ABSTRACT
Syndromic multiplex panel testing enables simultaneous detection of multiple respiratory pathogens, but limited data is available on the comparative diagnostic performance of different testing systems. In this multicenter prospective study, we aimed to compare the QIAstat‐Dx Respiratory Panel 2.0 (QIAstat‐Dx‐RP2.0) with the widely used BioFire‐RP2.1, using 269 respiratory clinical specimens. Concordant test results were obtained in 232 (86.3%) cases. Discordant test results included 33 BioFire‐RP2.1(+)/QIAstat‐Dx‐RP2.0(−) and 4 BioFire‐RP2.1(‐)/QIAstat‐Dx‐RP2.0(+) results. Discordant samples showed significantly lower pathogen loads than concordant ones (p < 0.01). Overall, the QIAstat‐Dx‐RP2.0 showed an analytical sensitivity of 50%–100% depending on the respiratory target, with an analytical specificity ≥ 99.0%. Most significant differences were found for the detection of adenovirus, human coronaviruses, respiratory syncytial virus, human rhinovirus/enterovirus and SARS‐CoV‐2 (kappa‐score: 0.67–0.91). Co‐detections of respiratory pathogens were identified in 47 cases by BioFire‐RP2.1 and 29 by QIAstat‐Dx‐RP2.0. Agreement rates between the two multiplex panel tests decreased from 91.8% for single pathogen detections to 65.0% and 42.9% for co‐detecting two and three pathogens, respectively. Pathogen loads were significantly lower in co‐detections compared to single pathogen detections (p < 0.01), potentially explaining the lower detection rates with the QIAstat‐Dx‐RP2.0 in cases of multiple pathogens. In conclusion, our prospective multicenter evaluation showed good diagnostic performance of the QIAstat‐Dx‐RP2.0 assay, but lower detection rates for some respiratory targets compared to BioFire‐RP2.1. As QIAstat‐Dx‐RP2.0 offers advantages in handling, noise emission, cost effectiveness, and provides semi‐quantitative results compared to BioFire‐RP2.1 an updated version with enhanced analytical sensitivity would be a viable alternative syndromic testing system for detecting respiratory pathogens.
Keywords: adenovirus, BioFire, influenza, QIAstat‐Dx, respiratory pathogen, respiratory tract infection, rhinovirus/enterovirus, RSV, SARS‐CoV‐2, syndromic multiplex panel testing
1. Introduction
Acute respiratory tract infections (RTIs) significantly contribute to morbidity and mortality, resulting in millions of deaths worldwide, particularly among vulnerable populations such as infants, older people, and immunocompromised individuals [1, 2]. Various pathogens, including SARS‐CoV‐2, other respiratory viruses and bacteria can cause RTIs. However, clinical presentation and symptoms often overlap for the different causative pathogens, posing a significant challenge to identify the infectious agent [3]. Despite bacterial infections being responsible for only a minority of RTIs, empiric antibiotic treatment targeting the most likely pathogens is commonly initiated by physicians due to diagnostic uncertainties [4]. Consequently, patients without confirmed bacterial infection receive unnecessary antibiotic treatment, leading to potential side effects and increased antibiotic resistance, underscoring the urgency for rapid diagnostics [5, 6].
Traditional methods such as bacterial culture and virus isolation by cell culture have been the gold standard for identifying bacterial and viral respiratory pathogens, respectively. However, these assays require skilled personnel and dedicated equipment, with a turn‐around‐time (TAT) of several days [3, 7]. In recent years, advances in molecular diagnostics with nucleic acid testing (NAT) have revolutionized the diagnosis of RTIs. The implementation of multiplex NAT assays has enabled the detection of multiple respiratory pathogens in a single test with high sensitivity and specificity [8, 9, 10], and with a significantly reduced TAT of approximately 6–8 h. However, a limitation remains in that a maximum of only five respiratory pathogens can be detected simultaneously in a single reaction. Therefore, multiple multiplex NAT assays must be performed on each respiratory sample to cover the wide variety of pathogens that can cause RTIs. Recently, several syndromic multiplex panel testing systems have become available that overcome this limitation. These systems can detect up to 23 respiratory pathogens in a single test with a very easy sample‐to‐answer workflow and a TAT of less than 2 h [11, 12]. Among those are the BioFire respiratory multiplex panel version 2.1 (BioFire‐RP2.1) and the QIAstat‐Dx respiratory panel version 2.0 (QIAstat‐Dx‐RP2.0), generating results from 23 different respiratory targets, including SARS‐CoV‐2 in approximately 1 h.
In this prospective multicenter study, we conducted a comparative evaluation of the BioFire‐RP2.1 and QIAstat‐Dx‐RP2.0 multiplex panel tests using 269 respiratory clinical specimens, prospectively collected from patients with RTIs.
2. Materials and Methods
2.1. Patients and Respiratory Samples
Patients presenting with RTIs at the outpatient clinics, emergency department, or who were hospitalized at the University Hospital Basel, the University of Basel Children's Hospital, or the University Hospital Bern were enrolled in this study (Table). Respiratory specimens were prospectively collected from November 2023 to February 2024 at the study sites, and were submitted in parallel for testing with the BioFire‐RP2.1 (Biomerieux, Marcy l’Étoile, France) and QIAstat‐Dx‐RP2.0 (Qiagen, Hilden, Germany) multiplex panel tests. The 269 respiratory specimens included 259 (96.3%) nasopharyngeal swabs, 8 (2.9%) bronchoalveolar lavages, and 2 (0.8%) tracheal secretions (Table S1).
2.2. Syndromic Multiplex Panel Testing
Respiratory specimens were tested in parallel with the two syndromic multiplex panel tests upon receipt in the diagnostic laboratory. The multiplex syndromic panel tests were performed according to the manufacturers' instructions.
For BioFire‐RP2.1, buffer and 0.3 mL respiratory sample were mixed in the injection vial and transferred to the pouch. The BioFire‐RP2.1 covers 23 respiratory targets (Supplementary Table 2), including human adenovirus (HAdV), human coronavirus (types ‐229E, ‐HKU1, ‐NL63 and ‐OC43), severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2), human metapneumovirus A and B (hMPV), human rhino‐/enterovirus (HRV/HEV), influenzavirus A (IV‐A), IV‐A/H1, IV‐A/H1‐2009, IV‐A/H3, influenzavirus B (IV‐B), Middle East respiratory syndrome (MERS) coronavirus, human parainfluenzavirus (HPIV) 1–4, respiratory syncytial virus A and B (RSV), Bordetella pertussis, Bordetella parapertussis, Chlamydophila pneumoniae, and Mycoplasma pneumoniae.
For QIAstat‐Dx‐RP2.0, 0.3 mL of sample was transferred to the main port of the QIAstat‐Dx cartridge. QIAstat‐Dx‐RP2.0 covers 23 targets (Table S2), including HAdV, HCoV (types ‐229E, ‐HKU1, ‐NL63 and ‐OC43), SARS‐CoV‐2, human bocavirus (HBoV), hMPV, HRV/HEV, IV‐A, IV‐A/H1, IV‐A/H1‐2009, IV‐A/H3, IV‐B, HPIV 1‐4, RSV, Bordetella pertussis, Chlamydophila pneumoniae, Legionella pneumophila, and Mycoplasma pneumoniae.
2.3. Pathogen‐Specific Nucleic Acid Testing
An aliquot of 500 μL was stored from respiratory specimens at ‐20°C for the study duration to be used for confirmation testing of samples with initial discordant results between the BioFire‐RP2.1 and QIAstat‐Dx‐RP2.0. Discordances in test results were resolved using laboratory‐developed NATs (LDTs) specific to the respective pathogen [13, 14, 15, 16, 17, 18, 19].
2.4. Viral Genotyping and Nanopore Sequencing
Genotyping of HRV‐positive clinical specimens was performed using NAT primers that match conserved sequences from the viral capsid regions 2 and 4 across different HRV genotypes [20]. NATs used the Iproof High Fidelity DNA Polymerase kit (BioRad, CA, USA) with 600 nM end concentration of the forward primer 5’‐CTACTTTGGGTGTCCGTGTTTC‐3’ and reverse primer 5’‐ATCHGGHARYTTCCAMCACCA‐3’. NATs had a reaction volume of 25 μL and 5 μL of reverse‐transcribed cDNA, and were run on Veriti Thermal Cyclers (Applied Biosystems, MA, USA) using the thermal cycling protocol specified in the Iproof High Fidelity DNA Polymerase kit. Library preparation was done from the full‐length amplicons using the Native Barcoding Kit (Oxford Nanopore Technologies, UK), following the manufacturers' instructions. Sequencing was performed on a GridION Mk1 platform (Oxford Nanopore Technologies). Raw FAST5 files produced were basecalled under high accuracy mode using the ONT basecaller Dorado (version 0.5.3; basecall_model_version_id “dna_r10.4.1_e8.2_400bps_hac@v4.2.0”). Basecalled FASTQ files were further processed using LORCAN (LOng Read Consensus ANalysis) pipeline [21, 22]. The LORCAN pipeline was developed for the analysis of barcoded amplicons sequenced with ONT‐based methods and was adapted for HRV‐based genotyping in this study. Briefly, the pipeline consists of the following main steps: Reads are filtered by length, keeping only those with a length of ±50 bp around the modal sequence length and a maximum of 3000 reads per barcode. Reads are mapped using minimap2 [23] to the closest reference and binned accordingly. Finally, a consensus sequence is derived using a 50% majority rule consensus. To enable the analysis of HRV sequences, a custom reference database was created by downloading 38 HRV reference sequences from GenBank using the following GenBank accession numbers (KC881035, KC881032, JN837694, JQ837724, D00239, MF973194, KF543906, FJ950838, OL133743, KF543907, OQ842402, FJ950829, OM001419, LC699424, OK539479, KF543904, MN749147, OK539497, OL961528, OM001457, OM001410, OM001389, OM001388, OL638423, OK539486, MH648026, OQ791565, MN749157, MN749156, MN749153, MH648063, MH648045, MH648033, OP886969, OM001345, OK539463, OK254859). Sequences from the database were further trimmed to the region of interest (i.e., the amplicon sequence excluding the primer binding sites). For HRV genotype identification, a minimum coverage of 100 for the target region was required, and a sequence similarity of 91.30% to 100% was allowed to account for the high genetic diversity found in human rhinoviruses.
2.5. Statistical Analyses
All statistical data analysis was done in R version 4.3.2 (https://cran.r-project.org), VassarStats (http://www.vassarstats.net/), and Prism version 10 (GraphPad Software, CA, USA) was used for data visualization. We evaluated the overall percent agreement, along with Cohen's kappa score, for detecting respiratory pathogens with the BioFire‐RP2.1 and the QIAstat‐Dx‐RP2.0 multiplex panel tests. The interpretation of the kappa statistic (k) was as follows: 0.81–1.00 indicated almost perfect agreement, 0.61–0.80 good agreement, 0.41–0.60 moderate agreement, 0.21–0.40 fair agreement, 0.00–0.20 minimal agreement, and less than 0.00 signified poor agreement between the assays [24].
Positive percent agreement, negative percent agreement, overall percent agreement, and k‐score were calculated for each respiratory pathogen on the QIAstat‐Dx‐RP2.0 system, using the BioFire‐RP2.1 as the reference. This comparison could not be conducted for MERS‐CoV, HBoV, and Legionella pneumophila, as these pathogens were only covered by one of the multiplex panel tests (Table S2).
3. Results
3.1. Respiratory Pathogen Molecular Epidemiology
The study included 269 respiratory clinical specimens submitted for routine syndromic multiplex panel testing from 259 patients with RTIs (female: 115, 44.4%; children ≤ 18 years: 103, 39.8%; Table S1 and Figure S1). Median patient age was 41 years (range: 1–93 years). At least one respiratory pathogen was detected in 181 of the 269 (67.3%) respiratory clinical specimens with the BioFire‐RP2.1. In those 181 respiratory specimens, 226 viral targets and 18 bacterial targets were identified with the BioFire‐RP2.1, with viruses being the predominantly detected species in > 92% cases. Most frequently detected viral pathogens were HRV/HEV (n = 70, 29.2%), followed by SARS‐CoV‐2 (n = 42, 17.5%), RSV (n = 33, 13.8%), IV‐A (n = 21, 8.8%) and HAdV (n = 16, 6.9%), while M. pneumoniae was detected as the most common bacterial agent (n = 16, 6.7%). All other respiratory pathogens were detected in less than 5% of tested clinical specimens (Figure 1A).
Figure 1.
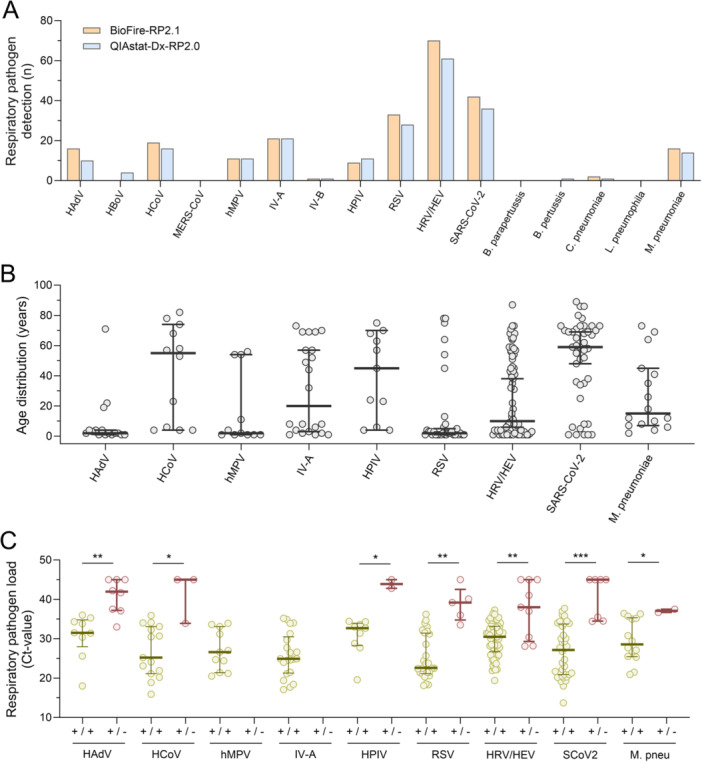
Respiratory pathogen detection by BioFire‐RP2.1 and QIAstat‐Dx‐RP2.0 multiplex panel tests. (A) Detection of respiratory pathogens in 269 respiratory samples from 259 patients using BioFire‐RP2.1 (orange) and QIAstat‐Dx‐RP2.0 (blue) between November 2023 and February 2024. (B) Age distribution of patients who tested positive for the respective respiratory pathogens (median, 25% and 75%). (C) Respiratory pathogen loads in patient samples that tested positive with both the BioFire‐RP2.1 and QIAstat‐Dx‐RP2.0 (yellow), or were detected exclusively in one of the multiplex panel tests (red; median, 25% and 75%). p values of the Mann‐Whitney U‐test, with a single asterisk (*) indicating a significance level of less than 0.05, a double asterisk (**) indicating a significance level of less than 0.01, and a triple asterisk (***) indicating a significance level of less than 0.001. HAdV, adenovirus (HAdV); HBoV, human bocavirus; HCoV, human coronavirus (types ‐229E, ‐HKU1, ‐NL63 and ‐OC43); hMPV, human metapneumovirus A and B; HPIV, human parainfluenzavirus; HRV/HEV, human rhino‐/enterovirus; IV‐A, influenzavirus A; IV‐B, influenzavirus B; M. pneu, Mycoplasma pneumoniae; MERS, Middle East respiratory syndrome coronavirus; RSV, respiratory syncytial virus A and B (RSV); SCoV2/SARS‐CoV‐2, severe respiratory syndrome coronavirus.
When comparing the epidemiology and prevalence of respiratory pathogen detections among adult and pediatric patients, we found similar detection rates among both for HRV/HEV and IV‐A. In contrast, most cases of RSV infections (78.8%, p < 0.0001) and HAdV infections (76.5%; p < 0.01) occurred in pediatric patients with a median patient age of 2 years (range: 1–78 years) and 2 years (range: 1–71 years), respectively (Figure 1B). SARS‐CoV‐2 infections (79.5%, p < 0.0001) and HCoV infections (69.2%; p < 0.01) were mainly found in adult individuals, with a median patient age of 59 years (range: 1–89 years) and 55 years (range: 4–82 years), respectively. Bacterial pathogens such as M. pneumoniae were predominantly detected in young adults, with a median patient age of 15 years (range: 2–73 years; Figure 1B).
3.2. Overall Agreement in Pathogen Detection Between the Two Respiratory Panel Tests
We compared the detection of respiratory pathogens among the BioFire‐RP2.1 and QIAstat‐Dx‐RP2.0 multiplex panel tests. Test results showed agreement in 232 out of 269 clinical specimens, with 148 (55.0%) samples testing concordant positive and 84 (31.2%) testing concordant negative. This resulted in an overall agreement of 86.3% and a Kappa coefficient of 0.71 (Table 1). Discordances were noted in 37 samples (13.8%), including 33 BioFire‐RP2.1(+)/QIAstat‐Dx‐RP2.0(−) and four BioFire‐RP2.1(−)/QIAstat‐Dx‐RP2.0(+) results. When comparing respiratory pathogen loads among clinical specimens with concordant positive test results (median cycle threshold (Ct)‐value 28; range: 14–38) and discordant ones (median Ct‐value 41; range: 28–45), significantly lower viral loads were detected in the former (p < 0.01). This could potentially explain the discordant test results at the detection limit of the QIAstat‐Dx‐RP2.0 multiplex panel test. When evaluating differences for individual respiratory pathogens, the most significant differences were found for HAdV, RSV, HRV/HEV, and SARS‐CoV‐2 (p < 0.01; Figure 1C).
Table 1.
Percent agreement and Cohen's kappa score for the comparison of BioFire‐RP2.1 and QIAstat‐Dx‐RP2.0 multiplex panel tests in 269 respiratory clinical specimens.
| QIAstat‐Dx‐RP2.0 | Agreement | Kappa | |||
|---|---|---|---|---|---|
| positive | negative | ||||
| BioFire‐RP2.1 | Positive | 148 | 33 | 86.3% | 0.71 |
| Negative | 4 | 84 | |||
3.3. Agreement for Detecting Specific Pathogens Between the Two Respiratory Panel Tests
Overall, respiratory pathogens were detected with good agreement and κ‐values between the BioFire‐RP2.1 and QIAstat‐Dx‐RP2.0 tests (Table 1). Based on these findings, we proceeded to assess whether similar patterns of detection would apply to specific respiratory pathogens. While most pathogens showed overall agreement rates over 99% between the BioFire‐RP2.1 and the QIAstat‐Dx‐RP2.0, differences were noted for the detection of HAdV (16 vs. 10; 62.5%), HRV/HEV (70 vs. 61; 87.1%), RSV (33 vs. 28; 84.8%), SARS‐CoV‐2 (42 vs. 36; 85.7%) and M. pneumoniae (16 vs. 14; 87.5%; Figure 1A, Table 2). These differences in respiratory pathogen detection led to lower overall agreement and κ‐values, resulting in lower positive percent agreement ranging from 50% to 100% for the QIAstat‐Dx‐RP2.0 multiplex panel test when compared to BioFire‐RP2.1 (Table 2). Differences in respiratory pathogen detection may be attributed to either insufficient coverage of genetic viral diversity or reduced analytical sensitivity of the QIAstat‐Dx‐RP2.0. Since the highest number of false negative test results by the QIAstat‐Dx‐RP2.0 were found for HRV (Table 2), we conducted HRV genotyping using Nanopore sequencing. The same HRV genotypes were identified in both BioFire‐RP2.1/QIAstat‐Dx‐RP2.0 concordant positive and BioFire‐RP2.1 only positive clinical specimens (Table S3). Furthermore, we retested respiratory specimens with initial discordant results using pathogen‐specific LTDs to evaluate the pathogen loads in samples missed by the QIAstat‐Dx‐RP2.0. Retesting confirmed six of nine (66.7%) positive HRV/HEV detections by BioFire‐RP2.1 with viral loads of 100, 33 700, 165 500, and 174 400 copies/mL. Additionally, Nanopore sequencing detected HRV‐specific reads in two more samples that tested negative with HRV‐specific LDT (Table S4). Furthermore, four of seven (66.7%) HAdV detections by BioFire‐RP2.1 were confirmed by LDT, although with low viral loads of 100, 100, 1200, and 1300 copies/mL. Moreover, BioFire‐RP2.1 tested an additional five samples positive for RSV, with viral loads of 700, 1300, 11 100, and 61 000 copies/mL, and two samples positive for M. pneumoniae, with loads of 1100 and 1900 copies/mL as determined by LDT (Table S4). Taken together, these findings suggests that limited analytical sensitivity of the QIAstat‐Dx‐RP2.0 rather than insufficient coverage of viral genetic diversity accounts for the observed differences in respiratory pathogen detection compared to BioFire‐RP2.1.
Table 2.
Diagnostic performance of the QIAstat‐Dx‐RP2.0 compared to the BioFire‐RP2.1 multiplex panel test.
| Viruses | Pathogen | TP | FP | TN | FN | PPA (95% CI) | NPA (95% CI) | OPA | κ–Score |
| Adenovirus | 9 | 1 | 252 | 7 | 56.3 (30.6–79.2) | 99.6 (97.4–100) | 97.0 | 0.68 | |
| Bocavirus1 | 4 | 0 | 265 | 0 | 100 (39.6–100) | 100 (98.2–100) | na | na | |
| HCoV‐229E | 2 | 0 | 267 | 0 | 100 (19.8–100) | 100 (98.3–100) | 100 | 1.00 | |
| HCoV‐HKU1 | 8 | 0 | 259 | 2 | 80.0 (44.2–96.5) | 100 (98.2–100) | 99.3 | 0.89 | |
| HCoV‐NL63 | 0 | 0 | 269 | 0 | na | 100 (98.3–100) | 100 | na | |
| HCoV‐OC43 | 6 | 0 | 262 | 1 | 85.7 (42.0–99.3) | 100 98.2–100) | 99.6 | 0.92 | |
| MERS‐CoV | 0 | 0 | 269 | 0 | na | 100 (98.3–100) | 100 | na | |
| hMPV | 11 | 0 | 258 | 0 | 100 (67.9–100) | 100 (98.2–100) | 100 | 1.00 | |
| IV‐A2 | 21 | 0 | 248 | 0 | 100 (80.8–100) | 100 (98.1–100) | 100 | 1.00 | |
| IV‐A/H1‐2009 | 19 | 0 | 250 | 0 | 100 (79.1–100) | 100 (98.1–100) | 100 | 1.00 | |
| IV‐A/H1 | 0 | 0 | 269 | 0 | na | 100 (98.3–100) | 100 | na | |
| IV‐A/H3 | 2 | 0 | 267 | 0 | 100 (19.8–100) | 100 (98.3–100) | 100 | 1.00 | |
| IV‐B | 1 | 0 | 268 | 0 | 100 (5.5–100) | 100 (98.2–100) | 100 | 1.00 | |
| HPIV‐1 | 0 | 0 | 269 | 0 | na | 100 (98.3–100) | 100 | na | |
| HPIV‐2 | 1 | 1 | 267 | 0 | 100 (5.4–100) | 99.6 (97.6–100) | 99.6 | 0.67 | |
| HPIV‐3 | 2 | 0 | 267 | 0 | 100 (39.6–100) | 100 (98.3–100) | 100 | 1.00 | |
| HPIV‐4 | 6 | 1 | 262 | 0 | 100 (51.7‐100) | 99.6 (97.6–100) | 99.6 | 0.92 | |
| RSV | 28 | 0 | 236 | 5 | 84.8 (67.3–94.3) | 100 (98.0–100) | 98.1 | 0.91 | |
| HRV/HEV | 61 | 0 | 199 | 9 | 87.1 (76.5–93.6) | 100 (98.0–100) | 96.7 | 0.91 | |
| SARS‐CoV‐2 | 36 | 1 | 226 | 6 | 85.7 (70.8–94.1) | 99.6 (97.2–100) | 97.4 | 0.90 | |
| Bacteria | B. parapertussis | 0 | 0 | 269 | 0 | na | 100 (98.3–100) | 100 | na |
| B. pertussis | 0 | 1 | 268 | 0 | na | 99.6 (98.2–100) | 99.6 | na | |
| C. pneumoniae | 1 | 0 | 267 | 1 | 50.0 (2.7–97.3) | 100 (98.2–100) | 99.6 | 0.67 | |
| L. pneumophila | 0 | 0 | 269 | 0 | na | 100 (98.3–100) | 100 | na | |
| M. pneumoniae | 14 | 0 | 253 | 2 | 87.5 (60.4–97.8) | 100 (98.1–100) | 99.3 | 0.93 |
Abbreviations: FN, false negative; FP, false positive; HAdV, human adenovirus; HCoV, human coronavirus; hMPV, human metapneumovirus A and B; HPIV, human parainfluenzavirus; HRV/HEV, human rhino‐/enterovirus; IV‐A, influenzavirus A; IV‐B, influenzavirus B; k, kappa score; MERS, Middle East respiratory syndrome coronavirus; NPA, negative percent agreement; OPA, overall percent agreement; PPA, positive percent agreement; RSV, respiratory syncytial virus A and B; TN, true negative; TP, true positive.
HBoV is not included in the BioFire‐RP2.1 multiplex panel test.
BioFire‐RP2.1 multiplex panel test is reporting influenza A subtypes.
3.4. Co‐Detection of Pathogens With the Two Respiratory Panel Tests
Next, we evaluated the diagnostic performance of the two multiplex panel tests for the co‐detection of respiratory pathogens. The BioFire‐RP2.1 identified 181 out of 269 samples (67.3%) as positive for at least one respiratory agent, including 134 (74.0%) single detections and 47 (26.0%) co‐detections. The co‐detections included 40 cases of dual and 7 cases of triple pathogen detections (Table 3). HRV/HEV was the most prevalent pathogen, identified in 29 co‐detections primarily with HAdV (n = 6), SARS‐CoV‐2 (n = 7), and RSV (n = 5). Other frequently detected pathogens were SARS‐CoV‐2 in 12 co‐detections, among others with IV‐A (n = 5) and HPIV (n = 2), and HAdV in 13 co‐detections, with RSV (n = 4) and hMPV (n = 2). IV‐A and different seasonal HCoV types were identified in 8 and 9 co‐detections, respectively, with various respiratory pathogens (Table 3). In contrast, the QIAstat‐Dx‐RP2.0 identified at least one respiratory pathogen in 152 out of 269 (56.5%) samples, resulting in 123 (80.9%) single detections and 29 (19.1%) co‐detections. The co‐detections included 26 cases of dual pathogen detections and 3 cases with three pathogens (Table 3). Taken together, both respiratory multiplex panel tests detected more than one pathogen in 51 samples (Table 3). However, the same pathogen was identified in only 24 out of 51 samples (47.1%). Moreover, the agreement rates for pathogen detections significantly decreased from 91.8% for single pathogen detections, to 65.0% for the detection of two pathogens, and 42.9% for the detection of three pathogens (Table 3; Figure 2A). When comparing the age distribution among single respiratory pathogen detections and cases of co‐detections, we found 36 of 51 co‐detections (70.6%) among pediatric patients (Figure 2B). Moreover, median patient age significantly decreased with the number of pathogens detected (p < 0.01; Figure 2B). The comparison of respiratory pathogen loads among clinical specimens with single pathogen detections (median Ct‐value 28; range: 14–38), two pathogens (median Ct‐value 31; range: 17–38) and three pathogens (median Ct‐value 32; range: 19–39), showed significantly lower viral loads for co‐detections (p < 0.01). This could potentially explain the lower detection rates of pathogens with the QIAstat‐Dx‐RP2.0 multiplex panel test in cases where multiple pathogens are present (Figure 2C).
Table 3.
Co‐detections of respiratory pathogens by BioFire‐RP2.1 and QIAstat‐Dx‐RP2.0.
| Co‐detections1 | Total | BioFire (+)/QIAstat (+) | BioFire (+)/QIAstat (−) | BioFire (−)/QIAstat (+) | |
|---|---|---|---|---|---|
| SARS‐CoV‐2 & | HRV/HEV | 3 | 3 | ||
| HRV/HEV & HBoV2 | 1 | 1 | |||
| IV‐A & RSV | 1 | 1 | |||
| HRV/HEV | 2 | 2 | |||
| IV‐A | 1 | 1 | |||
| HAdV | 1 | 1 | |||
| HPIV | 1 | 1 | |||
| IV‐A & HPIV | 1 | 1 | |||
| SARS‐CoV‐2 & | IV‐A | 2 | 2 | ||
| HCoV‐229E | 1 | 1 | |||
| HRV/HEV | 1 | 1 | |||
| IV‐A | RSV | 1 | 1 | ||
| HAdV | 1 | 1 | |||
| HPIV & HCoV | 1 | 1 | |||
| RSV & HRV/HEV & HBoV2 | 1 | 1 | |||
| HAdV | HRV/HEV | 3 | 3 | ||
| RSV | 1 | 1 | |||
| hMPV | 1 | 1 | |||
| HAdV | RSV | 2 | 2 | ||
| HRV/HEV | 2 | 2 | |||
| HRV/HEV & RSV | 1 | 1 | |||
| hMPV & C. pneumoniae 3 | 1 | 1 | 1 | ||
| HCoV | HRV/HEV | 2 | 2 | ||
| HCoV & HPIV | 1 | 1 | |||
| HPIV & HBoV2 | 1 | 1 | |||
| M. pneumoniae | 1 | 1 | |||
| HCoV | HPIV & HRV/HEV | 1 | 1 | ||
| RSV | HRV/HEV | 1 | 1 | ||
| HRV/HEV & HBoV2 | 1 | 1 | |||
| HRV/HEV & HCoV | 1 | 1 | |||
| HCoV | 1 | 1 | |||
| RSV | HRV/HEV | 1 | 1 | ||
| HRV/HEV | hMPV | 3 | 3 | ||
| M. pneumoniae | 2 | 2 | |||
| C. pneumoniae | 1 | 1 | |||
| HRV/HEV | M. pneumoniae | 1 | 1 | ||
| hMPV | 1 | 1 | |||
| M. pneumoniae & HPIV | 1 | 1 | |||
| B. pertussis | 1 | 1 | |||
| Total | 51 | 24 | 23 | 5 | |
Abbreviations: FN, false negative; FP, false positive; HAdV, human adenovirus; HCoV, human coronavirus; hMPV, human metapneumovirus A and B; HPIV, human parainfluenzavirus; HRV/HEV, human rhino‐/enterovirus; IV‐A, influenzavirus A; IV‐B, influenzavirus B; k, kappa score; MERS, Middle East respiratory syndrome coronavirus; NPV, negative predictive value; PPV, positive predictive value; RSV, respiratory syncytial virus A and B; TN, true negative; TP, true positive.
Respiratory pathogens missed by the respective other multiplex panel test are marked with an underline.
HBoV is not included in the BioFire‐RP2.1 multiplex panel test; rated as double infection in Figure 2.
BioFire‐RP2.1 multiplex panel test missed the HAdV detection, while QIAStat‐Dx‐RP2.0 missed the C. pneumoniae detection.
Figure 2.
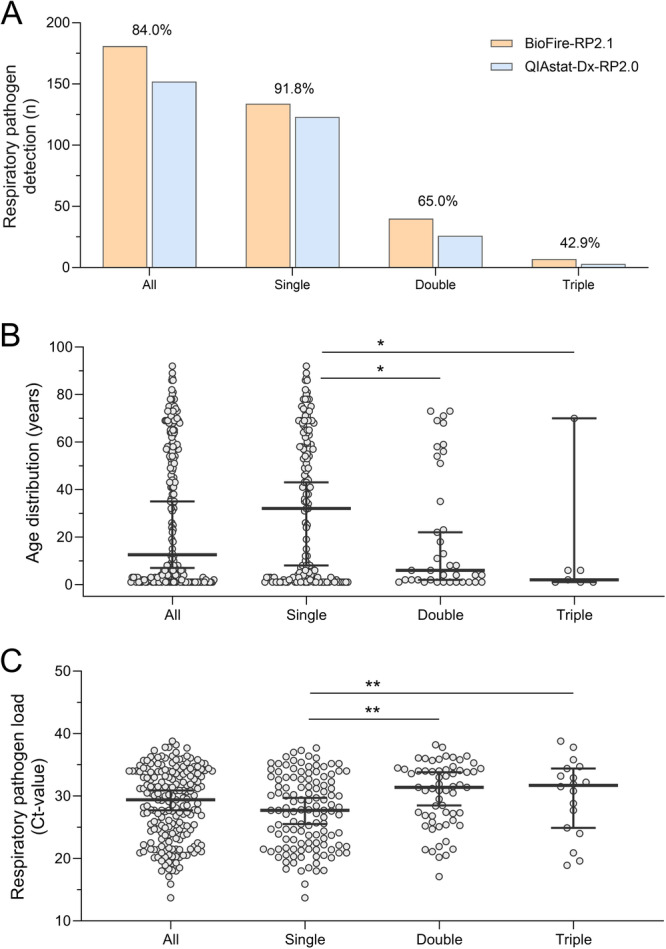
Co‐detection of respiratory pathogens by BioFire‐RP2.1 and QIAstat‐Dx‐RP2.0 multiplex panel tests. (A) Number of single detections, and the co‐detection of two or three respiratory pathogens in the same patient sample using BioFire‐RP2.1 (orange) and QIAstat‐Dx‐RP2.0 (blue). Comparison of co‐detection of respiratory pathogens by percent agreement. B. Age distribution of patients who tested positive for a single, two or three respiratory pathogens in the same patient sample (median, 25% and 75%). C. Respiratory pathogen loads in patient samples with a single detection, and the co‐detection of two or three respiratory pathogens (median, 25% and 75%). p values of the Mann‐Whitney U‐test indicate a statistically significant difference when compared to single respiratory pathogen detection. A single asterisk (*) denotes a significance level of less than 0.05, and a double asterisk (**) indicates a significance level of less than 0.01.
4. Discussion
This multicenter study prospectively evaluated the diagnostic performance characteristics of the QIAstat‐Dx‐RP2.0 compared to the BioFire‐RP2.1 in tertiary hospital settings using 269 respiratory clinical specimens collected during the 2023/2024 winter season.
When comparing the BioFire‐RP2.1 and QIAstat‐Dx‐RP2.0 multiplex panel tests, we observed an 86.3% agreement rate across all cases. Most instances of discordance in our study occurred in cases where BioFire‐RP2.1 tested positive and QIAstat‐Dx‐RP2.0 failed to detect the respective pathogen. Specifically, the QIAStat‐Dx‐RP2.0 identified 148 of the 181 (81.8%) pathogen‐positive clinical specimens detected by BioFire‐RP2.1 (Table 2). Confirmation testing of respiratory specimens with LDTs showed that discordant test results mainly occurred in samples with limited analyte, particularly those yielding positive LDT results with Ct‐values of ≥ 30. However, some cases involved higher viral loads of 11 100, and 61 000 copies/mL for RSV, and 33 700, 165 500, and 174 400 for HRV. A previous study suggested that sample inhibition could prevent the amplification of the respiratory target on the QIAstat‐Dx platform without the internal control indicating an invalid test result [11]. A review and comparison of internal control Ct‐values from discordant samples with concordant ones did not reveal a significant difference (p > 0.5). However, since respiratory targets are amplified in separate reaction chambers from the internal control in the QIAstat‐Dx system, the possibility of target amplification inhibition cannot be excluded. We furthermore assessed whether certain HRV genotypes might be missed by the QIAstat‐Dx‐RP2.0 due to incomplete coverage of viral genetic diversity by the primers and probes used in the multiplex panel test. Nanopore sequencing detected HRV genotypes A, A1, B3, and C1 in samples that tested positive in both QIAstat‐Dx‐RP2.0 and BioFire‐RP2.1, as well as in those not detected by QIAstat‐Dx‐RP2.0. This suggests that the differences in pathogen detection results between the QIAstat‐Dx‐RP2.0 and the BioFire‐RP2.1 are more likely due to lower analytical sensitivity rather than the omission of specific HRV genotypes.
Both multiplex panel tests detected viral pathogens in > 92% of assessed respiratory specimens, underscoring viruses as an important cause of RTIs in both pediatric and adult patients [25]. The predominant pathogens identified were HRV/HEV, SARS‐CoV‐2, and RSV. The pathogen‐specific results showed high positive percent agreement of 100% for the detection of HCoV‐229E, hMPV, IV‐A, IV‐B, HPIV‐2, HPIV‐3 and HPIV‐4. Lower pathogen detection rates were found for HAdV, RSV, HRV/HEV, SARS‐CoV‐2, and M. pneumoniae, resulting in low positive percent agreement ranging from 50% to 100% for the QIAstat‐Dx‐RP2.0 multiplex panel test compared to BioFire‐RP2.1. The lower detection rates were due to the QIAstat‐Dx‐RP2.0 multiplex panel test failing to detect respiratory pathogens at low viral loads, such as SARS‐CoV‐2 with 2900–5400 copies/mL. This reduced analytical sensitivity poses significant challenges for early pathogen detection, as viral loads are typically low during the initial stages of viral infection but can surge up to 10 000‐fold within a single day [26]. Thus, our data suggest that early viral infections with low viral loads may be missed by QIAstat‐Dx‐RP2.0.
Failing to detect respiratory pathogens by multiplex panel testing undermines containment strategies within hospital settings. It impedes the adequate identification of infected individuals and their proper isolation, thereby increasing transmission risks, particularly among vulnerable populations like patients with compromised immune systems and comorbid conditions. Additionally, false‐negative results can delay the initiation of appropriate medical interventions, exacerbating the risk of severe disease in children, older patients, and immunocompromised individuals. Therefore, enhancing the sensitivity of diagnostic testing is essential for accurate pathogen detection and timely administration of effective therapeutic measures, ultimately reducing hospital stays, morbidity and mortality rates [27, 28]. On the other hand, the identification of HRV does not significantly impact hygiene procedures and transmission precautions. However, it is important to note that identifying HRV, for example, could have clinical advantages. Specifically, it could lead to the discontinuation of empirically started antibiotic therapy, thereby reducing unnecessary antibiotic use and preventing antibiotic resistance development [5, 6]. Additionally, it could enable the de‐escalation of preemptive isolation strategies when another causative pathogen has been identified. These benefits underscore the importance of rapid and sensitive diagnostic methods, as they allow for more targeted and appropriate patient management decisions.
The observed differences in analytical sensitivity between the BioFire‐RP2.1 and the QIAstat‐Dx‐RP2.0 may be attributed to the former employing a nested PCR approach, which typically offers higher sensitivity, as well as potential variations in gene targets or cartridge chemistry. Both assays demonstrated high negative percent agreement, exceeding 99% for all assessed respiratory pathogens. High analytical specificity is crucial for diagnostic tests as false positive results can delay medical procedures such as surgeries and lead to unnecessary treatments and isolation.
Previous studies evaluating the diagnostic performance of the QIAstat‐Dx‐RP2.0 compared to other currently available multiplex panel testing systems include a multicenter, retrospective analysis of 287 clinical respiratory specimens using the Genmark ePlex Respiratory Pathogen Panel (ePlex RPP) [11], a single‐center, retrospective comparison of 65 clinical respiratory specimens using the ePlex RPP and BioFire‐RP2.1 [29], and a single‐center, retrospective comparison involving 137 patient nasopharyngeal swabs with the BioFire‐RP2.1 [12]. The findings from these studies align with the results of our study, demonstrating good diagnostic performance of the QIAstat‐Dx‐RP2.0, with high positive agreement rates of 92%–94% with the ePlex RPP [11, 29], and 82% with the BioFire‐RP2.1 [12]. The higher sensitivity of the BioFire‐RP2.1 for pathogen detections observed in our study has also been reported in previous comparative studies of older versions of the BioFire‐RP with the ePlex RPP [30], and QIAstat‐Dx‐RP2.0 [12, 31, 32, 33].
The findings from these studies emphasize the importance of understanding the strengths and limitations of different multiplex panel testing platforms to ensure high‐quality diagnostic testing for the diagnosis of RTIs. The QIAstat‐Dx‐RP2.0 demonstrated ease of handling by allowing direct sample transfer to a cartridge that is then loaded into the QIAstat‐Dx system. It also offers advantages in terms of noise emission, cost‐effectiveness, and environmental sustainability. Additionally, the QIAstat‐Dx‐RP2.0 provides Ct‐values as an automatic output result, offering valuable quantitative information that allows for further comparison with pathogen‐specific LDTs [18, 19]. Furthermore, respiratory pathogen loads have been reported to be prognostic for patient outcomes in hospital settings [34, 35, 36]. Therefore, pathogen quantification by QIAstat‐Dx‐RP2.0 may aid in estimating the viral or bacterial burden and potentially assess the risk of patients for more severe disease.
The QIAstat‐Dx‐RP2.0 allows for random‐access testing with a runtime of 70 min, resulting in a TAT of less than 2 h. While the BioFire‐RP2.1 offers relatively quicker results with a TAT of approximately 50 min, its workflow involves mixing buffer and sample in an injection vial before transferring to a pouch for loading. In contrast, the QIAstat‐Dx‐RP2.0 allows for direct introduction of the respiratory specimen into the cartridge, thereby reducing hands‐on time.
This study had some limitations. First, this evaluation was conducted from November 2023 to February 2024, resulting in a small data set for some respiratory targets.
Second, due to limited sample volumes, confirmatory testing was done with LDTs, with no repeat testing performed using the QIAstat‐Dx‐RP2.0 and BioFire‐RP2.1. Third, additional freezing and thawing of sample before retesting with LDTs could have degraded the nucleic acids of the pathogens in respiratory specimens, potentially leading to negative results in samples with low pathogen loads.
In conclusion, the QIAstat‐Dx‐RP2.0 assay provides a comprehensive panel for detecting twenty‐three viral and bacterial respiratory pathogens with minimal hands‐on time and a rapid turnaround. Our prospective multicenter evaluation showed good diagnostic performance of the QIAstat‐Dx‐RP2.0 assay, but lower detection rates for same respiratory targets compared to BioFire‐RP2.1. As QIAstat‐Dx‐RP2.0 offers advantages in handling, noise emission, cost effectiveness, and provides semi‐quantitative results (Ct‐values) compared to BioFire‐RP2.1, an updated version with increased analytical sensitivity would be a good alternative for the detection of respiratory pathogens using a syndromic approach.
Author Contributions
Karoline Leuzinger and Peter M. Keller designed the study. Rainer Gosert, Pascal Bittel and Roger Koller performed the laboratory analysis. Jakob Meyer, Sarah Dräger, Roland Bingisser, Christian H. Nickel, Stefano Bassetti and Sarah Tschudin Sutter reviewed and extracted data. Karoline Leuzinger, Rainer Gosert, Peter M. Keller, Pascal Bittel, Roger Koller and Alban Ramette performed the data analysis. Rainer Gosert and Karoline Leuzinger wrote the manuscript. All authors reviewed and provided feedback on the manuscript.
Ethics Statement
The study was conducted according to good laboratory practice and in accordance with the Declaration of Helsinki and national and institutional standards for laboratory quality control and was approved by the Ethical Committee of North‐Western and Central Switzerland (EKNZ 2024‐00212).
Conflicts of Interest
The authors declare no conflicts of interest.
Supporting information
Supporting information.
Supporting information.
Supporting information.
Supporting information.
Supporting information.
Acknowledgments
We kindly thank the biomedical analysts of the Clinical Virology unit, and the Clinical Bacteriology unit, University Hospital Basel, Basel, Switzerland and Alexandra Aerschmann, Institute for Infectious Diseases, University of Bern, Bern, Switzerland, for expert help and assistance.
Rainer Gosert and Roger Koller shared first authors.
Peter M. Keller and Pascal Bittel shared last authors.
Data Availability Statement
The sequencing data for human rhinovirus genotyping is available under the Sequence Read Archive Project ID PRJNA1190638. Additional data supporting the findings of this study can be obtained from the corresponding author upon reasonable request.
References
- 1. Ferkol T. and Schraufnagel D., “The Global Burden of Respiratory Disease,” Annals of the American Thoracic Society 11, no. 3 (2014): 404–406. [DOI] [PubMed] [Google Scholar]
- 2. Ison M. G. and Hirsch H. H., “Community‐Acquired Respiratory Viruses in Transplant Patients: Diversity, Impact, Unmet Clinical Needs,” Clinical Microbiology Reviews 32, no. 4 (2019): e00042, 10.1128/cmr.00042-00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nichols W. G., Peck Campbell A. J., and Boeckh M., “Respiratory Viruses Other Than Influenza Virus: Impact and Therapeutic Advances,” Clinical Microbiology Reviews 21, no. 2 (2008): 274–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ieven M., Coenen S., Loens K., et al., “Aetiology of Lower Respiratory Tract Infection in Adults in Primary Care: A Prospective Study in 11 European Countries,” Clinical Microbiology and Infection 24, no. 11 (2018): 1158–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gonzales R., Malone D. C., Maselli J. H., and Sande M. A., “Excessive Antibiotic Use for Acute Respiratory Infections in the United States,” Clinical Infectious Diseases 33, no. 6 (2001): 757–762. [DOI] [PubMed] [Google Scholar]
- 6. Kronman M. P., Zhou C., and Mangione‐Smith R., “Bacterial Prevalence and Antimicrobial Prescribing Trends for Acute Respiratory Tract Infections,” Pediatrics 134, no. 4 (2014): e956–e965. [DOI] [PubMed] [Google Scholar]
- 7. Melendez J. H., Frankel Y. M., An A. T., et al., “Real‐Time PCR Assays Compared to Culture‐Based Approaches for Identification of Aerobic Bacteria in Chronic Wounds,” Clinical Microbiology and Infection 16, no. 12 (2010): 1762–1769. [DOI] [PubMed] [Google Scholar]
- 8. Templeton K. E., Scheltinga S. A., Beersma M. F. C., Kroes A. C. M., and Claas E. C. J., “Rapid and Sensitive Method Using Multiplex Real‐Time Pcr for Diagnosis of Infections by Influenza A and Influenza B Viruses, Respiratory Syncytial Virus, and Parainfluenza Viruses 1, 2, 3, and 4,” Journal of Clinical Microbiology 42, no. 4 (2004): 1564–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Scheltinga S. A., Templeton K. E., Beersma M. F. C., and Claas E. C. J., “Diagnosis of Human Metapneumovirus and Rhinovirus in Patients With Respiratory Tract Infections by an Internally Controlled Multiplex Real‐Time Rna PCR,” Journal of Clinical Virology 33, no. 4 (2005): 306–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Templeton K. E., Scheltinga S. A., van den Eeden W. C. J. F. M., Graffelman W. A., van den Broek P. J., and Claas E. C. J., “Improved Diagnosis of the Etiology of Community‐Acquired Pneumonia With Real‐Time Polymerase Chain Reaction,” Clinical Infectious Diseases 41, no. 3 (2005): 345–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Boers S. A., Melchers W. J., Peters C. J., et al., “Multicenter Evaluation of Qiastat‐Dx Respiratory Panel V2 for Detection of Viral and Bacterial Respiratory Pathogens,” Journal of Clinical Microbiology 58, no. 6 (2020): e01793‐19, 10.1128/jcm.01793-01719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cassidy H., van Genne M., Lizarazo‐Forero E., Niesters H. G. M., and Gard L., “Evaluation of the QIAstat‐Dx RP2. 0 and the Biofire Filmarray RP2. 1 for the Rapid Detection of Respiratory Pathogens Including Sars‐CoV‐2,” Frontiers in Microbiology 13 (2022): 854209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gosert R., Naegele K., and Hirsch H. H., “Comparing the Cobas Liat Influenza A/B and Respiratory Syncytial Virus Assay With Multiplex Nucleic Acid Testing,” Journal of Medical Virology 91, no. 4 (2019): 582–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Beckmann C. and Hirsch H. H., “Comparing Luminex NxTAG‐Respiratory Pathogen Panel and RespiFinder‐22 for Multiplex Detection of Respiratory Pathogens,” Journal of Medical Virology 88, no. 8 (2016): 1319–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Beckmann C. and Hirsch H. H., “Diagnostic Performance of Near‐Patient Testing for Influenza,” Journal of Clinical Virology 67 (2015): 43–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Khanna N., Widmer A. F., Decker M., et al., “Respiratory Syncytial Virus Infection in Patients With Hematological Diseases: Single‐Center Study and Review of the Literature,” Clinical Infectious Diseases 46, no. 3 (2008): 402–412. [DOI] [PubMed] [Google Scholar]
- 17. Khanna N., Steffen I., Studt J. D., et al., “Outcome of Influenza Infections in Outpatients After Allogeneic Hematopoietic Stem Cell Transplantation,” Transplant Infectious Disease 11, no. 2 (2009): 100–105. [DOI] [PubMed] [Google Scholar]
- 18. Leuzinger K., Roloff T., Gosert R., et al., “Epidemiology of Severe Acute Respiratory Syndrome Coronavirus 2 Emergence Amidst Community‐Acquired Respiratory Viruses,” The Journal of Infectious Diseases 222, no. 8 (2020): 1270–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Leuzinger K., Gosert R., Søgaard K. K., et al., “Epidemiology and Precision of SARS‐CoV‐2 Detection Following Lockdown and Relaxation Measures,” Journal of Medical Virology 93, no. 4 (2021): 2374–2384. [DOI] [PubMed] [Google Scholar]
- 20. Linsuwanon P., Payungporn S., Samransamruajkit R., et al., “High Prevalence of Human Rhinovirus C Infection in Thai Children With Acute Lower Respiratory Tract Disease,” Journal of Infection 59, no. 2 (2009): 115–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Neuenschwander S. M., Terrazos Miani M. A., Amlang H., et al., “A Sample‐To‐Report Solution for Taxonomic Identification of Cultured Bacteria in the Clinical Setting Based on Nanopore Sequencing,” Journal of Clinical Microbiology 58, no. 6 (2020): e00060‐20, 10.1128/jcm.00060-00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Grädel C., Terrazos Miani M. A., Barbani M. T., Leib S. L., Suter‐Riniker F., and Ramette A., “Rapid and Cost‐Efficient Enterovirus Genotyping From Clinical Samples Using Flongle Flow Cells,” Genes 10, no. 9 (2019): 659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li H., “Minimap2: Pairwise Alignment for Nucleotide Sequences,” Bioinformatics 34, no. 18 (2018): 3094–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Landis J. R. and Koch G. G., “The Measurement of Observer Agreement for Categorical Data,” Biometrics 33 (1977): 159–174. [PubMed] [Google Scholar]
- 25. Ruuskanen O., Lahti E., Jennings L. C., and Murdoch D. R., “Viral Pneumonia,” The Lancet 377, no. 9773 (2011): 1264–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang S., Pan Y., Wang Q., Miao H., Brown A. N., and Rong L., “Modeling the Viral Dynamics of SARS‐CoV‐2 Infection,” Mathematical Biosciences 328 (2020): 108438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. van Rijn A. L., Nijhuis R., Bekker V., et al., “Clinical Implications of Rapid Eplex® Respiratory Pathogen Panel Testing Compared to Laboratory‐Developed Real‐Time PCR,” European Journal of Clinical Microbiology & Infectious Diseases: Official Publication of the European Society of Clinical Microbiology 37 (2018): 571–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rogers B. B., Shankar P., Jerris R. C., et al., “Impact of a Rapid Respiratory Panel Test on Patient Outcomes,” Archives of Pathology & Laboratory Medicine 139, no. 5 (2015): 636–641. [DOI] [PubMed] [Google Scholar]
- 29. Mills D. C., Huder J. B., Bloemberg G. V., and Huber M., “Comparison of Three Cartridge‐Based Platforms for Syndromic Testing for Respiratory Viruses,” Diagnostic Microbiology and Infectious Disease 109, no. 3 (2024): 116308. [DOI] [PubMed] [Google Scholar]
- 30. Babady N. E., England M. R., Jurcic Smith K. L., et al., “Multicenter Evaluation of the ePlex Respiratory Pathogen Panel for the Detection of Viral and Bacterial Respiratory Tract Pathogens in Nasopharyngeal Swabs,” Journal of Clinical Microbiology 56, no. 2 (2018): e01658‐17, 10.1128/jcm.01658-01617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Leber A. L., Lisby J. G., Hansen G., et al., “Multicenter Evaluation of the QIAstat‐Dx Respiratory Panel for Detection of Viruses and Bacteria in Nasopharyngeal Swab Specimens,” Journal of Clinical Microbiology 58, no. 5 (2020): e00155‐20, 10.1128/jcm.00155-00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Parčina M., Schneider U. V., Visseaux B., Jozić R., Hannet I., and Lisby J. G., “Multicenter Evaluation of the Qiastat Respiratory Panel—A New Rapid Highly Multiplexed PCR Based Assay for Diagnosis of Acute Respiratory Tract Infections,” PLoS One 15, no. 3 (2020): e0230183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gupta A., Soni A., Rooge S., et al., “Syndromic Approach to SARS‐CoV‐2 Detection Using QIAstat‐Dx SARS‐CoV‐2 Panel From Clinical Samples,” Journal of Virological Methods 298 (2021): 114300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Clark T. W., Ewings S., Medina M., et al., “Viral Load Is Strongly Associated With Length of Stay in Adults Hospitalised With Viral Acute Respiratory Illness,” Journal of Infection 73, no. 6 (2016): 598–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Magleby R., Westblade L. F., Trzebucki A., et al., “Impact of Severe Acute Respiratory Syndrome Coronavirus 2 Viral Load on Risk of Intubation and Mortality Among Hospitalized Patients With Coronavirus Disease 2019,” Clinical Infectious Diseases 73, no. 11 (2021): e4197–e4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pronier C., Gacouin A., Lagathu G., Le Tulzo Y., Tadié J.‐M., and Thibault V., “Respiratory Influenza Viral Load as a Marker of Poor Prognosis in Patients With Severe Symptoms,” Journal of Clinical Virology 136 (2021): 104761. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information.
Supporting information.
Supporting information.
Supporting information.
Supporting information.
Data Availability Statement
The sequencing data for human rhinovirus genotyping is available under the Sequence Read Archive Project ID PRJNA1190638. Additional data supporting the findings of this study can be obtained from the corresponding author upon reasonable request.


