Abstract
Although several reports have characterized the hematopoietic stem cell (HSC) transcriptome, the roles of HSC-specific genes in hematopoiesis remain elusive. To identify candidate regulators of HSC fate decisions, we compared the transcriptome of human umbilical cord blood and bone marrow CD34+CD33−CD38−Rholoc-kit+ cells, enriched for hematopoietic stem/progenitor cells with CD34+CD33−CD38−Rhohi cells, enriched in committed progenitors. We identified 277 differentially expressed transcripts conserved in these ontogenically distinct cell sources. We next performed a morpholino antisense oligonucleotide (MO)-based functional screen in zebrafish to determine the hematopoietic function of 61 genes that had no previously known function in HSC biology and for which a likely zebrafish ortholog could be identified. MO knock down of 14/61 (23%) of the differentially expressed transcripts resulted in hematopoietic defects in developing zebrafish embryos, as demonstrated by altered levels of circulating blood cells at 30 and 48 h postfertilization and subsequently confirmed by quantitative RT-PCR for erythroid-specific hbae1 and myeloid-specific lcp1 transcripts. Recapitulating the knockdown phenotype using a second MO of independent sequence, absence of the phenotype using a mismatched MO sequence, and rescue of the phenotype by cDNA-based overexpression of the targeted transcript for zebrafish spry4 confirmed the specificity of MO targeting in this system. Further characterization of the spry4-deficient zebrafish embryos demonstrated that hematopoietic defects were not due to more widespread defects in the mesodermal development, and therefore represented primary defects in HSC specification, proliferation, and/or differentiation. Overall, this high-throughput screen for the functional validation of differentially expressed genes using a zebrafish model of hematopoiesis represents a major step toward obtaining meaningful information from global gene profiling of HSCs.
Microarray studies are combined with a functional screen in zebrafish to identify new genes that are involved in the function and differentiation of hematopoietic stem cells.
Introduction
Hematopoiesis is the process by which hematopoietic stem cells (HSCs) give rise to all hematopoietic lineages during the lifetime of an individual. To sustain lifelong hematopoiesis, HSCs must self-renew to maintain or expand the HSC pool [1], and they must differentiate to form committed hematopoietic progenitor cells (HPCs) that progressively lose self-renewal potential and become increasingly restricted in their lineage potential. A combination of extrinsic and intrinsic signals are thought to converge to regulate HSC differentiation versus self-renewal decisions, but the molecular mechanisms that regulate these processes are poorly understood [2].
A multitude of cytokines have been cloned that affect HSCs and HPCs; however, to date none of these, alone or in combination, can induce the symmetrical, self-renewing HSC cell division in vitro that is required for HSC expansion. Recently, several novel regulators of HSC fate decisions have been identified. For instance, overexpression of Hoxb4 results in expansion of murine and human HSCs with an increased competitive repopulation potential [3–5]; novel extrinsic regulators implicated in self-renewal of HSCs include Notch [6], Wnt [7,8], and the morphogens, SHH (sonic hedgehog) [9] and BMP4 (bone morphogenetic protein 4) [9]. While the discovery of these novel regulators provides credence to the hypothesis that extrinsic and intrinsic signals can influence HSC fate, a more global gene and/or protein expression analysis of human HSC should provide additional insight into pathways that support HSC self-renewal.
Our current understanding of the expressed gene profile of HSCs comes primarily from murine HSCs that can be purified to near homogeneity [10–14]. The inability to purify human HSCs to similar degrees of homogeneity makes study of the transcriptome of human HSCs more difficult. Human HSCs and HPCs are CD34 positive, while cells that engraft in severe combined immunodeficiency (SCID) mice are enriched in the CD34+Lineage(Lin)−CD38− fraction [15]. As fewer than 1/500 CD34+Lin−CD38− cells can repopulate SCID mice [15], the expressed gene profile of CD34+Lin−CD38− cells is likely only partially enriched for HSC-specific genes [12,16]. We previously demonstrated that the rhodamine (Rho) 123− and c-kit+ subpopulation of CD34+Lin−CD38− cells (Rholo) cells are highly enriched for primitive HPCs with myeloid–lymphoid initiating cell (ML–IC) capacity relative to CD34+CD38−CD33−Rhohi (Rhohi) cells [17]. Thus, such selection separates CD34+Lin−CD38− cells into HSC-enriched and HSC-depleted populations.
We hypothesized that comparison of the transcriptome of Rholo and Rhohi cells from umbilical cord blood (UCB) and bone marrow (BM) should identify conserved genes and gene pathways that define the human HSC. Because of the inherent limitations of using gene expression data to infer biological gene function, we also assessed the hematopoietic role of these genes in a high-throughput in vivo functional genomics screen in the zebrafish. Using this strategy we have not only identified a series of genes that may represent novel regulators of human HSC fate decisions, but this work also represents, to our knowledge, the first example of a functional genetic screening strategy that is a critical step toward obtaining biologically relevant functional data from global gene-profiling studies.
Results/Discussion
ML–ICs Are Highly Enriched in Rholo Compared to Rhohi Cells
The study of human HSCs has been limited since the CD34+Lin−CD38− fraction of hematopoietic cells, commonly used as an HSC-enriched population, contains fewer than 0.2% SCID-repopulating cells [15], suggesting considerable heterogeneity. We have shown that ML–IC s, single hematopoietic cells that can generate several daughter cells capable of reinitiating long-term myeloid and long-term lymphoid cultures, are highly enriched by selecting the Rholo fraction of CD34+Lin−CD38− cells. While the Rholo population still only contains 15%–25% ML–ICs and therefore remains heterogeneous, the enrichment factor is 5- to 10-fold greater than CD34+Lin−CD38− cells [17]. Similar to our previous studies, the ML–IC frequency was greater than or equal to 10-fold higher in UCB Rholo compared to Rhohi cells (Figure S1).
Genes Differentially Expressed between Rholo and Rhohi Cells from Both UCB and BM
We hypothesized that comparing genes differentially expressed between Rholo and Rhohi cells from ontogenically distinct sources would identify conserved genes and gene pathways that govern self-renewal and differentiation of human HSCs. The experimental design used is illustrated in Figure 1. We defined differentially expressed probe sets as those with p < 0.05, using a paired Student's t-test. By taking into account the variability present in primary cell populations, this provides a more accurate analysis of differential gene expression compared with the commonly used fold change cutoff.
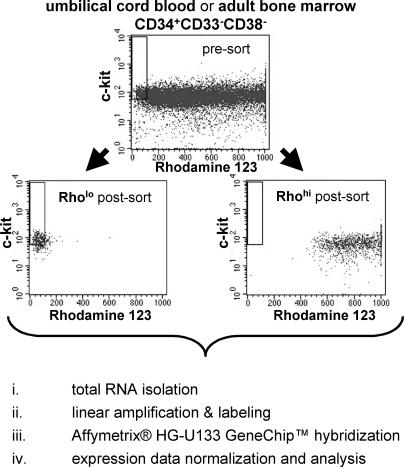
Figure 1. Fluorescence-Activated Cell Sorting and Gene Expression Analysis.
UCB or adult BM CD34+CD33−CD38−Rholoc-kit+ (Rholo; stem cell enriched) and CD34+CD33−CD38−Rhohi (Rhohi; stem cell depleted) cell populations were sorted for subsequent global gene expression profiling. Total RNA was isolated from Rholo and Rhohi cell populations prior to linear amplification and labeling for hybridization to the Affymetrix HG-U133 GeneChip set (approximately 45,000 probe sets) and subsequent data analysis.
We identified 2,707 and 4,667 probe sets differentially expressed between Rholo and Rhohi cells from UCB and BM, respectively (see Tables S1 and S2). The fidelity of our microarray results was confirmed using quantitative RT-PCR (Q-RT-PCR) (see Figure S2). We focused our further analysis on 277 unique transcripts, represented by 304 probe sets that were differentially expressed between Rholo and Rhohi cells from both UCB and BM with a fold change greater than 1.5 in either UCB or BM (Table S3).
Among the conserved genes enriched in Rholo cells, many have been implicated in early hematopoiesis, including CDKN1A, a cell cycle regulator required for maintenance of murine HSCs [18], and ABCB1, the ABC-transporter family member responsible for the Rholo phenotype [17]. Several transcription factors (TFs) known to play a role in early hematopoiesis or leukemogenesis were also identified, including HLF, involved in leukemogenic chromosomal translocations [19] and EVI1, a TF associated with myeloid leukemias [20]. Other TFs without a known role in hematopoiesis were also more highly expressed in Rholo cells, including HMGA2, a high-mobility group gene, and the zinc finger TFs ZNF165, ZNF331, and KLF5. All Rholo-enriched genes are listed in Table S3. As demonstrated in previous HSC gene-profiling studies [12–14], more than 40% of genes enriched in Rholo cells lack a functional annotation, are hypothetical proteins, or are expressed sequence tags, and thus may represent currently uncharacterized regulators of HSC fate decisions (Figure S3).
Some genes with well-established roles in HSC self-renewal and early differentiation are not present in the Rholo enriched gene list. However, most of these were differentially expressed in both datasets, but differences did not reach statistical significance. For instance, LMO2 [21] and GATA 2 [22], known to be involved in HSC development and self-renewal, were expressed significantly higher in BM Rholo than in Rhohi cells. Although similar trends were seen in UCB Rholo cells, these differences were not statistically significant (Table S4). Conversely, HOXB4 [4] expression was significantly higher in UCB Rholo than in Rhohi cells, but this difference was not statistically significant in BM. Although our stringent criteria for differential expression likely contribute to the omission of some genes that might be differentially expressed, another explanation might be that expression of these genes is maintained when cells differentiate from a Rholo to a Rhohi stage. The latter is consistent with the fact that most known HSC-associated genes were expressed at much higher levels than the normalized average microarray expression level in Rholo and in Rhohi cells from both UCB and BM (Table S4).
Conserved genes enriched in the Rhohi cells included LEF1, an effector of Wnt signaling expressed in pre-B and T cells [23], and NOTCH2, involved in hematopoietic differentiation cell-fate decision [24]. Several TFs known to play a role in hematopoietic cell differentiation were more highly expressed in Rhohi compared to Rholo cells, including HELLS and MAFB [25,26]. Additional TFs with no known role in hematopoietic development were also enriched in Rhohi cells, such as the zinc finger homeobox gene, ZFHX1B and the polycomb genes, EZH2 and SUZ12, the latter required for germ cell development [27]. Globin (Hb) gene family members were also more highly expressed in UCB and BM Rhohi than in Rholo cells. Consistent with the ontogenic expression patterns of fetal versus adult Hb genes, Hbγ genes were more highly expressed in perinatal UCB Rhohi cells, while Hbβ genes were more highly expressed in adult BM Rhohi cells. Additional genes enriched in Rhohi cells are listed in Table S3.
Because of functional redundancy among gene families, we examined the data for common differentially expressed gene family members. The Id family of transcriptional repressors [28] was enriched in the Rholo fraction, but was represented by different family members in UCB (ID4) and BM (ID1, ID2, and ID3). Similarly, various H1 and H2 histone genes were enriched in the Rholo fraction in both datasets, but were represented by distinct family members.
We also evaluated whether common differentially expressed genes were concentrated on specific chromosomes. We found that genes were not only concentrated on certain chromosomes, but at specific g-band addresses. Of the genes enriched in Rholo cells, 9% reside at 6p21, a region involved in recurrent chromosomal translocations in myeloid [29] and lymphoid [30] leukemias, and home to the PIM1 oncogene [31]. Six members of the H2B and one member of the H1 histone family, as well as CDKN1A, more highly expressed in Rholo than Rhohi cells, reside at 6p21. The remaining Rholo-enriched genes at 6p21 consist of six class II major histocompatibility complex (MHC) family members and a putative testis-specific zinc finger TF, ZNF165. H1 and H2 histone gene family members [11,13], class II MHC antigens [12,13], and CDKN1A [13] were also found among the genes identified in studies characterizing the transcriptome of murine HSC. The differential expression of such a large number of genes located at this chromosomal address suggests that, like CDKN1A, other genes located at 6p21 with as yet unknown hematopoietic function may play a role in HSC proliferation or differentiation.
We also compared the genes expressed more highly in Rholo versus Rhohi cells with published gene expression data. Comparison with the study by Ivanova et al. [12] that compared human CD34+Lin−CD38− with CD34+Lin−CD38+ cells, yielded only seven genes in common: ARMCX2, CRYGD, HLF, KIAA1102, RBPMS, SLCO3A1, and SSBP2. The lack of overlap may not be that surprising as Rholo and Rhohi cells are subpopulations of the CD34+Lin−CD38− population used by Ivanova et al. Comparison of genes expressed more highly in Rholo versus Rhohi cells with genes expressed more highly in murine side population/KLS/CD34− compared to total BM cells published by Ramalho-Santos et al. [13] identified 16 likely orthologs and 38 common gene family members (Table S5), suggesting that HSC-specific genes are conserved across species.
In vivo functional genomics screen in zebrafish
Because gene profiling per se does not prove functional importance, we developed an in vivo functional genomics screen in zebrafish (Figure 2). The zebrafish, Danio rerio, is an ideal organism for high-throughput genetic screens [32] as organogenesis is highly conserved from zebrafish to man [33]. There is abundant evidence that hematopoiesis in zebrafish occurs via a highly conserved genetic program. As in mammals, hematopoiesis in zebrafish occurs via specification of mesoderm to a hemangioblast stage that subsequently commits to either HSC or angioblasts [34], and genes and signals involved in specification (BMP signaling) and commitment (vascular endothelial growth factor signaling; flk1, also known as kdr; lmo2; scl, also known as tal1; gata2; gata1) are conserved from fish to man [35]. This high degree of homology in the genetic control of zebrafish and human hematopoietic development makes genetic screens in zebrafish a powerful tool to elucidate the role of genes in hematopoiesis. Additionally, rapid reverse genetic screens can be accomplished using morpholino antisense oligonucleotides (MOs) to knock down gene expression in the developing zebrafish embryo [36].
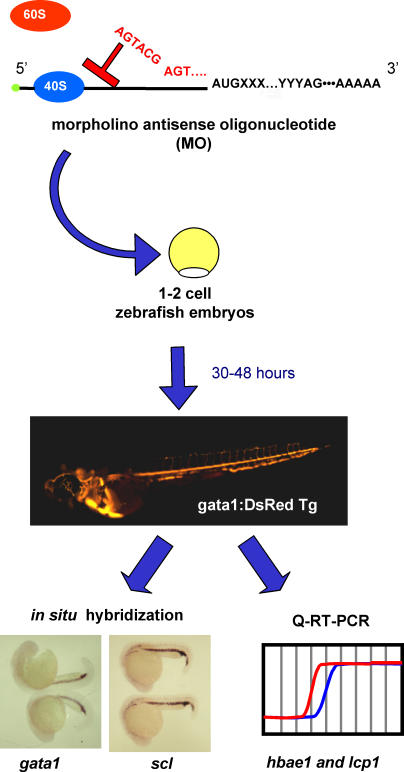
Figure 2. Functional Genomics Screen in Zebrafish.
The hematopoietic function of differentially expressed candidate genes was determined by injecting MOs into one- to two-cell embryos from gata1:DsRed Tg zebrafish to disrupt gene expression. Injected embryos were scored at 30–48 hpf for the presence of DsRed+ blood cells by fluorescence microscopy. Subsequently, MO-targeted embryos with gross hematopoietic defects were analyzed for the expression of the early hematopoietic markers gata1 and scl by whole-mount in situ hybridization, and the late hematopoietic markers hbae1 and lcp1 by Q-RT-PCR.
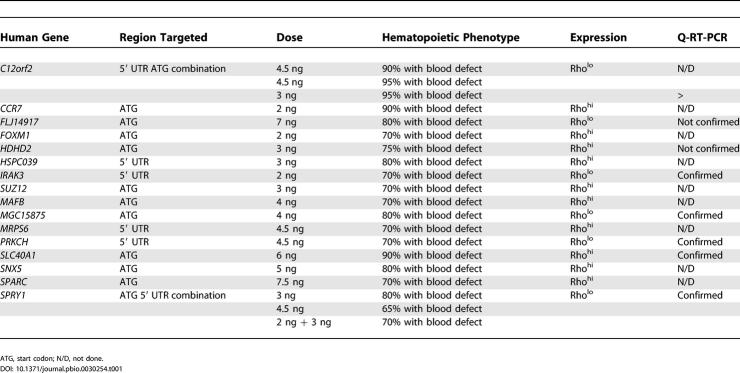
From the 277 unique transcripts that were differentially expressed between Rholo and Rhohi cells of both UCB and BM (Table S3), we eliminated genes with known function in hematopoiesis, MHC genes, histones, and genes that are known to play a role in glucose and protein metabolism and RNA and DNA synthesis, resulting in a final list of 158 genes. Of these, we identified a putative zebrafish ortholog for 86, and designed MOs against 61 (Table S6). The 61 MOs were injected in one- to two-cell zebrafish embryos and assayed for effects on blood development. Initial dosing studies identified 16/61 MOs that reduce blood cell production without confounding toxicities (Table 1). The 16 MOs induced a blood defect in more than 70% of embryos in two or more independent injections. Blood defects identified by gata1:DsRed transgenic (Tg) fluorescence microscopy were confirmed by Q-RT-PCR of the erythroid-specific hbae1 and myeloid-specific lcp1 transcripts in MO-targeted embryos compared to uninjected controls in three or more independent experiments of n = 5 embryos per experiment. A greater than 2-fold reduction in both erythroid and myeloid gene expression levels were seen for five of seven MOs analyzed (see Table 1; Figures 3 and 4). Thus, the addition of Q-RT-PCR to the screening process provides an independent confirmation and quantitation of the observed phenotypes, thereby limiting the false-positive rate, while maintaining the high-throughput nature of the screen. Additionally, the observed reduction of both erythroid and myeloid gene expression following knockdown of candidate genes is consistent with their presumed roles in HSC fate decisions prior to specification of the common myeloid progenitor. The validity of the Q-RT-PCR analysis was corroborated by analysis of hbae1 and lcp1 transcript levels in gata1 MO-targeted embryos, in which there was a virtually complete loss of hbae1 expression and an almost 2-fold increase in myeloid-specific lcp1 transcripts (Figure 4) consistent with the published expression patterns for these genes following loss of gata1 expression [37]. Analysis of vascular development by injecting MOs into fli1:enhanced green fluorescent protein (EGFP) Tg embryos, which express EGFP in endothelial cells, demonstrated no major abnormalities in vascular morphogenesis or remodeling that precludes the circulation of the remaining blood cells, indicating that the hematopoietic defect is not secondary to a vascular phenotype (Figure 4; Table S7).
Table 1. Genes Differentially Expressed between Human Rholo and Rhohi Cell Populations That Have a Functional Role in Zebrafish Hematopoietic Development.
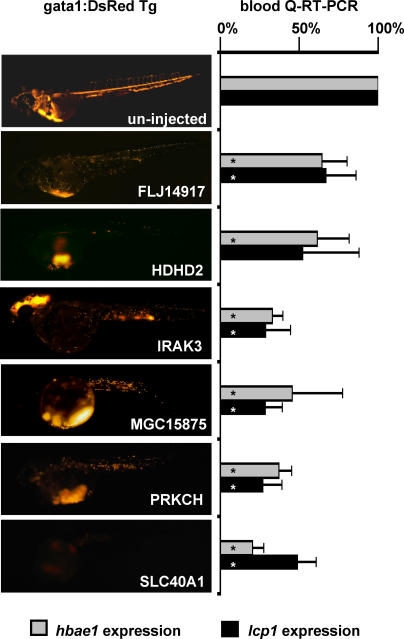
Figure 3. Representative Hematopoietic Phenotypes Observed in MO-Targeted Zebrafish Fluorescence microscopic images of gata1:DsRed Tg zebrafish embryos display the hematopoietic phenotypes observed for six MO-targeted zebrafish embryos compared to an uninjected control.
Phenotypes shown are representative of greater than 70% of injected embryos at 48 hpf (greater than or equal to three experiments of n > 40 embryos). Hematopoietic defects were quantified by Q-RT-PCR for the expression of erythroid-specific hbae1 and myeloid-specific lcp1 transcripts in MO-targeted embryos relative to uninjected clutchmate controls (greater than or equal to three experiments of n = 5 embryos; * p < 0.05)
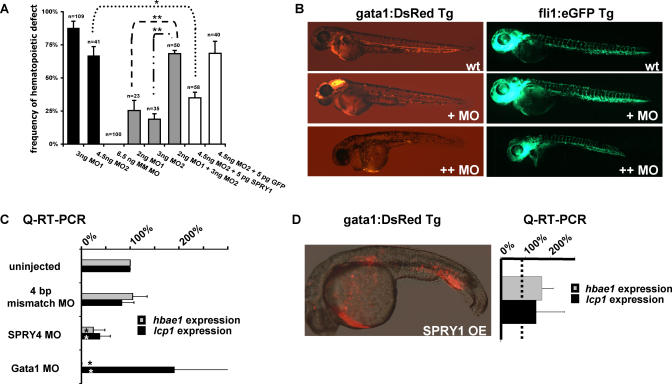
Figure 4. spry4 Is Required for Normal Hematopoietic Development in Zebrafish Embryos.
(A) The observed frequency of hematopoietic defects in gata1:DsRed Tg zebrafish embryos are indicated for two independent spry4-targeted MOs (MO1 and MO2; black bars), a four-base mismatched control MO (MM MO; no bar indicates a 0% frequency), a low-dose injection of MO1 and MO2 individually and in combination (gray bars), and MO2 coinjected with a human SPRY1 DNA expression vector or a GFP control (white bars) (error bars = standard deviation of the mean, *p < 0.05, **p ≤ 0.01).
(B) Representative pictures of the phenotypes seen with spry4 MO1 at 48 h using fli1:EGFP/gata1:DsRed double Tg zebrafish embryos that have EGFP+ vascular endothelial cells and DsRed+ erythroid cells. The embryos display a more drastic reduction in DsRed+ blood cells with some blood pooling and pericardial edema at higher MO doses (++MO) without major vasculature defects. The gata1:DsRed and fli1:EGFP Tg images are representative of three experiments of n > 40 embryos each.
(C) Q-RT-PCR analysis of hbae1 and lcp1 transcripts in spry4 MO1, control mismatch MO and a gata1 MO-injected zebrafish at 48 hpf (*p < 0.05).
(D) Injection of 30 pg of human SPRY1 DNA expression vector resulted in an expansion of DsRed+ hematopoietic cells in the posterior ICM in greater than 50% of successfully injected embryos at 32 hpf (three experiments of n = 30), and a representative bright-field image is pictured with the fluorescence micrograph overlaid (left). Q-RT-PCR analysis of hbae1 and lcp1 transcripts in SPRY1 overexpressing embryos relative to uninjected clutchmate controls (three experiments of n = 5 embryos) (right).
The greater than 20% frequency of blood defects seen in our screen compares very favorably with the 0.5%–1% frequency of hematopoietic phenotypes seen by ethylnitrosourea mutagenesis screens that mutate genes in a near random fashion [38] and the 4% of hematopoietic phenotypes seen in a morpholino-based functional screen of the zebrafish secretome [39] (S. C. Ekker, unpublished data). The high incidence of blood defects also demonstrates that the candidate genes identified by comparing the transcriptome of Rholo and Rhohi cells represent genes with important roles in HSC biology. We were unable to identify a candidate ortholog in zebrafish for 72/158 of the differentially expressed human genes. This may be because currently only one quarter of the zebrafish genome is high-quality finished sequence. Hence, some genes with important roles in hematopoiesis may have been untested. However, from a sample of ten genes that lacked a zebrafish match, only one has a likely ortholog in the Fugu or Medaka sequencing projects, and thus incomplete genome coverage provides only a partial explanation. Alternative possibilities include a reduced level of primary sequence conservation between functional orthologs that would have been missed using our simple comparative genomics criteria, or that a number of genes are not conserved between fish and man, and thus might be less important for the conserved processes of hematopoietic self-renewal and differentiation. Therefore, the relatively high incidence of blood defects in conserved genes may in part reflect “evolutionary filtering” in our screen. Consistent with this hypothesis, all zebrafish genes whose mutation resulted in a visible embryonic phenotype identified using a retroviral insertion strategy have a likely human ortholog [40].
Although the frequency of blood defects is high, the screen is not as well suited for the identification of knockdown phenotypes that result in increased HSC proliferation and/or differentiation. We and others have successfully shown that dramatically increased hematopoietic development can be modeled in zebrafish, as is the case for knockdown of the BMP-antagonist chd (chordin) and the corresponding dino mutant [41,42]; however, more modest increases in hematopoietic cell production or skewing of lineage differentiation may be undetectable. Although these caveats may lead to underestimation of the true frequency of genes with a role in early hematopoietic development and differentiation, the screening procedure used has proven effective for extracting functional information from a global gene expression profiling dataset in a high-throughput manner.
Of note, viable knockout mice exist for 4/14 genes identified in our functional screen (Sparc, Irak3, Ccr7, and Prkch) [43–46]. One could argue that if a viable knockout mouse exists, the gene of interest may not be important in hematopoiesis. However, lack of an overt hematopoietic phenotype does not preclude a role of a gene in HSC self-renewal and differentiation, as this may only be detectable under conditions where the hematopoietic system is stressed or in transplantation experiments. For example, Hoxb4 −/− mice develop normally and present with only subtle differences in spleen and BM cellularity [47]. However, the proliferative response of HSC in vitro and in vivo is decreased, consistent with the observation that overexpression of Hoxb4 supports expansion of competitive repopulating units and SCID-repopulating cells [4,5].
Characterization of Sprouty Family Members in Zebrafish Hematopoiesis
To further verify the hematopoietic role of genes identified by gene array, we performed a more extensive evaluation of the zebrafish targeted with an MO against spry4 (spry4 morphant or spry4MO). Although human SPRY1 was differentially expressed, an MO against zebrafish spry4 was used, as it is expressed in the region of the lateral plate mesoderm, the first site of zebrafish hematopoiesis [48], and it was the full-length zebrafish Sprouty gene with the greatest protein homology to human SPRY1. Recently the partial sequence of a potential zebrafish spry1 ortholog was predicted by Ensembl's gene prediction software based on genomic sequence information. However, the single exon that was predicted does not contain an ATG start codon, or a conserved splice donor or acceptor site, and the putative zebrafish spry1 sequence only partially covers the human SPRY1 gene. Moreover, the genomic location of Ensembl's putative zebrafish spry1 is currently not known, and therefore it is not possible to use syntenic relationships to determine the most likely zebrafish ortholog for human SPRY1. At present, there is not sufficient sequence data available to design gain- or loss-of-function experiments for the putative zebrafish spry1, thus precluding an analysis of hematopoietic function in the zebrafish model. Therefore, spry4 is currently the best full-length, MO-targetable candidate ortholog for human SPRY1, and based on our results, at the very least zebrafish spry4 and SPRY1 share a conserved function in hematopoiesis.
To confirm the specificity of MO targeting in the spry4MO, a second spry4 MO of independent sequence and a four-base mismatched spry4 MO were injected into zebrafish embryos. Injection of the independent spry4 MOs induced a hematopoietic phenotype in more than 65% of injected embryos, while the four-base mismatched MO did not induce any phenotypic changes (Figure 4). Q-RT-PCR for hbae1 and lcp1 transcripts also did not show changes in expression in four-base mismatched MO embryos. Furthermore, the two independent spry4 MOs acted synergistically when coinjected (Figure 4). In addition to the blood phenotype, a slight facial outgrowth was seen at a low frequency. Also, a weak dorsalization phenotype was seen at 2-fold higher MO doses based on a decreased somite size.
To rule out the possibility that the hematopoietic defect observed in the spry4MO was secondary to a vascular defect, we injected spry4 MO into fli1:EGFP Tg zebrafish. While the resulting embryos exhibited minor defects in cardinal vein remodeling and morphogenesis of intersegmental vessels in the posterior tail, there were no major defects in vascular development (Figure 4). The integrity of the vascular network in spry4MO fish was further demonstrated by the unimpeded circulation of the remaining DsRed+ blood cells in gata1:DsRed Tg spry4MO (data not shown).
Thisse et al. [35] have shown that overexpression of zebrafish spry4 mRNA leads to an expansion of the posterior intermediate cell mass (ICM) [48]. We overexpressed human SPRY1 in gata1:DsRed Tg zebrafish embryos, and observed a similar dose-dependent expansion of DsRed+ blood cells in the posterior ICM (Figure 4). Q-RT-PCR analysis of embryos overexpressing SPRY1 revealed a 1.7- and 1.4-fold increase hbae1 and lcp1 mRNA levels, respectively, confirming the observed expansion of blood cells in the ICM. Coinjection of human SPRY1 cDNA with spry4 MOs ameliorated the spry4MO phenotype, yet another confirmation of the specificity of MO targeting (Figure 4). The similar hematopoietic phenotypes observed following the overexpression of either human SPRY1 or zebrafish spry4 indicate that these genes encode proteins with a similar functional potential in blood development. In addition to the blood phenotype, the overexpression often caused a reduction and/or curve in the posterior tail (Figure 4) not seen in the overexpression of zebrafish spry4 [48].
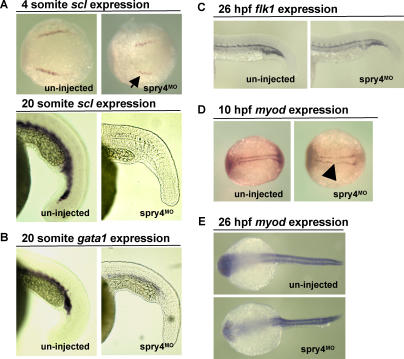
Characterization of hematopoietic gene expression in the spry4MO by whole-mount in situ hybridization revealed a reduction in scl expression at four somites (8/15), and virtually no scl (12/20) or gata1 (18/25) expression at 20 somites (Figure 5), consistent with a defect in mesodermal commitment to HSCs and/or HSC proliferation and differentiation. The few hematopoietic cells that are present in the morphant are hemoglobinized based on o-dianisidine staining (data not shown). To determine whether the hematopoietic defects observed in spry4MO were the result of a defect in mesoderm specification during development, we performed whole-mount in situ hybridization for the vasculature-specific flk1 and muscle-specific myod transcripts. At 10 h postfertilization (hpf) there was a slight defect in myod expression (5/10), while myod expression at 26 hpf was comparable to wild type (21/21) (Figure 5). This suggests that there were no major defects in mesodermal commitment in the spry4 morphants. Considering the nearly absent expression of the early hemangioblast and HSC cell marker scl, these results may suggest that the defect induced in the spry4MO occurs prior to HSC specification. However, the normal expression pattern of other mesodermal genes, such as flk1 and myod at 26 hpf in the spry4MO (Figure 5), indicate that the hematopoietic phenotype is not merely a consequence of defective specification of mesoderm. Finally, the hematopoietic gene cmyb, a presumed marker of definitive HSC in zebrafish [49], was absent at 38 hpf (10/12) (data not shown), suggesting that the spry4MO embryos are devoid of definitive HSC.
Figure 5. spry4 Is Required for Early Hematopoietic Development, but Not Mesodermal Commitment, in Developing Zebrafish Embryos.
(A) Expression of the early hematopoietic marker scl was reduced at the four-somite stage (arrow) and virtually absent at the 20-somite stage in spry4MO embryos compared to uninjected controls.
(B) The more mature hematopoietic marker gata1 was also reduced in spry4MO compared to uninjected zebrafish embryos at 20 somites.
(C and D) In contrast to hematopoietic genes, the mesodermal-specific flk1 (vasculature) transcripts were expressed at similar levels to uninjected controls at 26 hpf, and myod (muscle) had a slight defect (arrowhead) at 10 hpf, while expression was similar to controls at 26 hpf. All images are representative of greater than or equal to two experiments of n ≥ 8 embryos.
In vertebrates, Sprouty family members act as antagonists for fibroblast growth factor (FGF), vascular endothelial growth factor, and epidermal growth factor signaling, and they may be involved in feedback regulation, as Sprouty gene expression is induced by activation of these signaling pathways [50]. Sprouty genes antagonize receptor tyrosine kinase signaling at the level of the Ras/Raf/mitogen-activated protein kinase pathway; however, they also can serve as positive regulators of these pathways in some cell types [50]. Therefore, our current hypothesis is that SPRY1 may affect HSC by modulating FGF-mediated, perhaps in combination with other receptor tyrosine kinase-mediated, signaling. In fact, three of the 14 genes that induce a hematopoietic defect in the zebrafish screen, SPRY1,MAFB and SPARC, are all involved in FGF signaling [50–52], thus suggesting a role for FGF in hematopoiesis. Studies are ongoing to confirm a role of SPRY1 in mammalian hematopoiesis, by testing the effect of overexpression of SPRY1 on the repopulating ability of human and murine HSCs. Similar definitive studies in zebrafish and subsequent confirmation in mammalian HSC models are underway for genes that were functionally validated in our zebrafish screen. We believe that the sequential genetic screen in zebrafish followed by confirmation in mammalian models as described here will establish a hematopoietic function for genes identified by gene array analysis in a high-throughput and efficient manner.
Materials and Methods
Isolation of Rholo and Rhohi cell populations from UCB and BM
Human UCB from full-term delivered infants and BM from healthy donors were obtained after informed consent in accordance with guidelines approved by the University of Minnesota Committee on the Use of Human Subjects in Research. Each biologically distinct replicate was comprised of one to four donors for UCB and individual donors for BM samples. CD34+CD38−CD33−Rholoc-kit+ and CD34+CD38−CD33−Rhohi fractions were selected by sequential Ficoll-Hypaque separation, MACS column depletion, and fluorescence-activated cell sorting as previously described [17]. Postsort analysis demonstrated that sorted populations contained fewer than 1%–2% contaminating cells from the opposing population (see Figure 1).
Determination of ML–IC frequencies
ML–IC frequencies for UCB samples (n = 3) were determined as previously described [17]. An ML–IC was defined as a single cell that gave rise to at least one LTC–IC and one NK–IC. Results are presented as ML–IC frequency ± standard deviation of the mean.
Processing of RNA samples and oligonucleotide microarray analysis
Total cellular RNA was isolated from UCB (n = 5) and BM (n = 4) Rholo and Rhohi cells using the PicoPure RNA Isolation Kit (Arcturus, Mountain View, California, United States) per the manufacturer's instructions. Seven to 10,000 Rholo and Rhohi cells were sorted directly into 100 μl extraction buffer (XB) provided with the PicoPure RNA Isolation Kit prior to RNA isolation. Labeled cRNA was generated by one round of IVT-based, linear amplification using the RiboAmp OA RNA Amplification Kit (Arcturus) followed by labeling with the Enzo Bioarray HighYield RNA Transcript Labeling Kit (Enzo Life Sciences, Farmingdale, New York, United States) according to manufacturer's instructions. Samples were hybridized to Affymetrix HG-U133 A and B chips (Affymetrix Inc., Santa Clara, California, United States) were washed, and scanned at the University of Minnesota Affymetrix Microarray Core Facility as described in the Affymetrix GeneChip Expression Analysis Technical Manual.
Oligonucleotide microarray data analysis
Affymetrix HG-U133 GeneChips were processed using GeneData Refiner software (GeneData, San Francisco, California, United States) to assess overall quality. Feature intensities for each chip were condensed into a single intensity value per gene using the Affymetrix Statistical Algorithm (MAS 5.0) with τ = 0.015, α1 = 0.04, α2 = 0.06, and a scaling factor of 500. Expression data was analyzed using GeneData's Expressionist and Microsoft Excel (Microsoft, Redmond, Washington, United States). Differential gene expression for comparison of Rholo versus Rhohi cells was defined by using a paired Student's t-test with a threshold of p< 0.05, and a paired fold change was used to rank gene lists. Differentially expressed genes were classified according to their respective gene pathways and gene ontologies when available by using the Web-based Affymetrix NetAffx analysis tool (http://www.affymetrix.com) and the National Institutes of Allergy and Infectious Disease Database for Annotation, Visualization and Integrated Discovery analysis tool (DAVID) (http://apps1.niaid.nih.gov/david).
Microarray Q-RT-PCR confirmation
Labeled cRNA was reverse transcribed to generate cDNA using SuperScript II Reverse Transcriptase (Invitrogen, Carlsbad, California, United States) according to manufacturer's instructions. Q-RT-PCR was performed using the ABI Prism 7000 Sequence Detection System (Applied Biosystems, Foster City, California, United States). Briefly, 3 ng of cDNA was amplified by 40 cycles of a two-step PCR reaction (95 °C for 15 s denaturation and 60 °C for 1 min. annealing/elongation) containing 100 nM gene-specific primers (Table S8) and the SYBR Green PCR Master Mix (Applied Biosystems). Gene expression was normalized using human ACTB (also β-actin) expression levels. Results are presented as percent expression relative to uninjected clutchmate control embryos ± standard deviation of the mean.
High-throughput loss-of-function genetic screen in zebrafish
The likely zebrafish orthologs of differentially expressed human genes were identified using Ensembl's gene homology prediction program (http://www.ensembl.org, build Zv3) in combination with comparison of the human protein sequence to the Institute for Genomic Research zebrafish EST database (http://www.tigr.org, release 13–15). The criteria for a likely ortholog was ≥ 40% amino acid identity over the entire length of the protein or ≥ 50% if the fish protein sequence was only partial. MO (Gene Tools, Philomath, Oregon, United States) sequences were designed complimentary to the region of translational initiation of the zebrafish orthologs in order to inhibit protein translation (Table S6) and injected into one- to two-cell zebrafish embryos as previously described [36]. MOs were injected at 1.5, 3, 4.5, and 6 ng into 50–70 embryos derived from mating Tg zebrafish containing a fluorescent DsRed reporter cassette driven by the gata1 promoter (gata1:DsRed Tg) or a fluorescent EGFP reporter cassette driven by the fli1 promoter (fli1:EGFP Tg). MO-targeted (morphant) zebrafish were evaluated for defects in hematopoietic and vascular development compared to uninjected controls from the same embryo clutch at 30 and 48 hpf, using fluorescence microscopy for visualization of DsRed+ blood cells and EGFP+ vasculature.
Whole zebrafish Q-RT-PCR confirmation
Total RNA was isolated from five MO-injected zebrafish embryos at 48 hpf with hematopoietic defects, determined based on gata1:DsRed+ hematopoietic cell production, or five uninjected clutchmate controls using the RNeasy Mini Kit (Qiagen, Valencia, California, United States) according to the manufacturer's instructions. Total RNA was incubated with DNaseI (Invitrogen) to digest contaminating genomic DNA, and reverse transcribed to generate cDNA using SuperScript II Reverse Transcriptase (Invitrogen) according to the manufacturer's protocol. Q-RT-PCR was performed using the ABI Prism 7000 Sequence Detection System (Applied Biosystems). Briefly, 1/20 of the total cDNA from five zebrafish embryos was amplified by 40 cycles of a two-step PCR reaction (95 °C for 15 s denaturation and 60 °C for 1 min. annealing/elongation) containing 100 nM gene-specific primers (Table S8) and the SYBR Green PCR Master Mix (Applied Biosystems). Gene expression was normalized using a zebrafish gapd expression level. Results are presented as percentage expression relative to uninjected clutchmate control embryos ± standard deviation of the mean.
Whole-mount in situ hybridization
The scl, myod, flk1, gata1, and cmyb riboprobes were generated and whole-mount in situ hybridization of zebrafish embryos was conducted as previously described [41].
Overexpression of human and zebrafish Sprouty gene family members
The XhoI and KpnI fragment of the SPRY1 open reading frame (ORF) (Open Biosystems, Huntsville, Alabama, United States) was cloned into pENTR1A (Invitrogen) and subsequently transferred into a modified pFRM2.1 zebrafish expression vector using the Gateway cloning system (Invitrogen) to create the pFRM2.1–SPRY1 vector. pFRM2.1–SPRY1 was coinjected with pFRM2.1–EGFP at a 5:1 ratio into the yolk/cell interface of one-cell gata1:DsRed Tg zebrafish embryos as described for MO injections. Defects in hematopoietic development of EGFP+ embryos were analyzed by comparison to embryos from the same clutch injected with pFRM2.1–EGFP alone, using fluorescence microscopy to visualize DsRed+ blood cells.
Supporting Information
ML–ICs are highly enriched in Rholo (white bar) compared to Rhohi (black bar) cells from UCB (n = 3, *p < 0.05).
(3.6 MB TIF).
The Rholo to Rhohi fold change for DLK1, ABCB1, BMP6, HELLS, CDC25A, MAFB, and S100A8 was determined using Affymetrix GeneChip analysis (gray bars) and Q-RT-PCR (black bars) to confirm the fidelity of the microarray results for (A) adult BM (n ≥ 2) and (B) UCB (n ≥ 3).
(7.2 MB TIF).
Percentages of Gene Ontology classifications of the genes differentially expressed between Rholo and Rhohi cells from both UCB and adult BM (p < 0.05 and fold change > 1.5 in either UCB or BM).
(2.7 MB TIF).
(437 KB XLS).
(738 KB XLS).
(75 KB XLS).
(17 KB XLS).
(22 KB XLS).
(20 KB XLS).
(19 KB XLS).
(16 KB XLS).
Accession Numbers
The National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/) Entrez Gene accession numbers for the following genes are: ABCB1 (5243), ARMCX2 (9823), BMP4 (652), C12orf2 (11228), CCR7 (1236), Ccr7 (12775), CDKN1A (1026), chd (30161), cmyb (30519), CRYGD (1421), EVI1 (2122), EZH2 (2146), FLJ14917 (84947), flk1 (also known as kdr) (58106), FOXM1 (2305), gata1 (30481), GATA2 (2624), gata2 (30480), hbae1 (30597), HDHD2 (84064), HELLS (3070), HLF (3131), HMGA2 (8091), Hoxb4 (15412), HOXB4 (3214), HSPC039 (51124), IRAK3 (11213), Irak3 (73914), KIAA1102 (22998), KLF5 (688), lcp1 (30583), LEF1 (51176), LMO2 (4005), lmo2 (30332), MAFB (9935), MGC15875 (85007), MRPS6 (64968), myod (30513), NOTCH2 (4853), PIM1 (5292), PRKCH (5583), Prkch (18755), RBPMS (11030), scl (also known as tal1) (30766), SHH (6469), SLC40A1 (30061), SLCO3A1 (28232), SNX5 (27131), SPARC (6678), Sparc (20692), spry4 (114437), SPRY1 (10252), SSBP2 (23635), SUZ12 (23512), ZFHX1B (9839), ZNF165 (7718), and ZNF331 (55422).
The microarray data in this manuscript have been deposited in the GEO database (http://www.ncbi.nlm.nih.gov/geo/, and have been assigned the accession number GSE2666.
Acknowledgments
We thank Brent Bill, Aubrey Nielsen, and Shridhar Sivasubbu for zebrafish injections, Greg Veltri and Joel Sederstrom for cell sorting at the University of Minnesota Flow Cytometry Facility, Dr. Leonard Zon for the gata1:DsRed Tg zebrafish line, and Drs. Stephanie Salesse and David Largaespada for critical review of the manuscript. This research was supported by Medical Scientist Training Program (GM008244) and National Cancer Institute (NCI) (CA009138) training grants to CEE, a MinnCResT training fellowship to MAP sponsored by the National Institute of Dental and Craniofacial Research (DE07288), NCI Program Project Grant (CA065493) to CMV, and National Institute of General Medical Sciences R01 (GM63904) to SCE.
Competing interests. University of Minnesota and Stephen C. Ekker are cofounders and shareholders of a small biotechnology company called Discovery Genomics Inc. that has the exclusive license for the commercial use of morpholinos in zebrafish. Dr. Ekker is a consultant to this company.
Abbreviations
- BM
bone marrow
- EGFP
enhanced green fluorescent protein
- FGF
fibroblast growth factor
- HPC
hematopoietic progenitor cell
- hpf
hours postfertilization
- HSC
hematopoietic stem cell
- ICM
intermediate cell mass
- MHC
major histocompatibility complex
- ML–IC
myeloid–lymphoid initiating cell
- MO
morpholino antisense oligonucleotide
- Q-RT-PCR
quantitative RT-PCR
- Rho
rhodamine
- SCID
severe combined immunodeficiency
- TF
transcription factor
- Tg
transgenic
- UCB
umbilical cord blood
Author contributions. CEE, EMM, SCE, and CMV conceived and designed the experiments. CEE, EMM, CMF, and TFW performed the experiments. CEE, EMM, CMF, TFW, SMG, SCE, and CMV analyzed the data. MAP and SMG contributed reagents/materials/analysis tools. CEE, EMM, SCE, and CMV wrote the paper.
Citation: Eckfeldt CE, Mendenhall EM, Flynn CM, Wang TF, Pickart MA, et al. (2005) Functional analysis of human hematopoietic stem cell gene expression using zebrafish. PLoS Biol 3(8): e254.
References
- Morrison SJ, Uchida N, Weissman IL. The biology of hematopoietic stem cells. Annu Rev Cell Dev Biol. 1995;11:35–71. doi: 10.1146/annurev.cb.11.110195.000343. [DOI] [PubMed] [Google Scholar]
- Krause DS. Regulation of hematopoietic stem cell fate. Oncogene. 2002;21:3262–3269. doi: 10.1038/sj.onc.1205316. [DOI] [PubMed] [Google Scholar]
- Sauvageau G, Thorsteinsdottir U, Eaves CJ, Lawrence HJ, Largman C. Overexpression of HOXB4 in hematopoietic cells causes the selective expansion of more primitive populations in vitro and in vivo. Genes Dev. 1995;9:1753–1765. doi: 10.1101/gad.9.14.1753. [DOI] [PubMed] [Google Scholar]
- Antonchuk J, Sauvageau G, Humphries RK. HOXB4 overexpression mediates very rapid stem cell regeneration and competitive hematopoietic repopulation. Exp Hematol. 2001;29:1125–1134. doi: 10.1016/s0301-472x(01)00681-6. [DOI] [PubMed] [Google Scholar]
- Buske C, Feuring-Buske M, Abramovich C, Spiekermann K, Eaves CJ. Deregulated expression of HOXB4 enhances the primitive growth activity of human hematopoietic cells. Blood. 2002;100:862–868. doi: 10.1182/blood-2002-01-0220. [DOI] [PubMed] [Google Scholar]
- Varnum-Finney B, Xu L, Brashem-Stein C, Nourigat C, Flowers D. Pluripotent, cytokine-dependent, hematopoietic stem cells are immortalized by constitutive Notch1 signaling. Nat Med. 2000;6:1278–1281. doi: 10.1038/81390. [DOI] [PubMed] [Google Scholar]
- Reya T, Duncan AW, Ailles L, Domen J, Scherer DC. A role for Wnt signalling in self-renewal of haematopoietic stem cells. Nature. 2003;423:409–414. doi: 10.1038/nature01593. [DOI] [PubMed] [Google Scholar]
- Murdoch B, Chadwick K, Martin M, Shojaei F, Shah KV. Wnt-5A augments repopulating capacity and primitive hematopoietic development of human blood stem cells in vivo. Proc Natl Acad Sci U S A. 2003;100:3422–3427. doi: 10.1073/pnas.0130233100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj G, Murdoch B, Wu D, Baker DP, Williams KP. Sonic hedgehog induces the proliferation of primitive human hematopoietic cells via BMP regulation. Nat Immunol. 2001;2:172–180. doi: 10.1038/84282. [DOI] [PubMed] [Google Scholar]
- Terskikh AV, Miyamoto T, Chang C, Diatchenko L, Weissman IL. Gene expression analysis of purified hematopoietic stem cells and committed progenitors. Blood. 2003;102:94–101. doi: 10.1182/blood-2002-08-2509. [DOI] [PubMed] [Google Scholar]
- Park IK, He Y, Lin F, Laerum OD, Tian Q. Differential gene expression profiling of adult murine hematopoietic stem cells. Blood. 2002;99:488–498. doi: 10.1182/blood.v99.2.488. [DOI] [PubMed] [Google Scholar]
- Ivanova NB, Dimos JT, Schaniel C, Hackney JA, Moore KA. A stem cell molecular signature. Science. 2002;298:601–604. doi: 10.1126/science.1073823. [DOI] [PubMed] [Google Scholar]
- Ramalho-Santos M, Yoon S, Matsuzaki Y, Mulligan RC, Melton DA. “Stemness”: Transcriptional profiling of embryonic and adult stem cells. Science. 2002;298:597–600. doi: 10.1126/science.1072530. [DOI] [PubMed] [Google Scholar]
- Fortunel NO, Otu HH, Ng HH, Chen J, Mu X. Comment on “‘Stemness': Transcriptional profiling of embryonic and adult stem cells” and “a stem cell molecular signature.”. Science. 2003;302:393. doi: 10.1126/science.1082380. [DOI] [PubMed] [Google Scholar]
- Bhatia M, Wang JC, Kapp U, Bonnet D, Dick JE. Purification of primitive human hematopoietic cells capable of repopulating immune-deficient mice. Proc Natl Acad Sci U S A. 1997;94:5320–5325. doi: 10.1073/pnas.94.10.5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shojaei F, Gallacher L, Bhatia M. Differential gene expression of human stem progenitor cells derived from early stages of in utero human hematopoiesis. Blood. 2004;103:2530–2540. doi: 10.1182/blood-2003-09-3209. [DOI] [PubMed] [Google Scholar]
- Liu H, Verfaillie CM. Myeloid–lymphoid initiating cells (ML–IC) are highly enriched in the rhodamine-c-kit(+)CD33(−)CD38(−) fraction of umbilical cord CD34(+) cells. Exp Hematol. 2002;30:582–589. doi: 10.1016/s0301-472x(02)00796-8. [DOI] [PubMed] [Google Scholar]
- Cheng T, Rodrigues N, Shen H, Yang Y, Dombkowski D. Hematopoietic stem cell quiescence maintained by p21cip1/waf1. Science. 2000;287:1804–1808. doi: 10.1126/science.287.5459.1804. [DOI] [PubMed] [Google Scholar]
- Seidel MG, Look AT. E2A-HLF usurps control of evolutionarily conserved survival pathways. Oncogene. 2001;20:5718–5725. doi: 10.1038/sj.onc.1204591. [DOI] [PubMed] [Google Scholar]
- Buonamici S, Chakraborty S, Senyuk V, Nucifora G. The role of EVI1 in normal and leukemic cells. Blood Cells Mol Dis. 2003;31:206–212. doi: 10.1016/s1079-9796(03)00159-1. [DOI] [PubMed] [Google Scholar]
- Warren AJ, Colledge WH, Carlton MB, Evans MJ, Smith AJ. The oncogenic cysteine-rich LIM domain protein rbtn2 is essential for erythroid development. Cell. 1994;78:45–57. doi: 10.1016/0092-8674(94)90571-1. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Lim KC, Onodera K, Takahashi S, Ohta J. Rescue of the embryonic lethal hematopoietic defect reveals a critical role for GATA -2 in urogenital development. EMBO J. 1998;17:6689–6700. doi: 10.1093/emboj/17.22.6689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travis A, Amsterdam A, Belanger C, Grosschedl R. LEF-1, a gene encoding a lymphoid-specific protein with an HMG domain, regulates T-cell receptor alpha enhancer function [corrected] Genes Dev. 1991;5:880–894. doi: 10.1101/gad.5.5.880. [DOI] [PubMed] [Google Scholar]
- Bigas A, Martin DI, Milner LA. Notch1 and Notch2 inhibit myeloid differentiation in response to different cytokines. Mol Cell Biol. 1998;18:2324–2333. doi: 10.1128/mcb.18.4.2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashizume H, Hamalainen H, Sun Q, Sucharczuk A, Lahesmaa R. Downregulation of mafB expression in T-helper cells during early differentiation in vitro. Scand J Immunol. 2003;57:28–34. doi: 10.1046/j.1365-3083.2003.01181.x. [DOI] [PubMed] [Google Scholar]
- Geiman TM, Muegge K. Lsh, an SNF2/helicase family member, is required for proliferation of mature T lymphocytes. Proc Natl Acad Sci U S A. 2000;97:4772–4777. doi: 10.1073/pnas.97.9.4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birve A, Sengupta AK, Beuchle D, Larsson J, Kennison JA. Su(z)12 a novel Drosophila Polycomb group gene that is conserved in vertebrates and plants. Development. 2001;128:3371–3379. doi: 10.1242/dev.128.17.3371. [DOI] [PubMed] [Google Scholar]
- Ruzinova MB, Benezra R. Id proteins in development, cell cycle and cancer. Trends Cell Biol. 2003;13:410–418. doi: 10.1016/s0962-8924(03)00147-8. [DOI] [PubMed] [Google Scholar]
- Hoyle CF, Sherrington P, Hayhoe FG. Translocation (3;6)(q21;p21) in acute myeloid leukemia with abnormal thrombopoiesis and basophilia. Cancer Genet Cytogenet. 1988;30:261–267. doi: 10.1016/0165-4608(88)90193-8. [DOI] [PubMed] [Google Scholar]
- Sonoki T, Harder L, Horsman DE, Karran L, Taniguchi I. Cyclin D3 is a target gene of t(6;14)(p21.1;q32.3) of mature B-cell malignancies. Blood. 2001;98:2837–2844. doi: 10.1182/blood.v98.9.2837. [DOI] [PubMed] [Google Scholar]
- Nagarajan L, Louie E, Tsujimoto Y, ar-Rushdi A, Huebner K. Localization of the human pim oncogene (PIM) to a region of chromosome 6 involved in translocations in acute leukemias. Proc Natl Acad Sci U S A. 1986;83:2556–2560. doi: 10.1073/pnas.83.8.2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekker SC. Morphants: A new systematic vertebrate functional genomics approach. Yeast. 2000;17:302–306. doi: 10.1002/1097-0061(200012)17:4<302::AID-YEA53>3.0.CO;2-#. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackermann GE, Paw BH. Zebrafish: A genetic model for vertebrate organogenesis and human disorders. Front Biosci. 2003;8:d1227–1253. doi: 10.2741/1092. [DOI] [PubMed] [Google Scholar]
- Liao W, Ho CY, Yan YL, Postlethwait J, Stainier DY. Hhex and scl function in parallel to regulate early endothelial and blood differentiation in zebrafish. Development. 2000;127:4303–4313. doi: 10.1242/dev.127.20.4303. [DOI] [PubMed] [Google Scholar]
- Thisse C, Zon LI. Organogenesis—Heart and blood formation from the zebrafish point of view. Science. 2002;295:457–462. doi: 10.1126/science.1063654. [DOI] [PubMed] [Google Scholar]
- Nasevicius A, Ekker SC. Effective targeted gene “knockdown” in zebrafish. Nat Genet. 2000;26:216–220. doi: 10.1038/79951. [DOI] [PubMed] [Google Scholar]
- Galloway JL, Wingert RA, Thisse C, Thisse B, Zon LI. Loss of gata1 but not gata2 converts erythropoiesis to myelopoiesis in zebrafish embryos. Dev Cell. 2005;8:109–116. doi: 10.1016/j.devcel.2004.12.001. [DOI] [PubMed] [Google Scholar]
- Haffter P, Granato M, Brand M, Mullins MC, Hammerschmidt M. The identification of genes with unique and essential functions in the development of the zebrafish, Danio rerio . Development. 1996;123:1–36. doi: 10.1242/dev.123.1.1. [DOI] [PubMed] [Google Scholar]
- Pickart MA, Sivasubbu S, Nielsen AL, Shriram S, King RA. Functional genomics tools for the analysis of zebrafish pigment. Pigment Cell Res. 2004;17:461–470. doi: 10.1111/j.1600-0749.2004.00189.x. [DOI] [PubMed] [Google Scholar]
- Golling G, Amsterdam A, Sun Z, Antonelli M, Maldonado E. Insertional mutagenesis in zebrafish rapidly identifies genes essential for early vertebrate development. Nat Genet. 2002;31:135–140. doi: 10.1038/ng896. [DOI] [PubMed] [Google Scholar]
- Leung AY, Mendenhall EM, Kwan TT, Liang R, Eckfeldt C. Characterization of expanded intermediate cell mass in zebrafish chordin morphant embryos. Dev Biol. 2005;277:235–254. doi: 10.1016/j.ydbio.2004.09.032. [DOI] [PubMed] [Google Scholar]
- Hammerschmidt M, Pelegri F, Mullins MC, Kane DA, van Eeden FJ. Dino and mercedes, two genes regulating dorsal development in the zebrafish embryo. Development. 1996;123:95–102. doi: 10.1242/dev.123.1.95. [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Hernandez LD, Galan JE, Janeway CA, Medzhitov R. IRAK-M is a negative regulator of toll-like receptor signaling. Cell. 2002;110:191–202. doi: 10.1016/s0092-8674(02)00827-9. [DOI] [PubMed] [Google Scholar]
- Forster R, Schubel A, Breitfeld D, Kremmer E, Renner-Muller I. CCR7 coordinates the primary immune response by establishing functional microenvironments in secondary lymphoid organs. Cell. 1999;99:23–33. doi: 10.1016/s0092-8674(00)80059-8. [DOI] [PubMed] [Google Scholar]
- Gilmour DT, Lyon GJ, Carlton MB, Sanes JR, Cunningham JM. Mice deficient for the secreted glycoprotein SPARC/osteonectin/BM40 develop normally but show severe age-onset cataract formation and disruption of the lens. EMBO J. 1998;17:1860–1870. doi: 10.1093/emboj/17.7.1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chida K, Hara T, Hirai T, Konishi C, Nakamura K. Disruption of protein kinase Ceta results in impairment of wound healing and enhancement of tumor formation in mouse skin carcinogenesis. Cancer Res. 2003;63:2404–2408. [PubMed] [Google Scholar]
- Brun AC, Bjornsson JM, Magnusson M, Larsson N, Leveen P. Hoxb4-deficient mice undergo normal hematopoietic development but exhibit a mild proliferation defect in hematopoietic stem cells. Blood. 2004;103:4126–4133. doi: 10.1182/blood-2003-10-3557. [DOI] [PubMed] [Google Scholar]
- Furthauer M, Reifers F, Brand M, Thisse B, Thisse C. Sprouty4 acts in vivo as a feedback-induced antagonist of FGF signaling in zebrafish. Development. 2001;128:2175–2186. doi: 10.1242/dev.128.12.2175. [DOI] [PubMed] [Google Scholar]
- Thompson MA, Ransom DG, Pratt SJ, MacLennan H, Kieran MW. The cloche and spadetail genes differentially affect hematopoiesis and vasculogenesis. Dev Biol. 1998;197:248–269. doi: 10.1006/dbio.1998.8887. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Bar-Sagi D. Modulation of signalling by Sprouty A developing story. Nat Rev Mol Cell Biol. 2004;5:441–450. doi: 10.1038/nrm1400. [DOI] [PubMed] [Google Scholar]
- Brekken RA, Sage EH. SPARC, a matricellular protein: At the crossroads of cell-matrix communication. Matrix Biol. 2001;19:816–827. doi: 10.1016/s0945-053x(00)00133-5. [DOI] [PubMed] [Google Scholar]
- Marin F, Charnay P. Hindbrain patterning: FGFs regulate Krox20 and mafB/kr expression in the otic/preotic region. Development. 2000;127:4925–4935. doi: 10.1242/dev.127.22.4925. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
ML–ICs are highly enriched in Rholo (white bar) compared to Rhohi (black bar) cells from UCB (n = 3, *p < 0.05).
(3.6 MB TIF).
The Rholo to Rhohi fold change for DLK1, ABCB1, BMP6, HELLS, CDC25A, MAFB, and S100A8 was determined using Affymetrix GeneChip analysis (gray bars) and Q-RT-PCR (black bars) to confirm the fidelity of the microarray results for (A) adult BM (n ≥ 2) and (B) UCB (n ≥ 3).
(7.2 MB TIF).
Percentages of Gene Ontology classifications of the genes differentially expressed between Rholo and Rhohi cells from both UCB and adult BM (p < 0.05 and fold change > 1.5 in either UCB or BM).
(2.7 MB TIF).
(437 KB XLS).
(738 KB XLS).
(75 KB XLS).
(17 KB XLS).
(22 KB XLS).
(20 KB XLS).
(19 KB XLS).
(16 KB XLS).