Abstract
Background
Infections are common complications and causes of death during immunochemotherapy in diffuse large B-cell lymphoma (DLBCL). The gut microbiota plays a significant role in bacterial infection, but its relationship and predictive capacity with infectious complications in DLBCL are unknown.
Methods
We performed 16S rRNA gene sequencing of fecal samples collected from 41 patients with newly diagnosed DLBCL at baseline, after every two cycles of standard immunochemotherapy, during infection, and after infection recovery. Analysis of the diversity and species composition of these samples was used to evaluate the relationship between gut microbiota and bacterial infection.
Results
Our findings demonstrate the dynamic changes of Enterobacteriaceae in patients with DLBCL during immunochemotherapy. The abundance of Enterobacteriaceae was markedly higher at baseline in patients who subsequently developed bacterial infection during immunochemotherapy than in those who did not (P < 0.0001), and showed a further increase during infection (P < 0.01), after recovery from the infection, the Enterobacteriaceae was significantly decreased (P < 0.001). While there was no significant change in patients who did not develop bacterial infection. The univariate and multivariate analysis showed that baseline abundance of Enterobacteriaceae > 4.5% was independently associated with post-immunochemotherapy bacterial infection.
Conclusions
Our findings suggest that the gut microbiota signatures differ between patients with DLBCL who do and do not develop bacterial infection. The baseline abundance of Enterobacteriaceae is associated with the post-immunochemotherapy bacterial infection, and it has certain predictive value. Detecting the changes of gut microbiota can help predict the risk of bacterial infection after immunochemotherapy.
Keywords: Diffuse large B-cell lymphoma, Gut microbiota, Bacterial infection, Immunochemotherapy
Introduction
Diffuse large B-cell lymphoma (DLBCL) is the most common subtype of lymphoma, representing approximately 30%–40% of all non-Hodgkin's lymphoma [1]. Most patients are diagnosed at an advanced stage. With the emergence of new drugs, the survival of DLBCL patients has greatly improved. Combination immunochemotherapy with rituximab, cyclophosphamide, doxorubicin, vincristine, and prednisone (RCHOP) is the current standard treatment, over 60% patients being cured by this regimen [2]. However, high-dose combination immunochemotherapy can cause bone marrow suppression and immunosuppression [3, 4], making patients susceptible to infections. Randomized clinical trials have shown that the incidence of treatment-related infections with RCHOP immunochemotherapy is approximately 10%–20% [5], whereas real-world studies have reported incidences reaching 40%–60% [6, 7]. While mild infections prolong hospitalization and increase economic burden, severe infections can lead to treatment interruptions that impact treatment efficacy and even result in death. Currently, the process of diagnosing post-immunochemotherapy infections is time-consuming with low sensitivity, mainly relying on clinical symptoms, laboratory indicators, and pathogen test results. Furthermore, patients who develop and are then treated for infection may experience serious consequences, such as poor treatment effects and further disease progression. Therefore, it is necessary to strengthen the early identification and prediction of immunochemotherapy-related infections in patients with DLBCL.
A healthy gut microbiota is an important biological barrier that can prevent infection by inhibiting pathogenic species colonization [8–10]. Under normal circumstances, opportunistic pathogens do not infect hosts, but interventions such as chemotherapy, radiotherapy, and surgery can significantly disrupt the gut microecology and lead to dysbiosis of the intestinal microbiota. Such conditions allow opportunistic pathobionts to overgrow and expand, escape into the systemic circulation by translocation through damaged epithelial tissues, and induce infection [11–14]. Recent research has confirmed that the relative abundance of Enterobacteriaceae before treatment is associated with febrile neutropenia (FN) after RCHOP immunochemotherapy in patients with DLBCL [15]. Similar findings have been confirmed in patients with leukemia and those hematopoietic stem cell transplant(HSCT) patients [16–18]. For example, a low baseline Shannon diversity is associated with infection during neutropenia in patients with acute myeloid leukemia [16]. Therefore, pre-treatment characteristics of the gut microbiota could potentially serve as biomarkers to identify individuals at high risk of chemotherapy-related infections. However, most of these investigations have focused on infection during neutropenia. Considering that some infectious episodes occur in the absence of grade 4 neutropenia [19], we conducted this study to analyze the relationship between the gut microbiota and all bacterial infection during RCHOP immunochemotherapy in patients with DLBCL.
Methods
Cohort study
This study participants comprised 41 treatment-naïve, newly diagnosed patients with DLBCL undergoing RCHOP in the Second Affiliated Hospital of Nanchang University from September 2021 to July 2023. The study protocol was approved by the Institutional Ethics Committee of Second Affiliated Hospital of Nanchang University and was conducted in compliance with the Declaration of Helsinki. The inclusion criteria were as follows: 1) pathologic diagnosis of DLBCL without previous chemotherapy; and 2) absence of chronic inflammatory gastrointestinal diseases or other tumors. The exclusion criteria were as follows: 1) history of acute inflammation or infectious disease within the past month; 2) history of antibiotic use within the past month; 3) presence of autoimmune disease and long-term use of steroids or immunosuppressants; 4) history of gastrointestinal surgery; and 5) use of probiotic preparations within the previous 3 months.
The following clinical information was recorded: sex, age, immunotype, serum lactate dehydrogenase (LDH) and β−2 microglobulin levels, Eastern Cooperative Oncology Group (ECOG) performance status, body mass index (BMI), Ann Arbor stage and International Prognostic Index (IPI), and absolute neutrophil and lymphocyte counts, infectious episodes, neutropenia. The response evaluation was based on the Lugano response criteria, which include complete remission (CR), partial remission (PR), stable disease (SD), and progressive disease (PD). The follow up of each patient started at the beginning of the immunochemotherapy and ended 3 weeks after the last dose or patients occurred bacterial infection.
Patients were treated with first-line standard R-CHOP regimen, which includes rituximab (375 mg/m2, on day 0), cyclophosphamide (750 mg/m2, on day 1), doxorubicin (50 mg/m2, on day 1), vincristine (1.4 mg/m2, with a maximum dose of 2 mg, on day 1), and prednisone (100 mg, administered from day 1 to 5) every 21 days, and pegylated G-CSF was administered prophylactically every cycle. None of the patients received prophylactic antibiotic therapy prior to the initiation of immunochemotherapy.
Neutropenia is defined as an absolute neutrophil count < 500 cells/mm3 or that is expected to decrease to < 500 cells/mm3 during the next 48 h [20].
Bacterial infections were determined when microbiological evidence was present or clinically diagnosed [21]. Microbiologically defined bacterial infection was diagnosed when there are signs and symptoms of infection with microbiologic culture confirmation. Clinically defined bacterial infection was diagnosed when symptoms (e.g. fever, cough, purulent sputum, acute diarrhea, abdominal pain, pyuria, dysuria), signs (e.g. pulmonary rales, abdominal tenderness, tenderness in lumbosacral region), clinical test (e.g. increased peripheral blood leukocytes and neutrophils, elevated C-reactive protein and procalcitonin, increased white blood cells in urine) and medical imaging (e.g. lobar consolidation, multiple patchy consolidations, pleural effusion, localized lesion exudation, intestinal edema) of infection were evident.
Stool sample collection and DNA extraction
A total of 41 patients were enrolled in the study. Stool specimens were obtained for microbiota analysis at baseline, after every two cycles of standard immunochemotherapy, during infection and after infection recovery. And all samples were frozen at − 80 °C until DNA extraction.
Fecal DNA isolation
Fecal samples were weighed and total DNA was extracted with the DP328 Fecal Genome Extraction Kit (Tiangen Biotech) according to the manufacturer's instructions. DNA concentration and purity were measured with Nanodrop 2000.
-
2.
16S rRNA gene sequencing
The 16S rRNA gene regions V3-V4 was amplified with universal primers (Invitrogen, Carlsbad, CA, USA) 341F and 806R using the PCR reactions. The libraries for sequencing were prepared using the NEBNext® Ultra™ DNA Library Prep Kit designed for Illumina® sequencing (New England Biolabs, USA). The prepared DNA libraries was sequenced on an Illumina Nova6000 platform, generating paired-end reads with a length of 250 bp paired-end reads [22].
-
3.
Sequencing data processing
Paired-end raw reads quality control
Fastp (version 0.14.1, https://github.com/OpenGene/fastp) was used to control the quality of the Raw Data by sliding window (-W 4 -M 20). The primers were removed by using cutadapt software (https://github.com/marcelm/cutadapt/) according to the primer information at the beginning and end of the sequence to obtain the paired-end clean reads.
Paired-end clean reads assembly
Paired-end clean reads were merged using usearch-fastq_mergepairs (V10, http://www.drive5.com/usearch/)according to the relationship of the overlap between the paired-end reads, when at least 16 bp overlap the read generated from the opposite end of the same DNA fragment, the maximum mismatch allowed in overlap region was 5 bp, and the spliced sequences were called Raw Tags.
Raw tags quality control
Fastp (version 0.14.1, https://github.com/OpenGene/fastp) was used to control the quality of the raw Data by sliding window (-W 4 -M 20) to obtain the paired-end clean tags.
-
4.
OTU cluster and Species annotation
The usearch software (Version 10, http://www.drive5.com/usearch/) was employed to perform sequence analysis. Sequences that demonstrated a similarity of 97% or above were categorized into the same OTU. The representative sequence of each OTU was the most recurrent sequence, and was screened for subsequent annotation.
Statistical analysis
Differences between two groups were assessed using Student’s t-test or the Mann–Whitney U test. One-way analysis of variance (ANOVA) was used for multi-group (more than two groups) datasets. A receiver operating characteristic (ROC) curve was generated and the corresponding area under the curve (AUC) value was calculated for baseline Enterobacteriaceae abundance as a predictor of infection during immunochemotherapy. Kaplan–Meier curves were used to visualize time to infection based on “high” and “low” abundance, and were compared using log-rank tests. Cox proportional hazard models were used to investigate the univariate and multivariate analyses of the association between infection and gut microbiota data. P values < 0.05 were considered to indicate statistical significance. SPSS 27.0 and GraphPad Prism 9.0 software were used for statistical analysis.
Results
Patient characteristics and outcomes
The clinical characteristics of the 41 study patients are presented in Table 1. There were 23 male patients (56.0%) and 19 patients (46%) who were > 60 years of age. According to the Hans classification, 24 patients (59%) had the non-germinal center B cell (non-GCB) subtype of DLBCL. At the time of initial treatment, 18 patients (44%) had an ECOG score < 2, 16 patients (39%) were in Ann Arbor stages I–II, and 15 (37%) and 10 (24%) patients had low-risk and high-risk IPI scores, respectively. Thirteen patients (32%) had elevated LDH levels at initial treatment. There were 28 patients (68%) with a reduced absolute lymphocyte count, and three patients (7%) with a reduced absolute neutrophil count. Nineteen patients (46%) experienced at least one bacterial infection during the RCHOP immunochemotherapy period, 53% of which occurred in the absence of grade 4 neutropenia. The overall effectiveness of RCHOP immunochemotherapy was 83%: 26 patients (63%) achieved CR, eight patients (20%) had PR, six patients (15%) had PD, and one patient was not evaluated for treatment efficacy (Table 1).
Table 1.
Patients’ characteristics
| N = 41 | ||
|---|---|---|
| number (percent) | ||
| Baseline characteristics | ||
| Age, years | ≤ 60 | 22 (54%) |
| > 60 | 19 (46%) | |
| Sex | Male | 23 (56%) |
| Female | 18 (44%) | |
| ECOG PS | < 2 | 18 (44%) |
| ≥ 2 | 23 (56%) | |
| BMI, Kg/m2 | < 18.5 | 3 (7%) |
| 18.5–24.9 | 24 (59%) | |
| ≥ 25 | 14 (34%) | |
| Serum LDH | Normal | 28 (68%) |
| Elevated | 13 (32%) | |
| β2-microglobulin | Normal | 20 (49%) |
| Elevated | 21 (51%) | |
| Cell of origin | GCB | 17 (41%) |
| Non-GCB | 24 (59%) | |
| Stage | I-II | 16 (39%) |
| III-IV | 25 (61%) | |
| IPI | Low | 15 (37%) |
| Low-Intermediate | 10 (24%) | |
| High-Intermediate | 6 (15%) | |
| High | 10 (24%) | |
| Baseline neutrophil count | Normal | 38 (93%) |
| Reduced | 3 (7%) | |
| Baseline lymphocyte count | Normal | 13 (32%) |
| Reduced | 28 (68%) | |
| Patients with infections | Totol | 19 (46%) |
| Grade 4 neutropenia | 9 (47%) | |
| Without grade 4 neutropenia | 10 (53%) | |
| Treatment outcome after chemotherapy | ||
| Complete response | 26 (63%) | |
| Partial response | 8 (20%) | |
| Progression | 6 (15%) | |
| Not evaluated | 1 (2%) | |
Abbreviations: BMI body mass index, ECOG PS Eastern Cooperative Oncology Group Performance Status, GCB germinal center B cell, IPI International Prognostic Index
Baseline abundance of Enterobacteriaceae increased in patients who developed bacterial infection
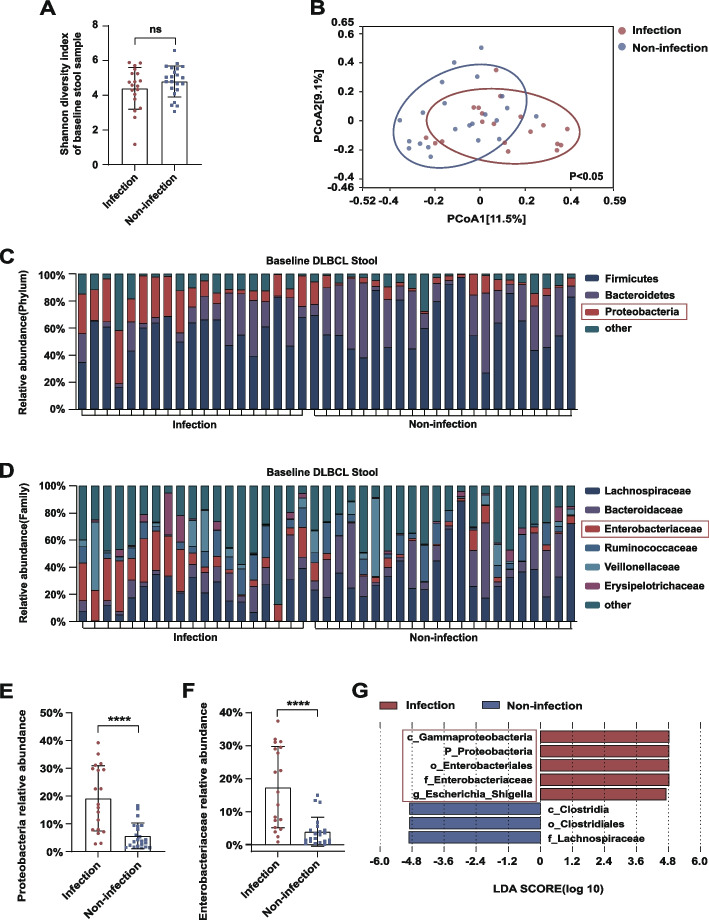
The 16S rRNA gene sequencing analysis showed a lower baseline level of alpha diversity of the gut microbiota in patients who developed bacterial infection than in those who did not; however, this difference did not reach statistical significance (P = 0.233; Fig. 1A). By contrast, there was a significant difference in beta diversity between the two groups (P < 0.05; Fig. 1B). In the taxonomic comparison of gut microbial composition at the phylum level, the baseline abundance of Proteobacteria was found to be significantly higher in the post-immunochemotherapy infection group than in the non-infection group (P < 0.0001; Fig. 1C, E). At the family level, patients who developed bacterial infection had a greater abundance of Enterobacteriaceae (P < 0.0001; Fig. 1D, F). Furthermore, linear discriminant analysis (LDA) effect size (LEfSe) analysis showed that, compared with those in the non-infection group, c_Gammaproteobacteria, p_Proteobacteria, o_Enterobacteriales, and Enterobacteriaceae with g_Escherichia_Shigella were higher in relative abundance in the infection group when the cutoff for the LDA score was set to 4.5 (Fig. 1G). These results suggested that the baseline abundance of Enterobacteriaceae differed between patients with DLBCL who did and did not develop bacterial infection.
Fig. 1.
A, B Shannon index of alpha diversity and Bray–Curtis index of beta diversity in baseline fecal samples of patients who did and did not develop bacterial infection during RCHOP. C, D Baseline relative abundance of bacteria at the phylum and family levels in stool samples of patients who did and did not develop bacterial infection during RCHOP. E, F Comparisons of Proteobacteria and Enterobacteriaceae abundance between patients who did and did not develop bacterial infection during RCHOP. G LEfSe analysis showing the higher baseline abundance of Proteobacteria and Enterobacteriaceae in patients who developed bacterial infection during RCHOP (LDA score cutoff: 4.5). Asterisks indicate significant differences identified using the two-tailed Mann–Whitney U test. *P < 0.05, **P < 0.01, ***P < 0.001. ns = not significant
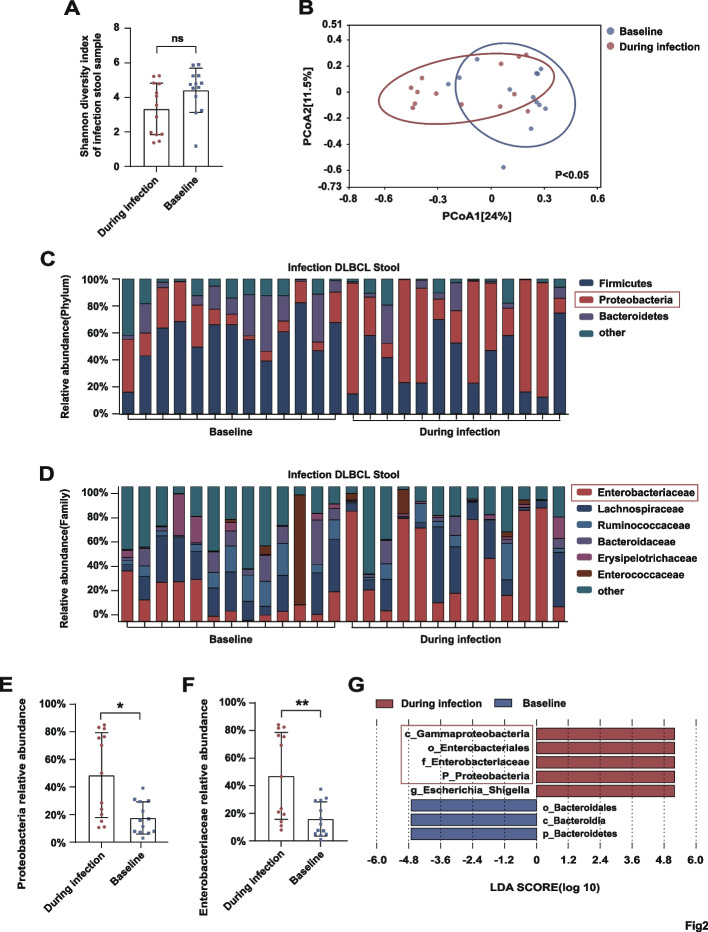
Abundance of Enterobacteriaceae further increased above baseline during infection
Although the alpha diversity of gut microbes in the infection group did not differ significantly between the baseline and during-infection collection points (P = 0.061; Fig. 2A), the beta diversity was lower during infection than at baseline (P < 0.05; Fig. 2B). At the phylum level, the abundance of Proteobacteria exhibited a further significant increase between baseline and during infection (P < 0.05; Fig. 2C, E). At the family level, patients had a greater abundance of Enterobacteriaceae during infection than at baseline (P < 0.01; Fig. 2D, F). LEfSe analysis identified higher abundances of c_Gammaproteobacteria, p_Proteobacteria, o_Enterobacteriales, and f_Enterobacteriaceae with g_Escherichia_Shigella in patients during infection compared with those at baseline when the LDA score cutoff was set to 4.5 (Fig. 2G).
Fig. 2.
A, B Shannon index of alpha diversity and Bray–Curtis index of beta diversity in fecal samples collected at baseline and during infection. C, D Relative abundance of bacteria at the phylum and family levels in stool samples collected at baseline and during infection. E, F Comparisons of Proteobacteria and Enterobacteriaceae abundance at baseline and during infection. G LEfSe analysis showing the higher abundance of Proteobacteria and Enterobacteriaceae during infection (LDA score cutoff: 4.5). Asterisks indicate significant differences identified using the two-tailed Mann–Whitney U test. *P < 0.05, **P < 0.01, ***P < 0.001. ns = not significant
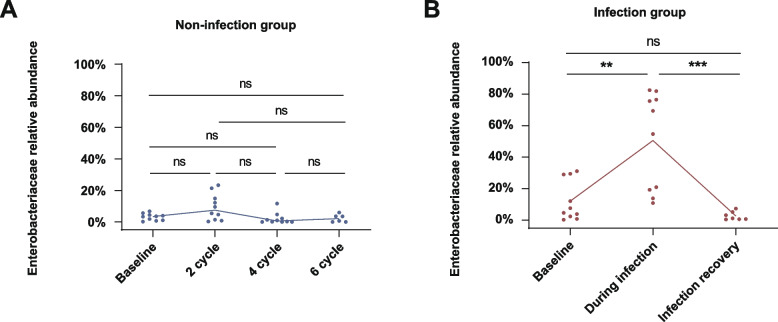
To further investigate the trend in Enterobacteriaceae abundance during immunochemotherapy, we analyzed fecal samples that had been collected at baseline, and after 2, 4, and 6 cycles of immunochemotherapy from 10 patients who did not develop bacterial infection. The results showed no significant change in the abundance of Enterobacteriaceae in these patients during RCHOP immunochemotherapy (Fig. 3A). Additionally, we analyzed fecal samples collected at baseline, during infection, and after infection recovery from 10 patients who developed bacterial infections. In these patients, the abundance of Enterobacteriaceae significantly increased between the baseline and infection periods, then significantly decreased following anti-infection treatment and recovery from the infection (Fig. 3B). These findings demonstrated that the gut microbiota of patients with DLBCL had a greater abundance of Enterobacteriaceae during infection compared with that at baseline and compared with that in DLBCL patients who did not develop bacterial infection on RCHOP.
Fig. 3.
A Change in Enterobacteriaceae abundance in fecal samples collected from 10 patients without bacterial infection during RCHOP for DLBCL at four time points: baseline, and after two cycles, four cycles, and six cycles. B Change in Enterobacteriaceae abundance in fecal samples collected from 10 patients who contracted a bacterial infection during RCHOP for DLBCL at three time points: baseline, during infection, and after infection recovery. Significant differences were identified using the two-tailed Mann–Whitney U test, Student’s t-test, or one-way ANOVA; *P < 0.05, **P < 0.01, ***P < 0.001. ns = not significant
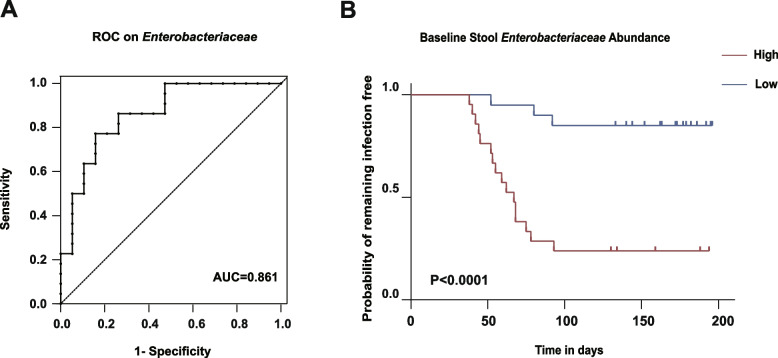
Predictive value of baseline Enterobacteriaceae abundance for bacterial infection
The finding of higher baseline Enterobacteriaceae abundance in stool samples from patients that developed bacterial infection during immuno chemotherapy led us to further examine the relationship between baseline Enterobacteriaceae abundance and post-immunochemotherapy bacterial infection. An ROC curve showed the strong predictive ability of baseline Enterobacteriaceae abundance for post-immunochemotherapy bacterial infection in DLBCL patients (AUC = 0.861; 95% confidence interval [CI]: 0.747–0.975; P < 0.0001). An Enterobacteriaceae abundance threshold of 4.5% exhibited a sensitivity of 84% and a specificity of 77% (Fig. 4A). The patient cohort was divided into high and low Enterobacteriaceae abundance groups based on the threshold value derived from ROC curve analysis. The Kaplan–Meier curve showed that the group with high baseline Enterobacteriaceae abundance had a significantly higher rate of bacterial infection compared with the low-abundance group (P < 0.0001; Fig. 4B).
Fig. 4.
A ROC curve of baseline Enterobacteriaceae abundance. B Kaplan‒Meier (KM) plot illustrating the difference in infection outcome between two groups stratified by low abundance (blue) and high abundance (red) of Enterobacteriaceae (abundance threshold: 4.5%)
To identify other baseline clinical indicators that might be predictive of the occurrence of bacterial infection, we used univariate and multivariate Cox proportional hazards models. In addition to the baseline abundance of Enterobacteriaceae, we incorporated age, sex, ECOG score, BMI, IPI and lymphocyte count. The univariate and multivariate Cox analyses showed that baseline Enterobacteriaceae abundance (hazard ratio: 11.307; 95% CI: 2.983–42.861; P < 0.001) was independently associated with the risk of post-immunochemotherapy bacterial infection (Table 2).
Table 2.
Univariate and multivariate analyses of infection complication
| Univariate | Multivariate | |||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P-value | HR | 95% CI | P-value | |
| Female | 1.327 | 0.538–3.271 | 0.539 | 0.748 | 0.271–2.064 | 0.575 |
| Age > 60 | 0.928 | 0.334–2.579 | 0.886 | 0.816 | 0.240–2.776 | 0.745 |
| ECOG ≥ 2 | 0.822 | 0.334–2.027 | 0.671 | 1.320 | 0.435–4.006 | 0.624 |
| BMI | 0.983 | 0.867–1.113 | 0.784 | 1.016 | 0.880–1.172 | 0.831 |
| IPI > 2 | 0.985 | 0.388–2.505 | 0.975 | 1.134 | 0.373–3.444 | 0.825 |
| Lymphocyte | 1.167 | 0.519–2.622 | 0.709 | 1.510 | 0.576–3.961 | 0.402 |
| Enterobacteriaceae > 4.5% | 9.015 | 2.591–31.359 | < .001 | 11.307 | 2.983–42.861 | < .001 |
Abbreviations: ECOG Eastern Cooperative Oncology Group Performance Status, BMI body mass index, IPI International Prognostic Index, HR hazard ratio, CI confidence interval
Discussion
The current RCHOP regimen has become the standard first-line treatment for DLBCL. However, while approximately 60% of patients are cured [2], infection remains a non-negligible complication and cause of death during immunochemotherapy [6, 7]. In this study, we used 16S rRNA gene sequence analysis of fecal samples to investigate the relationship between baseline characteristics of the gut microbiota and bacterial infection after RCHOP in 41 patients with DLBCL. We found that patients who developed bacterial infection after immunochemotherapy had a higher baseline abundance of Enterobacteriaceae compared with those who did not, and showed a further increase during infection. The baseline Enterobacteriaceae abundance > 4.5% was independently associated with bacterial infection and, after including potential clinical factors [3, 19], it remained an independent risk factor. Previous studies on DLBCL and gut microbiota have focused on disease characteristics and treatment outcomes, there are fewer studies on infectious complications. Currently, the means to predict infectious complications during immunochemotherapy before treatment are extremely limited. Our study for the first time demonstrates the trend of changes in Enterobacteriaceae in patients with DLBCL during immunochemotherapy by continuously collecting fecal samples, reveals its difference between infection and non-infection group. These findings show that baseline Enterobacteriaceae may have important predictive value for subsequent bacterial infection. Physicians treating DLBCL cases with high baseline abundance of Enterobacteriaceae may need to pay more attention to the risk of bacterial infection and take corresponding measures, as necessary.
The typical causative pathogens of infections in patients with malignant tumors mainly originate from the gut, such as Escherichia coli, Klebsiella, Enterococcus, Pseudomonas aeruginosa [23–25], most of these opportunistic pathogens belong to the Enterobacteriaceae. Mancini et al. found that the abundance of Enterobacteriaceae before transplantation is an independent risk factor for sepsis after allogeneic hematopoietic stem cell transplantation (allo-HSCT) [26], which is consistent with our findings. Another study discovered that the baseline abundance of Proteobacteria in the gut could predict FN after chemotherapy in children with acute lymphoblastic leukemia [27]. Recently, studies have also found that the baseline abundance of Enterobacteriaceae was related to FN after RCHOP in patients with DLBCL [15]. In line with the findings of previous report, our study found that the baseline abundances of Proteobacteria and Enterobacteriaceae were significantly increased in patients who later developed bacterial infection during immunochemotherapy. What’s more, our results have shown the dynamic changes of Enterobacteriaceae in patients who do or do not develop bacterial infection. And these studies mainly focused on infection during neutropenia or sepsis. However, more than half of the infectious episodes in our cohort occurred without grade 4 neutropenia, which has also been reported in the literature [19]. This implies that patients who do not develop innate immunity deficiencies through chemotherapy may still experience severe bacterial infection, and these patients may have been overlooked in scientific literature. Our findings show that the Enterobacteriaceae still has predictive value for those patients.
Previous studies have shown before the occurrence of an infection, a corresponding microbial community already dominates in the gut. Taur et al. studied the relationship between the gut microbiota and bacteremia during allo-HSCT and found that patients with a predominance of Enterococcus in the gut had a nine-fold increased risk of vancomycin-resistant Enterococcus bacteremia, and those with a predominance of Enterobacteriaceae had a five-fold increased risk of Gram-negative bacilli bacteremia, compared with other patients [28]. And there is another research discovered that colonization by Bacteroidetes, Lachnospiraceae, and Ruminococcaceae bacteria before transplantation has a protective effect against Clostridium difficile infection after allo-HSCT [29]. And it was also found that patients with a higher abundance of fecal butyrate-producing bacteria after allo-HSCT had a five-fold lower risk of respiratory viral infections compared with other patients [30]. In recent years, increasingly more evidence suggests that supplementation with probiotics may help prevent infections [31–34]. For example, both in vivo and in vitro experimental studies have shown that certain probiotics have a protective effect against C. difficile infections, such as yeasts, Bifidobacteria, and Lactobacillus [31]. Research by Piewngam et al. showed that Bacillus species can inhibit colonization by S. aureus in the human gut and nasal passages, and these results were validated in mouse models [32]. These studies reveal the efficacy of using probiotics for pathogen decolonization and suggest the potential for probiotics to prevent pathogen colonization. Whether probiotics can be safely used to prevent post-immunochemotherapy infections requires further in-depth research.
However, our study has some limitations. First, not all patients provided stool samples at all points, and it did not adjust for additional factors such as diet, BMI, age that might affect changes in gut microbiota. Secondly, its single-center design and a larger sample size is needed to confirm the current results. Thirdly, dynamic changes of the gut microbiota during treatment may impact the risk of acquiring infection. Previously, we have shown in mouse models that fluorouracil induces differential regulatory effects on the gut microbiota across individual mouse. Fluorouracil induced an increase and translocation of pathogenic bacteria in the intestines of in certain mice, leading to infection and mortality. Conversely, other mice did not exhibit a rise in intestinal pathogenic bacteria nor significant infection-related mortality [35]. In line with our mouse experiment findings, in the current study, we observed that in patients who did not develop bacterial infection, the abundance of Enterobacteriaceae did not significantly change post-immunochemotherapy. However, insufficient number of samples were available for an investigation on whether immunochemotherapy induced change in gut microbiota would impact on the risk of bacterial infection in the current study. In future, larger investigation into the differential responses and dynamic changes of individual gut microbiota to immunochemotherapy and its correlation with infection incidence will be of significant interest.
In summary, our study has revealed the predictive capability of the baseline abundance of Enterobacteriaceae for post-immunochemotherapy bacterial infections in DLBCL, and it may be a biomarker. Although we did not address the effect of chemotherapy on the change of the gut microbiota. we think further investigation into the dynamic alterations of the gut microbiota during chemotherapy is imperative, as it facilitates a deeper understanding of the processes and mechanisms underlying bacterial infection development. The influence of chemotherapy on the microbiome and its subsequent impact on bacterial infections represents a promising and valuable avenue for future research. We are engaged in further studies, with the aim of reducing the rate of post-immunochemotherapy infectious complications and thereby improving the prognosis for patients with DLBCL.
Acknowledgements
This research was supported by the Jiangxi Provincial Nature Science Foundation (20204BCJ22030 and 20204BCJ22027), and the National Natural Science Fund of China (NSFC-82160029).
Authors’ contributions
MS and DT collected samples and analyzed the majority of data. JJ, YW, CY, and RQ helped with sample collection and data recording. HW and ST conceived and designed the study. The manuscript was written by MS and ST and commented on by all other authors.
Data availability
No datasets were generated or analysed during the current study.
Declarations
Ethics approval and consent to participate
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the Declaration of Helsinki. The study protocol was approved by the Institutional Ethics Committee of Second Affiliated Hospital of Nanchang University (Review [2023] NO.098), Nanchang, China. All methods were carried out in accordance with approved rules and regulations. Informed consent was waived because of the retrospective nature of this study. No personal patient information was disclosed.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Man Sun and Duozhuang Tang contributed equally to this work.
Contributor Information
Hua Wang, Email: ndefy19002@ncu.edu.cn.
Si Tao, Email: ndefy11188@ncu.edu.cn.
References
- 1.Li S, Young KH, Medeiros LJ. Diffuse large B-cell lymphoma. Pathology. 2018;50:74–87. 10.1016/j.pathol.2017.09.006. [DOI] [PubMed] [Google Scholar]
- 2.Liu W, Ji X, Song Y, et al. Improving survival of 3760 patients with lymphoma: Experience of an academic center over two decades. Cancer Med. 2020;9:3765–74. 10.1002/cam4.3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Choi YW, Jeong SH, Ahn MS, et al. Patterns of Neutropenia and risk factors for febrile neutropenia of diffuse large B-cell lymphoma patients treated with rituximab-CHOP. J Korean Med Sci. 2014;29:1493. 10.3346/jkms.2014.29.11.1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van der Kolk LE, Baars JW, Prins MH, van Oers MHJ. Rituximab treatment results in impaired secondary humoral immune responsivenes s. Blood. 2002;100:2257–9. [PubMed] [Google Scholar]
- 5.Ohmoto A, Fuji S. Infection profiles of different chemotherapy regimens and the clinical feasibility of antimicrobial prophylaxis in patients with DLBCL. Blood Rev. 2021;46:100738. 10.1016/j.blre.2020.100738. [DOI] [PubMed] [Google Scholar]
- 6.Clausen MR, Ulrichsen SP, Juul MB, et al. Prognostic significance of infectious episodes occurring during first-line therapy for diffuse large B-cell lymphoma - A nationwide cohort study. Hematol Oncol. 2020;38:318–25. 10.1002/hon.2734. [DOI] [PubMed] [Google Scholar]
- 7.Dendle C, Gilbertson M, Spelman T, et al. Infection is an independent predictor of death in diffuse large B Cell lymphoma. Sci Rep. 2017;7:4395. 10.1038/s41598-017-04495-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pickard JM, Zeng MY, Caruso R, Núñez G. Gut microbiota: Role in pathogen colonization, immune responses, and inflammatory disease. Immunol Rev. 2017;279:70–89. 10.1111/imr.12567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bäumler AJ, Sperandio V. Interactions between the microbiota and pathogenic bacteria in the gut. Nature. 2016;535:85–93. 10.4049/jimmunol.1403169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sassone-Corsi M, Raffatellu M. No vacancy: how beneficial microbes cooperate with immunity to provide colonization resistance to pathogens. J Immunol. 2015;194:4081–7. 10.4049/jimmunol.1403169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galloway-Peña J, Brumlow C, Shelburne S. Impact of the microbiota on bacterial infections during cancer treatment. Trends Microbiol. 2017;25:992–1004. 10.1016/j.tim.2017.06.006. [DOI] [PubMed] [Google Scholar]
- 12.Finegold SM, Sutter VL, Boyle JD, Shimada K. The Normal Flora of Ileostomy and transverse colostomy effluents. J Infect Dis. 1970;122:376–81. 10.1093/infdis/122.5.376. [DOI] [PubMed] [Google Scholar]
- 13.Lu Q, Liang Y, Tian S, et al. Radiation-Induced Intestinal Injury: Injury Mechanism and Potential Treatment Strategies. Toxics. 2023;11:1011. 10.3390/toxics11121011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Vliet MJ, Harmsen HJM, de Bont ESJM, Tissing WJE. The role of intestinal microbiota in the development and severity of chemotherapy-induced mucositis. PLoS Pathog. 2010;6:e1000879. 10.1371/journal.ppat.1000879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoon SE, Kang W, Choi S, et al. The influence of microbial dysbiosis on immunochemotherapy-related efficacy and safety in diffuse large B-cell lymphoma. Blood blood. 2023;2022:018831. 10.1182/blood.2022018831. [DOI] [PubMed] [Google Scholar]
- 16.Galloway-Peña JR, Shi Y, Peterson CB, et al. Gut microbiome signatures are predictive of infectious risk following induction therapy for acute myeloid leukemia. Clin Infect Dis. 2020;71:63–71. 10.1093/cid/ciz777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galloway-Peña JR, Smith DP, Sahasrabhojane P, et al. The role of the gastrointestinal microbiome in infectious complications during induction chemotherapy for acute myeloid leukemia. Cancer. 2016;122:2186–96. 10.1002/cncr.30039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Montassier E, Al-Ghalith GA, Ward T, et al. Pretreatment gut microbiome predicts chemotherapy-related bloodstream infection. Genome Med. 2016;8:49. 10.1186/s13073-016-0301-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alonso JJ, Cánovas A, Barreiro JG, Aguirre C. Infectious complications of chemotherapy in clinically aggressive mature B and T cell lymphomas. Eur J Intern Med. 2012;23:255–60. 10.1016/j.ejim.2011.11.018. [DOI] [PubMed] [Google Scholar]
- 20.Freifeld AG, Bow EJ, Sepkowitz KA, et al. Clinical practice guideline for the use of antimicrobial agents in neutropenic patients with cancer: 2010 update by the infectious diseases Society of America. Clin Infect Dis. 2011;52:427–31. 10.1093/cid/ciq147. [DOI] [PubMed] [Google Scholar]
- 21.Pizzo PA, Armstrong D, Bodey G, et al. From the immunocompromised host society: the design, analysis, and reporting of clinical trials on the empirical antibiotic management of the neutropenic patient: report of a consensus panel. J Infect Dis. 1990;161:397–401. 10.1093/infdis/161.3.397. [DOI] [PubMed] [Google Scholar]
- 22.Tang D, Zeng T, Wang Y, et al. Dietary restriction increases protective gut bacteria to rescue lethal methotrexate-induced intestinal toxicity. Gut Microbes. 2020;12:1714401. 10.1080/19490976.2020.1714401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Velasco E, Byington R, Martins C, a. S, et al. Comparative study of clinical characteristics of neutropenic and non-neutropenic adult cancer patients with bloodstream infections. Eur J Clin Microbiol Infect Dis. 2006;25:1–7. 10.1007/s10096-005-0077-8. [DOI] [PubMed] [Google Scholar]
- 24.Lin L, Jia L, Fu Y, et al. A comparative analysis of infection in patients with malignant cancer: A clinical pharmacist consultation study. J Infect Public Health. 2019;12:789–93. 10.1016/j.jiph.2019.03.021. [DOI] [PubMed] [Google Scholar]
- 25.Yao D, Cao W, Qing Z. Analysis of pathogen distribution and drug resistance of nosocomial infections accompanied in patients with malignant tumor. Chin J Cancer Res. 2008;20:155–8. 10.1007/s11670-008-0155-4 [Google Scholar]
- 26.Mancini N, Greco R, Pasciuta R, et al. Enteric microbiome markers as early predictors of clinical outcome in allogeneic hematopoietic stem cell transplant: results of a prospective study in adult patients. Open Forum Infect Dis. 2017;4:ofx215. 10.1093/ofid/ofx215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hakim H, Dallas R, Wolf J, et al. Gut microbiome composition predicts infection risk during chemotherapy in children with acute lymphoblastic leukemia. Clin Infect Dis. 2018;67:541–8. 10.1093/cid/ciy153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taur Y, Xavier JB, Lipuma L, et al. Intestinal domination and the risk of bacteremia in patients undergoing allogeneic hematopoietic stem cell transplantation. Clin Infect Dis. 2012;55:905–14. 10.1093/cid/cis580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee YJ, Arguello ES, Jenq RR, et al. Protective factors in the intestinal microbiome against Clostridium difficile infection in recipients of allogeneic hematopoietic stem cell transplantation. J Infect Dis. 2017;215:1117–23. 10.1093/infdis/jix011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haak BW, Littmann ER, Chaubard J-L, et al. Impact of gut colonization with butyrate-producing microbiota on respiratory viral infection following allo-HCT. Blood. 2018;131:2978–86. 10.1182/blood-2018-01-828996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wei Y, Yang F, Wu Q, et al. Protective effects of bifidobacterial strains against toxigenic clostridium difficile. Front Microbiol. 2018;9:888. 10.3389/fmicb.2018.00888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Piewngam P, Zheng Y, Nguyen TH, et al. Pathogen elimination by probiotic Bacillus via signalling interference. Nature. 2018;562:532–7. 10.1038/s41586-018-0616-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boonma P, Spinler JK, Venable SF, et al. Lactobacillus rhamnosus L34 and Lactobacillus casei L39 suppress Clostridium difficile-induced IL-8 production by colonic epithelial cells. BMC Microbiol. 2014;14:177. 10.1186/1471-2180-14-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Geeraerts S, Ducatelle R, Haesebrouck F, Van Immerseel F. Bacillus amyloliquefaciens as prophylactic treatment for Clostridium difficile-associated disease in a mouse model. J Gastroenterol Hepatol. 2015;30:1275–80. 10.1111/jgh.12957. [DOI] [PubMed] [Google Scholar]
- 35.Tang D, Qiu R, Qiu X, et al. Dietary restriction rescues 5-fluorouracil -induced lethal intestinal toxicity in old mice by blocking translocation of opportunistic pathogens. Gut Microbes. 2024;16(1):2355693. 10.1080/19490976.2024.2355693. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No datasets were generated or analysed during the current study.