Abstract
Enzymatic hydrolysis of cellulosic biomass is a complex process involving many factors, including multiple enzymes, heterogeneous substrates, and multi-step enzyme reactions. Cellulase researchers have conventionally used a double-exponential equation to fit the experimental time course of product formation, but no theoretical basis for this has been established. Here we present a mechanism-based equation that fits well the progress curves of cellulase reaction, incorporating the concepts of non-productive and productive binding on the cellulose surface and processivity. The derived equation is double exponential. Our findings indicate that the reaction mechanism of cellulase itself can account for the double-exponential nature of the progress curve independently of other factors that may contribute, such as substrate heterogeneity and involvement of other enzymes.
Keywords: cellulase, glycoside hydrolase, progress curve, biomass utilization
Abbreviations
CBD, cellulose-binding domain; CBH, cellobiohydrolase; CD, catalytic domain; EG, endo-glucanase; GH, glycoside hydrolase; HS-AFM, high-speed atomic force microscopy.
INTRODUCTION
In progressing towards a carbon-neutral society, enzymatic hydrolysis of cellulose is a bottleneck in the production of soluble saccharides such as glucose and cellobiose from non-edible biomass [1]. Cellulase is an attractive option for converting cellulosic biomass because it can hydrolyze cellulose under mild conditions, and it is becoming more widely utilized as recent developments in biotechnology have reduced its cost [2]. Therefore, it is important to understand its mechanism of action in detail to maximize its utility.
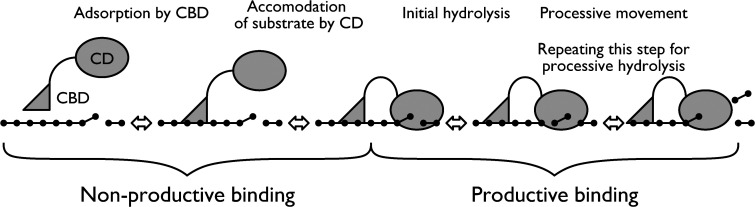
Cellulases that act on insoluble cellulose typically consist of two domains, the cellulose-binding (CBD) and catalytic (CD) domains [3, 4]. As shown in Fig. 1, cellulose hydrolysis is typically initiated by the adsorption of the enzyme on the cellulose surface, mediated by the CBD, though the resulting enzyme-substrate complex is “non-productive” because adsorption on cellulose itself does not produce any products [5, 6]. The following reaction is highly dependent on the characteristics of the CD for saccharide production [7, 8], i.e., the cellulose chain is retained after the initial hydrolysis and a further hydrolytic reaction occurs to produce soluble saccharide (typically cellobiose); this is called the “processive reaction” [9]. However, if the CD leaves the chain soon after the initial hydrolysis reaction, the reaction is called “non-processive”. Processivity is the key to the degradability of crystalline cellulose by the enzyme [10, 11]. We previously used high-speed atomic force microscopy (HS-AFM) to visualize the processive movement of microbial cellobiohydrolases (CBHs) [12, 13, 14] and we discussed the structure-function relationships of CBHs based on the idea of convergent evolution of cellulolytic bacteria and fungi [15]. These recent studies indicate that a combination of cutting-edge biophysical approaches with quantitative biochemical analysis is required to understand the reaction mechanism of the enzyme.
Fig. 1. Mechanism of action of cellulase with a dual domain structure, used to construct the theoretical equation.
CBD, cellulose-binding domain; CD, catalytic domain.
Many biochemical approaches have been used to examine the time course of product formation (the progress curve) by cellulases, but no consistent understanding has emerged, probably because of the complexity of enzymatic cellulose hydrolysis [11, 16]. In early investigations, it was suggested that the heterogeneity of cellulose, i.e., the presence of a mixture of crystalline and amorphous regions, might be important because of the endo-exo theory, i.e., that endo-glucanases (EGs) degrade amorphous regions of cellulose while crystalline cellulose is hydrolyzed by exo-type enzymes later called CBHs [17, 18, 19, 20, 21]. Since amorphous regions are quickly degraded by EGs while the degradation of crystalline part is slow, it seems reasonable to apply a two-term equation, such as a double exponential, to fit the progress curves. However, it was found that degradation was biphasic even when a single, highly purified enzyme was applied to highly crystalline cellulose [6, 11]. Thus, it appeared that the fitting of the observations to a double-exponential equation is not simply due to substrate heterogeneity or involvement of multiple enzymes.
In the present manuscript, we have developed a mechanism-based double-exponential equation that fits well the progress curve of the cellulase reaction, thereby providing a theoretical basis for the observation that the progress curve of cellulase shows a double-exponential character. This approach should be applicable not only to cellulases, but also to other enzymes that act on insoluble substrates with or without processivity.
MATERIALS AND METHODS
The theoretical development is presented step-by-step in the following Results and Discussion section. The kinetic data of glycoside hydrolase (GH) family 6 CBH from Phanerochaete chrysosporium (PcCel6A) [22] and GH7 CBH from Trichoderma reesei (TrCel7A) [6] towards highly crystalline cellulose from green algae Cladphora spp. used in this study are taken from the cited references. Global fitting of the data was done using Igor Pro (ver. 8.04, Wavemetric, Inc.)
RESULTS AND DISCUSSION
Recent biophysical experiments using HS-AFM indicate that cellulases, especially processive enzymes, move along the substrate chains of crystalline cellulose [12, 13, 14], and biochemical results need to be reanalyzed to take account of the dynamics of the enzyme. However, as regards the parameters, Väljamäe and coworkers' stated “It must be pointed out that we do not attach any physical interpretation to these constants since they are empirical” [11], and this situation has not changed in the past quarter of a century. In the present manuscript, we take a bottom-up approach based on the molecular mechanism of the enzyme.
As shown in Fig. 1, cellulase molecules may exist in three states: in solution, and on cellulose in a non-productive or productive manner. Chemical equations for the process are written as
where Es, Enp, Ep, and P are the concentrations of the enzyme molecule in solution, non-productively bound enzyme, productively bound enzyme, and the products, respectively. ν is the number of processive movements before the enzyme leaves from the cellulose surface. Note that we define the completion of productive binding when a cellulose chain is fully accommodated into the end of the tunnel in CD of the cellulase, simultaneously hydrolyzed and yielding a single cellobiose molecule [7]. Also, we assumed that the transition from non-productive binding to productive binding is reversible, given that this process is driven by the gradient of the potential energy, which could be perturbated by thermal fluctuations [23]. The following relationships hold, where E0 is the total amount of enzyme in the reaction mixture:
| (1) |
| (2) |
| (3) |
| (4) |
Note that in Eq. (2), the term for the transition from productive to non-productive binding was denoted as k−2Ep instead of k−2EpP because this process only involves mechanical detachment and does not require the hydrolyzed cellobiose molecule. From Eqs. (1) and (2), and (1) and (3), we obtain the following relationships:
| (5) |
| (6) |
The following relationships are defined:
| (7) |
| (8) |
From Eqs. (7) and (8),
Simplifying, we have
| (9) |
The eigenvalues of this second-order linear differential equation λ1 and λ2 are obtained as follows:
Given that Es is generally much larger than Enp and Ep (since only cellulases near the surface of cellulose contribute to the reaction), in the situation where k1 < k−2 (i.e., C2 < 0), both λ1 and λ2 become negative real numbers. This was confirmed by fitting to actual data as described later.
Thus, the solution of the differential Eq. (9) can be expressed as
and thus, we obtain
| (10) |
where ω1 and ω2 are constants satisfying the following relationships derived from x(0)=0
and (0)=k1E0 , respectively:
Similar equations for y can also be derived from Eqs. (7) and (8):
Simplifying, we have
| (11) |
Since Eq. (11) is similar to (9),
| (12) |
where ω3 and ω4 are constants satisfying the following relationships derived from (0) = 0 and y(0) = 0, respectively:
Finally, a differential equation for P is derived from Eqs. (4), (10) and (12);
where
The equation is then integrated as
where
and Cp is a constant,
Using the initial condition, P(t=0)=0,
| (13) |
Therefore, Eq. (13) can be simplified into the following double-exponential form with a linear term as follows:
| (14) |
where b < 0, d < 0 and f > 0.
In the derivation of Eq. (14), there is no assumption that heterogeneity, i.e. amorphous or crystalline form of cellulose, or involvement of other enzymes such as EGs and CBHs plays a role. Instead, the double exponential equation simply arises from the reaction mechanism of single enzyme. However, we should point out that this does not exclude contributions of other factors such as those mentioned above. Nevertheless, a double exponential equation can be expected simply on the basis of the substrate/enzyme combination alone.
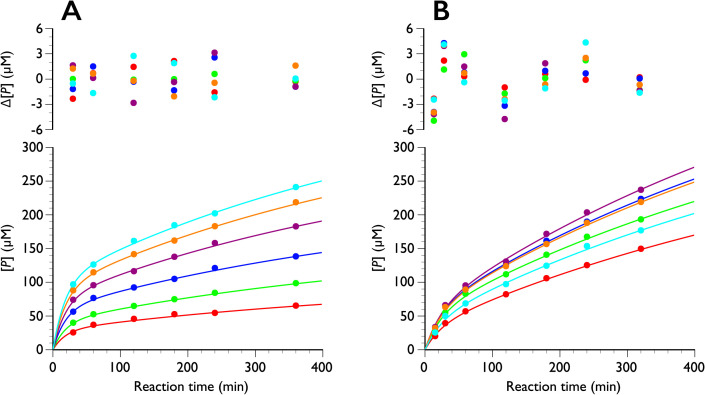
The progress curves of PcCel6A and TrCel7A were then re-analyzed using global fitting together with Eq. (14). Global fitting has recently been utilized to analyze enzyme kinetics because multiple parameters are optimized at once by non-linear regression. When the famous Michaelis-Menten study of sucrose hydrolysis by sucrase (invertase) was re-analyzed using this technique [24], it became apparent that substrate inhibition had not been considered in the initial kinetic study by Michaelis and Menten [25]. Moreover, we have applied global fitting to the complicated substrate loading system in processive cellulases [26]. Thus, global fitting is a powerful tool when multiple parameters must be considered. Since λ1 and λ2 (b and d in Eq. (14), respectively) are usually treated as rate constants and they do not include enzyme concentration (E0), these parameters were fixed at all enzyme concentrations tested, and other parameters were simulated from the data of PcCel6A [22] and TrCel7A [6] using Eq. (14) as shown in Fig. 2. The obtained parameters are listed in Tables 1, 2, 3.
Fig. 2. Time course of product formation from crystalline cellulose Iα at 30 ˚C.
The progress curves of PcCel6A (A) and TrCel7A (B) were globally fitted to Eq. (14). The residue values between the experimental data and the curve fitting are shown above the plots. Red, green, blue, purple, orange and cyan are 0.25, 0.51, 1.0, 2.2, 5.4, and 8.9 μM for PcCel6A in (A), and 0.40, 0.84, 1.3, 2.2, 4.3, and 8.6 μM for TrCel7A in (B), respectively.
Table 1. Parameters of crystalline cellulose hydrolysis by PcCel6A estimated using global fitting.
| E 0 | a | b | c | d | a/b | c/d |
| (μM) | (=λ1) | (=λ2) | ||||
| 0.25 | −1.55±0.12 | −0.0541 | −0.136±0.015 | −0.00180 | 28.6±2.0 | 75.5±11.0 |
| 0.51 | −2.21±0.14 | ±0.0040 | −0.215±0.021 | ±0.00037 | 40.8±2.3 | 120±16 |
| 1.0 | −3.23±0.18 | −0.297±0.027 | 59.6±2.8 | 165±21 | ||
| 2.2 | −4.01±0.20 | −0.410±0.035 | 74.0±3.4 | 228±29 | ||
| 5.4 | −4.84±0.24 | −0.478±0.041 | 89.4±3.8 | 265±34 | ||
| 8.9 | −5.47±0.27 | −0.525±0.045 | 101±4 | 291±37 |
Table 2. Parameters of crystalline cellulose hydrolysis by TrCel7A estimated using global fitting without f in Eq. (14).
| E 0 | a | b | c | d | a/b | c/d |
| (μM) | (=λ1) | (=λ2) | ||||
| 0.40 | −1.33±0.14 | −0.0412 | −0.455±0.053 | −0.00146 | 32.2±5.1 | 311±81 |
| 0.84 | −2.13±0.16 | ±0.0062 | −0.556±0.066 | ±0.00053 | 51.6±6.5 | 381±97 |
| 1.3 | −2.33±0.17 | −0.650±0.076 | 56.5±7.2 | 445±114 | ||
| 2.2 | −2.35±0.17 | −0.705±0.081 | 57.1±7.5 | 484±124 | ||
| 4.3 | −2.28±0.17 | −0.638±0.075 | 55.5±7.1 | 437±112 | ||
| 8.6 | −1.68±0.15 | −0.533±0.062 | 40.7±5.8 | 365±95 |
Table 3. Parameters of crystalline cellulose hydrolysis by TrCel7A estimated using global fitting including f in Eq. (14).
| E 0 | a | b | c | d | f | a/b | c/d |
| (μM) | (=λ1) | (=λ2) | |||||
| 0.40 | −1.31±0.15 | −0.0413 | −0.459±0.057 | −0.00158 | 0.0108 | 31.7±5.4 | 291±87 |
| 0.84 | −2.11±0.17 | ±0.0063 | −0.555±0.071 | ±0.00064 | ±0.0291 | 51.1±6.8 | 353±109 |
| 1.3 | −2.31±0.18 | −0.644±0.084 | 56.0±7.5 | 409±131 | |||
| 2.2 | −2.34±0.18 | −0.689±0.101 | 56.7±7.7 | 437±153 | |||
| 4.3 | −2.23±0.17 | −0.590±0.158 | 55.5±7.2 | 375±177 | |||
| 8.6 | −1.73±0.21 | −0.422±0.316 | 41.8±6.8 | 268±251 |
As shown in Fig. 2, the progress curves of both enzymes are well fitted by the equation. In the case of PcCel6A, all progress curves at various enzyme concentrations (0.25 to 8.9 μM) were well fitted by Eq. (14) and the f-value converged to almost 0 (<10−18). Therefore, the parameters were obtained using f = 0. From Eq. (13), this assumption implies that k2, the rate of forming a productive binding, is significantly smaller than k1 and k3, which is reasonable considering the limited access to the reaction. On the other hand, progress curves of TrCel7A seem to include an f-value, though the errors of each parameter were large, especially for the second exponential term, c and c/d, possibly because the second exponential term and linear term are quite similar to each other. Therefore, we also analyzed the data using Eq. (14) without f (i.e., f=0) for TrCel7A. The rate constants of the two enzymes are rather similar, though that of PcCel6A is slightly faster, in agreement with reported data.
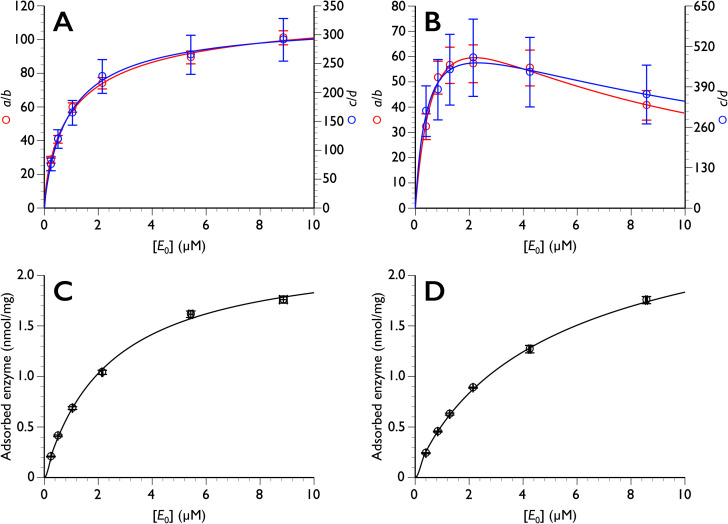
However, the enzyme concentration dependence of the amplitudes of the exponential terms, a/b and c/d values, are very different from each other. As shown in Fig. 3, the enzyme concentration dependence of both a/b and c/d was hyperbolic in the case of PcCel6A, while that of TrCel7A decreased with increasing enzyme concentration, which is similar to a typical substrate inhibition curve [27, 28, 29]. In a previous kinetic study of TrCel7A, we proposed possible crowding effects, and our hypothesis has been confirmed by biophysical HS-AFM and simulation studies [30]. Indeed, HS-AFM examination of processivity [15] indicated that the velocity of processive cellulase tends to decrease with increasing concentration of the enzyme, due to a “traffic jam” effect [13]. When we look at the construction of Eq. (14), most of the parameters except for λ1 and λ2 are dependent on E0, and it is difficult to clarify the relationship between each parameter and E0. However, it is clear that a/b and c/d are highly dependent on the amount of adsorbed enzyme, indicating that it is important to distinguish productive and non-productive adsorption.
Fig. 3. The enzyme concentration dependence of both a/b and c/d.
Top: enzyme concentration (E0) dependence of a/b and c/d for PcCel6A (A) and TrCel7A (B). Bottom: amount of adsorbed enzyme plotted versus E0 for PcCel6A (C) and TrCel7A (D). The data in A, C, and D were fitted to typical hyperbolic curve (y = (a×x) / (b+x) where a and b are constants) while inhibition is considered (y = (a×x) /{b+x+(x2/c)} where a, b and c are constants) in B. Note that the E0 dependence of adsorption (C and D) does not correspond to a typical Langmuir plot of ES versus adsorbed enzyme.
In this paper, we modeled the formation of a productive binding event with the assumption that one product is generated. Even if we adopted the formulation including this term in the processive reaction, the conclusion that the time course of the product follows a double exponential form remains valid. This is because, in this case, the first term on the right side of Eq. (4) would be eliminated, making P dependent solely on Ep, which itself has a double exponential form.
So far, progress curves of cellulase have been fitted conventionally by a single or double exponential equation, or a single exponential equation with a linear function [6, 11]. Equation (14) includes all terms in those equations, and therefore curve-fitting with this equation is reasonable. Looking at the Eq. (13), however, several values, such as λ and ω, remain undetermined because the experimental data is not accurate enough. Nevertheless, it should be possible to estimate them individually, for example, by adsorption experiments for k1/k−1 ratio, pre-steady-state kinetics for k2/k−2, processivity measurement by HS-AFM for ν, and so on. Recently we succeeded in simulating approximately 100 million cellulase molecules using a customized traffic simulator and demonstrated that consideration of the dual domain structure of cellulase is indispensable to explain the gap between single-molecule behavior and bulk biochemical results [30]. In the simulation study, we defined the parameters for adsorption, non-productive and productive enzymes, and processivity, and the obtained progress curve fitted well to the actual biochemical plots, supporting the idea that theoretical and simulated approaches can give insight into real biophysical and biochemical observations.
In conclusion, we have constructed a mechanism-based equation that for the first time provides a theoretical basis for the double-exponential character of the progress curve of cellulase reaction. Thus, it is not necessary to consider the heterogeneity of the substrate or the involvement of multiple enzymes in order to explain the double-exponential nature of the progress curve, although we cannot rule out a contribution of such effects. We believe this work represents an important step towards understanding the actual saccharification of cellulosic biomasses. Furthermore, since the developed equation describes a model consisting of a simple enzymatic reaction at a solid/liquid interface, a similar approach may be appliable to many other enzymatic reactions involving insoluble substrates. The origin of the double exponential time course lies in a two-step reaction process involving long timescale equilibration. While such a form can arise in general two-step enzyme reactions, if the relaxation rates of each process are sufficiently fast, the behavior would effectively follow a single exponential.
CONFLICTS OF INTEREST
The authors declare no conflict of interests.
ACKNOWLEDGEMENTS
The authors thank Mr. Sho Sakuma in the University of Tokyo, who initiated the mathematical modelling of cellulase during his undergraduate research. This work was supported in part by Grants-in-Aid for Scientific Research (A) 23H00341 and (B) 19H03013 (to K.I.) from the Japan Society for the Promotion of Science (JSPS) and a Grant-in-Aid for Innovative Areas 18H05494 (to K.I.) from the Japanese Ministry of Education, Culture, Sports, and Technology (MEXT), and by the Environment Research and Technology Development Fund No. JPMEERF21S11900 and JPMEERF21S11902 (to K.I.) from the Environmental Restoration and Conservation Agency of Japan (ERCA).
REFERENCES
- [1].Brandao AS, Goncalves A, Santos JMRCA. Circular bioeconomy strategies: From scientific research to commercially viable products. J Clean Prod. 2021; 295: 126407. [Google Scholar]
- [2].Olivieri G, Wijffels RH, Marzocchella A, Russo ME. Bioreactor and bioprocess design issues in enzymatic hydrolysis of lignocellulosic biomass. Catalysts. 2021; 11: 680. [Google Scholar]
- [3].Teeri TT, Koivula A, Linder M, Wohlfahrt G, Divne C, Jones TA. Trichoderma reesei cellobiohydrolases: why so efficient on crystalline cellulose? Biochem Soc Trans. 1998; 26: 173-8. [DOI] [PubMed] [Google Scholar]
- [4].Teeri TT. Crystalline cellulose degradation: New insight into the function of cellobiohydrolases. Trends Biotechnol. 1997; 15: 160-7. [Google Scholar]
- [5].Sugimoto N, Igarashi K, Wada M, Samejima M. Adsorption characteristics of fungal family 1 cellulose-binding domain from Trichoderma reesei cellobiohydrolase I on crystalline cellulose: negative cooperative adsorption via a steric exclusion effect. Langmuir. 2012; 28: 14323-9. [DOI] [PubMed] [Google Scholar]
- [6].78 Igarashi K, Wada M, Hori R, Samejima M. Surface density of cellobiohydrolase on crystalline celluloses. A critical parameter to evaluate enzymatic kinetics at a solid-liquid interface. FEBS J. 2006; 273: 2869-. [DOI] [PubMed] [Google Scholar]
- [7].Divne C, Ståhlberg J, Teeri TT, Jones TA. High-resolution crystal structures reveal how a cellulose chain is bound in the 50 Å long tunnel of cellobiohydrolase I from Trichoderma reesei. J Mol Biol. 1998; 275: 309-25. [DOI] [PubMed] [Google Scholar]
- [8].Divne C, Ståhlberg J, Reinikainen T, Ruohonen L, Pettersson G, Knowles JKC, et al. The 3-dimensional crystal-structure of the catalytic core of cellobiohydrolase I from Trichoderma reesei. Science. 1994; 265: 524-8. [DOI] [PubMed] [Google Scholar]
- [9].Wilson DB. Processive and nonprocessive cellulases for biofuel production-lessons from bacterial genomes and structural analysis. Appl Microbiol Biotechnol. 2012; 93: 497-502. [DOI] [PubMed] [Google Scholar]
- [10].Kipper K, Våljamåe P, Johansson G. Processive action of cellobiohydrolase Cel7A from Trichoderma reesei is revealed as ‘burst’ kinetics on fluorescent polymeric model substrates. Biochem J. 2005; 385: 527-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Våljamåe P, Sild V, Pettersson G, Johansson G. The initial kinetics of hydrolysis by cellobiohydrolases I and II is consistent with a cellulose surface - erosion model. Eur J Biochem. 1998; 253: 469-75. [DOI] [PubMed] [Google Scholar]
- [12].Igarashi K, Koivula A, Wada M, Kimura S, Penttilå M, Samejima M. High speed atomic force microscopy visualizes processive movement of Trichoderma reesei cellobiohydrolase I on crystalline cellulose. J Biol Chem. 2009; 284: 36186-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Igarashi K, Uchihashi T, Koivula A, Wada M, Kimura S, Okamoto T, et al. Traffic jams reduce hydrolytic efficiency of cellulase on cellulose surface. Science. 2011; 333: 1279-82. [DOI] [PubMed] [Google Scholar]
- [14].Igarashi K, Uchihashi T, Koivula A, Wada M, Kimura S, Penttilå M, et al. Visualization of cellobiohydrolase I from Trichoderma reesei moving on crystalline cellulose using high-speed atomic force microscopy. Methods Enzymol. 2012; 510: 169-82. [DOI] [PubMed] [Google Scholar]
- [15].Uchiyama T, Uchihashi T, Nakamura A, Watanabe H, Kaneko S, Samejima M, et al. Convergent evolution of processivity in bacterial and fungal cellulases. Proc Natl Acad Sci USA. 2020; 117: 19896-903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Sild V, Ståhlberg J, Pettersson G, Johansson G. Effect of potential binding site overlap to binding of cellulase to cellulose: A two-dimensional simulation. FEBS Lett. 1996; 378: 51-6. [DOI] [PubMed] [Google Scholar]
- [17].Eriksson KE, Pettersson B. Extracellular enzyme system utilized by fungus Sporotrichum pulverulentum (Chrysosporium lignorum) for breakdown of cellulose. 1. Separation, purification and physicochemical characterization of 5 Endo-1,4-β-glucanases. Eur J Biochem. 1975; 51: 193-206. [DOI] [PubMed] [Google Scholar]
- [18].Streamer M, Eriksson KE, Pettersson B. Extracellular enzyme-system utilized by fungus Sporotrichum pulverulentum (Chrysosporium lignorum) for breakdown of cellulose - Functional characterization of 5 endo-1,4-β-Glucanases and one exo-1,4-β-glucanase. Eur J Biochem. 1975; 59: 607-13. [DOI] [PubMed] [Google Scholar]
- [19].Almin KE, Eriksson KE, Pettersson B. Extracellular enzyme system utilized by fungus Sporotrichum pulverulentum (Chrysosporium lignorum) for breakdown of cellulose. 2. Activities of 5 endo-1,4-β-glucanases towards carboxymethylcellulose. Eur J Biochem. 1975; 51: 207-11. [DOI] [PubMed] [Google Scholar]
- [20].Coughlan MP. Mechanisms of cellulose degradation by fungi and bacteria. Anim Feed Sci Technol. 1991; 32: 77-100. [Google Scholar]
- [21].Jalak J, Kurasin M, Teugjas H, Våljamåe P. Endo-exo synergism in cellulose hydrolysis revisited. J Biol Chem. 2012; 287: 28802-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Igarashi K, Maruyama M, Nakamura A, Ishida T, Wada M, Samejima M. Degradation of crystalline celluloses by Phanerochaete chrysosporium cellobiohydrolase II (Cel6A) heterologously expressed in methylotrophic yeast Pichia pastoris. J Appl Glycosci. 2012; 59: 105-10. [Google Scholar]
- [23].Mulakala C, Reilly PJ. Hypocrea jecorina (Trichoderma reesei) Cel7A as a molecular machine: A docking study. Proteins. 2005; 60: 598-605. [DOI] [PubMed] [Google Scholar]
- [24].Johnson KA, Goody RS. The original Michaelis constant: translation of the 1913 Michaelis-Menten paper. Biochemistry-Us. 2011; 50: 8264-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Michaelis L, Menten ML. Die kinetik der invertinwirkung. Biochem Z. 1913; 49: 333-69. [Google Scholar]
- [26].Nakamura A, Kanazawa T, Furuta T, Sakurai M, Saloheimo M, Samejima M, et al. Role of tryptophan 38 in loading substrate chain into the active-site tunnel of cellobiohydrolase I from Trichoderma reesei. J Appl Glycosci. 2021; 68: 19-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Kawai R, Igarashi K, Kitaoka M, Ishii T, Samejima M. Kinetics of substrate transglycosylation by glycoside hydrolase family 3 glucan (1->3)-β-glucosidase from the white-rot fungus Phanerochaete chrysosporium. Carbohydr Res. 2004; 339: 2851-7. [DOI] [PubMed] [Google Scholar]
- [28].Igarashi K, Momohara I, Nishino T, Samejima M. Kinetics of inter-domain electron transfer in flavocytochrome cellobiose dehydrogenase from the white-rot fungus Phanerochaete chrysosporium. Biochem J. 2002; 365: 521-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Igarashi K, Yoshida M, Matsumura H, Nakamura N, Ohno H, Samejima M, et al. Electron transfer chain reaction of the extracellular flavocytochrome cellobiose dehydrogenase from the basidiomycete Phanerochaete chrysosporium. FEBS J. 2005; 272: 2869-77. [DOI] [PubMed] [Google Scholar]
- [30].Ezaki T, Nishinari K, Samejima M, Igarashi K. Bridging the micro-macro gap between single-molecular behavior and bulk hydrolysis properties of cellulase. Phys Rev Lett. 2019; 122: 098102. [DOI] [PubMed] [Google Scholar]