Abstract
Background
Cancer patients frequently suffer from pain, often managed with opioids. However, undertreated pain remains a significant concern. Opioid effectiveness varies due to genetic differences in how individuals metabolize some of these medications. While prior research suggests promise in tailoring opioid prescriptions based on CYP2D6 genetic makeup, its application in cancer pain management remains limited. This study investigates the potential benefits of preemptive CYP2D6 genotyping for cancer patients initiating opioid therapy, focusing on codeine, tramadol, and hydrocodone, whose efficacy is demonstrably impacted by CYP2D6 variations.
Methods
This is a randomized, prospective study to evaluate the effects of preemptive pharmacogenomic (PGx) testing on opioid dosing decisions/selections and composite pain score in oncology patients. Patients with metastatic solid tumors for whom near-future opioid therapy is anticipated will be randomized to PGx and control arms, stratified by the presence or absence of bony metastases and history of opioid use. In the PGx arm, patients will be preemptively tested using a panel of pharmacogenomic genetic variants, and providers will receive opioid dosing guidance via an electronic medical record-embedded clinical decision support tool. In the control arm, pain prescribing will occur per standard of care without genotype information.
Planned Outcome
The primary study outcome will be composite pain intensity during the first 45 days after an index opioid prescription for codeine, tramadol, or hydrocodone. Safety will be assessed by comparing opioid-related adverse event rates between the two study arms. Secondary outcomes will include rates of hospitalization/emergency room visits, cumulative morphine equivalents received, and type of first opioid prescribed.
Keywords: pharmacogenomics, CYP2D6, codeine, tramadol, hydrocodone, genetic testing, pain score
Introduction
Pain is a common and distressing symptom that can have a significant impact on the lives of people with cancer. A recent systemic review shows that about 39% of cancer patients experience pain after curative treatment, 55% during anticancer treatment, and 66% in advanced, metastatic, or terminal disease.1 Despite its high prevalence in the patient population, numerous studies indicate a problem of analgesic undertreatment in cancer.2 It is well documented that the effectiveness of pain therapies varies from patient to patient, even for those with similar pain levels or disease status.3 To safely and effectively control cancer pain, it is important to have a thorough knowledge of the pharmacokinetics and pharmacodynamics of opioids and utilize this understanding to guide selection and dosing.4
Nevertheless, current pain prescribing for oncology patients often follows a predictable pattern, in which oncologists typically use a codeine-containing medication, tramadol, or hydrocodone as the “first-line opioid analgesic.5” CYP2D6 is a major drug-metabolizing enzyme that is responsible for breaking down a quarter of all prescribed drugs.6 The corresponding gene is highly polymorphic, leading to different metabolizer phenotypes in patients, and it is thus one of the key pharmacokinetic biomarkers for predicting drug response in patients.7 Notably, numerous studies have indicated that these differences in CYP2D6 gene expression are responsible for the interindividual variability in safety and efficacy of different pain medications, including but not limited to codeine, tramadol, and hydrocodone.8
CYP2D6 activity is significantly reduced in CYP2D6 poor metabolizers (PMs – two non-functional alleles) and intermediate metabolizers (IMs – one nonfunctional and one reduced function allele, or one functional and one nonfunctional allele) compared to normal metabolizers (NMs – two functional alleles). This is of importance for opioids that rely on CYP2D6-mediated formation of an active metabolite to achieve full analgesic effects. Codeine is the classical prototype for dependence on CYP2D6, however other opioids also utilize CYP2D6-mediated biotransformation to produce metabolites with potent analgesic properties.8,9 For example, while tramadol has both opioid and non-opioid mechanisms of action, its opioid-mediated analgesia is enhanced by CYP2D6-catalyzed conversion to O-desmethyltramadol, particularly in situations requiring opioid-level pain relief.10,11 The CYP2D6 enzyme also converts hydrocodone into hydromorphone, which has a more potent binding affinity to μ-opioid receptors and is a predictive factor for hydrocodone pain relief.12 Consequently, PMs and IMs may have inadequate analgesic response to these pain medications because the resulting metabolites have reduced activity. Ultra-rapid metabolizers (UMs), who conversely demonstrate increased CYP2D6 activity, may instead have higher risk of toxicity from CYP2D6-dependent opioids at typical doses due to the higher levels of active metabolites.8 In contrast, opioids such as morphine, hydromorphone, and fentanyl are not metabolized via CYP2D6 and thus exert their analgesic effects independent of genetic variation in this gene.
Some of the existing guidelines and literature support the use of CYP2D6-guided opioid prescribing for better pain management.8,13,14 Our retrospective study showed an increased risk of hospitalization or pain-related encounters requiring non-CYP2D6 activated opioids in oncology patients.15 D’Agostino et al indicated that pharmacogenetic analysis of CYP2D6 polymorphisms can aid in predicting the safety and efficacy of opioid treatment in chronic low back pain patients, with reduced CYP2D6 activity being associated with therapeutic failure and increased risk of adverse reactions.16 A recent prospective study by Smith et al, found that chronic pain patients receiving tramadol/codeine with CYP2D6 dosing guidance had improved therapeutic composite outcomes.17 Thomas et al demonstrated that implementing CYP2D6 genotype-guided postoperative pain management is feasible and may result in reduced opioid consumption without adversely affecting pain control in adults undergoing total joint arthroplasty.18 A large-scale NIH-sponsored IGNITE study is currently ongoing to assess the impact of postoperative CYP2D6-guided opioid prescribing on pain control and opioid usage, aiming to provide evidence on the clinical utility of this approach for improving postoperative pain management and reducing opioid-related risks.19 The latest Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines give therapeutic recommendations on dosing and selection of opioids such as codeine and tramadol using CYP2D6 genotypes based on the current body of evidence.8
To our knowledge, only a few studies have explored the utilization of PGx in cancer palliative setting. Andreassen et al, focused solely on oxycodone prescribing with CYP2D6 genotypes and failed to find positive findings.20 Mosely et al and Patel et al both ran a PGx study including major opioid drugs metabolized by CYP2D6, but neither have generated results confirming clinical utility, potentially due to small sample size and actionable genotypes.21,22 Opioids undergo varying degrees of metabolism by the CYP2D6 enzyme. Unlike codeine, tramadol, and hydrocodone, oxycodone experiences moderately lower metabolism via CYP2D6, resulting in less direct influence by its enzyme activity.23 Evidence regarding CYP2D6 relevance for oxycodone is currently mixed, and CPIC recommendations presently do not support actionability for CYP2D6 and oxycodone. Consequently, for a comprehensive evaluation of the clinical efficacy of CYP2D6-guided opioid prescribing, it is prudent to prioritize opioids that are significantly impacted by CYP2D6 metabolism. However, Andreassen et al did not consider the inclusion of other opioids in their study design, and the study by Mosley et al consisted of 71% of study participants taking oxycodone at baseline, potentially affecting the effectiveness outcome. Furthermore, the non-randomized design and comparison of outcomes between a contemporary intervention group and historical controls in the study by Patel et al pose additional challenges for analysis. In light of the aforementioned gaps in the literature, our study will investigate the advantage of CYP2D6 preemptive genotyping in cancer opioid treatment within a prospective, randomized context with a focus on codeine, tramadol and hydrocodone. This research is purported to fulfill these unmet needs and adds to the current body of evidence to further support the routine use of genotyped guided dosing/selection in clinical oncology.
Methods
Design
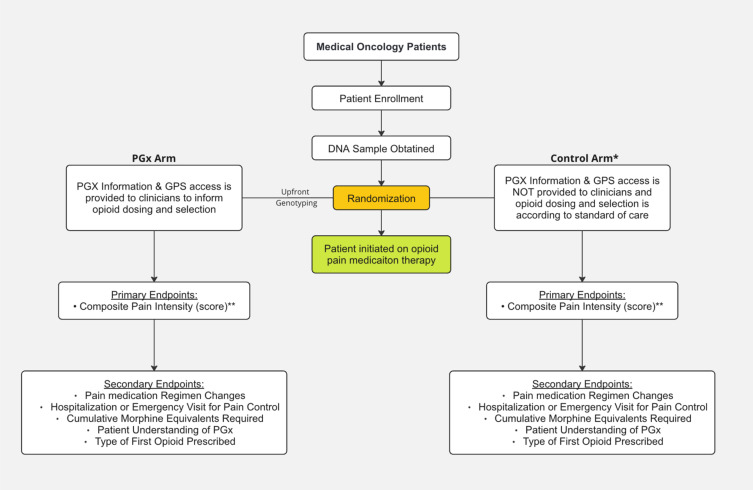
We here propose a prospective randomized study to assess the impact of preemptive pharmacogenomic (PGx) testing on opioid prescribing and pain management in cancer patients. In the study design, stage 4 cancer patients for whom near-future opioid use will likely be needed for treatment of cancer pain will be randomized to either a PGx guided group or a control group. The randomization will be carried out using two stratification factors: presence or absence of bone metastases; and prior opioid use (yes vs no) (Figure 1). The PGx guided group will undergo genetic testing at the start to identify potential medication response-influencing variations, with a genotyping turnaround time expected to be under 1 week. Healthcare providers will then have access to this genetic information and dosing guidance throughout the patient’s treatment course via electronic-medical record embedded pharmacogenomic clinical decision support (CDS) tools that have been developed as part of our institutional software system called the Genomic Prescribing System (GPS).24 GPS has been in use since 2012 at the University of Chicago and integrates laboratory-derived genomic data with evidence-based pharmacogenomic summaries to annotate a patient’s genetic profile for variants affecting drug metabolism or response. It provides interruptive, color-coded CDS alerts within the electronic health record (EHR) when clinicians prescribe medications that have actionable PGx information, indicating the level of gene–drug interaction risk and the potential clinical consequences. Clinicians can access detailed information and alternative prescribing recommendations by clicking on CDS-embedded related indicators. GPS has previously been deployed in several pharmacogenomic trials within and outside of the University of Chicago integrated care network, with demonstrations of its utility across various clinical contexts.25–27 Standard prescribing practices will be followed in the control group, without access to genotype information. The study will track various outcomes, including composite pain scores (primary), drug adverse event outcomes (co-primary), changes in pain medication regimens (secondary), hospitalization/emergency room visits (secondary), total morphine equivalents received (secondary) and type of first opioid prescribed (secondary). Additionally, it will explore the use of GPS for prescribing other supportive medications, patients’ quality of life, and their understanding of PGx. We hypothesize that providing clinicians with preemptive PGx information will lead to adjustments in opioid dosing or selection, potentially reducing inadequate pain relief and improving overall pain management for cancer patients.
Figure 1.
Study Schema for the C-PAIN Trial. Design of this prospective pharmacogenomics clinical trial, including patient allocation to pharmacogenomic (PGx) and control arms, and primary and secondary endpoints of the study.
*All patients in the control arm will be genotyped with results and GPS access provided to their clinical team approximately 3 months after index opioid prescription. **The primary endpoint comparison will focus solely on IM/PM patients within the PGx and control arms.
Abbreviations: GPS, Genomic Prescribing System; IM, Intermediate Metabolizer; PM, Poor Metabolizer.
Subjects
Adult oncology patients with a known diagnosis of a metastatic solid tumor who are receiving longitudinal care at the University of Chicago Medical Center and who are likely to require a near-future opioid medication for treatment of cancer-related pain will be enrolled. No specific threshold for baseline pain levels is required for inclusion. These broad inclusion criteria aim to capture a diverse range of stage 4 cancer patients who are likely to need opioid pain management specifically for cancer-related pain, regardless of their current pain levels, so as to increase the generalizability of the eventual results of this study. Patients likely to require near-future opioid treatment will be pre-screened by a dedicated clinical research coordinator prior to being approached for enrollment based on factors including current pain scores, existing non-opioid analgesic use, presence of bone metastases, signs of disease progression, medical record documentation of clinical discussions of new treatments, and treatment care team input. Exclusion criteria encompass individuals who fall into one or more of the following categories: 1) taking an opioid currently, or within the past 30 days, 2) who are currently undergoing palliative radiation, 3) who have undergone, or are being actively considered for, bone marrow, liver or kidney transplantation, 4) with a history of or active blood cancer (eg, leukemia), 5) with chronic kidney disease as defined by GFR < 30/mL/min/1.73m2, due to the risk of decreased drug excretion, 6) liver dysfunction defined by a total bilirubin level of ≥1.5 mg/dL and aspartate aminotransferase (AST) and alanine aminotransferase (ALT) levels ≥2.5 times the upper limit of normal (or ≥5 times the upper limit of normal if hepatic metastases are present) due to the risk of decreased drug metabolism, 7) inability to understand and give informed consent.
All participants will undergo pharmacogenomic testing at enrollment and be randomly assigned to either an PGx guided arm, where their clinicians will have real-time access to their genetic results through the GPS, or a control group.
Genotyping and Translation into Actionable Recommendations
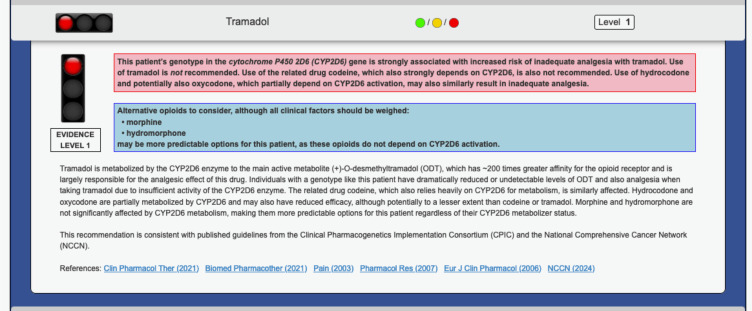
Timing of genotyping will vary between PGx guided arm and control arm. Genotyping will occur immediately for subjects randomized to PGx guided arm after sample collection in the Clinical Laboratory Improvement Amendments (CLIA)-certified and College of American Pathologists (CAP)-accredited laboratory at the University of Chicago. The enrollment sample from control arm will be held for genotyping until after 3 months of index opioid prescription. For comprehensive CYP2D6 genotyping, a custom panel powered by Hologic’s Invader technology will be utilized.28 Additionally, TaqMan qPCR will be employed to detect copy number variations within the CYP2D6 gene.29 An estimated turnaround time of 1 week is expected for CYP2D6 genotyping results. The results will be translated at a phenotype level, depicting metabolizer status through star allele-based activity score calculation.8 Star allele-based activity scores will be adjusted according to current guidelines for patients taking strong or moderate CYP2D6 inhibitors at the onset of opioid therapy.30 To accommodate this possibility of phenoconversion, if the patient is concurrently using a strong inhibitor, the genotype-based activity score will be multiplied by 0; if using a moderate inhibitor, the genotype-based activity score will be multiplied by 0.5 (Table 1). Recommendations will be delivered employing the GPS, a secure and password-protected online portal seamlessly integrated with our EMR (Figure 2).24,27
Table 1.
Impact of Strong and Moderate CYP2D6 Inhibitors on Genotype-Based Activity Scores
| Phenoconversion Table* | |
|---|---|
| Inhibitors | Genotype-Based Activity Score Adjustment |
| Strong: Bupropion, Fluoxetine, Paroxetine, Quinidine, Perhexiline | x 0 |
| Moderate: Abiraterone, Cinacalcet, Clobazam, Doxepin, Duloxetine, Halofantrine, Lorcaserin, Moclobemide, Rolapitant, Terbinafine | x 0.5 |
Note: *Adopted from the Indiana University School of Medicine Cytochrome P450 Drug Interaction Table Available at: https://drug-interactions.medicine.iu.edu/MainTableaspx (Accessed: 10.31.2024).
Figure 2.
Representative Pharmacogenomic Clinical Decision Support (CDS) Summary. Structure and key components of a representative pharmacogenomic clinical decision support summary that will be delivered to treating providers via the electronic medical record within prescribing workflows to inform opioid selection during the trial.
Intervention
This study will implement a novel PGx-guided approach to opioid therapy for patients randomized to the PGx-guided arm. Clinicians will access genotype-based/phenoconversion-adjusted therapeutic recommendations for opioids via the institutional pharmacogenomic portal GPS. The GPS portal will serve as a one-stop hub, displaying tailored drug options, supporting references, and a customized traffic light symbol (green/yellow/red) indicating PGx risk for each genomic result. This comprehensive information empowers clinicians to make informed treatment decisions, while patients in the control arm will receive treatment according to standard care procedures without access to genotype information. Ultimately, all treatment decisions will be made at the sole discretion of the treating clinician.
Endpoints
The primary objective of this study is to assess the impact of upfront pharmacogenomic testing on cancer pain control. Accordingly, the primary endpoint is composite pain intensity rating during the first 45 days after an index opioid prescription for codeine, tramadol, or hydrocodone. Additionally, the study will evaluate the safety of pharmacogenomic clinical decision support guidance by comparing severe opioid-related adverse event rates between the two study arms. Secondary endpoints are: 1) pain medication regimen changes, 2) rates of hospitalization or emergency room visits, 3) cumulative morphine dose equivalents received, 4) type of first opioid prescribed. As exploratory endpoints, 1) clinicians’ use and dosing of additional PGx informed supportive care medications, 2) subject reported quality of life, 3) subject understanding of pharmacogenomics are investigated. Furthermore, patients receiving oxycodone as the first-line medication will be analyzed post-hoc for exploratory purposes.
Assessment
Composite Pain Outcomes
The Composite Pain Intensity will represent a singular score that amalgamates the worst, least, average, and current pain scores, calculated by taking their mean. Each score will be assessed biweekly using numeric rating scale from Brief Pain Inventory-Short Form (BPI-SF) by patients during the first 45 days after an index opioid prescription of codeine, tramadol, or hydrocodone.31 The difference in Composite Pain Intensity between the PGx and control arms will be compared as the primary outcome of our study. Cancer pain will be differentiated from neuropathic pain using the modified Douleur Neuropathique 4 (DN4) questionnaire.32 This methodology aims to focus on pain directly arising from cancer tumor burden/cancer progression, as opposed to neuropathic pain, which is more commonly caused by chemotherapeutic effects in cancer patients and which likely has a different underlying pathophysiology and often follows different treatment approaches. The patient survey will importantly collect data on the use of non-opioid analgesics and other pain management substances, including acetaminophen, gabapentin, pregabalin, nonsteroidal anti-inflammatory drugs (NSAIDs), muscle relaxants, and cannabinoids, to account for adjunctive therapies commonly used in cancer pain management.
Drug Adverse Event Outcomes
Adverse events of special interest (AESIs) that are potentially related to opioid medication use will be recorded for all randomized subjects, with dates, and will be updated on a continual basis for each subject during the course of medical care from the time of first opioid prescription through 45 days of observation. These potentially opioid-related AESIs are defined below. AESIs will be identified by review of subject medical records by a member or members of the research team who is blinded to the patient’s study arm (assessor blinded methodology). The blinded study research staff will assign a grade based on Common Terminology Criteria for Adverse Events (CTCAE) v5.0, and will assign a corresponding attribution based on likelihood of relation of the AESI(s) to opioid medication(s). The following potentially opioid-related AESIs are designated for this study and will be tracked: nausea, constipation, somnolence, confusion, dry mouth, pruritus, vomiting, and dizziness.
Pain Medication Regimen Changes/Augmentations
To determine the extent to which providing comprehensive PGx information impacts opioid pain treatment, pain medication regimen changes will be assessed for patients who were taking an opioid metabolized primarily by CYP2D6 and were subsequently: 1) switched to a non-opioid analgesic, 2) switched to an opioid that is not metabolized as much by CYP2D6, and 3) augmented with additional opioid/non-opioid analgesic as combination therapy. The timing of subsequent changes in pain regimes after initial opioid selection will also be recorded.
Hospitalization or Emergency Visit for Pain Control
As a secondary endpoint of our study, we will examine the rates of hospitalization and emergency room visits for pain. Prior studies have shown that inadequate pain control in cancer patients is associated with increased utilization of health care resources, such as emergency department visits, hospitalizations, and pain consultations. This increased utilization can have a negative impact on the quality of life of cancer patients and can lead to increased health care costs.33,34 To fully examine the impact of pharmacogenomics (PGx) on pain management in cancer patients, we will capture the number and type of hospital visits related to pain, including hospitalizations for pain, pain clinic visits, pain-related procedures (such as joint and epidural injections), and emergency department visits for pain during the study observation period.
Cumulative Morphine Equivalents Required
Cumulative Morphine Equivalents will be defined as total amount of opioids taken by a patient that is converted into its morphine equivalent. Dose and frequency of each opioid given during 45 days after its index prescription will be considered during calculation to give final values, which will be compared to Cumulative Morphine Equivalents for control arms treated per standard of care.
Type of First Opioid Prescribed
Because the study allows (and in fact desires) enrollment of subjects who are likely to require near-future opioid use, in some cases it is possible that pharmacogenomic (PGx) information will be available for subjects in the intervention arm for treating providers prior to the time of deciding on a first opioid prescription. It is therefore possible that, based on this PGx information, providers may choose to avoid a given index opioid based on PGx guidance and instead prescribe a non-CYP2D6 metabolized opioid as the first opioid prescription. Such subjects, because of the design of the study as being triggered by the start of an “index opioid”, may thus never become part of the evaluable study population. These subjects, however, may nevertheless comprise a highly informative sub-population of this study regarding the potential influence of PGx on opioid prescribing. Therefore, to assess the influence of PGx information on first opioid prescription patterns, the type of first opioid prescribed will be recorded for all randomized subjects. We will compare the rates of individual opioid prescription types between the PGx and control arms to evaluate potential differences influenced by pharmacogenomic data.
Data Analysis
Data Collection
Patient data will be collected through electronic medical records (EMR) and a customized patient survey. The survey will include validated instruments like BPI-SF, EuroQoL 5-Dimension 3-Level (EQ-5D-3L), and a modified DN-4, along with additional internally and externally tested questionnaires.19,31,32,35 Surveys will be administered at baseline and every 14 days during the 45-day evaluation period (with a flexibility of ±7 days) either in person or via telephone or email, and they will be captured and managed using REDCap electronic data tools hosted at the University of Chicago.
Statistics
Primary Endpoint Analysis
The primary objective is the comparison of the composite pain intensities recorded over a 45-day evaluation period since index prescription, among CYP2D6 IM/PM patients. The longitudinal data will be analyzed by fitting mixed effects regression models to compare the changes over time between the PGx and control arms. These models allow for varying numbers and spacing of measurements and take into account correlation within subjects. Treatment arm, time of assessment, and treatment arm-by-time interaction terms will constitute the fixed effects, and subjects will form a random effect. Quadratic terms (in time) will be added if a linear model does not fit the data.
For the sample-size calculation, we consider the change from baseline (defined as time of index prescription) to 45 days. Smith et al found in their study of 51 CYP2D6-guided patients and 19 usual care patients a mean change over 3 months of −0.84 and −0.12 points, respectively, with a pooled standard deviation of 1.4.17 We regard a minimally meaningful difference between the two groups to be 1 point on the 10-point numeric rating scale (NRS), that is, a change from baseline that is 1 point greater in the PGx arm compared to the control group. While this effect size is slightly larger than that observed by Smith et al, it is plausible because our patient population (unlike the Smith et al study) will not have chronic pain syndromes and will be currently opioid-naive, allowing larger detectable effects on index prescriptions. A sample size of 64 patients (32 per group) will provide 80% power, based on a two-sided t-test at the 0.05 alpha level. Assuming a 10% IM/PM prevalence, a total of 640 evaluable subjects are needed for this aim. To achieve 640 evaluable subjects (those receiving index prescriptions of codeine, tramadol, or hydrocodone during the 5-year study period), we estimated based on preliminary data that 800 total cancer patients will need to be enrolled and randomized.
Safety Interim Analysis
From a safety standpoint for this study, while we hypothesize that the delivery of PGx information is most likely to improve patient care by promoting optimization and customization of medication choices, we nevertheless acknowledge that it is possible that PGx-guided clinical decision support could potentially influence providers to make medication choices that could result in unintended harm (eg, upfront use of more potent opioids with the potential for increased toxicity). Thus, to ensure that the use of PGx information for pain prescribing does not result in a significant increase in opioid-related AESIs, an interim analysis (IA) for the purpose of evaluating safety will be performed after the first 400 randomized subjects are enrolled. Based on a prior comprehensive review of opioid pain treatment studies, the expected rate of a “severe” opioid-related AESI (defined as a grade 3 or higher opioid-related AESI; or an AE resulting in opioid discontinuation) is approximately 15%.36,37 The IA will assess the severe AESI rate among the first 400 randomized subjects (approximately 200 subjects per arm), comparing the rate among intervention arm subjects with the rate among control arm subjects. If there is a significant difference in the severe AESI rate in the intervention arm (defined as an absolute increase of greater than or equal to 15%), then the research team will immediately halt enrollment to the study and convene a meeting of the entire study team (a quorum of all investigators named on the NIH award supporting this study, including mandatory participation in this meeting by the independent clinical study monitor who is the Associate Director for Clinical Investigation of the University of Chicago Cancer Center). The study PI will also consult with the Chair of the IRB. These meetings will provide a consensus recommendation to either (a) discontinue the study permanently due to unacceptable risk/benefit; or (b) implement additional modifications to the study protocol to mitigate the observed risks, changes, which would require IRB approval before the study may proceed. If, however, the IA reveals no significant difference in the severe AESI rates, the study will continue as planned.
Secondary Analysis
Finally, in addition to the above primary analyses, the study will evaluate several secondary endpoints to comprehensively assess the impact of PGx information on pain management strategies. First, we will evaluate the proportion and timing of any subsequent pain medication regimen changes/augmentations beyond the index prescription, compared between arms. We hypothesize that the average time to switching/dose altering the index medication will be shorter in the PGX Arm for IM/PMs (because of upfront PGx guidance recommending consideration of a non-CYP2D6 opioid), compared to IM/PMs in the control arm. With 32 evaluable patients in each arm, and an estimated standard deviation of 10 days, the study will also have 85% power to detect a difference of 14 vs 22 days in the time to switching to an alternative dose or a non-CYP2D6 opioid, using a two-sided t-test with alpha of 0.05. Also, we will capture the incidence (number of visits over 24 months of follow-up) of pain-related hospitalization/emergency visits for each group: PG IM/PM patients, control IM/PMs, PGx NMs, and control NMs. Incidence will be compared between IM/PM PGx Vs IM/PM controls, and NM PG vs NM controls, using Wilcoxon rank-sum tests. Poison regression analyses will be performed to evaluate the relationship between pain-related hospitalization/emergency visits and treatment arm, adjusting for demographic and clinical factors including sex. Separately, among all randomized subjects (including those who never start any index opioid), the type of first opioid prescribed will be analyzed to assess the potential influence of PGx information on initial opioid selection. We will formally compare the frequency of CYP2D6 substrate vs non-substrate opioids prescribed initially between the PGx and control arms. Finally, while provider contamination cannot be eliminated given randomization at the patient level (ie, treating providers may be nonspecifically influenced for controls because of the information they receive on PGx patients), the large number of providers being studied reduces this risk. Nevertheless, contamination or “spillover” effects will be measured by fitting a mixed effects model to include random provider effects and a “spillover effect” covariate equal to the number of prior actionable PGx Arm patients seen.
Conclusion
Opioids remain a mainstay for cancer pain management, yet individual responses and adverse effects may vary considerably.38 Emerging evidence implicates CYDP2D6 polymorphisms as key contributors to this variability for specific opioids (eg, codeine, tramadol, and hydrocodone).17,19,38 While preemptive CYDP2D6 genotyping has shown promise in different clinic settings for personalized opioid dosing and adverse events, its application in cancer-specific pain management remains underexplored. To our knowledge, there have been only three earlier studies that have examined the potential of PGx in cancer pain management, but their scopes have been limited to specific opioids or lacked robust methodologies.20–22 Our prospectively designed, randomized trial fills this gap by investigating the clinical efficacy of preemptive CYP2D6 genotyping with a wider range of opioids in a large cancer population. This comprehensive design will allow us to demonstrate the impact of CYP2D6 genotyping on pain control and opioid dosage needs in the context of cancer pain management. By providing actionable insights, our study aims to facilitate the integration of CYP2D6 genotyping into routine clinical practice, ultimately optimizing opioid therapy for cancer patients.
Acknowledgment
The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the NIH 1R01HG012273-01 (P. H. O’Donnell as PI), the University of Chicago Committee on Clinical Pharmacology and Pharmacogenomics T32 training grant NIH 5T32GM007019 (Y. Cho as trainee), and the Benjamin McAllister Research Fellowship Award (Y. Cho as trainee).
Ethics and Dissemination
This trial adheres to the principles of the Declaration of Helsinki and is registered with ClinicalTrials.gov (NCT06511401). The study has been approved by an independent review board within the University of Chicago Medicine (Date of Letter: 7/10/2024, Protocol Number: IRB24-0700). An independent review board within the University of Chicago Medicine will oversee the study’s procedures, ensuring participant safety and informed decision-making. The protocol complies with Good Clinical Practice, and all patients will provide written informed consent to participate in this trial.
Disclosure
Dr Russell Szmulewitz reports grants from Bayer, grants from Pfizer, personal fees from Novartis, outside the submitted work. Dr Everett Vokes reports personal fees from AbbVie, personal fees, non-financial support from AstraZeneca, personal fees from BioNTech, outside the submitted work. Dr Mark Ratain reports grants from National Institutes of Health, during the conduct of the study. Dr Peter O’Donnell reports grants from National Institutes of Health, during the conduct of the study; personal fees from National Institutes of Health, personal fees from O’Brien and Ryan LLP, personal fees from ISMIE, outside the submitted work. The authors report no other conflicts of interest in this work.
References
- 1.van den Beuken-van Everdingen MH, Hochstenbach LMJ, Joosten EAJ, et al. Update on prevalence of pain in patients with cancer: systematic review and meta-analysis. J Pain Symptom Manage. 2016;51(6):1070–1090e9. doi: 10.1016/j.jpainsymman.2015.12.340 [DOI] [PubMed] [Google Scholar]
- 2.Roberto A, Greco MT, Uggeri S, et al. Living systematic review to assess the analgesic undertreatment in cancer patients. Pain Pract. 2022;22(4):487–496. doi: 10.1111/papr.13098 [DOI] [PubMed] [Google Scholar]
- 3.Kleine-Brueggeney M, Musshoff F, Stuber F, et al. Pharmacogenetics in palliative care. Forensic Sci Int. 2010;203(1–3):63–70. doi: 10.1016/j.forsciint.2010.07.003 [DOI] [PubMed] [Google Scholar]
- 4.Bruera E, Paice JA. Cancer pain management: safe and effective use of opioids. Am Soc Clin Oncol Educ Book. 2015;e593–9. doi: 10.14694/EdBook_AM.2015.35.e593 [DOI] [PubMed] [Google Scholar]
- 5.Mestdagh F, Steyaert A, Lavand’homme P. Cancer pain management: a narrative review of current concepts, strategies, and techniques. Curr Oncol. 2023;30(7):6838–6858. doi: 10.3390/curroncol30070500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dean L. Codeine therapy and CYP2D6 genotype. In: Pratt VM, Scott SA, Pirmohamed M, Esquivel B, Kane, MA, editor. Medical Genetics Summaries. Bethesda (MD): National Center for Biotechnology Information (US); 2012. [PubMed] [Google Scholar]
- 7.Del Tredici AL, Malhotra A, Dedek M, et al. Frequency of CYP2D6 alleles including structural variants in the United States. Front Pharmacol. 2018;9:305. doi: 10.3389/fphar.2018.00305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crews KR, Monte AA, Huddart R, et al. Clinical Pharmacogenetics Implementation Consortium Guideline for CYP2D6, OPRM1, and COMT Genotypes and Select Opioid Therapy. Clin Pharmacol Ther. 2021;110(4):888–896. doi: 10.1002/cpt.2149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith HS. Opioid metabolism. Mayo Clin Proc. 2009;84(7):613–624. doi: 10.1016/S0025-6196(11)60750-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raffa RB. On subclasses of opioid analgesics. Curr Med Res Opin. 2014;30(12):2579–2584. doi: 10.1185/03007995.2014.952717 [DOI] [PubMed] [Google Scholar]
- 11.Subedi M, Bajaj S, Kumar MS, et al. An overview of tramadol and its usage in pain management and future perspective. Biomed Pharmacother. 2019;111:443–451. doi: 10.1016/j.biopha.2018.12.085 [DOI] [PubMed] [Google Scholar]
- 12.Stauble ME, Moore AW, Langman LJ, et al. Hydrocodone in postoperative personalized pain management: pro-drug or drug? Clin Chim Acta. 2014;429:26–29. doi: 10.1016/j.cca.2013.11.015 [DOI] [PubMed] [Google Scholar]
- 13.Cavallari LH, Johnson JA. A case for genotype-guided pain management. Pharmacogenomics. 2019;20(10):705–708. doi: 10.2217/pgs-2019-0068 [DOI] [PubMed] [Google Scholar]
- 14.Matic M, Nijenhuis M, Soree B, et al. Dutch Pharmacogenetics Working Group (DPWG) guideline for the gene-drug interaction between CYP2D6 and opioids (codeine, tramadol and oxycodone). Eur J Hum Genet. 2022;30(10):1105–1113. doi: 10.1038/s41431-021-00920-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reizine N, Danahey K, Schierer E, et al. Impact of CYP2D6 pharmacogenomic status on pain control among opioid-treated oncology patients. Oncologist. 2021;26(11):e2042–e2052. doi: 10.1002/onco.13953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dagostino C, Allegri M, Napolioni V, et al. CYP2D6 genotype can help to predict effectiveness and safety during opioid treatment for chronic low back pain: results from a retrospective study in an Italian cohort. Pharmgenomics Pers Med. 2018;11:179–191. doi: 10.2147/PGPM.S181334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith DM, Weitzel KW, Elsey AR, et al. CYP2D6-guided opioid therapy improves pain control in CYP2D6 intermediate and poor metabolizers: a pragmatic clinical trial. Genet Med. 2019;21(8):1842–1850. doi: 10.1038/s41436-018-0431-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thomas CD, Parvataneni HK, Gray CF, et al. A hybrid implementation-effectiveness randomized trial of CYP2D6-guided postoperative pain management. Genet Med. 2021;23(4):621–628. doi: 10.1038/s41436-020-01050-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cavallari LH, Cicali E, Wiisanen K, et al. Implementing a pragmatic clinical trial to tailor opioids for acute pain on behalf of the IGNITE ADOPT PGx investigators. Clin Transl Sci. 2022;15(10):2479–2492. doi: 10.1111/cts.13376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andreassen TN, Eftedal I, Klepstad P, et al. Do CYP2D6 genotypes reflect oxycodone requirements for cancer patients treated for cancer pain? A cross-sectional multicentre study. Eur J Clin Pharmacol. 2012;68(1):55–64. doi: 10.1007/s00228-011-1093-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mosley SA, Hicks JK, Portman DG, et al. Design and rational for the precision medicine guided treatment for cancer pain pragmatic clinical trial. Contemp Clin Trials. 2018;68:7–13. doi: 10.1016/j.cct.2018.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Patel JN, Boselli D, Hamadeh IS, et al. Pain management using clinical pharmacy assessments with and without pharmacogenomics in an oncology palliative medicine clinic. JCO Oncol Pract. 2020;16(2):e166–e174. doi: 10.1200/JOP.19.00206 [DOI] [PubMed] [Google Scholar]
- 23.Kane M. Oxycodone Therapy and CYP2D6 Genotype. In: Pratt VM, Scott SA, Pirmohamed M, Esquivel B, editor. Medical Genetics Summaries. Bethesda (MD): National Center for Biotechnology Information (US); 2012. [PubMed] [Google Scholar]
- 24.Danahey K, Borden BA, Furner B, et al. Simplifying the use of pharmacogenomics in clinical practice: building the genomic prescribing system. J Biomed Inform. 2017;75:110–121. doi: 10.1016/j.jbi.2017.09.012 [DOI] [PubMed] [Google Scholar]
- 25.Truong TM, Apfelbaum J, Shahul S, et al. The ImPre SS trial: implementation of point-of-care pharmacogenomic decision support in perioperative care. Clin Pharmacol Ther. 2019;106(6):1179–1183. doi: 10.1002/cpt.1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reizine N, Vokes EE, Liu P, et al. Implementation of pharmacogenomic testing in oncology care (PhOCus): study protocol of a pragmatic, randomized clinical trial. Ther Adv Med Oncol. 2020;12:1758835920974118. doi: 10.1177/1758835920974118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O’Donnell PH, Bush A, Spitz J, et al. The 1200 patients project: creating a new medical model system for clinical implementation of pharmacogenomics. Clin Pharmacol Ther. 2012;92(4):446–449. doi: 10.1038/clpt.2012.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fang H, Liu X, Ramírez J, et al. Establishment of CYP2D6 reference samples by multiple validated genotyping platforms. Pharmacogenomics J. 2014;14(6):564–572. doi: 10.1038/tpj.2014.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leung EKY, Agolini E, Pei X, et al. Validation of an extensive CYP2D6 assay panel based on invader and TaqMan copy number assays. J Appl Lab Med. 2017;1(5):471–482. doi: 10.1373/jalm.2016.021923 [DOI] [PubMed] [Google Scholar]
- 30.Nahid NA, Johnson JA. CYP2D6 pharmacogenetics and phenoconversion in personalized medicine. Expert Opin Drug Metab Toxicol. 2022;18(11):769–785. doi: 10.1080/17425255.2022.2160317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mathias SD, Crosby RD, Qian Y, et al. Estimating minimally important differences for the worst pain rating of the brief pain inventory-short form. J Support Oncol. 2011;9(2):72–78. doi: 10.1016/j.suponc.2010.12.004 [DOI] [PubMed] [Google Scholar]
- 32.Bouhassira D, Attal N, Alchaar H, et al. Comparison of pain syndromes associated with nervous or somatic lesions and development of a new neuropathic pain diagnostic questionnaire (DN4). Pain. 2005;114(1–2):29–36. doi: 10.1016/j.pain.2004.12.010 [DOI] [PubMed] [Google Scholar]
- 33.Whitney RL, Bell JF, Tancredi DJ, et al. Unplanned hospitalization among individuals with cancer in the year after diagnosis. J Oncol Pract. 2019;15(1):e20–e29. doi: 10.1200/JOP.18.00254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Whitney RL, Bell JF, Tancredi DJ, et al. Hospitalization rates and predictors of rehospitalization among individuals with advanced cancer in the year after diagnosis. J Clin Oncol. 2017;35(31):3610–3617. doi: 10.1200/JCO.2017.72.4963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rabin R, de Charro F. EQ-5D: a measure of health status from the EuroQol Group. Ann Med. 2001;33(5):337–343. doi: 10.3109/07853890109002087 [DOI] [PubMed] [Google Scholar]
- 36.Corli O, Santucci C, Corsi N, Radrezza S, Galli F, Bosetti C. The burden of opioid adverse events and the influence on cancer patients’ symptomatology. J Pain Symptom Manage. 2019;57(5):899–908e6. doi: 10.1016/j.jpainsymman.2019.02.009 [DOI] [PubMed] [Google Scholar]
- 37.Moore RA, McQuay HJ. Prevalence of opioid adverse events in chronic non-malignant pain: systematic review of randomised trials of oral opioids. Arthritis Res Ther. 2005;7(5):R1046–51. doi: 10.1186/ar1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bugada D, Lorini LF, Fumagalli R, et al. Genetics and opioids: towards more appropriate prescription in cancer pain. Cancers. 2020;12(7):1951. doi: 10.3390/cancers12071951 [DOI] [PMC free article] [PubMed] [Google Scholar]