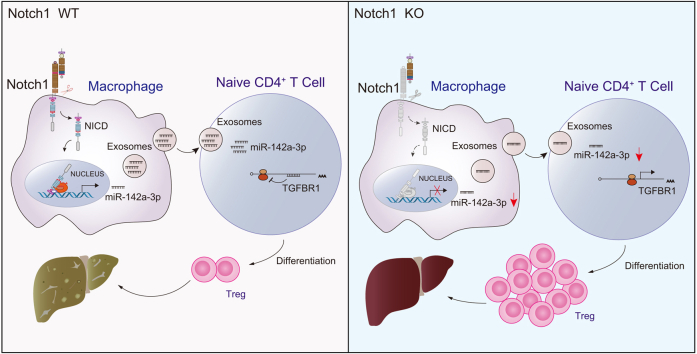
Abstract
Background & Aims
Hepatic immune imbalance is crucial for driving metabolic dysfunction-associated steatotic liver disease (MASLD) progression. However, the role of hepatic regulatory T cells (Tregs) in MASLD initiation and the mechanisms responsible for their change are not completely understood.
Methods
A mouse model subjected to a short-term high-fat diet (HFD) to mimic early steatosis, along with liver biopsy samples from patients with simple steatosis, and macrophage-specific Notch1-knockout mice (Notch1M-KO), were used to investigate the role of Tregs in early MASLD and the effect of hepatic macrophage Notch1 signaling on Treg frequency. The miRNAs correlated with Treg differentiation were analyzed using exosomal miRNA sequencing.
Results
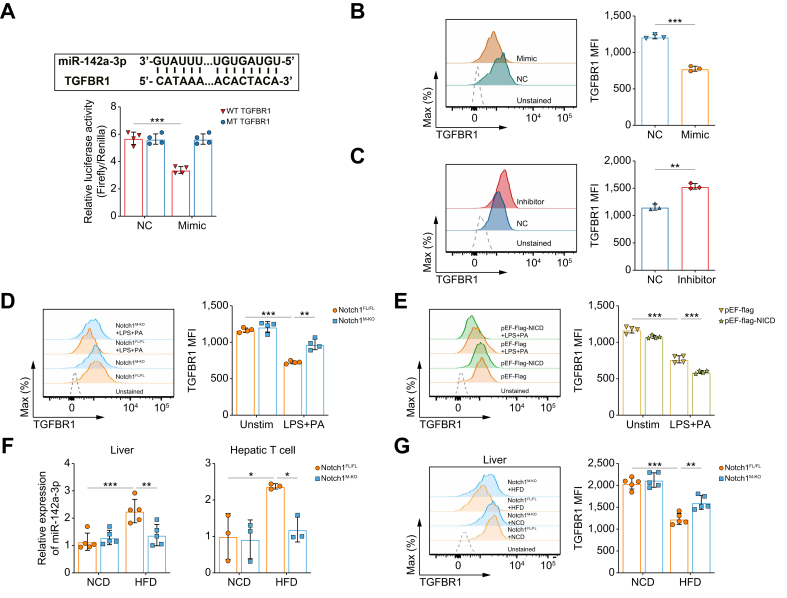
A decrease in Tregs contributed to HFD-induced hepatic steatosis and insulin resistance (five/group/time point, p <0.001). Remarkably, the frequency of Tregs was negatively correlated with Notch1 activation in hepatic macrophages during hepatic steatosis (38/group, r = -0.735, p <0.001). Furthermore, Notch1 deficiency attenuated hepatic lipid deposition and reversed Treg levels (five/group, p <0.01 and <0.05, respectively). Moreover, Treg depletion in Notch1M-KO mice greatly diminished the ameliorative effect of macrophagic Notch1 deletion on hepatic steatosis. Mechanistically, macrophage Notch1 activation increased the level of exosomal miR-142a-3p (by one- to two- fold), impairing Treg differentiation by targeting transforming growth factor beta receptor 1 (TGFBR1) on T cells. Consistently, HFD-fed Notch1M-KO mice exhibited reduced miR-142a-3p levels, elevated TGFBR1 expression on T cells, and increased Treg frequency in the liver.
Conclusions
These findings highlight the crucial role of hepatic Tregs during the early stage of MASLD and add a novel, non-negligible pathway for macrophage involvement in hepatic steatosis. We identify a previously unrecognized molecular mechanism involving the macrophage Notch1/exosomal miR-142a-3p/TGFBR1 pathway in regulating Treg differentiation, providing a rationale for refined therapeutic strategies for MASLD.
Impact and implications:
The immune mechanisms driving MASLD progression, particularly during the early stages of disease, are not fully understood, which limits the development of effective interventions. This study elucidated a novel mechanism by which hepatic macrophage Notch1 signaling modulated Tregs through the exosomal miR-142a-3p/TGFBR1 axis, contributing to the progression of MASLD. These findings provide a rationale for a potential immunological approach to treat MASLD in the future.
Keywords: Hepatic Tregs, Exosomes, Macrophages, Notch1, Hepatic steatosis
Graphical abstract
Highlights:
-
•
Decreased Tregs contribute to HFD-induced hepatic steatosis and insulin resistance.
-
•
Notch1 deficiency in hepatic macrophages reduces lipid accumulation and restores Tregs.
-
•
Notch1-regulated Exos-miR-142a-3p from macrophages hinders Treg production in hepatic steatosis.
Introduction
Metabolic dysfunction-associated steatotic liver disease (MASLD), formerly known as nonalcoholic fatty liver disease, initiating from simple steatosis to steatohepatitis (metabolic dysfunction-associated steatohepatitis; MASH), ultimately leading to fibrosis, and hepatocellular carcinoma, has emerged as the leading cause of gastrointestinal diseases, affecting ∼25% of the population worldwide.1 Nevertheless, there are currently no FDA-approved pharmacological therapies for MASLD, given that the molecular mechanisms involved have not been fully elucidated, highlighting the need for deeper insights into the mechanisms underlying MASLD and the identification of novel therapeutic strategies.
Several hypotheses have been proposed regarding the pathogenesis of MASLD, from ‘2-hit’ to ‘multi-hit’, which involve lipotoxicity, oxidative stress, mitochondrial injury, immune imbalance, and inflammatory cytokine production.2 Among these factors, immune imbalance is a crucial element in driving MASLD progression.3 While pronounced inflammation may only be observed during MASH, profound alterations of the hepatic immune system occurring even during steatosis, indicating the involvement of the immune system across the entire MASLD spectrum.4 Regulatory T cells (Tregs), a distinct lineage of CD4+ T lymphocytes, have a crucial role in peripheral immune tolerance, especially in the liver.5 The fine-tuning of Tregs is thought to contribute to the development of various liver diseases.6,7 Previous studies found that the frequency of hepatic Tregs is reduced during MASH,8 but increased during its premalignant stage.9 These findings suggested the Tregs undergo dynamic changes during MASLD progression. Recently, adipose Tregs were shown to directly modulate lipid metabolism.10 Hepatic steatosis is generally considered to be a key initiating event in MASLD;11 however, the role of hepatic Tregs in hepatic steatosis is not completely clear.
The liver is a vital place for peripherally induced Tregs because it has unique tolerogenic properties that favor the differentiation of antigen-specific Tregs.12 The production of hepatic Tregs depends on multiple cells, including macrophages.12 In particular, given the high plasticity of macrophages, different macrophage phenotypes exert diverse effects on Treg differentiation. Under homeostatic conditions, macrophages can induce CD4+ T cells to differentiate into Tregs to maintain hepatic immune tolerance. However, this differentiation is impaired when macrophages display a proinflammatory phenotype.13 Of note, previous studies demonstrated that hepatic macrophages preferentially polarize into a proinflammatory phenotype and recruit more proinflammatory cells into the liver during the initiation and development of MASLD.14,15 Nevertheless, whether and how macrophages regulate Tregs in initiating MASLD remain elusive.
Exosomes (Exos), which have emerged as crucial mediators of intercellular communication, carry and deliver miRNAs, proteins, and metabolites from host cells to recipient cells.16 Their contents are tightly regulated by physiological and pathological stimuli and highly dependent on the cell type.17 Remarkably, Notch1 signaling has been reported to regulate miRNA expression and Exo release.18,19 In addition, emerging reports indicate that Notch signaling has a crucial role in both innate and adaptive immunity,20,21 and the Notch1 activation-mediated proinflammatory transforming effect of macrophages is involved in multiple chronic inflammatory diseases.22,23 Thus, Notch1 signaling could be a potential modulating machinery for the crosstalk between macrophages and Tregs.
In the present study, we revealed the crucial role of hepatic Tregs and their correlation with Notch1 activation in hepatic macrophages during the early stages of MASLD. Using macrophage-specific Notch1-knockout (Notch1M-KO) mice, we identified that macrophage-derived Exo-miR-142a-3p regulated by Notch1 impedes Treg production during hepatic steatosis. Our findings provide novel molecular insights into macrophage–Treg interactions, targeting of which might offer a potential strategy for preventing MASLD.
Materials and methods
Animals
Floxed Notch1 (Notch1FL/FL) mice (Jackson Laboratory, Bar Harbor, ME, USA) and Lyz2-Cre mice (LysM-Cre; Jackson Laboratory) were used to generate Notch1M-KO mice. Mouse genotyping was performed by PCR of the tail DNA (Fig. S1). Male Notch1FL/FL and Notch1M-KO mice aged 8 weeks were used in the experiments.
Methodological details are provided in the Supplementary information online.
Isolation of Exos
Exo purification was achieved through differential ultracentrifugation. Briefly, the supernatant was collected after culturing bone marrow-derived macrophages (BMDMs) in Exo-free medium for 48 h. Exos were isolated via five sequential centrifugation steps at 4 °C: (1) 10 min at 300 × g to remove cells; (2) 10 min at 2,000 × g to remove cell debris; (3) 30 min at 10,000 × g to break organelles; (4) ultracentrifugation at 100,000 × g for 120 min to pellet exosomes; and (5) washing with a large amount of ice-cold PBS and ultracentrifuge at 100,000 g for 120 min. Exos were then resuspended in PBS and stored at −80 °C until use.
miRNA sequencing and data analysis
miRNA sequencing was done on RNA from BMDMs-Exos by Novogene (Beijing, China). Briefly, 2 μg of total RNA was used to prepare the miRNA library after checking the RNA quality and integrity. Then, miRNA sequencing was performed on TruSeq SR Cluster Kit v3-cBot-HS (Illumina) following the manufacturer’s instructions. Differential expression analysis of the 2 groups was performed using the DESeq R package (version 1.24.0; R Foundation for Statistical Computing, Vienna, Austria). Sequencing data have been deposited in the SRA database (www.ncbi.nlm.nih.gov/sra) with the following accession number: PRJNA1046700.
Details of other methods are described in the Supplementary information online.
Results
Reduction in hepatic Tregs contributes to HFD-induced hepatic steatosis and insulin resistance
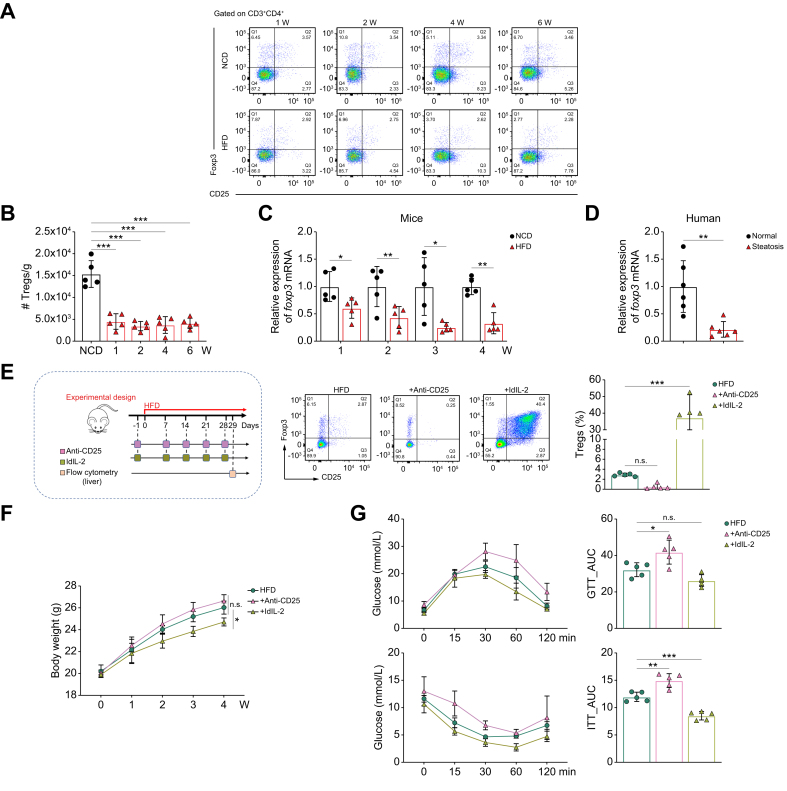
To evaluate the changes in Tregs during the initiation of MASLD, we analyzed the proportion and number of hepatic CD4+CD25+Foxp3+ Tregs and the mRNA level of foxp3, a specific marker of Tregs, in liver tissues from HFD-fed mice at 1, 2, 4, and 6 weeks after the start of HFD feeding. Compared with the normal chow diet (NCD) group, the decline in hepatic CD4+CD25+Foxp3+ Tregs was obvious, starting at 1 week and continuing until 6 weeks of HFD feeding, as determined by flow cytometry and quantitative reverse transcription (RT-qPCR) (Fig. 1A–C). As expected, multiple indices of HFD mice, including body weight, serum lipid levels, and intrahepatic lipid content, were higher than those of the NCD group (Fig. S2). Consistent with the murine model, fewer Tregs were observed in the steatotic human liver compared with the control group (Fig. 1D).
Fig. 1.
Frequency of Tregs is associated with HFD-induced hepatic steatosis and insulin resistance.
(A) Representative flow plots of the proportion of hepatic CD25+Foxp3+ Tregs gated on CD3+CD4+ T cells from mice fed a NCD or HFD (—four to five/group). (B) Number of hepatic CD4+CD25+Foxp3+ Tregs of mice fed a NCD or HFD for 1, 2, 4, and 6 weeks (five/group). (C, D) Foxp3 mRNA expression of liver tissues from (C) the NCD or HFD group (five/group), and (D) patients with or without hepatic steatosis (six/group). (E) Schematic of the construction of Treg-depleted mice (injected intraperitoneally with anti-CD25 antibodies) and Treg-expand mice (injected intraperitoneally with ldIL-2): representative dot plots of CD25+Foxp3+ Tregs gated on CD3+CD4+ T cells in the liver and a graph of the proportion of hepatic CD4+CD25+Foxp3+ Tregs (five/group). Comparison of (F) body weight (five/group), (G) GTT and ITT (five/group), (H) insulin-stimulated Akt and pAkt expression in the liver (three/group), (I) serum TC and total TG levels (five/group), and liver TC and TG levels (five/group) in Treg-depleted vs. Treg-expanded mice. (J) Representative images of H&E and Oil red O staining of liver sections (scale bars: 50 μm and 100 μm, respectively). (K) Representative images of Oil red O staining (scale bar: 100 μm) and TG content of primary hepatocytes co-cultured with Tregs or not (three/group). (L) Immunoblot analysis of insulin-stimulated Akt and pAkt in primary hepatocytes co-cultured with Tregs or not (three/group). Values represent means ± SD. Statistical analysis was performed by 2-tailed unpaired Student t test (B–D) or one-way ANOVA and Tukey's test (F–I,K,L): ∗p <0.05, ∗∗p <0.01, ∗∗∗p <0.001. Abbreviations: FFA, free fatty acid; GTT, glucose tolerance test; HFD, high-fat diet; ITT, insulin tolerance test; ldIL-2, low-dose IL-2; NCD, normal chow diet; pAkt, phosphorylated Akt; TC, total cholesterol; TG, triglyceride; Treg, regulatory T cell.
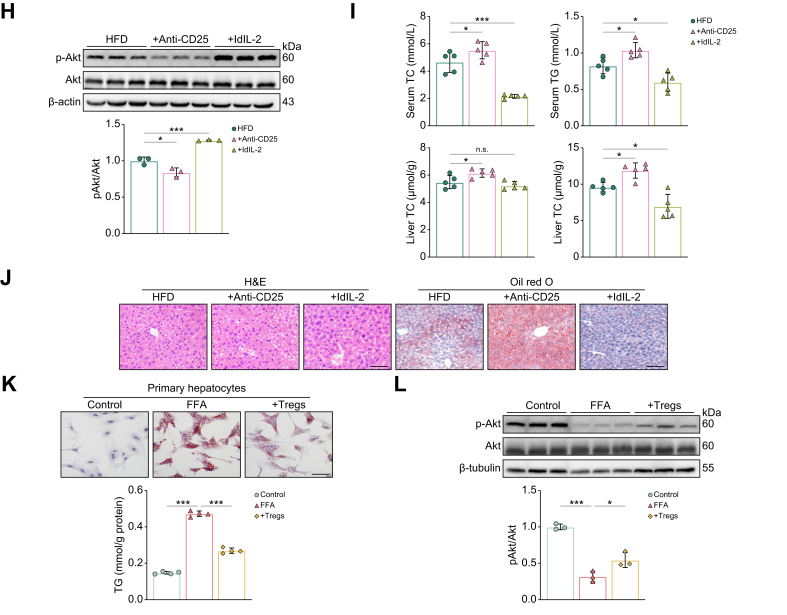
Next, to further clarify whether hepatic steatosis was directly correlated with the reduced Treg frequency, we constructed Treg-depleted mice and Treg-expanded mice by intraperitoneal injection of anti-CD25 antibodies24 and low-dose IL-2 (ldIL-2),25 respectively. There was a remarkable reduction in hepatic Tregs after anti-CD25 antibody administration, whereas ldIL-2 administration resulted in a significant increase in hepatic Tregs (Fig. 1E). After 4 weeks of HFD feeding, Treg-depleted mice showed higher body weight gain, whereas Treg-expanded mice showed lower body weight gain, although the difference was not significant (Fig. 1F). Glucose and insulin tolerance testing (GTT and ITT, respectively) showed that Treg-depleted mice had lower glucose tolerance and insulin sensitivity compared with HFD mice, whereas these phenomena were improved in Treg-expanded mice (Fig. 1G). Consistent with the systemic insulin resistance (IR), insulin signaling, as measured by the phosphorylation of Akt at S473 (pAkt), was impaired in the liver of Treg-depleted mice, but promoted in Treg-expanded mice (Fig. 1H). In addition, total cholesterol (TC) and triglyceride (TG) levels in the serum and liver were significantly decreased in Treg-expanded mice compared with HFD mice, whereas these indicators displayed the opposite trend in Treg-depleted mice (Fig. 1I). Accordingly, H&E and Oil red O staining of liver sections also reflected significant lipid accumulation in Treg-depleted mice, an effect that was markedly attenuated in Treg-expanded mice (Fig. 1J). These results suggested that Tregs influence both hepatic lipid homeostasis and IR.
Tregs are one of the major sources of IL-10 within the liver,26 which is an important protective factor against diet-induced hepatic steatosis and IR.27 ELISA results showed increased IL-10 production and reduced levels in Treg-depleted mice compared with HFD mice (Fig. S3A). Isolation of Tregs (Fig. S3B) and their co-culture assays with primary hepatocytes showed that Tregs significantly reduced free fatty acid (FFA)-induced lipid accumulation (Fig. 1K) and enhanced insulin-induced pAkt levels in hepatocytes (Fig. 1L). Neutralization of IL-10 confirmed that the ability of Tregs to ameliorate IR and reduce lipid deposition in hepatocytes was IL-10 dependent (Fig.S3C-F).
Aberrant activation of hepatic macrophage Notch1 signaling occurs in parallel with reduced hepatic Treg frequency in hepatic steatosis
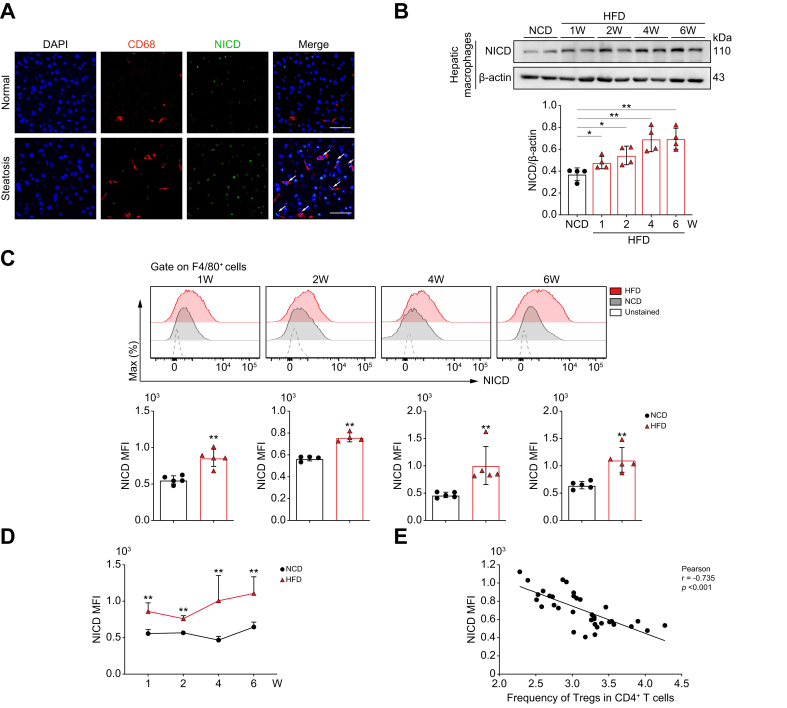
To address the reduction in hepatic Treg frequency, we interrogated the GEO database (GEO: GSE83452) and noted that the reduction in Tregs was accompanied by an increase in the proinflammatory macrophage population in patients with MASLD (Fig. S4A). Given the important role of Notch1 signaling in the proinflammatory transformation of macrophages, we examined the activation levels of Notch1 (Notch1 intracellular domain; NICD) in hepatic macrophages of patients with simple steatosis. Immunofluorescence (IF) staining of liver sections from patients with hepatic steatosis revealed that Notch1 activation was increased and predominantly located in macrophages compared with normal samples (Fig. 2A). Subsequently, we monitored the level of hepatic macrophage Notch1 activation during hepatic steatosis in mice by western blot and flow cytometry. Mice fed a HFD showed a significant and sustained increase in Notch1 activation in hepatic macrophages from 1 to 6 weeks of a HFD diet, compared to the NCD group (Fig. 2B–D). Interestingly, the level of macrophage Notch1 activation was negatively correlated with the frequency of CD4+CD25+Foxp3+ Tregs (Fig. 2E). These results suggested that Notch1 signaling activation in hepatic macrophages is involved in the reduced hepatic Treg frequency during the early stage of MASLD.
Fig. 2.
Hepatic macrophage Notch1 activation is negatively correlated with the frequency of hepatic CD4+CD25+Foxp3+ Tregs in hepatic steatosis.
(A) Representative pictures of IF staining for NICD in CD68+ cells from liver tissues of patients with or without hepatic steatosis (scale bar: 50 μm). WT C57BL/6 mice were fed with NCD or HFD. (B) Western blot analysis of NICD expression in hepatic macrophages isolated from mice fed a NCD or HFD (four 4/group). (C) Notch1 activation in hepatic macrophages was examined by flow cytometry at different time points (four or five/group). (D) Dynamic changes in the level of Notch1 activation in hepatic macrophages of mice fed a NCD or HFD for 1, 2, 4, and 6 weeks. (E) Relationship between the level of Notch1 activation in hepatic macrophages and frequency of hepatic CD4+CD25+Foxp3+ Tregs (38/group). Values represent means ± SD. Statistical analysis was performed by a 2-tailed unpaired Student t test: ∗p <0.05, ∗∗p <0.01. Abbreviations: HFD, high-fat diet; IF, immunofluorescence; MFI, mean fluorescence intensity; NCD, normal chow diet; NICD, Notch1 intracellular domain; Treg, regulatory T cell; WT, wild-type.
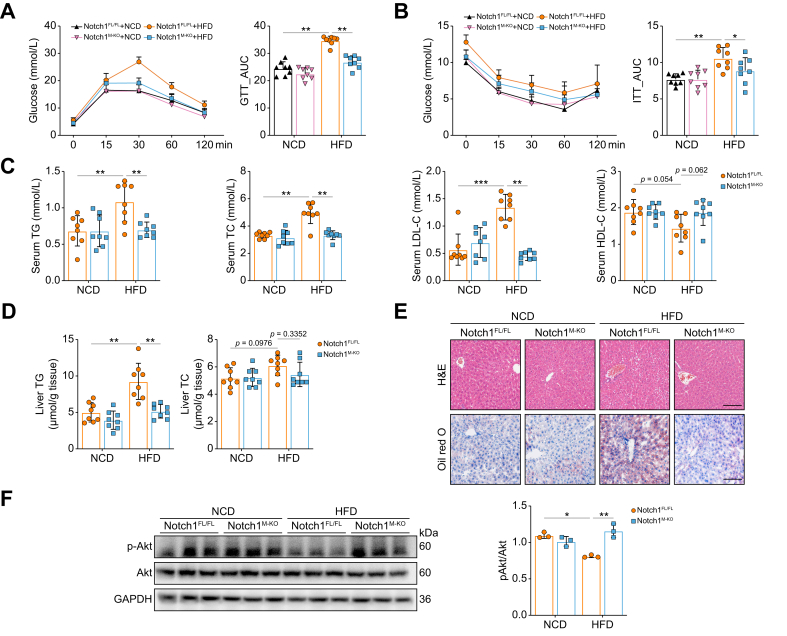
Deletion of Notch1 in macrophages ameliorates HFD-induced hepatic steatosis and insulin resistance
Next, we investigated the role of macrophage Notch1 in hepatic steatosis by feeding Notch1M-KO mice and Notch1FL/FL mice with HFD for 6 weeks. Notch1M-KO mice showed significantly slower body weight gain (Fig. S4C) compared with Notch1FL/FL mice. In addition, compared with Notch1FL/FL mice fed a HFD, Notch1 deficiency showed enhanced glucose tolerance and insulin sensitivity, as determined by GTT and ITT, respectively (Fig. 3A,B). In addition, serum TG, TC, and LDL cholesterol (LDL-C) were largely reduced, and HDL cholesterol (HDL-C) was slightly increased in Notch1M-KO mice (Fig. 3C). Consistent with these results, hepatic TG was significantly lower in Notch1M-KO mice, whereas there were no differences in hepatic TC (Fig. 3D). Similarly, H&E and Oil Red O staining of liver sections also confirmed less hepatic steatosis in Notch1M-KO mice compared with Notch1FL/FL mice (Fig. 3E). Moreover, Notch1 deficiency enhanced the insulin-stimulated hepatic pAkt level, revealing the improvement of hepatic IR in HFD-fed mice (Fig. 3F). Given that inflammation has a primary role in the etiology of hepatic IR, we measured the levels of proinflammatory (TNF-α and IL-1β) and anti-inflammatory [IL-10 and transforming growth factor (TGF)-β] cytokines in liver tissues, finding that Notch1 deficiency reduced the former and elevated the latter. Consistency changes were also found in the percentage of proinflammatory M1 (F4/80+CD11c+) macrophages and anti-inflammatory M2 (F4/80+CD206+) macrophages (Figs. S4D and E).
Fig. 3.
Deletion of Notch1 in macrophages ameliorates HFD-induced hepatic steatosis and insulin resistance.
Notch1FL/FL mice and Notch1M-KO mice were fed a NCD or HFD for 6 weeks. GTT (A), ITT (B), serum TG, TC, LDL-C, and HDL-C levels (C), and liver TG and TC levels (D) were measured (eight/group). (E) Representative images of H&E and Oil red O staining of liver sections (scale bar: 100 μm). (F) Western blot analyses of insulin-stimulated expression of Akt and pAkt in the liver (three/group). Values represent means ± SD. Statistical analysis was performed by one-way ANOVA and Tukey's test: ∗p <0.05, ∗∗p <0.01, ∗∗∗p <0.001. Abbreviations: GTT, glucose tolerance test; HDL-C, HDL cholesterol; HFD, high-fat diet; ITT, insulin tolerance test; LDL-C, LDL cholesterol; NCD, normal chow diet; Notch1FL/FL, floxed Notch1; Notch1M-KO, myeloid-specific Notch1-knockout; TC, total cholesterol; TG, triglyceride.
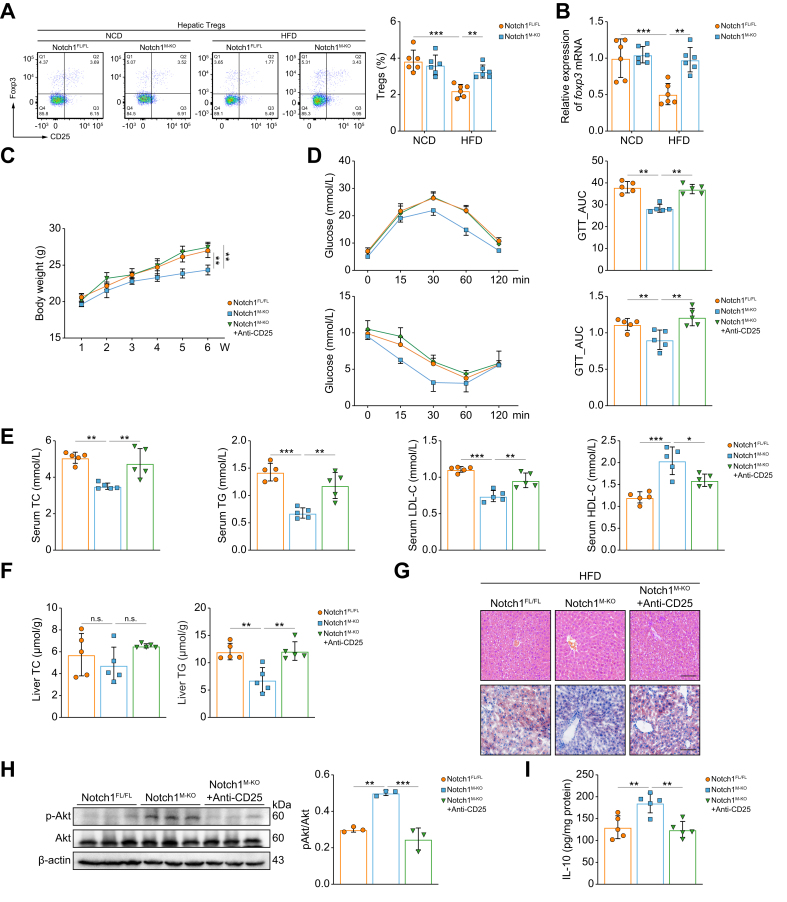
Tregs are required to reverse HFD-induced hepatic steatosis and insulin resistance in Notch1M-KO mice
Next, to confirm the correlation between the activation of hepatic macrophage Notch1 signaling and the Treg population, we analyzed the frequency of CD4+CD25+Foxp3+ Tregs and the mRNA expression of foxp3 in liver tissues of Notch1FL/FL and Notch1M-KO mice fed a HFD. Compared with Notch1FL/FL mice, Notch1 deficiency significantly reversed the HFD-induced reduction in Treg frequency (Fig. 4A,B, Fig. S5). Furthermore, we determined whether the ameliorative effects of macrophage Notch1 deficiency on MASLD were related to Treg frequency changes by abolishing Tregs in Notch1M-KO mice fed a HFD. Elimination of Tregs significantly increased the body weight (Fig. 4C), systemic IR (Fig. 4D), serum lipid levels (TC, TG, LDL, but not HDL) (Fig. 4E), and intrahepatic lipid content (Fig. 4F,G), but dramatically decreased pAkt and IL-10 (Fig. 4H,I) levels in the liver of Notch1M-KO mice, suggesting that these phenotypes associated with the deletion of Notch1 in macrophages were substantially offset by Treg elimination. Together, these results imply that the ameliorating effect of Notch1 depletion in macrophages on hepatic steatosis and IR is largely dependent on the increased frequency of Tregs.
Fig. 4.
Improvement effect of Notch1 deficiency in macrophages on hepatic steatosis and insulin resistance is mainly dependent on the increase in Treg frequency.
Notch1FL/FL mice and Notch1M-KO mice were fed a NCD or HFD for 6 weeks. (A) Proportion of hepatic CD25+Foxp3+ Tregs gated on CD3+CD4+ T cells was analyzed by flow cytometry in Notch1FL/FL mice and Notch1M-KO mice (six/group). (B) Foxp3 mRNA expression in the liver of Notch1FL/FL mice and Notch1M-KO mice (six/group). Notch1FL/FL mice, Notch1M-KO mice, and Notch1M-KO mice with Tregs eliminated (injected intraperitoneally with anti-CD25 antibodies) were fed a HFD for 6 weeks. The body weight (C), GTT, ITT (D), serum TG, TC, LDL-C, and HDL-C levels (E), and liver TG and TC levels (F) were measured (five/group). (G) Representative images of H&E and Oil red O staining of liver sections (scale bar: 100 μm). (H) Western blot analyses of insulin-stimulated expression of Akt and pAkt in the liver (three/group). (I) The level of IL-10 in the liver was measured by ELISA (five/group). Values represent means ± SD. Statistical analysis was performed by one-way ANOVA and Tukey's test. ∗p <0.05, ∗∗p <0.01, ∗∗∗p <0.001. Abbreviations: GTT, glucose tolerance test; HDL-C, HDL cholesterol; HFD, high-fat diet; ITT, insulin tolerance test; LDL-C, LDL cholesterol; NCD, normal chow diet; Notch1FL/FL, floxed Notch1; Notch1M-KO, myeloid-specific Notch1-knockout; TC, total cholesterol; TG, triglyceride; Treg, regulatory T cell.
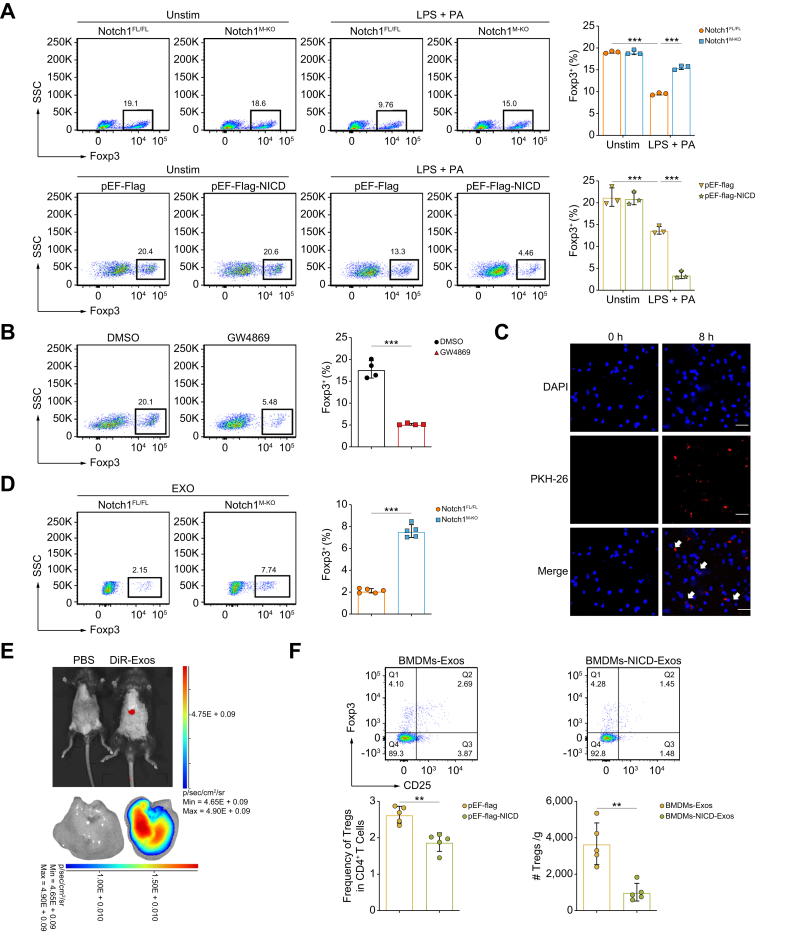
Exos from Notch1-activated macrophages impede Treg differentiation
Given that the liver is the main source of peripherally induced Tregs derived from naïve CD4+ T cell differentiation, we proposed the hypothesis that Notch1 activation in hepatic macrophages impedes Treg differentiation in hepatic steatosis. To test this hypothesis, an in vitro BMDM/naïve CD4+ T cell co-culture system was established (Fig. S6A). BMDMs were stimulated with lipopolysaccharide (LPS) + palmitic acid (PA) (the main stimulating factors of hepatic macrophages in hepatic steatosis) for 12 h, resulting in a significant increase in Notch1 activation (Fig. S6B). CD4+ T cells were harvested 3 days after the co-culture to analyze Foxp3+ Treg frequency by flow cytometry. After LPS+PA treatment, the induction of Foxp3+ Tregs was decreased significantly compared with those of unstimulated cells (Fig. 5A). Strikingly, BMDMs from Notch1M-KO mice exhibited higher induction of Foxp3+ Tregs. Subsequently, we overexpressed NICD in BMDMs through transfection of NICD overexpression plasmids (Fig. S5C). Induction of Foxp3+ Tregs was further reduced in response to overexpression of NICD in BMDMs (Fig. 5A). Consistent with this finding, IF staining also showed that the inducible Foxp3+ Tregs population was markedly enhanced following Notch1 KO and further reduced following NICD overexpression (Fig. S6D).
Fig. 5.
Exos from Notch1-activated macrophages impede Treg differentiation.
BMDMs from Notch1FL/FL and Notch1M-KO mice and pEF-Flag-NICD or pEF-Flag transfected BMDMs were stimulated with LPS (100 ng/ml) and PA (250 μM) or PBS for 12 h and then co-cultured with naïve CD4+ T cells for 3 days. (A) Induction of Foxp3+ Tregs was analyzed by flow cytometry (three/group). (B) Naïve CD4+ T cells were co-cultured with BMDMs treated with GW4869 (5 μM) for 3 days, and the induction of Foxp3+ Tregs was detected by flow cytometry (four/group). (C) BMDM-Exos were labeled with PKH26 (red) and then co-cultured with naïve CD4+ T cells for 8 h. The resulting T cells were collected for fluorescence confocal microscopy to detect Exo uptake and their nuclear location was determined by DAPI (blue) staining (scale bar: 25 μm). (D) BMDMs from Notch1FL/FL and Notch1M-KO mice were stimulated with LPS (100 ng/ml) and PA (250 μM) for 12 h. Naïve CD4+ T cells were co-cultured with BMDM-Exos for 3 days, and induction of Foxp3+ Tregs was detected by flow cytometry (five/group). (E) Small animal in vivo imaging was used to detect the distribution of DiR-labeled Exo fluorescence signals in vivo. (F) Exos were extracted from BMDMs transfected with pEF-Flag-NICD or pEF-Flag after LPS + PA stimulation, and their concentrations were measured using BCA. BMDM-Exos at a dose of 200 μg/mouse were injected via the tail vein into mice fed a HFD for 1 week, followed by another week of HFD feeding. Flow cytometry was used to detect the proportion and number of intrahepatic Tregs mice. Values represent means ± SD. Statistical analysis was performed by (A) one-way ANOVA and Tukey's test or (B,D) 2-tailed unpaired Student t test. ∗∗∗p <0.001. Abbreviations: BMDM, bone marrow-derived macrophage; Exo, exosome; LPS, lipopolysaccharide; NICD, Notch1 intracellular domain; Notch1FL/FL, floxed Notch1; Notch1M-KO, myeloid-specific Notch1-knockout; PA, palmitic acid; Treg, regulatory T cell.
Exos are an effective mechanism of intercellular communication in the liver.28 Consequently, we sought to determine whether Exos mediated the effect of macrophages on Treg differentiation. BMDMs were pretreated with GW4869, a commonly used chemical inhibitor that blocks the release of Exos. The inducible effect of BMDMs on Treg differentiation was significantly blunted after GW4869 treatment (Fig. 5B). To further define the molecular effects of BMDM-Exos on Treg differentiation, we isolated Exos from BMDMs and characterized them by transmission electron microscopy and flow nanoanalysis. The vesicles exhibited a cup- or sphere-shaped morphology with a 40–150 nm diameter (Figs. S7A and B). In addition, the Exo marker proteins CD63, CD9, and TSG101 were detected in the BMDM-Exos (Fig. S7C). These results confirmed that Exos were successfully extracted. Next, we examined whether these BMDM-Exos could be taken up by naïve CD4+ T cells. We labeled BMDM-Exos with the red fluorescent dye PKH26 and then added them into the culture medium of naïve CD4+ T cells. After 8 h, confocal microscopy analysis showed that the labeled Exos were efficiently taken up by naïve CD4+ T cells (Fig. 5C). Furthermore, we explored whether the Exos mediated the regulation of macrophagic Notch1 signaling on Treg differentiation. We incubated naïve CD4+ T cells with Exos isolated from LPS+PA-treated Notch1FL/FL-BMDMs and Notch1M-KO-BMDMs. Compared with the Notch1FL/FL group, the frequency of Tregs was increased significantly in the Notch1M-KO group (Fig. 5D).
To confirm the effect of macrophage-derived Exos on hepatic Tregs in vivo, we tracked the distribution of Exos after tail vein injection and found liver enrichment within 6 h (Fig. 5E). Exos from NICD-transfected BMDMs, stimulated by LPS + PA, were injected into HFD-fed mice, and flow cytometry showed a significant reduction in hepatic Tregs compared with controls (Fig. 5F). Collectively, these data indicated that Notch1 activation in macrophages impedes Treg production via Exos.
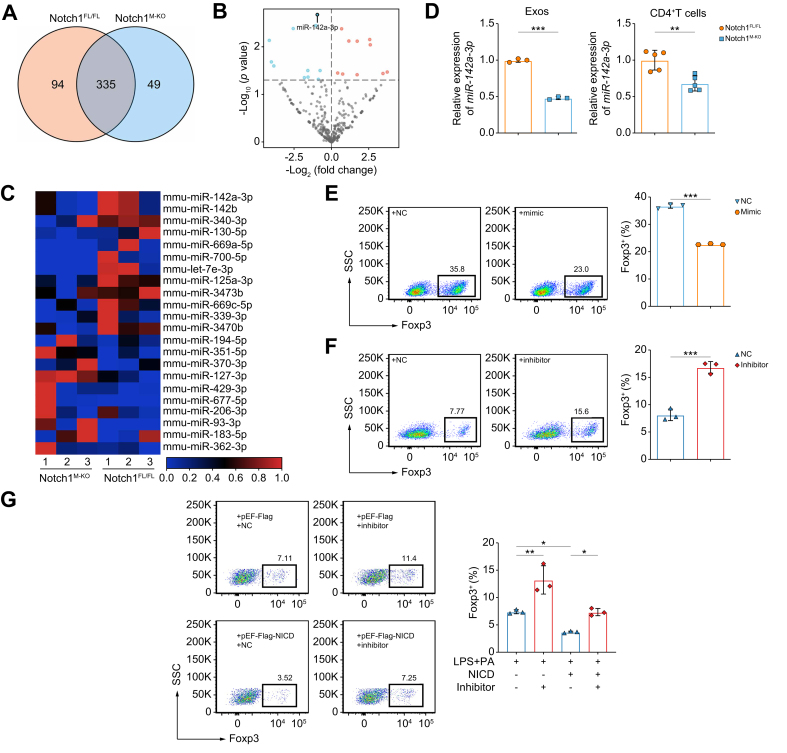
Notch1-activated macrophages impede Treg differentiation via exosomal miR-142a-3p
To decipher how the Exos from Notch1-activated macrophages affect Treg differentiation, we further analyzed the miRNA expression profiles in Exos from LPS+PA-treated Notch1M-KO-BMDMs compared with those in Exos from Notch1FL/FL-BMDMs exposed to the same treatment. miRNA sequencing results indicated that there were 335 miRNAs in common among the two groups (Fig. 6A). The volcano plots and heat maps displayed remarkable differences in miRNA profiles between Notch1M-KO-Exos and Notch1FL/FL-Exos (Fig. 6B,C). Among these differentially significantly expressed miRNAs, miR-142a-3p was reported to target TGF-β receptor 1 (TGFBR1) in both human and mouse,29,30 which is closely associated with Treg differentiation.31 Subsequently, we validated the change in miR-142a-3p in Exos by RT-qPCR, which was consistent with the miRNA sequencing results. Furthermore, we also found that miR-142a-3p was significantly decreased in CD4+ T cells co-cultured with Notch1M-KO-BMDM-Exos, compared with that in Notch1FL/FL-BMDM group (Fig. 6D). These results indicated that Notch1 activation in macrophage-modulated Exo-miR-142a-3p was transferred to CD4+ T cells.
Fig. 6.
Exo-miR-142a-3p from Notch1-activated macrophages impedes Treg differentiation.
BMDMs from Notch1FL/FL and Notch1M-KO mice were stimulated with LPS (100 ng/ml) and PA (250 μM) for 12 h. The miRNA expression profiles in BMDM-Exos were analyzed by miRNA sequencing (three/group). (A) Venn diagram showing miRNAs in both groups. (B) Volcano plot of differentially expressed miRNAs in both groups. (C) Heatmap of miRNAs that differed significantly (p <0.05) between the groups. (D) Validation of the expression of miR-142a-3p in BMDM-Exos (left panel) (three/group) and CD4+ T cells (right panel) (five/group) between the two groups by RT-qPCR analysis. (E) Induction of Foxp3+ Tregs was detected by flow cytometry after naïve CD4+ T cells transfected with NC or miR-142a-3p mimic were co-cultured with BMDMs (three/group). (F) Induction of Foxp3+ Tregs was detected by flow cytometry after naïve CD4+ T cells transfected with NC or miR-142a-3p-inhibitor were co-cultured with BMDMs pretreated with LPS (100 ng/ml) and PA (250 μM) for 12 h (three/group). (G) pEF-Flag-NICD or pEF-Flag transfected BMDMs were stimulated with LPS (100 ng/ml) and PA (250 μM) or PBS for 12 h, respectively, and then co-cultured with naïve CD4+ T cells transfected with NC or miR-142a-3p-inhibitor for 3 days. The induction of Foxp3+ Tregs was analyzed by flow cytometry (three/group). Values represent means ± SD. Statistical analysis was performed by (D–F) 2-tailed unpaired Student t test or (G) one-way ANOVA and Tukey's test: ∗p <0.05, ∗∗p <0.01, ∗∗∗p <0.001. Abbreviations: BMDM, bone marrow-derived macrophage; Exo, exosome; LPS, lipopolysaccharide; NC, negative control; NICD, Notch1 intracellular domain; Notch1FL/FL, floxed Notch1; Notch1M-KO, myeloid-specific Notch1-knockout; PA, palmitic acid; RT-qPCR, quantitative reverse transcription PCR; Treg, regulatory T cell.
To confirm the effect of miR-142a-3p on Treg differentiation, naïve CD4+ T cells were transfected with an miR-142a-3p inhibitor, mimic, or negative control (NC), and then all cells were co-cultured with BMDMs stimulated with LPS+PA or not. With confirmation of successful transfection (Fig. S8A), we found that the miR-142a-3p mimic significantly inhibited the induction of Tregs (Fig. 6E), whereas the miR-142a-3p inhibitor enhanced the induction of Tregs, compared with the corresponding NC (Fig. 6F). More importantly, the inhibition of Tregs induced by overexpressing NICD in macrophages was reversed when CD4+ T cells were transfected with the miR-142a-3p inhibitor (Fig. 6G). Overall, these results suggested that Notch1 activation in macrophages impedes Treg differentiation through Exo-miR-142a-3p.
Notch1-induced miR-142a-3p impedes Treg differentiation by targeting TGFBR1
To confirm the target gene of miR-142a-3p, we constructed a wild-type (WT) TGFBR1 3′-untranslated region (UTR) plasmid, as well as a control plasmid in which the miR-142a-3p binding site of the TGFBR1 3′UTR was mutated for use in a luciferase reporter assay (Fig. 7A, Fig. S8B). TGFBR1 3′UTR activities were significantly inhibited by miR-142a-3p-mimic transfection compared with cells transfected with NC, indicating that this 3′UTR is subject to miR-142a-3p regulation (Fig. 7A). Furthermore, we evaluated the effect of miR-142a-3p on the expression of TGFBR1 on CD4+ T cells co-cultured with BMDMs by flow cytometry. Overexpression of miR-142a-3p significantly decreased the expression of TGFBR1 on CD4+ T cells co-cultured with BMDMs (Fig. 7B), whereas miR-142a-3p inhibition increased TGFBR1 expression on CD4+ T cells co-cultured with LPS+PA-treated BMDMs compared with the NC group (Fig. 7C).
Fig. 7.
Notch1-induced miR-142a-3p impedes Treg differentiation by targeting TGFBR1.
(A) Prediction of the binding site of miR-142a-3p in TGFBR1 by miRWalk and miRDB (upper panel) and luciferase assay revealing binding of miR-142a-3p to the TGFBR1 3′UTR (lower panel). MFI of TGFBR1 in T cells was detected by flow cytometry after (B) naïve CD4+ T cells transfected with NC or a miR-142a-3p mimic were co-cultured with BMDMs (three/group), (C) naïve CD4+ T cells transfected with NC or miR-142a-3p inhibitor were co-cultured with BMDMs pretreated with LPS (100 ng/mL) and PA (250 μM) for 12 h (three/group), (D) naïve CD4+ T cells co-cultured with BMDMs from Notch1FL/FL and Notch1M-KO mice pretreated with LPS (100 ng/ml) and PA (250 μM) or PBS for 12 h (four/group), and (E) naïve CD4+ T cells co-cultured with LPS (100 ng/ml) and PA (250 μM) or PBS-pretreated BMDMs transfecting pEF-Flag-NICD or control plasmids for 12 h (four/group). The level of miR-142a-3p (F) and MFI of TGFBR1 in CD4+ T cells (G) in the liver tissues of Notch1FL/FL mice and Notch1M-KO mice fed with NCD or HFD for 6 weeks (five/group) were detected. Values represent means ± SD. Statistical analysis was performed by (A, D–G) one-way ANOVA and Tukey's test, or (B, C) 2-tailed unpaired Student t test. ∗∗p <0.01, ∗∗∗p <0.001. Abbreviations: BMDM, bone marrow-derived macrophage; HFD, high-fat diet; LPS, lipopolysaccharide; MFI, mean fluorescence intensity; NC, negative control; NCD, normal chow diet; NICD, Notch1 intracellular domain; Notch1FL/FL, floxed Notch1; Notch1M-KO, myeloid-specific Notch1-knockout; PA, palmitic acid; TGFBR1, transforming growth factor beta receptor 1; UTR, untranslated region.
Next, to further confirm that the effect of macrophage Notch1 signaling on Treg differentiation is mediated by TGFBR1, the expression of TGFBR1 in CD4+ T cells co-cultured with BMDMs was determined. The expression of TGFBR1 markedly increased in CD4+ T cells co-cultured with LPS+PA-treated Notch1M-KO-BMDMs compared with the Notch1FL/FL group, and was markedly reduced when Notch1 was overexpressed in BMDMs (Fig. 7D,E). In addition, in Notch1M-KO mice fed a HFD, macrophage Notch1 deficiency decreased miR-142a-3p levels in the liver and hepatic T cells and increased TGFBR1 expression in hepatic CD4+ T cells compared with Notch1FL/FL mice (Fig. 7F,G). Collectively, these results suggest that Notch1 signaling in macrophages regulates miR-142a-3p, which subsequently affects the expression of TGFBR1 on CD4+ T cells, leading to a decrease in Tregs.
Discussion
The role of Tregs in suppressing local inflammation has been extensively investigated,32 and emerging reports have highlighted their function in modulating non-immune lineages to maintain tissue homeostasis.33,34 In the present study, we identified that decreased Tregs contributed to HFD-induced hepatic steatosis and IR during the early stage of MASLD. Our results are consistent with prior work on hepatic Tregs, in which hepatic Treg frequency showed a significant decrease in mice after 1 week of a HFD and continued to decrease throughout the HFD feeding period.35 Furthermore, our results showed that hepatic Treg depletion significantly exacerbated lipid accumulation and IR. In agreement with this, a previous RNA-sequencing study demonstrated substantial changes in the expression of hepatic genes regulating lipid metabolism following Treg depletion.36 In addition, Tregs in visceral adipose tissue have been shown to have a clear insulin-sensitizing function,37 with adoptive Treg transfer also decreasing adipocyte size in mouse models of obesity.38 However, increasing the frequency of Tregs only in subcutaneous adipose tissue but not in the liver can exacerbate hepatic steatosis,39 a phenomenon that might result from organ crosstalk. In this study, ldIL-2 treatment was used to achieve hepatic Treg amplification, which significantly reduced hepatic steatosis. This suggests that attention should be paid to whether Tregs can be specifically enriched in liver tissue during the expansion of Tregs as part of in vivo therapy for liver diseases. Hepatic Tregs suppress MASH by alleviating intrahepatic inflammation,8 whereas they promote tumor development during the premalignant stage of MASH livers,9 which suggest that the effect of hepatic Tregs varies in response to various stimuli and the corresponding microenvironment during different stages of disease (early or late).
Environmental signals in peripheral tissues favor the development of antigen-specific Tregs. It is well documented that the activation status of macrophages in tissues is tightly connected to Treg homeostasis. For example, tumor-associated macrophages induce the imbalance of Tregs and T helper type 17 cells (Th-17),40 and the proinflammatory macrophages of visceral adipose tissue in obesity can cause Th17/Treg imbalance in visceral adipose tissue.41 In this study, increased Notch1 activation in hepatic macrophages was synchronized with the reduced hepatic Treg frequency in HFD-induced mice with hepatic steatosis. Additionally, our in vivo data revealed that deletion of Notch1 in macrophages reversed the HFD-induced reduction in hepatic Treg frequency, which was accompanied by improvements in hepatic steatosis and IR, largely attributed to changes in Treg frequency. Previous studies on Treg differentiation have primarily focused on the intrinsic role of Notch1 signaling in T cells;42,43 however, our study revealed that Notch1 signaling in macrophages could regulate macrophage–T cell interactions, highlighting the importance of cell-specific activities of Notch1 in different cell types.
Exos have been emerged as crucial mediators of intercellular communication and are involved in various pathological processes, including liver diseases, as well as carrying and delivering miRNAs, proteins, and metabolites from host to recipient cells.16 Previous work in MASH has noted the importance of Exo-mediated cellular communication between hepatic macrophages and other cells.44 The present study identified that macrophage-derived Exos could be taken up by naïve CD4+ T cells and perform a regulatory function on Treg differentiation. Notch1-deficient macrophages significantly improved the induction of Tregs by Exos secreted by LPS+PA-treated macrophages, suggesting that Exos are an important pathway for macrophage Notch1 signaling involved in regulating Treg differentiation. Exo-mediated miRNA transfer is important in various processes, including immune homeostasis.45 Multiple miRNAs have been identified to be involved in regulating Treg generation and function.46 In addition, Notch1 signaling has been reported to regulate miRNA expression.18,19 Further Exo-miRNA sequencing and functional studies demonstrated that macrophage Notch1 signaling regulating Treg differentiation might depend on an Exo-miR-142a-3p–TGFBR1 pathway. TGFBR1 serves as a crucial receptor in TGF-β signaling, which is required for the induction of Tregs.47 Kimura et al. revealed that the targeted silence of TGFBR1 in naïve CD4+ T cells inhibited the differentiation of Tregs.31 Our study demonstrated that Notch1 deletion in macrophages reduced the levels of miR-142a-3p, increased TGFBR1 expression on CD4+ T cells, and enhanced Treg frequency both in vitro and in vivo. These findings suggest that decreased Treg levels during the early stage of MASLD result from macrophages with aberrant Notch1 activation releasing miR-142a-3p, which inhibits the expression of TGFBR1 on CD4+ T cells, thereby impeding Treg production.
In conclusion, this study highlights the crucial role of hepatic Tregs during the early stage of MASLD and identifies a previously unrecognized molecular mechanism of a macrophage Notch1/exo-miR-142a-3p/TGFBR1 pathway in regulating Treg differentiation. Although we cannot rule out that macrophage Notch1 activation might regulate Treg production through pathways other than Exos, our findings provide evidence of the involvement of Exos in the crosstalk between macrophages and CD4+ T cells. However, because we focused exclusively on the early stage of MASLD, our findings might not be readily extended to MASH or fibrotic stages. Nevertheless, this study expands our current understanding of the role of crosstalk between immune cell populations in MASLD. Moreover, our findings provide rationale for a potential immunological approach for the treatment of MASLD by the induction of Tregs.
Abbreviations
BMDM, bone marrow-derived macrophage; Exo, exosome; FFA, free fatty acid; GTT, glucose tolerance test; HDL-C, HDL cholesterol; HFD, high-fat diet; IF, immunofluorescence; IR, insulin resistance; ITT, insulin tolerance test; ldIl-2, low-dose IL-2; LDL-C, LDL cholesterol; LPS, lipopolysaccharide; LysM-Cre, Lyz2-Cre mice; MASH, metabolic dysfunction-associated steatohepatitis; MASLD, metabolic dysfunction-associated steatotic liver disease; MFI, mean fluorescence intensity; NC, negative control; NCD, normal chow diet; NICD, Notch1 intracellular domain; Notch1FL/FL, floxed Notch1; Notch1M-KO, myeloid-specific Notch1-knockout; PA, palmitic acid; pAkt, phosphorylation of Akt at S473; RT-qPCR, quantitative reverse transcription PCR; TC, total cholesterol; TG, triglyceride; TGFBR1, transforming growth factor beta receptor 1; TGF-β, transforming growth factor-β; Th17, T helper type 17; Treg, regulatory T cell; UTR, untranslated region; WT, wild-type.
Financial support
This work was supported by the National Nature Science Foundation of China (82173872 and 81872663), the Natural Science Foundation of Hubei Province (2023AFB611), and the Basic-Clinical Medicine Joint Fund of Zhongnan Hospital of Wuhan University (ZNLH202207).
Authors’ contribution
Analyzed, interpreted the data, and drafted the manuscript: M-YZ, KL. Data collection: X-XH, D-QX, R-BZ, Q-TH, X-YD, Q-YZ, C-CJ, YG. Designed the original study, supervised research, and critically reviewed the paper: CYL, JP. Reviewed the draft for important intellectual content and approved the final article for submission: all authors.
Data availability statement
The miRNA-seq data have been deposited at GEO and are publicly available as of the date of publication. The accession number is PRJNA1046700.
Conflicts of interests
The authors declare no conflicts of interest that pertain to this work. Please refer to the accompanying ICMJE disclosure forms for further details.
Footnotes
Author names in bold designate share co-first authorship
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jhepr.2024.101242.
Contributor Information
Changyong Li, Email: lichangyong@whu.edu.cn.
Jie Ping, Email: pingjie@whu.edu.cn.
Supplementary data
The following are the Supplementary data to this article:
References
- 1.Huang D.Q., El-Serag H.B., Loomba R. Global epidemiology of NAFLD-related HCC: trends, predictions, risk factors and prevention. Nat Rev Gastroenterol Hepatol. 2021;18:223–238. doi: 10.1038/s41575-020-00381-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Febbraio M.A., Reibe S., Shalapour S., et al. Preclinical models for studying NASH-driven HCC: how useful are they? Cell Metab. 2019;29:18–26. doi: 10.1016/j.cmet.2018.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cai J., Zhang X.J., Li H. The role of innate immune cells in nonalcoholic steatohepatitis. Hepatology. 2019;70:1026–1037. doi: 10.1002/hep.30506. [DOI] [PubMed] [Google Scholar]
- 4.Peiseler M., Schwabe R., Hampe J., et al. Immune mechanisms linking metabolic injury to inflammation and fibrosis in fatty liver disease - novel insights into cellular communication circuits. J Hepatol. 2022;77:1136–1160. doi: 10.1016/j.jhep.2022.06.012. [DOI] [PubMed] [Google Scholar]
- 5.Allan S.E., Passerini L., Bacchetta R., et al. The role of 2 FOXP3 isoforms in the generation of human CD4+ Tregs. J Clin Invest. 2005;115:3276–3284. doi: 10.1172/JCI24685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hou X., Song J., Su J., et al. CD4(+)Foxp3(+) Tregs protect against innate immune cell-mediated fulminant hepatitis in mice. Mol Immunol. 2015;63:420–427. doi: 10.1016/j.molimm.2014.09.015. [DOI] [PubMed] [Google Scholar]
- 7.Wu K.J., Qian Q.F., Zhou J.R., et al. Regulatory T cells (Tregs) in liver fibrosis. Cell Death Discov. 2023;9:53. doi: 10.1038/s41420-023-01347-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roh Y.S., Kim J.W., Park S., et al. Toll-like receptor-7 signaling promotes nonalcoholic steatohepatitis by inhibiting regulatory T cells in mice. Am J Pathol. 2018;188:2574–2588. doi: 10.1016/j.ajpath.2018.07.011. [DOI] [PubMed] [Google Scholar]
- 9.Wang H., Zhang H., Wang Y., et al. Regulatory T-cell and neutrophil extracellular trap interaction contributes to carcinogenesis in non-alcoholic steatohepatitis. J Hepatol. 2021;75:1271–1283. doi: 10.1016/j.jhep.2021.07.032. [DOI] [PubMed] [Google Scholar]
- 10.Zhang Z., Salgado O.C., Liu B., et al. An OGT-STAT5 axis in regulatory T cells controls energy and iron metabolism. Front Immunol. 2022;13 doi: 10.3389/fimmu.2022.874863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loomba R., Friedman S.L., Shulman G.I. Mechanisms and disease consequences of nonalcoholic fatty liver disease. Cell. 2021;184:2537–2564. doi: 10.1016/j.cell.2021.04.015. [DOI] [PubMed] [Google Scholar]
- 12.Carambia A., Freund B., Schwinge D., et al. TGF-beta-dependent induction of CD4(+)CD25(+)Foxp3(+) Tregs by liver sinusoidal endothelial cells. J Hepatol. 2014;61:594–599. doi: 10.1016/j.jhep.2014.04.027. [DOI] [PubMed] [Google Scholar]
- 13.Heymann F., Peusquens J., Ludwig-Portugall I., et al. Liver inflammation abrogates immunological tolerance induced by Kupffer cells. Hepatology. 2015;62:279–291. doi: 10.1002/hep.27793. [DOI] [PubMed] [Google Scholar]
- 14.Stienstra R., Saudale F., Duval C., et al. Kupffer cells promote hepatic steatosis via interleukin-1beta-dependent suppression of peroxisome proliferator-activated receptor alpha activity. Hepatology. 2010;51:511–522. doi: 10.1002/hep.23337. [DOI] [PubMed] [Google Scholar]
- 15.Dong X., Feng Y., Xu D., et al. Targeting macrophagic 17beta-HSD7 by fenretinide for the treatment of nonalcoholic fatty liver disease. Acta Pharm Sin B. 2023;13:142–156. doi: 10.1016/j.apsb.2022.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sato K., Meng F., Glaser S., et al. Exosomes in liver pathology. J Hepatol. 2016;65:213–221. doi: 10.1016/j.jhep.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meng W., Hao Y., He C., et al. Exosome-orchestrated hypoxic tumor microenvironment. Mol Cancer. 2019;18:57. doi: 10.1186/s12943-019-0982-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao J.L., Huang F., He F., et al. Forced activation of Notch in macrophages represses tumor growth by upregulating miR-125a and disabling tumor-associated macrophages. Cancer Res. 2016;76:1403–1415. doi: 10.1158/0008-5472.CAN-15-2019. [DOI] [PubMed] [Google Scholar]
- 19.Boelens M.C., Wu T.J., Nabet B.Y., et al. Exosome transfer from stromal to breast cancer cells regulates therapy resistance pathways. Cell. 2014;159:499–513. doi: 10.1016/j.cell.2014.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Radtke F., MacDonald H.R., Tacchini-Cottier F. Regulation of innate and adaptive immunity by Notch. Nat Rev Immunol. 2013;13:427–437. doi: 10.1038/nri3445. [DOI] [PubMed] [Google Scholar]
- 21.Damle S.R., Martin R.K., Cockburn C.L., et al. ADAM10 and Notch1 on murine dendritic cells control the development of type 2 immunity and IgE production. Allergy. 2018;73:125–136. doi: 10.1111/all.13261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ballester-Lopez C., Conlon T.M., Ertuz Z., et al. The Notch ligand DNER regulates macrophage IFNgamma release in chronic obstructive pulmonary disease. EBioMedicine. 2019;43:562–575. doi: 10.1016/j.ebiom.2019.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.An L., Li Z., Shi L., et al. Inflammation-targeted celastrol nanodrug attenuates collagen-induced arthritis through NF-kappaB and Notch1 pathways. Nano Lett. 2020;20:7728–7736. doi: 10.1021/acs.nanolett.0c03279. [DOI] [PubMed] [Google Scholar]
- 24.Wang X., Sun L., Zhang L., et al. Effect of adoptive transfer or depletion of regulatory T cells on triptolide-induced liver injury. Front Pharmacol. 2016;7:99. doi: 10.3389/fphar.2016.00099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buitrago-Molina L.E., Pietrek J., Noyan F., et al. Treg-specific IL-2 therapy can reestablish intrahepatic immune regulation in autoimmune hepatitis. J Autoimmun. 2021;117 doi: 10.1016/j.jaut.2020.102591. [DOI] [PubMed] [Google Scholar]
- 26.Erhardt A., Biburger M., Papadopoulos T., et al. IL-10, regulatory T cells, and Kupffer cells mediate tolerance in concanavalin A-induced liver injury in mice. Hepatology. 2007;45:475–485. doi: 10.1002/hep.21498. [DOI] [PubMed] [Google Scholar]
- 27.Cintra D.E., Pauli J.R., Araujo E.P., et al. Interleukin-10 is a protective factor against diet-induced insulin resistance in liver. J Hepatol. 2008;48:628–637. doi: 10.1016/j.jhep.2007.12.017. [DOI] [PubMed] [Google Scholar]
- 28.Hou X., Yin S., Ren R., et al. Myeloid-cell-specific IL-6 signaling promotes microRNA-223-enriched exosome production to attenuate NAFLD-associated fibrosis. Hepatology. 2021;74:116–132. doi: 10.1002/hep.31658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen L., Xiong Y., Hu Y.Q., et al. Regulatory T cell-exosomal miR-142-3p promotes angiogenesis and osteogenesis via TGFBR1/SMAD2 inhibition to accelerate fracture repair. Chem Eng J. 2022;427 [Google Scholar]
- 30.Yang X., Dan X., Men R., et al. MiR-142-3p blocks TGF-beta-induced activation of hepatic stellate cells through targeting TGFbetaRI. Life Sci. 2017;187:22–30. doi: 10.1016/j.lfs.2017.08.017. [DOI] [PubMed] [Google Scholar]
- 31.Kimura K., Hohjoh H., Fukuoka M., et al. Circulating exosomes suppress the induction of regulatory T cells via let-7i in multiple sclerosis. Nat Commun. 2018;9:17. doi: 10.1038/s41467-017-02406-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Georgiev P., Charbonnier L.M., Chatila T.A. Regulatory T cells: the many faces of Foxp3. J Clin Immunol. 2019;39:623–640. doi: 10.1007/s10875-019-00684-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Panduro M., Benoist C., Mathis D. Tissue Tregs. Annu Rev Immunol. May 20 2016;34:609–633. doi: 10.1146/annurev-immunol-032712-095948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shao Q., Gu J., Zhou J., et al. Tissue Tregs and maintenance of tissue homeostasis. Front Cell Dev Biol. 2021;9 doi: 10.3389/fcell.2021.717903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ma X., Hua J., Mohamood A.R., et al. A high-fat diet and regulatory T cells influence susceptibility to endotoxin-induced liver injury. Hepatology. 2007;46:1519–1529. doi: 10.1002/hep.21823. [DOI] [PubMed] [Google Scholar]
- 36.Klingenberg R., Gerdes N., Badeau R.M., et al. Depletion of FOXP3+ regulatory T cells promotes hypercholesterolemia and atherosclerosis. J Clin Invest. 2013;123:1323–1334. doi: 10.1172/JCI63891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meng F., Hao P., Du H. Regulatory T cells differentiation in visceral adipose tissues contributes to insulin resistance by regulating JAZF-1/PPAR-gamma pathway. J Cell Mol Med. 2023;27:553–562. doi: 10.1111/jcmm.17680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eller K., Kirsch A., Wolf A.M., et al. Potential role of regulatory T cells in reversing obesity-linked insulin resistance and diabetic nephropathy. Diabetes. 2011;60:2954–2962. doi: 10.2337/db11-0358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Van Herck M.A., Vonghia L., Kwanten W.J., et al. Adoptive cell transfer of regulatory T cells exacerbates hepatic steatosis in high-fat high-fructose diet-fed mice. Front Immunol. 2020;11:1711. doi: 10.3389/fimmu.2020.01711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou J., Li X., Wu X., et al. Exosomes released from tumor-associated macrophages transfer miRNAs that induce a Treg/Th17 cell imbalance in epithelial ovarian cancer. Cancer Immunol Res. 2018;6:1578–1592. doi: 10.1158/2326-6066.CIR-17-0479. [DOI] [PubMed] [Google Scholar]
- 41.Zhang S., Gang X., Yang S., et al. The alterations in and the role of the Th17/Treg balance in metabolic diseases. Front Immunol. 2021;12 doi: 10.3389/fimmu.2021.678355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Radtke F., Fasnacht N., Macdonald H.R. Notch signaling in the immune system. Immunity. 2010;32:14–27. doi: 10.1016/j.immuni.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 43.Romero-Wolf M., Shin B., Zhou W., et al. Notch2 complements Notch1 to mediate inductive signaling that initiates early T cell development. J Cell Biol. 2020;219 doi: 10.1083/jcb.202005093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gao H., Jin Z., Bandyopadhyay G., et al. MiR-690 treatment causes decreased fibrosis and steatosis and restores specific Kupffer cell functions in NASH. Cell Metab. 2022;34:978–990. doi: 10.1016/j.cmet.2022.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu M.Y., Jia H.J., Zhang J., et al. Exosomal miRNAs-mediated macrophage polarization and its potential clinical application. Int Immunopharmacol. 2023;117 doi: 10.1016/j.intimp.2023.109905. [DOI] [PubMed] [Google Scholar]
- 46.Kunze-Schumacher H., Krueger A. The role of microRNAs in development and function of regulatory T Cells - lessons for a better understanding of microRNA biology. Front Immunol. 2020;11:2185. doi: 10.3389/fimmu.2020.02185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bronevetsky Y., Burt T.D., McCune J.M. Lin28b regulates fetal regulatory T cell differentiation through modulation of TGF-beta signaling. J Immunol. 2016;197:4344–4350. doi: 10.4049/jimmunol.1601070. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The miRNA-seq data have been deposited at GEO and are publicly available as of the date of publication. The accession number is PRJNA1046700.