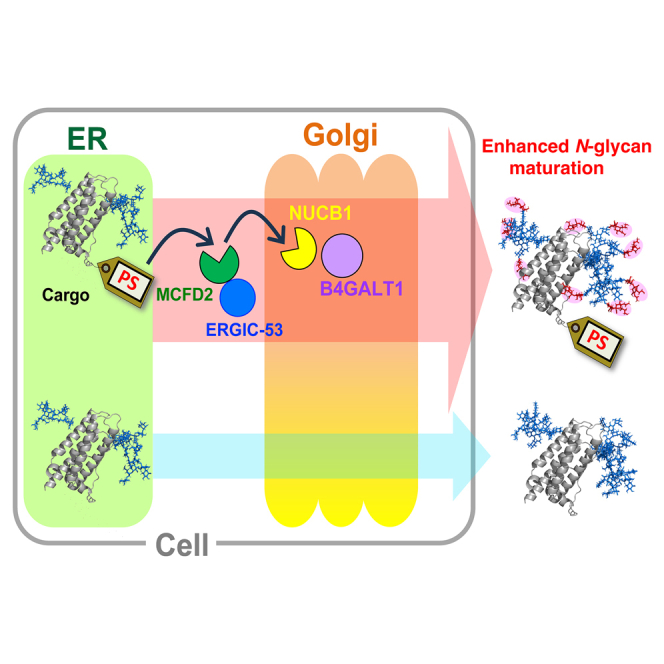
Summary
MCFD2 and ERGIC-53 form a cargo receptor complex that plays a crucial role in transporting specific glycoproteins, including blood coagulation factor VIII, from the endoplasmic reticulum to the Golgi apparatus. We have demonstrated that MCFD2 recognizes a 10-amino-acid sequence in factor VIII, thereby facilitating its efficient transport. Moreover, the secretion of biopharmaceutical recombinant glycoproteins, such as erythropoietin, can be enhanced by tagging them with this sequence, which we have termed the “passport sequence” (PS). Here, we found that the PS promotes the galactosylation and sialylation of N-glycans on glycoproteins. Furthermore, we discovered that glycoproteins tagged with the PS follow a unique route in the Golgi, where they encounter NUCB1. NUCB1 also recognizes the PS and mediates its interaction with the galactosylation enzyme B4GALT1. These findings offer a promising strategy for controlling the glycosylation of recombinant glycoproteins of biopharmaceutical interest.
Subject areas: Biochemistry, Protein, Properties of biomolecules, Glycobiology, Molecular biology
Graphical abstract
Highlights
-
•
The passport sequence promotes N-glycan maturation of recombinant glycoproteins
-
•
The passport sequence directs glycoproteins to interact with NUCB1 in Golgi
-
•
NUCB1 mediates the interaction of glycoproteins with galactosylation enzyme
-
•
This offers a novel strategy to control glycosylation of biopharmaceutical proteins
Biochemistry; Protein; Properties of biomolecules; Glycobiology; Molecular biology
Introduction
N-Glycosylation is a crucial process that most secretory and membrane proteins undergo within cells. Newly synthesized proteins are co-translationally modified with a tri-antennary, high-mannose-type tetradecasaccharide, which serves as a common precursor for N-glycans and undergoes sequential trimming by a series of glycosidases in the endoplasmic reticulum (ER).1 The resulting N-glycans, as processing intermediates, serve as quality control tags recognized by ER lectins, indicating the folding state of carrier proteins.2,3,4,5,6,7,8 Correctly folded glycoproteins are then transported from the ER to the Golgi apparatus, where the N-glycans are further trimmed and undergo diversification processes such as fucosylation, galactosylation, and sialylation, facilitated by specific glycosyltransferases.9 Through such maturation of the N-glycans, glycoproteins acquire diverse functions.10,11,12 Specifically, the affinity and specificity of glycoproteins for functionally interacting molecules are determined by their glycan structures. Furthermore, the in vivo dynamics and blood half-life of glycoproteins are governed by their glycans.13,14 This aspect is crucial in the development of glycoproteins as biopharmaceuticals.15
Certain glycoproteins require a specific cargo receptor for efficient translocation from the ER to the Golgi. MCFD2 and ERGIC-53 form a cargo receptor complex that plays a crucial role in transporting specific glycoproteins, including blood coagulation factor V (FV) and factor VIII (FVIII).16,17,18,19 ERGIC-53 is a lectin recognizing high-mannose-type N-glycans displayed on the glycoproteins as cargos, while MCFD2 is an EF-hand protein that is thought to recognize a specific sequence in the cargos.20,21,22 Genetic dysfunction of this cargo receptor complex causes combined deficiency of FV and FVIII (F5F8D).18
We have recently identified that MCFD2 recognizes a 10-amino-acid sequence in FVIII (SDLLMLLRQS at residues 807–816), thereby facilitating its efficient transport.21 Intriguingly, the secretion level of biopharmaceutical recombinant glycoproteins, such as erythropoietin (EPO), can be elevated by tagging them with this sequence. Hence, we named this sequence the “passport sequence” (PS) in the secretory pathway. These findings offer a potential approach for improvement in the production of recombinant glycoproteins of biopharmaceutical interest, simply by tagging them with the PS. In such applications, however, it is important not only to increase the production of biopharmaceutical glycoproteins but also to ensure their quality, including appropriate glycosylation.
In this study, we investigated the effect of PS tagging on N-glycosylation of secretory proteins primarily using recombinant EPO expressed in Expi293F cells as a model system. Consequently, we identified a previously unrecognized molecular mechanism by which the N-glycan maturation of secretory proteins is promoted in the MCFD2/ERGIC-53-mediated secretion pathway.
Results and discussion
Sialylation and galactosylation are promoted by PS tagging
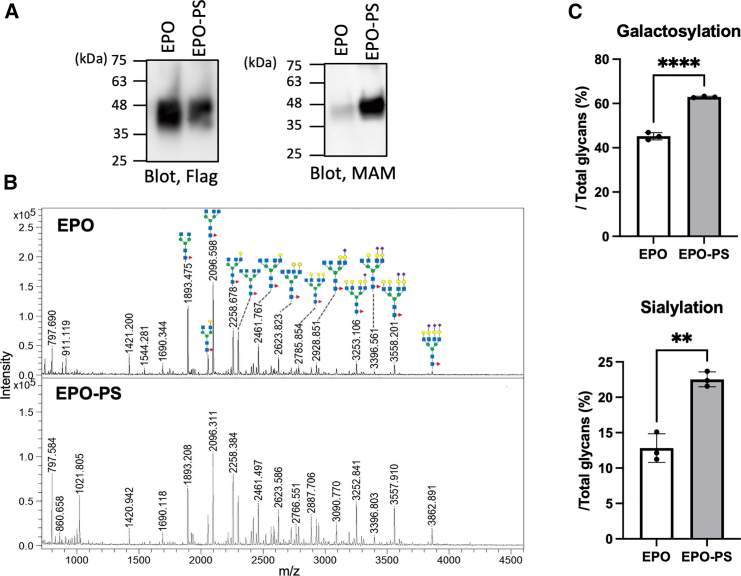
We first examined whether the C-terminal PS tagging, reported as the optimum method for enhancing secretion, influences the non-reducing terminal modifications of N-glycans of recombinant EPO secreted from Expi293F cells. Lectin blotting showed that the sialylation of EPO was enhanced by PS tagging (Figure 1A). Furthermore, N-glycosylation profiling using MALDI-TOF-MS revealed that the PS tagging promoted not only sialylation but also galactosylation, a prerequisite for the sialylation (Figures 1B and 1C). The levels of sialylation and galactosylation of soluble Fcγ receptor IIIa (sFcγRIII) were also elevated when expressed with PS tagging (Figure S1).
Figure 1.
The effect of passport sequence on the galactosylation and sialylation levels of recombinant EPO
(A) Tagging with the passport sequence increased the sialylation level of recombinant EPO produced in Expi293F cells. The sialylation level was determined through immunoblotting using an anti-FLAG antibody and lectin blotting using MAM.
(B) The N-glycans of recombinant EPO, with or without the passport sequence, were analyzed by MALDI-TOF-MS. The predicted structures, based on their mass values and the biosynthetic pathway, are shown above the profiles. The identities of sugar residues were inferred from the literature and are represented by symbols according to the Symbol Nomenclature for Glycans (SNFG) system. Details can be found at NCBI (http://www.ncbi.nlm.nih.gov/books/NBK310273/): GlcNAc, blue square; Man, green circle; Gal, yellow circle; Neu5Ac, magenta diamond; Fuc, red triangle.
(C) The galactosylation and sialylation levels of EPO, with or without the passport sequence, were calculated based on the peak intensities of MALDI-TOF-MS profiles. Error bars represent the standard error of the mean (SEM) from three independent transfections. Statistical significance was assessed by an unpaired t test: ∗∗∗∗p < 0.0001 and ∗∗p < 0.001.
In a previous study, we showed that FUT9, a human α1,3-fucosyltransferase, directly recognizes a specific sequence embedded within LAMP1, facilitating the LewisX modification of the N-glycans of recombinant glycoproteins tagged with this sequence.23 In human cells, the galactosylation of N-glycan outer branches is primarily catalyzed by B4GALT1, a β1,4-galactosyltransferase.24 Therefore, we tested the possibility that enhanced galactosylation in PS-tagged EPO is mediated through a direct interaction between the PS tag and B4GALT1. Recombinant EPO, produced by Expi293F cells and subjected to enzymatic treatment for de-sialylation and de-galactosylation, was used as a potential acceptor substrate in an in vitro enzymatic reaction with recombinant B4GALT1, both with and without the cleavage of the PS tag. The results indicated that B4GALT1 catalyzed galactosylation of EPO in a test tube, irrespective of PS tagging (Figure S2), suggesting that a certain intracellular condition is prerequisite for the PS-dependent promotion of N-glycan galactosylation.
PS defines cargo distribution in the Golgi
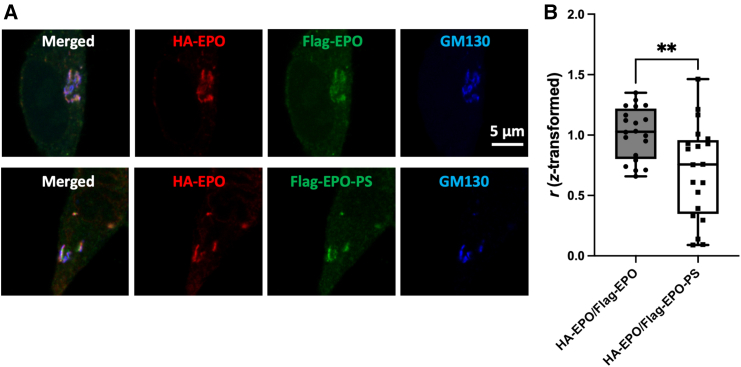
In the previous study, we demonstrated that interactions between the MCFD2/ERGIC-53 complex and the PS-tagged cargo glycoproteins enhance their trafficking efficiency from the ER to the ER-Golgi intermediate compartment (ERGIC), consequently increasing their expression level.21 We monitored the subsequent subcellular distribution of PS-tagged EPO (referred as EPO-PS) compared to EPO without PS (simply referred as EPO). Immunostaining revealed that both EPO and EPO-PS were co-localized with GM130, a cis-Golgi marker, suggesting that they were primarily distributed in the Golgi before being secreted to the extracellular space. Nevertheless, EPO-PS exhibited distinct distributions from EPO within the Golgi (Figure 2).
Figure 2.
The effect of the passport sequence on subcellular localization of recombinant EPO
(A) Immunocytochemistry of HA-EPO and FLAG-EPO-PS expressed in Expi293F cells. The transfected cells were treated with 4% paraformaldehyde solution in PBS, incubated with PBS containing 3% fetal bovine serum and 0.1% Triton X-100, then stained with anti-FLAG, anti-HA, and anti-GM130 antibodies as primary antibodies, and Alexa Fluor 488-conjugated anti-mouse IgG (green), Alexa Fluor 555-conjugated anti-rat IgG (red), and Alexa Fluor 350-conjugated anti-rabbit IgG (blue) as secondary antibodies. Scale bar: 5 μm.
(B) Quantification of colocalization based on Fisher Z-transformed values, which were reconverted to Pearson’s correlation coefficient values (r). Data represent the means of twenty cells. Error bars indicate SEM. Statistical significance was assessed by unpaired t test: ∗∗p < 0.001.
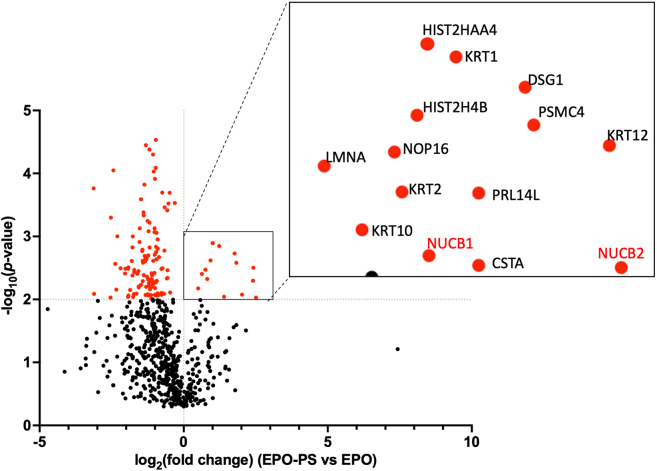
We used a proximity-dependent labeling technique in order to identify molecules localized in the sub-Golgi zone where EPO-PS is specifically distributed. EPO and EPO-PS, both fused with the biotin ligase TurboID, were expressed in Expi293F cells. The biotinylated proteins were then collected and subjected to liquid chromatography-mass spectrometry (LC-MS) analysis. The identified proteins included Golgi proteins specific to EPO-PS (Figure 3 and Table S2), indicating distinct sub-Golgi environments for PS-tagged cargo glycoproteins, which are transported into the Golgi through interactions with the MCFD2/ERGIC-53 complex.
Figure 3.
Volcano plot highlighting significantly enriched EPO-proximal proteins measured with LC-MS/MS
Expi293F cells expressing TurboID-EPO or TurboID-EPO-PS were incubated for 1 h with biotin to enrich biotinylated proteins in the secretory pathway. After pull down with streptavidin-Sepharose, the tryptic digested samples were analyzed using MS (n = 4).
NUCB1 mediates PS-dependent N-glycan maturation
The results obtained thus far suggest that a certain environmental factor in the sub-Golgi zone specific for the PS-tagged cargos enhances the enzymatic function of B4GALT1. We hypothesized that this enhancement involves a certain Golgi protein located in the zone where B4GALT1 and PS-tagged cargos co-exist. We attempted to capture this protein by proximity-dependent labeling. We identify Golgi proteins biotinylated with TurboID-fused B4GALT1 and compared them with the EPO-PS-specific proximal proteins (Table S3). As a result, NUCB1 and NUCB2 were found as common proximate proteins shared by B4GALT1 and EPO-PS.
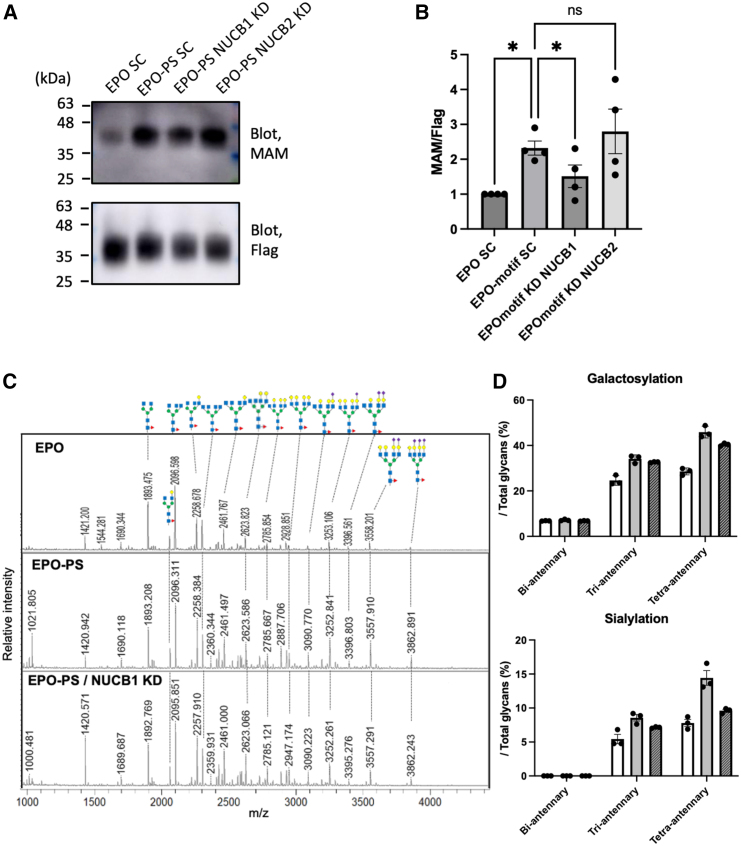
To determine whether these proteins are involved in the PS-dependent promotion of N-glycan maturation, EPO-PS was expressed in Expi293 cells with siRNA-mediated suppression of NUCB1 or NUCB2. Lectin blotting and MALDI-based glycosylation profiling revealed that the PS-dependent enhancement of galactosylation and sialylation was compromised by the knockdown of NUCB1 but not NUCB2 (Figure 4 and Figure S3). This indicates that NUCB1 is involved in PS-dependent N-glycan maturation of cargo glycoproteins. Knocking down NUCB1 did not affect secretory trafficking (Figure S4), suggesting that the increased secretion driven by the PS is likely due to efficient transport mediated by the MCFD2-ERGIC53 cargo receptor complex.
Figure 4.
The effect of NUCB1 on PS-dependent N-glycan maturation of recombinant EPO
(A) Knocking down NUCB1 decreased the PS-dependent sialylation level of EPO, as shown by lectin blotting with MAM. Expi293F cells expressing EPO or EPO-PS were treated with control-siRNA, NUCB1-siRNA, or NUCB2-siRNA. After 24 h of cultivation, EPO and EPO-PS were purified from the culture medium using anti-FLAG antibody-conjugated resin, followed by blotting with an anti-FLAG antibody and MAM lectin. See also Figure S3.
(B) The relative sialylation levels (MAM/FLAG) of EPO or EPO-PS in the culture medium of Expi293F cells treated with control-siRNA, NUCB1-siRNA, or NUCB2-siRNA were determined with immuno- and lectin-blotting. Error bars represent the SEM (n = 4 independent transfections). Significant differences (∗) were calculated compared with EPO expression levels using a one-way ANOVA with multiple comparisons (p < 0.05).
(C) The N-glycans of EPO and EPO-PS treated with control-siRNA and NUCB1-siRNA were observed using MALDI-TOF-MS analysis. The predicted structures based on their mass values and biosynthetic pathway are shown above the profile. Refer to the legend in Figure 1 for the keys.
(D) The galactosylation and sialylation levels of EPO (white) and EPO-PS (gray) treated with scrambled- and NUCB1-siRNA (striped) were calculated by the peak intensity of MALDI-TOF-MS profiles based on the structures. Error bars represent the SEM (n = 3).
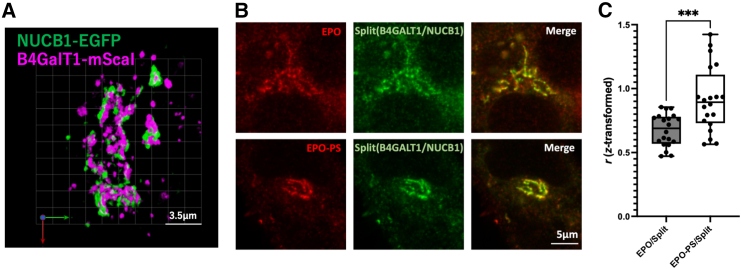
NUCB1 is a Ca2+-binding protein having an EF hand domain and located in the cis-Golgi, tightly associated with the luminal side of the membrane,25,26,27,28 in contrast to B4GALT1, which is a type-II membrane protein primarily localized in the trans-Golgi.29 Consistently, our simultaneous dual-color 3D observation by super-resolution confocal live imaging microscopy (SCLIM)30,31 confirmed localizations of these proteins are obviously different (Figure 5A). In addition, proximity ligation analysis with NUCB1 indicated that neither B4GALT1 nor other glycosyltransferases were significantly enriched among the identified proteins (Figure S5 and Table S4). However, it should be noted that significant fractions of NUCB1 and B4GALT were co-localized within the Golgi (Figure 5A). We further characterize their co-localization by proximity-dependent labeling using a split TurboID shared between B4GALT1 and NUCB1. Interestingly, the sub-Golgi zone mapped with biotinylation was considerably overlapped with the EPO-PS-specific zone (Figures 5B and 5C). These data indicate that the glycoproteins carrying PS are distributed in a unique sub-Golgi zone where NUCB1 and B4GALT1 co-exist.
Figure 5.
Fluorescence imaging of NUCB1 and B4GALT1 as well as EPO with or without passport sequence
(A) Dual-color 3D SCLIM images of the Golgi ribbon area in Expi293F cells expressing NUCB1-EGFP (green) and B4GALT1-mScarlet-I (magenta). Scale bars: 3.5 μm.
(B) Fluorescence imaging of split-TurboID shared by NUCB1 and B4GALT1. Expi293F cells transiently transfected with FLAG-EPO/FLAG-EPO-PS constructs and split-TurboID constructs (B4GALT1-TbN and NUCB1-TbC). The cells were treated with 50 μM biotin followed by 30 min of incubation. After fixations, the cells were treated with PBS containing 3% fetal bovine serum and 0.1% Triton X-100, then stained with an anti-FLAG as primary antibody, followed by Alexa Fluor 488-conjugated streptavidin (green) and Alexa Fluor 555-conjugated anti-mouse IgG (red) as secondary antibodies. Scale bar: 5 μm.
(C) Quantification of colocalization of EPO and EPO-PS with biotinylation by split-TurboID based on Fisher Z-transformed values, which were reconverted to Pearson’s correlation coefficient values (r). Data represent the means of at least twenty cells. Error bars indicate SEM. Statistical significance was assessed by unpaired t test: ∗∗∗p < 0.001.
Mechanistic insights into the PS functions
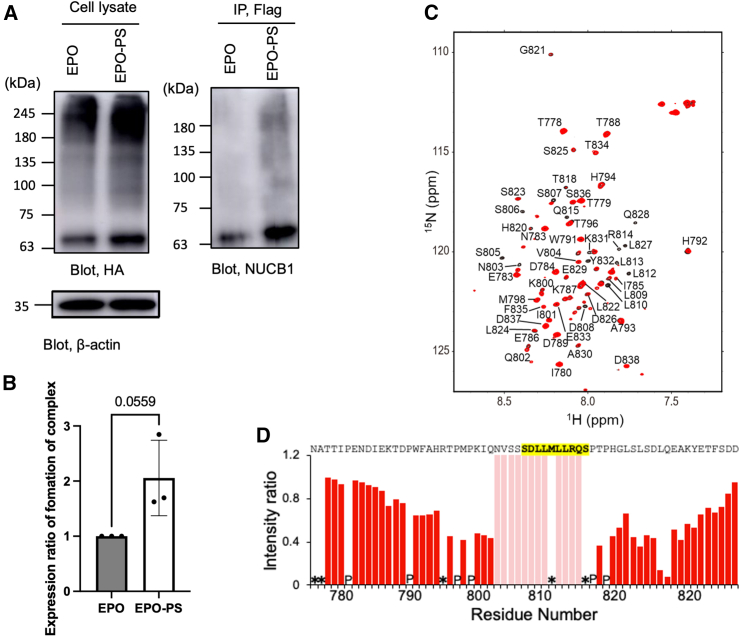
NUCB1 and MCFD2 are both EF-hand proteins. MCFD2 recognizes PS through its EF-hands. This prompted us to examine whether NUCB1 directly interacts with PS. For a pull-down assay, extracts of HCT116 cells co-expressing NUCB1-HA and FLAG-EPO with or without PS were subjected to chemical cross-linking and subsequent immunoprecipitation with an anti-FLAG antibody. The results showed that PS-tagged EPO was enriched in the fraction co-precipitating with NUCB1-HA compared to EPO without PS (Figures 6A and 6B). Furthermore, NMR analysis showed that NUCB1 interacts with PS in a 60-amino-acid segment corresponding to Asn776–Asp838 of FVIII (Figures 6C and 6D).
Figure 6.
Interaction of NUCB1 with PS
(A) FLAG-NUCB1 and EPO with or without passport sequence (EPO/EPO-PS) transiently expressed in HCT116 cells were homogenized and crosslinked with disuccinimidyl suberate. The cell lysates were subjected to immunoblotting with anti-HA and anti-β-actin antibodies and immunoprecipitation with anti-FLAG antibody. Immunoblot analysis of NUCB1 in the immunoprecipitates was performed using an anti-NUCB1 antibody.
(B) Density plot of relative chemical-linked NUCB1/EPO or NUCB1/EPO-PS expression levels normalized by NUCB1/EPO expression levels. Error bars represent the SEM (n = 3 independent experiments). Statistical significance was assessed by unpaired t test (p = 0.0559).
(C) 1H–15N heteronuclear single quantum coherence (HSQC) spectra of the 15N-labeled FVIII-derived peptide (Asn776–Asp838) alone (black) or in the presence of one molar equivalent of NUCB1 (red). The spectral assignments are based on our previous report.21.
(D) Plots of the intensity ratios of the backbone amide peaks of the FVIII-derived peptide with and without NUCB1. Pink bars indicate residues whose NMR peaks were undetectable due to extreme broadening upon addition of NUCB1. In the profiles of intensity ratios, residues for which the 1H–15N HSQC peaks could not be observed because of peak overlapping and/or broadening were denoted with asterisks, whereas proline residues were denoted with “P.”.
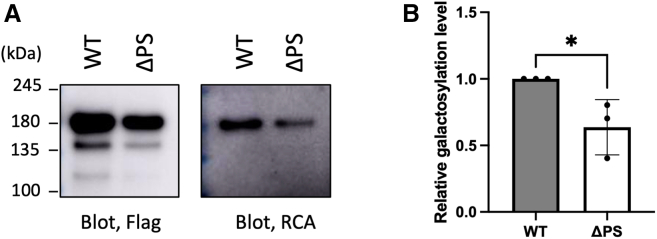
Since FVIII is a very large glycoprotein with a complicated structure, it requires the distinct, selective transport mechanism mediated by the MCFD2/ERGIC-53 cargo receptor complex.16,17,18,19 We previously demonstrated that FVIII harbors PS in the B domain and deletion of this sequence results in marked reduction in FVIII secretion.21 FVIII possesses multiple N-glycosylation sites, predominantly displaying bi-antennary complex-type oligosaccharides with sialylation, along with lower levels of high-mannose, tri-antennary, and tetra-antennary species.32 The N-glycosylation profile of recombinant FVIII depends on the production protocol. We examined the possible effect of PS deletion on N-glycosylation. The results revealed that the deletion of PS from FVIII led to a reduction not only in its secretion level but also in its galactosylation level (Figures 7 and S6). This suggests that the presence of PS contributes to the N-glycan maturation of FVIII, which is a native cargo in the MCFD2/ERGIC-53-mediated secretion pathway.
Figure 7.
PS-dependent N-glycan maturation of recombinant FVIII produced in Expi293F cells
(A) Blotting of FLAG-tagged wild-type FVIII (WT) and its PS-deleted mutant (ΔPS) treated with sialidase, utilizing RCA and anti-FLAG antibodies to assess galactosylation levels.
(B) Relative galactosylation levels of FVIIIΔPS to PS-harboring FVIII based on normalized RCA/FLAG ratios. Error bars denote the SEM (n = 3 independent experiments). Statistical significance was assessed by unpaired t test: ∗p < 0.05. See also Figure S6.
The MCFD2/ERGIC-53 cargo receptor complex shuttles between the ER and the ERGIC, where it releases its cargo glycoproteins.16,17,20 This study suggests that the cargo glycoproteins delivered by this complex subsequently traverse a specialized route through the Golgi apparatus (Figure 2). This Golgi trafficking pathway is mediated by NUCB1 as a secondary passport inspector and is equipped with the enzymes B4GALT1 and likely other N-glycan processing enzymes that are prerequisite for the galactosylation and subsequent sialylation of the cargo glycoproteins.
It has recently been reported that NUCB1 interacts with matrix metalloproteases (MMPs) and promotes their intra-Golgi trafficking.33 Intriguingly, ERGIC-53 has been identified as a potential cargo receptor for secretion of MMP-9.34 These data suggest that, in addition to the blood coagulation factors, other cargo glycoproteins may also utilize this pathway physiologically.
Applied perspectives
The N-glycan structure is a critical determinant of the circulatory half-life of glycoproteins, including FVIII.15,35,36,37 In particular, sialylation prolongs the in vivo survival of glycoproteins by masking galactose residues, thereby preventing their binding to the endocytic lectin asialoglycoprotein receptor.13,14,38 Most biopharmaceuticals currently in use are recombinant glycoproteins, such as FVIII and EPO.39 Extensive research has demonstrated that variations in the glycosylation profiles of these biopharmaceutical glycoproteins can significantly impact their efficacy including pharmacokinetic, pharmacodynamic, and immunogenic properties.15,35,40 Therefore, numerous attempts have been made to improve these properties by controlling glycosylation, with sialylation enhancement considered one of the most promising approaches.41
Accumulating evidence suggests that protein glycosylation is not a chaotic, stochastic process but is deterministic to some extent, depending on the interplay between a series of enzymes and glycoproteins as substrates.42,43 While the enzyme-substrate interactions or the accessibilities of the enzymes to the N-glycans are generally governed by their tertiary structures,44,45,46 certain glycosyltransferases directly recognize specific codes embedded in glycoproteins, facilitating targeted glycosylation.23,47,48 Incorporating such codes could potentially modify the glycosylation of biopharmaceutical glycoproteins.
In this study, we revealed the existence of another class of embeddable molecular codes that can determine the specific trafficking route of cargo glycoproteins in the secretory pathway, thereby controlling their interactions with glycosyltransferases and consequently modifying their glycosylation. Our findings offer a unique strategy for facilitating N-glycan maturation as well as the secretion of recombinant glycoproteins of biopharmaceutical interest. Recent studies have demonstrated that Golgi glycosyltransferases exhibit significantly different sub-Golgi localization, controlled by molecular codes embedded in the enzymes.49,50 Comprehensive understanding of the molecular codes that directly or indirectly control the interplay between cargo glycoproteins and glycosyltransferases will help decipher the putative blueprint of protein glycosylation in the secretory pathway, opening the possibility of tailored control of glycosylation in biopharmaceutical recombinant glycoproteins.
Limitations of the study
The mechanistic insights proposed in this study, particularly regarding the interactions among the cargo receptor, PS-tagged cargos, MCFD2, NUCB1, and B4GALT1, are based on qualitative observations, including proximity-dependent labeling and immunoprecipitation with chemical cross-linking. To enhance our understanding of these mechanisms, more quantitative data are necessary to elucidate the network of molecular interactions involved. This requires detailed information on the abundance of each molecule within the specific cellular microenvironment, as well as the environmental factors (such as local pH and metal ion concentrations) that may influence these interactions. Addressing these limitations will be crucial for future investigations.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Hirokazu Yagi (hyagi@phar.nagoya-cu.ac.jp).
Materials availability
Materials generated in this study will be made available on request, but we may require a completed Materials Transfer Agreement.
Data and code availability
-
•
The data on proximity-dependent labeling and protein identification using mass spectrometry have been deposited to jPOST under the ID JPST003247 and JPST003457. The microscopy images for the quantification analysis of colocalization have been deposited at Mendeley (http://www.doi.org/10.17632/przzrd7sb9.1) and are publicly available as of the date of publication.
-
•
This study did not produce any original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Acknowledgments
We are grateful to Prof. Kazuhiro Aoki (ExCELLS) for providing the expression vectors. We thank Ms. Kiyomi Senda, Ms. Kumiko Hattori (Nagoya City University), and Ms. Miho Waga (RIKEN) for help in constructing the expression vector and preparing the recombinant proteins. This work was supported in part by JST-CREST (grant number JPMJCR20E6 to S.A and JPMJCR21E3 to T.T. and K.K.), JST FOREST Program (grant number JPMJFR2255 to H.Y.), the Human Glycome Atlas Project, MEXT/JSPS KAKENHI (grant numbers JP22H02225 to S.A, JP22K06213 to T.T., JP17H06414 and JP21H02625 to H.Y. and JP20K21495 and JP24H00599 to K.K.), Grant-in-Aid for Scientific Research on Innovative Areas — Platforms for Advanced Technologies and Research Resources “Advanced Bioimaging Support,” MEXT Promotion of Development of a Joint Usage/ Research System Project: Coalition of Universities for Research Excellence Program (CURE) (grant number JPMXP1323015488 to K.K.), the Center for the Promotion of Integrated Sciences (CPIS) of SOKENDAI (to K.K.), the ExCELLS Advanced Co-creation Platform (Spatiotemporal atlas of dynamic structure and function of organelles, 23EXC601 to H.Y.), and ExCELLS “Golgi Atlas Project” (to K.K.). We are grateful to the Analytical Center, Meijo University for the MALDI-TOF/MS measurements of oligosaccharides, and to the Research Equipment Sharing Center at Nagoya City University for technical assistance. The NMR experiments were performed at ExCELLS of the NMR Platform supported by MEXT.
Author contributions
Conceptualization, H.Y. and K.K.; investigation, H.Y., R.Y, T.S., R.H., R.N, T.T., A.N., J.-i.F., H.K., K.N., S.A, K.I, S.Y., and M.Y.-U.; writing – original draft, H.Y. and K.K.; writing – review & editing, T.T., M.Y.-U., and K.K.; supervision, H.Y. and K.K.; project administration, H.Y. and K.K.; funding acquisition, H.Y., S.A., T.T., and K.K.
Declaration of interests
The authors declare no competing interests.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-Flag antibody (Clone M2) | Sigma-Aldrich | Cat# F1804; RRID: AB_262044 |
| anti-HA antibody (3F10) | Roche | Cat#12158167001; RRID: AB_390919 |
| GOLGA2/GM130 Polyclonal antibody | Proteintech | 11308-1-AP; RRID: AB_2919763 |
| Alexa Fluor 488-conjugated anti-mouse IgG | Thermo Fisher Scientific | Cat # A32723; RRID: AB_2633275 |
| Alexa Fluor 555-conjugated anti-rat IgG | Thermo Fisher Scientific | Cat # A-21434; RRID: AB_2535855 |
| Alexa Fluor 350-conjugated anti-rabbit IgG | Thermo Fisher Scientific | Cat # A-11046; RRID: AB_2534101 |
| anti-β-actin | Sigma-Aldrich | Cat #A2228; RRID: AB_476697 |
| Anti-Mouse IgG, HRP-Linked Whole Ab Sheep | Cytiva | Cat #NA931-1ML; RRID: AB_772210 |
| Anti-rabbit IgG, HRP-linked Antibody | Cell Signaling Technology | Cat #7074; RRID: AB_2099233 |
| Bacterial and virus strains | ||
| ECOSTM Competent E. coli DH5 α | Nippongene | Cat #310-06231 |
| E. coli BL21-CodonPlus (DE3) | Agilent Technologies | Cat #230245 |
| Chemicals, peptides, and recombinant proteins | ||
| DMEM, high glucose, GlutaMAX™ Supplement, pyruvate | Thermo Fisher Scientific | Cat# 10569010 |
| Expi293 Expression Medium | Thermo Fisher Scientific | Cat# A1435101 |
| NEBuilder® HiFi DNA Assembly Master Mix | NEW ENGLAND BioLabs | Cat# E2621S |
| KOD -Plus- Ver.2 | TOYOBO | Cat# KOD-211 |
| Polyethylenimine “Max” | Polysciences | Cat# 24765 |
| 293fectin™ Transfection Reagent | Thermo Fisher Scientific | Cat# 12347019 |
| poly-L-lysine hydrobromide | Sigma-Aldrich | Cat# P4832 |
| Lipofectamine 3000 | Thermo Fisher Scientific | Cat# L3000001 |
| isopropyl β-d-thiogalactopyranoside | WAKO | Cat# 096-05143 |
| cOmplete His-Tag Purification Resin | Roche | Cat# 08778850001 |
| Superdex™ 200 Increase 10/300 GL | Cytiva | Cat# 28990944 |
| anti-Flag M2 Affinity Gel | Sigma-Aldrich | Cat# A2220 |
| EZview™ Red anti-HA Affinity Gel | Sigma-Aldrich | Cat# E6779 |
| 3X FLAG tag peptide | Sigma-Aldrich | Cat# F4799 |
| HA Synthetic Peptide | Sigma-Aldrich | Cat# 11666975001 |
| Triton X-100 | Sigma-Aldrich | Cat#X-100 |
| Nα-((aminooxy)acetyl)tryptophanylarginine methyl ester (aoWR) | In house | https://www.sciencedirect.com/science/article/pii/S1535947620300347 |
| dihydrobenzoic acid (DHBA) | Sigma-Aldrich | Cat# 149357 |
| ethylenediaminetetraacetic acid (EDTA) | WAKO | Cat# 800682 |
| SoftLink™ Soft Release Avidin Resin | Promega | Cat# V2011 |
| 2-mercaptoethanol | WAKO | Cat# 131-14572 |
| 2-iodoacetamide | WAKO | Cat# 095-02151 |
| dithiothreitol | WAKO | Cat# 040-29223 |
| Sera-Mag™ SpeedBead Carboxylate-Modified [E3] Magnetic Particles | Cytiva | Cat#65152105050250 |
| Sera-Mag™ SpeedBead Carboxylate-Modified [E7] Magnetic Particles | Cytiva | Cat#45152105050250 |
| acetonitrile | WAKO | Cat# 014-00381 |
| lysylendopeptidase | Wako | Cat# 121-05063 |
| trypsin | Thermo Fisher Scientific | Cat# 90057 |
| Blocking One solution | Nacalai Tesque | Cat# 03953-95 |
| RCA-biotin | Vector | Cat# B-1085 |
| MAM biotin | MBL | Cat# J1001009 |
| HRP-conjugated streptavidin | Sigma-Aldrich | Cat# SA10001 |
| Immobilon Western Chemiluminescent HRP substrate solution | Millipore | Cat# WBKLS0500 |
| TRIzol reagent | Thermo Fisher Scientific | Cat#15596026 |
| sialidase from Arthrobacter ureafaciens | Nacalai Tesque | Cat# 24229-74 |
| β1-4galactosidase cloned from Streptococcus pneumoniae | New England Biolabs | Cat# P0745S |
| TEV protease | In house | N/A |
| uridine diphosphate-galactose | Sigma-Aldrich | Cat# 670111-M |
| Critical commercial assays | ||
| BlotGlyco beads | Sumitomo Bakelite | |
| PureLink™ RNA Mini Kit | Promega | Cat# 12183018A |
| Luna® universal qPCR master mix and LunaScript™ RT supermix kit | NEW ENGLAND BioLabs | Cat# M3003S |
| Deposited Data | ||
| LC-MS data | jPOST | ID: JPST003247 |
| LC-MS data | jPOST | ID: JPST003457 |
| Microscopy image Data | Mendeley Data | https://doi.org/10.17632/przzrd7sb9.2 |
| Experimental models: Cell lines | ||
| HCT116 | ATCC | CCL-247 |
| HeLa | RIKEN BRC | RCB0007: HeLa |
| Expi293F | Thermo Fisher Scientific | Cat# A14527 |
| Recombinant DNA | ||
| Plasmid | This Paper | Table S1 |
| C1(1–29)-TuroboID-V5 pCDNA3 | Addgene | #107173 |
| Software and algorithms | ||
| ImageJ | National Institutes of Health | https://imagej.net/ij/ |
| GraphPad Prism 9 | Dotmatics | RRID: SCR_002798; https://www.graphpad.com/ |
| SPARKY software | Goddard TD & Koeller DG51 | N/A |
| TopSpin | Bruker BioSpin | https://www.bruker.com/en/products-and-solutions/mr/nmr-software/topspin.html |
| Proteome Discoverer 2.2 with the SEQUEST search engine | Thermo Fisher Scientific | N/A |
Experimental model and study participant details
Microbe strains
We used Escherichia coli BL21(DE3) Codonplus strain (Agilent Technologies) for expression of recombinant proteins and DH5-alpha competent cells (Nippongene) for plasmid construction.
Cell lines
All cell lines used in this study for cellular assays are listed in the key resources table. HCT116 cells were obtained from ATCC and HeLa cells from RIKEN Cell bank. Expi293F cells was obtained from ThermoFisher Scientific. All cell lines were verified to be free of mycoplasma contamination.
Method details
Construction of plasmids
The plasmids used in this study are summarized in Table S1, which also includes Benchling links to the plasmid sequences and maps. The constructs for mammalian expression used in this study were inserted into pCAGGS or px3FlagCMV vectors, which included CAG or cytomegalovirus promoters and c-fos3′UTR, rabbit β-globin 3′-flanking sequence or growth hormone polyadenylation terminators. For bacterial expression, the pCold vector was used. Constructs were cloned using NEBuilder HiFi DNA Assembly (NEW ENGLAND BioLabs) with backbone PCR, or by ligation of double-digested DNA amplified by PCR. The TurboID gene was obtained from Addgene (C1(1–29)-TuroboID-V5 pCDNA3, #107173). The expression vector (NUCB1-HA) for human nucleobindin-1 (NUCB1) fused with an HA tag at the C-terminus is purchased from VecterBuilder.
Cell culture
HCT116 and HeLa cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum at 37°C in a 5% CO2 atmosphere. Expi293F cells (Thermo Fisher Scientific) were maintained in Expi293 Expression Medium (Thermo Fisher Scientific) under at 37°C with 8% CO2 on an orbital shaker platform.
Transfection
For transfection, HCT116 and HeLa cells were transfected using Polyethylenimine “Max” (PEI Max) (Polysciences) as previously described.52 Expi293F cells were transfected using 293fectin Transfection Reagent (Thermo Fisher Scientific) according to the manufacturer’s instruction. For the imagining analysis, cells were plated on a 35-mm glass-bottom dish (Iwaki) coated with poly-L-lysine (Sigma-Aldrich) the day before transfection with the expression plasmids using 293fectin Transfection Reagent or Lipofectamine 3000 (Thermo Fisher Scientific).
Preparation of proteins
The 13C-labeled FVIII fragment (Asn776–Asp838) was prepared using the protocol described previously.21 NUCB1 was expressed in the Escherichia coli BL21(DE3) Codonplus strain, induced with 0.5 mM isopropyl β-d-thiogalactopyranoside. The protein was purified using a cOmplete His-Tag Purification Resin (Roche), treated with TEV protease to cleave the hexahistidine tag, and further purified with a gel filtration column (Superdex 200 Increase; Cytiva).
For the expression of Flag- or HA-tagged soluble recombinant proteins (EPO-TEV-PS, FVIII-Flag, FVIII-ΔPS-Flag, NUCB1-HA) in mammalian cells, the tagged proteins were purified from the culture medium at 72 h post-transfection using anti-Flag M2 Affinity Gel (Sigma-Aldrich) or anti-HA Affinity Gel (Sigma-Aldrich). Proteins were eluted from each resin with 3X FLAG tag peptide (Sigma-Aldrich) or HA Synthetic Peptide (Sigma-Aldrich) in PBS. Recombinant full-length B4GALT1 was purified using affinity tags from cell lysate prepared by homogenizing cultured cells in lysis buffer (20 mM Tris-HCl, pH 7.6, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100), followed by centrifugation.
N-glycan structural analysis
The structural analysis of N-glycans was conducted following the methodologies outlined in previous studies.53,54 N-glycans were released from the proteins (100 μg) with 2 units of PNGase F (Roche) after reductive alkylation and trypsin digestion. The released N-glycans were then captured and labeled with Nα-((aminooxy)acetyl)tryptophanylarginine methyl ester (aoWR) using BlotGlyco beads (Sumitomo Bakelite).
After the removal of excess reagents via a HILIC column, matrix-assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF MS) analyses of aoWR-labeled glycans were carried out on an Autoflex Speed instrument (Bruker Daltonics), operated in positive-ion reflector mode. For MS acquisition, the aoWR-labeled glycans in water were mixed in a 1:1 ratio with dihydrobenzoic acid (10 mg/mL in 50% acetonitrile) and then spotted onto the target plate.
Microscopy
After overnight culture, cells expressing recombinant proteins were observed using florescence microscopes. Cell images were captured using the fluorescence microscope BZ-X700 and BZ-H3XF/Sectioning Module (Keyence), Leica TCS SP8 LIGHTNING (Leica Microsystems), and a super-resolution confocal live imaging microscope (SCLIM).30,31 For immunocytochemistry, primary antibodies used were anti-Flag (Sigma-Aldrich, Clone M2), anti-HA (Roche, 3F10), and anti-GM130 (Proteintech, 11308-1-AP). The secondary antibodies employed were Alexa Fluor 488-conjugated anti-mouse IgG (Thermo Fisher Scientific, Cat # A32723), Alexa Fluor 555-conjugated anti-rat IgG (Thermo Fisher Scientific, Cat # A-21434), and Alexa Fluor 350-conjugated anti-rabbit IgG (Thermo Fisher Scientific, Cat # A-11046).
TurboID sample preparation
Proximity labeling using the TurboID system was performed as previously described.55 Expi293F cells were allowed to adhere to the culture dish and then transfected with plasmids encoding TurboID-EPO, TurboID-EPO-PS, B4GALT-TurboID, B4GALT1-TbN, NUCB1-TbN and/or NUCB1-TbC. For proximity ligation and subsequent MS analysis, 50 μM biotin was added to the cells culture media 1 h after transfection, and the cells were incubated for an additional 1 h. For MS analysis, the cells were lysed in a buffer containing 25 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1 mM ethylenediaminetetraacetic acid (EDTA) and 1% Triton X-100. The cell lysates were homogenized by passing through a 26G syringe needle (Nipro). After centrifugation at 10,000 g for 2 min, the supernatant was collected as cell lysate. The biotinylated proteins in the cell lysates were captured using SoftLink Soft Release Avidin Resin (Promega). The resin was washed with a TBS buffer containing 1% Triton X-100, and the biotinylated proteins were eluted by incubation at 95°C for 2 min in a buffer containing 500 mM Tris-HCl, 10% glycerol, 2% SDS, and 5% 2-mercaptoethanol.
For the proximity ligation and subsequent microscopy imaging using split-TurboID constructs (B4GALT1-TbN and NUCB1-TbC), 50 μM biotin was added 12 h after transfection, and the cells were cultured for 5 h. The biotinylated proteins were then detected using a florescence microscope.
Mass spectrometry for protein identification
The peptides from biotinylated protein fractions were prepared for LC-MS analysis as described below: Eluted samples (10 μg) were adjusted to 100 μL by adding 1% SDS, then supplemented with 10 mM dithiothreitol and 10 mM 2-iodoacetamide. After a 30-min incubation at room temperature in the dark, magnetic beads (prepared by combining 10 μL each of Sera-Mag SpeedBead Carboxylate-Modified Magnetic Particles [E3 and E7] with 400 μL 100% acetonitrile (ACN)) were added to each sample to achieve a final concentration of 80% ACN. The mixture was allowed to bind for 5 min, followed by 2 min on a magnetic rack to immobilize the beads. The supernatant was then removed, and the beads were washed three times with 500 μL, 600 μL, and 800 μL of 80% ethanol, respectively. Next, the beads were resuspended in 50 μL of 1.4 M guanidine hydrochloride containing 100 ng of lysylendopeptidase (Wako) and incubated for 2 h at 37°C. Subsequently, 100 ng of trypsin (Thermo Fisher Scientific) was added to the samples, which were then incubated overnight at 37°C. The digested samples underwent purification using a magnetic rack to immobilize the beads. The generated peptides were examined utilizing an Evosep One LC system (EVOSEP) in conjunction with a Q-Exactive HF-X mass spectrometer (Thermo Scientific). The mobile phases consisted of 0.1% formic acid in water (solution A) and 0.1% formic acid in 99.9% acetonitrile (solution B). The analysis was performed in a data-dependent acquisition mode, selecting the top 25 MS spectra in the range of 380–1500 m/z for recording. All MS/MS spectra were queried against the protein sequences from the human Swiss-Prot database using Proteome Discoverer 2.2 with the SEQUEST search engine. The false discovery rate for peptide spectrum matches was set at 1%.
NMR spectroscopy
The 15N-labeled FVIII-derived peptide (0.1 mM) was dissolved in 20 mM MES (pH 6.0) containing 10 mM CaCl2, 150 mM NaCl, and 10% (v/v) D2O in the presence or absence of one molar equivalent of NUCB1. 1H–15N HSQC spectral measurements were conducted using AVANCE 800 spectrometer equipped with a 5-mm triple-resonance cryogenic probe (Bruker BioSpin). The NMR data were processed and analyzed with TopSpin (Bruker BioSpin) and SPARKY software.51
Immunoblotting and lectin blotting
Immunoblotting and lectin blotting were performed following established methods from previous studies.21,23 Purified proteins or cell lysates were resolved by 3–10% gradient SDS-polyacrylamide gel electrophoresis (PAGE) and subsequently transferred to a polyvinylidene difluoride (PVDF) membrane (Millipore). After blocking with Blocking One solution (Nacalai Tesque), membranes were incubated with monoclonal mouse anti-Flag M2 (Sigma-Aldrich, F1804) and anti-β-actin (Sigma-Aldrich, A2228) antibodies, followed by horseradish peroxidase (HRP)-conjugated secondary antibody (anti-mouse IgG antibody-HRP [Cytiva] and anti-rabbit Ig antibody-HRP [Cell Signaling Technology]) in 20 mM Tris–HCl (pH 7.6) containing 150 mM NaCl and 0.05% Tween 20. For lectin blotting, we used RCA and Maackia amurensis lectin (MAM), which specifically bind to non-reducing terminal Galβ1-4GlcNAc and Neu5Acα2-3Gal structures, respectively. Membranes were incubated with RCA-biotin (Vector) and MAM biotin (MBL), followed by HRP-conjugated streptavidin (Sigma-Aldrich, SA10001) in 20 mM Tris–HCl (pH 7.6) containing 150 mM NaCl, 1 mM CaCl2, and 0.05% Tween 20. The protein bands were visualized using Immobilon Western Chemiluminescent HRP substrate solution (Millipore).
Quantitative PCR analysis
Total RNAs were isolated from Expi293T cells using TRIzol reagent (Thermo Fisher Scientific) and an PureLink RNA Mini Kit (Thermo Fisher Scientific). Following extraction, one-step quantitative reverse transcription PCR (qRT-PCR) was performed using Luna universal qPCR master mix and LunaScript RT supermix kit (NEW ENGLAND BioLabs), following the recommended protocol. The primer sequences used were as follows (5′-3′): GAGGAAGCTGGAAGAGCAACA and CTCCCACACCTCCTTCAACTG for NUCB1; CTAAACCACCTGAATCCTGACAAG and ATAGTGTTCCAGATCACTTGTTGC for NUCB2; AACAGCGACACCCATCCTC and CATACCAGGAAATGAGCTTGACAA for GDPDH. Real-time PCR measurements were conducted using the Step One Real-Time PCR System (Applied Biosystems). The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene served as an internal control, and threshold cycle (Ct) values were calculated for each gene.
In vitro enzymatic activity assay of B4GALT1
For desialylation and degalactosylation, approximately 1 μg/mL of EPO-TEV-PS with or without the PS, was dissolved in 0.1 M MES buffer, pH 6.5, containing 150 mM NaCl. The solution was then incubated with 250 μunits/mL of sialidase from Arthrobacter ureafaciens (Nacalai Tesque) and 250 μunits/mL of β1-4galactosidase cloned from Streptococcus pneumoniae and expressed in E. coli (New England Biolabs) at 37°C for 3 h. Degalactosylation was confirmed via lectin blotting with RCA. A portion of the deglactosylated glycoproteins underwent TEV protease treatment to prepare PS-removed EPO. The degalactosyl glycoprotein solution was then mixed with an equal volume of buffer containing 2 μg/mL of uridine diphosphate-galactose (Sigma-Aldrich, approximately 2 μg/mL of B4GALT1, 40 mM MnCl2, and 0.02% Triton X-100. This mixture was incubated at 37°C for 1 h, and subsequently subjected to lectin blot and immunoblot analysis using RCA to detect galctosylation.
Quantification and statistical analysis
General data analysis
Data analyses in this study were conducted independently at least three times, and the results are presented as standard error of the mean (SEM). Statistical significance was assessed using analysis of one-way ANOVA with multiple comparisons and Student’s t test as appropriate, performed with GraphPad Prism 9 (Dotmatics). Statistical significance thresholds were set at p < 0.0001, p < 0.001, p < 0.01, or p < 0.05. Specific sample sizes are indicated in the figure legends.
Yagi et al. identify a "PS" enhancing glycoprotein secretion and N-glycan maturation. This sequence, recognized by MCFD2 and NUCB1, guides proteins through a specific Golgi route, promoting interactions with galactosylation enzyme B4GALT1. This discovery offers a novel strategy for controlling glycosylation of recombinant proteins, potentially improving biopharmaceutical production.
Published: November 22, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.111457.
Contributor Information
Hirokazu Yagi, Email: hyagi@phar.nagoya-cu.ac.jp.
Koichi Kato, Email: kkatonmr@ims.ac.jp.
Supplemental information
Two-sample t tests (EPO vs. EPO-PS)
Two-sample t tests (control vs. B4GALT1)
References
- 1.Kornfeld R., Kornfeld S. Assembly of asparagine-linked oligosaccharides. Annu. Rev. Biochem. 1985;54:631–664. doi: 10.1146/annurev.bi.54.070185.003215. [DOI] [PubMed] [Google Scholar]
- 2.Ellgaard L., Helenius A. Quality control in the endoplasmic reticulum. Nat. Rev. Mol. Cell Biol. 2003;4:181–191. doi: 10.1038/nrm1052. [DOI] [PubMed] [Google Scholar]
- 3.Kato K., Kamiya Y. Structural views of glycoprotein-fate determination in cells. Glycobiology. 2007;17:1031–1044. doi: 10.1093/glycob/cwm046. [DOI] [PubMed] [Google Scholar]
- 4.Lederkremer G.Z. Glycoprotein folding, quality control and ER-associated degradation. Curr. Opin. Struct. Biol. 2009;19:515–523. doi: 10.1016/j.sbi.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 5.Aebi M., Bernasconi R., Clerc S., Molinari M. N-glycan structures: recognition and processing in the ER. Trends Biochem. Sci. 2010;35:74–82. doi: 10.1016/j.tibs.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 6.Kamiya Y., Satoh T., Kato K. Molecular and structural basis for N-glycan-dependent determination of glycoprotein fates in cells. Biochim. Biophys. Acta. 2012;1820:1327–1337. doi: 10.1016/j.bbagen.2011.12.017. [DOI] [PubMed] [Google Scholar]
- 7.Satoh T., Yamaguchi T., Kato K. Emerging structural insights into glycoprotein quality control coupled with N-glycan processing in the endoplasmic reticulum. Molecules. 2015;20:2475–2491. doi: 10.3390/molecules20022475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Satoh T., Kato K. In: Glycobiophysics. Yamaguchi Y., Kato K., editors. Springer Singapore; 2018. Structural Aspects of ER Glycoprotein Quality-Control System Mediated by Glucose Tagging; pp. 149–169. [DOI] [PubMed] [Google Scholar]
- 9.Stanley P. Golgi glycosylation. Cold Spring Harbor Perspect. Biol. 2011;3 doi: 10.1101/cshperspect.a005199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Varki A. Biological roles of glycans. Glycobiology. 2017;27:3–49. doi: 10.1093/glycob/cww086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gabius H.-J., André S., Jiménez-Barbero J., Romero A., Solís D. From lectin structure to functional glycomics: principles of the sugar code. Trends Biochem. Sci. 2011;36:298–313. doi: 10.1016/j.tibs.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 12.Cummings R.D. The repertoire of glycan determinants in the human glycome. Mol. Biosyst. 2009;5:1087–1104. doi: 10.1039/b907931a. [DOI] [PubMed] [Google Scholar]
- 13.Morell A.G., Gregoriadis G., Scheinberg I.H., Hickman J., Ashwell G. The role of sialic acid in determining the survival of glycoproteins in the circulation. J. Biol. Chem. 1971;246:1461–1467. doi: 10.1016/s0021-9258(19)76994-4. [DOI] [PubMed] [Google Scholar]
- 14.Ashwell G., Harford J. Carbohydrate-specific receptors of the liver. Annu. Rev. Biochem. 1982;51:531–554. doi: 10.1146/annurev.bi.51.070182.002531. [DOI] [PubMed] [Google Scholar]
- 15.Solá R.J., Griebenow K. Effects of glycosylation on the stability of protein pharmaceuticals. J. Pharm. Sci. 2009;98:1223–1245. doi: 10.1002/jps.21504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nyfeler B., Zhang B., Ginsburg D., Kaufman R.J., Hauri H.-P. Cargo selectivity of the ERGIC-53/MCFD2 transport receptor complex. Traffic. 2006;7:1473–1481. doi: 10.1111/j.1600-0854.2006.00483.x. [DOI] [PubMed] [Google Scholar]
- 17.Zhang B., Cunningham M.A., Nichols W.C., Bernat J.A., Seligsohn U., Pipe S.W., McVey J.H., Schulte-Overberg U., de Bosch N.B., Ruiz-Saez A., et al. Bleeding due to disruption of a cargo-specific ER-to-Golgi transport complex. Nat. Genet. 2003;34:220–225. doi: 10.1038/ng1153. [DOI] [PubMed] [Google Scholar]
- 18.Zhang B., McGee B., Yamaoka J.S., Guglielmone H., Downes K.A., Minoldo S., Jarchum G., Peyvandi F., de Bosch N.B., Ruiz-Saez A., et al. Combined deficiency of factor V and factor VIII is due to mutations in either LMAN1 or MCFD2. Blood. 2006;107:1903–1907. doi: 10.1182/blood-2005-09-3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Appenzeller C., Andersson H., Kappeler F., Hauri H.P. The lectin ERGIC-53 is a cargo transport receptor for glycoproteins. Nat. Cell Biol. 1999;1:330–334. doi: 10.1038/14020. [DOI] [PubMed] [Google Scholar]
- 20.Nishio M., Kamiya Y., Mizushima T., Wakatsuki S., Sasakawa H., Yamamoto K., Uchiyama S., Noda M., McKay A.R., Fukui K., et al. Structural basis for the cooperative interplay between the two causative gene products of combined factor V and factor VIII deficiency. Proc. Natl. Acad. Sci. USA. 2010;107:4034–4039. doi: 10.1073/pnas.09085261070908526107. [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yagi H., Yagi-Utsumi M., Honda R., Ohta Y., Saito T., Nishio M., Ninagawa S., Suzuki K., Anzai T., Kamiya Y., et al. Improved secretion of glycoproteins using an N-glycan-restricted passport sequence tag recognized by cargo receptor. Nat. Commun. 2020;11:1368. doi: 10.1038/s41467-020-15192-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kamiya Y., Kamiya D., Yamamoto K., Nyfeler B., Hauri H.-P., Kato K. Molecular basis of sugar recognition by the human L-type lectins ERGIC-53, VIPL, and VIP36. J. Biol. Chem. 2008;283:1857–1861. doi: 10.1074/jbc.M709384200. [DOI] [PubMed] [Google Scholar]
- 23.Saito T., Yagi H., Kuo C.-W., Khoo K.-H., Kato K. An embeddable molecular code for Lewis X modification through interaction with fucosyltransferase 9. Commun. Biol. 2022;5:676. doi: 10.1038/s42003-022-03616-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shaper N.L., Charron M., Lo N.-W., Shaper J.H. β1,4-Galactosyltransferase and Lactose Biosynthesis. J. Mammary Gland Biol. Neoplasia. 1998;3:315–324. doi: 10.1023/a:1018719612087. [DOI] [PubMed] [Google Scholar]
- 25.Lin P., Le-Niculescu H., Hofmeister R., McCaffery J.M., Jin M., Hennemann H., McQuistan T., De Vries L., Farquhar M.G. The mammalian calcium-binding protein, nucleobindin (CALNUC), is a Golgi resident protein. J. Cell Biol. 1998;141:1515–1527. doi: 10.1083/jcb.141.7.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin P., Yao Y., Hofmeister R., Tsien R.Y., Farquhar M.G. Overexpression of CALNUC (nucleobindin) increases agonist and thapsigargin releasable Ca2+ storage in the Golgi. J. Cell Biol. 1999;145:279–289. doi: 10.1083/jcb.145.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lavoie C., Meerloo T., Lin P., Farquhar M.G. Calnuc, an EF-hand Ca(2+)-binding protein, is stored and processed in the Golgi and secreted by the constitutive-like pathway in AtT20 cells. Mol. Endocrinol. 2002;16:2462–2474. doi: 10.1210/me.2002-0079. [DOI] [PubMed] [Google Scholar]
- 28.Tulke S., Williams P., Hellysaz A., Ilegems E., Wendel M., Broberger C. Nucleobindin 1 (NUCB1) is a Golgi-resident marker of neurons. Neuroscience. 2016;314:179–188. doi: 10.1016/j.neuroscience.2015.11.062. [DOI] [PubMed] [Google Scholar]
- 29.Roth J., Berger E.G. Immunocytochemical localization of galactosyltransferase in HeLa cells: codistribution with thiamine pyrophosphatase in trans-Golgi cisternae. J. Cell Biol. 1982;93:223–229. doi: 10.1083/jcb.93.1.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tojima T., Miyashiro D., Kosugi Y., Nakano A. Super-Resolution Live Imaging of Cargo Traffic Through the Golgi Apparatus in Mammalian Cells. Methods Mol. Biol. 2023;2557:127–140. doi: 10.1007/978-1-0716-2639-9_10. [DOI] [PubMed] [Google Scholar]
- 31.Kurokawa K., Nakano A. Live-cell Imaging by Super-resolution Confocal Live Imaging Microscopy (SCLIM): Simultaneous Three-color and Four-dimensional Live Cell Imaging with High Space and Time Resolution. Bio. Protoc. 2020;10:e3732. doi: 10.21769/BioProtoc.3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Canis K., McKinnon T.A.J., Nowak A., Panico M., Morris H.R., Laffan M., Dell A. The plasma von Willebrand factor O-glycome comprises a surprising variety of structures including ABH antigens and disialosyl motifs. J. Thromb. Haemostasis. 2010;8:137–145. doi: 10.1111/j.1538-7836.2009.03665.x. [DOI] [PubMed] [Google Scholar]
- 33.Pacheco-Fernandez N., Pakdel M., Blank B., Sanchez-Gonzalez I., Weber K., Tran M.L., Hecht T.K.-H., Gautsch R., Beck G., Perez F., et al. Nucleobindin-1 regulates ECM degradation by promoting intra-Golgi trafficking of MMPs. J. Cell Biol. 2020;219 doi: 10.1083/jcb.201907058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Duellman T., Burnett J., Shin A., Yang J. LMAN1 (ERGIC-53) is a potential carrier protein for matrix metalloproteinase-9 glycoprotein secretion. Biochem. Biophys. Res. Commun. 2015;464:685–691. doi: 10.1016/j.bbrc.2015.06.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Teare J.M., Kates D.S., Shah A., Garger S. Increased branching and sialylation of N-linked glycans correlate with an improved pharmacokinetic profile for BAY 81-8973 compared with other full-length rFVIII products. Drug Des. Dev. Ther. 2019;13:941–948. doi: 10.2147/DDDT.S188171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sodetz J.M., Pizzo S.V., McKee P.A. Relationship of sialic acid to function and in vivo survival of human factor VIII/von Willebrand factor protein. J. Biol. Chem. 1977;252:5538–5546. doi: 10.1016/s0021-9258(19)63384-3. [DOI] [PubMed] [Google Scholar]
- 37.Bovenschen N., Rijken D.C., Havekes L.M., van Vlijmen B.J.M., Mertens K. The B domain of coagulation factor VIII interacts with the asialoglycoprotein receptor: Factor VIII binding to asialoglycoprotein receptor. J. Thromb. Haemostasis. 2005;3:1257–1265. doi: 10.1111/j.1538-7836.2005.01389.x. [DOI] [PubMed] [Google Scholar]
- 38.Varki A. Sialic acids in human health and disease. Trends Mol. Med. 2008;14:351–360. doi: 10.1016/j.molmed.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Walsh G., Walsh E. Biopharmaceutical benchmarks 2022. Nat. Biotechnol. 2022;40:1722–1760. doi: 10.1038/s41587-022-01582-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yagi H., Yanaka S., Kato K. Comprehensive Glycoscience. Elsevier; 2021. Structural and functional roles of the N-glycans in therapeutic antibodies; pp. 534–542. [DOI] [Google Scholar]
- 41.Chia S., Tay S.J., Song Z., Yang Y., Walsh I., Pang K.T. Enhancing pharmacokinetic and pharmacodynamic properties of recombinant therapeutic proteins by manipulation of sialic acid content. Biomed. Pharmacother. 2023;163 doi: 10.1016/j.biopha.2023.114757. [DOI] [PubMed] [Google Scholar]
- 42.Moremen K.W., Tiemeyer M., Nairn A.V. Vertebrate protein glycosylation: diversity, synthesis and function. Nat. Rev. Mol. Cell Biol. 2012;13:448–462. doi: 10.1038/nrm3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schjoldager K.T., Narimatsu Y., Joshi H.J., Clausen H. Global view of human protein glycosylation pathways and functions. Nat. Rev. Mol. Cell Biol. 2020;21:729–749. doi: 10.1038/s41580-020-00294-x. [DOI] [PubMed] [Google Scholar]
- 44.Thaysen-Andersen M., Packer N.H. Site-specific glycoproteomics confirms that protein structure dictates formation of N-glycan type, core fucosylation and branching. Glycobiology. 2012;22:1440–1452. doi: 10.1093/glycob/cws110. [DOI] [PubMed] [Google Scholar]
- 45.Lee L.Y., Lin C.-H., Fanayan S., Packer N.H., Thaysen-Andersen M. Differential site accessibility mechanistically explains subcellular-specific N-glycosylation determinants. Front. Immunol. 2014;5:404. doi: 10.3389/fimmu.2014.00404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Suga A., Nagae M., Yamaguchi Y. Analysis of protein landscapes around N-glycosylation sites from the PDB repository for understanding the structural basis of N-glycoprotein processing and maturation. Glycobiology. 2018;28:774–785. doi: 10.1093/glycob/cwy059. [DOI] [PubMed] [Google Scholar]
- 47.Kanagawa M., Saito F., Kunz S., Yoshida-Moriguchi T., Barresi R., Kobayashi Y.M., Muschler J., Dumanski J.P., Michele D.E., Oldstone M.B.A., Campbell K.P. Molecular recognition by LARGE is essential for expression of functional dystroglycan. Cell. 2004;117:953–964. doi: 10.1016/j.cell.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 48.Foley D.A., Swartzentruber K.G., Colley K.J. Identification of sequences in the polysialyltransferases ST8Sia II and ST8Sia IV that are required for the protein-specific polysialylation of the neural cell adhesion molecule. J. Biol. Chem. 2009;284:15505–15516. doi: 10.1074/jbc.M809696200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yagi H., Tateo S., Saito T., Ohta Y., Nishi E., Obitsu S., Suzuki T., Seetaha S., Hellec C., Nakano A., et al. Deciphering the sub-Golgi localization of glycosyltransferases via 3D super-resolution imaging. Cell Struct. Funct. 2024;49 doi: 10.1247/csf.24008. [DOI] [PubMed] [Google Scholar]
- 50.Welch L.G., Peak-Chew S.-Y., Begum F., Stevens T.J., Munro S. GOLPH3 and GOLPH3L are broad-spectrum COPI adaptors for sorting into intra-Golgi transport vesicles. J. Cell Biol. 2021;220 doi: 10.1083/jcb.202106115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee W., Tonelli M., Markley J.L. NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy. Bioinformatics. 2015;31:1325–1327. doi: 10.1093/bioinformatics/btu830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Longo P.A., Kavran J.M., Kim M.-S., Leahy D.J. Transient mammalian cell transfection with polyethylenimine (PEI) Methods Enzymol. 2013;529:227–240. doi: 10.1016/B978-0-12-418687-3.00018-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Uematsu R., Furukawa J.-I., Nakagawa H., Shinohara Y., Deguchi K., Monde K., Nishimura S.-I. High Throughput Quantitative Glycomics and Glycoform-focused Proteomics of Murine Dermis and Epidermis. Mol. Cell. Proteomics. 2005;4:1977–1989. doi: 10.1074/mcp.M500203-MCP200. [DOI] [PubMed] [Google Scholar]
- 54.Furukawa J.-I., Shinohara Y., Kuramoto H., Miura Y., Shimaoka H., Kurogochi M., Nakano M., Nishimura S.-I. Comprehensive approach to structural and functional glycomics based on chemoselective glycoblotting and sequential tag conversion. Anal. Chem. 2008;80:1094–1101. doi: 10.1021/ac702124d. [DOI] [PubMed] [Google Scholar]
- 55.Branon T.C., Bosch J.A., Sanchez A.D., Udeshi N.D., Svinkina T., Carr S.A., Feldman J.L., Perrimon N., Ting A.Y. Efficient proximity labeling in living cells and organisms with TurboID. Nat. Biotechnol. 2018;36:880–887. doi: 10.1038/nbt.4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Two-sample t tests (EPO vs. EPO-PS)
Two-sample t tests (control vs. B4GALT1)
Data Availability Statement
-
•
The data on proximity-dependent labeling and protein identification using mass spectrometry have been deposited to jPOST under the ID JPST003247 and JPST003457. The microscopy images for the quantification analysis of colocalization have been deposited at Mendeley (http://www.doi.org/10.17632/przzrd7sb9.1) and are publicly available as of the date of publication.
-
•
This study did not produce any original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.