Abstract
Background
Periodontitis is considered to be one of the major risk factors associated with cancers of the oral cavity. Periodontogenic pathogens such as Porphyromonas gingivalis, Treponema denticola, Tannerella forsythia, Fusobacterium nucleatum and Aggregatibacter actinomycetemcomitans are the important pathogens associated with periodontitis. Chronic exposure to bacterial components induces changes in the nearby cells. Hence, the present study has been designed to identify the molecular mechanisms that could be associated with the two disease conditions viz., periodontitis and head and neck cancer.
Objective
The present study investigated the differential gene expression profile in human gingival keratinocytes treated with P. gingivalis (Pg), a bacterium associated with periodontal disease, and its possible association with the development of Head and Neck Squamous Cell Carcinoma (HNSCC).
Methods
The study followed a computational design. Multiple tools and databases, such as GEOmnibus, STRING, Metascape, PANTHER, and UALCAN, cBioportal, were used to derive an association between gene expression during infection with P. gingivalis, and the resulting gene expression profiles were analyzed in the HNSCC dataset.
Results
The study revealed 29 genes from a pool of transcripts acquired after comparing the Pg-HIGK and Sham-HIGK. Among them, 3 genes i.e., FST, VRK3, and SGK1, were found to be overexpressed and significantly influenced patient survival. The upregulation of FST was found to correlate with poor prognosis in HNSCC patients.
Conclusion
The study provided insights into the possible association of FST, VRK3 and SGK1 in the development of HNSCC. Further investigations are warranted to confirm the functional role of these genes in establishing the cancer phenotype in patients with chronic infection with Pg.
Keywords: Microbiome, Head and neck cancer, Carcinoma, Genotoxic, Diagnosis, Prognosis
Graphical abstract

1. Introduction
The human microbiome, especially the oral microbiome, is vital in maintaining good health.1 The Human Oral Microbiome Database identifies over 700 bacterial species in this complex ecosystem. The advancements in sequencing technology have revealed a symbiotic relationship between the microbiome, host, and environment, with dysbiosis contributing to various diseases.2 Dysbiotic oral microbiomes have been linked to conditions such as periodontitis and systemic diseases, including cardiovascular disease and cancer.3 Oral squamous cell carcinoma (OSCC) and gastrointestinal cancers are connected to the oral microbiome through the “oral-gut axis”.4 While many studies focused on individual bacterial species, a growing understanding emphasizes the importance of interactions within multi-species microbial communities in cancer development. An extensive review of literature has recently focused on the connection between two established periodontal pathogens, Porphyromonas gingivalis (Pg) and Fusobacteria nucleatum, and their potential roles in the development of oral squamous cell carcinoma (OSCC), colorectal cancer (CRC), and pancreatic ductal adenocarcinoma (PDAC). The studies investigated the prevalence of keystone bacteria in multiple studies and explored the clinical and experimental evidence of their collaboration's impact. Although there is insufficient experimental evidence of synergy, the studies hypothesized to explain the underlying mechanisms, which could pave the way for further research in this evolving field.5 A recent study by Saba and team investigated the link between the anaerobic bacterium P. gingivalis and pancreatic ductal adenocarcinoma (PDAC). Administering Pg to mice, the bacteria that translocated from the oral cavity to the pancreas were detected in human PDAC lesions. In wild-type mice, P. gingivalis induced pancreatic changes, and in a PDAC model, it accelerated progression. In vitro, P. gingivalis infection induced markers of pancreatic changes and protected PDAC cells from stress. The findings revealed a causal role for Pg in pancreatic cancer development in mice, highlighting potential mechanistic pathways.6
Several pathways have been explored to identify the possible relationship between bacterial proteins and the development of tumors. The Pg induces chronic inflammation, which can turn into a tumor-promoting inflammation.7 The modulation of the immune response, production of gingipains that can damage host tissues,8 dysbiosis of oral microbiome triggered by complex interactions with other bacterial species, and activation of signaling pathways leading to cell proliferation are other mechanisms associated with cancer development. A supportive study demonstrated the relationship between bacterial lipopolysaccharide (LPS), programmed death ligand-1 (PD-L1) expression, and partial epithelial-mesenchymal transition (pEMT) in oral squamous cell carcinoma (OSCC). TLR4-expressing OSCC cell lines exhibited PD-L1 expression and pEMT when exposed to LPS from Pg or Escherichia coli (E. coli). While LPS was found to have no impact on cell morphology, migration, or proliferation, it was found to enhance the invasive capacity. Additionally, higher PD-L1 expression was observed in OSCC extracellular vesicles (EXOs) with LPS, suggesting a potential role of EXOs in mediating PD-L1 induction after LPS exposure.9 The periodontal pathogen has been associated with several forms of cancer, such as colorectal cancer,10 gastrointestinal cancer,11 colon cancer12 and many more.
The present study aimed to investigate the relationship between P. gingivalis infection fueling the development of head and neck carcinoma. The research is focused on analyzing the changes in gene expression that occur in the immortalized human gingival keratinocytes after they are treated with P. gingivalis. By examining these changes and their potential connection to the development of HNSCC, we demonstrated the key mechanisms that underlie the pathogenicity of P. gingivalis. The knowledge about the candidate genes expressed in both datasets would provide insights into the molecular pathways that may be involved in the initiation and progression of HNSCC and advance our understanding of the complex relationship between gum disease and oral cancer.
2. Materials and methods
2.1. Selection of pathogen
Extensive text mining was conducted before the pathogen for this study was selected. The process revealed that Porphyromonas gingivalis plays a crucial role in oral and orodigestive squamous cell carcinoma. Hence, the dataset for the same was queried in GEOmnibus. The strength of the present study lies in using computational tools to arrive at a preliminary result that can be further validated using experimental approaches. Since high throughput methodologies and large sample sizes are used to derive the results, the study design provides a reliable way to satisfy the aim of the study.
2.2. Sample dataset
The GEOmnibus dataset GSE12121 was used for the study. This series comprised 4 samples of HIGK (Human Immortalized Gingival Keratinocytes) infected with Porphyromonas gingivalis (Pg) (GSM305596, GSM305597, GSM305598, GSM305599), 4 Sham-infected HIGK cells (GSM305588, GSM305589, GSM305590, GSM305591). Sham-infected HIGK cells served as the control. The HIGK cells infected with Pg were taken as the test group (https://www.ncbi.nlm.nih.gov/geo/geo2r/?acc=GSE12121).13 The GEO2R analysis was performed to curate the differentially expressed genes (DEGs) from the pool of transcriptome based on the adjusted p-value and fold changes.
2.3. Protein-protein interaction analysis
We analyzed the top 29 differentially expressed genes (DEGs) using the STRING (Search Tool for the Retrieval of Interacting Genes/Proteins) tool, version 12.0. This bioinformatics resource provides information about direct physical interactions and functional associations between proteins. In the network, the nodes represent proteins, and the edges indicate the type of interaction, such as physical, enzymatic, or genetic. The analysis was conducted at a median confidence score of 0.400.14 Metascape, a gene annotation and analysis resource, was used to identify the gene enrichment pattern and possible association of the queried DEGs with disease conditions. The DisGENET database was also queried for this purpose.15
2.4. Gene ontology analysis
The gene ontology analysis was performed using the PANTHER database (v16.0; Protein Analysis Through Evolutionary Relationships). This analysis provided information on the molecular pathways and functions, biological processes, and sub-cellular localization of gene products. A user-defined query of the top 29 genes was fed as a batch to identify the functional classification of genes. Additionally, classification based on pathways was conducted to identify potential pathways associated with the genes.16,17
2.5. Gene expression and survival analysis
The information about the top 29 differentially expressed genes in the Pg-treated HIGK group, as assessed using the GEO2R program, was further investigated in the HNSCC dataset by employing the UALCAN database (http://ualcan.path.uab.edu/cgi-bin/TCGA-survival). The study analyzed 520 samples from patients with HNSCC primary tumors and 44 paired normal samples. The expression profile was measured in transcripts per million (TPM), a standard unit for normalizing RNA-seq data. The significance between different groups was determined by creating box-whisker plots using the TPM values. The study also demonstrated the overall survival of patients with HNSCC using Kaplan-Meier plots. The high-expression and low/medium-expression groups were compared to illustrate the effect of gene expression changes on patients' overall survival.18
2.6. Statistical analysis
The gene expression data from multiple datasets were analyzed using the GEO2R tool. GEO2R uses R packages from Limma to compare groups of samples in a GEO series to identify differentially expressed genes. It presents results as a table and graphic plots.19 The UALCAN portal analyzed the gene expression profile by comparing the expression levels between groups using the PERL script accompanied by the Comprehensive Perl Archive Network (CPAN) module. The survival plots were generated using “survival” and “survminer” R packages, which were compared using a log-rank test. Survival was exclusively used to perform survival analysis, including survival curves, hypothesis tests, and models, whereas Survminer improved the visualization of Kaplan-Meier and forest plots for clear representation and interpretation of complex survival data.20
2.7. Mutation profiling
The cBioPortal is an open-access platform for visualizing and analyzing cancer genomics data. It provides a user-friendly interface for researchers to explore multidimensional cancer genomics datasets, access various cancer studies, perform interactive analyses, and visualize results through customizable plots and charts. The mutation profiling of the 3 DEGs viz., FST, VRK3, and SGK1 was analyzed using the cBioportal site. Over 504 samples were queried for the presence of gross genetic abnormalities or point mutations.21,22
3. Results
3.1. Curation of DEGs
The GEO2R analysis of the transcriptome data derived from the comparison between Pg-treated HIGK and Sham-HIGK revealed 29 differentially expressed genes (DEGs) on comparison of gene expression profiles were identified, among which two were found to be downregulated, and 27 were upregulated (Table 1). The non-coding RNA and repetitive genes were excluded from further analysis. The KRTAP2 gene showed the highest fold change value (4.15) with an adjusted p-value of 0.0023. The gene RBSN exhibited a 3-fold change with an adjusted p-value of 0.0506. The adjusted p-values above 0.0506 were not significant (Fig. 1).
Table 1.
List of differentially expressed genes of the GSE12121 dataset (Sham-HIGK vs PG treated -HIGK).
| Gene symbol | Protein encoded | Adj.P.Val | LogFC |
|---|---|---|---|
| KRTAP2 | Keratin associated protein 2-3///keratin associated protein 2-4 | 0.00229 | 4.15 |
| EDN1 | Endothelin 1 | 0.00229 | 3.80 |
| PHLDB2 | Pleckstrin homology like domain family B member 2 | 0.00386 | 3.03 |
| PLK2 | Polo like kinase 2 | 0.00386 | 2.51 |
| PTGS2 | Prostaglandin-endoperoxide synthase 2 | 0.00386 | 3.06 |
| KLF6 | Kruppel like factor 6 | 0.0096 | 2.16 |
| TNFAIP3 | TNF alpha induced protein 3 | 0.01526 | 2.09 |
| IL6 | Interleukin 6 | 0.02675 | 1.93 |
| CTGF | Connective tissue growth factor | 0.02675 | 1.87 |
| FST | Follistatin | 0.02689 | 1.89 |
| NUAK2 | NUAK family kinase 2///A-kinase interacting protein 1 | 0.02689 | 1.76 |
| OVOL1 | Ovo like transcriptional repressor 1 | 0.02689 | 2.03 |
| GADD45A | Growth arrest and DNA damage inducible alpha | 0.02708 | 1.80 |
| ARRDC3 | Arrestin domain containing 3 | 0.02725 | −2.38 |
| PPP1R3B | Protein phosphatase 1 regulatory subunit 3B | 0.02725 | 1.70 |
| CYP1B1 | Cytochrome P450 family 1 subfamily B member 1 | 0.02725 | 1.65 |
| C1orf74 | Chromosome 1 open reading frame 74 | 0.02725 | 1.98 |
| SLC35E4 | Solute carrier family 35 member E4 | 0.02725 | 2.14 |
| CXCL8 | C-X-C motif chemokine ligand 8 | 0.02787 | 2.38 |
| TJP2 | Tight junction protein 2 | 0.03135 | 3.51 |
| KLF5 | Kruppel like factor 5 | 0.03182 | 1.59 |
| CYR61 | Cysteine rich angiogenic inducer 61 | 0.03942 | 1.68 |
| HES1 | Hes family bHLH transcription factor 1 | 0.03942 | 1.62 |
| CYP24A1 | Cytochrome P450 family 24 subfamily A member 1 | 0.03942 | 1.97 |
| VGLL3 | Vestigial like family member 3 | 0.04108 | 2.36 |
| VRK3 | Vaccinia related kinase 3 | 0.04451 | 2.37 |
| SGK1 | Serum/glucocorticoid regulated kinase 1 | 0.04698 | 1.42 |
| RORA | RAR related orphan receptor A | 0.04995 | −1.53 |
| RBSN | Rabenosyn, RAB effector | 0.05058 | 3.28 |
Fig. 1.
Volcano plot demonstrating differentially expressed genes (DEGs) that are upregulated (red) and downregulated (blue) in treated human immortalized gingival keratinocytes (HIGK). A p-value less than 0.05 is considered significant.
3.2. Gene ontology analysis
The gene ontology analysis revealed the following genes in each of the pathways described, PTGS2, CXCL8 - CCKR signaling, PHLDB2 - EGF receptor signaling, PTGS2, EDN1 - Endothelin signaling, FST - Gonadotrophin-releasing hormone receptor pathway, PTGS2, CXCL8, IL6 - Inflammation mediated by chemokine and cytokine signaling pathway, CXCL8, IL6 - Interleukin signaling pathway, HES1 - Notch signaling pathway, GADD45A - PI3 Kinase pathway, TNFAIP3, PTGS2 - Toll receptor signaling pathway, CYP24A1 - Vitamin D metabolism and pathway, EDN1 - Wnt signaling pathway, GADD45A - P38 MAPK pathway, GADD45A - P53 pathway (Fig. 2). About sixteen genes were categorized as non-panther genes since they did not fall under the predetermined category as classified by the PANTHER tool.
Fig. 2.
Gene ontology analysis using Panther (http://www.pantherdb.org/) revealed the molecular pathways associated with the differentially expressed genes.
∗ 16 genes were clustered into the unclassified group.
3.3. Protein Protein network interactions
The protein-protein interaction of the 29 DEGs identified one preliminary cluster and 12 other independent proteins. This large cluster accommodated PLK2, GADD45A, RORA, IL6, EDN1, FST, KLF6, CXCL8, CCN1, CCN2, PTGS2, CYP1B1, CYP24A1, TNFAIP3, SGK1, and PPP1R3. The KEGG pathway analysis demonstrated major signaling pathways associated with carcinogenesis such as IL17, TNF, NF-Kappa B etc., (Fig. 3a). The gene enrichment and DisGENET analysis revealed the association of the 29 DEGs with various forms of cancer such as mammary neoplasms, meningioma, ductal carcinoma, invasive carcinoma of the breast, etc (Fig. 3b).
Fig. 3.
a) The protein-protein interaction network derived from STRING version 12.0 demonstrates protein clusters interacting with each other and independent proteins showing no interactions in the network; b) DisGENET analysis demonstrates the association of DEGs with different types of carcinomas, including carcinoma of the breast and mammary neoplasms.
3.4. Gene expression and survival analysis
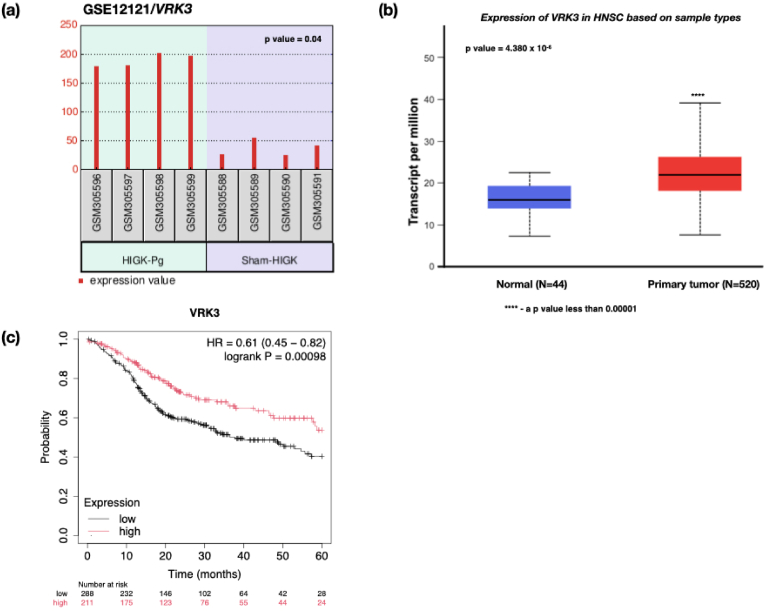
The genes FST, VRK3, and SGK1 were among the 29 differentially expressed genes that presented with an identical gene expression pattern in the Pg-treated HIGK and HNSCC datasets (Table 2). All 3 genes in the HNSCC group showed a statistically significant upregulation. These genes were found to influence the survival of HNSCC patients significantly. The upregulation of the FST gene (1.624 × 10−12) correlated well with the survival of HNSCC patients (p-value = 0.0026), wherein increased expression of the FST gene resulted in poor prognosis (Fig. 4). The VRK3 (4.380 × 10−6) (Fig. 5) and SGK1 (9.099 × 10−3) (Fig. 6) genes were significantly upregulated, exhibiting differential expression compared to the normal tissues. Interestingly, the overexpression of VRK3 (p-value = 0.00098) and SGK1 (p-value = 0.056) genes presented increased survival of HNSCC patients. The reason for such a presentation is worth investigating.
Table 2.
Expression profile of DEGs in PG treated-HIGK and HNSCC dataset (TCGA).
| Gene symbol | Gene expression in PG treated HIGK | Gene expression in HNSCC | P value | Survival (p value) |
|---|---|---|---|---|
| KRTAP2/KRTAP2-2 | Upregulated | Insignificant | Insufficient data | Insufficient data |
| EDN1 | Upregulated | Insignificant | 4.941 x 10-01 | 0.92 |
| PHLDB2 | Upregulated | Upregulated | <10−12 | 0.85 |
| PLK2 | Upregulated | Insignificant | 4.941 x 10-01 | 0.92 |
| PTGS2 | Upregulated | Upregulated | 1.679 × 10−12 | 0.69 |
| KLF6 | Upregulated | Downregulated | 1.105 x 10-2 | 0.78 |
| TNFAIP3 | Upregulated | Upregulated | 1.093 × 10−12 | 0.43 |
| IL6 | Upregulated | Insignificant | 6.393 x 10-02 | 0.047 |
| CTGF | Upregulated | Insignificant | 6.598 x 10-01 | 0.16 |
| FST | Upregulated | Upregulated | 1.624 x 10–12 | 0.021 |
| NUAK2 | Upregulated | Downregulated | 1.624 × 10−2 | 0.59 |
| OVOL1 | Upregulated | Insignificant | 9.050 × 10−2 | 0.58 |
| GADD45A | Upregulated | Insignificant | 3.559 × 10−1 | 0.36 |
| ARRDC3 | Downregulated | Insignificant | 5.383 × 10−2 | 0.31 |
| PPP1R3B | Upregulated | Insignificant | 7.617 × 10−2 | 0.45 |
| CYP1B1 | Upregulated | Insignificant | 5.186 × 10−1 | 0.55 |
| C1orf74 | Upregulated | Upregulated | 1.887 × 10−15 | 0.45 |
| SLC35E4 | Upregulated | Upregulated | 2.975 × 10−10 | 0.40 |
| CXCL8/IL8 | Upregulated | Upregulated | 7.281 × 10−09 | 0.057 |
| TJP2 | Upregulated | Insignificant | 7.120 × 10−1 | 0.55 |
| KLF5 | Upregulated | Insignificant | 6.545 × 10−2 | 0.49 |
| CYR61 | Upregulated | Insignificant | 2.696 × 10−1 | 0.65 |
| HES1 | Upregulated | Insignificant | 9.141 × 10−1 | 0.56 |
| CYP24A1 | Upregulated | Upregulated | 3.661 × 10−5 | 0.32 |
| VGLL3 | Upregulated | Insignificant | 9.000 × 10−2 | 0.14 |
| VRK3 | Upregulated | Upregulated | 4.380 x 10–6 | 0.0092 |
| SGK1 | Upregulated | Upregulated | 9.099 x 10–3 | 0.036 |
| RORA | Downregulated | Downregulated | 1.016 × 10−6 | 0.092 |
| RBSN | Upregulated | Insignificant | 6.267 × 10−1 | 0.006 |
Fig. 4.
(a) Histogram demonstrating the gene expression in HIGK cells infected with Pg. A significant fold change of 1.89 (p-value = 0.026) was observed, (b) Box-Whisker plot showing the gene expression profile of the FST gene in HNSCC datasets. The gene expression between the normal and the HNSCC primary tumor group demonstrated significant upregulation in transcript levels (p-value = 1.624 × 10−12), (c) Kaplan Meier plot demonstrating survival probability of HNSCC patients with a differential expression profile. A statistically significant change in survival was observed with the expression of FST (p-value = 0.0026). The patients with increased expression of FST presented with poor prognosis.
Fig. 5.
(a) Histogram demonstrating the VRK3 expression of the gene in HIGK cells infected with Pg, A significant fold change of 2.37 (p-value = 0.04) was observed, (b) Box-Whisker plot showing the gene expression profile of the VRK3 gene in HNSCC datasets. The gene expression between the normal and the HNSCC primary tumor group demonstrated significant upregulation in transcript levels (p-value = 4.380 × 10−6), (c) Kaplan Meier plot demonstrating survival probability of HNSCC patients with a differential expression profile. A statistically significant change in survival was observed with the expression of VRK3 (p-value = 0.00098). The patients with low/medium expression of VRK3 presented with poor prognosis.
Fig. 6.
(a) Histogram demonstrating the SGK1 gene expression in HIGK cells infected with Pg, A significant fold change of 1.42 (p-value = 0.04) was observed, (b) Box-Whisker plot showing the gene expression profile of the SGK1 gene in HNSCC datasets. The gene expression between the normal and the HNSCC primary tumor group demonstrated significant upregulation in transcript levels (p-value = 9.099 × 10−3), (c) Kaplan Meier plot demonstrating survival probability of HNSCC patients with a differential expression profile. A statistically significant change in survival was observed with the expression of SGK1 (p-value = 0.056). The patients with low/medium expression of SGK1 presented with poor prognosis.
3.5. Mutation profiling
Mutation profiling of FST, VRK3, and SGK1 genes revealed one to less than 1 % alterations in the queried genes (Fig. 7). The FST gene showed deep deletion and amplification in 6 patients, the VRK3 gene exhibited amplification and missense mutations (R461H, F296L, E299D), with a somatic mutation frequency of 0.6 %, and the SGK1 gene showed amplification, deep deletion, and a missense mutation (H365D).
Fig. 7.
Mutation profiling of the genes FST, VRK3, and SGK1 showing gross chromosomal abnormalities and point mutations.
4. Discussion
The present study investigated the possible association of genes differentially expressed during infection with Porphyromonas gingivalis and their potential role in developing head and neck cancers. Twenty-nine DEGs were curated based on the adjusted p-value from the transcriptome profile when comparing the Pg-HIGK and Sham-HIGK treated cells. The DEGs were investigated for molecular interactions, and pathway analysis revealed that most genes were involved in the inflammatory and cancer-related pathways. The DEGs were further checked for their gene expression in the HNSCC (TCGA dataset), which revealed increased expression of three genes, FST, VRK3, and SGK1, exhibiting similar expression patterns to that of Pg-treated HIGK cells (Fig. 4, Fig. 5, Fig. 6b). Among the 3 genes, FST was found to be oncogenic, wherein the increased expression of this gene resulted in poor prognosis (Fig. 4c). In contrast, the VRK3 and SGK1 genes presented with good prognosis. The upregulation of VRK3 and SGK1 improved HNSCC patient survival rates. The decrease in gene expression can be attributed primarily to the action of non-coding RNAs, such as miRNAs, that can target gene transcripts and inhibit the translation process. Further investigations are warranted to provide substantial evidence on the role of these genetic and epigenetic factors in precipitating cancer phenotypes.
A study by Oriuchi and their team investigated the connection between Pg LPS and the development of gastric cancer. The biopsy samples taken from patients with gastric cancer showed that when exposed to Pg lipopolysaccharide, they had reduced mucosal impedance and an increased expression of toll-like receptor 2 (TLR2), tumor necrosis factor α (TNFα), and apoptotic markers. The experiments conducted in vitro showed that during oral administration of Pg in Gan mice, ROS-related apoptosis and TLR2-β-catenin signaling activation occurred in gastric cell lines. The LPS led to tumor enlargement and β-catenin signaling activation. The findings suggest local exposure to Pg lipopolysaccharide may contribute to gastric cancer through apoptosis-related barrier dysfunction and TNFα secretion from activated macrophages, activating TLR2-β-catenin signaling.23 Thus, reports from experimental studies showed that the LPS-promoted tumor via TLR2-β-catenin signaling pathways. The present study supported the fact that inflammatory pathways can give rise to tumor-promoting inflammation.
Follistatin encoded by the FTS gene is a type of glycoprotein expressed in most tissues. Elevated levels of FTS have been implicated in type 2 diabetes (T2D).24 The protein has two major mature forms: follistatin 315 and 288.25 The alternative splicing of the same precursor gene produces these forms. Follistatin-like 3, which is similar in structure to follistatin, has also been identified. This gene is expressed in most tissues where activin mRNAs are found. Follistatin has a high affinity for activins and can block their effects by preventing them from binding to their receptors.26 A study by Oyelakin demonstrated the role of p63, an oncogenic transcription factor, in head and neck squamous cell carcinoma (HNSCC) progression. Through integrative analysis of multiple datasets, FST, encoding the glycoprotein Follistatin, was identified as a direct transcriptional target of p63. The FST, influenced by TGF-β and EGFR pathways, promoted tumor growth and migration in HNSCC cells. Single-cell RNA sequencing data link FST to cancer cells within the tumor microenvironment, suggesting its potential as a prognostic biomarker and mediator of p63 effects on both tumor and microenvironment in HNSCC.27 Another study revealed the role of syndecan-1 (SDC1) in oral cancer and its interaction with follistatin-related protein 1 (FSTL1). The study investigated their associations in oral squamous cell carcinoma models and revealed that SDC1 forms complexes with FSTL1, influencing tumor events. Knocking down FSTL1 or both FSTL1 and SDC1 resulted in less aggressive tumors with altered characteristics. The study thus identified a relationship between SDC1 and FSTL1 in cell signaling events, shedding light on their impact on cancer development.28 A recent study identified therapeutic targets for squamous cell carcinoma of the tongue by analyzing the functional gene expression datasets. The protocol revealed 154 genes that differed in expression between SCC and normal tissues. Bioinformatics analysis suggested that high levels of follistatin expression could lead to a poor prognosis. Experimental validation was conducted to confirm these findings through colony formation assays and molecular analyses, where they successfully demonstrated that reducing follistatin levels inhibits the proliferation of tongue SCC cells.29 These results revealed the potential of targeting Follistatin in SCC treatment to improve patient outcomes. A similar kind of observation was obtained in the present study, where the expression of the FST gene was overexpressed, leading to a decline in the survival of HNSCC patients.
Vaccinia-related kinase 1 (VRK1) and VRK3 are serine/threonine kinases belonging to the VRK family. These proteins are mainly located in the nucleus and have been found to play important roles in various biological processes, such as cell cycle regulation, DNA repair, chromatin assembly, and RNA processing. Recent studies have shown that VRK1/VRK3 interact with other proteins involved in these processes. Biochemical assays have confirmed these interactions, and in liver cancer cells, upregulation of VRK1/VRK3 has been observed to affect cell cycle progression. This highlights their regulatory roles in diverse biological processes.30 A clinical study on lung squamous cell carcinoma (LUSC) documented the elevated bovine pox virus-associated kinase 1 (VRK1) expression in both LUSC tissues and cells. Survival analysis revealed a lower overall survival in LUSC patients with high VRK1 expression. VRK1 downregulation through shRNA interference significantly impacted LUSC cell viability, migration, cell cycle progression, and apoptosis. The study suggests VRK1 as a potential therapeutic target for LUSC, emphasizing its role in DNA damage response.31 A similar study investigated the role of VRK1 in various cancers. Utilizing public databases and experimental analyses, VRK1 upregulation is identified across multiple cancer types, associating with worse prognosis in specific cases. The correlations with immune factors and promoter methylation variations were positive. Experimental results in hepatocellular carcinoma (HCC) revealed elevated VRK1 expression, establishing its role as an oncogene in HCC progression.32 The study on HNSCC cases was found to agree with the observations of the present study. However, the reduced expression of VRK1 inevitably resulted in a worse prognosis, an important fact to be noted. The interaction of the VRK1 gene products with that of specific non-coding RNA populations can result in a marked reduction in the transcript levels. Hence, more research is warranted in this area to explore and understand the role of VRK1 in head and neck carcinogenesis.
SGK1 is crucial in cellular stress response, promoting cell survival under various extracellular stress conditions. Factors like osmolarity, cell volume changes, and cellular stressors such as hyperosmotic stress, oxidative stress, heat shock, and UV irradiation influence the expression. The modulation of Sgk-1 promoter activity by specific response elements in response to glucocorticoids, p53, and hyperosmotic stress contributes to increased transcript expression in different cell systems. The kinetics, magnitude, and duration of induced Sgk-1 expression vary across stimuli, indicating its diverse role in maintaining physiological homeostasis.33 In non-small cell lung cancer (NSCLC), the PI3K signaling pathway is frequently altered and often associated with drug-induced resistance. This prompted the exploration of glucocorticoid-induced kinase 1 (SGK1) as a potential target. Inhibiting SGK1 alongside PI3K/AKT showed synergistic anticancer activity, inducing apoptosis, DNA damage, and G1 phase cell cycle arrest. High SGK1 expression correlated with increased resistance to PI3K inhibitors, suggesting its potential as a predictive marker. The study revealed that PI3K/AKT and SGK-1 inhibition eventually lead to anticancer activity.34
Although all three genes identified from the pool of DEGs demonstrated increased expression, the prognosis for the genes varied widely. This variation in the prognosis can be attributed to epigenetic mechanisms such as promoter methylation, histone modification, and regulation of gene expression via non-coding RNAs. Several studies have focused on microRNAs, regulating the expression of target genes,35,36 and methyltransferases serving as prognostic markers.37 Also, the dysbiosis of the microbiome in the oral cavity in response to metabolic disorders or habitual practices can invariably result in the deviation of gene expression, contributing to variation in patient prognosis. The genes identified can be targeted by small molecule inhibitors or non-coding RNAs to improve anti-cancer effects, thereby controlling the proliferation and promoting the death of the cancerous cells. Combined with all the data obtained through computational analysis, FST was considered a vital prognostic marker for Pg-mediated head and neck carcinogenesis. Nevertheless, the statement has to be validated further by performing experimental and clinical research to enhance our understanding of the functions and carcinogenic potential of Pg-induced genes in HNSCC. Cancer, being a multifactorial disorder with a complex interplay between genes and environment, underscores the need for prompt diagnosis and treatment. Recently, microbes have also been found to trigger pathways associated with carcinogenesis. Early treatment options for chronic inflammatory conditions directed towards eradicating pathogenic microorganisms could avoid the malignant transformation of cells nearby.
5. Limitations
The current study has several limitations that need to be addressed in addition to the strengths discussed earlier: (a) the study design uses a computational approach that needs validation through in vitro and in vivo models, (b) the study can be replicated in normal cells to rule out the influence of the immortalization process on gene expression, although the comparison group included sham-treated HIGK, (c) while the study demonstrated the influence of candidate gene expression on patient survival, it did not explore the potential impact of epigenetic mechanisms on the prognosis of HNSCC patients, which warrants further investigation, (d) also the gene expression profile is influenced by various factors such as exposure to carcinogens, habits, infectious diseases, inflammatory conditions, age, gender, stage, ethnic population, etc., requiring a more controlled environment to provide conclusive evidence about the association of the microbial pathogen with cancer development. A more detailed analysis of the exposure of the HNSCC group to periodontal pathogens will provide conclusive evidence about their role in cancer development.
6. Conclusion
Although the association between Pg and oral cancer has been established previously, the molecular mechanisms underlying the process have not been revealed. The observations from the present study provided preliminary evidence on the hub of DEGs identically expressed during two disease conditions. Future prospects of the study lie in experimental validation of the FST gene in animal models infected with Pg, which could provide more insights about this marker and its role in a biological environment. Additionally, knockout studies can unravel the role of these candidate gene markers in carcinogenesis. The observations from other markers, such as VRK1 and SGK1, are worth investigating due to their inverse relationship with the patient's survival. This study has opened novel avenues for clinical researchers to reveal the causal link between chronic infection, inflammation, and cancer.
Patient's/Guardian's consent
Not required - Computational study.
Ethical clearance
Not required - Computational analysis.
Sources of funding
Not Available.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
The authors are grateful to all the cohorts and groups involved in the compilation of data from patients for public use. Our sincere thanks also go to all the patients who have indirectly contributed to the scientific community by providing consent to share their data for research use.
References
- 1.Relman D.A. The human microbiome and the future practice of medicine. JAMA. 2015;314(11):1127–1128. doi: 10.1001/jama.2015.10700. [DOI] [PubMed] [Google Scholar]
- 2.Ogunrinola G.A., Oyewale J.O., Oshamika O.O., et al. The human microbiome and its impacts on health. Internet J Microbiol. 2020;2020 doi: 10.1155/2020/8045646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Giordano-Kelhoffer B., Lorca C., March Llanes J., et al. Oral microbiota, its equilibrium and implications in the pathophysiology of human diseases: a systematic review. Biomedicines. 2022;10(8):1803. doi: 10.3390/biomedicines10081803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li Z., Liu Y., Zhang L. Role of the microbiome in oral cancer occurrence, progression and therapy. Microb Pathog. 2022;169 doi: 10.1016/j.micpath.2022.105638. [DOI] [PubMed] [Google Scholar]
- 5.Wang B., Deng J., Donati V., et al. The roles and interactions of Porphyromonas gingivalis and Fusobacterium nucleatum in oral and gastrointestinal carcinogenesis: a narrative review. Pathogens. 2024;13(1):93. doi: 10.3390/pathogens13010093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saba E., Farhat M., Daoud A., et al. Oral bacteria accelerate pancreatic cancer development in mice. Gut. 2024 doi: 10.1136/gutjnl-2023-330941. gutjnl-2023-330941. [DOI] [PubMed] [Google Scholar]
- 7.Guo Z.C., Jing S.L., Jia X.Y., et al. Porphyromonas gingivalis promotes the progression of oral squamous cell carcinoma by stimulating the release of neutrophil extracellular traps in the tumor immune microenvironment. Inflamm Res. 2023 doi: 10.1007/s00011-023-01822-z. [DOI] [PubMed] [Google Scholar]
- 8.Kylmä A.K., Sorsa T., Jouhi L., et al. Prognostic role of Porphyromonas gingivalis gingipain rgp and matrix metalloproteinase 9 in oropharyngeal squamous cell carcinoma. Anticancer Res. 2022;42(11):5415–5430. doi: 10.21873/anticanres.16046. [DOI] [PubMed] [Google Scholar]
- 9.Omori Y., Noguchi K., Kitamura M., et al. Bacterial lipopolysaccharide induces PD-L1 expression and an invasive phenotype of oral squamous cell carcinoma cells. Cancers. 2024;16(2):343. doi: 10.3390/cancers16020343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kerdreux M., Edin S., Löwenmark T., et al. Porphyromonas gingivalis in colorectal cancer and its association to patient prognosis. J Cancer. 2023;14(9):1479–1485. doi: 10.7150/jca.83395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nasiri K., Amiri Moghaddam M., Etajuri E.A., et al. Periodontitis and progression of gastrointestinal cancer: current knowledge and future perspective. Clin Transl Oncol. 2023;25(10):2801–2811. doi: 10.1007/s12094-023-03162-0. [DOI] [PubMed] [Google Scholar]
- 12.Lu Y., Huang R., Zhang Y., et al. Porphyromonas gingivalis induced UCHL3 to promote colon cancer progression. Am J Cancer Res. 2023;13(12):5981–5995. [PMC free article] [PubMed] [Google Scholar]
- 13.Handfield M., Mans J.J., Zheng G., et al. Distinct transcriptional profiles characterise oral epithelium-microbiota interactions. Cell Microbiol. 2005;7(6):811–823. doi: 10.1111/j.1462-5822.2005.00513.x. [DOI] [PubMed] [Google Scholar]
- 14.Szklarczyk D., Gable A.L., Lyon D., et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou Y., Zhou B., Pache L., et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10(1):1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mi H., Thomas P. PANTHER pathway: an ontology-based pathway database coupled with data analysis tools. Methods Mol Biol. 2009;563:123–140. doi: 10.1007/978-1-60761-175-2_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mi H., Ebert D., Muruganujan A., et al. PANTHER version 16: a revised family classification, tree-based classification tool, enhancer regions and extensive API. Nucleic Acids Res. 2021;49(D1):D394–D403. doi: 10.1093/nar/gkaa1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chandrashekar D.S., Karthikeyan S.K., Korla P.K., et al. UALCAN: an integrated cancer data analysis platform update. Neoplasia. 2022;25:18–27. doi: 10.1016/j.neo.2022.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barrett T., Wilhite S.E., Ledoux P., et al. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res. 2013;41(Database issue):D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chandrashekar D.S., Bashel B., Balasubramanya S.A.H., et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19(8):649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cerami E., Gao J., Dogrusoz U., et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao J., Aksoy B.A., Dogrusoz U., et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:l1–pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oriuchi M., Lee S., Uno K., et al. Porphyromonas gingivalis lipopolysaccharide damages mucosal barrier to promote gastritis-associated carcinogenesis. Dig Dis Sci. 2024;69(1):95–111. doi: 10.1007/s10620-023-08142-6. [DOI] [PubMed] [Google Scholar]
- 24.Pan J., Nilsson J., Engström G., et al. Elevated circulating follistatin associates with increased risk of mortality and cardiometabolic disorders. Nutr Metabol Cardiovasc Dis. 2023;S0939–4753(23):371. doi: 10.1016/j.numecd.2023.09.012. X. [DOI] [PubMed] [Google Scholar]
- 25.Pierre A., Pisselet C., Monget P., et al. Testing the antagonistic effect of follistatin on BMP family members in ovine granulosa cells. Reprod Nutr Dev. 2005;45(4):419–425. doi: 10.1051/rnd:2005031. [DOI] [PubMed] [Google Scholar]
- 26.Wu C., Borné Y., Gao R., et al. Elevated circulating follistatin is associated with an increased risk of type 2 diabetes. Nat Commun. 2021;12(1):6486. doi: 10.1038/s41467-021-26536-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oyelakin A., Sosa J., Nayak K.B., et al. An integrated genomic approach identifies follistatin as a target of the p63-epidermal growth factor receptor oncogenic network in head and neck squamous cell carcinoma. NAR Cancer. 2023;5(3) doi: 10.1093/narcan/zcad038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zandonadi F.S., Yokoo S., Granato D.C., et al. Follistatin-related protein 1 interacting partner of Syndecan-1 promotes an aggressive phenotype on Oral Squamous cell carcinoma (OSCC) models. J Proteonomics. 2022;254 doi: 10.1016/j.jprot.2021.104474. [DOI] [PubMed] [Google Scholar]
- 29.Yu M., Xiao L., Chen Y., et al. Identification of a potential target for the treatment of squamous cell carcinoma of the tongue: follistatin. Br J Oral Maxillofac Surg. 2020;58(4):437–442. doi: 10.1016/j.bjoms.2020.01.028. [DOI] [PubMed] [Google Scholar]
- 30.Lee N., Kim D.K., Han S.H., et al. Comparative interactomes of VRK1 and VRK3 with their distinct roles in the cell cycle of liver cancer. Mol Cell. 2017;40(9):621–631. doi: 10.14348/molcells.2017.0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Du N., Zhang B., Zhang Y. Downregulation of VRK1 inhibits lung squamous cell carcinoma progression through DNA damage. Cancer Res J. 2023;2023 doi: 10.1155/2023/4533504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen D., Zhou W., Chen J., et al. Comprehensively prognostic and immunological analysis of VRK Serine/Threonine Kinase 1 in pan-cancer and identification in hepatocellular carcinoma. Aging (Albany NY) 2023;15(24):15504–15524. doi: 10.18632/aging.205389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O'Keeffe B.A., Cilia S., Maiyar A.C., et al. The serum- and glucocorticoid-induced protein kinase-1 (Sgk-1) mitochondria connection: identification of the IF-1 inhibitor of the F(1)F(0)-ATPase as a mitochondria-specific binding target and the stress-induced mitochondrial localisation of endogenous Sgk-1. Biochimie. 2013;95(6):1258–1265. doi: 10.1016/j.biochi.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kale R., Samant C., Bokare A., et al. Inhibition of SGK1 potentiates the anticancer activity of PI3K inhibitor in NSCLC cells through modulation of mTORC1, p-ERK and β-catenin signaling. Biomed Rep. 2023;19(6) doi: 10.3892/br.2023.1676. 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jayaseelan V.P., Arumugam P. Exosomal microRNAs targeting TP53 gene as promising prognostic markers for head and neck squamous cell carcinoma. Glob Med Genet. 2022;9(4):277–286. doi: 10.1055/s-0042-1758204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gopalakrishnan S., Pandi A., Arumugam P., Jayaseelan V.P. MicroRNAs targeting CDKN2A gene as a potential prognostic marker in head and neck squamous cell carcinoma. Mol Biol Res Commun. 2024;13(1):21–27. doi: 10.22099/mbrc.2023.48081.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muthumanickam P., Ramasubramanian A., Pandi C., et al. The novel m6A writer methyltransferase 5 is a promising prognostic biomarker and associated with immune cell infiltration in oral squamous cell carcinoma. J Oral Pathol Med. 2024;53(8):521–589. doi: 10.1111/jop.13568. [DOI] [PubMed] [Google Scholar]