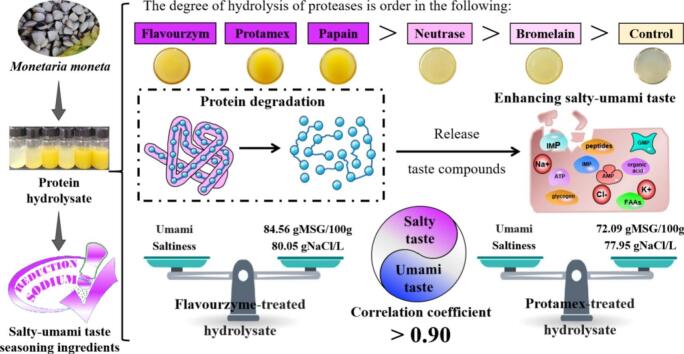
Abstract
To prepare dual-functional seasoning ingredients with a salty-umami taste, five proteases were applied to hydrolyze Monetaria moneta proteins, preparing enzymatic hydrolysates. Their taste compounds along with the salty-umami taste, were investigated. The results revealed that enzymatic hydrolysis facilitated the release of taste compounds from M. moneta. The whiteness and < 3 kDa peptides of enzymatic hydrolysates significantly increased. Moreover, flavorzyme and protamex, with high DHs, could thoroughly hydrolyze the proteins, generating the enzymatic hydrolysates abundant in taste compounds (e.g., amino acids, nucleotides) that synergistically provided a strong salty-umami taste. Saltiness and umami posed a strong positive correlation, with a correlation coefficient exceeding 0.90, resulting in the highest levels of equivalent salty intensity (ESI = 80.05 gNaCl/L) and equivalent umami concentration (EUC = 84.56 gMSG/100 g) in the flavorzyme-treated hydrolysate, followed by the protamex-treated hydrolysate. In summary, these findings offer novel insights into preparing dual-functional seasoning ingredients with a salty-umami taste, ideal for use in low-salt food production.
Keywords: Monetaria moneta, Protein hydrolysate, Taste compounds, Synergy, Salty-umami taste
Graphical abstract
Highlights
-
•
Enzymatic hydrolysis facilitates the release of salty and umami taste ingredients.
-
•
The salty intensity is evaluated by an innovative equivalent salty intensity.
-
•
The correlation coefficient between saltiness and umami is greater than 0.90.
-
•
The hydrolysates of Monetaria moneta can serve as dual-functional seasoning ingredients.
1. Introduction
Table salt is a principal ingredient in seasonings because it uniquely enhances the taste and aroma of food by deodorizing, degreasing, and amplifying their aromas (He & Tan, 2024; Taladrid et al., 2020). Currently, the excessive reuse of diverse seasoning ingredients has led to a dangerously high daily sodium intake, with worldwide averages falling between 3.54 and 4.72 g/day, which significantly exceeds the daily limit of 2.0 g/day recommended by the World Health Organization (WHO). This excessive intake poses a severe global public health crisis, underscoring the need to curb sodium consumption. In response, the WHO has proposed a strategy to reduce salt intake by 30 % before 2025 (Le et al., 2022; Wang, Huang, et al., 2023). However, the trend towards “salt reduction without loss of salty taste” profoundly impacts seasoning ingredients worldwide and causes to a huge impact on the food industry. Moreover, the rise of “civilization diseases” and “wealth diseases” directly threaten human health. Consequently, the demand for salty and umami flavors is reaching new heights, increasingly prioritizing “foods' taste” (Hu et al., 2021; Song et al., 2023).
In taste perception, salty and umami compounds interact complementary, and thus they establish a new platform that produces a composite sensation known as the “salty-umami taste” (Hu et al., 2021; Song et al., 2023). Furthermore, umami substances can enhance the perception of saltiness, reducing sodium intake, and thus synergistic umami effect represents a potential approach to salt reduction (Li et al., 2024). Recent studies have identified several salty peptides, such as Ala-Val, Asn-Ser-Glu, Asn-Glu, Lys-Ser-Ala-Glu-Asn, Arg-Glu-Glu, and Lys-Glu-Arg (Shen et al., 2022; Wang et al., 2024; Zhang et al., 2018). These peptides are rich in acidic amino acids like aspartic and glutamic acids, along with their sodium salts, which also exhibit a pronounced umami taste. This dual functionality of peptides underscores their ability to impart a strong salty-umami taste to food (Song et al., 2023). Consequently, an increasing trend in the seasoning industry is developing dual-functional seasoning ingredients with a strong salty-umami taste. Bio-enzymatic technology is widely used to produce natural seasoning ingredients because it can offer significant advantages such as mild reaction conditions, controllable reaction directions, and low energy consumption (Feng et al., 2024; Ildephonse et al., 2023). During enzymatic hydrolysis, proteins are cleaved and dissociated into a variety of free amino acids, peptides, and other taste compounds that are readily digestible and absorbable by the human body (Feng et al., 2024; Ildephonse et al., 2023). These compounds synergistically enhance the salty-umami flavor of food, allowing for reduced sodium intake without compromising the intensity of salty and umami tastes (Feng et al., 2024; Ildephonse et al., 2023; Zhang et al., 2018). Additionally, these taste compounds can interact with each other and even modify other taste formations, thereby creating a synergistic effect that intensifies the salty-umami taste of food (Shen et al., 2022; Wang et al., 2024; Zhang et al., 2018).
Monetaria moneta, a delicious edible shellfish, is a low-value marine species abundant in the South China Sea. Its small size and complex processing requirements often hinder its full utilization (Yuan et al., 2018). In the southern coastal regions of Guangdong and Guangxi, M. moneta is commonly used in soups, prized for its tasty meat and abundant nutritional content. Notably, the edible parts of M. moneta are high in proteins which can offer a complete amino acid profile, resulting in its inclusion in the first classic work of traditional Chinese medicine, “Shen Nong's Herbal Classic,” almost two thousand years ago (Yuan et al., 2018). However, there are relatively few publications about M. moneta. Specifically, its high protein content renders it suitable for preparing dual-functional seasoning ingredients with a robust salty-umami taste.
As a result, we systematically evaluated the effects of different proteases on the hydrolysis efficiency and taste compounds derived from M. moneta and analyzed the salty and umami tastes of protein hydrolysates. Based on the characteristics of taste compounds in protein hydrolysates, we also investigated the relationship between salty and umami tastes. This study offers a novel perspective on the relationship between salty and umami tastes, aiming to provide dual-functional seasoning ingredients with a robust salty-umami taste for use in low-salt food production.
2. Materials and methods
2.1. Materials and chemical reagents
Fresh M. moneta was purchased from a local aquatic market in Zhanjiang, Guangdong, China. After being washed and de-shelled, the meat was removed, drained, and packed in sample bags. It was then stored in a refrigerator at −20 °C.
Bromelain (300 U/mg), papain (10 U/mg), neutrase (100 U/mg), flavourzyme (20 U/mg), protamex (120 U/mg), and nucleotide standards were purchased from Yuanye Bio-Technology Co., Ltd. (Shanghai, China). Organic acids standards were obtained from Maclean Biochemical Technology Co., Ltd. (Shanghai, China), while amino acid standards were purchased from Sigma-Aldrich Co. Ltd. (St. Louis, MO, USA). The remaining chemical reagents used in the experiments were of analytical grade and sourced from the Chemical Reagent Factory in Guangzhou, China.
2.2. Preparation of protein hydrolysates
The preparation of protein hydrolysates was carried out according to a modified version of the method described by Wang et al. (2022). Specifically, 600 g of M. moneta meat was mixed with deionized water at a mass-to-volume ratio of 1:3. After homogenization at a speed of 10,000 rpm for 5 min, the pH of the mixture was adjusted to 7.0. Subsequently, the mixture was divided into six equal parts and subjected to hydrolysis at 50 °C for 4 h with various proteases (bromelain, papain, neutrase, flavourzyme, and protamex), and the proteases were added at an enzyme-to-protein ratio of 3000 U/g. Following complete enzymatic hydrolysis, the hydrolysates were immediately placed in boiling water for 15 min to deactivate the enzymes. They were then centrifuged at 8000 g for 20 min and stored at −40 °C for further analysis. Control experiments were conducted by following the same procedure but omitting the proteases.
2.3. Determinations of the degree of hydrolysis (DH)
The content of α-amino nitrogen was determined through formaldehyde titration, while the protein content was assayed using an automatic nitrogen analyzer (Kjeltec 8200, FOSS, Germany). This approach was based on a method described by Wang et al. (2022) with modifications. The protein content of M. moneta was found to be 10.54 g/100 g (wet weight). The total nitrogen content is typically estimated by dividing the protein content by a conversion factor of 6.25 (Yang et al., 2024). Subsequently, DH was calculated using formula (1), which determines the percentage of α-amino nitrogen content relative to the total nitrogen content.
| (1) |
In formula (1), T1 and T2 represent the α-amino nitrogen contents of M. moneta and its protein hydrolysate, respectively; while P1 denotes the total nitrogen content of M. moneta.
2.4. Whiteness evaluation
After equilibrating at room temperature (25 °C) for 30 min, the whiteness of protein hydrolysates was evaluated using a colorimeter (Model NS800; 3NH Technology Co., Ltd., Shenzhen, China) (Zhang, Liu, et al., 2024). The parameters L⁎ (lightness), a⁎ (red-green), and b⁎ (yellow-blue) were measured and the whiteness was calculated following the formula (2).
| (2) |
2.5. Determinations of molecular weight (MW) distribution
The MW distribution of protein hydrolysates was determined by high-performance liquid chromatography (HPLC) (Wang et al., 2022). After being filtered through a 0.22-μm membrane filter, the samples were introduced into a chromatographic column [TSKgel G2000SWXL(7.8 mm I·D × 30 cm, C—No.003HA02325H)]. The detection wavelength was established at 220 nm, and a mixed solution (Acetonitrile:water:trifluoroacetic acid = 20:80:0.1) was used as a mobile phase. Protein standards of varying molecular weights [Insulin (5807.58 Da), Bacitracin (1421.69 Da), Glycine-Glycine-Tyrosine-Arginine (451.48 Da), and Glycine-Glycine-Glycine (181.19 Da)] were utilized to generate a standard curve which was plotted as y = −0.0763× + 4.3866 (R2 = 0.9944). From this data, the MW distribution of the protein hydrolysates was calculated based on the retention times of the samples and ultimately expressed as percentages.
2.6. Determination of free amino acid (FAA) concentrations
The concentration of FAAs was analyzed based on a previous method with some modifications (Chen et al., 2022). The supernatants obtained in section 2.2 were filtered through a 0.45-μm membrane filter and then separated by using a ZORBAX Eclipse-AAA column (4.6 × 150 mm, 3.5 μm) with detection wavelengths set at 338 and 266 nm. Additionally, 40 mM NaH2PO4 (pH 7.8) and a mixed solvent (Acetonitrile:methanol:water = 45:45:10) were used as mobile phases A and B, respectively, in the HPLC analysis. The amino acid contents were quantified using calibration curves generated from authentic compounds relative to external standards.
2.7. Determination of nucleotide and organic acid concentrations
The concentrations of nucleotides and organic acids were measured using the methods described by Wang, Li, et al. (2023). After passing through a 0.45-μm membrane filter, the solutions were introduced into a COSMOSIL 5C18-MS-II column using HPLC (Agilent 1200, Agilent Technologies, Santa Clara, CA, USA). The detection wavelength was set at 220 nm, and mobile phases A and B consisted of 5 % of 0.1 % phosphoric acid and 95 % methanol solutions, respectively. The identity and quantity of the compounds were finally determined by comparing the retention times with those of standards and through calibration using the standard curves.
2.8. Determination of betaine, glycogen, and small peptide concentrations
The concentration of betaine was measured using the Reinecke salt crystallographic colorimetric method (Liu et al., 2024). The detection wavelength was established at 525 nm using a spectrophotometer (Thermo Spectronic Genesys 10UV, Thermo Fisher Scientific Inc., Waltham, MA). The betaine concentration was determined by referencing a standard curve, represented by the equation y = 11.024×-0.0056 (R2 = 0.9984). Besides, the glycogen concentration was determined using the phenol-sulphuric acid method (Liu et al., 2024). Absorbance measurement was taken at 470 nm using a spectrophotometer, and a standard curve was plotted by using glucose as a standard for measuring glycogen concentration, given by y = 0.0035× + 0.0663 (R2 = 0.9990).
Furthermore, to eliminate proteins, 20.0 mL of protein hydrolysates were blended with an equivalent volume of a 10 % trichloroacetic acid solution. After allowing the mixture to stand for 30 min, it was centrifuged at 4000 g for 15 min. Subsequently, the small peptide concentration was determined using Lowry's method. A standard curve was established using bovine serum albumin, plotted as y = 0.5474× + 0.0096 (R2 = 0.9995).
2.9. Determination of inorganic ion concentrations
Sodium (Na+) and potassium (K+) in protein hydrolysates were determined using inductively coupled plasma-optical emission spectroscopy (Thermo Fisher Scientific Inc., Cambridge, UK) (Liu et al., 2024). Additionally, chlorides (Cl−) were determined using the molybdenum blue spectrophotometric method reported in the study (Liu et al., 2024).
2.10. Taste activity value (TAV) of taste compounds
Taste compounds with a TAV (taste activity value) ≥ 1 are defined as key taste compounds that can contribute significantly to the taste of protein hydrolysates, while taste compounds with 0.1 ≤ TAV < 1 exhibit a modifying effect (Wang, Li, et al., 2023). The TAV is calculated using the following equation. In formula (3), Ci represents the concentration of taste compounds (mg/L), and Ti represents their corresponding threshold value in water (mg/L).
| (3) |
2.11. Sensory evaluation of salty and umami tastes
Quantitative descriptive analysis (QDA) was employed to conduct the sensory evaluation of protein hydrolysates (Yang et al., 2022). The intensities of salty and umami tastes were evaluated using a ten-point scoring system, structured as follows: very weak (scoring between 0 and 2), weak (scoring between 2 and 4), medium (scoring between 4 and 6), strong (scoring between 6 and 8), and very strong (scoring between 8 and 10). This scoring rubric facilitated a precise quantification of taste intensities. The salty/umami taste intensity of a 0.1 g/L NaCl/MSG standard solution was recorded as a “0 score”, while that of a 5.0 g/L NaCl/MSG standard solution was recorded as a “10 score”. Besides, 20.0 mL of protein hydrolysates were diluted tenfold, poured into clean and transparent cups, and randomly assigned with a three-digit code. After equilibrating at room temperature (25 °C) for 30 min, the tastes of the protein hydrolysates was evaluated by 20 trained food professionals aged between 20 and 30, who had experience in assessing the tastes of protein hydrolysates. They were required to conduct a sensory evaluation of the salty and umami tastes of the protein hydrolysates and assigned scores based on the scoring rubric. All protein hydrolysates were evaluated under consistent conditions.
2.12. The analysis of salty and umami tastes, equivalent salty intensity (ESI) and equivalent umami concentration (EUC)
A volume of 20.0 mL of protein hydrolysates was taken and diluted tenfold. After filtering through two layers of filter paper, the salty and umami tastes of the protein hydrolysates were analyzed using an electronic tongue equipped with CTO and AAE taste sensors (Liu et al., 2024). Additionally, a standard curve for salty intensity was plotted as y = 2.0420× + 19.9310 (R2 = 0.9660) using NaCl solutions. Based on the salty intensity recorded by the electronic tongue, we tentatively define the Equivalent Salty Intensity (ESI) as the salty intensity elicited by the protein hydrolysate that corresponds to the salty intensity elicited by a specific concentration of NaCl. Therefore, it is denoted as ESI (gNaCl/L) and can be calculated using the following equation. In formula (4), P represents the salty intensity elicited by the protein hydrolysate, and T represents the diluted ratio of the protein hydrolysate.
| (4) |
The term “equivalent umami concentration” refers to the combined effect of certain umami amino acids (Asp, Glu) and nucleotides (IMP, GMP, and AMP). The EUC is defined as a cumulative effect of amino acids (Asp, Glu) and nucleotides (IMP, GMP, and AMP) that produces an umami intensity equivalent to a specific concentration of monosodium glutamate (MSG) per 100 g (Liu, Li et al., 2024). This parameter can be calculated using the following equation. In formula (5), αi and αj represent the concentrations (g/100 g) of the amino acids and nucleotides, respectively. Additionally, βi denotes the relative umami concentrations (RUC) of amino acids (Glu (1.00) and Asp (0.077)) compared to MSG, while βj signifies the RUC of nucleotides (IMP (1.00), GMP (2.30), and AMP (0.18)) relative to MSG. The synergistic constant is fixed at 1218.
| (5) |
2.13. Statistical analysis
All experiments were independently replicated three times using various samples. Statistical analysis, including ANOVA and Duncan's Multiple Range tests, was conducted using SPSS software (version 19.0; SPSS Inc., Chicago, IL, USA). All data were presented as mean ± standard deviation (SD).
3. Results and discussion
3.1. Effects of different proteases on the DH of protein hydrolysates
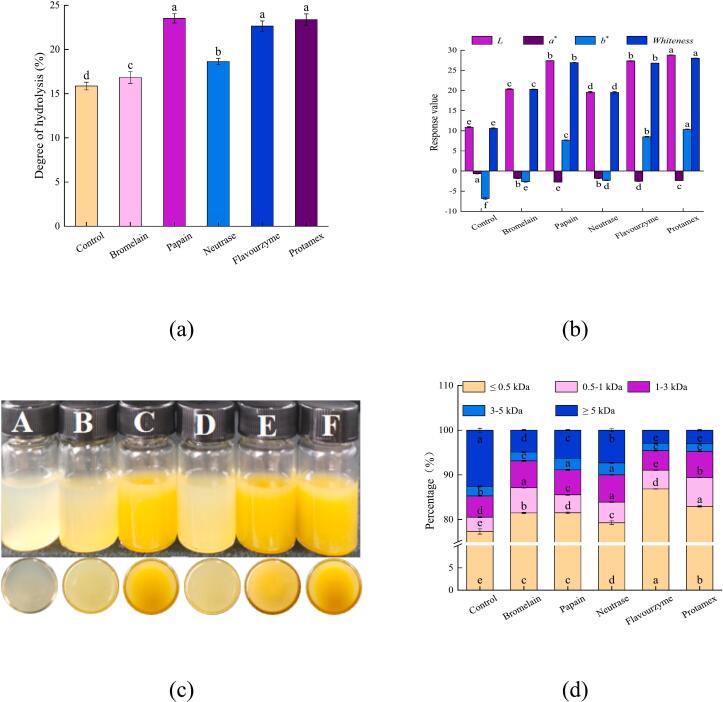
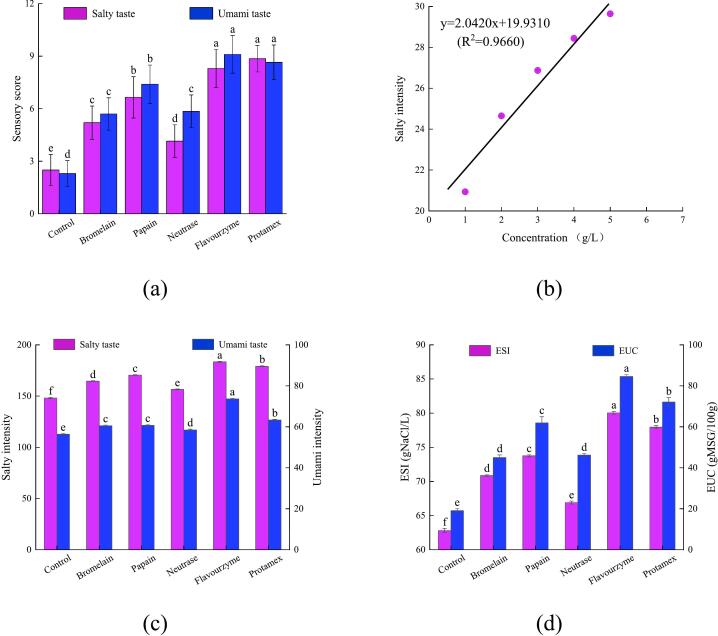
DH is a crucial indicator for assessing proteases' ability to hydrolyze proteins, which directly correlates with the efficiency of protein hydrolysis (Zhang, Tu, et al., 2024). As illustrated in Fig. 1 (a), there is a significant difference in the efficiency of enzymatic hydrolysis between the control and five types of proteases. Notably, the DH of the control was the lowest, at just 15.86 % (P < 0.05). In contrast, the DH of the proteases showed a marked increase compared to the control. This increase can be attributed to the ease with which M. moneta proteins are hydrolyzed by proteases, resulting in the generation of diverse amino acids and peptides (Zhang, Tu, et al., 2024). Furthermore, the DHs of the proteases exhibited substantial variation, arranged in the following order: papain, flavourzyme, and protamex > neutrase > bromelain > control. This variation may stem from the specificity of the proteases (Zhang, Liu, et al., 2024). Specifically, papain, flavourzyme, and protamex exhibited high hydrolysis efficiencies, with their DHs exceeding 22.64 %.
Fig. 1.
Effects of different proteases on the physicochemical properties of protein hydrolysates of Monetaria moneta. (a) the degree of hydrolysis, (b) whiteness, (c) protein hydrolysates, (d) molecular weight distribution, A-F represent control, protein hydrolysates of bromelain, papain, neutrase, flavorzyme, and protamex, respectively.
The muscles of shellfish, both striated and smooth, are abundant in myofibrillar, sarcoplasmic, and matrix proteins (Li et al., 2022). Specifically, matrix protein belongs to a class of insoluble proteins rich in collagen and elastin (Li et al., 2022). Consequently, these proteins were not extracted during the water extraction process, leading to the lowest DH in the control. However, M. moneta proteins underwent facile hydrolysis during the enzymatic process and converted proteins into various small molecule compounds, thereby significantly enhancing their DHs (Yang et al., 2024). Seven enzyme groups were applied to hydrolyze broken fruiting bodies of morel mushroom (Morchella sextelata), and the morel hydrolysate obtained from Neutrase-Flavourzyme combination contained the most contents of taste-activity compounds, which resulted from the highest DH of them, up to 36.64 %, thereby giving the best overall flavor properties (Gao et al., 2021).
Under the same conditions of enzymatic hydrolysis, various proteases could exert different extents of enzymatic hydrolysis on M. moneta proteins, which could potentially be determined by the specificity of the enzyme. According to their cleavage sites, proteolytic enzymes can be divided into exo-type proteinases and endo-type proteinases (peptidases) (Kieliszek et al., 2021). The first group cleave the peptide bond proximal to the amino or carboxy terminals, whereas endo-type proteinases cleave peptide bonds distant from the termini of the proteins (Kieliszek et al., 2021). Specifically, bromelain and papain are kinds of cysteine proteases, also known as thiol proteases. These enzymes can act on the carboxyl terminals of Arg and Lys within peptide chains, preferentially hydrolyzing amino acids with two carboxyl groups at the N-terminal (Veymar et al., 2021; Veymar et al., 2023). Nevertheless, the DH of bromelain was notably lower than that of papain. This disparity might be attributed to bromelain's susceptibility to metal ions, which can readily diminish its activity (Shukor et al., 2008; Veymar et al., 2021; Veymar et al., 2023). Neutrase, an endo-type protease derived from Bacillus subtilis, degrades peptide bonds between hydrophobic amino acids. It tends to cleave the peptide bond between Leu and Phe at the C-terminal, contributing to a lower DH (Chen et al., 2023; Zheng et al., 2024). Additionally, flavourzyme, an exo-type protease, cleaves peptide chains from both the C and N-terminals. Protamex, possessing dual characteristics of both endo- and exo-type proteases, can cleave proteins at multiple sites (Chen et al., 2023). Given the analysis of the DH and cleavage sites of various proteases, it was observed that flavourzyme and protamex exhibited proficiency in generating salty and umami ingredients, thereby imparting a distinct salty-umami taste to their protein hydrolysates.
3.2. Effects of different proteases on the whiteness of protein hydrolysates
Whiteness serves as a crucial indicator which can reflect both the color and quality of protein hydrolysates, and it also directly influences consumer preferences (Zhang, Liu, et al., 2024). Changes in whiteness offer a direct window into the internal transformations occurring within protein hydrolysates, which attributes to their dissolved compound components and the surface's optical properties. Essentially, the whiteness of protein hydrolysates is closely correlated with the DH of proteases and their cleavage sites (Zhang, Liu, et al., 2024).
As illustrated in Fig. 1 (b-c), the results demonstrated that, following enzymatic hydrolysis, the L⁎ and b⁎ values of all protein hydrolysates experienced a notable increase (P < 0.05), but the a⁎ value decreased significantly (P < 0.05). Most importantly, the protein hydrolysates of papain, flavourzyme, and protamex exhibited a more pronounced improvement in L⁎ and b⁎ values (P < 0.05), ultimately leading to a substantial boost in their whiteness (P < 0.05). In response, the protein hydrolysates were observed to be a yellow hue as a result. Combined with their DHs, it could be speculated that these proteases effectively hydrolyze M. moneta proteins, facilitating the release of taste compounds from M. moneta and thereby enriching the taste of the protein hydrolysates. Consequently, protein hydrolysate appearing in a yellow hue is likely to exhibit a pronounced salty-umami taste.
3.3. Effects of different proteases on the MW distribution of protein hydrolysates
To further investigate the salty-umami taste of protein hydrolysates produced by different proteases, the MW distribution was determined. This was because peptides with a small MW have been found to significantly affect taste (Yang et al., 2024). It is widely recognized that peptides weighing less than 3 kDa exhibit a strong taste, whereas those exceeding 3 kDa are typically weak or tasteless (Li et al., 2023; Song et al., 2023). Peptides under 1 kDa can considerably enhance the umami and salty tastes of protein hydrolysates (Wang et al., 2022; Zhang et al., 2022).
The results presented in Fig. 1 (d) reveal that the MW distribution of the protein hydrolysates primarily falls into five ranges, with the majority concentrated in the under 3 kDa range. In our study, the control contained a significant proportion of peptides exceeding 3 kDa, accounting for 14.70 % (P < 0.05). However, following hydrolysis with various proteases, the content of peptides over 3 kDa decreased remarkably (P < 0.05), while the proportion of peptides under 1 kDa increased significantly (P < 0.05). Furthermore, the flavourzyme-treated hydrolysate exhibited the highest content of peptides under 1 kDa (91.06 %), closely followed by the protamex-treated hydrolysate (89.41 %). This suggests that both proteases are capable of thoroughly hydrolyzing M. moneta proteins. Flavourzyme, an exo-type protease, demonstrates a strong propensity to produce amino acids. Meanwhile, protamex, possessing dual characteristics, efficiently cleaves proteins to generate a diverse array of small peptides and amino acids. These results account for the elevated levels of peptides under 1 kDa in their respective protein hydrolysates (Chen et al., 2023; Yang et al., 2024). Consequently, compared to the control and other proteases, the flavourzyme- and protamex-treated hydrolysates are likely to exhibit distinctive taste profiles, characterized by a pronounced salty-umami taste.
3.4. Effects of different proteases on the concentrations of FAAs in protein hydrolysates
As shown in Table 1, 24 varieties of FAAs were detected in both the control and protein hydrolysates, including 9 essential amino acids (EAAs) and 15 non-essential amino acids. The trends for total FAAs and EAAs were comparable. In the control, the concentrations of FAAs and EAAs were only 16.93 and 6.94 g/L, respectively. However, the concentrations of FAAs and EAAs in the protein hydrolysates rose obviously (P < 0.05). Interestingly, the flavourzyme-treated hydrolysate exhibited the highest concentrations of FAAs and EAAs, reaching 28.08 and 11.55 g/L, respectively (P < 0.05), closely followed by the protamex-treated hydrolysate. This difference could be attributed to the DHs and cleavage sites of the proteases involved (Fang et al., 2022; Zhang, Liu, et al., 2024). As a result, the nutritional value of protein hydrolysates treated by flavourzyme and protamex remained impressively intact.
Table 1.
Effects of different proteases on the concentration of free amino acids and nucleotides in protein hydrolysates of Monetaria moneta.
| Taste substance name | Protein hydrolysates of different proteases(g/L) |
|||||
|---|---|---|---|---|---|---|
| Control | Bromelain | Papain | Neutrase | Flavourzyme | Protamex | |
| Asp | 0.58 ± 0.01e | 0.68 ± 0.01c | 0.66 ± 0.02c | 0.63 ± 0.02d | 0.89 ± 0.03a | 0.82 ± 0.03b |
| Glu | 1.94 ± 0.02e | 2.62 ± 0.01c | 2.85 ± 0.04b | 2.46 ± 0.03d | 3.30 ± 0.05a | 2.91 ± 0.04b |
| Gly | 0.86 ± 0.03f | 1.78 ± 0.02d | 1.85 ± 0.01c | 1.67 ± 0.02e | 1.89 ± 0.02b | 2.11 ± 0.02a |
| Thr* | 0.76 ± 0.02e | 1.01 ± 0.02d | 1.06 ± 0.01c | 1.14 ± 0.03b | 1.31 ± 0.02a | 0.79 ± 0.03e |
| Ala | 2.07 ± 0.05e | 3.82 ± 0.01b | 3.73 ± 0.02c | 3.37 ± 0.01d | 3.76 ± 0.02c | 4.33 ± 0.02a |
| Pro | 0.31 ± 0.01d | 0.36 ± 0.01c | 0.38 ± 0.01c | 0.30 ± 0.01d | 0.61 ± 0.03a | 0.48 ± 0.05b |
| Ser | 0.42 ± 0.01d | 0.51 ± 0.01c | 0.61 ± 0.02b | 0.81 ± 0.02a | 0.82 ± 0.02a | 0.28 ± 0.02e |
| Arg | 1.44 ± 0.03e | 2.19 ± 0.01c | 2.32 ± 0.01b | 2.02 ± 0.02d | 2.53 ± 0.02a | 2.34 ± 0.02b |
| Tyr | 1.14 ± 0.03a | 1.03 ± 0.02c | 0.96 ± 0.02d | 0.63 ± 0.02e | 0.95 ± 0.02d | 1.07 ± 0.03b |
| His* | 0.28 ± 0.01c | 0.46 ± 0.01a | 0.48 ± 0.01a | 0.43 ± 0.02b | 0.48 ± 0.01a | 0.47 ± 0.01a |
| Val* | 0.92 ± 0.01d | 1.33 ± 0.02b | 1.31 ± 0.02b | 1.06 ± 0.03c | 1.44 ± 0.02a | 1.34 ± 0.03b |
| Met* | 0.53 ± 0.02d | 0.88 ± 0.01a | 0.83 ± 0.02b | 0.68 ± 0.02c | 0.89 ± 0.02a | 0.85 ± 0.02b |
| Trp* | 0.17 ± 0.01d | 0.28 ± 0.01ab | 0.27 ± 0.01b | 0.23 ± 0.02c | 0.30 ± 0.01a | 0.27 ± 0.02b |
| Phe* | 0.83 ± 0.01e | 1.22 ± 0.01a | 1.11 ± 0.02c | 0.93 ± 0.02d | 1.14 ± 0.01b | 1.14 ± 0.02b |
| Ile* | 0.85 ± 0.02e | 1.29 ± 0.01b | 1.20 ± 0.01c | 0.91 ± 0.02d | 1.34 ± 0.02a | 1.27 ± 0.02b |
| Leu* | 1.47 ± 0.02e | 2.57 ± 0.01a | 2.26 ± 0.03c | 1.68 ± 0.02d | 2.48 ± 0.01b | 2.47 ± 0.02b |
| Lys* | 1.12 ± 0.01f | 1.66 ± 0.01d | 2.04 ± 0.02b | 1.62 ± 0.02e | 2.17 ± 0.02a | 1.89 ± 0.02c |
| Asn | 0.32 ± 0.01e | 0.41 ± 0.01d | 0.45 ± 0.02c | 0.21 ± 0.01f | 0.57 ± 0.02a | 0.50 ± 0.02b |
| Gln | 0.25 ± 0.01e | 0.41 ± 0.01b | 0.38 ± 0.01c | 0.32 ± 0.02d | 0.46 ± 0.02a | 0.46 ± 0.01a |
| Cys | 0.20 ± 0.00a | 0.17 ± 0.01b | 0.17 ± 0.01b | 0.20 ± 0.01a | 0.21 ± 0.02a | 0.21 ± 0.02a |
| Cit | 0.12 ± 0.01c | 0.17 ± 0.01b | 0.07 ± 0.01d | 0.15 ± 0.03b | 0.23 ± 0.02a | 0.18 ± 0.01b |
| Nva | 0.05 ± 0.00d | 0.07 ± 0.01b | 0.07 ± 0.01b | 0.11 ± 0.01a | 0.06 ± 0.00c | 0.06 ± 0.01c |
| Hyp | 0.26 ± 0.01a | 0.13 ± 0.01d | 0.17 ± 0.01c | 0.20 ± 0.03bc | 0.21 ± 0.01b | 0.19 ± 0.01bc |
| Sar | 0.03 ± 0.00b | 0.02 ± 0.00b | 0.01 ± 0.00c | 0.02 ± 0.00b | 0.04 ± 0.00a | 0.02 ± 0.00b |
| FAAs | 16.93 ± 0.35e | 25.08 ± 0.26c | 25.22 ± 0.36c | 21.76 ± 2.37d | 28.08 ± 0.42a | 26.47 ± 0.27b |
| EAAs | 6.94 ± 0.13d | 10.71 ± 0.12b | 10.55 ± 0.15b | 8.67 ± 0.14c | 11.55 ± 0.15a | 10.49 ± 0.16b |
| SAAs | 2.52 ± 0.02f | 3.30 ± 0.02d | 3.52 ± 0.06c | 3.10 ± 0.05e | 4.18 ± 0.08a | 3.73 ± 0.07b |
| UAAs | 5.45 ± 0.09e | 8.90 ± 0.05c | 9.09 ± 0.09c | 8.13 ± 0.03d | 9.83 ± 0.12b | 10.18 ± 0.03a |
| TAAs | 5.85 ± 0.14f | 9.67 ± 0.08d | 9.94 ± 0.09c | 9.31 ± 0.07e | 10.91 ± 0.12a | 10.35 ± 0.11b |
| BAAs | 7.15 ± 0.13e | 10.45 ± 0.11b | 10.18 ± 0.14c | 7.92 ± 0.17d | 10.90 ± 0.13a | 10.50 ± 0.17b |
| AAAs | 2.80 ± 0.03f | 3.76 ± 0.03d | 3.99 ± 0.07c | 3.53 ± 0.08e | 4.67 ± 0.09a | 4.21 ± 0.08b |
| ATP | 0.01 ± 0.00d | 0.14 ± 0.00a | 0.14 ± 0.01a | 0.01 ± 0.00c | 0.02 ± 0.00b | 0.02 ± 0.00bc |
| ADP | 0.00 ± 0.00d | 0.04 ± 0.00a | 0.05 ± 0.00a | 0.01 ± 0.00d | 0.02 ± 0.00b | 0.01 ± 0.00c |
| AMP | 2.41 ± 0.10e | 5.03 ± 0.09d | 5.26 ± 0.10c | 4.99 ± 0.06d | 6.02 ± 0.13a | 5.46 ± 0.06b |
| IMP | 0.20 ± 0.01e | 0.28 ± 0.01d | 0.54 ± 0.03b | 0.37 ± 0.03c | 0.71 ± 0.02a | 0.70 ± 0.02a |
| HxR | 0.52 ± 0.05e | 0.79 ± 0.05c | 1.14 ± 0.07a | 0.64 ± 0.08d | 1.01 ± 0.04b | 0.98 ± 0.05b |
| Hx | 0.15 ± 0.01c | 0.44 ± 0.03a | 0.39 ± 0.01b | 0.40 ± 0.01b | 0.42 ± 0.02ab | 0.42 ± 0.02ab |
| GMP | 0.06 ± 0.01e | 0.08 ± 0.01d | 0.11 ± 0.01bc | 0.10 ± 0.00c | 0.12 ± 0.01b | 0.13 ± 0.01a |
| Total nucleotides | 3.36 ± 0.12f | 7.03 ± 0.12d | 7.39 ± 0.19c | 6.52 ± 0.09e | 8.32 ± 0.08a | 7.72 ± 0.14b |
Note: The data are expressed in the form of mean ± standard deviations (n = 3). Different letters (a-f) within the same row indicate significant differences (P < 0.05) between mean values. “*” represents essential amino acids; FAA represents total free amino acids; EAA represents total essential amino acids; SAA represents total salty amino acids (sum of Glu and Asp); UAA represents total umami amino acids (sum of Glu, Asp, Gly, and Ala); TAA represents total sweet amino acids (sum of Gly, Ala, Ser, Thr, Pro, and Arg); BAA represents total bitter amino acids (sum of Lys, Met, Val, Ile, Leu, Tyr, His, and Phe); AAA represents total acids amino acids (sum of Glu, Asp, and His).
Apart from their nutritional importance, free amino acids play a pivotal role in shaping the unique taste profiles of protein hydrolysates (Chen et al., 2022; Yang et al., 2024). Amino acids are categorized into five groups based on their taste attributes: salty amino acids (SAAs), umami amino acids (UAAs), sweet amino acids (TAAs), bitter amino acids (BAAs), and acidic amino acids (AAAs) (Wang et al., 2022; Zhang, Tu, et al., 2024). Reports have indicated that SAAs and UAAs predominantly influenced the salty-umami taste, especially Glu and Asp, which could contribute strongly to this taste profile in protein hydrolysates (Wang et al., 2022; Zhang, Tu, et al., 2024). In the control, SAAs and UAAs concentrations were limited to 2.52 and 5.45 g/L, respectively (P < 0.05). However, enzymatic treatment could elevate amino acid levels, which increased these concentrations significantly in the protein hydrolysates (P < 0.05), thereby enriching the salty-umami taste of the hydrolysates. This phenomenon aligned with trends observed in DH and MW distribution (referring to Fig. 1). Additionally, both flavourzyme and protamex exhibit a profound ability to hydrolyze and disrupt protein structures (Chen et al., 2023; Yang et al., 2024). Consequently, the concentrations of SAAs and UAAs surged significantly in the flavourzyme- and protamex-treated hydrolysates (P < 0.05).
Furthermore, Gly, Arg, and Ala are exemplary sweet amino acids abundantly found in protein hydrolysates. Apart from contributing sweetness, Gly and Arg can also mitigate the bitterness of protein hydrolysates and amplify their umami flavor (Yang et al., 2024; Zhu et al., 2023). For instance, when Ala is present alongside umami amino acids (such as Glu), they can create a synergistic effect, ultimately imparting a pronounced umami taste to protein hydrolysates (Liu et al., 2024). A diverse range of BAAs were also identified in protein hydrolysates, with Leu emerging as the most prevalent. Remarkably, even minute concentrations of BAAs have the potential to amplify the umami and sweet flavors imparted by other amino acids (Du et al., 2024; Zhu et al., 2023). As a result, protein hydrolysates processed by flavourzyme and protamex comprised a notable amount of taste-associated amino acids, ultimately endowing them with a distinct salty-umami taste in comparison to the control and other proteases.
3.5. Effects of different proteases on the concentrations of nucleotides in protein hydrolysates
Nucleotides and their related compounds constitute a class of taste compounds that can synergize with amino acids to not only enhance saltiness and umami but also improve the overall taste of protein hydrolysates (Liu et al., 2024; Wang et al., 2022). GMP primarily originates from RNA, while ADP, AMP, IMP, inosine (HxR), and hypoxanthine (Hx) are related products derived from ATP decomposition in shellfish after death (Chen et al., 2022; Liu et al., 2024). Previous studies have demonstrated that ATP decomposition can occur via two primary pathways: (1) ATP → ADP → AMP → IMP→HxR → Hx and (2) ATP → ADP → AMP → AdR → HxR → Hx (Chen et al., 2022; Liu et al., 2024). Among these nucleotides and their related compounds, four primarily taste-related compounds are AMP (2.41–6.02 g/L), IMP (0.20–0.71 g/L), HxR (0.52–1.14 g/L), and Hx (0.15–0.44 g/L), which account for 92.97–99.10 % of the total detected nucleotides in protein hydrolysates. These results confirm that pathway (2) is the primary pathway of ATP decomposition during the enzymatic hydrolysis process. Specifically, AMP has the highest content among the total nucleotides, and its concentration is much higher than that of IMP. This further supports the hypothesis that ATP is primarily degraded via pathway (2), with pathway (1) being secondary. These findings are also consistent with previous studies (Chen et al., 2022; Du et al., 2024; Yang et al., 2024), which observed that AMP is abundant in protein hydrolysates and pathway (2) dominates ATP decomposition during scallop adductors processing (Chen et al., 2022).
Moreover, the variation in total nucleotides and AMP in protein hydrolysates exhibited a similar trend. The highest concentrations of total nucleotides (8.32 g/L) and AMP (6.02 g/L) were detected in the flavourzyme-treated hydrolysate, closely followed by the protamex-treated hydrolysate (7.72 and 5.46 g/L), respectively. Notably, the concentration of AMP in protein hydrolysates was significantly higher than that of the control. These results aligned with previous studies where reported high levels of AMP in the trypsin-treated and flavourzyme-treated hydrolysates, specifically 1.302 and 1.084 mg/g, respectively (Yang et al., 2024). Such a high concentration of AMP could play a role in augmenting the umami taste of protein hydrolysates (Yang et al., 2024). Additionally, high concentrations of IMP and GMP were also observed in the flavourzyme- and protamex-treated hydrolysates (P < 0.05), echoing the trends seen in DH [referring to Fig. 1 (a)]. This was attributed to proteases promoting the release of non-volatile taste compounds, thereby elevating nucleotide concentrations (Yang et al., 2024). Gao et al. (2021) also highlighted that various enzymatic hydrolysis systems could boost nucleotide concentrations in protein hydrolysates. Based on this analysis, flavourzyme and protamex effectively promote the release of non-volatile taste compounds and elevate nucleotide concentrations during the enzymatic hydrolysis process, imparting their protein hydrolysates with a pronounced salty-umami taste.
3.6. Effects of different proteases on the concentrations of organic acids, betaine, and glycogen in protein hydrolysates
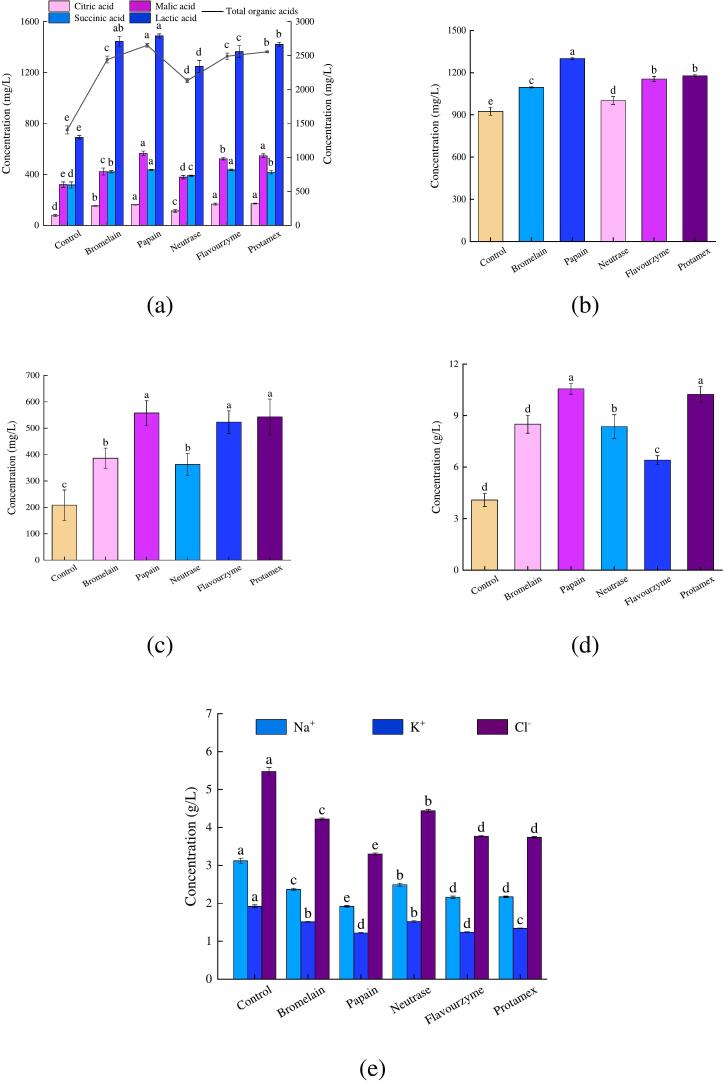
As illustrated in Fig. 2(a), citric, malic, succinic, and lactic acids were identified in both the control sample and the protein hydrolysates. These acids are metabolites of M. moneta, primarily derived from glycogen. Throughout the hydrolysis process, glycogen undergoes anaerobic degradation catalyzed by endogenous enzymes, resulting in elevated levels of organic acids in protein hydrolysates (Liu et al., 2024). Consequently, the concentrations of organic acids in protein hydrolysates were treated with different proteases ranged from 2.13 g/L to 2.65 g/L, significantly higher than the control's concentration of 1.41 g/L (P < 0.05). Most importantly, citric and lactic acids can impart a slightly fruity and sour taste while malic and succinic acids contribute a mild and crisp acidity, further enhancing the salty-umami taste of protein hydrolysates (Du et al., 2024; Wang, Huang, et al., 2023). Additionally, succinic acid and its sodium salt are crucial umami compounds among organic acids, since they synergize with inorganic ions and umami or sweet-free amino acids to amplify the umami taste (Bai et al., 2022; Liu et al., 2024). Hence, the abundance of malic and succinic acids in the flavorzyme- and protamex-treated hydrolysates contributes to the salty-umami taste.
Fig. 2.
Effects of different proteases on the concentration of taste compounds in protein hydrolysates of Monetaria moneta. (a) organic acids, (b) betaine, (c) glycogen, (d) peptides, (e) inorganic ions.
Betaine is a primary alkaloid compound that gives a unique sweetness to seafood and contributes to its taste (Chen et al., 2022; Wang, Li et al., 2023). Moreover, although glycogen is not directly linked to taste, it still holds a vital importance in the overall enhancement and modulation of flavor (Liu et al., 2024). As depicted in Fig. 2 (b-c), the protein hydrolysates demonstrated a comparable trend in the fluctuations of betaine and glycogen concentrations. The lowest concentrations of betaine and glycogen were observed in the control, measuring only 924.80 and 208.41 mg/L, respectively (P < 0.05). On the contrary, the concentrations of both betaine and glycogen in the protein hydrolysates increased significantly after enzymatic hydrolysis (P < 0.05). Additionally, high levels of betaine and glycogen were detected in the papain-, flavorzyme-, and protamex-treated hydrolysates, which also correlated positively with the high DHs [referring to Fig. 1(a)]. During the enzymatic hydrolysis process, proteases can efficiently hydrolyze proteins and disrupt muscle structure, releasing numerous taste compounds (Gao et al., 2021; Yang et al., 2024). Especially, glycoproteins undergo hydrolysis to release glycogen, significantly increasing the concentration of betaine and glycogen (Gao et al., 2021; Yang et al., 2024). According to variations in organic acids, betaine, and glycogen, it could be inferred that the salty-umami taste in the flavorzyme- and protamex-treated hydrolysates was extremely stronger compared to other hydrolysates (P < 0.05).
3.7. Effects of different proteases on the concentrations of peptide and inorganic ions in protein hydrolysates
Peptides are easier to digest than proteins composed of the same amino acids and taste better than a single amino acid (Wang et al., 2022). Hence, they are considered as crucial taste compounds that can impart a unique flavor to protein hydrolysates. The results presented in Fig. 2 (d) indicate that the concentrations of peptides in the protein hydrolysates produced by different proteases ranged from 6.41 g/L to 10.56 g/L, significantly exceeding the concentration of the control (4.09 g/L) (P < 0.05). Furthermore, the concentration of peptides in protein hydrolysates was closely correlated with the DH and enzyme cleavage site, which could explain the strong salty-umami taste observed in the flavourzyme- and protamex-treated hydrolysates. Papain and protamex, with their high DHs, efficiently hydrolyze proteins and convert them into peptides, elevating the peptide concentration in their protein hydrolysates. However, despite flavourzyme's high DH, as an exo-type protease, it cleaves peptide chains to produce free amino acids, leading to a lower peptide concentration in the flavourzyme-treated hydrolysates (Chen et al., 2023). On the contrary, the control, bromelain, and neutrase yielded the protein hydrolysates with low peptide concentrations owing to their low DHs. In addition, a series of taste-active peptides have been isolated and identified from enzymatic hydrolysates, and these taste-active peptides are all oligopeptides which can enrich the taste of enzymatic hydrolysates. For example, four decapeptides were found as taste-active peptides, and EDEGEQPRPF was the most potent taste-active compounds, and 0.4 mg/mL of the taste-active peptide in 50 mmol/L NaCl solution could increase its salty perception equivalent to the salt level of 63 mmol/L NaCl reference solution (Chen et al., 2021). The remaining peptides (VGPDDDEKSW, DEDEQPRPIP, and DEGEQPRPFP) also could give umami and kokumi tastes to food as well as show a weak saltiness-enhancing sensation (Chen et al., 2021). Therefore, the peptides with low molecular weights significantly contributed to the salty-umami taste, which resulted in the protamex-treated hydrolysate with obvious salty-umami flavor.
As shown in Fig. 2 (e), the primary taste-affecting inorganic ions detected in the protein hydrolysates were Na+, K+, and Cl−, and Cl− comprised over 50 % of the total inorganic ions. Moreover, the variations in inorganic ion concentration exhibited an inverse relationship with DH. Specifically, the highest concentration of inorganic ions was observed in the control, peaking at 10.52 g/L, but it plummeted after enzymatic hydrolysis. Additionally, the bromelain- and neutrase-treated hydrolysates contained higher concentrations of inorganic ions than those treated with other proteases. The flavourzyme- and protamex-treated hydrolysates showed lower concentrations of inorganic ions. Proteases with high DHs could proficiently hydrolyze M. moneta proteins, thereby releasing a substantial number of peptides, amino acids, polysaccharides, and other organic compounds (Gao et al., 2021; Yang et al., 2024). Because these compounds possess a strong affinity for chelating with inorganic ions, the concentrations of free inorganic ions became into decreasing as a result (Adelnia et al., 2023; Nickerson et al., 2008). Based on the aforementioned observations, it was further determined that flavourzyme and protamex exhibited high efficacy in hydrolyzing proteins, ultimately conferring a distinct salty-umami flavor to their protein hydrolysates.
3.8. Effects of different proteases on the TAV of taste compounds
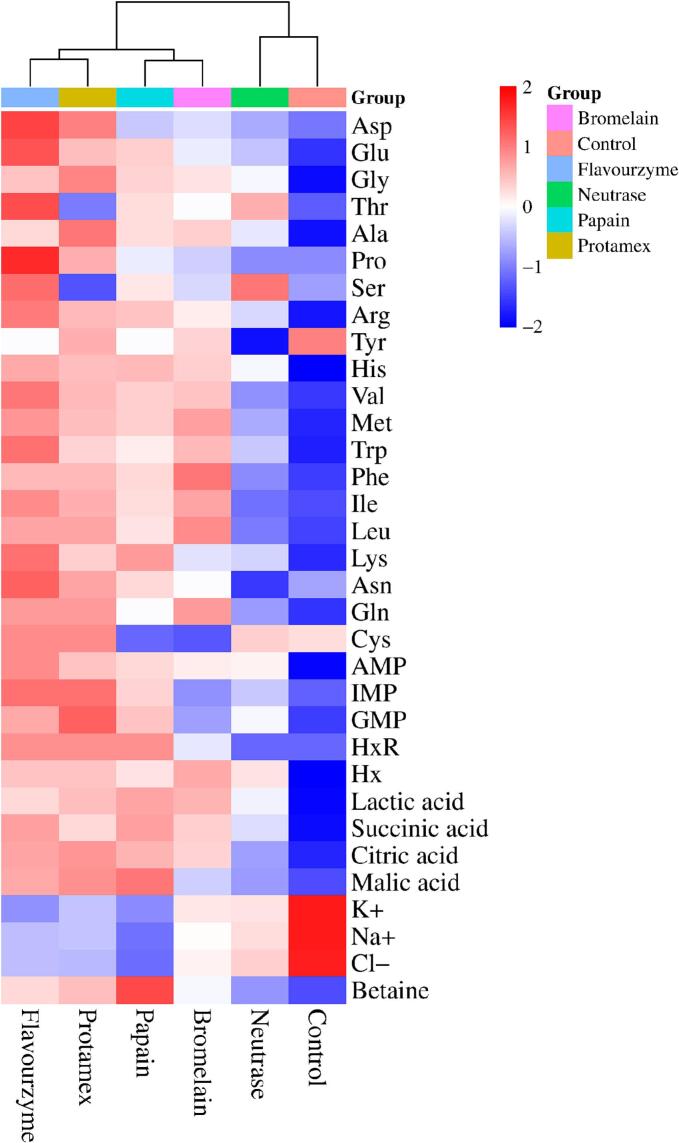
As shown in Table 2, the taste compounds with a TAV greater than 1.0 in the control and protein hydrolysates of various proteases were predominantly amino acids (Asp, Glu, Ala, Arg, Tyr, His, Val, Met, and Lys), AMP, succinic acid, and inorganic ions. Notably, the TAV of Glu and AMP reached impressive levels of 10.98 and 12.04, respectively, in the flavourzyme-treated hydrolysate. Essentially, these key compounds played a pivotal role in shaping the overall taste profiles of the protein hydrolysates. Compared to the control, the protein hydrolysates exhibited a significant increase in the TAV of amino acids, nucleotides, and organic acids (P < 0.05), but the TAV of Na+, K+, and Cl− decreased significantly (P < 0.05). Furthermore, the most substantial variations in the TAV of taste compounds were observed in the flavourzyme- and protamex-treated hydrolysates. Remarkably, both protein hydrolysates contained more than 20 types of taste compounds with a TAV greater than 1.0, imparting a rich salty-umami taste. This result can be attributed to the high DHs of flavourzyme and protamex, which allows for more thorough protein hydrolysis. Afterwards, this facilitates the release and concentration increase of non-volatile taste compounds, thereby elevating their TAV (Gao et al., 2021; Yang et al., 2024). Referring to Fig. 3, based on the variations in key taste compounds and their associated taste characteristics, the control and different protein hydrolysates can be categorized into four groups: Group I (control and neutrase-treated hydrolysate), Group II (bromelain- and papain-treated hydrolysates), and Group III (flavourzyme- and protamex-treated hydrolysates). Notably, Group III exhibited a significantly stronger (P < 0.05) salty-umami taste compared to the other groups.
Table 2.
Effects of different proteases on the taste compounds' TAV of protein hydrolysates of Monetaria moneta.
| Taste substance name | Taste threshold (g/L) |
TAV of taste substances in different protein hydrolysates |
|||||
|---|---|---|---|---|---|---|---|
| Control | Bromelain | Papain | Neutrase | Flavourzyme | Protamex | ||
| Asp | 0.53 | 1.10 ± 0.01e | 1.28 ± 0.01c | 1.25 ± 0.03c | 1.19 ± 0.03d | 1.67 ± 0.05a | 1.56 ± 0.06b |
| Glu | 0.30 | 6.47 ± 0.06e | 8.73 ± 0.05c | 9.52 ± 0.14b | 8.21 ± 0.12d | 10.98 ± 0.15a | 9.70 ± 0.13b |
| Gly | 1.30 | 0.66 ± 0.02f | 1.37 ± 0.02d | 1.42 ± 0.01c | 1.28 ± 0.01e | 1.45 ± 0.01b | 1.62 ± 0.01a |
| Thr | 2.60 | 0.29 ± 0.01e | 0.39 ± 0.01d | 0.41 ± 0.01c | 0.44 ± 0.01b | 0.50 ± 0.01a | 0.31 ± 0.01e |
| Ala | 0.60 | 3.46 ± 0.08e | 6.37 ± 0.02b | 6.21 ± 0.03c | 5.61 ± 0.02d | 6.26 ± 0.04c | 7.22 ± 0.04a |
| Pro | 3.00 | 0.10 ± 0.00d | 0.12 ± 0.00c | 0.13 ± 0.00c | 0.10 ± 0.01d | 0.20 ± 0.01a | 0.16 ± 0.02b |
| Ser | 1.50 | 0.28 ± 0.01d | 0.34 ± 0.00c | 0.41 ± 0.01b | 0.54 ± 0.01a | 0.55 ± 0.02a | 0.19 ± 0.02e |
| Arg | 0.50 | 2.88 ± 0.06e | 4.38 ± 0.03c | 4.64 ± 0.03b | 4.04 ± 0.04d | 5.06 ± 0.03a | 4.69 ± 0.04b |
| Tyr | 0.72 | 1.58 ± 0.04a | 1.42 ± 0.03c | 1.33 ± 0.03d | 0.87 ± 0.02e | 1.32 ± 0.03d | 1.49 ± 0.04b |
| His | 0.20 | 1.40 ± 0.05c | 2.31 ± 0.06a | 2.38 ± 0.06a | 2.14 ± 0.12b | 2.42 ± 0.06a | 2.36 ± 0.06a |
| Val | 0.40 | 2.31 ± 0.03d | 3.32 ± 0.04b | 3.27 ± 0.04b | 2.65 ± 0.07c | 3.60 ± 0.04a | 3.35 ± 0.07b |
| Met | 0.30 | 1.78 ± 0.06d | 2.93 ± 0.04a | 2.76 ± 0.06b | 2.27 ± 0.08c | 2.97 ± 0.06a | 2.82 ± 0.08b |
| Trp | – | – | – | – | – | – | – |
| Phe | 0.90 | 0.93 ± 0.01e | 1.36 ± 0.01a | 1.23 ± 0.02c | 1.03 ± 0.02d | 1.27 ± 0.01b | 1.27 ± 0.03b |
| Ile | 0.90 | 0.94 ± 0.02e | 1.44 ± 0.01b | 1.34 ± 0.01c | 1.01 ± 0.02d | 1.49 ± 0.03a | 1.42 ± 0.02b |
| Leu | 1.90 | 0.77 ± 0.01e | 1.35 ± 0.01a | 1.19 ± 0.02c | 0.88 ± 0.01d | 1.30 ± 0.01b | 1.30 ± 0.01b |
| Lys | 0.50 | 2.23 ± 0.03f | 3.33 ± 0.03d | 4.08 ± 0.04b | 3.24 ± 0.04e | 4.33 ± 0.05a | 3.77 ± 0.04c |
| Asn | – | – | – | – | – | – | – |
| Gln | – | – | – | – | – | – | – |
| Cys | 0.24 | 0.82 ± 0.04a | 0.70 ± 0.05b | 0.71 ± 0.05b | 0.84 ± 0.04a | 0.88 ± 0.07a | 0.88 ± 0.07a |
| AMP | 0.50 | 4.82 ± 0.21e | 10.07 ± 0.18d | 10.52 ± 0.21c | 9.99 ± 0.11d | 12.04 ± 0.25a | 10.91 ± 0.13b |
| IMP | 0.25 | 0.80 ± 0.06e | 1.12 ± 0.05d | 2.15 ± 0.13b | 1.49 ± 0.11c | 2.82 ± 0.07a | 2.81 ± 0.09a |
| GMP | 0.125 | 0.51 ± 0.05e | 0.66 ± 0.07d | 0.89 ± 0.08bc | 0.79 ± 0.03c | 0.93 ± 0.07b | 1.04 ± 0.06a |
| HxR | – | – | – | – | – | – | – |
| Hx | – | – | – | – | – | – | – |
| Lactic acid | 1.26 | 0.55 ± 0.01e | 1.15 ± 0.03ab | 1.18 ± 0.01a | 0.99 ± 0.04d | 1.08 ± 0.04c | 1.13 ± 0.01b |
| Succinic acid | 0.10 | 3.18 ± 0.24d | 4.21 ± 0.09b | 4.36 ± 0.05a | 3.91 ± 0.06c | 4.36 ± 0.06a | 4.17 ± 0.13b |
| Citric acid | 0.45 | 0.17 ± 0.02d | 0.34 ± 0.01b | 0.36 ± 0.00a | 0.25 ± 0.02c | 0.37 ± 0.02a | 0.38 ± 0.01a |
| Malic acid | 0.50 | 0.64 ± 0.04e | 0.85 ± 0.05c | 1.13 ± 0.04a | 0.76 ± 0.03d | 1.05 ± 0.02b | 1.09 ± 0.03a |
| K+ | 1.30 | 1.48 ± 0.03a | 1.16 ± 0.01b | 0.94 ± 0.01d | 1.17 ± 0.02b | 0.95 ± 0.01d | 1.03 ± 0.01c |
| Na+ | 1.80 | 1.73 ± 0.04a | 1.32 ± 0.02c | 1.07 ± 0.01e | 1.38 ± 0.02b | 1.20 ± 0.02d | 1.21 ± 0.01d |
| Cl− | 1.30 | 4.22 ± 0.08a | 3.25 ± 0.03c | 2.54 ± 0.03e | 3.42 ± 0.03b | 2.90 ± 0.01d | 2.88 ± 0.01d |
| Betaine | 2.50 | 0.37 ± 0.01e | 0.44 ± 0.00c | 0.52 ± 0.00a | 0.40 ± 0.01d | 0.46 ± 0.01b | 0.47 ± 0.00b |
Note: The data are expressed in the form of mean ± standard deviations (n = 3). Different letters (a-f) within the same row indicate significant differences (P < 0.05) between mean values. Taste recognition threshold value (g/L) of free amino acids (Bai et al., 2022; Wang et al., 2022), nucleotides (Du et al., 2024; Wang et al., 2022), organic acids (Zhu et al., 2023), inorganic ions and betaine (Chen et al., 2022).
Fig. 3.
Effects of different proteases on the dynamic heatmap of the taste compounds' TAV of protein hydrolysates of Monetaria moneta.
3.9. Effects of different proteases on salty and umami tastes of protein hydrolysates
Based on the analysis of E-tongue data and sensory scores shown in Fig. 4 (a-d), it is evident that the intensities of salty and umami tastes were the weakest in the control. In other words, the control exhibited the lowest levels of ESI and EUC with values of only 62.82 gNaCl/L and 19.08 gMSG/100 g, respectively (P < 0.05). Following enzymatic hydrolysis, the sensory scores for salty and umami tastes in the protein hydrolysates were significantly higher compared to the control (P < 0.05). As a result, the intensities of saltiness and umami were notably enhanced (P < 0.05). Additionally, their ESI and EUC varied in the range of 66.90–80.05 gNaCl/L and 44.97–84.56 gMSG/100 g, respectively. Most importantly, the flavourzyme-treated hydrolysate exhibited the highest levels of ESI (80.05 gNaCl/L) and EUC (84.56 gMSG/100 g) (P < 0.05), closely followed by the protamex-treated hydrolysate (77.95 gNaCl/L and 72.09 gMSG/100 g) (P < 0.05). These results aligned with the analysis presented in Table 1 and Table 2. Furthermore, similar observations were reported in a study by Li et al. (2023), where the salty and umami tastes in low-value red swamp crayfish hydrolysates increased significantly after hydrolysis with flavourzyme and protamex. Zhang, Tu, et al. (2024) also highlighted that saltiness and umami were the predominant characteristic tastes in the oyster hydrolysates.
Fig. 4.
Effects of different proteases on the salty and umami intensities of protein hydrolysates of Monetaria moneta. (a) sensory scores of salty and umami tastes, (b) the standard curve for the salty intensity, (c) salty and umami intensities, (d) equivalent salty intensity and equivalent umami concentration.
3.10. Interaction between umami and salty tastes in the protein hydrolysates
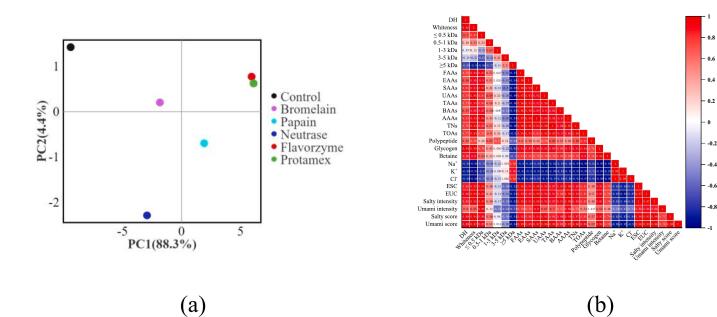
Based on the PCA analysis conducted using sensory scores for saltiness and umami, as shown in Fig. 5 (a), a significant disparity is evident between the control and the protein hydrolysates. Notably, the flavourzyme- and protamex-treated hydrolysates exhibited the most analogous salty-umami flavor characteristics. In conclusion, flavourzyme and protamex demonstrated exceptional protein hydrolysis abilities when compared to the control and other proteases. As shown in Fig. 5 (b), saltiness and umami also exhibited a positive correlation with the DH. Proteases could hydrolyze proteins more thoroughly, releasing numerous taste compounds and enhancing the whiteness of the protein hydrolysate (Gao et al., 2021; Yang et al., 2024). This resulted in a positive correlation between saltiness, umami and taste compounds (such as amino acids, nucleotides, organic acids, peptides) in the protein hydrolysates. Additionally, saltiness and umami also demonstrated a positive correlation with <3 kDa peptides but a negative correlation with >3 kDa peptides. In other words, there was a strong positive correlation between saltiness and umami in the protein hydrolysates, with a correlation coefficient exceeding 0.90. This further supported the idea that saltiness and umami could mutually reinforce each other. Collectively, our findings aligned with previous reports. For instance, under low-concentration ion conditions (such as Na+ and K+), Glu can behave similarly to MSG, a typical umami taste compound (Liu et al., 2024; Yang et al., 2024). Simultaneously, Glu, being a typical acidic amino acid, can ionize hydrogen ions in protein hydrolysates akin to organic acids. These hydrogen ions can convert chemical signals into molecular second messengers via a gustatory pathway, finally triggering the perception of a salty taste in the brain (Le et al., 2022; Song et al., 2023). Besides, organic acid anions exhibit a stronger affinity for cell membranes, facilitating ion penetration into taste cells and enhancing the salty taste perception as a result (Le et al., 2022; Song et al., 2023). Since sourness can enhance the perception of saltiness to a certain extent, umami peptides with sour taste characteristics may play a significant role in saltiness enhancement (Zhang, He, et al., 2024).
Fig. 5.
Effects of different proteases on the correlation analysis of protein hydrolysates of Monetaria moneta. (a) PCA analysis, (b) matrix heatmap analysis.
Furthermore, umami substances have the potential to enhance the perception of saltiness, reducing sodium intake, and thus synergistic umami effect represents a potential approach to salt reduction (Li et al., 2024). Higher concentrations of umami hydrolysates exhibit a greater enhancement effect on salty taste, which is likely due to MSG's ability to compensate for NaCl, optimizing palatability (Yang et al., 2022). Selamat et al. (2016) explored the acceptability of chicken soup with varying NaCl and MSG concentrations, revealing that the addition of 0.7 % MSG allowed for a 32.5 % reduction in NaCl without compromising taste. Salty-enhancing peptides, rich in arginine such as Arg-Arg, Arg-Val, and Ala-Arg, augment the frequency of sodium-induced responses, stimulating the α-subunit of the epithelial sodium channel (ENaC) (Song et al., 2023). These findings further confirm that the protein hydrolysates were enriched with salty and umami compounds which collectively impart a pronounced salty-umami taste to their protein hydrolysates (Li et al., 2024; Selamat et al., 2016; Zhang, He, et al., 2024). As a result, the flavourzyme- and protamex-treated hydrolysates were suitable as seasoning bases for producing dual-functional seasoning ingredients with a salty-umami taste, ideal for use in low-salt food production.
4. Conclusion
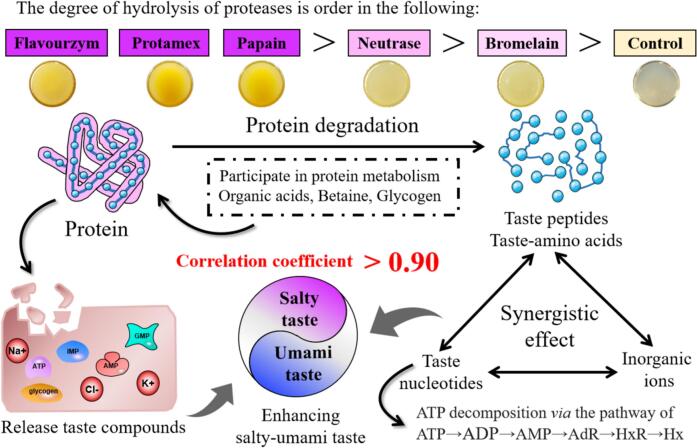
In conclusion, enzymatic hydrolysis proved beneficial for releasing taste compounds from M. moneta, enhancing the whiteness of protein hydrolysates, and significantly increasing the amount of <3 kDa peptides (referring to Fig. 6). Flavourzyme and protamex could hydrolyze M. moneta proteins more thoroughly, which resulted in various kinds of taste compounds serving as key contributors to the salty-umami taste, thereby imparting a distinct salty-umami taste to their hydrolysates. The taste compounds also worked synergistically to deliver a potent salty-umami taste, and a positive correlation was observed between saltiness and umami in the protein hydrolysates. Consequently, the flavourzyme-treated hydrolysate exhibited the highest levels of ESI and EUC, followed by the protamex-treated hydrolysates. Collectively, these results provide invaluable insights for augmenting the salty-umami taste of protein hydrolysates. Further investigation into the synergistic reactions among taste compounds to improve the salty-umami taste would be beneficial for utilizing protein hydrolysates as dual-functional seasoning ingredients, imparting a salty-umami taste to low-salt foods.
Fig. 6.
Effects of different proteases on the formation of salty-umami taste of protein hydrolysates of Monetaria moneta.
Ethical statement
We had confirmed that the appropriate protocols for protecting the rights and privacy of all participants were utilized in the execution of the sensory evaluation.
CRediT authorship contribution statement
Chunyong Song: Writing – review & editing, Writing – original draft, Methodology, Investigation, Data curation. Yaofang Yang: Methodology, Investigation, Formal analysis, Data curation, Conceptualization. Zhihang Zhao: Validation, Supervision, Software, Methodology, Investigation. Mingtang Tan: Software, Methodology, Investigation, Data curation, Conceptualization. Zhongqin Chen: Methodology, Investigation, Formal analysis, Data curation, Conceptualization. Huina Zheng: Visualization, Validation, Supervision, Software, Methodology. Jialong Gao: Visualization, Validation, Supervision, Software, Conceptualization. Haisheng Lin: Visualization, Validation, Supervision, Software, Conceptualization. Guoping Zhu: Visualization, Validation, Supervision, Software, Methodology. Wenhong Cao: Writing – review & editing, Resources, Project administration, Funding acquisition, Data curation.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by the Shenzhen Science and Technology Program (JCYJ20230807120316033) and the Modern Agricultural Industry Technology Research System of China (CARS-49). We gratefully acknowledge the anonymous referees for the comments and constructive suggestions provided for improving the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.fochx.2024.102056.
Appendix A. Supplementary data
Supplementary material
Data availability
Data will be made available on request.
References
- Adelnia H., Sirous F., Blakey I., Ta H.T. Metal ion chelation of poly (aspartic acid): From scale inhibition to therapeutic potentials. International Journal of Biological Macromolecules. 2023;229:974–993. doi: 10.1016/j.ijbiomac.2022.12.256. [DOI] [PubMed] [Google Scholar]
- Bai J., Fan Y., Zhu L.L., Wang Y.C., Hou H. Characteristic flavor of Antarctic krill (Euphausia superba) and white shrimp (Penaeus vannamei) induced by thermal treatment. Food Chemistry. 2022;378 doi: 10.1016/j.foodchem.2022.132074. [DOI] [PubMed] [Google Scholar]
- Chen J.H., Zhu L., Wu Q.M., Chen Y.L., Wu G.C., Zhang H. Structure characterization and bioactivities of protein hydrolysates of chia seed expeller processed with different proteases in silico and in vitro. Food Bioscience. 2023;55 doi: 10.1016/j.fbio.2023.102781. [DOI] [Google Scholar]
- Chen Y.P., Wang M.N., Blank I., Xu J.J., Chung H.Y. Saltiness-enhancing peptides isolated from the Chinese commercial fermented soybean curds with potential applications in salt reduction. Journal of Agricultural and Food Chemistry. 2021;69(35):10272–10280. doi: 10.1021/acs.jafc.1c03431. [DOI] [PubMed] [Google Scholar]
- Chen Z.Q., Zhu Y.H., Cao W.H., Zhou L.J., Zhang C.H., Qin X.M., Zheng H.N., Lin H.S., Gao J.L. Novel insight into the role of processing stages in nutritional components changes and characteristic flavors formation of noble scallop Chlamys nobilis adductors. Food Chemistry. 2022;378 doi: 10.1016/j.foodchem.2022.132049. [DOI] [PubMed] [Google Scholar]
- Du J.X., Xi J.P., Chen X., Sun H.L., Zhong L., Zhan Q.P., Zhao L.Y. Effects of different extraction methods on the release of non-volatile flavor components in shiitake mushroom (Lentinus edodes) Journal of Food Composition and Analysis. 2024;128 doi: 10.1016/j.jfca.2024.106001. [DOI] [Google Scholar]
- Fang X.J., Chen Z.Q., Wu W.J., Chen H.J., Nie S.P., Gao H.Y. Effects of different protease treatment on protein degradation and flavor components of Lentinus edodes. eFood. 2022;3(6) doi: 10.1002/EFD2.41. [DOI] [Google Scholar]
- Feng L., Wu Y.M., Han Y.T., Yao X.Q., Li Q.Q., Liu M.M., Cao Y.G. Structural characteristics, functional properties and nutritional value of walnut protein by limited enzymatic hydrolysis. LWT- Food Science and Technology. 2024;197 doi: 10.1016/j.lwt.2024.115923. [DOI] [Google Scholar]
- Gao J., Fang D.L., Kimatu B.M., Chen X., Wu X., Du J.X., Yang Q., Chen H., Zheng H.N., An X.X., Zhao L.Y., Hu Q.H. Analysis of umami taste substances of morel mushroom (Morchella sextelata) hydrolysates derived from different enzymatic systems. Food Chemistry. 2021;362 doi: 10.1016/j.foodchem.2021.130192. [DOI] [PubMed] [Google Scholar]
- He M., Tan M.Q. Hollow salt for sodium reduction in foods: Mechanisms, influence factors, applications and challenges. Trends in Food Science & Technology. 2024;147 doi: 10.1016/j.tifs.2024.104451. [DOI] [Google Scholar]
- Hu L.T., Elam E., Ni Z.J., Shen Y., Xia B., Thakur K., Jiang L., Zhang J.G., Wei Z.J. The structure and flavor of low sodium seasoning salts in combination with different sesame seed meal protein hydrolysate derived Maillard reaction products. Food Chemistry: X. 2021;12 doi: 10.1016/J.FOCHX.2021.100148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ildephonse H., Daniel N., Bertrand M., Falade E., Afusat Y.A., Marc A.N. Recent and novel processing technologies coupled with enzymatic hydrolysis to enhance the production of antioxidant peptides from food proteins: A review. Food Chemistry. 2023;423 doi: 10.1016/j.foodchem.2023.136313. [DOI] [PubMed] [Google Scholar]
- Kieliszek M., Pobiega K., Piwowarek K., Kot A.M. Characteristics of the proteolytic enzymes produced by lactic acid bacteria. Molecules. 2021;26(7):1858. doi: 10.3390/molecules26071858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le B., Yu B.B., Amin M.S., Liu R.X., Zhang N., Soladoye O.P., Aluko R.E., Zhang Y.H., Fu Y. Salt taste receptors and associated salty/salt taste-enhancing peptides: A comprehensive review of structure and function. Trends in Food Science and Technology. 2022;129:657–666. doi: 10.1016/J.TIFS.2022.11.014. [DOI] [Google Scholar]
- Li C.J., Tu Z.C., Liu W.Y., Wu C.L., Hu Y.M., Wang H. Flavor substances of low-valued red swamp crayfish (Procambarus clarkii) hydrolysates derived from double enzymatic systems. Food Research International. 2023;165 doi: 10.1016/j.foodres.2023.112461. [DOI] [PubMed] [Google Scholar]
- Li J.Y., Zhong F., Spence C., Xia Y.X. Synergistic effect of combining umami substances enhances perceived saltiness. Food Research International. 2024;189 doi: 10.1016/j.foodres.2024.114516. [DOI] [PubMed] [Google Scholar]
- Li W., Du R., Majura J.J., Chen Z.Q., Cao W.H., Zhang C.H., Zheng H.N., Gao J.L., Lin H.S., Qin X.M. The spatial distribution patterns, physicochemical properties, and structural characterization of proteins in oysters (Crassostrea hongkongensis) Foods. 2022;11(18):2820. doi: 10.3390/foods11182820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z.Y., Wei S., Xiao N.Y., Liu Y., Sun Q.X., Zhang B., Jin H.W., Cao H., Liu S.C. Insight into the correlation of key taste substances and key volatile substances from shrimp heads at different temperatures. Food Chemistry. 2024;450 doi: 10.1016/j.foodchem.2024.139150. [DOI] [PubMed] [Google Scholar]
- Nickerson M.T., Paulson A.T., Hallett F.R. Pre-gel solution properties of gellan polysaccharides: Effect of potassium and calcium ions on chain associations. Food Research International. 2008;41(5):462–471. doi: 10.1016/j.foodres.2007.12.009. [DOI] [Google Scholar]
- Selamat J., Parvaneh H., Roslina K., Sarian N., Simayi Y., Razak A.K. Reduction of sodium content in spicy soups using monosodium glutamate. Food & Nutrition Research. 2016;60(0):30463. doi: 10.3402/fnr.v60.30463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen D.Y., Pan F., Yang Z.C., Song H.L., Zou T.T., Xiong J., Li K., Li P., Hu N., Xue D.D. Identification of novel saltiness-enhancing peptides from yeast extract and their mechanism of action for transmembrane channel-like 4 (TMC4) protein through experimental and integrated computational modeling. Food Chemistry. 2022;388 doi: 10.1016/j.foodchem.2022.132993. [DOI] [PubMed] [Google Scholar]
- Shukor M.Y., Masdor N., Baharom N.A., Jamal J.A., Abdullah M.P.A., Shamaan N.A., Syed M.A. An inhibitive determination method for heavy metals using bromelain, a cysteine protease. Applied Biochemistry and Biotechnology. 2008;144(3):283–291. doi: 10.1007/s12010-007-8063-5. [DOI] [PubMed] [Google Scholar]
- Song C.Y., Wang Z.J., Li H.Q., Cao W.H., Chen Z.Q., Zheng H.N., Gao J.L., Lin H.S., Zhu G.P. Recent advances in taste transduction mechanism, analysis methods and strategies employed to improve the taste of taste peptides. Critical Reviews in Food Science and Nutrition. 2023;1-20 doi: 10.1080/10408398.2023.2280246. [DOI] [PubMed] [Google Scholar]
- Taladrid D., Laguna L., Bartolomé B., Moreno-Arribas M.V. Plant-derived seasonings as sodium salt replacers in food. Trends in Food Science & Technology. 2020;99:194–202. doi: 10.1016/j.tifs.2020.03.002. [DOI] [Google Scholar]
- Veymar G.T.P., Daniel C.V., Olga T., Ángel B.M., Beatriz T.S., Roberto F.L. Peptides with biological and technofunctional properties produced by bromelain hydrolysis of proteins from different sources: A review. International Journal of Biological Macromolecules. 2023;253(P5) doi: 10.1016/j.ijbiomac.2023.127244. [DOI] [PubMed] [Google Scholar]
- Veymar G.T.P., Daniel C.V., Roberto M.S., Olga T., Ángel B.M., Gilber V.G., Irfan A.R., Roberto F.L. Bioactive peptides from fisheries residues: A review of use of papain in proteolysis reactions. International Journal of Biological Macromolecules. 2021;184:415–428. doi: 10.1016/j.ijbiomac.2021.06.076. [DOI] [PubMed] [Google Scholar]
- Wang H.Y., Chen D., Lu W.J., Dang Y.L., Liu Z.M., Chen G.Y., Wang B., Zhang C., Xiao C.G. Novel salty peptides derived from bovine bone: Identification, taste characteristic, and salt-enhancing mechanism. Food Chemistry. 2024;447 doi: 10.1016/j.foodchem.2024.139035. [DOI] [PubMed] [Google Scholar]
- Wang J., Huang X.H., Zhang Y.Y., Li S.J., Dong X.P., Qin L. Effect of sodium salt on meat products and reduction sodium strategies-a review. Meat Science. 2023;205 doi: 10.1016/j.meatsci.2023.109296. [DOI] [PubMed] [Google Scholar]
- Wang Y.Y., Tang X.H., Luan J.J., Zhu W.H., Xu Y.X., Yi S.M., Li J.R., Wang J.X., Li X.P. Effects of ultrasound pretreatment at different powers on flavor characteristics of enzymatic hydrolysates of cod (Gadus macrocephalus) head. Food Research International. 2022;159 doi: 10.1016/J.FOODRES.2022.111612. [DOI] [PubMed] [Google Scholar]
- Wang Z.J., Li H.Q., Cao W.H., Chen Z.Q., Gao J.L., Zheng H.N., Lin H.S., Qin X.M. Effect of drying process on the formation of the characteristic flavor of oyster (Crassostrea hongkongensis) Foods. 2023;12(11) doi: 10.1016/J.FBIO.2024.103736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F., Fu A.Z., Meng H.Y., Liu Y., Bi S. Non-volatile taste active compounds and umami evaluation of Agrocybe aegerita hydrolysates derived using different enzymes. Food Bioscience. 2024;58 doi: 10.1016/j.fbio.2024.103772. [DOI] [Google Scholar]
- Yang F., Lv S., Liu Y., Bi S., Zhang Y. Determination of umami compounds in edible fungi and evaluation of salty enhancement effect of antler fungus enzymatic hydrolysate. Food Chemistry. 2022;387 doi: 10.1016/j.foodchem.2022.132890. [DOI] [PubMed] [Google Scholar]
- Yuan J., Chen S.X., Wang L.P., Xu T.T., Shi X., Jing Y., Zhang H.J., Huang Y.G., Xu Y., Li D., Chen X., Chen J.H., Xiong Q.P. Preparation of purified fractions for polysaccharides from Monetaria moneta Linnaeus and comparison their characteristics and antioxidant activities. International Journal of Biological Macromolecules. 2018;108:342–349. doi: 10.1016/j.ijbiomac.2017.12.023. [DOI] [PubMed] [Google Scholar]
- Zhang H., Liu Y., Gao L., Wang J.H. Analysis of flavor changes in Huangshan floral mushroom hydrolysates obtained by different enzyme treatments. Food Chemistry. 2024;443 doi: 10.1016/j.foodchem.2024.138554. [DOI] [PubMed] [Google Scholar]
- Zhang J.C., He W., Liang L., Sun B.G., Zhang Y.Y. Study on the saltiness-enhancing mechanism of chicken-derived umami peptides by sensory evaluation and molecular docking to transmembrane channel-like protein 4 (TMC4) Food Research International. 2024;182 doi: 10.1016/j.foodres.2024.114139. [DOI] [PubMed] [Google Scholar]
- Zhang J.N., Zhao M.M., Su G.W., Lin L.Z. Identification and taste characteristics of novel umami and umami-enhancing peptides separated from peanut protein isolate hydrolysate by consecutive chromatography and UPLC–ESI–QTOF–MS/MS. Food Chemistry. 2018;278:674–682. doi: 10.1016/j.foodchem.2018.11.114. [DOI] [PubMed] [Google Scholar]
- Zhang J.W., Tu Z.C., Hu Z.Z., Hu Y.M., Wang H. Efficient preparation of oyster hydrolysate with aroma and umami coexistence derived from ultrasonic pretreatment assisted enzymatic hydrolysis. Food Chemistry. 2024;437(2) doi: 10.1016/j.foodchem.2023.137881. [DOI] [PubMed] [Google Scholar]
- Zhang W.W., Shi K.X., Han Y.Q., Wang J.M., Yang C., Xu X., Li B.Y. Characterization of Pleurotus citrinopileatus hydrolysates obtained from Actinomucor elegans proteases compared with that by commercial proteases. Journal of Food Science. 2022;87(9):3737–3751. doi: 10.1111/1750-3841.16256. [DOI] [PubMed] [Google Scholar]
- Zheng Y., Yang F.F., Yuan X.W., Ji Y.Q., Li H.J., Li H.B., Yu J.H., Zulewska J. Enzymatic hydrolysis of whey proteins by the combination of alcalase and neutrase: Kinetic model and hydrolysis control. International Dairy Journal. 2024;151 doi: 10.1016/j.idairyj.2023.105867. [DOI] [Google Scholar]
- Zhu S.C., Zhu L., Ke Z.G., Chen H., Zheng Y.D., Yang P.…Liu S.L. A comparative study on the taste quality of Mytilus coruscus under different shucking treatments. Food Chemistry. 2023;412 doi: 10.1016/j.foodchem.2023.135480. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
Data will be made available on request.