Abstract
Species classified in Cordycipitaceae (Hypocreales) include multiple entomopathogenic fungi. Numerous changes have recently occurred in the nomenclature of cordycipitaceous fungi due to the single naming system proposed for pleomorphic fungi in 2011. Species of Cordycipitaceae are widely applied as herbal medicines, especially in Asian cultures. However, the diversity of Cordycipitaceae in Taiwan is based on relatively few literature records. Here we conducted a comprehensive survey of this family throughout the island of Taiwan and provided a glimpse of the diversity and distribution patterns. In addition, the present study reassesses the generic and species boundaries of Cordycipitaceae and finally provides an updated phylogenetic overview of Cordyceps and allied genera. Phylogenetic reconstructions using combined ITS, nrLSU, tef1-α, rpb1, and rpb2 sequence data, along with morphological features, resulted in 10 novel species: Akanthomyces taiwanicus sp. nov., Blackwellomyces taiwanensis sp. nov., Cordyceps hehuanensis sp. nov., C. locastrae sp. nov., C. malleiformis sp. nov., C. pseudorosea sp. nov., C. siangyangensis sp. nov., Samsoniella lasiocampidarum sp. nov., S. yuanzuiensis sp. nov., and Simplicillium salviniae sp. nov.; and nine new records for Taiwan: A. kanyawimiae, A. muscarius, S. cardinalis, S. hepiali, B. lii, B. medogensis, C. lepidopterorum, C. neopruinosa, and Si. chinense. Furthermore, we provided DNA sequence data of the ex-type strains of C. ninchukispora for the first time and determined the species limits of the taxon. In addition, the present study proposed to synonymize B. staphylinidicola and C. jakajanicola under B. bassiana and C. lepidopterorum, respectively. Moreover, three species, C. roseostromata, C. kyushuensis, and C. shuifuensis, that clustered within the species clade of C. militaris are proposed to be synonymized under the latter taxon. To maintain the monophyly of Cordyceps, we propose to classify Parahevansia koratensis in Cordyceps, which makes the genus Parahevansia obsolete.
Taxonomic novelties: New species: Akanthomyces taiwanicus W.Y. Chuang, B. Shrestha & H.A. Ariyaw., Blackwellomyces taiwanensis W.Y. Chuang & H.A. Ariyaw., Cordyceps hehuanensis W.Y. Chuang & H.A. Ariyaw., C. locastrae W.Y. Chuang & H.A. Ariyaw., C. malleiformis W.Y. Chuang & H.A. Ariyaw., C. pseudorosea W.Y. Chuang & H.A. Ariyaw., C. siangyangensis W.Y. Chuang & H.A. Ariyaw., Samsoniella lasiocampidarum W.Y. Chuang & H.A. Ariyaw., S. yuanzuiensis W.Y. Chuang & H.A. Ariyaw., Simplicillium salviniae W.Y. Chuang & H.A. Ariyaw. New combination: Cordyceps koratensis (Hywel-Jones) H.A. Ariyaw., M. Stadler & Luangsa-ard. New synonyms: Beauveria bassiana (Bals.-Criv.) Vuill., Cordyceps lepidopterorum Mongkols. et al., C. militaris (L.) Fr.
Citation: Chuang WY, Lin YC, Shrestha B, Luangsa-ard JJ, Stadler M, Tzean SS, Wu S, Ko CC, Hsieh SY, Wu ML, Wang SC, Shen TL, Ariyawansa HA (2024). Phylogenetic diversity and morphological characterization of cordycipitaceous species in Taiwan. Studies in Mycology 109: 1–56. doi: 10.3114/sim.2024.109.01
Keywords: Cordycipitaceae, entomopathogenic fungi, new species, phylogenetic analyses
INTRODUCTION
Hypocrealean fungi (Sordariomycetes, Ascomycota) include a wide range of entomopathogens (Sung et al. 2007a, Vega et al. 2009a, Vega et al. 2012, Araujo & Hughes 2016) and were historically all placed within a single family, Clavicipitaceae (Kobayasi 1941, 1982, Samson 1974, Luangsa-ard et al. 2004). However, phylogenetic studies have shown that entomopathogenic fungi within Hypocreales are polyphyletic and as such are now classified in several families including Bionectriaceae, Clavicipitaceae, Cocoonihabitaceae, Cordycipitaceae, Hypocreaceae, Ophiocordycipitaceae, and Polycephalomycetaceae (Sung et al. 2007a, b, Chen et al. 2016, Shrestha et al. 2017, Zhuang & Zeng 2017, Xiao et al. 2023). The majority of Cordycipitaceae species produce stalked, erect stromatic ascomata that are fleshy in texture, but some are characterized by reduced stipes or subiculate stromata (Kobayasi & Shimizu 1982, Sung et al. 2007a). In addition, several species of Cordyceps s.s. are now nomenclaturally synonymized with various asexual morphs (Kepler et al. 2017).
In agreement with the reframing of Article 59 of the International Code of Nomenclature for algae, fungi, and plants (ICN) (McNeill et al. 2012), Cordyceps was recommended to be conserved against Isaria by Kepler et al. (2017). Accordingly, Kepler et al. (2017) recognized nine genera in Cordycipitaceae, including Akanthomyces, Ascopolyporus, Beauveria, Cordyceps, Engyodontium, Gibellula, Hyperdermium, Parengyodontium, and Simplicillium and erected two new genera, Blackwellomyces and Hevansia. According to the outline of fungi and fungi-like organisms, 17 genera are currently accepted in the Cordycipitaceae (Wijayawardene et al. 2020). In addition, in recent studies, six new genera were introduced, including Bhushaniella, Gamszarea, Jenniferia, Neohyperdermium, Parahevansia, Pleurodesmospora, and Polystromomyces (Zhang et al. 2021, Chen et al. 2021, Thanakitpipattana et al. 2022, Mongkolsamrit et al. 2022, Mongkolsamrit et al. 2023).
Taiwan, a geographically isolated island, is home to multiple unique fungal species including cordycipitaceous fungi (Tzean et al. 1997, Ke & Ju 2015, Ariyawansa et al. 2018, Tsai et al. 2018, Chen et al. 2020, Yang et al. 2020). Knowledge of the cordycipitaceous taxa in Taiwan has a long history. For example, Sawada (1922) reported 40 species of entomopathogenic fungi including Beauveria, Gibellula, and Isaria. A comprehensive study done by Tzean et al. (1997) described over 831 entomopathogenic fungi in Taiwan and introduced four new species, one new variety, and 37 new records of entomopathogenic fungi in Taiwan. Tzean et al. (1997) also concluded that Paecilomyces is the most prevalent entomopathogenic fungal group in Taiwan followed by Aschersonia, Beauveria, Gibellula, and Nomuraea. In addition to these predominant genera, several genera in Cordycipitaceae such as Akanthomyces and Cordyceps were also reported. Unfortunately, most of the above-mentioned studies were done in the absence of DNA sequence data and fungal strain identifications were based only on the phenotypes of the fungal groups. However, many recent studies have suggested that natural classification of cordycipitaceous taxa based solely on morphological characters does not comply with molecular phylogenetic positions (Sung et al. 2007a, Chen et al. 2016, Kepler et al. 2017). Therefore, the objective of this study is to investigate the diversity, taxonomy, and molecular phylogeny of cordycipitaceous taxa in Taiwan and provide their phylogenetic classification.
MATERIALS AND METHODS
Specimen collection and isolation
Cordycipitaceous specimens were collected in forests of Taiwan (Fig. 1) during 2017–2019, ranging in altitude from ten to 3 952 meters above sea level. Sample preparations and morphological examinations were performed as described by Ariyawansa et al. (2018) and Yang et al. (2020). In brief, spores were mounted in one mL of sterile distilled water on a flame-sterilized cavity slide, and were tweaked and mixed to create a uniform suspension. The spore suspension was then diluted in five mL of sterile distilled water, spread on 2 % water agar (WA, BioShop®) and incubated at 25 °C in the dark for 12 h or further until spore germination was observed. Germinated spores were individually isolated and transferred to malt extract agar (MEA; HIMEDIA®) with a sterile syringe needle and then incubated at 25 °C for 7–14 d. All the single spore cultures were transferred to potato dextrose agar (PDA; HIMEDIA®) slants and preserved at 4 °C.
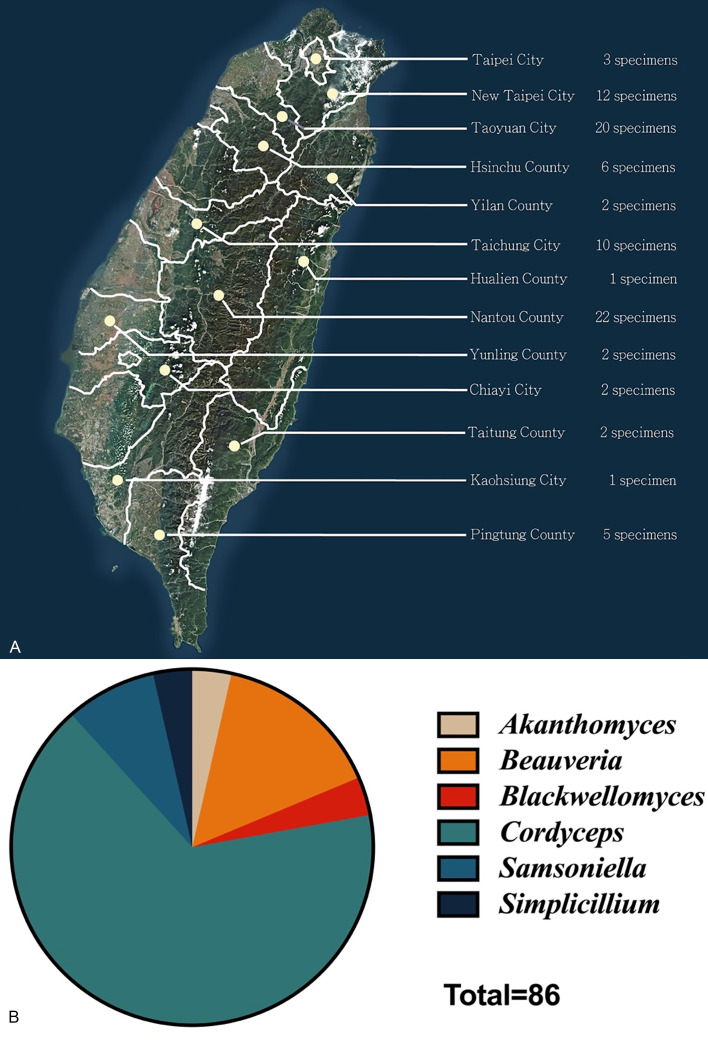
Fig. 1.
Overall information of the 86 cordycipitaceous fungi collected in this study. A. Geographic distribution of cordycipitaceous fungi collected in this study. (Satellite imagery source: FORMOSAT-5 (FS-5) satellite, National Space Organization, Hsinchu City, Taiwan.) B. Proportions of the genera of 86 cordycipitaceous fungi collected in this study.
Voucher specimens were deposited at the fungarium of the Department of Plant Pathology and Microbiology, National Taiwan University (NTUPPMH) in Taipei, Taiwan and the National Museum of Natural Science (NMNS) in Taichung, Taiwan. Ex-type cultures were deposited at the Department of Plant Pathology and Microbiology, National Taiwan University Culture Collection (NTUPPMCC) and the Bioresource Collection and Research Center (BCRC) in Hsinchu, Taiwan.
Morphological observations
To describe the sexual morphs, specimens were first observed and photographed using a Canon PowerShot SX730 HS camera. Hand sections of the perithecia were mounted in distilled water for microscopic observations. The micro-morphological characteristics such as shape and size of perithecia, asci, ascus caps, ascospores, part-spores, phialides, and conidia were examined and recorded using a compound microscope (Olympus® BX51) with differential interference contrast (DIC) illumination.
To describe asexual morphs in culture, single spore cultures were grown on 2 % PDA at 25 °C in the dark for 7–21 d. Sporulation structures were first observed under a dissecting microscope (Hamlet® SEM-HB) and transferred to a slide for morphological observation using a compound microscope (Olympus® BX51) with DIC illumination. For each micro-morphological structure such as conidia, phialides, etc., 30 measurements were taken and recorded using cellSens Standard software (Olympus®).
DNA extraction and PCR amplification
Pure cultures were grown on 2 % PDA at 25 °C for 7–21 d. Scraped mycelia were transferred to a 1.5 mL tube and homogenized with glass beads using a 2010 Geno homogenizer (Grinder® SPEX SamplePrep, USA). Then, genomic DNA was extracted using an EasyPure Genomic DNA Spin Kit (Bioman®, New Taipei, Taiwan) according to the manufacturer’s instructions. PCR amplification was carried out in a 25 μL reaction containing 1X Taq PCR Mix-RED (Bioman®, New Taipei, Taiwan) and 10 μM of each forward and reverse primer.
Five genomic regions (ITS, internal transcribed spacer region; nrLSU, large subunit ribosomal DNA; tef1-α, translation elongation factor 1-alpha gene; rpb1, RNA polymerase II large subunit 1; and rpb2) were selected for genotyping following recent publications (Tasanathai et al. 2016, Kepler et al. 2017, Mongkolsamrit et al. 2018). Primer sequences and PCR conditions are listed in Supplementary Table S1. PCR products were visualized on 1.5 % agarose electrophoresis gels. Purification and sequencing of PCR products were carried out at Genomics Company, New Taipei, Taiwan using the Sanger sequencing method and the obtained sequences were deposited in NCBI GenBank.
Phylogenetic analyses
DNA sequences were first analysed via BioEdit v. 7.2.5 software (Informer Technologies, Inc.) to check the quality of the sequences before submission to BLASTn on the NCBI website. Sequences of related species and outgroups were selected based on previous studies (Sung et al. 2007a, Kepler et al. 2017, Zhou et al. 2018, Mongkolsamrit et al. 2018, Vinit et al. 2018, Chen et al. 2019, Wei et al. 2019, Mongkolsamrit et al. 2020, Aini et al. 2020, Kondo et al. 2020, Wang et al. 2020, Chen et al. 2022, Dong et al. 2022) and downloaded from GenBank to construct multiple sequence alignments (Table 1). Multiple sequence alignments were performed via MAFFT v. 7.409 (http://mafft.cbrc.jp/alignment/server/index.html) with default settings. Alignments were manually improved using BioEdit v. 7.2.5 software and can be accessed under 10.6084/m9.figshare.25075217. Single gene alignment data matrixes for each locus and the combined gene datasets were analysed using different tree inference methods as described in Supplementary Table S2.
Table 1.
List of species and GenBank accession numbers of sequences used in this study
The accession numbers marked with T refer to sequences from type species. The sequences generated in this study are in bold.
Phylogenetic analyses were based on maximum likelihood (ML) for all single and combined loci while Bayesian inference (BI) was used only for the combined gene analyses. For the ML analyses, the best-fit substitution models were executed for each gene region under the Akaike Information Criterion (AIC) with the nexus-formed partition file using the Model Selection criteria provided in the IQ-TREE v. 1.6.12; the best fitting models for each gene partition are provided in Supplementary Table S2 (Kalyaanamoorthy et al. 2017). ML trees were inferred with 1 000 bootstrap replicates using the ultrafast algorithm in IQ-TREE with the concatenated data matrix, and the resulting maximum likelihood bootstrap (MLB) values ≥ 70 % were indicated at each node (Nguyen et al. 2015, Chernomor et al. 2016).
For the BI analyses, the best evolutionary model was decided under the AIC via MrModeltest v. 2.3 (Nylander 2004) and is given in Supplementary Table S2. MrBayes v. 3.2.7 (Ronquist et al. 2012) was used to generate BI trees under optimal criteria per partition. The Markov Chain Monte Carlo (MCMC) analysis used six chains following the method described by Tsai et al. (2018). The distribution of log-likelihood scores was examined to determine the stationary phase for each search and to decide if extra runs were required to achieve convergence using the program Tracer 1.7.2 (Rambaut & Drummond 2009). All sampled topologies beneath the asymptote (20 %) were discarded as part of the burn-in procedure, and the remaining trees were used for calculating posterior probabilities (PP) in the majority rule consensus tree. PP equal to or greater than 0.95 are given below or above each node (as seen in Figs 2–6). FigTree v. 1.4 (Rambaut & Drummond 2012) was used to view the phylogenetic trees and data files.
Fig. 2.
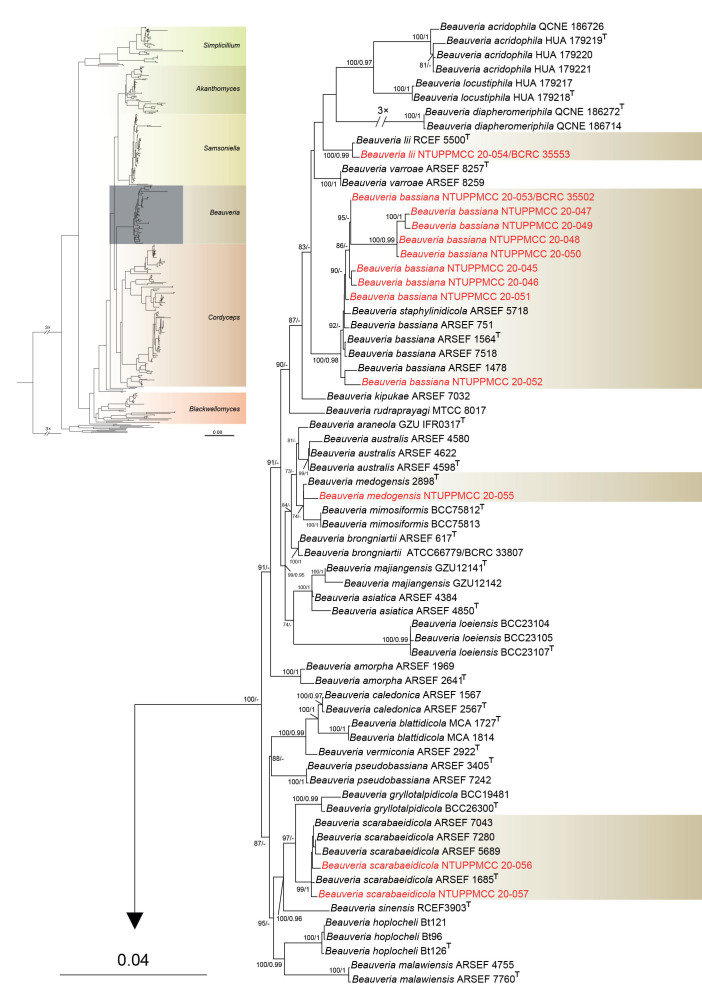
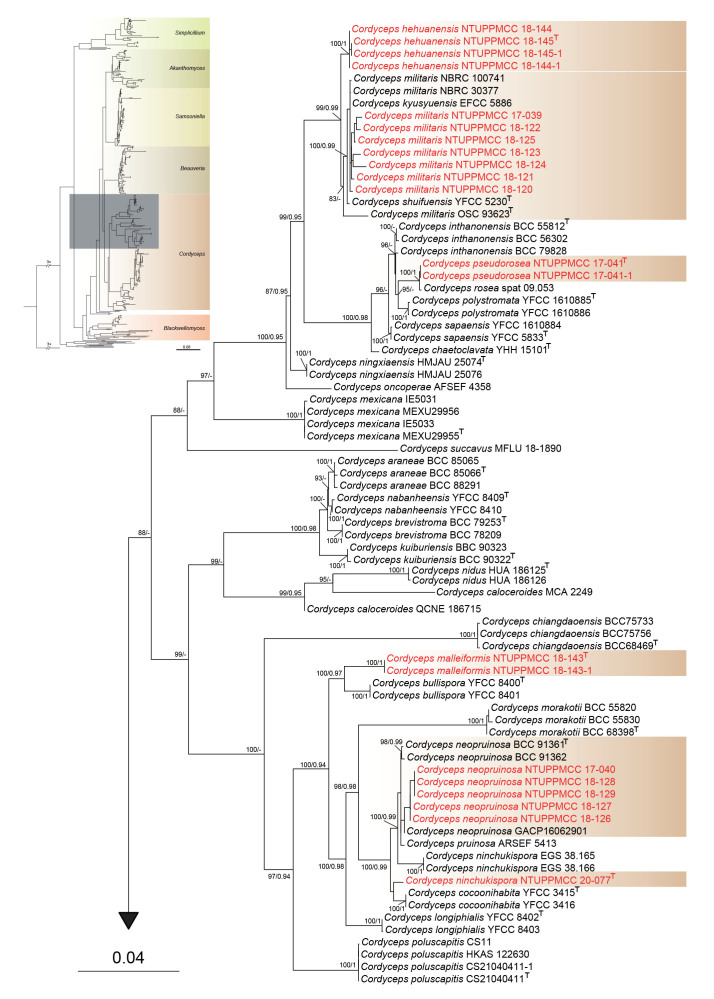
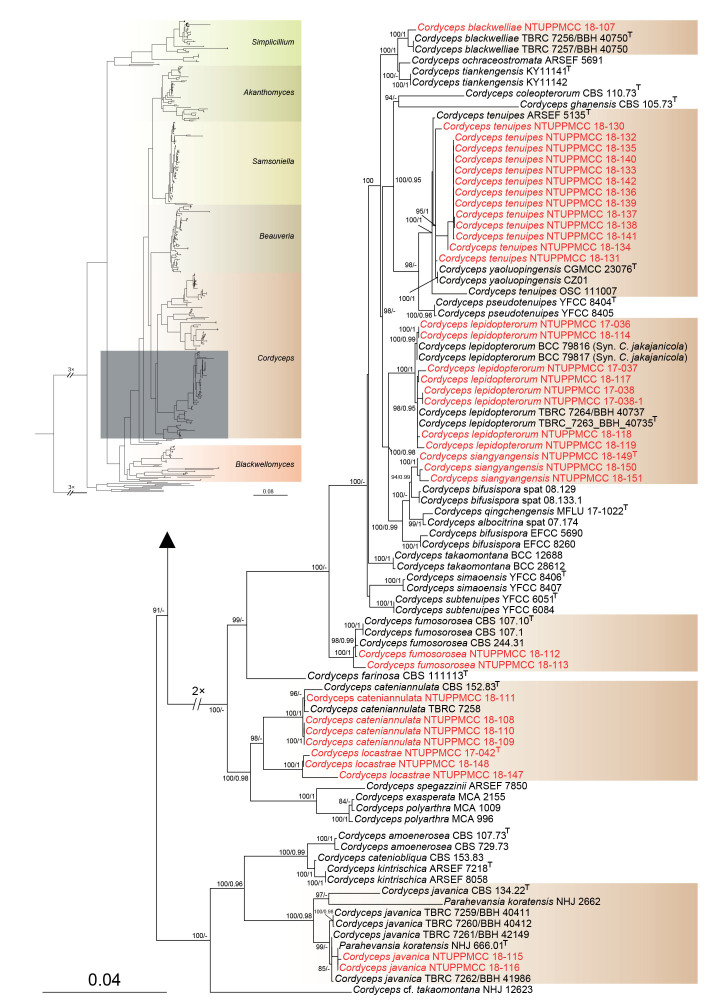
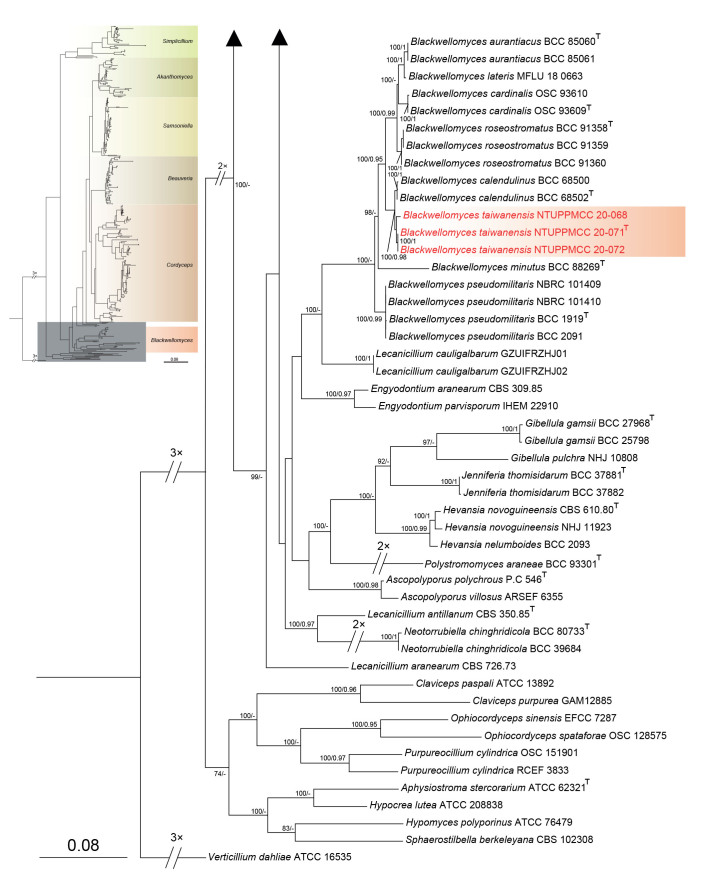
Maximum likelihood phylogenetic tree of Cordycipitaceae and related families based on a combined dataset of ITS, nrLSU, tef1-α, rpb1, and rpb2 gene regions. Verticillium dahliae (ATCC 16535) was selected as the outgroup taxon. ML bootstrap values (BS) ≥ 70 % and Bayesian posterior probabilities (PP) ≥ 0.95 are given at the nodes. Strains obtained in the present study are marked in red and type strains are indicated with a superscript T.
Fig. 6.
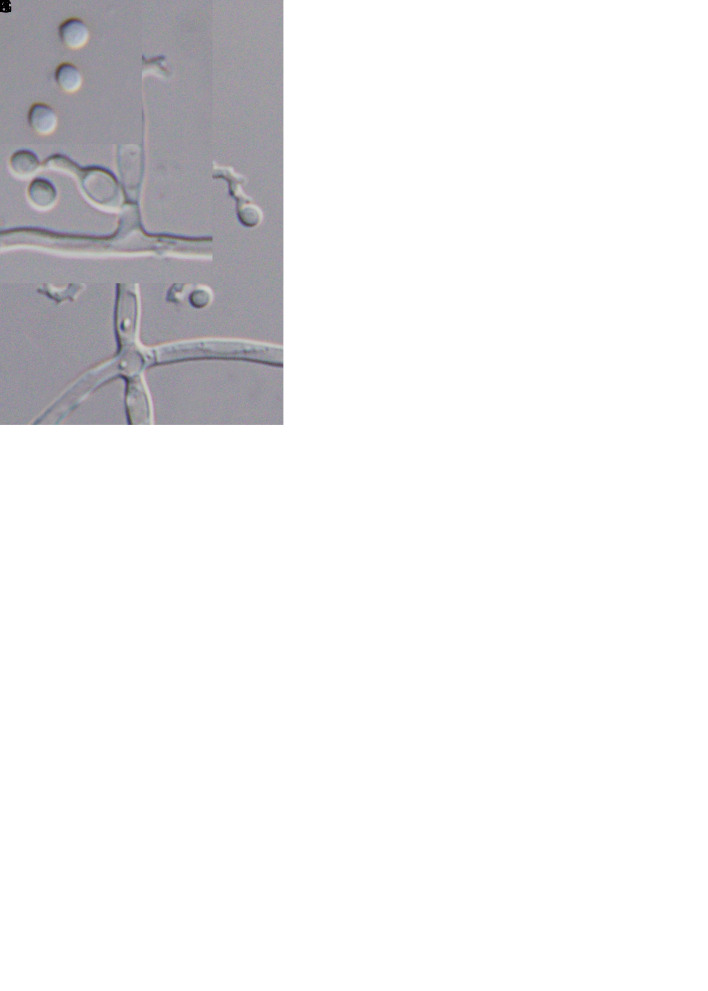
Morphology of Cordyceps locastrae (holotype NTUPPMH 17-042, ex-type culture NTUPPMCC 17-042). A, B. Synnemata on Locastra muscosalis pupa. C. Conidiophore with phialides and conidia. D, E. Conidia on the host. F. Obverse and reverse of colonies on PDA in 14 d. G, H. Evlachovaea-like conidiogenesis. I–K Phialides on PDA. L. Conidia on PDA culture. Scale bars = 5 μm.
RESULTS
Distribution and fungal isolations
During 2017–2019, 86 cordycipitaceous fungal specimens were collected from different forests throughout Taiwan, most of which were found in the mountainous areas of Taoyuan City and Nantou County as indicated in Fig. 1. Out of 86 specimens, 57 belonged to Cordyceps followed by 13 Beauveria, seven Samsoniella, three Blackwellomyces, three Akanthomyces, and three Simplicillium specimens. In addition, we obtained various strains of cordycipitaceous fungi from the BCRC, which were originally described from Taiwan and identified by previous scholars based only on morphological features.
Among the specimens in the present study, the majority were collected from the central and northern parts of Taiwan while a few were collected from the southern part of the Island. Most of the specimens with sexual morphs were collected at mountains above 1500 m altitude but asexual morphs were found irrespective of the altitude in the present study. The sample collection was carried out throughout the whole year; however, the majority (77 %) of the samples were obtained during the rainy season in Taiwan, specifically from April to October.
Phylogenetic analyses
Alignments of all single genes, ITS, nrLSU, tef1-α, rpb1, and rpb2, were initially analysed using ML. Congruence between individual genes made it possible to reconstruct the phylogenies by concatenating these genes as it provided a guarantee of gene orthology. The final phylogenetic analysis was based on the combined ITS-nrLSU-tef1-α-rpb1-rpb2 sequences of 475 strains representing 205 species of four families (Cordycipitaceae, Ophicordycipitaceae, Clavicipitaceae, Hypocreaceae) of the Hypocreales.
The concatenated sequence dataset is composed of 3 959 characters (ITS 519, nrLSU 788, tef1-α 926, rpb1 679, and rpb2 1047 bp). Phylogenetic trees obtained from the concatenated gene datasets using both ML and BI for the genera within Cordycipitaceae are shown in Fig. 2. A comparison of the alignment properties and nucleotide substitution models is shown in Supplementary Table S2. All methods resulted in a similar topology with high support for most branches in the ML and BI analyses and gave similar overall topologies of genus and species relationships as in previous studies based on ML and BI analyses (Sung et al. 2007a, Kepler et al. 2017, Mongkolsamrit et al. 2018, Mongkolsamrit et al. 2020).
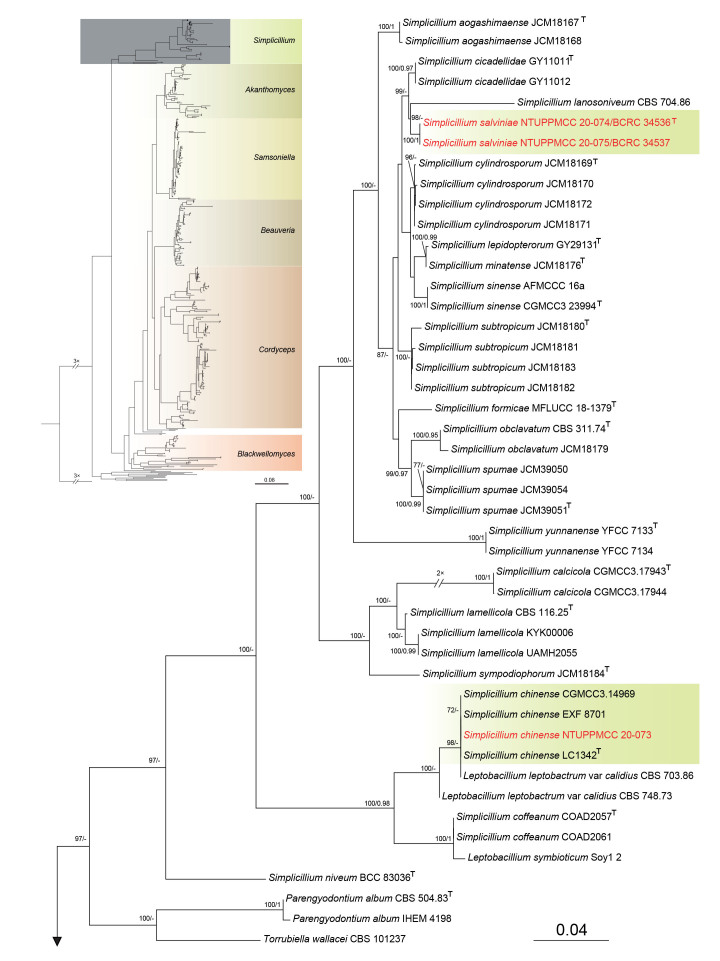
In total, three strains used in the present study clustered within Simplicillium. Among the three strains, Si. chinense (NTUPPMCC 20-073) isolated in this study clustered in a well-supported clade with another isolate of Si. chinense (CGMCC 3.14969) from submerged wood during freshwater fungi isolation. Si. chinense (CGMCC 3.14969) was used by Liu & Cai (2012) to describe the species, hence confirming the identity of the studied species. The remaining two strains (NTUPPMCC 20-074/BCRC 34536, NTUPPMCC 20-075/BCRC 34537) formed a distinct clade basal to the clade containing Si. cicadellidae (GY11011 and GY11012) and Si. lanosoniveum (CBS 704.86). Hence, the new lineage is herein regarded as the novel species Si. salviniae.
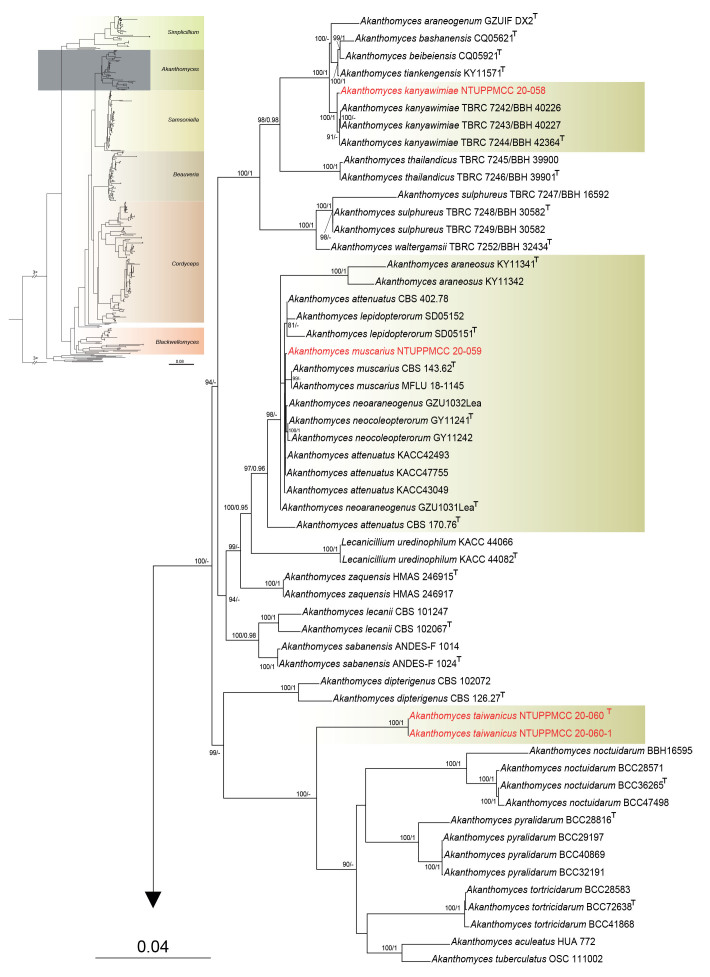
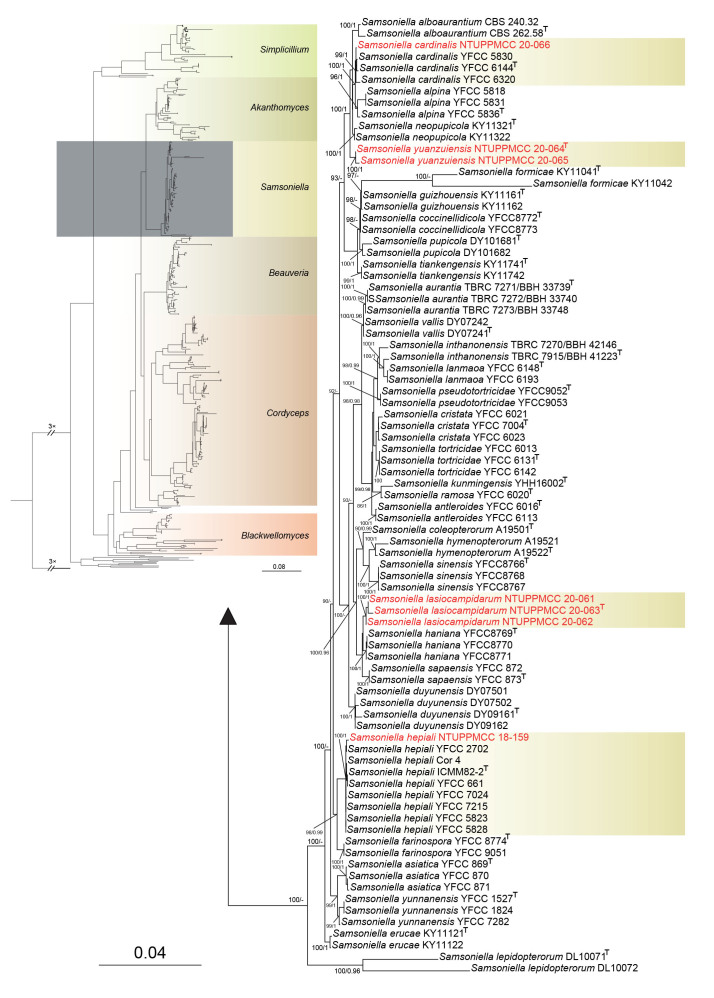
Furthermore, among the eleven strains used in the present study, five and seven strains were clustered within clades representing genera Akanthomyces and Samsoniella, respectively. Two newly collected Akanthomyces strains did not cluster with any known species, and two distinct clades were independently formed that did not group with any known species in Samsoniella. Hence, three novel species were proposed as Akanthomyces taiwanicus, Samsoniella lasiocampidarum, and S. yuanzuiensis to represent these distinct phylogenetic clades. The remaining strains clustered within the clades of S. cardinalis, S. hepiali, A. kanyawimiae, and A. muscarius, which are new records in Taiwan.
In total, 13 newly collected strains clustered with the Beauveria clade. The two new strains of B. scarabaeidicola (NTUPPMCC 20-056, NTUPPMCC 20-057) collected in this study formed a distinct clade with the ex-type strain of B. scarabaeidicola (ARSEF 1685). The B. medogensis (NTUPPMCC 20-055) strain included in this study clustered with the ex-type strain of B. medogensis 2898 that was examined by Imoulan et al. (2016) to describe the taxon, thus verifying the identity of the studied species. Furthermore, strain NTUPPMCC 20-054 (BCRC 35553) clustered with the ex-type strain of B. lii (RCEF 5500), and was thus identified as B. lii in the present study. Beauveria bassiana strains (NTUPPMCC 20-045 to NTUPPMCC 20-053) isolated in this study clustered in a well-supported clade containing the ex-type strain of B. bassiana (ARSEF 1564), therefore confirming the identification of the studied species. B. staphylinidicola ARSEF 5718 also clustered within the clade we identified as B. bassiana in the present study. Thus, we proposed to synonymize B. staphylinidicola ARSEF under B. bassiana (taxonomic treatments are explained below).
Among the strains clustered within Cordyceps, 28 clades of known species and five clades of unidentified species were resolved in the phylogenetic tree, each of which was used for the delimitation of species. In total, five distinct monophyletic clades were independently generated that did not cluster with any known species in Cordyceps. Hence, five novel species are proposed to accommodate these distinct lineages.
In addition, out of 52 strains, seven (NTUPPMCC 17-039, NTUPPMCC 18-120 to NTUPPMCC 18-125) were nested within the clade containing the ex-type strain (OSC 93623) plus strains of C. militaris and strains representing C. roseostromata, C. shuifuensis, and C. kyusyuensis. All these strains showed little variation in their morphology and molecular sequence data of ITS, nrLSU, tef1-α, rpb1, and rpb2 gene regions, thus we identified our strains as C. militaris and proposed to synonymize C. roseostromata, C. shuifuensis, and C. kyusyuensis under C. militaris. In addition, five strains (NTUPPMCC 17-040, NTUPPMCC 18-126 to NTUPPMCC 18-129) generated in this study were nested within the clade containing the ex-type strain of C. neopruinosa (BCC 91361), and therefore identified as C. neopruinosa in the present study. The ex-type strain of C. ninchukispora (NTUPPMCC 20-077/BCRC 31900) used in this analysis formed a basal clade to the clade comprising C. cocoonihabita, hence validating the phylogenetic placement of the studied species within Cordyceps. Furthermore, two strains (NTUPPMCC 18-115 and 116) clustered within the species clade containing ex-type strain of C. javanica (CBS 124.22), and was thus identified as C. javanica in this study. The two strains of Parahevansia koratensis (NHJ 2662 and NHJ 666.01) which were used by Mongkolsamrit et al. (2022) to introduce the genus Parahevansia are nested in Cordyceps. The strain NTUPPMCC 18-107 produced in this study was nested with the same clade containing the ex-type strain of C. blackwelliae (TBRC 7256), and is hence identified as C. blackwelliae.
In addition, eight strains included in the present clustered within the clades containing C. lepidopterorum and C. jakajanicola, which were initially introduced by Mongkolsamrit et al. (2018) and Crous et al. (2019) to accommodate species found on larvae of Lepidoptera and a cicada nymph from Thailand, respectively. However, strains representing these two species plus eight strains used in the present study nested in the same clade in both single and multi-gene phylogeny. Therefore, we recommend synonymizing C. jakajanicola under C. lepidopterorum by giving the priority of the oldest name and treated strains as C. lepidopterorum in the present study. Out of 52 strains clustered within Cordyceps, 13 were nested within the species clade of C. tenuipes, and hence identified as C. tenuipes in the current study. In total, two strains (NTUPPMCC 18-112 and NTUPPMCC 18-113) and four strains (NTUPPMCC 18-108, NTUPPMCC 18-109, NTUPPMCC 18-110 and NTUPPMCC 18-111), clustered within the species clades of C. fumosorosea and C. cateniannulata, respectively, and are therefore identified as C. fumosorosea and C. cateniannulata in the present study.
Three strains (NTUPPMCC 20-071, NTUPPMCC 20-072 and NTUPPMCC 20-068) used in the present study formed a distinct clade with high support (100/0.98) that is sister to the clade representing Blackwellomyces calendulinus (BCC 68500 and BCC 68502) within the genus Blackwellomyces in both single gene and concatenated gene analyses. Thus, the lineage is identified here as the new species Bl. taiwanensis.
Taxonomy of cordycipitaceous species
Based on the phylogenetic inference of concatenated sequences of ITS, nrLSU, tef1-α, rpb1, and rpb2 combined with morphological observations, 10 novel species were recognized within Cordycipitaceae. These novel species are described and illustrated below. Additionally, nine cordycipitaceous species have been recorded in Taiwan for the first time.
Taxonomic novelties
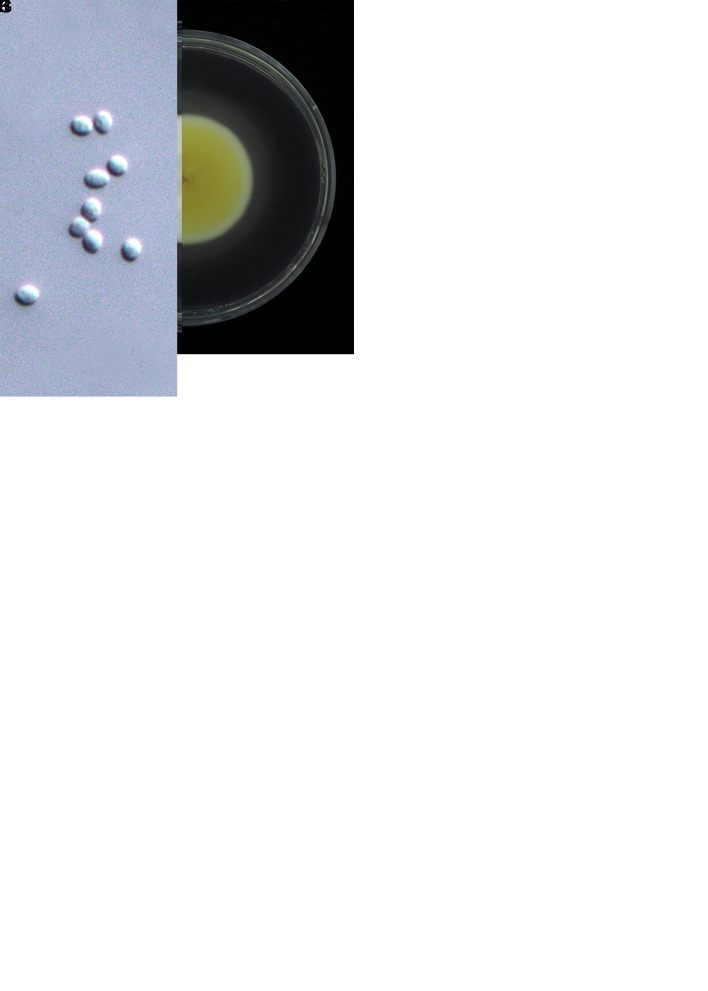
Akanthomyces taiwanicus W.Y. Chuang, B. Shrestha & H.A. Ariyaw., sp. nov. MycoBank MB 851850. Fig. 3.
Fig. 3.
Morphology of Akanthomyces taiwanicus (holotype NTUPPMH 20-060, ex-type culture NTUPPMCC 20-060). A. Synnemata arising from an adult Agrius convolvuli. B. The host adult A. convolvuli. C. Proboscis of A. convolvuli. D, E. Magnified synnemata. F. Phialides on the host. G. Conidia on the host. H. Obverse and reverse of colonies on PDA at 14 d. I, J. Phialides on PDA. K. Conidia from PDA culture. Scale bars: F, G = 10 μm; I, J = 10 μm; K = 5 μm.
Typus: Taiwan, New Taipei City, Xizhi Wujhihshan Scenic Area, on an adult of Agrius convolvuli (Lepidoptera), 1 Mar. 2018, Y.-T. Chu & W.-Y. Chuang (holotype NTUPPMH 20-060, ex-type culture NTUPPMCC 20-060).
Etymology: Referring to the Taiwanese Island from where the specimen was collected.
Description from specimen NTUPPMH 20-060: Sexual morph: Not observed. Asexual morph: Synnemata numerous, simple, or branched at the apex, white to light yellow, erect, bearing a powdery, white mass of conidia at the apex of synnemata, 3.1–7.3 mm long, arising from the adult of A. convolvuli on Cinnamomum camphora. Conidiophores produced along the synnemata on the specimen, monophialidic, consisting of solitary phialides. Phialides (12.5–)14.5–19(–20) × 2–3(–3.5) μm, subcylindrical, tapering into a distinct neck, 1–1.5(–2) μm in width. Conidia (3.5–)4–4.5(–5) × 2–2.5 μm, unicellular, hyaline, ovoid to ellipsoidal.
Culture characteristics: Colony on PDA (NTUPPMCC 20-060) white, convex, entire edge, moderately slow growing, 3.2 cm diam in 21 d; reverse cream white to light yellow. Synnemata formed 15 d after inoculation. Conidiophores produced along the synnemata in culture, monophialidic, consisting of solitary phialides. Phialides (12.5–)19.5–28.5(–35.5) × (2–)2.5–3 μm subcylindrical, tapering into a distinct neck, 1 μm in width. Conidia 4–5(–5.5) × 2–2.5 μm unicellular, hyaline, ovoid to ellipsoidal.
Distribution: New Taipei City; Taiwan.
Additional materials examined: Taiwan, New Taipei City, Xizhi Wujhihshan Scenic Area, on an adult of Agrius convolvuli (Lepidoptera), 1 Mar. 2018, Y.-T. Chu & W.-Y. Chuang, NTUPPMH 20-060-1, living culture NTUPPMCC 20-060-1.
Notes: In the multi-locus phylogeny, Akanthomyces taiwanicus (NTUPPMCC 20-060 and NTUPPMCC 20-060-1) forms a distinct clade basal to the clades representing A. aculeatus, A. noctuidarum, A. pyralidarum, A. tortricidarum and A. tuberculatus is thus introduced here as a novel taxon.
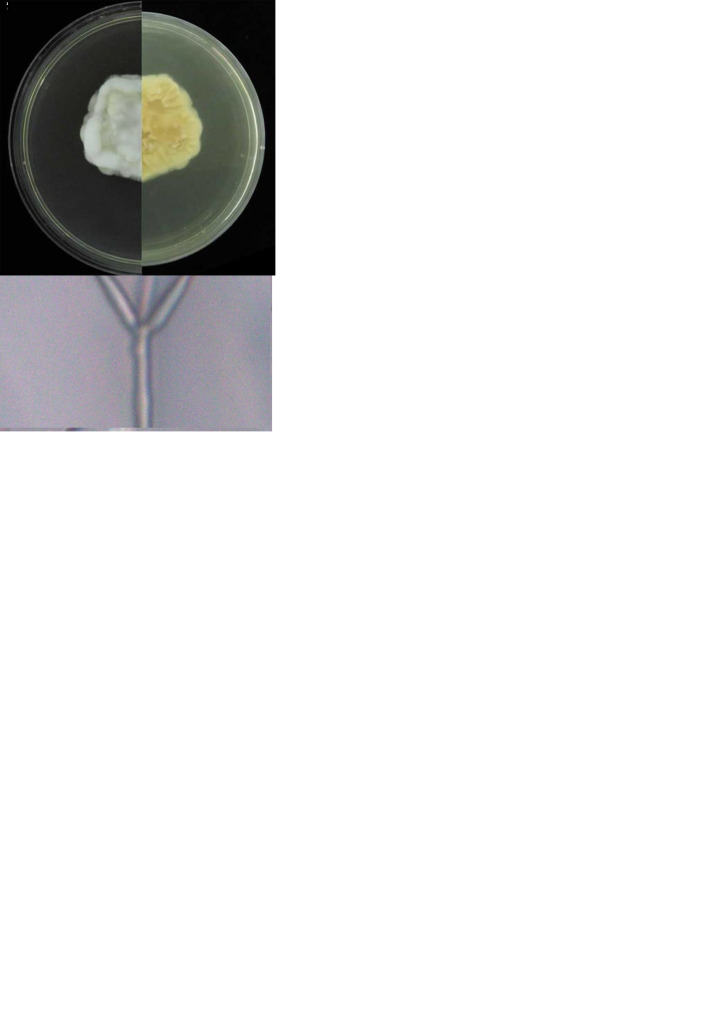
Blackwellomyces taiwanensis W.Y. Chuang & H.A. Ariyaw., sp. nov. MycoBank MB 839343. Fig. 4.
Fig. 4.
. Morphology of Blackwellomyces taiwanensis (holotype NTUPPMH 20-071, ex-type culture NTUPPMCC 20-071). A. Stromata on Tenebrionidae pupa. B. Stroma. C. Section of stroma. D. Section of perithecium. E–G. Immaure asci. H. Mature ascus. I. Ascus cap. J. Part-spores. K. Obverse and reverse of colonies on PDA. L, M. Phialides on PDA. N. Conidia from PDA culture. Scale bars: C = 200 μm; D = 40 μm; E–H = 20 μm; I, J = 5 μm; L–N = 10 μm.
Typus: Taiwan, Taoyuan City, Dongyan Mountain Forest Recreational Area, 24°49’49.5”N 121°24’32.1”E, on a tenebrionid larva (Coleoptera), 1 Jun. 2018, C.-Y. Chen, Y.-T. Wei (holotype NTUPPMH 20-071, ex-type culture NTUPPMCC 20-071).
Etymology: Referring to the Taiwanese Island from where the specimen was collected.
Description from specimen NTUPPMH 20-071: Sexual morph: Stromata solitary or multiple, arising from the head of the tenebrionid larva buried inside wood under the ground. The exposed part is composed of a fleshy stromata, with an orangish red stipe and orange fertile part. Perithecia numerous, gregarious, immersed to partially erumpent, flask-shaped, orangish red, (143–)173–238(–249) × (65–)83–132(–165) μm. Asci cylindrical, hyaline, 8-spored, (105–)106–145(–173) × (3.5–)4–5 μm with a prominent cap, 1.5–2 × 3–3.5(–4) μm at the apex. Ascospores filiform, septate, (97–)123–160(–183) ×1–1.5 μm, disarticulating into cylindrical part-spores, (2.5–)3–4.5(–5.5) ×1–1.5(–2) μm. Asexual morph: Not observed.
Culture characteristics: Colony on PDA (NTUPPMH 20-071) white, flat, concentric, moderately fast growing, 4.5 cm diam in 14 d at 25 °C; reverse beige to light yellow. Colony margin light pinkish. Conidiophores arising from the prostrate or aerial hyphae. Phialide single, hyaline, subcylindrical, (15–)17–26(–32) × (1.5–)2–2.5 μm, tapering towards the neck, 1–1.5(–2) μm in width. Conidia unicellular, sometimes forming chains, hyaline, oblong-elliptical, (4–)4.5–5.5(–6.5) × 2–2.5(–3) μm.
Distribution: New Taipei City, Taoyuan City; Taiwan.
Additional materials examined: Taiwan, Taoyuan City, Dongyan Mountain Forest Recreational Area, on a tenebrionid larva buried in wood, 1 Jun. 2018, C.-Y. Chen, Y.-T. Wei, NTUPPH 20-072, living culture NTUPPMCC 20-072; New Taipei City, Wulai District, on an unknown host destroyed by the fungus buried in wood, 15 Oct. 2018, C.-Y. Lee, NTUPPH 20-068, living culture NTUPPMCC 20-068; ibid., NTUPPH 20-069, living culture NTUPPMCC 20-069; ibid., NTUPPH 20-070, living culture NTUPPMCC 20-070.
Notes: Blackwellomyces taiwanensis exhibits typical Blackwellomyces morphology by producing orange to red stromata and disarticulating ascospores. Blackwellomyces taiwanensis is phylogenetically close to Bl. calendulinus. However, Bl. taiwanensis produces part-spores, which is not observed in Bl. calendulinus. In addition, the length of Bl. taiwanensis asci (106–145 μm) is shorter than that of Bl. calendulinus (220 × 4–5 μm). Moreover, Bl. taiwanensis morphologically resembles Cordyceps rubricapitata (Kobayasi & Shimizu 1983). Both Bl. taiwanensis and C. rubricapitata parasitize coleopteran larvae and show similar micro-morphological features (Kobayasi & Shimizu 1983). However, the new species can be differentiated from C. rubricapitata by the perithecial position (semi-immersed vs superficial) and the geographical location (Taiwan vs Japan).
Cordyceps hehuanensis W.Y. Chuang & H.A. Ariyaw., sp. nov. MycoBank MB 839345. Fig. 5.
Fig. 5.
Morphology of Cordyceps hehuanensis (holotype NTUPPMH 18-145, ex-type culture NTUPPMCC 18-145). A. Stromata arising from a lepidopteran adult. B, C. Enlarged fertile part of stroma showing semi-immersed perithecia. D. Section of perithecia. E–G. Ascus. H. Ascus cap. I. Part-spores. J. Obverse and reverse of colonies on PDA at 14 d. K, L Phialides on PDA. M, N. Conidia from PDA culture. Scale bars: D = 200 μm; E–G = 50 μm; H, I = 10 μm; K–N = 10 μm.
Typus: Taiwan, Nantou County, Ren’ai Township, Mt. Hehuan, 24°10’26.5”N 121°17’13.0”E, on a lepidopteran adult, 3 Sep. 2018, C.-M. Hu (holotype NTUPPMH 18-145, ex-type culture NTUPPMCC 18-145).
Etymology: Referring to Mt. Hehuan where the specimen was collected.
Description from specimen NTUPPMH 18-145: Sexual morph: Stromata simple, two, clavate, yellow to orangish, arising from the lateral side of the lepidopteran adult buried under leaf litter. Perithecia numerous, covering the ascomatal heads, semi-immersed, pyriform, orange to brown, (379–)382–443(–466) × (215–)223–287(–294) μm. Asci cylindrical, hyaline, 8-spored, (255–)319–410(–458) × (4–)4.5–5(–5.5) μm, with a hemispherical cap, 4 × 2.5 μm at the apex. Ascospores filiform, septate, (282– )303–390(–415) × 1–1.5 μm, disarticulating into part-spores. Part-spores cylindrical, with truncate ends, hyaline, (2.5–)3–3.5(–4) × 1 μm. Asexual morph: Not observed.
Culture characteristics: Colony on PDA (NTUPPMH 18-145) reaching 2.5 cm diam in 14 d at 25 °C, light yellow, reverse yellow to brown, flat, entire edge. Conidiophores arising perpendicularly from the aerial hyphae in culture, single, hyaline, subcylindrical, (11–)12–19(–23) × 2–2.5(–3) μm, slightly tapering into the neck, 1 μm in width. Conidia unicellular, hyaline, ovoid to globose, sometimes in a chain, (3.5–)4–5(–6) × 2.5–3 μm.
Distribution: Nantou County; Taiwan.
Additional materials examined: Taiwan, Nantou County, Ren’ai Township, Mt. Hehuan, on lepidopteran adult, 4 Sep. 2018, C.-M. Hu, NTUPPMH 18-144, living culture NTUPPMCC 18-144; ibid., NTUPPMH 18-145-1, living culture NTUPPMCC 18-145-1. Ren’ai Township, Mt. Hehuan, on lepidopteran adult, 3 Sep. 2018, C.-M. Hu, NTUPPMH 18-145-1, living culture NTUPPMCC 18-145-1.
Notes: Based on multi-locus phylogeny (Fig. 2), C. hehuanensis forms a well-supported clade sister to C. militaris. However, C. hehuanensis differs from C. militaris in producing smaller perithecia (382–443 × 223–287 μm vs 500–720 × 300–480 μm).
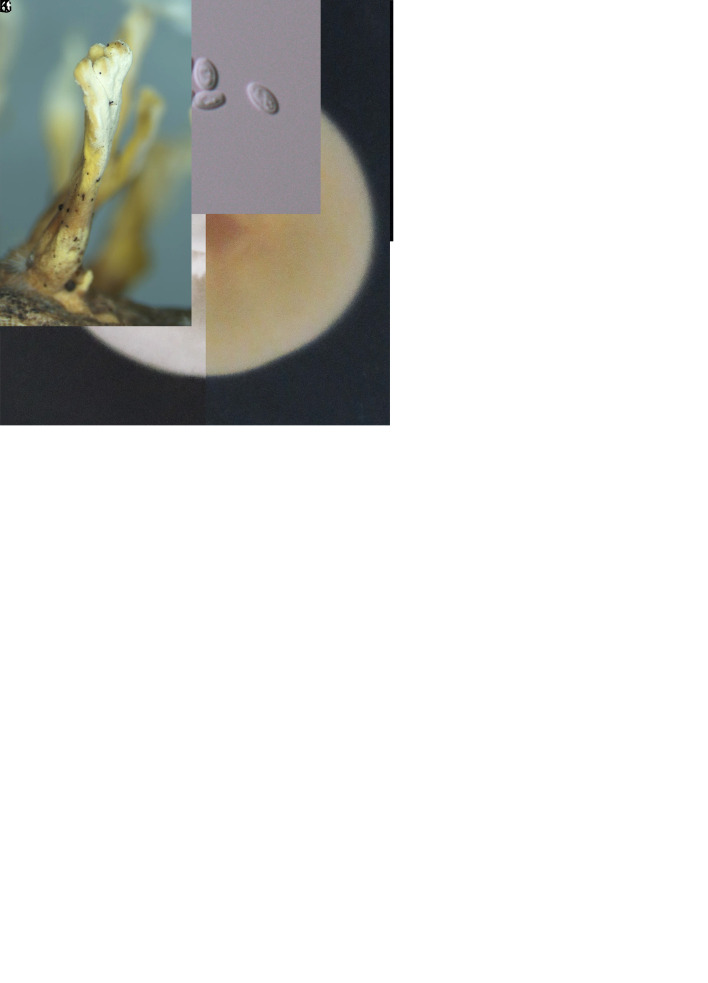
Cordyceps locastrae W.Y. Chuang & H.A. Ariyaw., sp. nov. MycoBank MB 839344. Fig. 6.
Typus: Taiwan, Taoyuan City, Taoyuan District, Mt. Hutou, 25°00’02.5”N 121°19’44.7”E, on Locastra muscosalis (Lepidoptera) pupa, 11 Nov. 2017, M.-L. Lo (holotype NTUPPMH 17-042, ex-type culture NTUPPMCC 17-042).
Etymology: Referring to the insect host genus, Locastra muscosalis (Lepidoptera).
Description from specimen NTUPPMH 18-149: Sexual morph: Not observed. Asexual morph: Synnemata numerous, simple, white, erect, buried under soil, bearing a powdery, white mass of conidia at the apex of the synnemata (head), 1–3 mm. Conidiophores arising from the host, consisting of solitary phialides along verticillate branches with two to three phialides in whorls. Phialides cylindrical, (3–)3.5–7.5(–10.5) × (1.5–)2.5–3(–3.5) μm, with a flask-shaped basal portion tapering into a neck, 1 μm in width. Conidia unicellular, hyaline, subglobose to globose, sometimes in a chain, 2.5–3 × 2.–2.5(–3) μm.
Culture characteristics: Colony on PDA (NTUPPMCC 18-149) reaching 2.5 cm diam in 14 d at 25 °C, white, reverse white to light yellow, convex, condensed, entire edge. Conidiophores arising from the host or on aerial hyphae in culture, consisting of solitary phialides along the hyphae or verticillate branches with two to three phialides in whorls. Phialides cylindrical, (6–)7–10(–13) × 2.5–3.5(–4) μm, with a flask-shaped basal portion tapering into a neck, 1 μm in width. Conidia unicellular, hyaline, subglobose to globose, sometimes in a chain, 3–3.5(–4) × (2–)2.5–3 μm.
Distribution: Taoyuan City; Taiwan.
Additional materials examined: Taiwan, Taoyuan City, Mt. Hutou, on Lepidoptera pupa, 10 Mar. 2018, W.-Y. Chuang, NTUPPMH 18-147, living culture NTUPPMCC 18-147; ibid., NTUPPMH 18-148, living culture NTUPPMCC 18-148.
Notes: Cordyceps locastrae is phylogenetically close to C. cateniannulata (Liang et al. 1981). However, C. locastrae produces subglobose to globose conidia (3–3.5 × 2.5–3 μm), which are slightly larger than those of C. cateniannulata (2–3.5 × 1–1.5 μm). Furthermore, C. locastrae produces larger phialides compared to C. cateniannulata (7–10 × 2.5–3.5 μm vs 3–8 ×1.5–3 μm) (Liang et al. 1981).
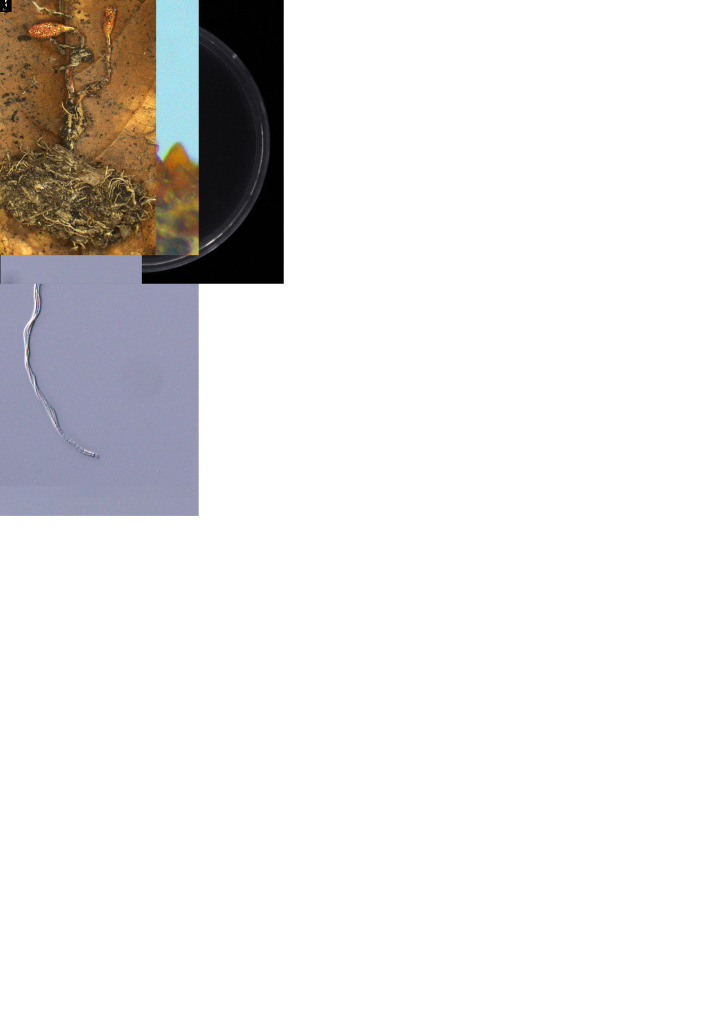
Cordyceps malleiformis W.Y. Chuang & H.A. Ariyaw., sp. nov. MycoBank MB 839348. Fig. 7.
Fig. 7.
Morphology of Cordyceps malleiformis (holotype NTUPPMH 18-143, ex-type culture NTUPPMCC 18-143). A, B. Stromata arising from a lepidopteran pupa. C. Part of stroma showing semi-immersed perithecia. D. Perithecium. E. Ostiole of perithecia. F. Perithecia cell wall. G–I. Ascus. J, K. Ascus cap L. Obverse and reverse of colonies on PDA at 14 d. M. Evlachovaea-like conidiogenesis. N. Phialides on PDA. O, P. Conidia from PDA culture. Scale bars: D = 50 μm; E, F = 20 μm; G–I = 50 μm; J, K = 5 μm; M = 20 μm; N–P = 10 μm.
Typus: Taiwan, Hsinchu County, Jianshi Township, Mt. Ptlaman, 24°44’27.6”N 121°16’43.1”E, on a lepidopteran pupa, 20 Aug. 2018, L.-J. Chu (holotype NTUPPMH 18-143, ex-type culture NTUPPMCC 18-143).
Etymology: Referring to the shape of the stromata – “mace-shaped”.
Description from specimen NTUPPMH 18-143: Sexual morph: Stromata simple, multiple, clavate, producing white rhizomorph-like hyphae below the ground (Fig. 7B), 1–1.5 cm long. Stromata light orange at the base becoming bright orange to red in terminal fertile head. Perithecia semi-immersed, pyriform, red, (195–)225–353 × 114–178 μm. Asci cylindrical, hyaline, 8-spored, 158–259 × 3–3.5 μm, with an oblate spheroidal or hemispherical cap, 1.5–2 × 3–3.5 μm at the apex. Ascospores smooth, filiform, aseptate, (153–)180– 277(–312) × 1–1.5 μm. Part-spores not observed.
Culture characteristics: Colony on PDA (NTUPPMCC 18-143) reaching 2.5 cm diam in 14 d at 25 °C, white, reverse yellow to yellow, dense, low convex, entire edge with slight furrows. Synnemata not observed. Conidiophores arising from the prostrate hyphae or on aerial hyphae in culture, mostly single, rarely branched, hyaline, subcylindrical, (10–)12–17(–20) × (1.5–)2–2.5 μm, tapering into the neck, 1 μm in width. Conidia unicellular, hyaline, cylindrical to ellipsoidal or sometimes bean or kidney-shaped, sometimes in a chain, (3–)3.5–4.5(–5.5) × 2 μm.
Distribution: Hsinchu County; Taiwan.
Additional materials examined: Taiwan, Hsinchu County, Jianshi Township, Mt. Ptlaman, on a lepidopteran pupa, 20 Aug. 2018, L.-J. Chu, NTUPPMH 18-143-1, living culture NTUPPMCC 18-143-1.
Notes: Cordyceps malleiformis is typical of Cordyceps in having brightly pigmented, fleshy stromata, hyaline, cylindrical asci with thickened ascus caps and long, filiform ascospores. Based on multi-locus phylogenetic trees, C. malleiformis forms a well-supported clade sister to the recently introduced species C. bullispora (holotype: YFCC 8400) in China. However, C. malleiformis can be easily distinguished from C. bullispora in having mace-shaped stromata and shorter conidia (3.5–4.5 vs 5–11 μm) (Dong et al. 2022). C. malleiformis shares similar morphological features with C. longiphialis by producing prominent superficial perithecia. Nevertheless, C. malleiformis can be easily differentiated from C. longiphialis by its longer and fatter asci (158–259 × 3–3.5 vs 113– 200 × 1–2.5 μm) and shorter phialide (12–17 vs 7– 8 μm) (Dong et al. 2022).
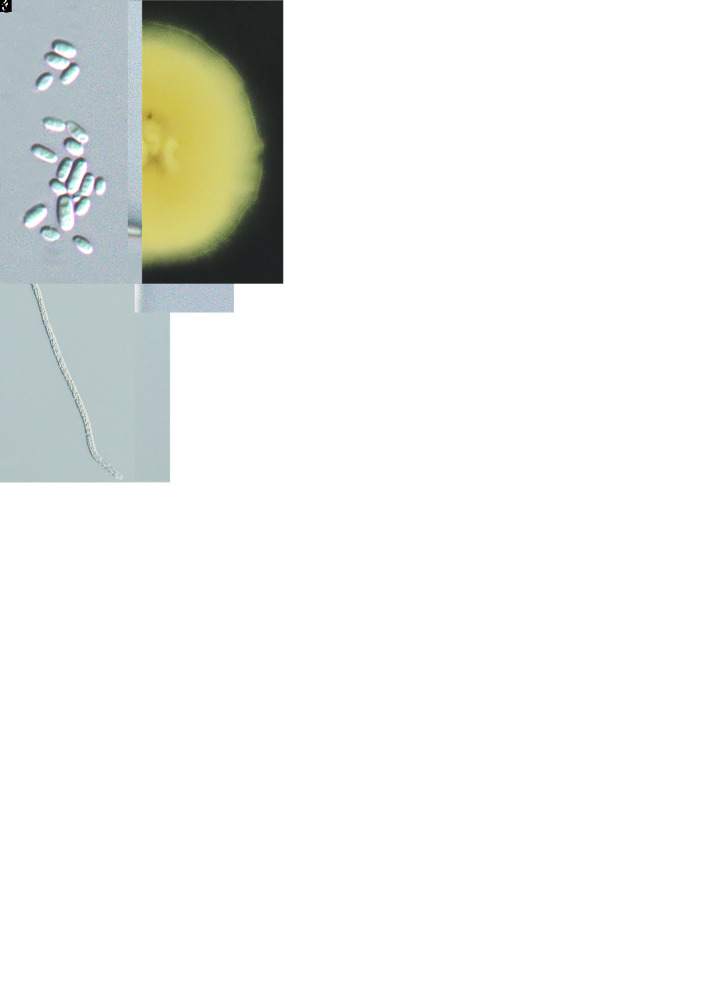
Cordyceps pseudorosea W.Y. Chuang & H.A. Ariyaw., sp. nov. MycoBank MB 839341. Fig. 8.
Fig. 8.
. Morphology of Cordyceps pseudorosea (holotype NTUPPMH 17-041, ex-type culture NTUPPMCC 17-041). A. Stromata arising from the ventral side of the anterior end of the erebid adult. B. Erebid adult host. C, D. Sections of perithecia. E–G. Asci. H. Ascus cap. I. Part-spores. J. Obverse and reverse of colonies on PDA. K, L. Evlachovaea-like conidiogenesis. M. Conidia from PDA culture. Scale bars: C = 200 μm; D = 40 μm; E–G = 50 μm; H, I = 10 μm; K–M = 10 μm.
Typus: Taiwan, Taichung City, Heping District, 710 Forest trail, 24°21’42.1”N 121°21’09.5”E, on an erebid adult (Lepidoptera) attached to the upperside of the leaf, 11 Nov. 2017, G. Shang, C.-K. Fan & D.-L. You (holotype NTUPPMH 17-041, ex-type culture NTUPPMCC 17-041).
Etymology: Referring to the morphological resemblance to C. rosea.
Description from specimen NTUPPMH 17-041: Sexual morph: Stromata simple, slightly curved, multiple, orangish red, arising from the dorsal side of the anterior part of the Erebidae adult. Perithecia produced sparsely from the middle of the stalk, becoming more crowded towards the apex, superficial, pyriform, orange to red, (407–)481–664(–727) × 341–374 μm. Asci cylindrical, hyaline, 8-spored, (172–)205–372(–425) × (4–)4.5–5(–5.5) μm, with a hemispherical cap, (3–)3.5–4(–4.5) × 2–2.5(–3) μm at the apex. Ascospores filiform, septate, (220–)230–383(–392) × (1–)1.5–2 μm, disarticulating into part-spores, cylindrical, (3.5–)4–5(–6) ×1.5– 2(–2.5) μm. Asexual morph: Not observed.
Culture characteristics: Colony on PDA (NTUPPMCC 17-041) reaching 3.0 cm diam in 14 d at 25 °C, white to light yellow, reverse beige to light yellow, flat. Synnemata not observed. Conidiophores arising from the sterile hyphae or on aerial hyphae, single, hyaline, subcylindrical, (9–)12.5–19.5(–26) × 2.5 μm, tapering towards the neck, 1–1.5(–2) μm in width. Conidia unicellular, hyaline, oblong-elliptical, sometimes in chain, (4–)4.5–6(–7) × 2–2.5(–3) μm.
Additional materials examined: Taiwan, Taichung City, Heping District, 710 Forest trail, on an erebid adult, 11 Nov. 2017, G. Shang, C. Kai, F. Li, D. Li & You, NTUPPMH 17-041-1, living culture NTUPPMCC 17-041-1.
Distribution: Taichung City; Taiwan.
Notes: Cordyceps pseudorosea forms a separate clade sister to tentatively named strain C. rosea (spat 09-053). Cordyceps rosea was described by Kobayasi & Shimizu (1982) from Japan. However, the description of C. rosea was based only on the sexual morph and was reported to be parasitizing a geometrid larva (Lepidoptera). The sequence of C. rosea in this study was used by Kepler et al. (2017) but it is not clear that these sequences were generated from the type material or epitype material. Furthermore, we did not find any morphological description corresponding to the particular strain. According to the original description by Kobayasi & Shimizu (1982), C. rosea possessed 100 × 3–4 μm asci and the ascospores did not disarticulate into part-spores. Our strains showed a few deviations in morphological features compared to the description of C. rosea in having larger asci (205–372 × 4.5–5 μm vs 100 × 3–4 μm). Moreover, ascospores of C. pseudorosea disarticulated into part-spores when mature while ascospores of C. rosea showed only septation (Kobayasi & Shimizu 1982). Therefore, considering the lack of DNA sequence data from the type material of C. rosea and the variation in the morphological features plus the difference in the location where the strains were collected (Japan vs Taiwan) we introduced our strains as a new species. Furthermore, considering the high similarity of phylogeny as shown in Fig. 2, we tentatively identify the putative strain of C. rosea (spat 09-053) as C. pseudorosea (spat 09-053) in the present study.
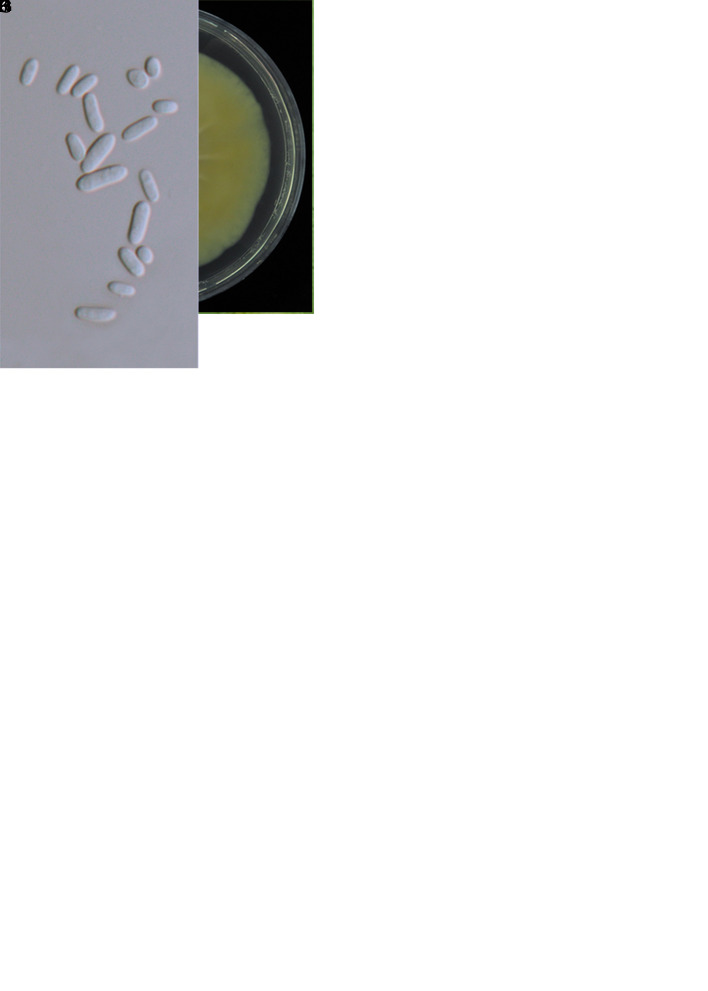
Cordyceps siangyangensis W.Y. Chuang & H.A. Ariyaw., sp. nov. MycoBank MB 839342. Fig. 9.
Fig. 9.
. The morphology of Cordyceps siangyangensis (holotype NTUPPMH 18-149, ex-type culture NTUPPMCC 18-149). A, B. Fungus on host. C. Magnified synnemata. D, E. Host. F. Phialides on the host. G. Conidia on the host. H. Obverse and reverse of colonies on PDA at 14 d. I, J. Isaria-like conidiogenesis. K, L. Phialides with conidia on PDA. M. Conidia from PDA. Scale bars: F, G = 5 μm; I, J = 20 μm; K–M = 10 μm.
Typus: Taiwan, Taitung County, Siangyang Forest Recreation, 23°15’01.8”N 120°58’59.6”E, on an unidentified adult insect, 28 Nov. 2019, W.-Y. Chuang (holotype NTUPPMH 18-149, ex-type culture NTUPPMCC 18-149).
Etymology: Referring to the location where the specimen was collected.
Description from specimen NTUPPMH 18-149: Sexual morph: Not observed. Asexual morph: Synnemata formed simple, single to multiple, light yellow, erect, arising from the host head, buried in soil, bearing a powdery isaria-like white mass of conidia on the apex of the branches, 1.8–2.2 cm long. Phialides cylindrical, (8–)8.5–14.5(–18) × (2–)3–4 μm, with globose basal portion tapering into a neck, 1–1.5 μm in width. Conidia unicellular, hyaline, cylindrical to ellipsoidal, sometimes forming chains, 5–6.5(–7.5) × 2.5–3.5(–4) μm.
Culture characteristics: Colony on PDA (NTUPPMCC 18-149) reaching 4 cm diam in 14 d at 20 °C, white, reverse white to pale yellow, flat, raised at the centre after 30 d, entire edge with slight furrows in the middle and concentric rings on the margin. Conidiogenous cells arising from the sterile hyphae or on aerial hyphae, consisting of solitary phialides along the hyphae or verticillate branches with two to four phialides in whorls. Phialides cylindrical, (7.5–)9–14.5(–18) × (2.5–)3–4(–4.5) μm, with globose basal portion tapering into a neck, 1–1.5 μm in width. Conidia unicellular, hyaline, cylindrical to ellipsoidal, sometimes forming chains, 6.5–8(–9) × (3–)3.5–4(–4.5) μm.
Distribution: New Taipei City, Taitung County; Taiwan.
Additional materials examined: Taiwan, Taitung County, Siangyang Forest Recreation, on unknown insect buried in moss, 28 Nov. 2019, W.-Y. Chuang, NTUPPMH 18-150, living culture NTUPPMCC 18-150; New Taipei City, Sanxia District, Manyueyuan Forest Recreation, on a Limacodidae cocoon underground, 6 Aug. 2018, W.-Y. Chuang, NTUPPMH 18-151, living culture NTUPPMCC 18-151.
Notes: Cordyceps siangyangensis is typical of Cordyceps (isaria-like asexual morphs) in having mono- or synnematous conidiophores with verticillate, globose phialides in whorls (Luangsa-Ard et al. 2005). Based on multi-gene phylogenetic trees, C. siangyangensis forms a well-supported clade (BS = 100 %, 1.00 PP), sister to the putative strain of C. bifusispora (spat 08-133.1) (Eriksson 1982). However, C. siangyangensis has shorter and swollen phialides compared to the original description of C. bifusispora (9–15 × 3–4 μm vs 9–30 × 1.5–2 μm) (Liu et al. 1996). Additionally, C. siangyangensis produces longer conidia than C. bifusispora (6.7–8.1 μm vs 2.5–3.5 μm) (Liu et al. 1996). Moreover, the conidiogenesis patterns between C. siangyangensis and C. bifusispora are different. The former species produces successive conidia in end-to-end orientations resulting in straight chains while the latter species produces successive conidia in alternate orientations resulting in zipper-like chains (Liu et al. 1996).
Samsoniella lasiocampidarum W.Y. Chuang & H.A. Ariyaw., sp. nov. MycoBank MB 839347. Fig. 10.
Fig. 10.
Morphology of Samsoniella lasiocampidarum (holotype NTUPPMH 20-063, ex-type culture NTUPPMCC 20-063). A. Synnemata on a lasiocampid larva. B, C. Synnemata and conidial mass. D. Phialide on lasiocampid host. E. Conidia on lasiocampid host. F. Obverse and reverse of colonies on PDA. G. Isaria-like conidiogenesis. H, I. Conidiophore on PDA. J. Conidia from PDA culture. Scale bars: D = 10 μm; E = 5 μm; G–J =10 μm.
Typus: Taiwan, Yilan County, Datong Township, Mingchih Forest Recreation Area, 24°39’03.0”N 121°28’24.7”E, on a lasiocampid larva (Lepidoptera) on the ground, 9 Jun. 2018, Y.-Y. Tang (holotype NTUPPMH 20-063, ex-type culture NTUPPMCC 20-063)
Etymology: Referring to the insect host family – Lasiocampidae.
Description from specimen NTUPPMH 20-063: Sexual morph: Not observed. Asexual morph: Synnemata arising from multiple parts of the host, erect, multiple, light yellow to orange, irregularly branched at the apex, bearing a powdery, white mass of conidia, 2.5–4.5 cm long. Conidiophores erect, mononematous, consisting of solitary phialides along the hyphae or verticillate branches with two to three phialides in whorls in culture. Phialides hyaline, subcylindrical, (4.5–)5–7(–7.5) × (1–)1.5–2.5(–3) μm, with a flask-shaped basal portion tapering into a distinct neck, 0.5–1 μm in width. Conidia unicellular, hyaline, fusiform or ovoid, sometimes in a chain, 2.5 × 1.5–2 μm.
Culture characteristics: Colony on PDA (NTUPPMCC 20-063) reaching 2.6 cm diam in 14 d at 25°C, white, reverse cream white to light yellow, convex, entire edge. Conidiophores erect, mononematous, consisting of solitary phialides along the hyphae or verticillate branches in culture. Phialides hyaline, subcylindrical, (5.5–)6.5–9.5(–11.5) × 2.5–3 μm, with a flask-shaped basal portion tapering into a distinct neck, 1 μm in width. Conidia unicellular, hyaline, fusiform or ovoid, sometimes in a chain, 3 × 2 μm.
Distribution: Pingtung County, Yilan County; Taiwan.
Additional materials examined: Taiwan, Pingtung County, Chunrih Township, Dahan Forest Road, on a lasiocampid larva, 20 May 2018, W.-Y. Chuang, NTUPPMH 20-062, living culture NTUPPMCC 20-062; ibid., 7 Oct. 2018, W.-Y. Chuang, NTUPPMH 20-061, living culture NTUPPMCC 20-061.
Notes: Samsoniella lasiocampidarum is similar to its phylogenetically closely related taxon S. aurantia in having yellow to light orange synnemata on the hosts bearing a powdery conidia mass at the branches along the synnemata. However, S. lasiocampidarum differs from S. aurantia by the size of conidia (3 × 2 μm vs 2.5–3.5 × 1.5 μm) and the size of phialides (6.5–9.5 × 2.5–3 μm vs 5.5–8.5 × 2–3 μm) (Mongkolsamrit et al. 2018). In addition to their morphological variations, S. lasiocampidarum can be distinguished from S. aurantia based on location (Taiwan vs Thailand) (Mongkolsamrit et al. 2018).
Samsoniella yuanzuiensis W.Y. Chuang & H.A. Ariyaw., sp. nov. MycoBank MB 840828. Fig. 11.
Fig. 11.
The morphology of Samsoniella yuanzuiensis (holotype NTUPPMH 20-064, ex-type culture NTUPPMCC 20-064). A, B. Synnemata growing on a lepidopteran pupa. C. Conidial mass on the apex of synnemata. D. Phialides on the host. E. Conidia on the host. F. Obverse and reverse of colonies on PDA. G, H. Isaria-like conidiogenesis. I, J. Conidiophore on PDA. K. Conidia from PDA culture. Scale bars =10 μm.
Typus: Taiwan, Taichung City, Mt. Yuanzui, 24°14’05.2”N 120°57’15.5”E, on a lepidopteran pupa buried under the ground, 1 Apr. 2018, W.-Y. Chuang (holotype NTUPPMH 20-064, ex-type culture NTUPPMCC 20-064).
Etymology: Referring to Mount Yuanzui where the specimen was collected.
Description from specimen NTUPPMH 20-064: Sexual morph: Not observed. Asexual morph: Synnemata white, numerous, irregularly branched or unbranched, 0.4–0.8 cm long, bearing a powdery, white mass of conidia at the apex, arising from an exarate lepidopteran pupa. Conidiophores consisting of two to three phialides in whorls. Phialides hyaline, 5–7(–8.5) × (1.5–)2–2.5(–3) μm, with a flask-shaped basal portion tapering into a distinct neck, 1 μm in width. Conidia unicellular, hyaline, ovoid to fusiform, sometimes in a chain, 2 × 1.5 μm.
Culture characteristics: Colony on PDA (NTUPPMCC 20-064) moderately slow growing, reaching 4.1 cm diam in 21 d at 25 °C, white, flat, entire edge, reverse light yellow to yellow. Conidiophores erect, consisting of solitary phialides along the hyphae or verticillate branches with two to three phialides in whorls in culture. Phialides hyaline, (6–)7–10.5(–14) × (2–)2.5–3(–3.5) μm, with a flask-shaped basal portion tapering into a distinct neck, 1 μm in width. Conidia unicellular, hyaline, ovoid to fusiform, sometimes in a chain, 3 × 2–2.5(–3) μm.
Distribution: Taichung City, Taiwan.
Additional materials examined: Taiwan, Taichung City, Mt. Yuanzui, on a pupa, 1 Apr. 2018, W.-Y. Chuang, NTUPPMH 20-065, living culture NTUPPMCC 20-065.
Notes: Samsoniella yuanzuiensis closely resembles S. alpina in having similar phialides. Both species produce an isaria-like asexual morph. However, S. yuanzuiensis differs from S. alpina in having larger conidia (2.5–3 × 2–2.5 vs 2–3.1 × 1.3–2.1 μm) (Wang et al. 2020). Moreover, synnemata of S. yuanzuiensis were found arising from the pupa while synnemata of S. alpina arose from the whole larval body (Wang et al. 2020).
Simplicillium salviniae W.Y. Chuang & H.A. Ariyaw., sp. nov. MycoBank MB 839349. Fig. 12.
Fig. 12.
Morphology of Simplicillium salviniae (ex-type culture NTUPPMCC 20-074/BCRC 34536). A, B. Obverse and reverse of colonies on PDA. C–E. Conidial mass on the top of a conidiophore. F–H. Conidiophore on PDA. I. Conidia from PDA culture. Scale bars = 10 μm.
Typus: Taiwan, Chiayi County, on Salvinia auriculata (a floating fern), Dec. 2006, Chen et al. (holotype NTUPPMH 20-074 preserved in a metabolically inactive state, ex-type culture NTUPPMCC 20-074/BCRC 34536).
Etymology: Referring to the plant host genus – Salvinia auriculata.
Description: See Chen et al. (2008).
Culture characteristics: Colony on PDA (NTUPPMCC 20-074) reaching 2.6 cm diam in 14 d at 25 °C, white, reverse yellow to light brown, convex, entire edge. Synnemata not observed. Phialides arising from the prostrate hyphae or on aerial hyphae in culture, single, solitary, hyaline, subcylindrical, (21–)27–45(–53) × 2 μm, tapering into the neck, 1 μm in width. Conidia produced in small globose heads at the apex of the phialides, unicellular, hyaline, subglobose to ellipsoidal, 3–4(–4.5) × 1.5–2(–2.5) μm.
Distribution: Chiayi County; Taiwan.
Additional materials examined: Taiwan, Chiayi County, on Salvinia auriculata, Dec. 2006, Chen et al., NTUPPMH 20-075, living culture NTUPPMCC 20-075/BCRC 34537.
Notes: The two strains used in this study, namely NTUPPMCC 20-074 (BCRC 34536) and NTUPPMCC 20-075 (BCRC 34537), were initially isolated from a brown spot on a Salvinia species in Taiwan and identified as Si. lanosoniveum based on the asexual reproduction structures and sequence data of the ITS region by Chen et al. (2008). These strains were published as a disease note to report Si. lanosoniveum for the first time in Taiwan (Chen et al. 2008). In the present study, we obtained these two strains from BCRC (34536 and 34537), the culture collection where the original authors deposited the strain and generated DNA sequence data for ITS, nrLSU, tef1-α, rpb1, and rpb2. Remarkably, both NTUPPMCC 20-074 (BCRC 34536) and NTUPPMCC 20-075 (BCRC 34537) clustered in the same clade distant from Si. lanosoniveum in our multigene phylogenetic analyses. Therefore, we proposed Si. salviniae sp. nov. to accommodate NTUPPMCC 20-074 (BCRC 34536) and NTUPPMCC 20-075 (BCRC 34537) in the genus Simplicillium. Si. salviniae can be easily separated from its phylogenetically closely related taxon, Si. lanosoniveum in having larger conidia (3–4 × 2 μm vs 1.5–3 × 0.7–1.3 μm) (Zare & Gams 2001). Moreover, the phialides of Si. salviniae were also larger than those of Si. lanosoniveum (27– 45 × 2 μm vs 15–35 × 0.7–1.5 μm) (Zare & Gams 2001).
New records for Taiwan
Akanthomyces kanyawimiae Mongkols. et al., Mycologia 110: 237. 2018. Fig. 13.
Fig. 13.
Morphology of Akanthomyces kanyawimiae (NTUPPMH 20-058, living culture NTUPPMCC 20-058). A. Synnemata of fungus arising from host (Araneae). B, C. Magnified synnemata. D. Phialides on host (Araneae). E, F. Conidia from host (Araneae). G. Obverse and reverse of colonies on PDA at 14 d. H. Isaria-like conidiogenesis. I, J. Phialides on PDA. K. Conidia from PDA culture. Scale bars = 10 μm.
Typus: Thailand, Phetahabun Province, Nam Nao National Park, on spider (Araneae), on the stem of dicotyledonous plant, 24 Nov. 2016, K. Tasanathai, S. Mongkolsamrit, R. Promharn (holotype BBH 42364, ex-type culture TBRC 7244).
Description: See Mongkolsamrit et al. (2018). The description below is based on specimen NTUPPMH 20-058 and living culture NTUPPMCC 20-058.
Material examined: Taiwan, Nantou City, Shuishe Great Mountain Nature Trail on Araneae host, 6 Apr. 2018, W.-Y. Chuang, NTUPPMH 20-058, living culture NTUPPMCC 20-058.
Description from specimen NTUPPMH 20-058: Sexual morph: Not observed. Asexual morph: Araneae host covered by dense white to light yellow mycelia, buried inside the leaves. Synnemata single, branched at the apex, light yellow, bearing powdery conidia, arising from the edge of the leaves, 0.1–0.4 mm long. Conidiophores arising from prostrate hyphae or on aerial hyphae in culture, consisting of solitary phialides along the hyphae or verticillate branches with two to three phialides in whorls. Phialides (7–)7.5–9.5(–11.5) × 2–3(–3.5) μm, subcylindrical to flask–shaped, slightly tapering into a neck, 1–1.5 μm in width. Conidia unicellular, hyaline, cylindrical to ellipsoidal, sometimes in a chain, 2.5–3(–3.5) × 2–2.5 μm.
Culture characteristics: Colony on PDA (NTUPPMCC 20-058) white, fluffy, entire edge, moderate growth, 4.2 cm diam in 21 d at 25 °C; reverse light yellow to brown. Conidiophores arising from prostrate hyphae or on aerial hyphae in culture, consisting of solitary phialides along the hyphae or verticillate branches with two to three phialides in whorls. Phialides (7–)10–17(–22) × 2–2.5 μm, subcylindrical to flask-shaped, slightly tapering into a neck, 1–1.5 μm in width. Conidia unicellular, hyaline, cylindrical to ellipsoidal, sometimes in a chain, 3.5 × 2.5 μm.
Distribution: Thailand, Taiwan.
Notes: Akanthomyces kanyawimiae was recently described by Mongkolsamrit et al. (2018) from Thailand. The fresh collection in the present study resembles A. kanyawimiae, and the multigene phylogenetic analysis showed that our strain NTUPPMCC 20-058 clustered with the ex-type strain (TBRC 7244) of A. kanyawimiae (Fig. 2).
Akanthomyces muscarius (Zare & Gams) Spatafora et al., IMA Fungus 8: 342. 2017. Fig. 14.
Fig. 14.
Morphology of Akanthomyces muscarius (NTUPPMH 20-059, living culture NTUPPMCC 20-059). A. Mycelium covering Araneae host. B. Phialides on the host. C, D. Conidia on the host. E. Obverse and reverse of colonies on PDA at 14 d. F. Conidiophores arising on aerial hyphae. G. Conidial mass on the top of a conidiophore. H–J. Phialides on PDA. K. Conidia from PDA culture. Scale bars: B = 20 μm; C, D = 10 μm; G = 20 μm; H–K = 10 μm.
Basionym: Cephalosporium muscarium Petch, The Naturalist London 102. 1931.
Synonyms: Lecanicillium muscarium Zare & Gams, Nova Hedwigia 73: 13. 2001.
See MycoBank (2023) for further synonyms.
Typus: UK, on fly (Diptera), 1931, collector unknown (holotype 156599); on Trialeurodes vaporariorum (Hemiptera), 1957, N.W. Hussey (epitype CBS H-7268, ex-epitype culture CBS 143.62 = IMI 068689 = ATCC 28300 = MUCL 9713).
Description: See Zare & Gams (2001). The description below is based on specimen NTUPPMH 20-059 and living culture NTUPPMCC 20-059.
Material examined: Taiwan, Hsinchu City, Mt. Ptlaman, on Araneae host, 29 Oct. 2019, L.-J. Chu, NTUPPMH 20-059, living culture NTUPPMCC 20-059.
Description from specimen NTUPPMH 20-059: Sexual morph: Not observed. Asexual morph: Araneae host covered by dense white to light yellow mycelia, attached on the underside of the leaves. Conidiophores consisting of solitary phialides along the hyphae or verticillate branches with two to four phialides in whorls. Phialides 16.5–26.5(–33.5) × 2–2.5(–3) μm, subcylindrical to flask–shaped, slightly tapering into a neck, 1 μm in width. Conidia 3–5.5(–6.5) × 2 μm, unicellular, hyaline, cylindrical to oblong-elliptical.
Culture characteristics: Colony on PDA (NTUPPMCC 20-059) white, flat, entire edge, moderately growing, 4 cm diam in 21 d at 25 °C; reverse light yellow. Conidiophores arising from prostrate hyphae or on aerial hyphae in culture, consisting of solitary phialides along the hyphae or verticillate branches with two to four phialides in whorls. Phialides (14.5–)17–25.5(–32) × (1.5–)2–2.5(–3) μm, subcylindrical to flask–shaped, slightly tapering into a neck, 1 μm in width. Conidia (3.5–)4–5(–5.5) × 2 μm, cylindrical to oblong-elliptical, unicellular, hyaline.
Distribution: Taiwan, UK.
Notes: Akanthomyces muscarius was initially described as Lecanicillium muscarium on a larva of Trialeurodes vaporariorum (Hemiptera) by Zare & Gams (2001). However, this species was later transferred to Akanthomyces by Kepler et al. (2017). In our phylogeny, strain NTUPPMCC 20-059 formed clade sister to A. muscarius (CBS 143.62T and MFLU 18-1145) with relatively well statistical support (97 % ML, 0.96 PP). The strain NTUPPMCC 20-059 is morphologically comparable with the original description provided by (Zare & Gams 2001) showing similar size in phialides (17–25.3 × 1.9–2.2 μm vs 20–35 × 1–1.7 μm) and conidia (3.8–4.8 × 1.8–2.2 μm vs 2.5–5.5 × 1–1.5 μm). Therefore, based on their similarity in phylogeny and morphology, we tentatively identified our specimen as A. muscarius, marking a new report for Taiwan.
Beauveria lii S.L. Zhang & B. Huang, Mycotaxon 121: 203. 2012. Fig. 15.
Fig. 15.
Mycelial colony and conidiation of Beauveria lii (NTUPPMCC 20-054). A, B. Obverse and reverse of colonies on PDA at 21 d. C–E. Conidiogenesis pattern and phialides on PDA. F–H. Conidia from PDA culture. Scale bars = 10 μm.
Typus: China, Shaanxi Province, Xunyi County, on the larva of Henosepilachna vigintioctopunctata (Coleoptera), 23 Sep. 2010, L.-M. He (holotype RCEF5500, ex-type culture ARSEF 11741).
Description: See Zhang et al. 2012. The description below is based on living culture NTUPPMCC 20-054.
Material examined: Taiwan, Yunlin City, Kukeng Township, on ladybird beetle. Living culture NTUPPMCC 20-054/BCRC 35553.
Culture characteristics: Colony on PDA (NTUPPMCC 20-054) white, dense, convex, entire edge, fast-growing, 6.1 cm diam in 21 d at 25 °C; reverse white to light yellow. Conidiophores developing on aerial hyphae, solitary, base flask-shaped, (4–)5–9(–11) × 0.5–1.5 μm, apex with an indeterminate, denticulate rachis. Conidia unicellular, hyaline, usually cylindrical to ellipsoidal, occasionally obovoid (4.5–)5.5–7(–9) × (2–)2.5–3(–3.5) μm.
Distribution: China, Taiwan.
Notes: Beauveria lii was isolated from larva of Henosepilachna vigintioctopunctata and introduced by Zhang et al. 2012 from China. The strain identified as B. lii (NTUPPMCC 20-054) in the present study was also isolated from H. vigintioctopunctata and shares similar morphology with the original description of B. lii. In addition, the multigene phylogenetic tree generated in the present study clearly exhibited that our B. lii strains clustered within the species clade of B. lii. Therefore, we identified NTUPPMCC 20-054 as B. lii, marking the first record of B. lii in Taiwan.
Beauveria medogensis Imoulan & Y.J. Yao, J. Invert. Pathol. 139: 79. 2016. Fig. 16.
Fig. 16.
Morphology of Beauveria medogensis (NTUPPMH 20-055, living culture NTUPPMCC 20-055). A. Mycelium arising from Formicidae host. B. Formicidae host. C. Phialide and conidia on the host. D. Obverse and reverse of colonies on PDA at 14 d. E. Conidiogenesis pattern. F–H. Phialides on PDA. I–K. Conidia from PDA culture. Scale bars: C = 10 μm, E–H = 10 μm, I–K = 5 μm.
Typus: China, Tibet, Mêdog County, from soil sample, 23 Apr. 2014, W.-L. Lu (holotype CGMCC 3.15617, ex-type culture 2898).
Description: See Imoulan et al. (2016). The description below is based on specimen NTUPPMH 20-055 and living culture NTUPPMCC 20-055.
Material examined: Taiwan, Taoyuan City, Mt. Hutou, on Formicidae host, 10 Mar. 2018, W.-Y. Chuang, NTUPPMH 20-055, living culture NTUPPMCC 20-055.
Description from specimen NTUPPMH 20-055: Sexual morph: Not observed. Asexual morph: Formicidae host covered by dense white mycelia with powdery conidia. Conidiophores solitary, base sometimes flask-shaped, produced laterally on aerial hyphae, (8.5–)10.5–19.5(–22) × (1.5–)2–3(–3.5) μm, apex with an indeterminate, denticulate rachis, 1 μm wide. Conidia (2–)2.5–3.5(–4.5) × (1.5–)2–3(–3) μm, subglobose to globose, hyaline, aseptate, scattered or gregarious in clusters.
Culture characteristics: Colony on PDA (NTUPPMCC 20-055) white, dense, low convex, entire edge, fast-growing, 4.7 cm diam in 14 d at 25 °C; reverse light yellow to orangish yellow. Conidiophores solitary, base sometimes flask-shaped, produced laterally on aerial hyphae, (9–)10.5–16(–20) × (2–)2.5–3(–3.5) μm, apex with an indeterminate, denticulate rachis, 1–1.5 μm wide. Conidia 2–3.5(–4.5) × (2–)2.5–3 μm, subglobose to globose, hyaline, aseptate, scattered or gregarious in clusters.
Distribution: China, Taiwan.
Notes: Beauveria medogensis was originally isolated from soil and described by Imoulan et al. (2016). Our strain NTUPPMCC 20-055 (Fig. 16) is similar to B. medogensis based on both morphology (in producing undetermined denticulate rachis with globose to subglobose conidia) and DNA sequences (Fig. 2). The pathogenicity of B. medogensis has been confirmed using Helicoverpa armigera and Tenebrio molitor larvae (Imoulan et al. 2016). Therefore, this is the first report of B. medogensis isolated as a pathogen on a formicid host under natural conditions and it is a new record in Taiwan.
In addition to these new records in Taiwan, based on the results of the multigene phylogenetic data coupled with morphological data, we proposed to synonymize Beauveria staphylinidicola under Beauveria bassiana.
Beauveria bassiana (Bals.-Criv.) Vuill., Bull. Soc. Bot. Fr. 59: 40. 1912.
Basionym: Botrytis bassiana Bals.-Criv., Linnaea 10: 611. 1835.
Synonyms: Beauveria staphylinidicola (Kobayasi & Shimizu) B. Shrestha et al., IMA Fungus 8: 345. 2017.
Cordyceps staphylinidicola Kobayasi & Shimizu [as ‘staphylinidaecola’], Bull. Nat. Sci. Mus. Tokyo B 8: 88. 1982.
See MycoBank (2023) for further synonyms.
Cordyceps lepidopterorum Mongkols. et al., Mycologia 110: 247. 2018. Fig. 17.
Fig. 17.
Morphology of Cordyceps lepidopterorum (NTUPPMH 18-118 and living culture NTUPPMCC 18-118). A, B. Synnemata arising from a Cicadidae nymph. C. Synnemata. D. Phialides on the host. E. Conidia on the host. F. Obverse and reverse of colonies on PDA at 14 d. G, H. Isaria-like conidiogenesis. I. Phialides on PDA. J. Conidia from PDA culture. Scale bars: D–E = 10 μm; G = 50 μm; H–I = 10 μm; J = 5 μm.
Synonym: Cordyceps jakajanicola Luangsa-ard et al., Persoonia 43: 357. 2019.
Typus: Thailand, Chiang Mai Province, Kanlayaniwatthana District, on lepidopteran larva in leaf litter, 22 Nov. 2015, K. Tasanathai, S. Mongkolsamrit, D. Thanakitpipattana, W. Noisripooma, R. Promharn, P. Srikitikulchai & S. Wongkanoun (holotype BBH 40735, ex-type culture TBRC 7263).
Description: See Mongkolsamrit et al. (2018). The description below is based on specimen NTUPPMH 18-118 and living culture NTUPPMCC 18-118.
Material examined: Taiwan, Nantou County, Mt. Nandongyan, on Cicadidae nymph, 29 Jul. 2018, C.-M. Hu, NTUPPMH 18-117, living culture, NTUPPMCC 18-117; Taichung City, Wufong district, on Cicadidae nymph, 19 Aug. 2018, C.-M. Hu, NTUPPMH 18-118, living culture, NTUPPMCC 18-118; New Taipei City Manyueyuan Forest Recreation, on Cicadidae nymph, 7 Oct. 2018, W.-Y. Chuang, NTUPPMH 17-038, NTUPPMCC 17-038; ibid., NTUPPMH 17-038-1, living culture NTUPPMCC 17-038-1; Nantou City, Huisun experimental forest, on Cicadidae nymph, 7 Sep. 2017, W.-Y. Chuang, NTUPPMH 17-037, living culture NTUPPMCC 17-037; Taoyuan City, Mt. Hutou, on Cicadidae nymph, 10 Mar. 2018, W.-Y. Chuang, NTUPPMH 18-114, living culture NTUPPMCC 18-114; Taipei City, Fuyang Eco Park, on Cicadidae nymph, 11 Nov. 2017, W.-Y. Chuang, NTUPPMH 17-036, living culture NTUPPMCC 17-036.
Description from specimen NTUPPMH 18-118: Sexual morph: Undetermined. Asexual morph: Synnemata numerous, light yellow, erect, arising from the head of the Cicadidae nymph, buried under soil, 7 cm long. Conidiophores arising from prostrate hyphae or on aerial hyphae, consisting of solitary phialides along the hyphae or verticillate branches with three to four phialides in whorls. Phialides (4.5–)5–6(–7) × (2.5–)3–4 μm, flask-shaped, with ellipsoidal basal portion abruptly tapering into a thick neck, 1 μm in width. Conidia (6.5–)7.5–12(–13.5) × (2.5–)3–3.5 μm, unicellular, hyaline, cylindrical to ellipsoidal, sometimes in a chain.
Culture characteristics: Colony on PDA (NTUPPMCC 18-118) white, entire edge, moderately fast growing, 4 cm diam in 14 d at 25 °C; reverse white to light yellow. Conidiophores arising from prostrate hyphae or on aerial hyphae, consisting of solitary phialides along the hyphae or verticillate branches with three to four phialides in whorls. Phialides (6.5–)8–11(–13) × (3.5–)4–4.5(–5) μm, flask-shaped, with ellipsoidal basal portion abruptly tapering into a thick neck, 1–1.5 μm in width. Conidia (7–)7.5–9(–10) × (3–)3.5–4(–4.5) μm, unicellular, hyaline, cylindrical to ellipsoidal, sometimes in a chain.
Distribution: Thailand, Taiwan.
Notes: Based on the multigene phylogeny, all strains (NTUPPMCC 17-036 to NTUPPMCC 17-038-1, NTUPPMCC 18-114, NTUPPMCC 18-117 to NTUPPMCC 18-119) recognized in the present study as C. lepidopterorum nested with the clade containing the ex-type strain of C. lepidopterorum (TBRC 7263). Our fresh collections show overlapping morphologies with the type specimen of C. lepidopterorum except for the host species from which they were isolated (Mongkolsamrit et al. 2018). All strains used in the present study parasitized Cicadidae nymphs while C. lepidopterorum (TBRC 7263) was identified as a pathogen on lepidopteran larvae. However, strains NTUPPMCC 18-114 and NTUPPMCC 17-036 identified as C. lepidopterorum also show overlapping morphologies with the phylogenetically closely related species C. jakajanicola, which was initially introduced by Luangsa-ard et al. 2019 to accommodate Cordyceps species parasitizing Cicadidae nymphs. In fact, while introducing C. jakajanicola, Luangsa-ard et al. 2019 considered the host specificity of the species as the main character to separate C. jakajanicola from C. lepidopterorum (Cicadidae nymphs vs lepidopteran larvae). Despite this difference, both species have similar morphological characters such as an isaria-like conidiogenesis pattern and conidial size and they showed little variation in their molecular data. Thus, our finding further indicated that host-based classification cannot be used as a key character to separate C. jakajanicola from C. lepidopterorum, and thus we propose to synonymize C. jakajanicola under C. lepidopterorum in the present study. According to our best knowledge, this is the first report of C. lepidopterorum from a Cicadidae nymph in Taiwan (Tzean et al. 1997).
Cordyceps neopruinosa Mongkols. et al., Mycol. Prog. 19: 976. 2020. Fig. 18.
Fig. 18.
Morphology of Cordyceps neopruinosa (NTUPPMH 18-128, living culture NTUPPMCC 18-128). A. Stromata growing on a limacodid cocoon. B. Ascomata of specimen. C. Section of perithecium. D. Cell wall of perithecium. E, F. Ascus. G. Obverse and reverse of colonies on PDA. H. Evlachovaea-like conidiogenesis. I, J. Phialides on PDA. K. Conidia from PDA culture. Scale bars: C = 50 μm, D = 20 μm, E = 40 μm, F = 30 μm, H = 30 μm. I, J = 10 μm, K, L = 5 μm.
Typus: Thailand, Chiang Mai Province, Kanlayaniwatthana District, 18°58′42.62″N, 98°17′13.01″E, on a pupa of Limacodidae (Lepidoptera) inside a cocoon buried in soil, 6 Nov. 2019, S. Mongkolsamrit, A. Khonsanit, W. Noisripoom, P. Khamsuntorn & S. Sommai (holotype BBH 47573, ex-type culture BCC 91361).
Description: See Mongkolsamrit et al. (2020). The description below is based on specimen NTUPPMH 18-128 and living culture NTUPPMCC 18-128.
Material examined: Taiwan, New Taipei City, Manyueyuan Forest Recreation, on a Limacodidae cocoon, 20 Sep. 2017, M.-J. Chen, NTUPPMH 17-040, living culture NTUPPMCC 17-040; Taoyuan City, Dongyan Mountain Forest Recreational Area, on a Limacodidae cocoon, 10 Sep. 2018, W.-Y. Chuang, NTUPPMH 18-126, living culture NTUPPMCC 18-126; Taoyuan City, Dongyan Mountain Forest Recreational Area, on a Limacodidae cocoon, 30 Sep. 2018, W.-Y. Chuang, NTUPPMCC 18-127, living culture NTUPPMCC 18-127; ibid., NTUPPMH 18-128, living culture NTUPPMCC 18-128; ibid., NTUPPMCC 18-129, living culture NTUPPMCC 18-129.
Description from specimen NTUPPMH 18-128: Sexual morph: Stromata arising from the upwards regions of the Limacodidae cocoon, buried under the soil. Stromata simple, solitary, clavate, light yellow to orange at the basal region. Terminal part of the stromata fertile, orange to red. Perithecia semi-immersed, ovoid, (265–)289–362 × (138–)145–195 μm. Asci cylindrical, hyaline, 8-spored, (94–)107–168(–203) × (2.5–)3–4(–4.5) μm with a hemispherical cap, (2.5–)3–3.5 × (2.5–)3–3.5 μm at the apex. Ascospores smooth, filiform, septate, (120–)148–182(–201) × 0.5 μm. No part-spores observed. Asexual morph: Not observed.
Culture characteristics: Colony on PDA (NTUPPMCC 18-128) white, flat, entire edge, moderately fast growing, 6 cm diam in 21 d at 25 °C; reverse amber to orange. Synnemata awl-shaped, white, scattered around the colony. Conidiophores arising from prostrate hyphae or on aerial hyphae, single, hyaline, navicular, (10–)11–23.5(–33) × 2–2.5(–3) μm, tapering into the neck, 1 μm in width. Conidia 5–7(–9) × 2–2.5(–3) μm, unicellular, hyaline, shape irregular, mostly cylindrical to ellipsoidal, sometimes in a chain. Chlamydospores and synnemata not observed.
Distribution: Thailand, Taiwan.
Notes: Cordyceps neopruinosa was introduced by Mongkolsamrit et al. (2020) to accommodate a fungal strain grown on a pupa of Limacodidae (Lepidoptera) inside a cocoon buried in soil in Thailand. Phenotypically (producing orange to red stromata and hyaline ascospores with septa), our fresh collection fits well with C. neopruinosa, and phylogenetic analysis showed that strains used in this study clustered in the same clade with the ex-type strain of C. neopruinosa (BCC 91361) (Fig. 2). Furthermore, our fresh collections as well as the original specimen of C. neopruinosa were collected as a parasite on Limacodidae cocoon. Thus, we recognized our specimen as C. neopruinosa, which is a new report for Taiwan.
Samsoniella cardinalis H. Yu et al., Fungal Diversity 103: 27. 2020. Fig. 19.
Fig. 19.
Mycelium of S. cardinalis (D–I) growing on fruiting body of cordyceps-like taxon (A–C) (NTUPPMH 20-066, living culture NTUPPMCC 20-066). A. Cordyceps sp. on cicada. B, C. Conidia of Cordyceps sp. D. Obverse of colonies on PDA at 21 d. E. Reverse of colonies on PDA at 21 d. F–H. Conidiogenesis pattern and phialides on PDA. I, J. Conidia from PDA culture. Scale bars: B, C = 5 μm; F–J = 10 μm.
Typus: China, Yunnan Province, Kunming City, Wild Duck Lake Forest Park, on the pupa of Limacodidae in a cocoon buried in soil, 12 Aug. 2017, H. Yu (holotype YHH 15732, ex-type living culture YFCC 6144).
Description: See Wang et al. (2020). The description below is based on specimen NTUPPMH 20-066 and living culture NTUPPMCC 20-066.
Material examined: Taiwan, Taoyuan City, Mt. Hutou, on Cordyceps sp. sporocarp, 3 Mar. 2018, M.-L. Lo, NTUPPMH 20-066, living culture NTUPPMCC 20-066.
Description from specimen NTUPPMH 20-066: Asexual morph: Mycelium growing on fruiting body of cordyceps-like taxon. Description of the cordycipitaceous fungi: Synnemata numerous, yellow to light brown, arising from the head of the nymph. Conidia unicellular, hyaline, cylindrical to ellipsoidal, (6.5–)7.5–8.5(–9.5) × 3.5 μm.
Culture characteristics: Colony on PDA (NTUPPMCC 20-066) white, fluffy, convex, entire edge, moderately fast-growing, 5.5 cm diam in 21 d at 25 °C; reverse light yellow to orangish yellow. Conidiophores erect, consisting of solitary phialides along the hyphae or verticillate branches with two to five phialides in whorls, (3–)4.5–10.5(–16) × 1.5–2.5(–3) μm. Phialides hyaline, (5.5–)6–13(–26) × 1.5–2.5 μm, with a flask-shaped basal portion tapering into a distinct neck, 1 μm in width. Conidia unicellular, hyaline, ovoid, (3–)3.5–4 × (2–)2.5–3 μm.
Distribution: China, Taiwan.
Notes: Samsoniella cardinalis was initially introduced by Wang et al. (2020) from Kunming City, China. In our phylogeny, strain NTUPPMCC 20-066 clustered within the clade containing S. cardinalis strains (YFCC 5830, YFCC 6144, YFCC 6320) with high statistical support. Furthermore, our strain shares similar phenotypic characters with S. cardinalis (YHH 15732) in having isaria-like conidiogenesis and similar-sized phialides and conidia. Thus, we identified our strains as S. cardinalis, and this is the first report of S. cardinalis in Taiwan.
Samsoniella hepiali (Q.T. Chen & R.Q. Dai ex R.Q. Dai et al.) H. Yu et al., Fungal Diversity 103: 31. 2020. Fig. 20.
Fig. 20.
Morphology of Samsoniella hepiali (NTUPPMH 18-159, living culture, NTUPPMCC 18-159). A. Synnemata arising from lepidopteran insect. B. Phialide on the host. C. Conidia on the host. D. Obverse and reverse of colonies on PDA at 14 d. E–G. Isaria-like conidiogenesis. H–J. Phialides on PDA. K. Conidia from PDA culture. Scale bars: B, C = 10 μm; E–K = 10 μm.
Basionym: Paecilomyces hepiali Q.T. Chen & R.Q. Dai ex R.Q. Dai et al., Mycosystema 27: 642. 2008.
Synonyms: Paecilomyces hepiali Chen & Dai, Acta Agric. Univ. Pekin. 6: 223. 1989.
Typus: China, Yunnan Province, Diqing Tibetan Autonomous Prefecture, Deqin County, Baima Snow Mountain, isolated from the larva of Hepialus armoricanus parasitized by Ophiocordyceps sinensis, Jun. 1982, R.-Q. Dai (holotype IMM 82–2 = CHICMM 82–2, ex-type living culture ICMM 82–2).
Description: See Wang et al. (2020). The description below is based on specimen NTUPPMH 18-159 and living culture NTUPPMCC 18-159.
Material examined: Taiwan, Taichung City, Mt. Yuanzui, 9 Jun. 2018, on lepidopteran larvae, W.-Y. Chuang, NTUPPMH 18-159, living culture, NTUPPMCC 18-159.
Description from specimen NTUPPMH 18-159: Sexual morph: Undetermined. Asexual morph: Synnemata arising from the whole body of lepidopteran insects, irregularly branched or unbranched at the apex, white, bearing a powdery, white mass of conidia. Conidiophores erect, consisting of two to three phialides in whorls. Phialides hyaline, 7.5–11(–12.5) × 2–3 μm, with a flask-shaped basal portion tapering into a distinct neck, 1 μm in width. Conidia unicellular, hyaline, ovoid, sometimes in a chain, (2–)2.5–3(–3.5) × (1.5)2–2.5 μm.
Culture characteristics: Colony on PDA (NTUPPMH 18-159) white, flat, entire edge, moderately slow growing, 3.9 cm diam in 21 d at 25 °C; reverse light yellow to yellow. Media around colony turned white. Conidiophores erect, consisting of solitary phialides along the hyphae or verticillate branches with two to three phialides in whorls. Phialides hyaline, (7.5–)9–12.5(–15) × 2.5 μm, with a flask-shaped basal portion tapering into a distinct neck, 1 μm in width. Conidia unicellular, hyaline, ovoid, sometimes in a chain, 3–3.5(–4) × 2–2.5(–3) μm.
Distribution: China, Taiwan.
Notes: Samsoniella hepiali (NTUPPMCC 18-159) identified in the present study is morphologically similar to S. hepiali in having an isaria-like conidiogenesis pattern and similar sized phialides and conidia (Wang et al. 2020). Moreover, our strain forms a monophyletic clade with the ex-type strain of S. hepiali (ICMM 82–2) which was used to introduce the species (Wang et al. 2020). Hence, we acknowledge our collection as S. hepiali, which is a new record for Taiwan.
Simplicillium chinense F. Liu & L. Cai, Cryptog. Mycol. 33: 139. 2012. Fig. 21.
Fig. 21.
Morphology of Beauveria brongniartii (A–J) and Simplicillium chinense (K–O) (NTUPPMH 20-073, living culture NTUPPMCC 20-073). A. Beauveria brongniartii on Locastra muscosalis cocoon. B. Locastra muscosalis. C. Stromata of cordycipitaceous fungi. D. Ascomata of cordycipitaceous fungi. E, F. Section of perithecium. G, H. Ascus. I. Ascus cap. J. Part-spores. K. Obverse and reverse of colonies on PDA. L. Conidiogenesis pattern. M, N. Phialide on PDA. O. Conidia from PDA culture. Scale bars: E = 100 μm; F–H = 50 μm; I, J = 10 μm; L = 20 μm; M–O = 10 μm.
Typus: China, 17 Mar. 2011, F. Liu (holotype HMAS 243490, ex-type living culture CGMCC 3.14970).
Description: See Liu & Cai (2018). The description below is based on specimen NTUPPMH 20-073 and living culture NTUPPMCC 20-073.
Material examined: Taiwan, Taoyuan City, Mt. Hutou, on Beauveria brongniartii, 10 Mar. 2018, W.-Y. Chuang, NTUPPMH 20-073, living culture NTUPPMCC 20-073.
Description from specimen NTUPPMH 20-073: Sexual morph: Undetermined. Asexual morph: Mycelium growing on cordycipitaceous fungal fruiting body. Description of the cordycipitaceous fungi: Stromata simple, solitary, brown, arising from the cocoon of Locastra muscosalis host, buried under soil. Perithecia numerous, gregarious at the apex of stromata, superficial, ovoid, (332–)400–552(–578) × (257–)282–351(–370) μm. Asci cylindrical, hyaline, 8-spored, (259–) 324–429(–484) × (5.5–)6.5–9.5(–11.5) μm with a hemispherical cap, 5.5–7(–8.5) × (4.5–)5–5.5(–6) μm at the apex. Ascospores filiform, septate, disarticulating into part-spores, cylindrical, 5.5–7(–8.5) × (2–)2.5–3.5 μm.
Culture characteristics: Colony on PDA (NTUPPMCC 20-073) white, low convex, radially striated with lobate edge, moderately growing, 3.0 cm diam in 14 d at 25 °C; reverse pale brown. Conidiophores arising from the prostrate hyphae or on aerial hyphae, single, hyaline, subcylindrical, (17–)18.5–25.5(–32.5) × 2 μm, tapering into the neck, 1 μm in width. Conidia (4–)4.5–5 × 1–1.5 μm, produced in branched or unbranched chains, unicellular, hyaline, cylindrical.
Distribution: China, Taiwan.
Notes: Simplicillium chinense was introduced by Liu & Cai (2012) and was isolated from submerged wood in China. This fungus was characterized with conidial chains. Our strain (NTUPPMCC 20-073) had similar morphological characters as Si. chinense but was isolated from the cordycipitaceous fungal fruiting body. Therefore, combining morphology and the results of phylogenetic analyses, our strains are identified as a new record of Si. chinense on cordycipitaceous fungi in Taiwan.
Cordyceps javanica (Bally) Kepler et al., IMA Fungus 8: 347. 2017. Fig. 22.
Fig. 22.
Morphology of C. javanica (NTUPPMH 18-115, living culture NTUPPMCC 18-115). A. Fungus on unknown insect host. B. Obverse and reverse of colonies on PDA at 14 d. C. Conidia on the host. D–F. Isaria-like conidiogenesis. G, H. Phialides on PDA. I, J. Conidia from PDA culture. Scale bars: C–E = 20 μm; F–H = 10 μm; I–J = 5 μm.
Basionym: Spicaria javanica Bally, Meded. Koffiebessenboeboek-Fonds, Malang 6: 146. 1923.
Synonyms: Paecilomyces javanicus (Bally) A.H.S. Br. & G. Sm., Trans. Brit. Mycol. Soc. 40: 65. 1957.
Isaria javanica (Bally) Samson & Hywel-Jones, Mycol. Res. 109: 588. 2005.
Typus: Indonesia, Java, on Stephanoderes hampei (Coleoptera), 1923, F.W. Bally (ex-lectotype CBS 134.22).
Description: See Samson (1974). The description below is based on specimen NTUPPMH 18-115 and living culture NTUPPMCC 18-115.
Material examined: Taiwan, Hsinchu County, Jianshi Township, Mt. Ptlaman, on unknown host, 8 Aug. 2018, L.-J. Chu, NTUPPMH 18-115, living culture NTUPPMCC 18-115; Nantou County, Xinyi township, on whitefly, 1996, collector unknown, living culture NTUPPMCC 18-116/BCRC FU30878.
Description from specimen NTUPPMH 18-115: Sexual morph: Not observed. Asexual morph: Synnemata numerous, simple, white, erect, arising from Erebidae (Lymantriinae), bearing powdery, white mass of conidia along the branches, 7.7–9.2 mm long. Conidiophores not observed. Conidia 3–4(–4.5) × 2–2.5 μm, hyaline, cylindrical to ellipsoidal.
Culture characteristics: Colony on PDA (NTUPPMCC 18-115) reaching 4 cm diam in 14 d at 25 °C, white, reverse white to pale yellow, fluffy, undulate. Conidiophores arising from the prostrate hyphae or on aerial hyphae, consisting of solitary phialides along the hyphae or verticillate branches with two to four phialides in whorls. Phialides (8–)9–13(–16) × (2–)2.5–3 μm, flask–shaped, with ovoid basal portion slightly tapering into a neck, 1 μm in width. Conidia produced in chains, unicellular, hyaline, cylindrical to ellipsoidal, 4.5–5.5(–6.5) × 2–2.5(–3) μm.
Distribution: Ghana, Indonesia, Taiwan.
Notes: The genus Parahevansia was initially introduced by Mongkolsamrit et al. (2022) to accommodate Hevansia koratensis in the family Cordycipitaceae. In their study, Mongkolsamrit et al. (2022) concluded that the two strains of P. koratensis (NHJ 2662 and NHJ 666.01) form an independent clade with strong support out of the Hevansia clade and in the proximity of Cordyceps species but the boundaries of the genus Cordyceps was determined by using few species in their phylogenetic analysis. However, our multilocus phylogenetic analysis indicated that these two strains are clearly clustered within the species clade of C. javanica after including all introduced Cordyceps species. Even though P. koratensis nested in the C. javanica clade in the present phylogeny, the morphology of P. koratensis (syn. Hevansia koratensis, Akanthomyces koratensis) described in Hywel-Jones (1996) showed huge variation in conidiogenesis compared to C. javanica (Fig. 22). Parahevansia koratensis is morphologically characterized by white to greyish cylindrical, attenuated synnematal growth covered by a hymenium-like layer of obovoid to ellipsoid phialides producing catenulate, clavate conidia while C. javanica produces isaria-like phialides (flask-shaped with long necks) and divergent long, dry chains of fusoid conidia. Thus, in the present study we tentatively propose to transfer P. koratensis in Cordyceps to maintain the monophyly of Cordyceps and treat it as C. koratensis without synonymizing under C. javanica considering the morphological variation of these two taxa.
Cordyceps koratensis (Hywel-Jones) H.A. Ariyaw., M. Stadler & Luangsa-ard, comb. nov. MycoBank 851851
Basionym: Akanthomyces koratensis Hywel-Jones, Mycol. Res. 100: 1067. 1996.
Synonyms: Parahevansia koratensis (Hywel-Jones) Mongkols. & Noisrip., MycoKeys 91: 138. 2022.
Hevansia koratensis (Hywel-Jones) Luangsa-ard et al., IMA Fungus 8: 349. 2017.
Cordyceps ninchukispora (C.H. Su & H.H. Wang) G.H. Sung et al., Stud. Mycol. 57: 49. 2007. Fig. 23.
Fig. 23.
Morphology of C. ninchukispora (ex-type culture NTUPPMCC 20-077/BCRC 31900). A, B. Obverse and reverse of colonies on PDA at 21 d. C–E. Phialides on PDA. F. Conidia from PDA. Scale bars: C–F = 10 μm.
Basionym: Phytocordyceps ninchukispora C.H. Su & H.H. Wang, Mycotaxon 26: 338. 1986.
Typus: Taiwan, Nantou County, Xitou Nature Education Area, on seed of Beilschmiedia erythrophloia, Jan. 1985, C.H. Su & HH. Wang (holotype NTU 850106, ex-type culture NTUPPMCC 20-077/BCRC 31900).
Description: Refer to Su & Wang (1986). The description below is based on ex-type culture NTUPPMCC 20-077:
Culture characteristics: Colony on PDA (NTUPPMCC 20-077/BCRC 31900) white to light yellow, fluffy, entire edge, moderately slow growing, 4.6 cm diam in 21 d at 25 °C; reverse light yellow to orange. Sexual morph: Not observed. Asexual morph: Synnemata not observed. Conidiophores arising from prostrate hyphae or on aerial hyphae, consisting of solitary phialides along the hyphae or verticillate branches with two to three phialides in whorls. Phialides (12.5–)14.5–23.5(–28) × (1.5–)2–2.5(–3) μm, subcylindrical to flask-shaped with basal portion tapering into a distinct neck, 1 μm in width. Conidia unicellular, hyaline, oblong-elliptical, (3.5–)5–7.5(–9) × 1.5–2(–2.5) μm.
Distribution: Taiwan.
Notes: Cordyceps ninchukispora was initially introduced as Phytocordyceps ninchukispora in Taiwan (Su & Wang 1986) considering bola-ascospores and its host association as a pathogen of Beilschmiedia erythrophloia Hayata (Lauraceae) plant seeds. However, Sung et al. (2007a) later placed P. ninchukispora in Cordyceps based on the results of their multigene phylogeny. Sung et al. (2007) used DNA sequence data generated from the authentic material of E.G.S. 38.165 and E.G.S. 38.166. However, many recent studies have doubted the placement of this species within the genus due to the lack of DNA sequence data from type material. Thus, in the present study we generated DNA sequence data from the ex-type strain of C. ninchukispora (NTUPPMCC 20-077/BCRC 31900) obtained from BCRC where the ex-type strain of the species was originally deposited by Su & Wang (1986). Based on our phylogenetic analysis, the ex-type strain (NTUPPMCC 20-077/BCRC 31900) obtained from BCRC used by Su & Wang (1986) to illustrate C. ninchukispora formed a basal clade to the clades representing C. cocoonihabita (YFCC 3415 and 3416), putatively named C. ninchukispora strains (E.G.S 38-125 and 126) and C. pruinosa (ARSEF 5413). Furthermore, the conidia morphology as well as conidiogenesis pattern description provided by Su & Wang (1986) are identical to the morphology of NTUPPMCC 20-077/BCRC 31900 provided in the present study. Therefore, we propose the natural classification of C. ninchukispora based on DNA sequences together with morphological features and resolve the correct phylogenetic placement of the species in the genus Cordyceps.
Cordyceps militaris (L.) Fr., Observ. Mycol., Cancellans Edn (Havniae) 2: 317. 1818. Fig. 24.
Fig. 24.
Morphology of Cordyceps militaris (NTUPPMH 18-120, living culture NTUPPMCC 18-120). A. Fungus on Notodontidae pupa. B. Ascomata of specimen. C. Notodontidae pupa. D. Section of perithecium. E. Cell wall of perithecium. F, G. Asci. H. Ascus cap. I. Part-spores. J. Obverse and reverse of colonies on PDA (NTUPPMCC 18-120). K. Isaria-like conidiogenesis. L, M. Phialides on PDA. N. Conidia from PDA culture. Scale bars: D–G = 50 μm; H, I = 10 μm; K–N = 10 μm.
Basionym: Clavaria militaris L., Sp. pl. 2: 1182. 1753.
Synonyms: Cordyceps roseostromata Kobayasi & Shimizu, Bull. Nat. Sci. Mus. Ser. B 9: 10. 1983.
Cordyceps kyushuensis A. Kawam., Icones of Japanese fungi 8: 841. 1955.
Cordyceps shuifuensis H. Yu et al., Fungal Diversity 103: 17. 2020. See MycoBank (2023) for further synonyms.
Typus: ex-type culture OSC 93623
Description: See Link (1833) and Pennycook (2013). The description below is based on specimen NTUPPMH 18-120 and living culture NTUPPMCC 18-120.
Materials examined: Taiwan, Hualien County, Sioulin Township, Mt. Yangtou, on lepidopteran pupa, 25 Sep. 2017, W.-Y. Chuang, NTUPPMH 17-039, living culture NTUPPMCC 17-039; Taoyuan City, Fuxing District, on Notodontidae pupa, 5 May 2018, C.-Y. Chen & Y.-T. Wei, NTUPPMH 18-120, living culture NTUPPMCC 18-120.
Description from specimen NTUPPMH 18-120: Stromata simple, clavate, solitary, arising from Notodontidae pupa, buried in decayed wood or soil. Terminal part of the stromata fertile, orange to red. Perithecia semi-immersed, ovoid, (220–)226–254(–250) × 88–129(–140) μm. Asci cylindrical, hyaline, 8-spored, (206–) 308–452(–493) × (2.5–)3.5–5.5(–6) μm, with an oblate spheroidal or hemispherical cap, (3–)3.5–5(–6) × 2–3(–3.5) μm, at the apex. Ascospores smooth, filiform, septate, disarticulating into part-spores, cylindrical, (2.5–)3–3.5(–4.5) × 1–1.5 μm.
Culture characteristics: Colony on PDA (NTUPPMCC 18-120) reaching 2.5 cm diam in 14 d at 25 °C, white, reverse yellow to chrome yellow, dense, low convex, entire edge. Synnemata not observed. Conidiophores arising from the prostrate hyphae or on aerial hyphae, single, hyaline, navicular, (6–)7–14(–23) × (1.5–) 2–2.5(–3) μm, tapering into the neck, 1–1.5 μm in width. Conidia produced in heads or in unbranched chains, unicellular, hyaline, subglobose to globose, 3–4(–5.5) × 2.5–3 μm.
Distribution: China, Denmark, England, Germany, Luxembourg, Netherlands, Taiwan.
Notes: In the present study, all strains recognized as C. militaris were clustered with the clade comprising the ex-type strain of C. militaris (OSC 93623) plus several representative strains (NBRC 30377 and NBRC 100741), together with C. roseostromata (ARSEF 4871), the type strain of C. shuifuensis (YFCC 5230), and C. kyusyuensis (EFCC 5886). C. shuifuensis (YFCC 5230) was introduced by Wang et al. (2020) to accommodate species that were morphologically identical to C. militaris, C. kyusyuensis, and C. roseostromata within Cordyceps even after resolving poor branch lengths with relatively low statistical support in their phylogeny. In fact, these particularly short overall branch lengths with relatively low statistical support may be due to poorly resolved branches that suggest populations rather than species. Furthermore, in the present study, all strains named C. militaris did not show notable differences in their morphology even though they showed small variations in the phylogenetic trees shown in Fig. 2. Cordyceps militaris has a broad host range and has been reported from various parts of the world (Shrestha et al. 2012,Shrestha et al. 2016). Therefore, considering the phenotypic variations, phylogeny, distribution, and host range of the species, we identified our strains as C. militaris rather than introducing it as a new taxon. Furthermore, we propose to synonymize C. roseostromata, C. kyushuensis, and C. shuifuensis under C. militaris considering their morphological and phylogenetic similarity.
DISCUSSION
An increasing number of cordycipitaceous fungi have been discovered, prompting a new phylogenetic classification of Cordycipitaceae, in which multiple new genera are established in this family including Blackwellomyces, Samsoniella, Flavocillium, and Liangia (Kepler et al. 2017, Mongkolsamrit et al. 2018, Mongkolsamrit et al. 2020, Wang et al. 2020). The majority of the cordycipitaceous fungi isolated in the present study were collected from the northern and central part of Taiwan. The main reason for this finding is due to the optimal macro- and micro-climate conditions plus altitudes reaching from sea level to about 1 500 meters high that are suitable for the growth of the entomopathogenic fungi classified in Cordycipitaceae. For example, 22 samples were collected in Nantou County, and the average temperature (19.3 °C), rain fall (2 343 mm), and relative humidity (81.7 %) are suitable for the growth of the entomopathogenic fungi classified in Cordycipitaceae. In addition, Fuxing District in Taoyuan (northern Taiwan) is identified as a hotspot for Cordycipitaceae collected in this study, which also has suitable average temperature (20.3 °C), rain fall (3 113 mm), and relative humidity (82.1 %).
In the current study, we conducted a phylogenetic analysis of Cordycipitaceae family members throughout Taiwan. We discovered 10 novel species of Cordycipitaceae, including Akanthomyces taiwanicus sp. nov., Bl. taiwanensis sp. nov., C. siangyangensis sp. nov., C. pseudorosea sp. nov., C. locastrae sp. nov., C. hehuanensis sp. nov., C. malleiformis sp. nov., S. lasiocampidarum sp. nov., S. yuanzuiensis sp. nov., and Si. salviniae sp. nov., in addition to ten newly reported species across genera of Akanthomyces, Beauveria, Cordyceps, Samsoniella, and Simplicillium.
Blackwellomyces was introduced to accommodate Bl. cardinalis and is characterized by the unique characters of the ascospore, which have irregularly spaced septa and do not disarticulate into part-spores at maturity (Kepler et al. 2017, Mongkolsamrit et al. 2020). Remarkably, we noticed that Bl. taiwanensis introduced in this study produces septate ascospores that are disarticulated into part-spores while maturing, which is not observed in other Blackwellomyces species (Mongkolsamrit et al. 2020). However, Bl. taiwanensis displays a red pigment diffusion around its colony, which was also reported in Bl. cardinalis, Bl. aurantiacus, and Bl. roseostromatus (Kepler et al. 2017, Mongkolsamrit et al. 2020).
The overall topology of our Cordyceps multigene phylogenetic tree is similar to that of previous studies (Tasanathai et al. 2016, Kepler et al. 2017, Mongkolsamrit et al. 2018, Wang et al. 2020). Nevertheless, a few confusing clades were identified in the phylogenies. For instance, C. exasperata (MCA2288 & MCA2155) grouped with C. polyarthra (MCA996 & MCA1009), which was observed by Kepler et al. (2017) with absolute bootstrap support (BP = 100 %, PP = 1). However, these strains do not represent the ex-type strains of these two species and we did not find any morphological data attached to these strains. Hence, epitypification is vital for these taxonomically important species, or else the evolutionary relationships of these taxa may remain unclear or overlooked in future publications. In addition, phylogenetic analyses of concatenated sequences indicate that the fungal strains showing an evlachovaea-like or isaria-like conidiogenesis pattern do not form a monophyletic clade within the generic clade of Cordyceps. This finding is consistent with previous studies of Mongkolsamrit et al. (2018, 2020).
In a recent study, Mongkolsamrit et al. (2020) introduced C. neopruinosa from Thailand, which is morphologically similar to C. ninchukispora and C. pruinosa and formed a sister clade to the putative strains of C. ninchukispora (E.G.S 38-125 and 126). Cordyceps ninchukispora was initially reported in Taiwan and was described based on whole, bola-shaped ascospores (Su & Wang 1986). However, the phylogenetic placement of C. ninchukispora and C. pruinosa was somewhat debatable due to the lack of DNA sequence data generated from ex-type strains (Mongkolsamrit et al. 2020). In order to clarify phylogenetic placement, we generated DNA sequence data from the ex-type strain of C. ninchukispora (NTUPPMCC 20-077/BCRC 31900), obtained from the BCRC in Taiwan. In our multigene phylogeny (Fig. 2), the ex-type strain of C. ninchukispora formed a basal clade to the clades containing C. cocoonihabita (YFCC 3415 and 3416), putatively named C. ninchukispora (strains E.G.S 38-125 and E.G.S 38-126) and C. pruinosa (ARSEF 5413) confirming correct phylogenetic placement of the species. However, in the present study putatively named strains of C. ninchukispora (E.G.S 38-125 and E.G.S 38-126) did not cluster with the ex-type strain of C. ninchukispora but formed a separate phylogenetic clade basal to species clade of C. neopruinosa, and thus should be treated as a new fungal species. However, we could not obtain any morphological data linked to these strains to compare with the morphology of the ex-type strain of C. ninchukispora (NTUPPMCC 20-077/BCRC 31900) nor for the other taxa that clustered with these two strains. Therefore, we tentatively recognize these strains as Cordyceps sp. without introducing it as a novel taxon.
In this study, we expanded the general knowledge of taxa classified in Cordycipitaceae and laid preliminary groundwork for the natural classification of cordycipitaceous fungi in Taiwan. The present study provided a glimpse into the diversity of cordycipitaceous fungi in Taiwan and supplemented the molecular data of cordycipitaceous fungal species recorded in the study of Tzean et al. (1997).
ACKNOWLEDGEMENTS
We appreciate the support given by M.-J. Chen, L.-J. Chu, C.-M. Hu, C.-Y. Lee, L.-H. Liao, M.-L. Lo, G. Shang, Y.-Y. Tang, and Y.-T. Wei. H.A. Ariyawansa is grateful to A.D Ariyawansa, D.M.K Ariyawansa, R. Ariyawansa, and A. Gunasekara for their valuable suggestions. Y.C. Lin appreciates the support from the German Academic Exchange Service (DAAD) for scholarship funding (Program number: 57650361).
DECLARATION ON CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
Funding information: This study was funded by the Ministry of Science and Technology, Taiwan (MOST project ID 106-2621-B-002-005-MY2 and 112-2313-B-002-027-MY3).
Supplementary Material: https://studiesinmycology.org/
Primer sets and PCR conditions used to amplify the five gene loci (nrLSU, ITS, tef1-α, rpb1 and rpb2).
Parameters for phylogenetic analysis. Nucleotide substitution models were determined by MrModeltest for both maximum likelihood and Bayesian analyses.
REFERENCES
- Aini AN, Mongkolsamrit S, Wijanarka W. et al. (2020). Diversity of Akanthomyces on moths (Lepidoptera) in Thailand. MycoKeys 71: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araújo JP, Hughes DP. (2016). Diversity of entomopathogenic fungi: which groups conquered the insect body? Advances in Genetics 94: 1–39. [DOI] [PubMed] [Google Scholar]
- Ariyawansa HA, Hyde KD, Jayasiri SC. et al. (2015). Fungal diversity notes 111–252—taxonomic and phylogenetic contributions to fungal taxa. Fungal Diversity 75: 27–274. [Google Scholar]
- Ariyawansa HA, Phillips AJ, Chuang WY. et al. (2018). Tzeananiaceae, a new pleosporalean family associated with Ophiocordyceps macroacicularis fruiting bodies in Taiwan. MycoKeys 37: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CH, Hsieh SY, Yeh YH. et al. (2020). Cladocillium musae, a new genus and species of cercosporoid fungi (Mycosphaerellaceae) on wild banana in Taiwan. Mycological Progress 19: 837–843. [Google Scholar]
- Chen WH, Han YF, Liang JD. et al. (2016). A new araneogenous fungus of the genus Clonostachysv. Mycosystema 35: 1061–1069. [Google Scholar]
- Chen WH, Han YF, Liang JD. et al. (2021). Multi-gene phylogenetic evidence indicates that Pleurodesmospora belongs in Cordycipitaceae (Hypocreales, Hypocreomycetidae) and Pleurodesmospora lepidopterorum sp. nov. on pupa from China. MycoKeys 80: 45–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WH, Liang JD, Ren XX. et al. (2022). Species diversity of cordyceps-like fungi in the Tiankeng karst region of China. Microbiology Spectrum 10: e01975-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WH, Liu C, Han YF. et al. (2019). Three novel insect-associated species of Simplicillium (Cordycipitaceae, Hypocreales) from Southwest China. MycoKeys 58: 83–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen RS, Huang CC, Li JC. et al. (2008). First report of Simplicillium lanosoniveum causing brown spot on Salvinia auriculata and S. molesta in Taiwan. Plant Disease 92: 1589–1589. [DOI] [PubMed] [Google Scholar]
- Chen ZC. (1978). Note on new formosan forest fungi VI. Genus Cordyceps and their distribution in Taiwan. Taiwania 23:153–162. [Google Scholar]
- Chernomor O, Von Haeseler A, Minh BQ. (2016). Terrace aware data structure for phylogenomic inference from supermatrices. Systematic Biology 65: 997–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous P, Wingfield M, Lombard L. et al. (2019). Fungal Planet description sheets: 951–1041. Persoonia 43: 223–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong QY, Wang Y, Wang ZQ. et al. (2022). Morphology and phylogeny reveal five novel species in the genus Cordyceps (Cordycipitaceae, Hypocreales) from Yunnan, China. Frontiers in Microbiology 13: 846909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson O. (1982). Cordyceps bifusispora spec. nov. Mycotaxon 15:185–188. [Google Scholar]
- Fries EM. (1818). Observationes mycologicae praecipue ad illustrandam Floram Suecicam. Pars secunda (Cancellans issue). Copenhagen: G. Bonnieri. [Google Scholar]
- Gomes AA, Pinho DB, Cardeal ZL. et al. (2018). Simplicillium coffeanum, a new endophytic species from Brazilian coffee plants, emitting antimicrobial volatiles. Phytotaxa 333: 188–198. [Google Scholar]
- Hywel-Jones NL. (1996). The genus Akanthomyces on spiders in Thailand. Mycological Research 100: 1065–1070. [Google Scholar]
- Imoulan A, Wu HJ, Lu WL. et al. (2016). Beauveria medogensis sp. nov., a new fungus of the entomopathogenic genus from China. Journal of Invertebrate Pathology 139: 74–81. [DOI] [PubMed] [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TKF. et al. (2017). Modelfinder: fast model selection for accurate phylogenetic estimates. Nature Methods 14: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke Y-H, Ju Y-M. (2015). Two rare ophiocordycipitaceous fungi newly recorded in Taiwan. Botanical Studies 56: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepler RM, Luangsa-Ard JJ, Hywel-Jones NL. et al. (2017). A phylogenetically-based nomenclature for Cordycipitaceae (Hypocreales). IMA Fungus 8: 335–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayasi Y. (1941). The genus Cordyceps and its allies. Science Reports of the Tokyo Bunrika Daigaku 5: 53–260. [Google Scholar]
- Kobayasi Y. (1982). Keys to the taxa of the genera Cordyceps and Torrubiella. Transactions of the Mycological Society of Japan 23: 329–364. [Google Scholar]
- Kobayasi Y, Shimizu D. (1982). Cordyceps Species from Japan 5. Bulletin of the National Science Museum Series B 8: 112–113. [Google Scholar]
- Kobayasi Y, Shimizu D. (1983). Cordyceps species from Japan. 6. Bulletin of the National Science Museum Tokyo 9:1–21. [Google Scholar]
- Kondo N, Iwasaki H, Tokiwa T. et al. (2020). Simplicillium spumae (Cordycipitaceae, Hypocreales), a new hyphomycetes from aquarium foam in Japan. Mycoscience 61: 116–121. [Google Scholar]
- Liu F, Cai L. (2012). Morphological and molecular characterization of a novel species of Simplicillium from China. Cryptogamie, Mycologie 33: 137–144. [Google Scholar]
- Liang Z. (1981). Two new species of Paecilomyces from insects. Acta Microbiologica Sinica 21: 31–34. [Google Scholar]
- Liu Z, Liang Z, Liu A. (1996). Identification of anamorph of Cordyceps bifusispora. Acta Mycologica Sinica 15: 210–214. [Google Scholar]
- Luangsa-Ard JJ, Hywel-Jones NL, Manoch L. et al. (2005). On the relationships of Paecilomyces sect. Isarioidea species. Mycological Research 109: 581–589. [DOI] [PubMed] [Google Scholar]
- Luangsa-ard JJ, Hywel-Jones NL, Samson RA. (2004). The polyphyletic nature of Paecilomyces sensu lato based on 18S-generated rDNA phylogeny. Mycologia 96: 773–780. [DOI] [PubMed] [Google Scholar]
- McNeill J, Barrie FR, Buck WR. et al. (2012). International Code of Nomenclature for algae, fungi, and plants (Melbourne Code). Koeltz Scientific Books, Königstein. [Google Scholar]
- Mongkolsamrit S, Noisripoom W, Thanakitpipattana D. et al. (2018). Disentangling cryptic species with isaria-like morphs in Cordycipitaceae. Mycologia 110: 230–257. [DOI] [PubMed] [Google Scholar]
- Mongkolsamrit S, Noisripoom W, Tasanathai K. et al. (2020). Molecular phylogeny and morphology reveal cryptic species in Blackwellomyces and Cordyceps (Cordycipitaceae) from Thailand. Mycological Progress 19: 957–983. [Google Scholar]
- Mongkolsamrit S, Noisripoom W, Tasanathai K. et al. (2022). Comprehensive treatise of Hevansia and three new genera Jenniferia, Parahevansia and Polystromomyces on spiders in Cordycipitaceae from Thailand. MycoKeys 91: 113–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongkolsamrit S, Sandargo B, Ebada SS. et al. (2023). Bhushaniella gen. nov. (Cordycipitaceae) on spider eggs sac: a new genus from Thailand and its bioactive secondary metabolites. Mycological Progress 22: 64. [Google Scholar]
- Nguyen LT, Schmidt HA, von Haeseler A. et al. (2015) IQ-TREE: A fast and effective stochastic algorithm for estimating maximum likelihood phylogenies. Molecular Biology and Evolution 32: 268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonaka K, Kaifuchi S, Ōmura S. et al. (2013). Five new Simplicillium species (Cordycipitaceae) from soils in Tokyo, Japan. Mycoscience 54: 42–53. [Google Scholar]
- Nylander JAA. (2004). MrModeltest 2: program distributed by the author. Evolutionary Biology Centre, Uppsala University. [Google Scholar]
- Rambaut A, Drummond AJ. (2009) Tracer v1.5.0. http://beast.bio.ed.ac.uk.
- Rambaut A. (2012) FigTree v1.4. http://tree.bio.ed.ac.uk/software/figtree/. [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P. et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systems Biology 61: 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada K. (1922). Descriptive catalogue of the Formosan Fungi. 1st edn.Department of Agriculture Government Research Institute Formosa. [Google Scholar]
- Samson RA. (1974). Paecilomyces and some allied hyphomycetes. Studies in Mycology 6: 1–119. [Google Scholar]
- Shrestha B, Sung GH, Sung JM. (2017). Current nomenclatural changes in Cordyceps sensu lato and its multidisciplinary impacts. Mycology 8: 293–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha B, Tanaka E, Hyun MW. et al. (2016). Coleopteran and lepidopteran hosts of the entomopathogenic genus Cordyceps sensu lato. Journal of Mycology 2016: 7648219. [Google Scholar]
- Shrestha B, Zhang W, Zhang Y. et al. (2012). The medicinal fungus Cordyceps militaris: research and development. Mycological Progress 11: 599–614. [Google Scholar]
- Su CH, Wang HH. (1986). Phytocordyceps, a new genus of the Clavicipitaceae. Mycotaxon 26: 337–344. [Google Scholar]
- Sung GH, Hywel-Jones NL, Sung JM. et al. (2007a). Phylogenetic classification of Cordyceps and the clavicipitaceous fungi. Studies in Mycology 57: 5–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung GH, Sung JM, Hywel-Jones NL. et al. (2007b). A multi-gene phylogeny of Clavicipitaceae (Ascomycota, Fungi): Identification of localized incongruence using a combinational bootstrap approach. Molecular Phylogenetics and Evolution 44: 1204–1223. [DOI] [PubMed] [Google Scholar]
- Tasanathai K, Thanakitpipattana D, Noisripoom W. et al. (2016). Two new Cordyceps species from a community forest in Thailand. Mycological Progress 15: 28. [Google Scholar]
- Thanakitpipattana D, Mongkolsamrit S, Khonsanit A. et al. (2022). Is Hyperdermium congeneric with Ascopolyporus? Phylogenetic relationships of Ascopolyporus spp. (Cordycipitaceae, Hypocreales) and a new genus Neohyperdermium on scale insects in Thailand. Journal of Fungi 8: 516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai I, Maharachchikumbura SS, Hyde KD. et al. (2018). Molecular phylogeny, morphology and pathogenicity of Pseudopestalotiopsis species on Ixora in Taiwan. Mycological Progress 17: 941–952. [Google Scholar]
- Tzean SS, Hsieh LS, Wu WJ. (1997). Atlas of entomopathogenic fungi from Taiwan. 1st edn. Council of Agriculture, Executive Yuan, Taiwan. [Google Scholar]
- Vega FE, Goettel MS, Blackwell M. et al. (2009). Fungal entomopathogens: new insights on their ecology. Fungal Ecology 2: 149–159. [Google Scholar]
- Vega FE, Meyling NV, Luangsa-ard JJ. et al. (2012). Fungal entomopathogens. Insect Pathology 2: 171–220. [Google Scholar]
- Vinit K, Doilom M, Wanasinghe DN. et al. (2018). Phylogenetic placement of Akanthomyces muscarius, a new endophyte record from Nypa fruticans in Thailand. Current Research in Environment & Applied Mycology 8: 404–417. [Google Scholar]
- Wang YB, Wang Y, Fan Q. et al. (2020). Multigene phylogeny of the family Cordycipitaceae (Hypocreales): new taxa and the new systematic position of the Chinese cordycipitoid fungus Paecilomyces hepiali. Fungal Diversity 103: 1–46. [Google Scholar]
- Wei DP, Wanasinghe DN, Hyde KD. et al. (2019). The genus Simplicillium. MycoKeys 60: 69–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijayawardene NN, Hyde KD, Al-Ani LKT. et al. (2020). Outline of Fungi and fungus-like taxa. Mycosphere Online: Journal of Fungal Biology 11: 1060–1456. [Google Scholar]
- Xiao YP, Wang YB, Hyde KD. et al. (2023). Polycephalomycetaceae, a new family of clavicipitoid fungi segregates from Ophiocordycipitaceae. Fungal Diversity 120:1–76. [Google Scholar]
- Yang JI, Stadler M, Chuang WY. et al. (2020). In vitro inferred interactions of selected entomopathogenic fungi from Taiwan and eggs of Meloidogyne graminicola. Mycological Progress 19: 97–109. [Google Scholar]
- Zare R, Gams W. (2001). A revision of Verticillium section Prostrata. IV. The genera Lecanicillium and Simplicillium gen. nov. Nova Hedwigia 73: 1–50. [Google Scholar]
- Zhang ZF, Zhou SY. et al. (2021). Culturable mycobiota from Karst caves in China II, with descriptions of 33 new species. Fungal Diversity 106: 29–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang WY, Zeng ZQ. (2017). Cocoonihabitus sinensis gen. et sp. nov. on remaining leaf veins of cocoons in a new family (Cocoonihabitaceae fam. nov.) of Hypocreales. Mycosystema 36: 1591–1598. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primer sets and PCR conditions used to amplify the five gene loci (nrLSU, ITS, tef1-α, rpb1 and rpb2).
Parameters for phylogenetic analysis. Nucleotide substitution models were determined by MrModeltest for both maximum likelihood and Bayesian analyses.