Abstract
Purpose
Integrated MRI and linear accelerator systems (MR-Linacs) provide superior soft tissue contrast, and the capability of adapting radiotherapy plans to changes in daily anatomy. In this dataset, serial MRIs of the abdomen of patients undergoing radiotherapy were collected and the luminal gastro-intestinal tract was segmented to support an online segmentation algorithm competition. This dataset may be further utilized by radiation oncologists, medical physicists, and data scientists to further improve auto segmentation algorithms.
Acquisition and validation of methods
Serial 0.35T MRIs from patients who were treated on an MR-Linac for tumors located in the abdomen were collected. The stomach, small intestine and large intestine were manually segmented on all MRIs by a team of annotators under the supervision of a board-certified radiation oncologist. Annotator segmentations were validated on 4 representative abdominal MRIs by comparing to the radiation oncologist's contours using 3D Hausdorff distance and 3D Dice coefficient metrics.
Data format and usage notes
The dataset includes 467 de-identified scans and their contours from 107 patients. Each patient underwent 1–5 MRI scans of the abdomen. Most of the scans consisted of 144 axial slices with a pixel resolution of 1.5 × 1.5 × 3 mm, leading to 67,248 total slices in the dataset. Images in DICOM format were converted into Portable Graphics Format (PNG) files. Each Portable Graphics Format (PNG) image file stored a slice of the scan, with pixels recorded in 16 bits to cover the full range of intensity values. DICOM-RT segmentations were converted into Comma-Separated Values (CSV) format. Data including images and the annotations is publicly available at https://www.kaggle.com/ds/3577354.
Potential applications
While manual segmentations are subject to bias and inter-observer variability, the dataset has been used for the UW-Madison GI Tract Image Segmentation Challenge hosted by Kaggle and may be used for ongoing segmentation algorithm development and potentially for dose accumulation algorithms.
Keywords: MR-Linac, Automatic contouring, Adaptive radiotherapy, Oncology
Specifications Table
| Subject | Medical Imaging |
| Specific subject area | Serial MRIs of the abdomen of patients undergoing radiotherapy were collected and the luminal gastro-intestinal tract was segmented. |
| Type of data | Raw MRIs, organ segmentation |
| Data collection | Serial 0.35T MRIs from patients who were treated on an MR-Linac for tumors located in the abdomen were collected. The stomach, small intestine and large intestine were manually segmented on all MRIs by a team of annotators under the supervision of a board-certified radiation oncologist. Annotator segmentations were validated on 4 representative abdominal MRIs by comparing to the radiation oncologist's contours using 3D Hausdorff distance and 3D Dice coefficient metrics. |
| Data source location | Department of Human Oncology, University of Wisconsin-Madison, 600 Highland Ave, Madison, Wisconsin 53,792, USA. |
| Data accessibility | Repository name: Kaggle Data identification number: DOI: 10.34740/kaggle/ds/3577354 Direct URL to data: https://www.kaggle.com/ds/3577354 |
| Related research article | None |
1. Value of the Data
-
•
To our knowledge, this is the only publicly available dataset of serial abdominal MRIs of patients undergoing abdominal radiotherapy.
-
•
The manual segmentation of the gastrointestinal tract can be used in the development of automatic segmentation algorithms and dose accumulation algorithms.
-
•
Automatic segmentation of the gastrointestinal tract can improve the efficiency of delivering adaptive radiotherapy and thereby increase patient access to personalized radiotherapy with an improved therapeutic ratio.
-
•
Automatic segmentation is the first step for artificial intelligence and radiomics analysis and can be used to move radiology from one-dimensional measurements to volumetric measurements.
-
•
The dataset has been used for the UW-Madison GI Tract Image Segmentation Challenge hosted by Kaggle.
2. Background
Randomized controlled trials show that stereotactic ablative body radiotherapy (SABR) can improve survival in patients with oligometastatic disease although at a possibly higher risk of treatment-related toxicity and deaths [1]. The treatment of malignancies in the upper abdomen with SABR is particularly challenging due to respiratory and bowel motion, poor visualization of anatomy with cone-beam CT on most conventional linear accelerators, and the risk of complications to the gastrointestinal (GI) tract.
The introduction of integrated MRI and linear accelerator systems, also known as MR-linacs, enables the delivery of higher radiation doses to abdominal malignancies with SABR while limiting treatment-related toxicity [2]. Due to the enhanced soft-tissue contrast, MR-linacs provides the ability to adapt the treatment based on daily changes in shape, size and position of the tumor and surrounding normal tissues to increase the accuracy of treatment delivery [2]. In combination with advanced online motion-compensation, SABR delivered on an MR-linac can improve tumor targeting accuracy by allowing for smaller planning target volume margins. However, MR-linac based adaptive radiotherapy treatments are much more labour-intensive [3,4].
3. Data Description
Here we present the curation of the Serial Abdominal MRI Collection, a single-institution collection of 107 patients treated with radiotherapy on a 0.35 T MRIdian MR-linac (ViewRay, Oakwood Village, OH) at the University of Wisconsin Carbone Cancer Center from 2015 to 2019. The Serial Abdominal MRI Collection contains 1-5 de-identified serial volumetric abdominal MRIs of each patient acquired on separate days during their radiotherapy treatment and manual segmentations of the gastrointestinal tract. This data has been uploaded onto the online machine learning community and competition platform, Kaggle, and was used in an online competition to find the best machine learning algorithm to automatically segment the gastrointestinal tract. The full dataset is available for open access for interinstitutional comparisons and ongoing evaluation of new segmentation algorithms.
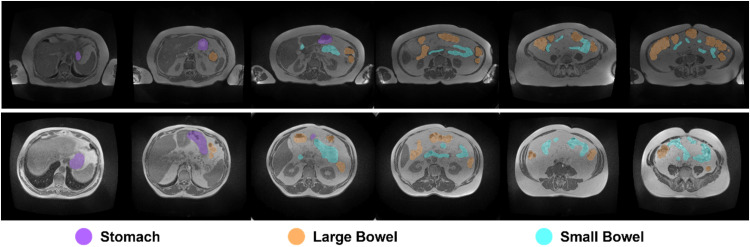
The final dataset contains 467 de-identified scans and their contours from 107 patients from 2015 to 2019. The dataset has been used for the UW-Madison GI Tract Image Segmentation Challenge hosted by Kaggle (https://www.kaggle.com/competitions/uw-madison-gi-tract-image-segmentation), and is publicly available at https://www.kaggle.com/ds/3577354 [5]. Sample slices from two scans along with their annotated contours are shown in Fig. 1.
Fig. 1.
Example images and their corresponding annotated contours from the dataset. Each row displays sample slices from a single scan, with contours color-coded according to the organ.
Data from each patient are organized into folders named “caseX”, where X is a randomly generated patient ID. Each folder might contain multiple scans from the same patient acquired at different time intervals, with each scan saved in a separate sub-folder named “caseX_dayY”, where X is the same patient ID and Y is the time relative to the first scan with day 0 indicating the first scan, the simulation scan. For example, “case10_day12” means that the scan was from patient ID 10 and captured 12 days after the simulation scan. Each sub-folder contains a set of Portable Graphics Format (PNG) image files storing the slices in the scan with 16-bit pixel data. The image file names follow “slice_[ID]_[slice width (in pixels)]_[slice height (in pixels)]_[spacing width (in mm)]_[spacing in height (in mm)]”, e.g., slice_0001_276_276_1.63_1.63.png. SliceID indexes the slices in their axial order, and the rest of the 4 numbers indicate slice height and width (integers in pixels), and height and width pixel spacing (floating points in mm). The axial spacing is 3 mm for all scans and thus is not encoded in the file name.
All contours from the scan are stored in a single CSV file with 3 data fields (columns): (slice) ID, class, and segmentation. “ID” follows “caseX_dayY_sliceID” (e.g., case123_day20_slice_0001), and points to one slice indexed by “sliceID” within a case indicated by “caseX_dayY”. “class” records the type of organ and must be one of the following: large bowel, small bowel, and stomach. “segmentation” is the run-length encoded mask [6] on the target slice of the particular organ. If an organ is not presented at the slice, the “segmentation” field is kept empty.
3.1. Usage notes
The Serial Abdominal MRI Collection is provided as a collection of Portable Graphics Format (PNG) files with accompanying CSV files. The dataset can be downloaded as a single compressed file in ZIP format by clicking the “Download” button on the Kaggle webpage. This file can be further decompressed into data folders. To facilitate access to the dataset, we have also provided sample code (sample_loader.py) as part of our dataset. Images and their annotations can be accessed using existing tools. PNG files storing individual slices can be viewed using an image viewer that supports 16-bit pixel values, such as ImageJ [7]. CSV files storing the annotations can be viewed with a spreadsheet editor, such as Microsoft Excel. However, decoding the contours stored in the CSV files requires a specialized program, which is included in the sample code.
Use of this data set is open to all researchers in accordance with the Kaggle usage policies. When citing this collection, both the DOI (10.34740/kaggle/ds/3577354) for this collection and this dataset manuscript should be cited.
3.2. Code availability
Python code for reading the imaging data and visualizing the masks are provided to facilitate the use of the data. The visualization code superimposes color-coded masks of organs on top of an intensity image of a slice. We briefly summarize the code. The code is made available at https://github.com/happyharrycn/UW-Madison-GI-Tract-Segmentation-Data-Tools.
4. Experimental Design, Materials and Methods
4.1. Patient selection
To develop this dataset, the records of consecutive patients with malignancies located in the upper abdomen treated on a single 0.35T MR-linac at the University of Wisconsin Carbone Cancer Center from 2015to 2019, were screened. After the exclusion of patients with prior GI tract resection, tumors greater than 10 cm causing deformations of the GI tract, or large image artifacts from metal or motion that overlaps with the GI tract, a total of 107 patients were included. This study was approved by the institutional review board of the University of Wisconsin-Madison. Clinical data were stored in and retrieved from EPIC (EPIC Systems, Verona, WI), Viewray treatment planning system (Viewray, Oakwood Village, OH), and ARIA (Varian Medical Systems, Palo Alto, CA).
35 of 107 patients had the most common primary histology of pancreatic adenocarcinoma (Table 1). 79 of the 107 patients had treatment to the primary site of disease while the remaining 28 were treated to a metastatic lesion. The most common anatomic site of treatment was the pancreas with 41 patients, followed by the liver with 38 patients.
Table 1.
Characteristics including primary histology, body site and whether the tumor was a metastatic site for patients in this data set.
| Characteristics | N = 107 | % |
|---|---|---|
| Primary Cancer | ||
| Adrenal | 1 | 1% |
| Anal | 1 | 1 % |
| Biliary Tree | 12 | 11 % |
| Breast | 2 | 2 % |
| Colorectal | 7 | 7 % |
| Esophagus | 9 | 8 % |
| HCC | 9 | 8 % |
| Hematologic | 7 | 7 % |
| Lung | 9 | 8 % |
| Pancreas | 35 | 33 % |
| Prostate | 2 | 2 % |
| Renal | 3 | 3 % |
| Skin | 2 | 2 % |
| Stomach | 4 | 4 % |
| Thyroid | 1 | 1 % |
| Uterine | 3 | 3 % |
| Body Site | ||
| Adrenal | 4 | 4 % |
| Esophagus | 7 | 7 % |
| Kidney | 5 | 5 % |
| Liver | 38 | 35 % |
| Lymph Node | 6 | 5 % |
| Pancreas | 41 | 37 % |
| Stomach | 6 | 7 % |
| Primary versus Metastasis | ||
| Metastasis | 28 | 26 % |
| Primary | 79 | 74 % |
Because patients with large tumors and those that underwent gastrointestinal surgery were excluded, these patients had relatively normal anatomy. The presence of small tumors increases the variability in the anatomy compared to a dataset of healthy volunteers or patients without cancer. Therefore, this dataset with a representative sample of patients with cancer can be used for diverse research purposes on cancer patients. Table 1: Characteristics including primary histology, body site and whether the tumor was a metastatic site for patients in data set.
4.2. Image acquisition
All images in this dataset were acquired at the University of Wisconsin Carbone Cancer Center. 0.35T abdomen MRIs were obtained at the time of radiotherapy simulation and daily during radiation delivery on an integrated MR-linac system, which started about 2 weeks after the radiotherapy simulation. The 0.35T MRI underwent regular imaging quality assurance. Weekly and monthly imaging quality assurance was performed to test the geometric accuracy, high contrast, spatial resolution, slice thickness accuracy, slice position accuracy, image intensity uniformity, and distortion. There were no major variations in quality over time. Each daily radiotherapy fraction was delivered at least 1 day apart. The abdominal MRI at simulation and up to 4 additional MRIs taken during radiotherapy scheduled daily or every other day were included in the dataset. MRIs were acquired prior to online adaptive replanning, approximately 10–20 min prior to the delivery of radiotherapy. There were approximately 7–14 days between simulation scan and the first fraction. There were between approximately 7–14 days between the first and last fraction. Overall, the serial MRIs for each patient were taken over a period of approximately 3–4 weeks. Patients were positioned and immobilized in head-first supine position on a thin mattress, head rest, with both arms up using a wing board and a triangular knee support. Gadoxetate disodium (Bayer, Leverkusen, Germany) was used for liver metastases cases, and 3–4 h of fasting prior to simulation scans and radiation delivery was required for any patients with metastases near gastrointestinal luminal organs. True fast imaging with a steady state precession (TrueFISP) was used for the simulation and daily scans. The planning MRIs were captured with torso receiver coils placed on patients. Most of the MRIs (>95 %) have a field of view of either 40×40×43 cm (with pixel size 0.15×0.15×0.3 cm) or 54×47×43 cm (with pixel size 0.16×0.16×0.3 cm). Most scans covered the diaphragm superiorly to the L3-4 vertebra inferiorly. MR scans were acquired in the maximum inhale breathe hold phase.
4.3. Dataset curation
10 annotators manually contoured the abdominal MRIs. All annotators were actively involved in the treatment of patients on a single Viewray system and included 4 radiation therapists, 4 radiation planners, and 2 radiation physicists (P.Y., J.M., J.S., D.H., A.A., S.O., T.K., J.S., K.B., B.L.). All contours were manually reviewed by a board-certified radiation oncologist specializing in GI malignancies (S.L.L.). 467 abdominal MRIs from 107 patients were manually contoured.
MIM (version 6.6.11, MIM Software Inc., Cleveland, OH) was used for contouring of the GI tract, and Matlab (version R2021a, The MathWorks, Inc., Natick MA) was used for DICOM-to-PNG conversion on clinical Mac desktop computers (Apple Inc., Cupertino, CA). No routine preprocessing steps of the MRIs were required, although individual annotators were allowed to change the window levels to improve the visualization of the GI tract. The 2D Brush tool was used for manual contouring. In cases where there was sufficient contrast between the organ of interest and surrounding adipose tissue, the use of the Dynamic Brush, an automatic thresholding tool, was acceptable. The Smooth Tool, was used to smooth out the contours prior to interpolation. Interpolation of contours was allowed for every other slice but had to be checked for accuracy. The GI tract from the gastro-esophageal junction to the large intestines (stomach, small intestines, large intestines) were contoured de novo using standard contouring guidelines [8,9]. The original contours used for the delivered radiotherapy plans were not included in this dataset due to inconsistencies between physicians and incomplete contouring. For example, in many cases, the radiation oncologist only contoured the GI tract in the transverse slices within 3 cm of the PTV [10]. In order to contour the organs and landmarks consistently and completely, specific instructions were followed by all annotators (see supplementary material). The transition from the esophagus to the stomach was specified as the MRI slice where the left and right diaphragmatic crura surround the GI lumen by 180-degrees. The duodenum was included with the small intestines due to the relative difficulty in finding a consistent border between the duodenum and the jejunum [11]. To minimise any ambiguity between organs, annotators were instructed to contour the stomach first, then the large bowel, then the small bowel, as the small bowel had the least predictable position. The GI luminal organs were selected for contouring in this dataset as high performing auto-contouring solutions exist for solid GI organs [11,12]. Annotators had no time limit for completing the contours on each abdominal MRI.
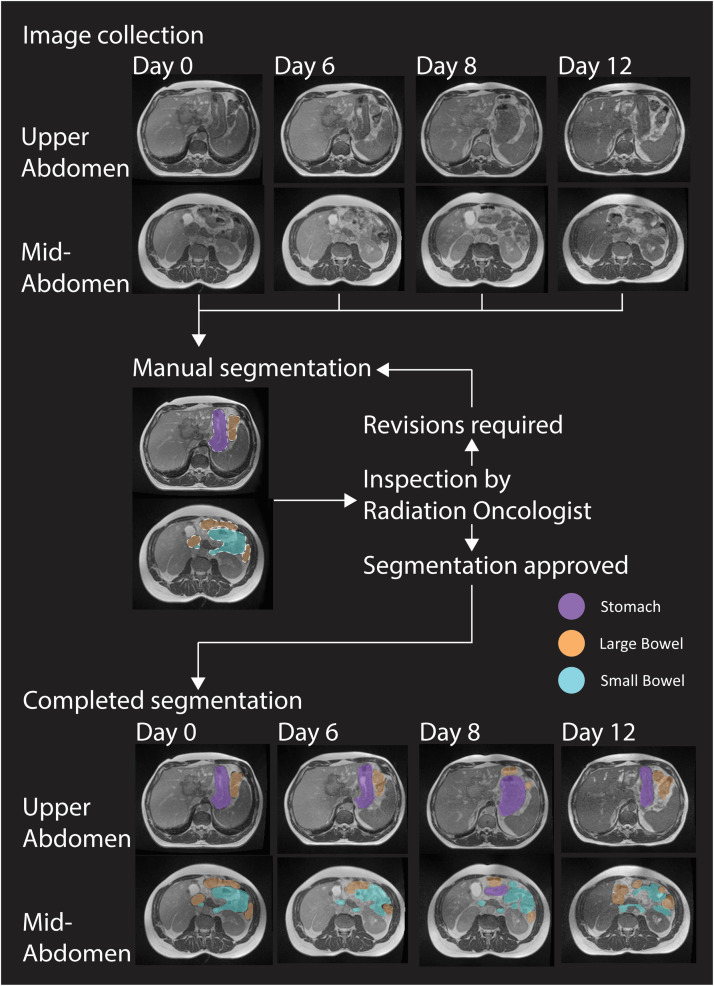
Annotators were given a list of patients to contour. Each MRI was contoured by a single annotator. A supervising radiation oncologist (S.L.L.) instructed that the annotators make corrections where necessary until the contours were of sufficient accuracy. The radiation oncologist reviewed all the contours and gave feedback for any necessary corrections iteratively before the annotator moved onto the next patient on their respective lists (Fig. 2). While no quantitative criteria were used for quality control, the contours were required to be sufficiently accurate to be used in a clinical radiotherapy planning scenario.
Fig. 2.
Schema representation of image collection and manual segmentation of dataset. Day 0 is the MRI simulation and Days 6, 8 and 12 are the MRIs taken before the delivery of each fraction in this example.
The mean contouring time per MRI for all annotators was 31.5 min. The mean contouring time per MRI was 23 minutes for the fastest annotator and 38 min for the slowest annotator. On average each annotator contoured 46.7 MRIs, with a range of 19–91 MRIs.
Overall, 47.5 % of the MRIs required revisions before they were approved by the radiation oncologist. Reasons for revision included mislabelling pancreas and blood vessels as bowel, confusing small bowel with large bowel, missing bowel loops, including fat as small bowel loops, incorrectly drawing edges at the interface between gas and liquids, and not contouring the full superior to inferior extent of the organs.
The method of contouring used in this study can be applied to other imaging modalities such as higher-field MRI and CT scans. However, the accuracy of the contouring would depend on factors such as slice thickness, soft tissue contrast and the type of pulse sequence used.
4.4. Data validation
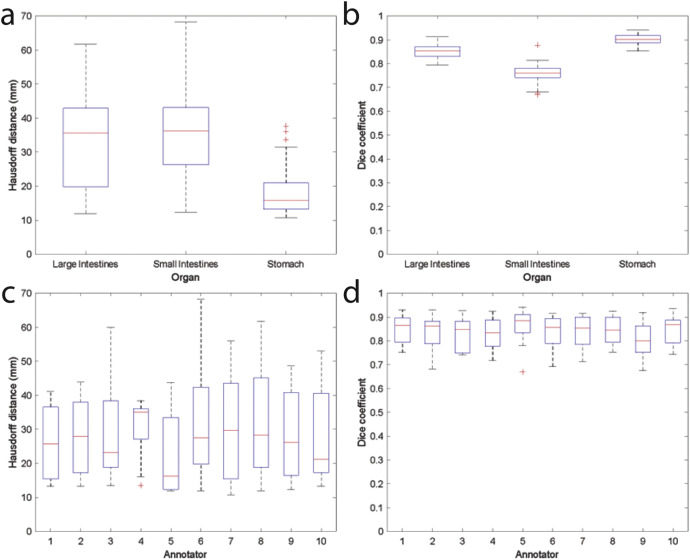
All annotators were asked to contour the same 4 representative abdominal MRIs to ensure accuracy before moving onto their assigned patients. The contours on the 4 representative MRIs were compared to the radiation oncologist's contours using 3D Hausdorff distance and 3D Dice coefficient metrics (Fig. 3). The stomach had the lowest Hausdorff distance and highest Dice coefficient while the small intestines had the highest Hausdorff distance and lowest Dice coefficient, reflecting the anatomic complexities of each organ (Table 2). The inter-observer variability was in line with that observed for CT based contours [13]. In general, a Dice coefficient above 0.9 and a Hausdorff distance of less than 10 mm could be considered as excellent. However, the applicability and acceptability of these quantitative metrics in radiotherapy is context specific. For example, the Hausdorff distance may be important for ensuring no ablative radiation hotpots fall on the organ at risk, while the Dice coefficient may be more important for assessing contour adaptation time and time-saving [14].
Fig. 3.
Comparison of each annotator's contours to those of a radiation oncologist's on a common set of 4 MRIs. (a) Hausdorff distance for each organ. (b) Dice coefficient for each organ. (c) Hausdorff distance of the stomach, large intestine and small intestine in aggregate for each annotator. (d) Dice coefficient of the stomach, large intestine and small intestine in aggregate for each annotator. On each box, the central mark indicates the median, and the bottom and top edges of the box indicate the 25th and 75th percentiles, respectively. The whiskers extend to the most extreme data points not considered outliers, and the outliers are plotted individually using the '+' symbol. The whiskers correspond to approximately +/– 2.7σ and 99.3 percent coverage if the data are normally distributed.
Table 2.
Comparison of each non-radiation oncologist annotator's contours for each organ to those of a radiation oncologist's on a common set of 4 MRIs.
| Hausdorff distance mean ± standard deviation (mm) | Dice coefficient mean ± standard deviation | |
|---|---|---|
| Large Intestines | 32.3 ± 12.3 | 0.85 ± 0.03 |
| Small Intestines | 35.5 ± 12.6 | 0.76 ± 0.04 |
| Stomach | 18.0 ± 6.9 | 0.90 ± 0.02 |
4.5. Data anonymization and transmission to Kaggle
Custom code in Matlab (version R2021a, The MathWorks, Inc., Natick MA) was developed to convert a DICOM scan [15] into a set of Portable Graphics Format (PNG) files. Conversion to Portable Graphics Format (PNG) format ensured that all patient identifying information was removed and that the public dataset would be more accessible to a wider audience including computer scientists without a medical imaging background. Each Portable Graphics Format (PNG) image file stored a slice of the scan, with pixels recorded in 16 bits. The DICOM files are also stored pixels in 16 bits. Therefore, the converted PNG files cover the full range of intensity values. Information in the DICOM header was discarded during the conversion to de-identify the imaging data, with the exception of the 3D spacing, which was recorded in the image file names. The same Matlab code also exported DICOM-RT contours into binary Matlab format (MAT) files, where each contour was stored as a 3-dimensional binary mask. Similarly, information in the DICOM-RT [16] header was removed.
A custom Python (version 3.7, Python Software Foundation, DE) program was further developed to organize the Portable Graphics Format (PNG) and MAT files into the data format ready for release. Portable Graphics Format (PNG) files of the same scan were grouped into a separate folder. MAT files were also converted into Comma-Separated Values (CSV) format using the Python code. Masks of one scan were stored in a single CSV file.
Folders including all scans, together with CSV files of all contours, were packed into a zip file and released to Kaggle. The Kaggle team further repackaged the data to facilitate the integration with the platform.
4.6. Data reusability
Segmentation is often the initial step in a biomedical imaging analysis pipeline. By automatically segmenting organs of interest, researchers can then focus on applying artificial intelligence algorithms on those organs or look at radiomic features. In radiation oncology, automatic segmentation can help guide and evaluate deformable registration algorithms which are a prerequisite for any dose accumulation calculations. By tracking the position and shape of the GI tract over time, one could calculate the optimal planning organ at risk (PRV) margins. Automatic segmentation allows radiologists to make more objective anatomic measurements. Instead of relying on one-dimensional measurements, more robust volumetric measurements can highlight changes in anatomy. For example, instead of measuring the diameter of the small bowel on one slice of a volumetric image, determining the overall volume of the whole small bowel may be more sensitive in detecting bowel obstruction.
Limitations
A limitation of this dataset is that manual segmentations are subject to bias and inter-observer variability. Due to the time-consuming nature of manually contouring the GI tract, the majority of the MRIs were only contoured by a single annotator and therefore inter-observer variability of the segmentation on every MRI could not be determined. We acknowledge that the inter-observer variability of any data set can limit the performance of the automatic segmentation algorithms derived from that data. The images were contoured on the axial slices and therefore segmentation algorithms based off this data set may perform better on individual axial slices compared to other orientations. Further refinements in the segmentations could possibly be made to improve the dataset. The Hausdorff distance metric used in the validation is sensitive to outliers but is appropriate as the maximum point dose can vary greatly due to outlier contours with adaptive SABR. A Hausdorff distance > 10–30 mm for the small intestines is in line with other studies on interobserver contouring variation [13]. Data was compiled from a single institution in the US which could limit its generalizability to other populations. The ethnicities of the patient population included in this data set, while not specifically analysed, likely represents the demographics of Madison, Wisconsin, with 69 % Caucasian, 9 % Asian, and 7 % Black [17]. Despite these limitations, this is the first publicly available dataset of gastrointestinal tract segmentations on MRIs to the authors’ knowledge.
Ethics Statement
This study (Project ID AAH4984) was approved by the institutional review board of the University of Wisconsin-Madison and follows the ethical requirements in accordance with the Declaration of Helsinki. Given the retrospective nature of the study and lack of identifiable patient information in the abdominal MRIs, the requirement for patient consent was waived.
CRediT Author Statement
Sangjune L. Lee: Conceptualization, Methodology, Data curation, Writing- Original draft preparation, Funding acquisition. Poonam Yadav: Conceptualization, Methodology, Data curation, Writing- Original draft preparation. Yin Li: Conceptualization, Methodology, Writing- Original draft preparation, Visualization, Software. Jason J. Meudt: Data curation. Jessica Strang: Data curation. Dustin Hebel: Data curation. Alyx Alfson: Data curation. Stephanie J. Olson: Data curation. Tera R. Kruser: Data curation. Jennifer B. Smilowitz: Data curation. Kailee Borchert: Data curation. Brianne Loritz: Data curation. Laila Gharzai: Data curation. Shervin Karimpour: Data curation. John Bayouth: Supervision. Michael F. Bassetti: Supervision, Funding acquisition.
Acknowledgments
This research was supported by the University of Wisconsin Carbone Cancer Center Pancreas Pilot Grant. We thank Janet Bresnahan from the University of Wisconsin for her assistance in establishing a data sharing agreement. We thank Maggie Demkin and Phil Culliton from Kaggle for their assistance in successfully executing the Kaggle data science competition.
Declaration of Competing Interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: SL reports educational funding on behalf of the radiation oncology division from AstraZeneca, Bayer, Merck, Pfizer, not related to this work. PY reports research funding from Viewray, not related to this work. MB reports non-financial support from Viewray Inc, outside the submitted work, and MERCK Clinical Trial Support, Astra Zeneca Clinical Trial Support, EMD Serono Clinical Trial Support.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.dib.2024.111159.
Appendix. Supplementary materials
Data Availability
References
- 1.Chalkidou A., Macmillan T., Grzeda M.T., Peacock J., Summers J., Eddy S., et al. Stereotactic ablative body radiotherapy in patients with oligometastatic cancers: a prospective, registry-based, single-arm, observational, evaluation study. Lancet Oncol. 2021;22:98–106. doi: 10.1016/S1470-2045(20)30537-4. [DOI] [PubMed] [Google Scholar]
- 2.Henke L., Kashani R., Robinson C., Curcuru A., DeWees T., Bradley J., et al. Phase I trial of stereotactic MR-guided online adaptive radiation therapy (SMART) for the treatment of oligometastatic or unresectable primary malignancies of the abdomen. Radiother. Oncol. 2018;126:519–526. doi: 10.1016/j.radonc.2017.11.032. [DOI] [PubMed] [Google Scholar]
- 3.Shelley C.E., Bolt M.A., Hollingdale R., Chadwick S.J., Barnard A.P., Rashid M., et al. Implementing cone-beam computed tomography-guided online adaptive radiotherapy in cervical cancer. Clin. Transl. Radiat. Oncol. 2023;40 doi: 10.1016/j.ctro.2023.100596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bertelsen A.S., Schytte T., Møller P.K., Mahmood F., Riis H.L., Gottlieb K.L., et al. First clinical experiences with a high field 1.5 T MR linac. Acta Oncol. 2019;58:1352–1357. doi: 10.1080/0284186X.2019.1627417. [DOI] [PubMed] [Google Scholar]
- 5.Lee SL, Yadav P, Li Y, Meudt JJ, Strang J, Hebel D, et al. UW Madison GI tract image segmentation dataset 2023. 10.34740/KAGGLE/DS/3577354.
- 6.Golomb S. Run-length encodings (Corresp.) IEEE Trans. Inf. Theory. 1966;12:399–401. doi: 10.1109/TIT.1966.1053907. [DOI] [Google Scholar]
- 7.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jabbour S.K., Hashem S.A., Bosch W., Kim T.K., Finkelstein S.E., Anderson B.M., et al. Upper abdominal normal organ contouring guidelines and atlas: a radiation therapy oncology group consensus. Pract. Radiat. Oncol. 2014;4:82–89. doi: 10.1016/j.prro.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lukovic J., Henke L., Gani C., Kim T.K., Stanescu T., Hosni A., et al. MRI-based upper abdominal organs-at-risk atlas for radiation oncology. Int. J. Radiat. Oncol. Biol. Phys. 2020;106:743–753. doi: 10.1016/j.ijrobp.2019.12.003. [DOI] [PubMed] [Google Scholar]
- 10.Bohoudi O., Bruynzeel A.M.E., Senan S., Cuijpers J.P., Slotman B.J., Lagerwaard F.J., et al. Fast and robust online adaptive planning in stereotactic MR-guided adaptive radiation therapy (SMART) for pancreatic cancer. Radiother. Oncol. 2017;125:439–444. doi: 10.1016/j.radonc.2017.07.028. [DOI] [PubMed] [Google Scholar]
- 11.Fu Y., Mazur T.R., Wu X., Liu S., Chang X., Lu Y., et al. A novel MRI segmentation method using CNN-based correction network for MRI-guided adaptive radiotherapy. Med. Phys. 2018;45:5129–5137. doi: 10.1002/mp.13221. [DOI] [PubMed] [Google Scholar]
- 12.Liang F., Qian P., Su K-H., Baydoun A., Leisser A., Van Hedent S., et al. Abdominal, multi-organ, auto-contouring method for online adaptive magnetic resonance guided radiotherapy: An intelligent, multi-level fusion approach. Artif. Intell. Med. 2018;90:34–41. doi: 10.1016/j.artmed.2018.07.001. [DOI] [PubMed] [Google Scholar]
- 13.Kim H., Jung J., Kim J., Cho B., Kwak J., Jang J.Y., et al. Abdominal multi-organ auto-segmentation using 3D-patch-based deep convolutional neural network. Sci. Rep. 2020;10:6204. doi: 10.1038/s41598-020-63285-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vaassen F., Hazelaar C., Vaniqui A., Gooding M., van der Heyden B., Canters R., et al. Evaluation of measures for assessing time-saving of automatic organ-at-risk segmentation in radiotherapy. Phys. Imaging Radiat. Oncol. 2020;13:1–6. doi: 10.1016/j.phro.2019.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mildenberger P., Eichelberg M., Martin E. Introduction to the DICOM standard. Eur. Radiol. 2002;12:920–927. doi: 10.1007/s003300101100. [DOI] [PubMed] [Google Scholar]
- 16.Law M.Y.Y., Liu B. DICOM-RT and its utilization in radiation therapy. RadioGraphics. 2009;29:655–667. doi: 10.1148/rg.293075172. [DOI] [PubMed] [Google Scholar]
- 17.Madison, Wisconsin. Wikipedia 2024.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.