Abstract
Introduction
Chronic alcohol consumption and tobacco usage are major risk factors for esophageal squamous cell carcinoma (ESCC). Excessive tobacco and alcohol consumption lead to oxidative stress and the generation of reactive carbonyl species (RCS) which induce DNA damage and cell apoptosis. This phenomenon contributes to cell damage and carcinogenesis in various organs including ESCC. However, it also raises an important question on how ESCC cells evade RCS-induced apoptosis and grow rapidly under these conditions. Therefore, we hypothesize that some enzymes produced by ESCC cells are capable of catabolizing RCS, preventing ESCC neoplastic cells from undergoing RCS-induced apoptosis, potentially contributing to ESCC progression.
Methods
To identify significant gene clusters involved in the metabolism of RCS in ESCC, we used an Agilent SurePrint G3 Human V2 GE 8 × 60 K microarray kit to analyze differentially expressed genes between nine paired ESCC tissues and adjacent normal esophageal tissues taken from areas distant from the tumor site. Bioinformatics analysis using gene ontology (GO) was performed to categorize these genes. To validate the findings, immunohistochemical staining in specimens from 169 surgically resected ESCC patients was performed and then correlated with treatment outcomes. Furthermore, the identified signaling pathway and its biological effects were investigated in ESCC cell lines in vitro and 4-nitroquinoline 1-oxide (4-NQO)-induced-ESCC murine model in vivo.
Results
Interestingly, we found that one of the significantly altered 57 GO molecular function domain terms (GO:0004033 aldo-keto reductase activity; P = 0.021) between nine paired ESCC tumors and adjacent normal tissue specimens was associated with the RCS metabolism. Among significant genes within this domain, AKR1B10 (aldo-keto reductase family 1 member B10; P = 0.006) was identified as the most significantly altered gene. Immunohistochemical analysis revealed that AKR1B10 expression was higher in ESCC cells than in adjacent normal esophageal epithelium. In addition, AKR1B10 expression was independently significantly associated with a poorer prognosis in 169 ESCC patients. Enzyme-linked immunosorbent assay results further demonstrated that blood AKR1B10 concentrations were significantly higher in 72 ESCC patients than in 24 healthy controls. In vitro experiments revealed that inhibiting endogenous AKR1B10 enhanced the cytotoxicity of 4-hydroxy trans-2-nonenal, a type of RCS. In a 4-NQO-induced-ESCC murine model, oleanolic acid, an AKR1B10 inhibitor, significantly reduced the incidence of esophageal tumors.
Conclusions
Our findings suggested that AKR1B10 is an independent adverse prognosticator for patients with ESCC, and could prevent ESCC neoplastic cells from undergoing RCS-induced apoptosis, and promote ESCC progression. Therefore, AKR1B10 signaling could be a potential therapeutic strategy for ESCC.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12935-024-03623-8.
Keywords: AKR1B10, ESCC, RCS
Introduction
Esophageal cancer is the sixth leading cause of cancer-related death worldwide in 2018 [1]. The primary two histological types of esophageal cancer are squamous cell carcinoma and adenocarcinoma. Unlike adenocarcinomas, which often arise from Barrett’s esophagus in Western countries, esophageal squamous cell carcinoma (ESCC) is the predominate type in the Asia-Pacific region [2]. In particular, ESCC is more prevalent in Taiwan, accounting for more than 90% of esophageal cancer cases. Despite substantial improvement in surgical techniques, chemotherapy, and radiotherapy, the prognosis of patients with ESCC remains unfavorable [3], with a 5-year survival of approximately 20 ~ 40%.3 Therefore, identifying new targets for ESCC is crucial, necessitating a comprehensive understanding of its underlying signaling pathways.
Constant exposure of cells to oxygen could generate highly reactive molecules known as free radicals or reactive oxygen species (ROS) [4]. These unstable free radicals or oxidants include hydroxyl radicals (OH−), singlet oxygen, superoxide anion (O2−) and hydrogen peroxide (H2O2) [4]. Under physiological conditions, the antioxidant system within cells mitigates the effects of ROS. However, when ROS levels exceed the capacity of cellular antioxidants, oxidative stress is induced. Accumulating evidence suggests that persistent oxidative stress can trigger chronic inflammation, which predisposes to several chronic diseases including infection, autoimmune disease, and cancer [5–12]. Low levels of ROS can be effectively neutralized by both enzymatic and nonenzymatic processes; however, excessive ROS production overwhelms the intrinsic antioxidant defenses of the tissue leading to cell injury and apoptosis. Tobacco and alcohol are major risk factors for ESCC, and patients with ESCC often have a history of long-term use of these substances. Overconsumption of tobacco and alcohol has been associated with increasing ROS production leading to oxidative stress [13].
A notable characteristic of ROS is the ability to propagate and exacerbate the oxidative damage by interacting with various cellular components. Among these, membrane lipids, particularly polyunsaturated fatty acids, are primary targets because they can readily stabilize free radicals such as ROS. Consequently, when attacked by free radicals, they initiate a cascade of autocatalytic reactions known as “lipid peroxidation”. ROS-triggered lipid-peroxidation reactions generate a variety of reactive carbonyl species (RCS), which further intensify ROS-related injury by reacting with nucleophilic cell constituents such as the sulfhydryl side chain of polypeptides and the amine groups in proteins, DNA and lipids [14, 15]. Similar to ROS, RCS can react with multiple cell components, forming covalent adducts that alter their structure, and induce DNA damage and cell apoptosis. Excessive production and accumulation of RCS are hallmark features of ROS production. ROS-triggered lipid-peroxidation reactions generate a range of RCS, and these RCS propagate ROS-related injury. Notably, the RCS, 4-hydroxy trans-2-nonenal (4-HNE), acrolein and malondialdehyde (MDA) have been studied extensively and are recognized as general indicators of oxidative stress resulting from excessive tobacco smoking or alcohol consumption [16–18].
Previous studies reported that oxidative stress and these RCS cause DNA damage and carcinogenesis in various organs including ESCC [18–21]. However, it also raises an important question on how ESCC cells evade RCS-induced apoptosis, and grow rapidly under these conditions. For example, although high concentrations of RCS such as HNE, acrolein, or MDA can induce cell apoptosis [21], ESCC neoplastic cells seem to evade the RCS-induced apoptosis and exhibit rapid growth. Therefore, we hypothesized that some enzymes produced by ESCC cells catabolize RCS, which prevent ESCC neoplastic cells from undergoing RCS-induced apoptosis, and promote ESCC progression.
Methods
Microarray analysis
Fresh human ESCC tumor specimen and adjacent normal esophageal tissues taken from sites distant from the tumor site were collected from the tissue bank at the Kaohsiung Chang Gung Memorial Hospital. Total RNA was extracted from tissue samples using the TRIzol method following the manufacturer’s protocol. To identify important gene clusters involved in the metabolism of RCS in ESCC, Agilent SurePrint G3 Human V2 GE 8 × 60 K microarray kit (Agilent Technologies, USA) was used to analyze the differentially expressed genes between nine paired ESCC tumors and adjacent normal esophageal tissues. Briefly, 0.2 µg of total RNA were amplified using a Low Input Quick-Amp Labeling kit (Agilent Technologies, USA) and then labeled with Cy3 (CyDye, Agilent Technologies, USA) during the in vitro transcription process. Then, 0.6 µg of Cy3-labled cRNA were fragmented to an average size of about 50–100 nucleotides by incubating with fragmentation buffer at 60 °C for 30 min. The fragmented and labeled cRNA was then pooled and hybridized using an Agilent SurePrint G3 Human V2 GE 8 × 60 K Microarray kit at 65 °C for 17 h. Following washing and drying with a nitrogen gun, the microarrays were scanned using an Agilent microarray scanner (Agilent Technologies, USA) at 535 nm for Cy3. Subsequently, The scanned images were analyzed using Feature extraction10.5.1.1 software (Agilent Technologies, USA), an image analysis and normalization tool, to quantify signal and background intensity for each feature [22]. To identify differentially expressed genes, the raw signal data were subjected to quantile normalization [23]. Subsequently, enrichment analyses were performed using the cluster Profiler, which identifies differentially expressed genes enriched in specific gene ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes pathways [24, 25].
Patient population
This study was approved by the Chang Gung Medical Foundation Institutional Review Board (approval number: 201601568B0 and 202100927B0). For immunohistochemistry, data from ESCC patients who had undergone esophagectomy at Kaohsiung Chang Gung Memorial Hospital were retrospectively reviewed. Patients who were treated with preoperative chemotherapy, radiotherapy, or chemoradiotherapy, and those with synchronous cancers in other organs were excluded. Finally, 169 patients were identified. Patients who underwent surgery received either a radical esophagectomy with cervical esophagogastric anastomosis (McKeown procedure) or an Ivor Lewis esophagectomy with intrathoracic anastomosis, reconstruction of the digestive tract with a gastric tube, and pylorus drainage procedures. All the patients underwent two-field lymph node dissection. The pathological TNM stage was determined according to the eighth edition of American Joint Committee on Cancer (AJCC) staging system [26]. Overall survival (OS) was calculated from the date of surgery to death from any cause, and disease-free survival (DFS) was calculated from the date of surgery to recurrence or death from any cause without evidence of recurrence.
Immunohistochemistry was performed using an immunoperoxidase technique, as previously described [27]. Staining was performed on slides (4 mm) of formalin-fixed, paraffin-embedded tissue sections with primary antibodies against aldo-keto reductase family 1 member B10 (AKR1B10) (H00057016-M01, 1:80, Abnova Corporation). Briefly, after deparaffinization and rehydration, antigen retrieval was performed by treating the slides in 10 mmol/L citrate buffer (pH 6.0) in a hot water bath (95°C) for 20 mins. Endogenous peroxidase activity was blocked with 0.3% hydrogen peroxide for 15 mins, followed by blocking with 1% goat serum for 1 h at room temperature. The sections were then incubated with primary antibodies for at least 18 hours at 4°C overnight. Immunodetection was performed using the LSAB2 kit (Dako, Carpinteria, CA) followed by 3–3’-diaminobenzidine for color development and hematoxylin for counterstaining. Negative controls were prepared by removing primary antibodies. Normal human colon mucosa was used as a positive control. Immunohistochemical staining was assessed using a semi-quantitative immunoreactive score (IRS) [27]. The IRS was calculated by multiplying the staining intensity (graded as: 0: no staining, 1: weak staining, 2: moderate staining, and 3: strong staining) and the percentage of positively stained cells (0: no stained cell, 1: <10% of stained cells, 2: 10–50% of stained cells, 3: 51–80% of stained cells, and 4: >80% of stained cells). A specimen with an IRS is ≧ 6 was considered to have overexpression.
AKR1B10 levels in peripheral blood samples from patients with ESCC and healthy controls without a history of malignancy were collected, and analyzed using enzyme-linked immunosorbent assay (ELISA). Serum AKR1B10 levels were was measured using commercially available ELISAs kits (MyBioSource, San Diego, CA, USA) following the manufacturer’s instructions.
Cell culture, transient transfection, and cell viability assay
Human ESCC cell lines, KYSE70 and TE11 were purchase from the American Type Culture Collection. All cells were cultured in Dulbecco’s modified eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 100 µg/mL of penicillin, and 100 U/mL of streptomycin (Gibco) in a 37 ℃ humidified incubator with 5% CO2. The cells were regularly tested for mycoplasma contamination. For transfection, siAKR1B10 and the negative control were transfected into ESCC cell lines using Lipofectamine 3000 for 48 h following the manufacture’s protocol. For viability assay, cells were seeded into 96-well plates at a density of 1 × 104/well in triplicate and incubated for an additional 24 h. After transfection for 24 h, 10 µl of CCK-8 solution was added to each well, followed by incubation for 3 h at 37 ℃. The optical density at 450 nm was measured by microplate reader (Bio-Rad).
Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was extracted from the cell samples by using TRIzol reagent according to the standard procedure. The isolated RNA was reverse transcribed into cDNA using a Reverse Transcription System Kit (Takara) according to the manufacturer’s instruction. AKR1B10 expression was analyzed using SYBR Green I (Takara) on the ABI7500 system. The relative gene expression level was normalized to β-actin and calculated using the 2-ΔΔCT method. The primer sequences are presented. AKR1B10 forward: 5’-GCTGAGCTATCTGGACGTCT-3’; AKR1B10 reverse: 5’-CGTTACAGGCCCTCCAGTTT-3’; β-actin forward 5’-ACCCTGAAGTACCCCATCGAG-3’ β-actin reverse 5’-AGCACAGCCTGGATAGCAAC-3’.
4-nitroquinoline 1-oxide (4-NQO)-induced murine ESCC model
The 4-NQO-induced murine ESCC model was established as previously described [28]. The carcinogen, 4-NQO (Sigma Aldrich, St Louis, MO, USA) stock, was first dissolved in DMSO at 50 mg/mL as a stock solution and stored at -20 °C until future use. On the days of 4-NQO administration, the stock solution was dissolved in propylene glycol (Sigma Aldrich, St Louis, MO, USA) and added to the drinking water bottles containing autoclaved tap water to obtain a final concentration of 100 µg/m. The 60 six-week-old C57Bl/6 mice received 100 µg/ml 4-NQO in drinking water for 16 weeks followed by normal autoclaved drinking water for another 16 weeks (total 32 weeks). The mice were divided into two groups (n = 30 in each group): the AKR1B10 inhibitor, oleanolic acid [29] (75 mg/kg/day for 10 weeks intraperitoneally started 16 weeks after initiation of 4-NOQ treatment) and the vehicle control group. Mice that died before the end of the experiment were excluded from the analysis. At the end of the experiment, the mice were euthanized, and the entire esophagus and stomach were opened longitudinally, and macroscopic lesions were observed and identified carefully. For histological examination, the esophagus and gross lesions were fixed in 10% buffered formalin, embedded in paraffin blocks and stained with hematoxylin and eosin. Histological determination was performed by a pathologist (W.T.H) according to the criteria described previously [28]. ESCC was defined as the presence of neoplastic cells invading the subepithelial tissues.
Statistical analysis
All patient data were analyzed using SPSS software (version 17.0; IBM Corp., Armonk, NY, USA). Chi-square and Fisher’s exact tests were used to compare the two groups. Serum AKR1B10 levels between controls and ESCC patients were evaluated using the t-test. Survival analysis was performed using the Kaplan–Meier method for univariate analysis.Differences between survival curves were assessed using a log-rank test. Statistically significant variables in the univariate analysis were then assessed using a Cox regression model to evaluate their relative prognostic importance. For cell line and mice experiments, all statistical analyses were performed using a t-test, and a P value < 0.05 indicates statistical significance.
Results
Aldo-keto reductase signaling was significantly altered between nine paired ESCC tumors and adjacent normal tissue specimens and involved in RCS metabolism
To identify key gene clusters involved in the metabolism of RCS in ESCC, microarray analysis was performed using Agilent SurePrint G3 Human V2 GE 8 × 60 K microarray to analyze the differentially expressed genes between nine paired ESCC tumor samples and adjacent normal esophageal tissue specimens. The Venn diagram plot revealed that 3406 transcripts (P < 0.05, fold change > 1.5) were differentially expressed between ESCC tumor samples and adjacent normal esophageal tissue specimens. Compared with the adjacent normal esophageal tissues, 3406 differentially expressed genes, including 1785 upregulated transcripts and 1621 downregulated transcripts, were identified in the paired nine paired ESCC tissues. Bioinformatic analysis using gene ontology (GO) was performed, and we try to find the potential gene clusters involved in the metabolism of RCS in ESCC. Among the 57 GO molecular function domain terms that were significantly altered between tumors and adjacent normal tissue specimens (Table 1), one GO terms (GO:0004033 aldo-keto reductase activity; P = 0.021, Table 1; Fig. 1A) was associated with the RCS metabolism. Previous studies described that aldo-keto reductase catalyzes the reduction of a carbonyl moiety of RCS to an alcohol using NADPH as a cofactor [30, 31].
Table 1.
Significantly up-regulated and down-regulated 57 gene ontology (GO) molecular function domain terms (p < 0.05) in 9 paired human esophageal squamous cell carcinoma and adjacent normal tissue specimens
| Term | Description | p-value | Term | Description | P-value |
|---|---|---|---|---|---|
| GO:0008092 | cytoskeletal protein binding | 2.35E-11 | GO:0008093 | cytoskeletal adaptor activity | 0.010957 |
| GO:0003779 | actin binding | 7.95E-07 | GO:0003690 | double-stranded DNA binding | 0.011835 |
| GO:0004714 | transmembrane receptor protein tyrosine kinase activity | 9.06E-06 | GO:0003678 | DNA helicase activity | 0.011911 |
| GO:0004672 | protein kinase activity | 1.34E-05 | GO:0000217 | DNA secondary structure binding | 0.012965 |
| GO:0005524 | ATP binding | 4.94E-05 | GO:0019838 | growth factor binding | 0.013504 |
| GO:0032559 | adenyl ribonucleotide binding | 7.56E-05 | GO:0008307 | structural constituent of muscle | 0.016790 |
| GO:0032403 | protein complex binding | 1.50E-04 | GO:0005200 | structural constituent of cytoskeleton | 0.017610 |
| GO:0001883 | purine nucleoside binding | 1.57E-04 | GO:0004033 | aldo-keto reductase activity | 0.021094 |
| GO:0030554 | adenyl nucleotide binding | 2.10E-04 | GO:0019902 | phosphatase binding | 0.023008 |
| GO:0001882 | nucleoside binding | 2.38E-04 | GO:0005539 | glycosaminoglycan binding | 0.025147 |
| GO:0004713 | protein tyrosine kinase activity | 4.24E-04 | GO:0003677 | DNA binding | 0.026861 |
| GO:0008094 | DNA-dependent ATPase activity | 4.29E-04 | GO:0019903 | protein phosphatase binding | 0.027569 |
| GO:0032553 | ribonucleotide binding | 6.38E-04 | GO:0019207 | kinase regulator activity | 0.031196 |
| GO:0032555 | purine ribonucleotide binding | 6.38E-04 | GO:0005158 | insulin receptor binding | 0.033105 |
| GO:0005516 | calmodulin binding | 9.00E-04 | GO:0060090 | molecular adaptor activity | 0.036026 |
| GO:0004674 | protein serine/threonine kinase activity | 0.001092 | GO:0019955 | cytokine binding | 0.036901 |
| GO:0003682 | chromatin binding | 0.001340 | GO:0019887 | protein kinase regulator activity | 0.037270 |
| GO:0017076 | purine nucleotide binding | 0.001375 | GO:0019201 | nucleotide kinase activity | 0.038960 |
| GO:0003777 | microtubule motor activity | 0.002028 | GO:0008201 | heparin binding | 0.040527 |
| GO:0046870 | cadmium ion binding | 0.002870 | GO:0001871 | pattern binding | 0.040875 |
| GO:0003774 | motor activity | 0.004732 | GO:0030247 | polysaccharide binding | 0.040875 |
| GO:0004385 | guanylate kinase activity | 0.004774 | GO:0019901 | protein kinase binding | 0.041954 |
| GO:0017124 | SH3 domain binding | 0.004951 | GO:0005509 | calcium ion binding | 0.042079 |
| GO:0005178 | integrin binding | 0.005317 | GO:0004386 | helicase activity | 0.043019 |
| GO:0043566 | structure-specific DNA binding | 0.006323 | GO:0019904 | protein domain specific binding | 0.043117 |
| GO:0019899 | enzyme binding | 0.006538 | GO:0030695 | GTPase regulator activity | 0.047138 |
| GO:0003697 | single-stranded DNA binding | 0.007353 | GO:0016887 | ATPase activity | 0.048814 |
| GO:0015631 | tubulin binding | 0.007920 | GO:0060589 | nucleoside-triphosphatase regulator activity | 0.049010 |
| GO:0000166 | nucleotide binding | 0.008806 |
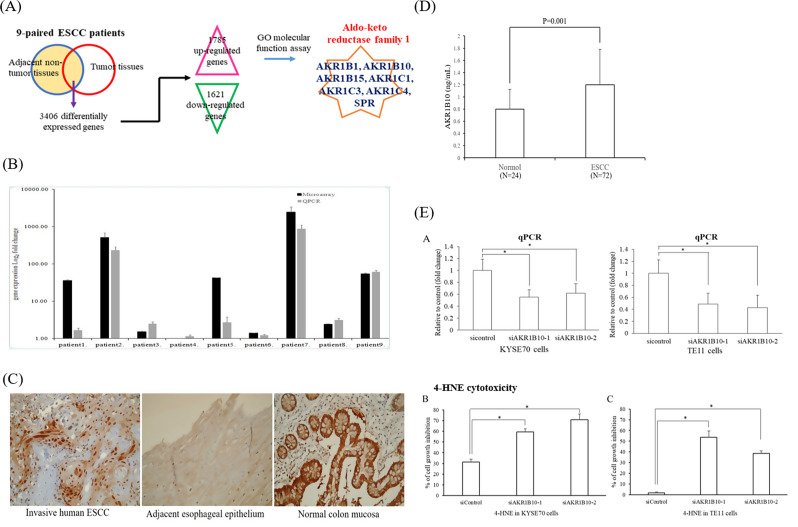
Fig. 1.
(A) Upper: Venn diagram analysis showed 3406 significant (> 1.5-fold) transcripts including 1785 upregulated and 1621 downregulated transcripts in nine-paired ESCC tissues in comparison with adjacent normal esophageal tissues. Bioinformatics analysis using gene ontology (GO) revealed 57 GO molecular function domain, and one domain, GO:0004033 (aldo-keto reductase activity; P = 0.021), was associated with the RCS metabolism. Among the significant genes in aldo-keto reductase activity (GO:0004033), AKR1B10 was the most significant gene identified between ESCC tumors and adjacent normal esophageal tissue specimens, followed by AKR1B15, AKR1B1, AKR1C4, AKR1C3, SPR, and AKR1C1.(B) The differentially expressed gene, AKR1B10, between nine paired ESCC tumors and adjacent normal tissue specimens identified by microarray was validated by q-RT-PCR. (C) Representative immunohistochemical staining of AKR1B10 in esophageal squamous cell carcinoma and adjacent normal esophageal epithelium. Immunohistochemistry of AKR1B10 in esophagectomy samples showed higher AKR1B10 protein expression in ESCC tissues than in adjacent normal esophageal epithelium. Positive AKR1B10 immunoreactivity was observed in the normal human colon mucosa, which served as a positive control. Original magnification: ×200. (D) AKR1B10 concentration was significantly higher in 22 ESCC patients than in 22 healthy controls. ELISA of AKR1B10 concentration of blood samples from ESCC patients and healthy controls. The results are presented as the mean ± SD of triplicate wells in three independent experiments. *statistically significant (P = 0.001). (E) The 4-HNE, a reactive carbonyl species (RCS), is recognized as a general marker of oxidative stress induced by excessive tobacco smoking or alcohol consumption. AKR1B10 silencing by small-interfering RNA (left panel) significantly enhance 4-HNE cytotoxicity in ESCC cell lines, KYSE70 and TE11, suggesting that AKR1B10 can prevent ESCC cells from undergoing RCS-induced apoptosis. The percentage (%) of cell growth inhibition = (viable cell number under 0µM 4-HNE - viable cell number under 40µM 4-HNE)/ viable cell number under 0µM 4-HNE. *Significantly different from the control (P < 0.05); ESCC, esophageal squamous cell carcinoma; RCS, reactive carbonyl species; AKR1B10, aldo-keto reductase family 1 member B10; qRT-PCR, quantitative real-time polymerase chain reaction; 4-HNE, 4-hydroxy trans-2-nonenal
AKR1B10 is the most significant gene among GO:0004033 aldo-keto reductase activity and prevent ESCC cells from undergoing RCS-induced apoptosis
Among the significant genes associated with GO:0004033 aldo-keto reductase activity, AKR1B10 (aldo-keto reductase family 1 member B10; P = 0.006, Fig. 1A) was the most significant gene between ESCC tumors and adjacent normal esophageal tissue specimens, followed by AKR1B15 (P = 0.007), AKR1B1 (P = 0.008), AKR1C4 (P = 0.013), AKR1C3 (P = 0.015), sepiapterin reductase (SPR; P = 0.02), and AKR1C1 (P = 0.047). To validate the microarray findings, q-RT-PCR of AKR1B10 was performed in these nine paired ESCC tumor samples and adjacent normal tissue specimens, which revealed that AKR1B10 expression was higher in the ESCC tumor specimens than in adjacent normal esophageal tissues. (Fig. 1B). Immunohistochemistry of AKR1B10 in esophagectomy samples from ESCC patients showed that AKR1B10 protein expression was higher in ESCC tumors than in adjacent normal esophageal epitheliums (Fig. 1C). Furthermore, we measured serum AKR1B10 level in 72 patients with ESCC and 24 healthy controls, and found that serum AKR1B10 levels were significantly higher in patients with ESCC than in controls (P = 0.001, Fig. 1D). These findings suggest that AKR1B10 expression is higher in ESCC than in normal esophageal epitheliums. We then investigated whether AKR1B10 prevents ESCC cells from undergoing RCS-induced apoptosis using ESCC cell lines. The 4-hydroxy trans-2-nonenal (4-HNE), an RCS, is recognized as a general indicator of oxidative stress resulting from excessive tobacco smoking or alcohol consumption. We found that the inhibition of endogenous AKR1B10 by small-interfering RNA (Fig. 1E, left panel) significantly enhanced 4-HNE cytotoxicity in the ESCC cell lines, KYSE70 and TE11 (Fig. 1E, middle and right panels), suggesting that AKR1B10 can prevent ESCC cells from undergoing 4-HNE-induced cell death.
AKR1B10 expression and its correlation with clinicopathologic variables in ESCC patients
To further explore the clinical significance of AKR1B10, we performed AKR1B10 immunohistochemical analysis in tissue samples from 169 patients with ESCC who had undergone esophagectomy. The demographic characteristics of the patients are presented in Table 2. The median follow-up duration was 67 months (range, 60–238 months) for 60 survivors and 34 months (range, 1-238 months) for all 169 patients.
Table 2.
Baseline characteristics of 169 patients with esophageal squamous cell carcinoma receiving esophagectomy
| Age | ||
| median | 55 | |
| mean | 56.3 | |
| range | 29 ~ 80 | |
| Sex | ||
| male | 163 (96%) | |
| female | 6 (4%) | |
| Primary tumor location | ||
| Upper | 27 (16%) | |
| Middle | 63 (37%) | |
| Lower | 79 (47%) | |
| Pathological T classification | ||
| T1 | 56 (33%) | |
| T2 | 34 (20%) | |
| T3 | 64 (38%) | |
| T4 | 15 (9%) | |
| Pathological N classification | ||
| N0 | 115 (68%) | |
| N1 | 33 (20%) | |
| N2 | 14 (8%) | |
| N3 | 7 (4%) | |
| Pathological 8th AJCC Stage | ||
| IA | 5 (3%) | |
| IB | 46 (27%) | |
| IIA | 41 (24%) | |
| IIB | 22 (13%) | |
| IIIA | 9 (5%) | |
| IIIB | 26 (16%) | |
| IVA | 20 (12%) | |
| Histological grading | ||
| Grade 1 | 15 (9%) | |
| Grade 2 | 110 (65%) | |
| Grade 3 | 44 (26%) | |
| Surgical margin | ||
| Negative | 150 (89%) | |
| Positive | 19 (11%) | |
| AKR1B10 expression | ||
| Low expression | 81 (48%) | |
| Overexpression | 88 (52%) |
AJCC, American Joint Committee on Cancer; AKR1B10, Aldo-keto reductase 1B10; SREBP1, Sterol regulatory element-binding protein 1
Of the 169 patients analyzed, 88 (52%) patients exhibited AKR1B10 overexpression. Correlations between clinicopathological parameters and immunohistochemical expressions were outlined in Table 3. AKR1B10 overexpression showed a significant association with advanced pathological N classification N1/2/3 (P = 0.023), and advanced pathological 8th AJCC stage II/III/IVA (P = 0.011).
Table 3.
Associations between AKR1B10 expression and clinicopathological parameters in 169 patients with esophageal squamous cell carcinoma receiving esophagectomy
| Parameters | AKR1B10 expression | |||
|---|---|---|---|---|
| Low | Over | P value | ||
| Age (years) | <55 | 38 | 45 | 0.58 |
| ≧ 55 | 43 | 43 | ||
| Sex | Male | 78 | 85 | 0.92 |
| Female | 3 | 3 | ||
| Primary tumor location | U/M | 46 | 44 | 0.38 |
| L | 35 | 44 | ||
| Pathological T classification | T1/T2 | 47 | 43 | 0.23 |
| T3/T4 | 34 | 45 | ||
| Pathological N classification | N0 | 62 | 53 | 0.023* |
| N1/2/3 | 19 | 35 | ||
| Pathological 8th AJCC Stage | I/II | 60 | 54 | 0.078 |
| III/IVA | 21 | 34 | ||
| Pathological 8th AJCC Stage | I | 32 | 19 | 0.011* |
| II/III/IVA | 49 | 69 | ||
| Histological grading | Grade 1/2 | 61 | 64 | 0.70 |
| Grade 3 | 20 | 24 | ||
| Surgical margin | Negative | 72 | 78 | 0.96 |
| Positive | 9 | 10 | ||
AJCC, American Joint Committee on Cancer; AKR1B10, Aldo-keto reductase 1B10; SREBP1, Sterol regulatory element-binding protein 1. *Statistically significant
Survival analyses of patients with ESCC receiving esophagectomy
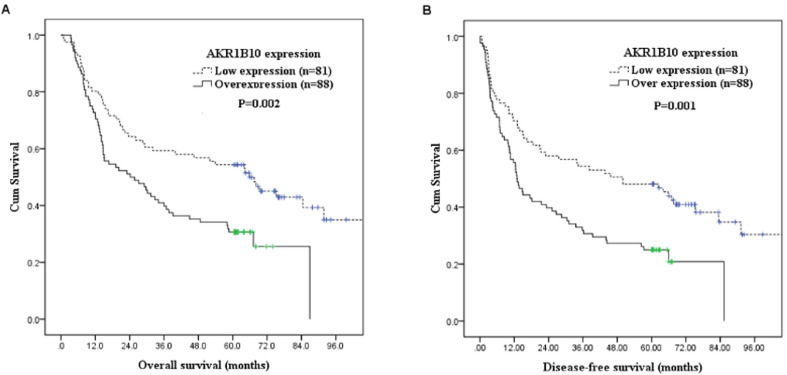
Associations between survival and clinicopathological factors, along with AKR1B10 expression were listed in Table 4. AKR1B10 overexpression (P = 0.002, Fig. 2A), positive surgical margin (P = 0.01), pathological T classification, T3/4 (P < 0.001), pathological N classification, N1/2/3 (P < 0.001), and pathological 8th AJCC stage III/IVA (P < 0.001) were significantly correlated with poor OS. Moreover, AKR1B10 overexpression (P = 0.001, Fig. 2B), positive surgical margin (P = 0.02), pathological T classification, T3/4 (P < 0.001), pathological N classification, N1/2/3 (P < 0.001), and pathological 8th AJCC stage III/IVA (P < 0.001) were associated with worse DFS.
Table 4.
Results of univariate log-rank analysis of prognostic factors for overall survival and disease-free survival in 169 patients with esophageal squamous cell carcinoma receiving esophagectomy
| Factors | No. of patients |
Overall survival (OS) | Disease-free survival (DFS) | |||
|---|---|---|---|---|---|---|
| 5-yr OS rate (%) | P value | 5-yr DFS rate (%) | P value | |||
| Age (years) | ||||||
| < 55 | 83 | 48% | 0.35 | 43% | 0.15 | |
| ≧ 55 | 86 | 36% | 29% | |||
| Location | ||||||
| U/M | 90 | 43% | 0.98 | 37% | 0.70 | |
| L | 79 | 41% | 36% | |||
| Pathological T classification | ||||||
| T1/2 | 90 | 58% | < 0.001* | 48% | < 0.001* | |
| T3/4 | 79 | 24% | 23% | |||
| Pathological N classification | ||||||
| N0 | 115 | 53% | < 0.001* | 45% | < 0.001* | |
| N1/2/3 | 54 | 19% | 17% | |||
| Pathological 8th AJCC stage | ||||||
| I/II | 114 | 54% | < 0.001* | 46% | < 0.001* | |
| III/IVA | 55 | 16% | 16% | |||
| Pathological 8th AJCC stage | ||||||
| I | 51 | 67% | < 0.001* | 59% | < 0.001* | |
| II/III/IVA | 118 | 31% | 26% | |||
| Histological grading | ||||||
| Grade 1/2 | 125 | 46% | 0.088 | 38% | 0.18 | |
| Grade 3 | 44 | 32% | 30% | |||
| Surgical margin | ||||||
| Negative | 150 | 45% | 0.01* | 38% | 0.02* | |
| Positive | 19 | 21% | 21% | |||
| AKR1B10 expression | ||||||
| Low expression | 81 | 54% | 0.002* | 48% | 0.001* | |
| Overexpression | 88 | 31% | 25% | |||
AJCC, American Joint Committee on Cancer; AKR1B10, Aldo-keto reductase 1B10; SREBP1, Sterol regulatory element-binding protein 1 *Statistically significant
Fig. 2.
Kaplan–Meier curves showed that patients with AKR1B10 overexpression had significantly worse overall survival and disease-free survival than those with low AKR1B10 expression. (A) Overall survival according to AKR1B10 expression. (B) Disease-free survival according to AKR1B10 expression. AKR1B10, Aldo-keto reductase 1B10
Multivariate survival analysis identified AKR1B10 overexpression (P = 0.029, hazard ratio = 1.579, 95% confidence interval: 1.047 ~ 2.382) as an independent factor associated with poor OS, along with pathological T classification, T3/4 (P = 0.007, hazard ratio = 1.956, 95% confidence interval: 1.198 ~ 3.193) and pathological N classification, N1/2/3 (P = 0.002, hazard ratio = 1.950, 95% confidence interval: 1.274 ~ 2.984). Moreover, AKR1B10 overexpression (P = 0.011, hazard ratio = 1.668, 95% confidence interval: 1.126 ~ 2.472), pathological T classification, T3/4 (P = 0.012, hazard ratio = 1.821, 95% confidence interval: 1.143 ~ 2.898), and pathological N classification, N1/2/3 (P = 0.007, hazard ratio = 1.767, 95% confidence interval: 1.170 ~ 2.670) were identified as independent poor prognostic factors for DFS. The 5-year OS and DFS rates were 31% and 25% in patients with AKR1B10 overexpression, and 54% and 48% in patients with low AKR1B10 expression, respectively.
AKR1B10 inhibitor, oleanolic acid, considerably decreased the incidence of esophageal tumor in the 4-NQO-induced ESCC murine model
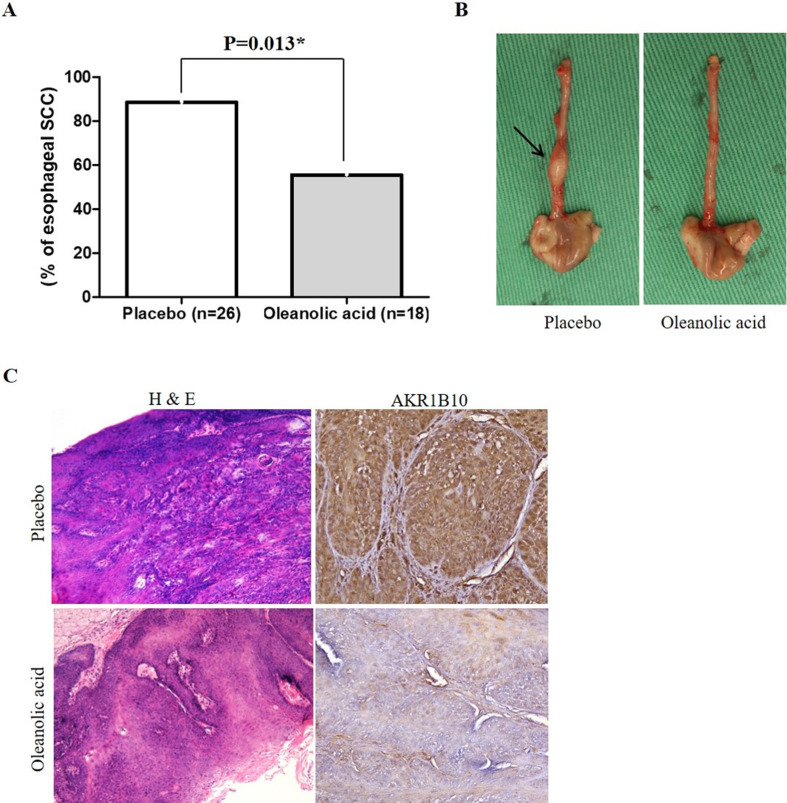
We investigated the potential inhibitory effect of the AKR1B10 inhibitor, oleanolic acid, in a 4-NQO-induced ESCC murine model. Among the mice administered with vehicle control, 23 (88%) out of 26 mice developed ESCC. Conversely, among the mice treated with oleanolic acid, 10 (56%) out of 18 mice developed ESCC. This demonstrated that the incidence of ESCC was significantly lower in mice treated with oleanolic acid than in those treated with vehicle control (56% versus 88%; P = 0.013; Fig. 3A and B). Furthermore, immunohistochemistry of the murine esophagus revealed that AKR1B10 expression (Fig. 3C) was lower in mice treated with oleanolic acid than in those treated with vehicle control.
Fig. 3.
AKR1B10 inhibitor, oleanolic acid, significantly decrease the incidence of ESCC in 4-NQO-induced ESCC murine model. (A) The incidence of ESCC in mice treated with oleanolic acid was significantly lower than that in mice treated with placebo control (56% versus 88%; P = 0.013). (B) Left: Gross appearance of the esophagus from representative mice treated with placebo control showed enlargement of the esophageal tumor indicated by the arrow. Right: Gross appearance of the esophagus from representative mice treated with oleanolic acid revealed no enlargement of the esophageal tumor. (C) Hematoxylin and eosin stained sections from representative mice treated with placebo control showed ESCC with stromal invasion, whereas sections from representative mice treated with oleanolic acid displayed only esophageal dysplasia. Original magnification ×100. Immunohistochemistry revealed lower AKR1B10 expression in the oleanolic acid group than in the placebo group. Original magnification ×200. AKR1B10, Aldo-keto reductase 1B10; ESCC, esophageal squamous cell carcinoma; 4-NQO, 4-nitroquinoline 1-oxide *statistically significant
Discussion
Major risk factors for ESCC include chronic tobacco use and alcohol consumption [2]. Excessive tobacco and alcohol consumption leads to production of ROS, generation of RCS, and persistent oxidative stress. This phenomenon can induce DNA damage and esophageal epithelial cell apoptosis [13, 31]. Continued oxidative stress can lead to chronic inflammation, which exacerbates esophageal epithelial cell damage and ESCC carcinogenesis [19]. However, the ability of ESCC tumor cells to evade RCS-induced apoptosis, and proliferate rapidly under these conditions remains elusive. Therefore, we hypothesized that ESCC tumor cells produce enzymes capable of catabolizing RCS, thereby preventing these ESCC tumor cells from undergoing RCS-induced apoptosis, and promoting ESCC progression. To align with our hypothesis, the expression levels of these enzymes should be higher in ESCC tumor cells than in normal esophageal epithelial cells. Therefore, we used the Agilent SurePrint G3 Human V2 GE 8 × 60 K microarray kit to analyze differentially expressed genes between nine paired tumors and adjacent normal esophageal tissue specimens. Among the 57 GO terms that were significantly altered between tumors and adjacent normal tissue specimens, we identified that GO:0004033 aldo-keto reductase activity molecular functional domain was significantly altered (P = 0.021), which was correlated with RCS metabolism. The aldo-keto reductases superfamily comprises monomeric, soluble, NADPH-dependent oxidoreductases. They play a crucial role in modifying the carbonyl groups present on aldehydes and ketones, facilitating their conversion into alcohols by eliminating reactions [32]. Given its functionality, it is plausible to suggest that these enzymes can catabolize RCS and may prevent ESCC tumor cells from RCS-induced apoptosis. Kazemi-Noureini et al. [33]. reported that AKR1C2 is up-regulated in ESCC compared to normal esophageal epithelium. Zhang et al. [34]. also found that AKR1C2 is up-regulated in ESCC tissues. Miller et al. [35]. revealed strong AKR1C3 immunoreactivity in columnar epithelium, with only mild immunoreactivity in squamous epithelium of the gastrointestinal junction. Additionally, both adenocarcinoma and squamous cell carcinoma originating from the gastroesophageal junction exhibited extensive AKR1C3 immunoreactivity. These studies further supported our findings.
Among the significant genes in GO:0004033 aldo-keto reductase activity, AKR1B10 was the most significant gene between ESCC tumors and adjacent normal esophageal tissue specimens. However, studies exploring the role of AKR1B10 in ESCC carcinogenesis are lacking. Using ESCC cell lines, we found that inhibiting endogenous AKR1B10 enhanced the cytotoxicity of 4-HNE, an RCS. In a 4-NQO-induced-ESCC murine model, oleanolic acid, an AKR1B10 inhibitor, significantly decreased the incidence of esophageal tumors. These results further support the hypothesis that AKR1B10 prevents ESCC tumor cells from undergoing RCS-induced apoptosis and promotes ESCC progression. Among the 169 patients with ESCC, 88(52%) exhibited AKR1B10 overexpression. However, 81 (48%) patients showed low AKR1B10 expression, suggesting that enzymes other than AKR1B10 may also be involved. In addition to AKR1B10, RCS can be metabolized by several oxidoreductases, including other members of the aldo-keto reductase superfamily, aldehyde dehydrogenases, and alcohol dehydrogenases [31].
The prognostic role of AKR1B10 in various cancers has been investigated, with conflicting results. Liu et al. [36]. reported that AKR1B10 overexpression predicts poor prognosis in gastric cancer patients undergoing surgical resection. Conversely, Yao et al. [37]. suggested that AKR1B10 may serve as a favorable prognostic indicator in patients with gastric cancer. Qu et al. [38]. demonstrated that AKR1B10 expression was positively correlated with lymph node metastasis, tumor size, and Ki67 expression, whereas it was negatively correlated with OS and DFS in patients with breast cancer. Hojnik et al. [39]. showed that AKR1B1 and AKR1B10 play protective roles in endometrioid endometrial cancer and are potential prognostic biomarkers. Ko et al. [40]. identified AKR1B10 overexpression as an independent poor prognostic biomarker for patients with oral squamous cell carcinoma. Hung et al. [41]. showed that AKR1B10 overexpression was a significant prognostic factor for poor recurrence-free survival in patients with resected lung adenocarcinoma. In contrast, Ha et al. [42]. reported contrasting findings in hepatocellular carcinoma, suggesting that high AKR1B10 protein expression may indicate a favorable prognosis in patients who underwent curative hepatectomy. Similarly, Schmitz et al. [43]. presented that high AKR1B10 expression reflected less aggressive tumor behavior in patients with hepatocellular carcinoma. The prognostic significance of AKR1B10 in ESCC has not been previously reported. Our findings indicated that AKR1B10 overexpression was significantly correlated with advanced pathological N classification and advanced pathological 8th AJCC Stage. Moreover, AKR1B10 overexpression was independently significantly associated with poor OS and DFS in ESCC patients. Our findings further supported the oncogenic role of AKR1B10 in ESCC.
The most abundant RCS are derived from oxidation of carbohydrates, lipids, and amino acids [44, 45]. Over 20 RCS have been identified in biological samples. RCS has the ability to modulate homeostasis at various levels, probably by damaging biological molecules, or participating in signaling/transcription regulation. This dual nature of RCS biological event appears to be dose- and time-dependent manner. The toxicity of RCS can be attributed to the inactivation of certain target enzymes. Accumulating evidence has shown that RCS such as 4-HNE, and MDA can function as messengers that activate or inhibit signaling pathways under physiologic or pathologic conditions. For example, low level of 4-HNE promotes proliferation, but at higher concentrations it induces differentiation and apoptosis [46, 47]. In the current study, the mechanism of how AKR1B10 protects RCS-induced apoptosis will be further investigated.
Using a 4-NQO-induced-ESCC murine model, we found that the AKR1B10 inhibitor significantly decreased the incidence of ESCC, suggesting that AKR1B10 may be a therapeutic target for ESCC. To the best of our knowledge, the therapeutic role of AKR1B10 inhibitor in ESCC has not been explored before. However, Suvilesh et al. [48]. used patient-derived tumor organoids in non-small cell lung cancer and found that AKR1B10 could be targeted by repurposing the AKR1B10 inhibitor, epalrestat, to overcome chemoresistance. Kong et al. [49]. reported that pharmaceutical inhibition of AKR1B10 with oleanolic acid in vitro and in a xenograft model significantly induced tumor regression and sensitized pancreatic adenocarcinoma cells to gemcitabine. Jin et al. [50]. showed that AKR1B10 inhibitor enhanced the inhibitory effect of sorafenib on hepatocellular carcinoma xenografts. These studies highlight the therapeutic potential of targeting AKR1B10 in malignancies, underscoring the need for further clinical trials in ESCC.
Our study had several limitations. First, among the significant genes in GO:0004033 aldo-keto reductase activity, cell line and mouse experiments were performed only for the most significant gene, AKR1B10. Second, the sample size was relatively small. There were only nine paired ESCC tumor samples and adjacent normal tissues for microarray analysis. In addition, our findings were based on a retrospective analysis from a single hospital. Third, several potential confounding factors may have influenced gene expression and serum concentrations of AKR1B10. Previous studies revealed that AKR1B10 expression has been observed in non-neoplastic diseases, such as diabetes nephropathy [51], atopic dermatitis [52], ulcerative colitis [53], and leprosy [54]. Park et al. [55]. also reported that blood AKR1B10 is elevated in people with nonalcoholic steatohepatitis and hepatic fibrosis. Fourth, our study found that AKR1B10 prevents ESCC neoplastic cells from undergoing RCS-induced apoptosis, but specific molecular mechanism was not deeply delved.
In conclusion, AKR1B10 expression was higher in human ESCC tumors than in adjacent normal esophageal epithelium, and independently significantly associated with poorer prognosis in 169 ESCC patients receiving esophagectomy. In vitro experiments revealed that inhibiting endogenous AKR1B10 enhanced the cytotoxicity of 4-HNE, a type of RCS, indicating that AKR1B10 prevents ESCC neoplastic cells from undergoing RCS-induced apoptosis. In a 4-NQO-induced-ESCC murine model, oleanolic acid, an AKR1B10 inhibitor, significantly decreased the incidence of ESCC. These findings suggest that AKR1B10 is involved in ESCC progression and may be a potential therapeutic target for ESCC.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Material 1: Supplemental Fig. 1: Left: Heat map and hierarchical clustering of genes in GO:0004033 aldo-keto reductase activity between nine paired ESCC tumors and adjacent normal tissue specimens. Right: Among the genes in GO:0004033 aldo-keto reductase activity, AKR1B10, AKR1B15, AKR1B1, AKR1C4, AKR1C3, AKR1C, and sepiapterin reductase (SPR) were significantly differentially expressed genes between nine paired ESCC tumors and adjacent normal tissue specimens. Each row represents one gene, and each column represents one paired patient sample (value = gene expression of tumor/gene expression of adjacent normal tissue). Relative gene expression was visualized using a color scale, where red indicates up-regulation and green represents down-regulation. ESCC, esophageal squamous cell carcinoma.
Acknowledgements
We thank Chang Gung Medical Foundation Kaohsiung Chang Gung Memorial Hospital Biobank & Research Specimen Processing Lab (CLRPG8L0084) for excellent technical support.
Abbreviations
- ESCC
Esophageal Squamous Cell Carcinoma
- RCS
Reactive Carbonyl Species
- GO
Gene Ontology
- 4-NQO
4–Nitroquinoline 1-Oxide
- AKR1B10
Aldo-Keto Reductase Family 1 Member B10
- ROS
Reactive Oxygen Species
- 4-HNE
4-Hydroxy Trans-2-Nonenal
- MDA
Malondialdehyde
- AJCC
American Joint Committee on Cancer
- OS
Overall Survival
- DFS
Disease-Free Survival
- IRS
Immunoreactive Score
- ELISA
Enzyme-Linked Immunosorbent Assay
- DMEM
Dulbecco’s Modified Eagle Medium
- FBS
Fetal Bovine Serum
- qRT-PCR
quantitative Real-Time Polymerase Chain Reaction
- SPR
Sepiapterin Reductase
Author contributions
SH Li and CH Chen designed the study and drafted the manuscript. WT Huang, YH Chen, HI Lu and CM Lo conserved tissue samples and collected the clinical data. HT Tsai performed experiments. All authors read and approved the final manuscript.
Funding
This work was supported in part by grants from the National Science Council, Taiwan (NSTC 112-2314-B-182 A-029-MY3), and Chang Gung Memorial Hospital (CMRPG8M0903, CMRPG8K1293, CORPG8L1271, and CORPG8P0491). This work was supported by grants from the National Science Council, Taiwan (NSTC 112-2320-B-075 A-003), and by grant TCVGH-1127319C, TCVGH-1137309C, TCVGH-1143914C, and TCVGH-1143904D from Taichung Veterans General Hospital Research Program, and by grant TCVGH-NCH U1137620 from Taichung Veterans General Hospital and National Chung Hsing University. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Data availability
The datasets supporting the conclusions of this article are included within the article.
Declarations
Ethics approval and consent to participate
This study was approved by the Chang Gung Medical Foundation Institutional Review Board (approval number: 201601568B0 and 202100927B0). All the methods were carried out in accordance with the approved guidelines, and written informed consent of the patients or their families was not judged necessary for this kind of retrospective study by the Chang Gung Medical Foundation Institutional Review Board. All animal experiments were approved by the Institutional Animal Care and Use Committee of the Chang Gung Memorial Hospital (2016112302).
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Chung CS, Lee YC, Wang CP et al. Secondary prevention of esophageal squamous cell carcinoma in areas where smoking, alcohol, and betel quid chewing are prevalent. J Formos Med Assoc. 2010;109(6):408 – 21. (In eng). DOI: S0929-6646(10)60072-1 [pii] 10.1016/S0929-6646(10)60072-1 [DOI] [PubMed]
- 3.Lo CM, Wang YM, Chen YH, et al. The impact of Radiotherapy Dose in patients with locally advanced esophageal squamous cell carcinoma receiving Preoperative Chemoradiotherapy. Curr Oncol. 2021;28(2):1354–65. 10.3390/curroncol28020129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Trachootham D, Alexandre J, Huang P. Targeting cancer cells by ROS-mediated mechanisms: a radical therapeutic approach? Nat Rev Drug Discovery. 2009;8(7):579–91. 10.1038/nrd2803. [DOI] [PubMed] [Google Scholar]
- 5.Thanan R, Oikawa S, Hiraku Y, et al. Oxidative stress and its significant roles in neurodegenerative diseases and cancer. Int J Mol Sci. 2015;16(1):193–217. 10.3390/ijms16010193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khan TH, Srivastava N, Srivastava A, et al. SHP-1 plays a crucial role in CD40 signaling reciprocity. J Immunol. 2014;193(7):3644–53. 10.4049/jimmunol.1400620. [DOI] [PubMed] [Google Scholar]
- 7.Zhang X, Kiapour N, Kapoor S, et al. IL-11 induces encephalitogenic Th17 cells in multiple sclerosis and experimental autoimmune encephalomyelitis. J Immunol. 2019;203(5):1142–50. 10.4049/jimmunol.1900311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tarique M, Naz H, Suhail M, et al. Differential expression of programmed death 1 (PD-1) on various immune cells and its role in human leprosy. Front Immunol. 2023;14:1138145. 10.3389/fimmu.2023.1138145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seyedsadr M, Wang Y, Elzoheiry M, et al. IL-11 induces NLRP3 inflammasome activation in monocytes and inflammatory cell migration to the central nervous system. Proc Natl Acad Sci U S A. 2023;120(26):e2221007120. 10.1073/pnas.2221007120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hisamuddin M, Rizvi I, Malik A, et al. Characterization of pH-induced conformational changes in recombinant DENV NS2B-NS3pro. Int J Biol Macromol. 2023;253(Pt 3):126823. 10.1016/j.ijbiomac.2023.126823. [DOI] [PubMed] [Google Scholar]
- 11.Das S, Khan TH, Sarkar D. Comprehensive Review on the Effect of Stem cells in Cancer Progression. Curr Tissue Microenvironment Rep. 2024;5(2):39–59. 10.1007/s43152-024-00053-6. [Google Scholar]
- 12.Khan TH, Muhammad N, Tarique M, Usmani D, Naz H, Sarode A. The role of Cancer-Specific Target antigens in CAR T Cell Therapy in Hematological Malignancies. Curr Tissue Microenvironment Rep. 2024;5(2):61–7. 10.1007/s43152-024-00055-4. [Google Scholar]
- 13.Mena S, Ortega A, Estrela JM. Oxidative stress in environmental-induced carcinogenesis. Mutat Res. 2009;674(1–2):36–44. 10.1016/j.mrgentox.2008.09.017. [DOI] [PubMed] [Google Scholar]
- 14.Marnett LJ, Riggins JN, West JD. Endogenous generation of reactive oxidants and electrophiles and their reactions with DNA and protein. J Clin Invest. 2003;111(5):583–93. 10.1172/JCI18022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Esterbauer H, Schaur RJ, Zollner H. Chemistry and biochemistry of 4-hydroxynonenal, malonaldehyde and related aldehydes. Free Radic Biol Med. 1991;11(1):81–128. http://www.ncbi.nlm.nih.gov/pubmed/1937131. [DOI] [PubMed] [Google Scholar]
- 16.Csala M, Kardon T, Legeza B, et al. On the role of 4-hydroxynonenal in health and disease. Biochim Biophys Acta. 2015;1852(5):826–38. 10.1016/j.bbadis.2015.01.015. [DOI] [PubMed] [Google Scholar]
- 17.Pizzimenti S, Ciamporcero E, Daga M, et al. Interaction of aldehydes derived from lipid peroxidation and membrane proteins. Front Physiol. 2013;4:242. 10.3389/fphys.2013.00242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Y, Chen H, Sun Z, Chen X. Molecular mechanisms of ethanol-associated oro-esophageal squamous cell carcinoma. Cancer Lett. 2015;361(2):164–73. 10.1016/j.canlet.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haas SL, Ye W, Lohr JM. Alcohol consumption and digestive tract cancer. Curr Opin Clin Nutr Metab Care. 2012;15(5):457–67. 10.1097/MCO.0b013e3283566699. [DOI] [PubMed]
- 20.Mansoori AA, Jain SK. Molecular links between Alcohol and Tobacco Induced DNA damage, Gene Polymorphisms and patho-physiological consequences: a systematic review of hepatic carcinogenesis. Asian Pac J Cancer Prev. 2015;16(12):4803–12. http://www.ncbi.nlm.nih.gov/pubmed/26163595. [DOI] [PubMed] [Google Scholar]
- 21.Barrera G. Oxidative stress and lipid peroxidation products in cancer progression and therapy. ISRN Oncol. 2012;2012:137289. 10.5402/2012/137289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Watson A, Mazumder A, Stewart M, Balasubramanian S. Technology for microarray analysis of gene expression. Curr Opin Biotechnol. 1998;9(6):609–14. http://www.ncbi.nlm.nih.gov/pubmed/9889134. [DOI] [PubMed] [Google Scholar]
- 23.Ritchie ME, Phipson B, Wu D, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.clusterProfiler. An R package for comparing biological themes among gene clusters.OMICS. 2012;16(5):284–7. 10.1089/omi.2011.0118. Epub 2012 Mar 28. [DOI] [PMC free article] [PubMed]
- 25.Pathview. An R/Bioconductor package for pathway-based data integration and visualization.Bioinformatics, 29(14):1830–1, 10.1093/bioinformatics/btt285 [DOI] [PMC free article] [PubMed]
- 26.D’Journo XB. Clinical implication of the innovations of the 8(th) edition of the TNM classification for esophageal and esophago-gastric cancer. J Thorac Disease. 2018;10(Suppl 22):S2671–81. 10.21037/jtd.2018.03.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li SH, Qian L, Chen YH, et al. Targeting MYO1B impairs tumorigenesis via inhibiting the SNAI2/cyclin D1 signaling in esophageal squamous cell carcinoma. J Cell Physiol. 2022;237(9):3671–86. 10.1002/jcp.30831. [DOI] [PubMed] [Google Scholar]
- 28.Li SH, Lu HI, Chang AY, et al. Angiotensin II type I receptor (AT1R) is an independent prognosticator of esophageal squamous cell carcinoma and promotes cells proliferation via mTOR activation. Oncotarget. 2016;7(41):67150–65. 10.18632/oncotarget.11567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang X, Bai H, Zhang X, et al. Inhibitory effect of oleanolic acid on hepatocellular carcinoma via ERK-p53-mediated cell cycle arrest and mitochondrial-dependent apoptosis. Carcinogenesis. 2013;34(6):1323–30. 10.1093/carcin/bgt058. [DOI] [PubMed] [Google Scholar]
- 30.Penning TM. The aldo-keto reductases (AKRs): overview. Chem Biol Interact. 2015;234:236–46. 10.1016/j.cbi.2014.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Singh M, Kapoor A, Bhatnagar A. Oxidative and reductive metabolism of lipid-peroxidation derived carbonyls. Chem Biol Interact. 2015;234:261–73. 10.1016/j.cbi.2014.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Penning TM. The aldo-keto reductases (AKRs): overview. Chem Biol Interact. 2014. 10.1016/j.cbi.2014.09.024. In Eng. [DOI] [PMC free article] [PubMed]
- 33.Kazemi-Noureini S, Colonna-Romano S, Ziaee AA, et al. Differential gene expression between squamous cell carcinoma of esophageus and its normal epithelium; altered pattern of mal, akr1c2, and rab11a expression. World J Gastroenterol. 2004;10(12):1716–21. 10.3748/wjg.v10.i12.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang ZF, Huang TJ, Zhang XK, et al. AKR1C2 acts as a targetable oncogene in esophageal squamous cell carcinoma via activating PI3K/AKT signaling pathway. J Cell Mol Med. 2020;24(17):9999–10012. 10.1111/jcmm.15604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller VL, Lin HK, Murugan P, et al. Aldo-keto reductase family 1 member C3 (AKR1C3) is expressed in adenocarcinoma and squamous cell carcinoma but not small cell carcinoma. Int J Clin Exp Pathol. 2012;5(4):278–89. https://www.ncbi.nlm.nih.gov/pubmed/22670171. [PMC free article] [PubMed] [Google Scholar]
- 36.Liu YY, Liu YW, Huang GK, et al. Overexpression of AKR1B10 predicts poor prognosis in gastric Cancer patients undergoing Surgical Resection. Curr Oncol. 2022;30(1):85–99. 10.3390/curroncol30010007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yao HB, Xu Y, Chen LG, et al. AKR1B10, a good prognostic indicator in gastric cancer. Eur J Surg Oncol. 2014;40(3):318–24. 10.1016/j.ejso.2013.12.014. [DOI] [PubMed] [Google Scholar]
- 38.Qu J, Li J, Zhang Y, et al. AKR1B10 promotes breast cancer cell proliferation and migration via the PI3K/AKT/NF-kappaB signaling pathway. Cell Bioscience. 2021;11(1):163. 10.1186/s13578-021-00677-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hojnik M, Frkovic Grazio S, Verdenik I, Rizner TL. AKR1B1 and AKR1B10 as prognostic biomarkers of endometrioid endometrial carcinomas. Cancers (Basel). 2021;13(14). 10.3390/cancers13143398. [DOI] [PMC free article] [PubMed]
- 40.Ko HH, Cheng SL, Lee JJ, Chen HM, Kuo MY, Cheng SJ. Expression of AKR1B10 as an independent marker for poor prognosis in human oral squamous cell carcinoma. Head Neck. 2017;39(7):1327–32. 10.1002/hed.24759. [DOI] [PubMed] [Google Scholar]
- 41.Hung JJ, Yeh YC, Hsu WH. Prognostic significance of AKR1B10 in patients with resected lung adenocarcinoma. Thorac Cancer. 2018;9(11):1492–9. 10.1111/1759-7714.12863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ha SY, Song DH, Lee JJ, Lee HW, Cho SY, Park CK. High expression of aldo-keto reductase 1B10 is an independent predictor of favorable prognosis in patients with hepatocellular carcinoma. Gut Liver. 2014;8(6):648–54. 10.5009/gnl13406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schmitz KJ, Sotiropoulos GC, Baba HA, et al. AKR1B10 expression is associated with less aggressive hepatocellular carcinoma: a clinicopathological study of 168 cases. Liver Int. 2011;31(6):810–6. 10.1111/j.1478-3231.2011.02511.x. [DOI] [PubMed] [Google Scholar]
- 44.Alessandra A, Giovanna B, Erica G, et al. Lipid peroxidation derived reactive carbonyl species in free and conjugated forms as an index of lipid peroxidation: limits and perspectives. Redox Biol. 2021;42. 10.1016/j.redox.2021.101899. Epub 2021 Feb 17.53. [DOI] [PMC free article] [PubMed]
- 45.Halyna MS. eCollection, Reactive carbonyl species in vivo: generation and dual biological effects. ScientificWorldJournal. 2014:2014:417842. 10.1155/2014/417842. 2014. [DOI] [PMC free article] [PubMed]
- 46.E E D VAD. Role of 4-hydroxy-trans-2-nonenal in cell function. Biochem (Mosc). 2010;75(9):1069–87. 10.1134/s0006297910090014.55. [DOI] [PubMed] [Google Scholar]
- 47.Yogesh CA, Rajendra S. Role of 4-hydroxynonenal in stress-mediated apoptosis signaling. Mol Aspects Med. 2003;24(4–5):219–30. 10.1016/s0098-2997(03)00017-7. [DOI] [PubMed] [Google Scholar]
- 48.Suvilesh KN, Manjunath Y, Nussbaum YI, et al. Targeting AKR1B10 by drug repurposing with epalrestat overcomes chemoresistance in non-small cell lung cancer patient-derived tumor organoids. Clin Cancer Res. 2024. 10.1158/1078-0432.CCR-23-3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kong D, Wu Y, Tong B, et al. CHES1 modulated tumorigenesis and senescence of pancreas cancer cells through repressing AKR1B10. Biochim Biophys Acta Mol Basis Dis. 2024;1870(6):167214. 10.1016/j.bbadis.2024.167214. [DOI] [PubMed] [Google Scholar]
- 50.Jin YY, Han C, Geng N, et al. [AKR1B10 inhibitor enhances the inhibitory effect of sorafenib on liver cancer xenograft]. Zhonghua Gan Zang Bing Za Zhi. 2019;27(1):39–44. 10.3760/cma.j.issn.1007-3418.2019.01.009. [DOI] [PubMed] [Google Scholar]
- 51.Shaw N, Yang B, Millward A, Demaine A, Hodgkinson A. AKR1B10 is induced by hyperglycaemia and lipopolysaccharide in patients with diabetic nephropathy. Cell Stress Chaperones. 2014;19(2):281–7. 10.1007/s12192-013-0455-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ghosh D, Ding L, Sivaprasad U, et al. PLoS ONE. 2015;10(12):e0144316. 10.1371/journal.pone.0144316. Multiple Transcriptome Data Analysis Reveals Biologically Relevant Atopic Dermatitis Signature Genes and Pathways. [DOI] [PMC free article] [PubMed]
- 53.Shen Y, Ma J, Yan R, et al. Impaired self-renewal and increased colitis and dysplastic lesions in colonic mucosa of AKR1B8-deficient mice. Clin Cancer Res. 2015;21(6):1466–76. 10.1158/1078-0432.CCR-14-2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Soares CT, Fachin LRV, Trombone APF, Rosa PS, Ghidella CC, Belone AFF. Potential of AKR1B10 as a biomarker and therapeutic target in type 2 Leprosy reaction. Front Med (Lausanne). 2018;5:263. 10.3389/fmed.2018.00263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Park A, Choi SJ, Park S, et al. Plasma Aldo-Keto Reductase Family 1 Member B10 as a Biomarker performs well in the diagnosis of nonalcoholic steatohepatitis and fibrosis. Int J Mol Sci. 2022;23(9). 10.3390/ijms23095035. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Ghosh D, Ding L, Sivaprasad U, et al. PLoS ONE. 2015;10(12):e0144316. 10.1371/journal.pone.0144316. Multiple Transcriptome Data Analysis Reveals Biologically Relevant Atopic Dermatitis Signature Genes and Pathways. [DOI] [PMC free article] [PubMed]
Supplementary Materials
Supplementary Material 1: Supplemental Fig. 1: Left: Heat map and hierarchical clustering of genes in GO:0004033 aldo-keto reductase activity between nine paired ESCC tumors and adjacent normal tissue specimens. Right: Among the genes in GO:0004033 aldo-keto reductase activity, AKR1B10, AKR1B15, AKR1B1, AKR1C4, AKR1C3, AKR1C, and sepiapterin reductase (SPR) were significantly differentially expressed genes between nine paired ESCC tumors and adjacent normal tissue specimens. Each row represents one gene, and each column represents one paired patient sample (value = gene expression of tumor/gene expression of adjacent normal tissue). Relative gene expression was visualized using a color scale, where red indicates up-regulation and green represents down-regulation. ESCC, esophageal squamous cell carcinoma.
Data Availability Statement
The datasets supporting the conclusions of this article are included within the article.