (3c7r), (N) MAPK8 (3elj), (O) PGR (2w8y), (P) PPARG (lzgy), (Q) MAPK14 (3fmh) and (R) MET (5ya5) by molecular docking.
Cytotoxicity of Cur on OS cells
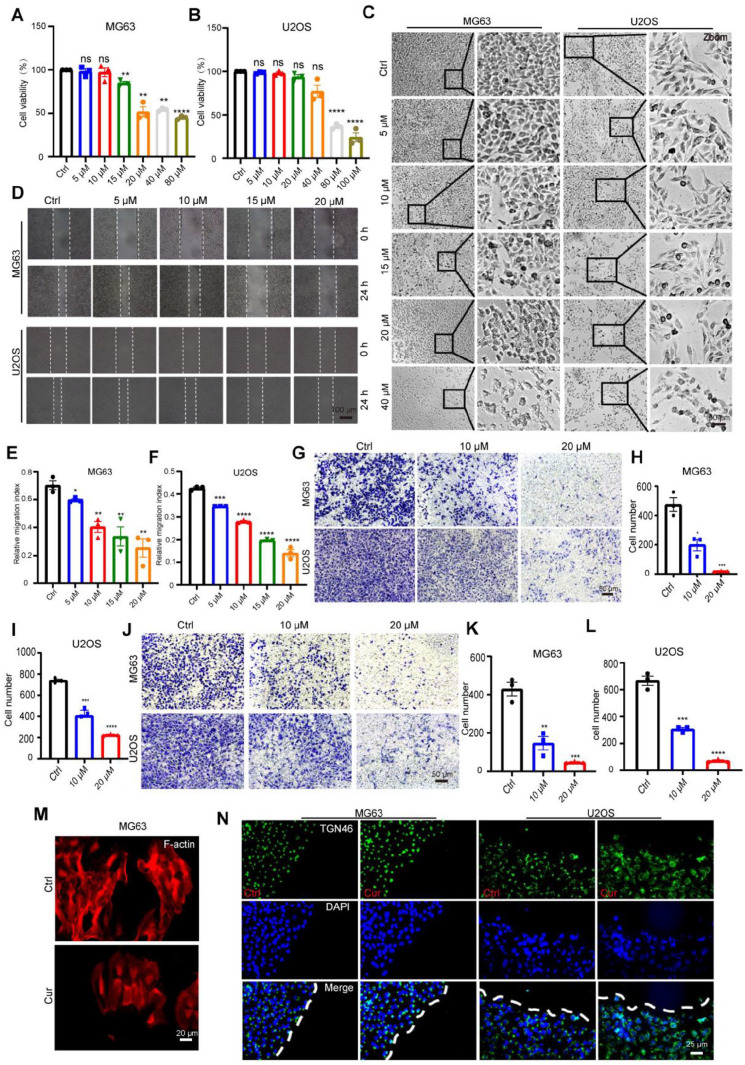
We first evaluated the toxicity of Cur on MG63 and U2OS cells using the CCK-8 assay. As shown in Figure. 4A-C, Cur exposure exhibited a dose-dependent inhibitory effect on cell survival. Cur (above 15 μM) inhibited the cell viability of MG63 cells, and at a dose of 20 μM, the cells exhibited morphological shrinkage. In U2OS cells, inhibition of cell viability was observed only when the concentration reached 80 µM, and morphological changes in these cells could be seen under bright field microscopy.
Figure 4.
Cur inhibits the migration and invasion of MG63, U2OS cells. (A,B) Cell viability of MG63, U2OS cells treated with different concentrations of Cur for 24 h, respectively; (C) Morphological changes of cells under different concentrations (0 μM, 5 μM, 10 μM, 15 μM, 20 μM, 40 μM) of Cur treatment about 24 h, Bar=100 um;(D) Wound healing distance of MG63, U2OS cells after 24 h of different concentrations Cur treatment;(E, F). Relative quantification of wound healing distance in Fig. 4D; (G) The number of cells transferred into lower chamber after 24 h of Cur treatment, Bar=50 um; (H, I) Relative quantification of cell number in Fig. 4G; (G) The number of cells invaded into lower chamber after 24 h of Cur treatment (0 μM, 10 μM, 20 μM), Bar=50 um; (K. L) Relative quantification of cell number in Fig. 4J. (M) F-actin labeled skeletal changes in cells treated with Cur (10 μM) treatment, Bar=20 um; (N) Golgi reorientation ability in Ctrl, Cur treatment group during migration, respectively, Bar=25 um.
Inhibition of Cur on migration and invasion of OS cells
We further performed wound healing experiments to determine the regulatory effect of Cur on the migration of OS cells. The results (Figure. 4D) show that OS cells treated with Cur exhibit a reduced migration distance, indicating a diminished migratory capacity. The Transwell migration experiment further confirmed this finding (Figure. 4G). F-actin, labeled with phalloidin, displayed more active membrane ruffling structures in the Cur-treated group (Figure. 4M), a phenomenon that facilitates cell migration. However, Figure. 4N shows that Cur does not inhibit the reorientation of the Golgi apparatus during cell migration (labeled with TGN46). Meanwhile, results from the Matrigel invasion assay (Figure. 4J) showed that Cur treatment reduces the number of cells in the Matrigel-precoated Transwell. This suggests that Cur also weakens the invasion capacity of the cells. However, further functional experiments are still needed to fully explore these effects.
Cur inhibits the proliferation OS cells
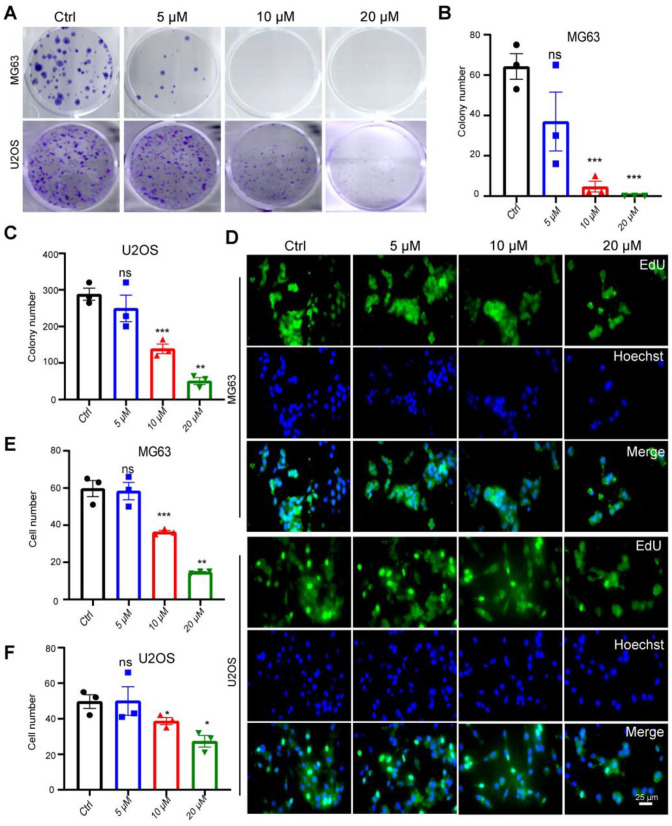
The regulatory effect of Cur on the proliferation of OS cells was determined by a colony formation assay. The results, as depicted in Figure. 5A, demonstrate a decrease in the number of colonies following Cur treatment in both MG63 and U2OS cells, indicating a reduction in the proliferation capacity of the cells. This finding was further confirmed using EdU proliferation staining. The outcomes presented in Figure. 5D suggest that higher Cur concentrations correspond to lower levels of cell proliferation.
Figure 5.
Cur inhibits the proliferation of MG63, U2OS cells. (A) colony number of MG63, U2OS cells after 2 weeks of Cur treatment (0 μM, 5 μM, 10 μM, 15 μM, 20 μM); (B,C) Relative quantification of colony number in figure A; (D) Edu Fluorescence positive cells after 48 h of Cur treatment (0 μM, 5 μM, 10 μM, 15 μM, 20 μM) in MG63, U2OS cells, Bar=25 um; (E,F) Relative quantification of cells in Fig. 5D.
The anti-OS activity of Cur depends on the EGFR/Src signaling axis
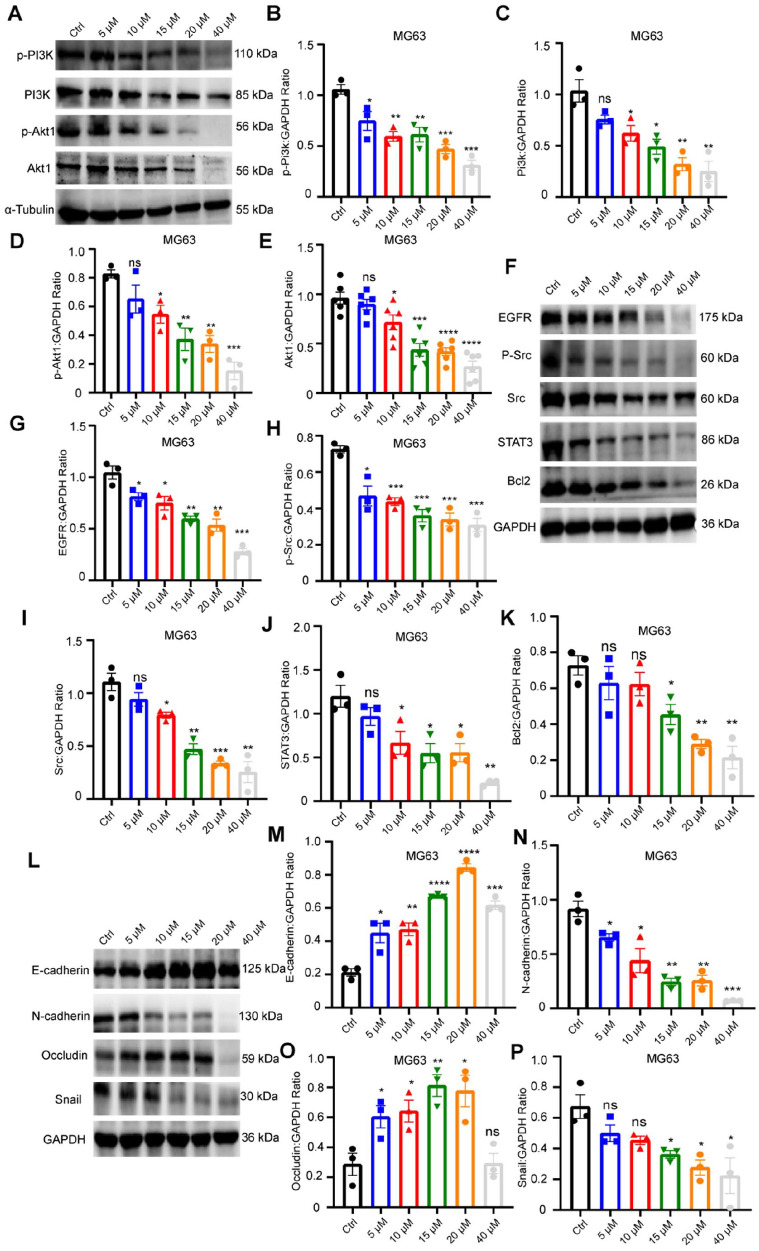
The enrichment analysis of Cur based on network pharmacology suggests that Cur may exert its anti-OS effects via the PI3K/Akt signaling pathway. This pathway administers various cellular processes, including proliferation, differentiation, and apoptosis. As depicted in Figure. 6A, the levels of PI3K, as well as the phosphorylated and total protein forms of Akt, were notably reduced after treatment with varying concentrations of Cur.
Figure6.
Cur inhibits metastasis of OS dependent on EGFR/Src pathway. (A) The PI3K, Akt1, p-Akt1 protein expression in MG63 cells; (B-E) Relative quantification of protein level in Fig. 6A; (F) The STAT3, Bcl2, p-Src, Src, EGFR protein expression in MG63 cells; (G-K) Relative quantification of protein level in Fig. 6F; (L) The EMT protein (E-cadherin, N-cadherin, Occludin, Snail ) expression in MG63 cells; (M-P) Relative quantification of protein level in Fig. 6L.
Based on our network drug target prediction, we further explored the regulatory role of the EGFR/Src signaling pathway in Cur’s tumor-inhibiting function. The results, as illustrated in Figure. 6F, show that following treatment with varying concentrations of Cur, the protein expression of EGFR, Src and phosphorylated Src (Tyr416) gradually decreased. In addition, the downstream regulators, including ERK1/2, STAT3, and Akt also decreased after Cur treatment. Src, as an ancient proto-oncogene, is involved in numerous processes related to tumorigenesis and development. The reduction in activated Src protein is likely to affect its downstream substrate, given that Src regulates the downstream substrate E-cadherin. We also detected the expression of EMT protein markers. As shown in Figure. 6L, Cur treatment significantly reduced the N-cadherin and Sail expression, while increasing the protein levels of E-cadherin and Occludin. These findings suggest that Cur inhibits EMT, probably contributing to its anti-OS effects.
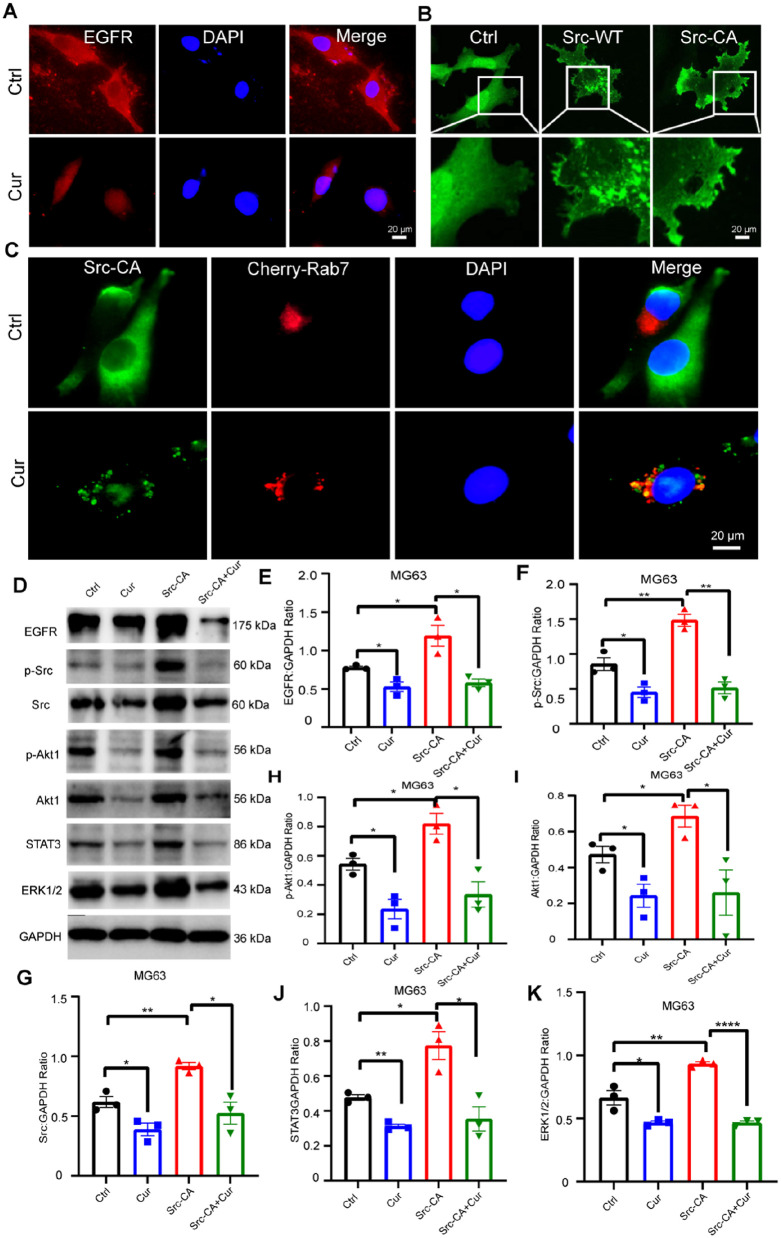
Consistent with the partially localized EGFR signal on the cell membrane, we observed that EGFR protein expression decreased following Cur treatment (Figure. 7A). We then focused on understanding the role of Src in Cur’s anti-tumor activity in OS cells. Src is localized on the cell membrane and regulates the formation of adhesion plaques and dynamic pseudopodia during tumor cell migration. In this study, we constructed a constitutively activated Src plasmid (Src-CA) and found that Src-CA was predominantly localized on the cell membrane, as previously reported (Figure. 7B). Rab7, a regulatory protein involved in vesicular transport, is mainly located on late endosomes and lysosomes. Interestingly, we discovered a strong colocalization between phosphorylated Src and the exogenously expressed Cherry-Rab7 protein in OS cells. This suggests that Src-CA is degraded into Rab7-positive vesicles after Cur treatment in MG63 cells (Figure. 7C). We then conducted further exploration to determine whether the downstream substrates are affected by this process. Our results revealed that overexpression of Src-CA significantly enhanced downstream pathways involving Akt, ERK1/2, and STAT3. Remarkably, this influence was reversed by Cur treatment (Figure. 7D). Notably, the expression of EGFR was also found to be upregulated.
Figure 7.
Cur inhibits metastasis of OS dependent on EGFR/Src pathway. (A) In MG63 cells, EGFR intracellular location under Cur treatment or not, Bar=20 um; (B) Construction and identification of Ctrl, Src wild type (SRC-WT)and persistently active plasmid (Src-CA), Bar=20 um; (C) colocation of Exogenous p-Src (Tyr416) and Rab7-positive vesicles after Cur treatment or not in MG63 cells, Bar= 20 um; (D) Relative expression of protein level (PI3K, Akt1, p-Akt1, STAT3, ERK1/2, p-Src, Src, EGFR) in treat with Ctrl, Cur (10 μM), Over-expression of Src-CA and Cur (10 μM) + Over-expression of Src-CA; (E-K) Relative quantification of protein level in Fig 7D.
Discussion
OS is a systemic bone tumor that predominantly affects adolescents and children and often metastasizes to the lungs. 1 Cur is a yellow compound derived from turmeric, a natural spice. Despite its demonstrated anti-cancer properties and high safety profile in clinical trials, the clinical application of Cur is limited, likely due to its poor water solubility, stability, and consequently low bioavailability. 21 However, potential analogs of Cur and nanoparticle-based Cur delivery systems are being progressively developed to overcome these challenges. 22 Existing reports indicate that Cur can effectively inhibit OS cell migration, invasion, and proliferation, while promoting cell death and apoptosis. However, the mechanisms through which Cur regulates the development and progression of OS are complex and remain to be explored.
Research has shown that Cur triggers apoptosis by generating ROS, which activate the mitochondrial apoptosis pathway via the ROS/Cyto-C/Caspase-3 signaling pathway in OS cells. 23 Additionally, Cur inhibits OS cell proliferation and invasion under both normal and hypoxic conditions by down-regulating Notch-1 and p-JAK2/p-STAT3 signaling.24,25 Notably, Cur also affects miRNA-138, thereby regulating its target genes Smad4, NF-κB p65 and cyclin D3, which results in tumorigenesis inhibition. 26 Similarly, overactivation of the miR-125a/ERRα signaling pathway has been found to promote OS cell death. 14 Despite these findings, the role of Cur in regulating EMT in OS remains unexplored.
In this study, we utilized network pharmacology analysis to look for potential targets of Cur, and performed target-pathway network analyses to explore its mechanisms against OS. Among the identified targets, AKT1, CASP3, and EGFR emerged as the most probable candidates for alteration according to the topological analysis. AKT1 is a member of a family of closely related serine/threonine-protein kinases (AKT1, AKT2, and AKT3), collectively referred to as the AKT kinase. They regulate multifarious biological processes, including metabolism, proliferation, cell survival, growth, and angiogenesis.27,28 CASP3 is involved in the activation cascade of caspases responsible for executing apoptosis, 29 and it cleaves and inhibits AKT1 in response to oxidative stress. 30 The impacts of Cur on CASP3 and Akt1 have been reported in previous studies.21,31,32 Our functional enrichment analysis revealed that Cur might inhibit OS progression via the PI3K/Akt signaling pathway. Typically, The PI3K/Akt pathway was activated in cancer and contributing to tumor progression and anticancer resistance. The PI3K pathway plays a pivotal role in numerous cellular processes and is frequently altered in cancer, contributing to tumor growth and survival. 33 Additionally, the importance of PI3K signaling in various cellular processes has been gradually revealed. 34 Cur’s potential to target the MAPK/ERK, PI3K/Akt, microRNA, Notch pathways has been evaluated to enhance lifetime in patients with OS. 32 In this work, we performed cell experiments to further corroborate the reliability of our computational predictions.
We verified the expression of potential targets and further elaborated the mechanisms of Cur. Our study demonstrated that Cur exerted anti-tumor effects in a concentration-dependent manner, effectively inhibiting the migration, invasion, and proliferation of MG63 and U2OS cells. Based on network pharmacology data analysis, we identified potential Cur targets, including EGFR, Src, and Akt1. Notably, Cur treatment resulted in the downregulation of EGFR, CDK2 in MG63 cells, suggesting an EGFR-mediated regulatory role in OS cells.
Additionally, it remains unclear whether Cur regulates central kinase signaling, such as EGFR/Src, to inhibit downstream substrates involved in migration, invasion, proliferation, death, and apoptosis. EGFR, a tyrosine kinase receptor frequently overexpressed and/or mutated in many tumors, regulates tumor growth through signal transduction. Src, the first non-receptor tyrosine kinase discovered, is commonly implicated in cell morphology, motility, proliferation, and survival across various cancer types. It also phosphorylates EGFR on tyrosine 845 (Y845) within the Src-EGFR complex. 35 Nevertheless, EGFR can function as an upstream activator of Src phosphorylation. 36 Interestingly, we observed that Cur significantly decreased Src protein levels, suggesting that Cur may effectively disrupt EGFR/Src interaction. Previous studies indicate that Src serves as an upstream kinase of Akt1, STAT3, Bcl2, and ERK1/2, which are involved in tumor metastasis, angiogenesis, and other processes. In our study, we found Cur effectively inhibited EGFR/Src signaling axis. Previous studies have highlighted Src’s role in E-cadherin regulation and EMT. 37 Our investigation revealed that Cur induced the epithelialization of cytoskeleton, upregulated epithelial markers E-cadherin, Occludin, and downregulated mesenchymal markers N-cadherin, snail, suggesting that Cur might regulate EMT by inhibiting Src signaling. Finally, Cur suppressed PI3K/Akt signaling, a finding consistent with previous reports, thereby further substantiating the reliability of our network pharmacology-based findings.
Moreover, to further investigate Cur’s regulatory influence on Src, we engineered constitutively active (Src-CA) and wild-type (Src-WT) mutants featuring a phosphomimetic mutation at Tyr416. Typically, inactive Src is primarily concentrated in perinuclear endosomes. EHD1 recruits MICAL-L1-positive structures to promote Src’s release from endosomes, enabling Src endosomal trafficking to the membrane where it regulates cell adhesion, pseudopodia formation, and downstream processes with FAK, integrins, and EGFR.38,39 Our findings showed that the introduction of active Src activated downstream signaling pathways such as Akt, ERK1/2, and STAT3. Interestingly, EGFR protein levels also increased, which may be attributed to Src’s role as an upstream regulator upregulating EGFR, 40 or it could result from active Src restoring EGFR expression through compensatory mechanisms. These changes, in turn, could enhance tumor migration and invasion through these interactions. Cur may reduce EGFR levels, limit EGFR/Src interactions, accelerate Src protein turnover, and further trigger the inhibition of Src-mediated downstream signaling. Collectively, these effects could enable Cur to exert anti-OS activity by modulating EGFR/Src signaling.
To sum up, in this study, we utilized bioinformatic approaches to uncover the targets and signaling pathways of Cur in the treatment of OS. Key targets and related signaling pathways were validated through subsequent in vitro experiments. However, more in vitro techniques are needed to demonstrate the direct interaction of Cur with these targets. Furthermore, we conducted focused investigations on the core gene Src, revealing its pivotal role in Cur-based therapy for OS. Additionally, we analyzed the biological mechanisms by which Cur regulates the migration, invasion, and proliferation of OS cells through the EGFR/Src signaling axis. However, the anti-OS effect of Cur and the role of Src kinase in this process still need to be further verified in vivo. Our research elucidates the mechanistic role of Cur in treating OS, providing valuable insights into its therapeutic targets and signaling pathways, particularly emphasizing the crucial role of Src. These findings not only deepen our understanding of Cur’s biological effects on OS cells, but also pave the way for potential therapeutic strategies targeting the EGFR/Src signaling axis in OS treatment.
Supplemental Material
Supplemental material, sj-doc-1-iji-10.1177_03946320241308082 for Curcumin suppresses metastasis, invasion, and proliferation in osteosarcoma cells by regulating the EGFR/Src signaling axis by Huiying Liu, Zhiqiang Li, Binwu Xu, Zhipeng Li, Xili Yang and Jun Luo in International Journal of Immunopathology and Pharmacology
Footnotes
Abbreviations: OS Osteosarcoma
Cur Curcumin
EMT epithelial-mesenchymal transition
Akt AKT serine/threonine kinase 1
Src SRC proto-oncogene, non-receptor tyrosine kinase
EGFR epidermal growth factor receptor
CASP3 caspase 3
HSP90A1 heat shock protein 90-A1
ESR1 estrogen receptor 1
IGF1 insulin like growth factor 1
ANXA5 annexin A5
RHOA ras homolog family member A
MAPK14 mitogen-activated protein kinase 14
PGR progesterone receptor
KDR kinase insert domain receptor
AR androgen receptor
PPARG peroxisome proliferator activated receptor gamma
MAPK8 mitogen-activated protein kinase 8
CDK2 cyclin dependent kinase 2
CCNA2 cyclin A2
MET MET proto-oncogene, receptor tyrosine kinase
STAT3 signal transducer and activator of transcription 3
IGF1R insulin like growth factor 1 receptor
OCLN occludin
Bcl2 BCL2 apoptosis regulator
ERK1/2 mitogen-activated protein kinase
E-cadherin cadherin 1
N-cadherin cadherin 2
Src-CA Persistent active of Src
Src-WT wild type of Src
ROS reactive oxygen species
EHD1 EH domain containing 1
MICAL-L1 MICAL like 1
FAK Focal adhesion kinase
Notch-1 notch receptor 1
JAK2 Janus kinase 2
Smad4 SMAD family member 4
NFκB nuclear factor kappa B subunit 1
ERRα estrogen related receptor alpha
Author contribution: Jun Luo: Conceptualization; resources; supervision. Huiying Liu: Data curation; methodology; Resources; project administration; writing-original draft; writing-review and editing. Zhiqiang Li: Formal analysis; project administration. Binwu Xu: Data curation; formal analysis; software; Resources. Zhipeng Li : writing-review and editing; Xili Yang: writing-review and editing.
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported Project of department of Education science and technology research of Jiangxi Province, China (GJJ2200229); National Natural Science Foundation incubation project of the Second Affiliated Hospital of Nanchang University (2022YNFY12038); Jiangxi Provincial Health Commission science and technology project (202410028); Project of the Science and Technology Department of Jiangxi Province (20202BABL206083).
Ethics approval: Ethical approval was not sought for the present study because we have not involved animal experiments in this study.
Informed consent: Not applicable.
Trial registration: Not applicable.
ORCID iD: Jun Luo  https://orcid.org/0000-0002-3016-0086
https://orcid.org/0000-0002-3016-0086
Supplemental material: Supplemental material for this article is available online.
References
- 1. Czarnecka AM, Synoradzki K, Firlej W, et al. (2020) Molecular biology of osteosarcoma. Cancers (Basel) 12(8): 2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Luetke A, Meyers PA, Lewis I, et al. (2014) Osteosarcoma treatment - where do we stand? A state of the art review. Cancer Treatment Reviews 40(4): 523–532. [DOI] [PubMed] [Google Scholar]
- 3. Bacci G, Rocca M, Salone M, et al. (2008) High grade osteosarcoma of the extremities with lung metastases at presentation: Treatment with neoadjuvant chemotherapy and simultaneous resection of primary and metastatic lesions. Journal of Surgical Oncology 98(6): 415–420. [DOI] [PubMed] [Google Scholar]
- 4. Hattinger CM, Pasello M, Ferrari S, et al. (2010) Emerging drugs for high-grade osteosarcoma. Expert Opinion on Emerging Drugs 15(4): 615–634. [DOI] [PubMed] [Google Scholar]
- 5. Schwarz R, Bruland O, Cassoni A, et al. (2009) The role of radiotherapy in oseosarcoma. Cancer Treatment and Research 152: 147–164. [DOI] [PubMed] [Google Scholar]
- 6. Mahajan A, Woo SY, Kornguth DG, et al. (2008) Multimodality treatment of osteosarcoma: Radiation in a high-risk cohort. Pediatric Blood & Cancer 50(5): 976–982. [DOI] [PubMed] [Google Scholar]
- 7. DeLaney TF, Park L, Goldberg SI, et al. (2005) Radiotherapy for local control of osteosarcoma. International Journal of Radiation Oncology, Biology, Physics 61(2): 492–498. [DOI] [PubMed] [Google Scholar]
- 8. Errani C, Longhi A, Rossi G, et al. (2011) Palliative therapy for osteosarcoma. Expert Review of Anticancer Therapy 11(2): 217–227. [DOI] [PubMed] [Google Scholar]
- 9. Zhou Y, Kong Y, Jiang M, et al. (2022) Curcumin activates NLRC4, AIM2, and IFI16 inflammasomes and induces pyroptosis by up-regulated ISG3 transcript factor in acute myeloid leukemia cell lines. Cancer Biology & Therapy 23(1): 328–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bhatia M, Bhalerao M, Cruz-Martins N, et al. (2021) Curcumin and cancer biology: Focusing regulatory effects in different signalling pathways. Phytotherapy Research 35(9): 4913–4929. [DOI] [PubMed] [Google Scholar]
- 11. Prasad S, Gupta SC, Tyagi AK, et al. (2014) Curcumin, a component of golden spice: From bedside to bench and back. Biotechnology Advances 32(6): 1053–1064. [DOI] [PubMed] [Google Scholar]
- 12. Leow PC, Tian Q, Ong ZY, et al. (2010) Antitumor activity of natural compounds, curcumin and PKF118-310, as Wnt/β-catenin antagonists against human osteosarcoma cells. Investigational New Drugs 28(6): 766–782. [DOI] [PubMed] [Google Scholar]
- 13. Lee DS, Lee MK, Kim JH. (2009) Curcumin induces cell cycle arrest and apoptosis in human osteosarcoma (HOS) cells. Anticancer Research 29(12): 5039–5044. [PubMed] [Google Scholar]
- 14. Chen P, Wang H, Yang F, et al. (2017) Curcumin promotes osteosarcoma cell death by activating miR-125a/ERRα signal pathway. Journal of Cellular Biochemistry 118(1): 74–81. [DOI] [PubMed] [Google Scholar]
- 15. Shang W, Zhang J, Song H, et al. (2021) Mechanism of tetrandrine against endometrial cancer based on network pharmacology. Drug Design, Development and Therapy 15: 2907–2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Poornima P, Kumar JD, Zhao Q, et al. (2016) Network pharmacology of cancer: From understanding of complex interactomes to the design of multi-target specific therapeutics from nature. Pharmacological Research 111: 290–302. [DOI] [PubMed] [Google Scholar]
- 17. Wang J, Li Y, Yang Y, et al. (2017) Systems pharmacology dissection of multiscale mechanisms of action for herbal medicines in treating rheumatoid arthritis. Molecular Pharmaceutics 14(9): 3201–3217. [DOI] [PubMed] [Google Scholar]
- 18. Wang X, Shen Y, Wang S, et al. (2017) PharmMapper 2017 update: A web server for potential drug target identification with a comprehensive target pharmacophore database. Nucleic Acids Research 45(W1): W356–w360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shen J, Cheng F, Xu Y, et al. (2010) Estimation of ADME properties with substructure pattern recognition. Journal of Chemical Information and Modeling 50(6): 1034–1041. [DOI] [PubMed] [Google Scholar]
- 20. Lin X, Zhang J, Chen L, et al. (2017) Tyrosine phosphorylation of Rab7 by Src kinase. Cell Signalling 35: 84–94. [DOI] [PubMed] [Google Scholar]
- 21. Yang ZJ, Huang SY, Zhou DD, et al. (2022) Effects and mechanisms of curcumin for the prevention and management of cancers: An updated review. Antioxidants (Basel): 11(8): 1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhu X, Yu Z, Feng L, et al. (2021) Chitosan-based nanoparticle co-delivery of docetaxel and curcumin ameliorates anti-tumor chemoimmunotherapy in lung cancer. Carbohydrate Polymers 268: 118237. [DOI] [PubMed] [Google Scholar]
- 23. Chang Z, Xing J, Yu X. (2014) Curcumin induces osteosarcoma MG63 cells apoptosis via ROS/Cyto-C/Caspase-3 pathway. Tumor Biology 35(1): 753–758. [DOI] [PubMed] [Google Scholar]
- 24. Sun Y, Liu L, Wang Y, et al. (2019) Curcumin inhibits the proliferation and invasion of MG-63 cells through inactivation of the p-JAK2/p-STAT3 pathway. OncoTargets and Therapy 12: 2011–2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang Z, Zhang K, Zhu Y, et al. (2017) Curcumin inhibits hypoxia-induced proliferation and invasion of MG-63 osteosarcoma cells via downregulating Notch1. Molecular Medicine Reports 15(4): 1747–1752. [DOI] [PubMed] [Google Scholar]
- 26. Yu D, An F, He X, et al. (2015) Curcumin inhibits the proliferation and invasion of human osteosarcoma cell line MG-63 by regulating miR-138. International Journal of Clinical and Experimental Pathology 8(11): 14946–14952. [PMC free article] [PubMed] [Google Scholar]
- 27. Nicholson KM, Anderson NG. (2002) The protein kinase B/Akt signalling pathway in human malignancy. Cell Signalling 14(5): 381–395. [DOI] [PubMed] [Google Scholar]
- 28. Hers I, Vincent EE, Tavaré JM. (2011) Akt signalling in health and disease. Cell Signalling 23(10): 1515–1527. [DOI] [PubMed] [Google Scholar]
- 29. Nicholson DW, Ali A, Thornberry NA, et al. (1995) Identification and inhibition of the ICE/CED-3 protease necessary for mammalian apoptosis. Nature 376(6535): 37–43. [DOI] [PubMed] [Google Scholar]
- 30. Sen T, Sen N, Noordhuis MG, et al. (2012) OGDHL is a modifier of AKT-dependent signaling and NF-κB function. PLoS One 7(11): e48770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yavuz Türel G, Şahin Calapoğlu N, Bayram D, et al. (2022) Curcumin induces apoptosis through caspase dependent pathway in human colon carcinoma cells. Molecular Biology Reports 49(2): 1351–1360. [DOI] [PubMed] [Google Scholar]
- 32. Farnood PR, Pazhooh RD, Asemi Z, et al. (2023) Targeting signaling pathway by curcumin in osteosarcoma. Current Molecular Pharmacology 16(1): 71–82. [DOI] [PubMed] [Google Scholar]
- 33. LoRusso PM. (2016) Inhibition of the PI3K/AKT/mTOR pathway in solid tumors. Journal of Clinical Oncology 34(31): 3803–3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Martini M, De Santis MC, Braccini L, et al. (2014) PI3K/AKT signaling pathway and cancer: An updated review. Annals of Medicine 46(6): 372–383. [DOI] [PubMed] [Google Scholar]
- 35. Sato K. (2013) Cellular functions regulated by phosphorylation of EGFR on Tyr845. International Journal of Molecular Sciences 14(6): 10761–10790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chen Z, Oh D, Dubey AK, et al. (2018) EGFR family and Src family kinase interactions: Mechanics matters? Current Opinion in Cell Biology 51: 97–102. [DOI] [PubMed] [Google Scholar]
- 37. Nagathihalli NS, Merchant NB. (2012) Src-mediated regulation of E-cadherin and EMT in pancreatic cancer. Frontiers in Bioscience-Landmark 17(6): 2059–2069. [DOI] [PubMed] [Google Scholar]
- 38. Reinecke JB, Katafiasz D, Naslavsky N, et al. (2014) Regulation of Src trafficking and activation by the endocytic regulatory proteins MICAL-L1 and EHD1. Journal of Cell Science 127(Pt 8): 1684–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tai YL, Chu PY, Lai IR, et al. (2015) An EGFR/Src-dependent β4 integrin/FAK complex contributes to malignancy of breast cancer. Scientific Reports 5: 16408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li J, Li Y, He H, et al. (2016) Csk/Src/EGFR signaling regulates migration of myofibroblasts and alveolarization. American Journal of Physiology-Lung Cellular and Molecular Physiology 310(6): L562–571. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-doc-1-iji-10.1177_03946320241308082 for Curcumin suppresses metastasis, invasion, and proliferation in osteosarcoma cells by regulating the EGFR/Src signaling axis by Huiying Liu, Zhiqiang Li, Binwu Xu, Zhipeng Li, Xili Yang and Jun Luo in International Journal of Immunopathology and Pharmacology