Abstract
Background
Epidemiologic researches show that short sleep duration may affect feeding behaviors resulting in higher energy intake and increased risk of obesity, but the further mechanisms that can interpret the causality remain unclear. The circadian rhythm is fine-tuned by the suprachiasmatic nucleus (SCN) as the master clock, which is essential for driving rhythms in food intake and energy metabolism through neuronal projections to the arcuate nucleus (ARC) and paraventricular nucleus (PVN).
Results
We showed that chronic SD-induced aberrant expressions of AgRP/NPY and POMC attributed to compromised JAK/STAT3 signals and reduced energy expenditure in the mice, which can be rescued with AAV-genetic overexpression of BMAL1 into SCN. The potential mechanism may be related to the disruptions of SCN efferent mediated by BMAL1.
Conclusions
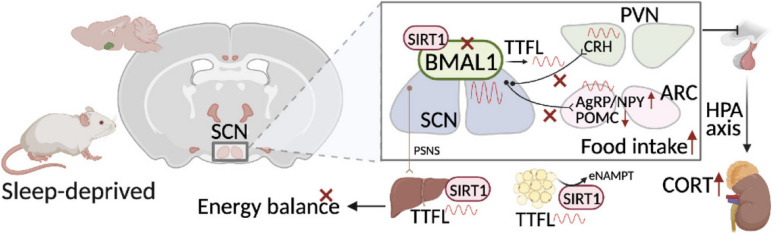
Chronic SD impairs energy balance through directly dampening BMAL1 expression, probably in the transcription level, in the SCN, which in turn affects the neuron projections to ARC and PVN. Remarkably, we provide evidence that may explain the causal mechanisms associated with sleep curtailment and obesity in adolescents.
Graphical Abstract
Supplementary Information
The online version contains supplementary material available at 10.1186/s12915-024-02097-4.
Keywords: Chronic sleep deprivation, SCN, Food intake, Energy expenditure, BMAL1
Background
Adequate sleep during adolescence is crucial for mental and physiological development, including growth, maturation, and neuroendocrine function [1]. Insufficient sleep and social jetlag in adolescents are common in modern society due to social and family obligations, stress, sleep disorders, and other medical illnesses [2]. It has been well established from epidemiological and experimental studies that chronic sleep loss may increase the risk of obesity and weight gain during adolescence [3–6]. Adolescents are developmentally and socially vulnerable to unhealthy diets. Insufficient sleep duration might affect adolescents’ self-control when making food choices, resulting in increased intake of unhealthy foods and increased risk of overeating [7–10]. The latest research revealed increased response to food stimuli in the brain reward system, such as left ventral tegmental area and substantia nigra in adolescents after sleep restriction [11]. A recent review found that the aberrant appetite regulation in young adults caused by sleep deprivation may be driven by augmented or reduced gut hormones (such as leptin and ghrelin) [12]. However, little evidence illustrates the effect of sleep deprivation on hypothalamic food desire, and the underlying mechanisms need further exploration, given that the hypothalamus plays an essential role in the modulation of appetite and energy expenditure and a dysfunctional hypothalamus is involved in the development of obesity [13–15].
Alqaderi et al. evaluated the relationship between the sleep variables and salivary and serum biomarkers in adolescents; the results indicated that the inflammatory and metabolic biomarkers had significant associations with sleep debt and social jetlag, which could disrupt circadian rhythm [16]. It is widely acknowledged that circadian rhythm disruption could be the major risk of energy metabolic disorders via affecting metabolic and appetite-related hormones, and energy expenditure [17, 18]. Circadian rhythm can influence subjective hunger and appetite independent of the fasting/feeding cycle, with peaks in the biological evening and troughs in the biological morning [19]. In mammals, the circadian rhythm is timed by the suprachiasmatic nucleus (SCN) as the master clock located in the hypothalamus, and the daily rhythms are driven by a transcriptional feedback loop formed by core clock genes including Clock, Bmal1, Per1, Per2, Cry1, and Cry2 [20]. The cycle is stabilized by accessory loops in which CLOCK and BMAL1 drive E-box-mediated circadian expression of the nuclear receptors RORα and REV-ERBα which in turn act via REV response element (RRE) sequences to activate and suppress BMAL1 transcription, respectively [20]. SCN can make projections to different types of cell bodies in the hypothalamus, including the arcuate nucleus (ARC) and paraventricular nucleus (PVN). It is essential for driving rhythms in food intake, with efferent fibers co-localized with neurons in ARC [21–23]. ARC, adjacent to the third ventricle, is a central area in the regulation of feeding behavior. There are two main neuron types that co-expressed anorexigenic or orexigenic neuropeptide in the ARC: proopiomelanocortin (POMC)/CART neurons that are related to inhibition of food intake and neuropeptide Y (NPY)/AgRP neurons that are associated with energy need. They are also called the first-order neurons that respond to circulating nutrients and hormones such as glucose, insulin, leptin, and ghrelin [24, 25]. In addition, GABAergic projections from AgRP neurons also suppress POMC neurons in ARC and the activity of MC3R/4R neurons in the PVN, which plays a pivot role in energy metabolism as extinguished representing the HPA axis [26]. PVN also can receive direct SCN input through different neurotransmitters and indirect SCN input via the ARC [27, 28].
BMAL1, as a transcription factor, has been demonstrated a role in controlling the expression of orexigenic neuropeptide expression in the hypothalamus [18]. Loss of BMAL1 also altered the time-dependent expression of NPY and POMC and disrupted AgRP rhythmicity [29]. Depletion of BMAL1 in PVN neurons reduces diurnal rhythmicity in metabolism and causes obesity [30]. Previous studies have demonstrated that cortical and hypothalamic expressions of several clock genes are affected by sleep deprivation [5, 31]. However, the role of BMAL1 on the effects of sleep deprivation on appetite regulation in the hypothalamus is not entirely clear.
The cytoplasmic tyrosine kinase Janus kinase 2 (JAK2)/signal transducer and activator of transcript 3 (STAT3) signals of sensory neurons in ARC can be phosphorylated by circulating leptin via the long form of the leptin receptor (LepRb). Phosphorylated STAT3 translocates into the nucleus and binds to the promoter of the POMC gene, which will reduce food intake and increase energy expenditure [32]. It has been demonstrated that chronic sleep deprivation can attenuate the phosphorylation of STAT3 in the hypothalamus of mice [32]. Our previous study also observed that chronic sleep deprivation could attenuate the JAK2/STAT3 signal pathway and expression of POMC in young adult rats, with compromised BMAL1 expression at the transcriptional levels. Still, the mechanisms of the role of BMAL1 require further exploration [5]. Therefore, our present study was firstly designed to investigate the role of BMAL1 on the effects of chronic sleep deprivation on food intake and energy metabolism in adolescent mice; secondly, the modification of temporal code emitted by the SCN under chronic sleep deprivation is unknown, we tried to establish the neural connections of SCN efferent mechanisms underlying disrupted food intake and energy expenditure due to chronic sleep deprivation, illustrating the causal mechanisms associated with insufficient sleep and obesity in adolescents.
Results
Chronic sleep deprivation-induced aberrant food intake and weight gain
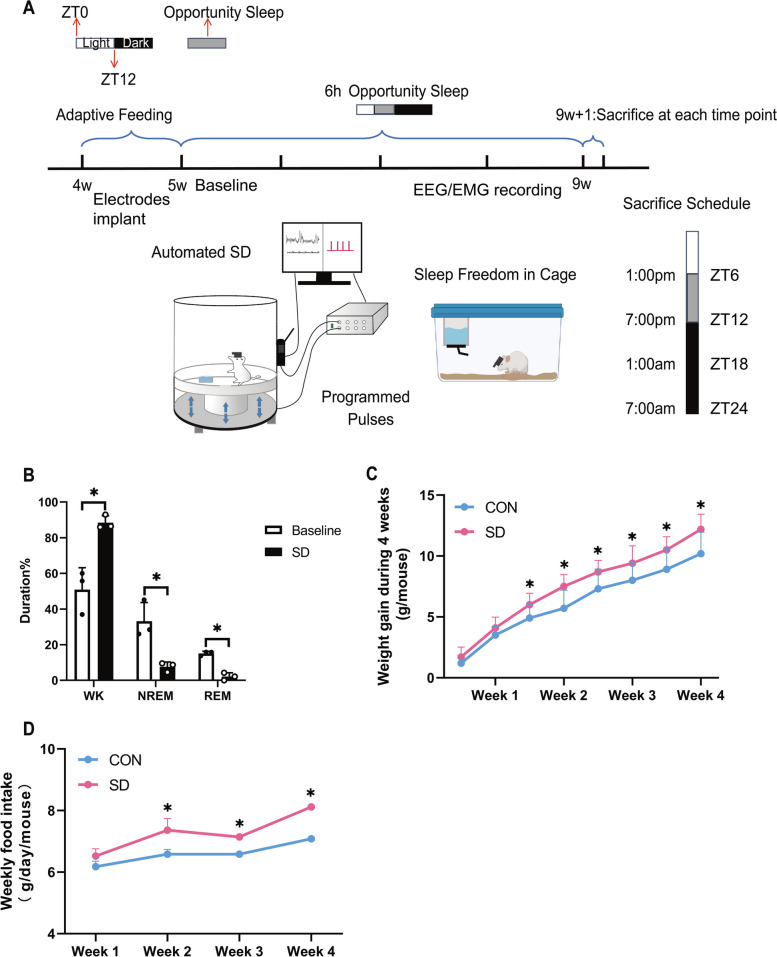
In the first part of the present study, mice were randomly divided into two groups: the control group (CON) and the chronic sleep deprivation group (SD) and the brain tissues and serum were taken immediately at ZT6, ZT12, ZT18, and ZT24 on the last sleep deprivation day after 4 weeks procedure. EEG/EMG electrodes were implanted 1 week before the sleep deprivation process (as shown in Fig. 1A). Sleep duration in WK (wakefulness) stage of mice was significantly increased in SD groups, compared to the baseline, whereas the durations of NREM (non-rapid eye movement) and REM (rapid eye movement) stage of mice were significantly decreased (p < 0.05, shown in Fig. 1B). Our preliminary analysis of sleep-deprived mice revealed a trend toward increased food intake and body weight reached statistical significance in the chronic SD group, compared to the control group from 2 to 4 weeks of sleep deprivation (p < 0.05; shown in Fig. 1C and D). The relationship between circadian rhythm and metabolic disease has recently drawn wide attention. SCN in the hypothalamus acts as an oscillation pacemaker of circadian rhythm with extra-SCN in the brain and peripheral organs [33]. Moreover, our previous publication provided a clue that the increased weight gain and food intake in chronic sleep-deprived rats might be attributed to the dampened expressions of circadian clock genes of the hypothalamus, and the expressions of master regulators, BMAL1 and CLOCK, were downregulated in the transcriptional level [5]. CLOCK usually acts as a histone acetyltransferase in the clockwork, binding to BMAL1 as a heterodimer to drive the system [34]. Therefore, we next mainly focus on BMAL1 in subsequent explorations of our chronic sleep deprivation model.
Fig. 1.
Chronic sleep deprivation-induced aberrant feeding behavior. A Experimental design and timeline. B Changes of different sleep stages duration after sleep deprivation for 4 weeks (n = 3) and presented as bar graphs. EEG data were recorded on the last sleep deprivation day. Significant difference (single asterisk) was defined as p < 0.05 by a two-sided unpaired t-test. C Line chart, weekly body weight gain during chronic sleep deprivation (n = 10). Significant difference (single asterisk) was defined as p < 0.05 by two-way repeated-measures ANOVA and pairwise comparisons. D Line chart, weekly food intake during chronic sleep deprivation (n = 12, 3 mice per cage for distribution). Significant difference (single asterisk) was defined as p < 0.05 by two-way repeated-measures ANOVA and pairwise comparisons. Data were represented as means + standard deviations for B, C, and D. Data were presented as the means ± standard deviations. NREM, non-rapid eye movement; REM, rapid eye moment; SD, sleep deprivation group, mice received 4 weeks of sleep deprivation (18 h/day); CON, control group. ZT, Zeitgeber time
Changes in circadian clock expression patterns in the hypothalamus induced by chronic sleep deprivation
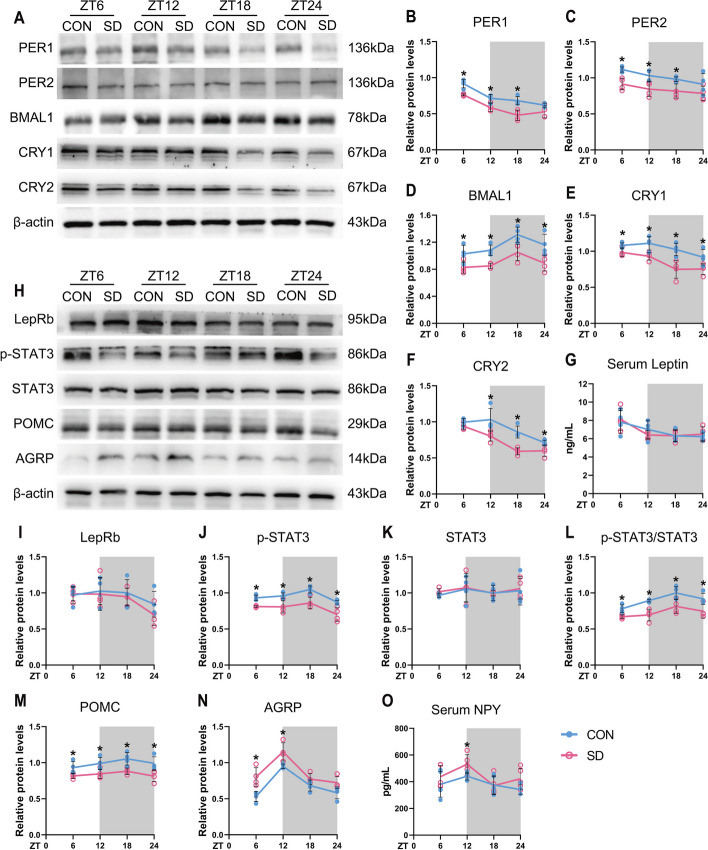
We firstly explored oscillation changes of core circadian genes in the established chronic sleep-deprived mice model; the entire hypothalamus tissues were taken every 6 h starting at ZT6 throughout a 12-h light/dark cycle, and the expressions of core circadian clocks were measured. As shown in Fig. 2A and Additional file 1: Fig. S3, the protein levels of BMAL1 in the control groups were lower in the light phase (ZT6 and ZT12) and higher in the dark phase (ZT18 and ZT24); in contrast, the expressions of PER1 and CRY1 protein were higher in the light phase and lower in the dark phase. Moreover, the circadian amplitudes of key circadian gene expressions, BMAL1, PER2, and CRY1, in the hypothalamus of chronic sleep-deprived mice after sleep deprivation for 4 weeks were shrunken.
Fig. 2.
Chronic sleep deprivation changed expressions of circadian clock genes and appetite-related peptides in the hypothalamus. A Western blot analysis, the typical blots of circadian clocks in the entire hypothalamus. B–F Relative protein levels for circadian clocks in the entire hypothalamus. β-actin was used as an internal control. The band intensities were quantified using Image J software and presented as line charts to show relative quantification (n = 4 mice/group/time point). G Line chart, serum leptin concentrations in CON and SD mice at 6-h intervals after 4-week sleep deprivation (n = 4 mice/group/time point). H Western blot analysis, the typical blots of appetite-related peptides in the entire hypothalamus. I-N. Relative protein levels for appetite-related peptides in the entire hypothalamus. β-actin was used as an internal control. The band intensities were quantified using Image J software and presented as line graphs to show relative quantification (n = 4 mice/group/time point). O Line chart, serum NPY concentrations in CON and SD mice at 6-h intervals after 4-week sleep deprivation (n = 5 mice/group/time point). The white part represented the light phase and the gray part represented the dark phase. Significant difference (single asterisk) was defined as p < 0.05 by two-way repeated-measures ANOVA and pairwise comparisons. Data were presented as the means ± standard deviations for the line charts. Mice were sacrificed at 6-h intervals across the 12:12 light/dark cycle (ZT0, 7:00 am). CON, control group; SD, sleep deprivation group, mice received 4 weeks of sleep deprivation (18 h/day); ZT, Zeitgeber
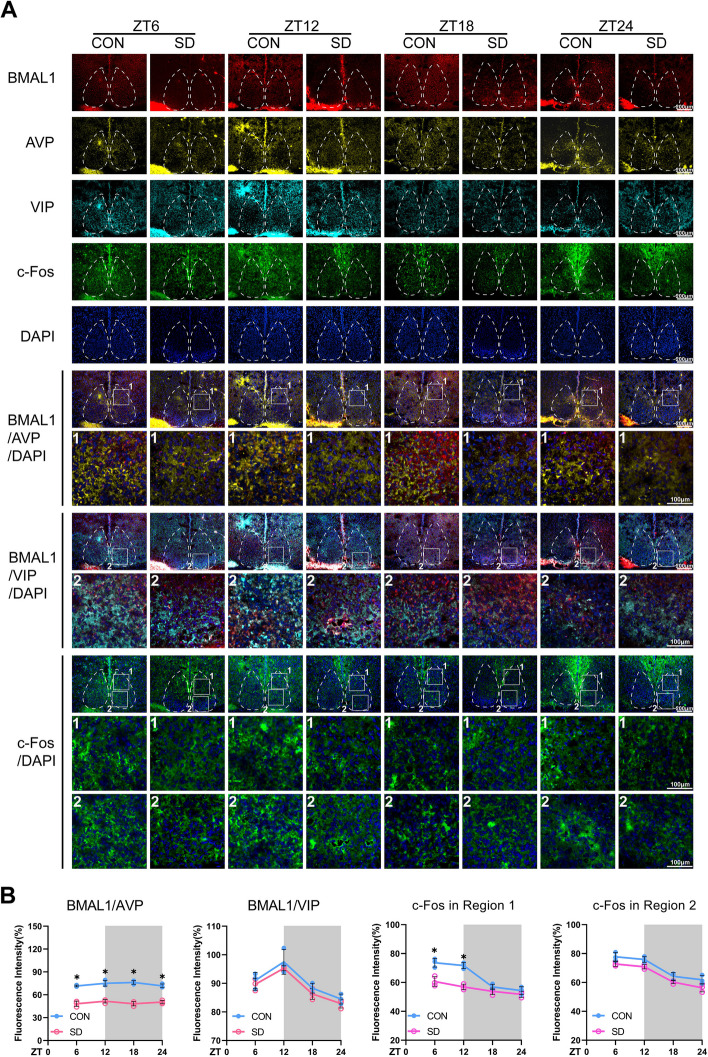
It is well established that there are two anatomic subdivisions in the SCN of the mouse, a ventral “core” region receives retinal input and is partially enveloped by a dorsal “shell” region. Neurons in core and shell sub-regions are distinguished by neuropeptides. Within the shell are mainly the neurons containing arginine vasopressin (AVP), while neurons within the core are mainly containing vasoactive intestinal polypeptide (VIP) [33]. Confocal images (Fig. 3) presented the changes in immunostaining for BMAL1 in the SCN; the average fluorescence intensities of BMAL1 in the control group were lower in the light phase (ZT6 and ZT12) and higher in the dark phase (ZT18 and ZT24). Furthermore, the reductions of BMAL1 occurred in the “shell” region of the SCN since the average fluorescence intensities of BMAL1 co-stained with AVP were decreased at each time point, instead of VIP (p < 0.05), as we expected (Fig. 3B). The neural activities, characterized by c-Fos staining, in the SCN of chronic sleep-deprived mice were significantly dampened in the light phase (p < 0.05, shown in Fig. 3B), especially in the shell region. AVP is also a downstream target of BMAL1, the neuronal connectivity of the SCN network in AVP-Bmal1−/− mice could be restored by compensation of BMAL1 in AVP neurons [35]. Therefore, we could speculate that chronic sleep deprivation might directly cause the depletion of BMAL1 in AVP neurons and subsequently impact the pacemaker function of the SCN.
Fig. 3.
Chronic sleep deprivation changed the in situ expressions of BMAL1 and c-Fos in the SCN. A Representative immunofluorescence (IF) staining for BMAL1 (red), AVP (yellow), VIP (cyan), c-Fos (green), and DAPI (blue) (scale bars = 500 μm) and merged images of BMAL1/AVP/DAPI, BMAL1/VIP/DAPI, and c-Fos/DAPI (scale bars, upper, 500 μm; lower, 100 μm) in the SCN of the mice in CON and SD group at each time point. The number 1 represented the shell region, and the number 2 represented the core region of the SCN. B The average IF intensities of BMAL1 co-stained with AVP or VIP, and the average IF intensities of c-Fos were determined by Image J software and presented as line charts (n = 3). Mean intensity = integrated density in co-staining area/area of AVP (or VIP). The white part represented the light phase, and the gray part represented the dark phase. Data were presented as the means ± standard deviations. Significant difference (single asterisk) was defined as p < 0.05 by two-way repeated-measures ANOVA and pairwise comparisons. Mice were sacrificed at 6-h intervals across the 12:12 light/dark cycle (ZT0, 7:00 am). CON, control group; SD, sleep deprivation group, mice received 4 weeks of sleep deprivation (18 h/day); ZT, Zeitgeber time
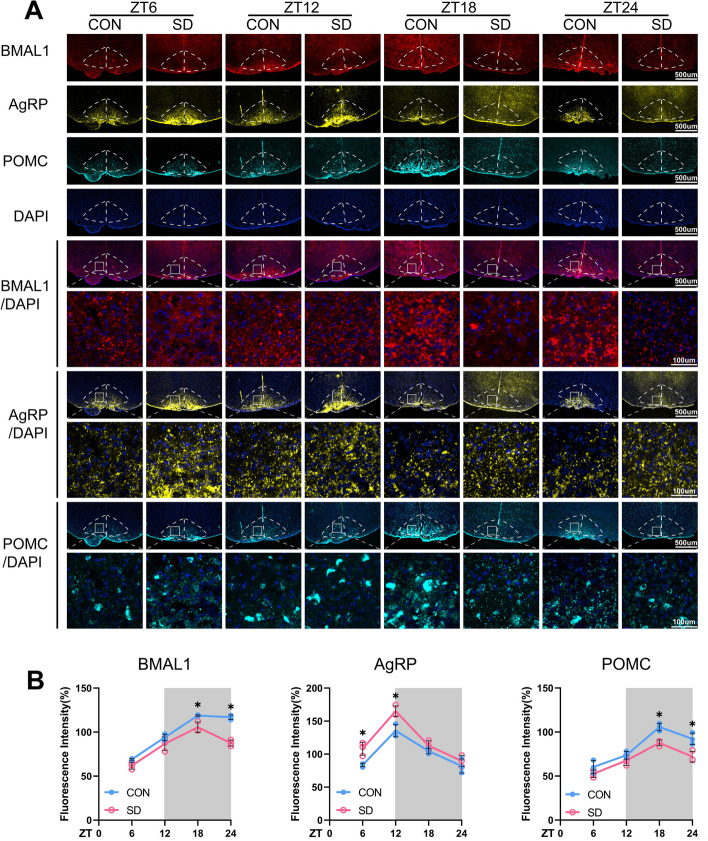
ARC can receive direct projections from the SCN with the same neuronal firing patterns that are coordinated with daily waves of SCN input [36]. The present results of confocal images also showed the same oscillation expressions of BMAL1 in ARC as the SCN in the control group; the expressions of BMAL1 in ARC were significantly decreased in chronic SD groups at ZT18 and ZT24 as compared to controls (Fig. 4).
Fig. 4.
Chronic sleep deprivation changed in situ expressions of AgRP and POMC in the ARC. A Representative immunofluorescence (IF) staining for BMAL1 (red), AgRP (yellow), POMC (cyan), and DAPI (blue) (scale bars = 500 μm), and merged images of BMAL1 and DAPI, AgRP and DAPI, POMC and DAPI (scale bars, upper, 500 μm; lower, 100 μm) in the ARC of the mice in CON and SD group at each time point. B The average IF intensities of BMAL1, AgRP, and POMC were determined by Image J software and presented as bar graphs (n = 3). Mean intensity = integrated density/area. The white part represented the light phase and the gray part represented the dark phase. Data were presented as the means ± standard deviations. Significant difference (single asterisk) was defined as p < 0.05 by two-way repeated-measures ANOVA and pairwise comparisons. Mice were sacrificed at 6-h intervals across the 12:12 light/dark cycle (ZT0, 7:00 am). CON, control group; SD, sleep deprivation group, mice received 4 weeks of sleep deprivation (18 h/day); ZT, Zeitgeber time
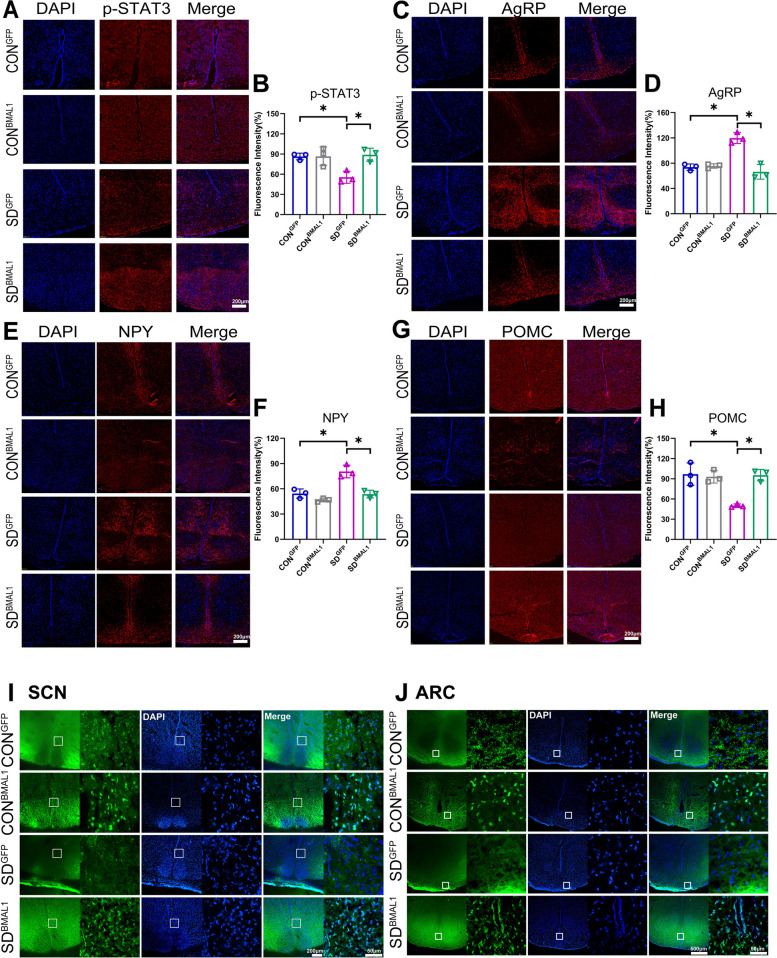
Impact of chronic sleep deprivation on AgRP and POMC expressions in ARC
ARC is regarded as the “first-order” nuclei of appetite regulation and an important sensor of nutrient signals coming from the blood, with its anatomical mapping in the hypothalamus located close to fenestrated capillaries [23]. AgRP neurons and POMC neurons are the two populations mainly expressed in ARC, which exert opposite functions on appetite regulation. The activation of AgRP neurons promotes food seeking and taking, whose firing rate is high at the light/fasted phase in mice. In contrast, POMC neurons express anorexigenic peptides with higher neural activities in the dark/feed phase in mice [37]. Western blots of the entire hypothalamus were performed to detect the changes in the circadian patterns of AgRP and POMC expressions at the protein level; the results showed that the expressions of POMC were decreased in both the light and dark phases, but the expressions of AgRP were upregulated only in the light phase (p < 0.05, Fig. 4). We also tried to illustrate the changes in diurnal rhythmicity of AgRP and POMC expressions examined by IF in ARC; the changes of AgRP protein expression was in accordance with the Western blots, but POMC protein staining was only significantly decreased in the dark phase (p < 0.05). Interestingly, the same diurnal expression rhythms of POMC and BMAL1 in ARC were observed, and they exhibited higher levels of protein expression during the dark phase, compared to the light (shown in Fig. 2H, D, M, N and Additional file 2: Fig. S3). Whereas control mice exhibited a robust diurnal rhythm, chronic sleep-deprived mice showed a dramatic reduction in POMC and BMAL1 expressions during the dark phase (ZT18 and ZT24) as shown in Fig. 4 (p < 0.05). On the other hand, AgRP expressions were significantly upregulated in the chronic SD group, compared to controls, especially at the light phase (ZT6 and ZT12) as shown in Fig. 4, which were in accordance with the result of Western blot shown in Fig. 2H and N (p < 0.05).
POMC and AgRP in the ARC neurons can be activated by the circulating leptin through the transmembrane receptor, LepRb. LepRb binds to a cytoplasmic tyrosine kinase JAK2 to acquire enzymatic activity. Then JAK2 phosphorylates LepR on Tyr1138 in response to leptin and following recruits the SH2 domain of STAT3 [38, 39]. The phosphorylated STAT3 subsequently translocates into nuclear and acts as a transcription factor to regulate the expression of POMC and AgRP. Activation of STAT3 promotes POMC and suppresses AgRP and NPY expressions at the transcriptional level [40]. Our present results indicated that the protein level of p-STAT3 in the hypothalamus also showed a robust diurnal rhythm with the same pattern as BMAL1 in the control group (shown in Fig. 2H and J). Whether sleep deprivation could reduce the circulating leptin is still controversial [41]. The convey results seem to show that leptin level is apparently decreased in the acute sleep deprivation model, whereas there are no consistent results observed from chronic sleep-deprived rodents [42]. In our chronic sleep deprivation model, leptin level exhibited a circadian rhythm through daytime and night; interestingly, there was no difference between controls and sleep deprivation groups at any time point nor the expression of LepRb in the hypothalamus (shown in Fig. 2H, G, and I). Based on these results, we speculated that the compromised STAT3 signal pathway might be due to the reduction of BMAL1 expression rather than the changes in circulating leptin level.
Evidence has consistently shown that SCN efferent projects to the ARC and synchronizes the circadian clocks of ARC; hence, SCN-ARC axis is essential for daily food seeking and intake [23]. Therefore, we next explored whether overexpression of BMAL1 in SCN could rescue the dysregulation of feeding behavior in chronic sleep-deprived mice by stereotaxic injection of AAV1-hSyn-Bmal1 into the shell region of the SCN.
Compensation of BMAL1 in SCN shapes sleep behavior
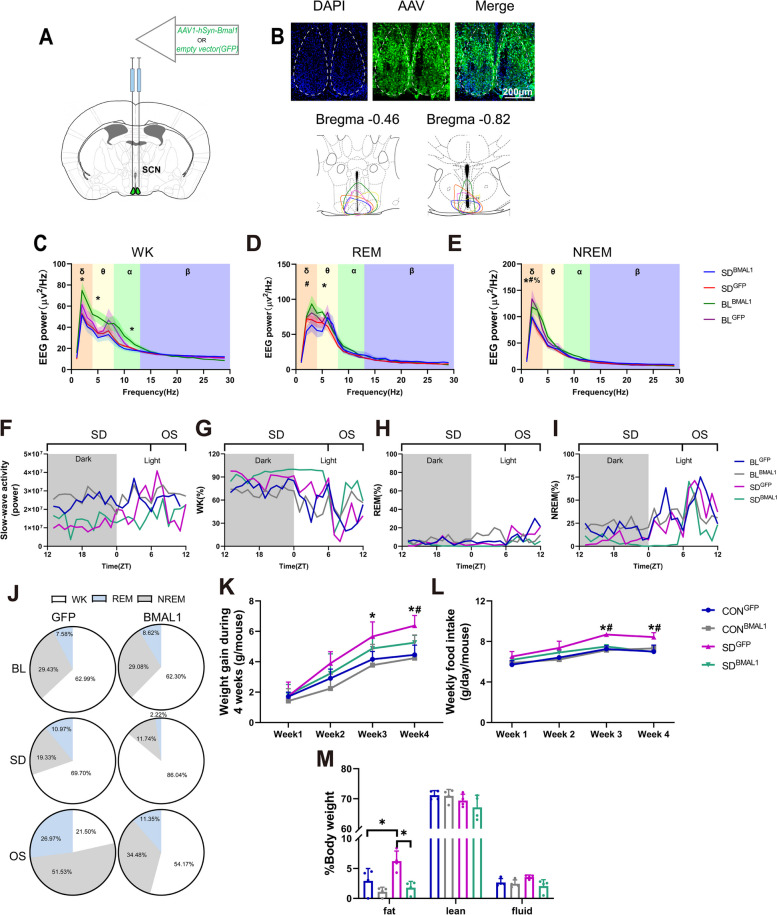
In the following intervention experiments, mice were randomly divided into four groups: the control group with empty vector (CONGFP), the control group with AAV-genetic overexpression of BMAL1 (CONBMAL1), the chronic SD group with empty vector (SDGFP), and the chronic SD with AAV-genetic overexpression of BMAL1 group (SDBMAL1). EEG/EMG electrodes and viral injection (as shown in Fig. 5A) were performed 1 week before the chronic sleep deprivation procedure (4 weeks). As shown in Fig. 4C, the injection site was targeted almost in the shell region of SCN where chronic sleep deprivation resulted in the depletion of BMAL1 as we illustrated above. EEG/EMG data (presented in Fig. 5C–I) were adopted on the last day of the chronic sleep deprivation procedure. Figure 5C–E shows the comparisons of power in different sleep stages. Animal behavior is visually classified as “WK,” “REM,” or “NREM” for consecutive 4-s epochs based on the EEG and EMG signals. WK is characterized by EEG activity of mixed frequency and low amplitude, and muscle tone is present and variable [43]. NREM is accompanied by large-amplitude, low-frequency activity in the EEG, with a spectral peak around the delta band (0.5–4 Hz), whereas REM sleep shows regular EEG activity in the theta band (4–9 Hz), together with muscle atonia [43]. Power spectral analysis of delta EEG activity in NREM, also known as SWA (slow-wave activity), is considered a proxy for sleep homeostasis, with the initial SWA accumulation after sleep onset reflecting sleep drive or sleep need, and the dissipation of SWA reflecting the progression of recovery from the sleep debt that accumulated over the day. SWA increases proportionally with the duration of WK, with longer time awake, homeostatic sleep pressure builds, resulting in higher SWA [44].
Fig. 5.
Chronic sleep deprivation changed sleep behavior. A Viral injection, empty vector/AAV1-hSyn-Bmal1 was injected at the shell of SCN bilaterally in the mice. B Schematic timeline. C Representative images showing GFP, DAPI, and merged images in the SCN, and schematic showing dissection neurons. D–F. EEG power during wakefulness (WK), rapid eye movement (REM), and non-rapid eye movement (NREM) (n = 4 for BLGFP, n = 5 for BLBMAL1, n = 6 for SDGFP and SDBMAL1). Single asterisk, BLBMAL1 vs. SDBMAL1 p < 0.05; percent, BLGFP vs. SDGFP p < 0.05; number sign, SDGFP vs. SDBMAL1 p < 0.05 by one-way ANOVA. G A representative 24-h recording of slow-wave activity (SWA), and SWA is plotted as the power of the mean activity during NREM over 24 h. H–J Representative hourly amounts of sleep during a constant 24-h. K Representative pie chart for sleep distribution during 24-h baseline (BL), 18-h sleep deprivation (SD), and 6-h opportunity sleep (OS). L Weight gain during 4 weeks of sleep deprivation (18 h/day) (n = 8). Single asterisk was defined as p < 0.05 by two-way repeated-measures ANOVA with Tukey’s post hoc test. Data were represented as means + standard deviations for L, M, and N. M Weekly food intake during 4 weeks of sleep deprivation (18 h/day) (n = 8, 2 or 3 mice per cage for distribution). Single asterisk was defined as p < 0.05 by two-way repeated-measures ANOVA with Tukey’s post hoc test. N Body composition after chronic sleep deprivation (n = 4). BLGFP, mice received injection of AAV-GFP before sleep deprivation; BLBMAL1, mice received injection of AAV-BMAL1 before sleep deprivation; CONGFP, the control group with empty vector; CONBMAL1, the control group with AAV-genetic overexpression of BMAL1; SDGFP, the chronic SD group with empty vector; SD.BMAL1, the chronic SD with AAV-genetic overexpression of BMAL1
We performed baseline recordings for 24 h, including an 18-h SD and 6 h of opportunity sleep (termed recovery recording) on the last day of sleep deprivation for 4 weeks. The average power spectra during different sleep stages in baseline and sleep deprivation were obtained. Power spectra displayed peaks at low frequencies during NREM and WK and medium frequencies during REM, corresponding to δ and θ frequency bands, respectively. When analyzing the baseline, EEG power showed no difference in δ and θ power of WK, REM, and NREM in mice of BLBMAL1 relative to the BLGFP (shown in Fig. 5C–E). After chronic sleep deprivation, mice in SDGFP and SDBMAL1 exhibited decreased δ power of NREM relative to the baseline (p < 0.05) (shown in Fig. 5E). EEG measures in SDBMAL1 mice exhibited decreased δ, θ, and α power relative to the BLBMAL1 (baseline) during WK (p < 0.05) (shown in Fig. 5C). Mice in SDBMAL1 exhibited decreased θ power of REM relative to the BLBMAL1 (p < 0.05, shown in Fig. 5D). SDBMAL1 mice showed a significant decrease in θ power during REM, compared to BLBMAL1 (p < 0.05, shown in Fig. 5D), indicating that mice need more sleep duration after chronic sleep deprivation since a REM sleep deficit must be repaid in the hard currency of time, regardless compensation of BMAL1. SWA, a quantitative measure of the amplitude and prevalence of EEG slow waves, tracks sleep needs over time. Homeostatic sleep pressure accumulates during WK and is thus higher following sleep deprivation. Representative EEG recordings were shown in Fig. 5F, mice in BLGFP and BLBMAL1 exhibit higher SWA than the SDGFP and SDBMAL1 group at the dark phase showing that chronic sleep deprivation disrupted sleep homeostasis. During sleep deprivation, the SWA of mice in the SDGFP group showed a gradual ascend but below the baseline and reached the highest level at ZT6, then gradually descended, whereas this change was not observed in SDBMAL1 mice. The SWA of SDBMAL1 mice was lower than SDGFP indicating that compensation of BMAL1 in the SCN exhibited less sleep debt after chronic sleep deprivation than the SDGFP group. Representative duration recordings were shown in Fig. 5G–I; the measurements of the time spent in WK, NREM, and REM indicated that compensation of BMAL1 caused a different sleep–wake distribution after chronic sleep deprivation. During 6-h opportunity sleep, we observed a reduced time spent on WK and an increased time spent in NREM and REM in SDGFP mice, compared to that in the baseline (BLGFP), and mice in the SDBMAL1 group showed increased time spent on WK stage. We further investigated the effect of chronic sleep deprivation on the sleep–wake distribution of different stages by computing the time course of the phase during 24 h. The distributions of mice in the BLGFP and BLBMAL1 groups are nearly the same during different sleep stages. During baseline, the typical sleep–wake distribution of the mice in the BLGFP and BLBMAL1 groups was observed with similar time spent in different sleep stages. As shown in Fig. 5J, the comparison between data of BLGFP and SDGFP indicated a different distribution of these states across the period on NREM, REM, or WK; mice in the SDGFP group exhibited more durations of WK and REM and less NREM. During sleep deprivation, mice in the SDBMAL1 group exhibited progressively more durations of WK and less NREM and REM than the SDGFP group. The mice in both SDGFP and SDBMAL1 groups exhibited less WK and more NREM and REM durations than that in baseline during sleep deprivation. These results suggest that δ power in the overall spectrum decreases in all sleep periods caused by sleep deprivation, and δ power in NREM is of great significance for evaluating the establishment of sleep deprivation models and sleep homeostasis. Therefore, we continued to analyze the changes in SWA with ZT, and the results showed that SWA during sleep deprivation was lower than the baseline level. It was gradually increased and reached a peak at ZT6 when mice were restored to sleep and gradually decreased during the recovery period, indicating that this model could increase the sleep homeostatic pressure of mice. Although SWA was increased at ZT6 after mice re-entered sleep, it was still less than 200% indicating the sleep could not be fully compensated. Moreover, overexpression of BMAL1 could reduce sleep homeostasis pressure, suggesting that circadian clock genes might be involved in the regulation of sleep homeostasis. The results in the sleep stages of mice in each group showed that sleep deprivation reduced the sleep duration of mice in each period, and the sleep duration of opportunity sleep in the BMAL1 overexpression group was less than that in the GFP-carrier control group, suggesting that BMAL1 overexpression could reduce the sleep debt caused by sleep deprivation, which was consistent with the SWA results shown above.
Overexpression of BMAL1 rescues chronic sleep deprivation-induced aberrant weight gain and food intake
The weight gains of mice in the SDGFP group were significantly increased after sleep deprivation for 3 and 4 weeks, compared to controls (p < 0.05), and SDBMAL1 showed a relative reduction to the higher level of the SDGFP group (p < 0.05, shown in Fig. 5K and L). Similar results were observed on regular chow intake. Furthermore, we also found that body composition seemed to be a significant increase in fat tissue of mice in the SDGFP group (p < 0.05, shown in Fig. 5M).
Compensation of BMAL1 in SCN rescues molecular clocks in the hypothalamus
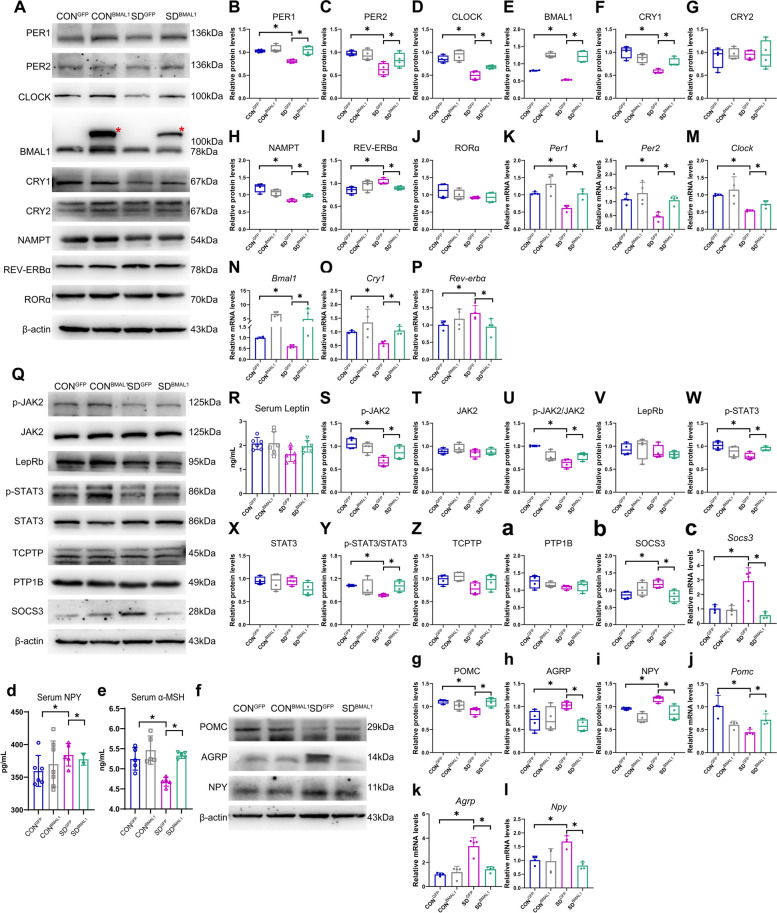
Given our findings mentioned above, the neural activities of the shell region in the SCN were significantly decreased in the light phase, probably caused by the depletion of BMAL1, then mice were sacrificed in the light phase immediately after sleep deprivation in our procedure (ZT6) to explore the role of BMAL1 in fine-tuning circadian rhythms. Representative protein blots of circadian clock proteins were shown in Fig. 6A and Additional file 3: Fig.S5. We found that BMAL1, CLOCK, CRY1, PER1, and PER2 protein levels in the hypothalamus were significantly reduced in the SDGFP group after chronic sleep deprivation compared to controls (p < 0.05, Fig. 6B–F and Additional file 4: Fig.S6). Compensation of BMAL1 in SCN could rescue the depletion of these protein expressions. The mRNA levels of BMAL1, CLOCK, CRY1, PER1, and PER2 in the hypothalamus of the chronic SD group were also significantly dampened, which could be rescued by injection of exogenous AAV-genetic BMAL1 (p < 0.05, Fig. 6K–O). To examine the further mechanism underlying the depletion of BMAL1 expression, the regulators of TTFL including other CCGs and kinase pathways were also detected. The transcriptional suppressor of BMAL1, REV-ERBα, was significantly upregulated in the SDGFP group, compared to the CONGFP group (p < 0.05, Fig. 6I and P), and slightly decreased in the SDBMAL1 group, failing to fall to the baseline. However, the activator, RORα, did not show any change in the SDGFP group (Fig. 6J). NAD+-dependent deacetylase sirtuin 1 (SIRT1) is a major influencer on the circadian machinery; it can directly deacetylate PER2 to promote degradation [45] and BMAL1 to relieve suppression of CRY1 [46]. The activity of SIRT1 depends on NAD+ oscillating levels and nicotinamide phosphoribosyl transferase (NAMPT), the rate-limiting enzyme in the NAD+ salvage pathway, which is also an output target of CLOCK:BMAL1 [47]. As shown in Fig. 6H, NAMPT in the SDGFP group was significantly downregulated as compared to the CONGFP group (p < 0.05), and the protein level of NAMPT could be rescued in the SDBMAL1 group. SIRT1 can also directly activate the transcription of Bmal1 via recruiting its co-activator PGC-1α to bind with RORα [48]. Our results showed that the expression of SIRT1 in the protein level was compromised in the hypothalamus of chronic sleep-deprived mice at each detected time point (shown in Fig. 8G), providing us a clue that the depletion of SIRT1 might be the cause of inhibition in the transcriptional level of BMAL1. Collectively, these results supported the notion that rescuing the depletion of BMAL1 might be the potential intervention target for chronic sleep deprivation.
Fig. 6.
Compensation of BMAL1 in SCN rescued compromised molecular clocks induced by chronic sleep deprivation. A The typical blots of circadian clocks in the entire hypothalamus. The red asterisk represented as AAV-genetic compensation of BMAL1 in the SCN. B–J. Relative protein levels for circadian clocks in the entire hypothalamus. β-actin was used as an internal control (n = 4). The band intensities were quantified using Image J software and presented as box graphs to show relative quantification for B–J, S–Z, a–b, and g–i. Data were presented as the means ± standard deviations. K–P. Relative mRNA levels for circadian clocks in the entire hypothalamus. β-actin was used as an internal control (n = 4). Data were represented as means + standard deviations for K–P and j–l and presented as bar graphs to show relative quantification. Q The typical blots of JAK2/STAT3 signal pathway in the hypothalamus. R Serum leptin concentrations (n = 6). The serum indicators were presented as bar graphs with means + standard deviations. S–Z and a–b. Relative protein levels for JAK2/STAT3 signal pathway in the entire hypothalamus. β-actin was used as an internal control (n = 4). c Relative mRNA levels for SOCS3 in the entire hypothalamus (n = 4). Data were represented as means + standard deviations. d, e Serum NPY and α-MSH concentrations (n = 5). f–i The typical blots of POMC, AgRP, and NPY in the entire hypothalamus and relative protein levels in the entire hypothalamus. β-actin was used as an internal control (n = 4). j–l. Relative mRNA levels for POMC, AgRP, and NPY in the entire hypothalamus. β-actin was used as an internal control (n = 4). CONGFP, the control group with empty vector; CONBMAL1, the control group with AAV-genetic overexpression of BMAL1; SDGFP, the chronic SD group with empty vector; SDBMAL1, the chronic SD with AAV-genetic overexpression of BMAL1. Samples were taken at ZT6 (1:00 pm). Significant difference (single asterisk) was defined as p < 0.05 by one-way ANOVA
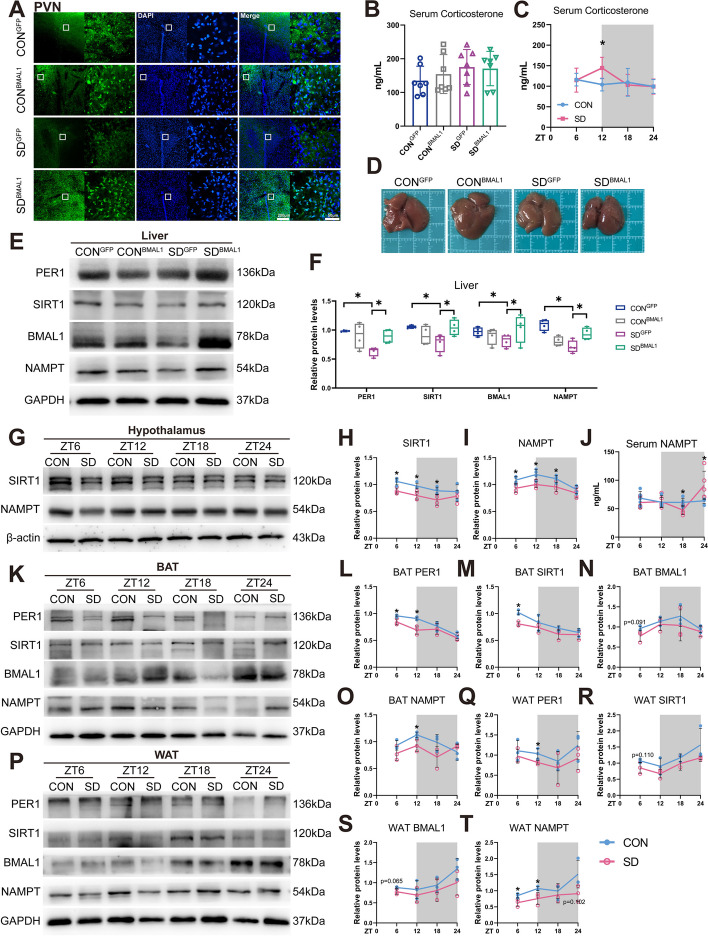
Fig. 8.
Chronic sleep deprivation-induced disruption of energy homeostasis and metabolic disorder. A Representative autofluorescence, DAPI, and merged images in the PVN. B Serum CORT concentrations (n = 7) in the intervention experiments and presented as the bar graph. Significant difference (single asterisk) was defined as p < 0.05 by one-way ANOVA. C Serum CORT concentrations in wild-type mice of CON and SD groups at 6-h intervals after 4-week sleep deprivation and presented as the line chart (n = 6 mice/group/time point). D Representative photograph of the autopsy of the liver in different groups. E Western blot detects changes in circadian clock expression in the liver. F Relative protein levels for circadian clock expression in the liver and presented as box graphs (n = 4). Significant difference (single asterisk) was defined as p < 0.05 by one-way ANOVA. G Representative Western blot for changes in SIRT1 and NAMPT protein levels in the entire hypothalamus. H, I Relative protein levels for SIRT1 and NAMPT in the entire hypothalamus and presented as line charts (n = 4 mice/group/time point). J Serum NAMPT concentrations in CON and SD mice at 6-h intervals after 4-week sleep deprivation (n = 5 mice/group/time point). K–O Representative Western blots and relative protein levels for PER1, SIRT1, BMAL1, and NAMPT in brown adipose tissue and presented as line charts (BAT) (n = 3 mice/group/time point). P–T Representative Western blots and relative protein levels for PER1, SIRT1, BMAL1, and NAMPT in white adipose tissue and presented as line charts (WAT) (n = 3 mice/group/time point). Significant difference between CON and SD at each time point was calculated by two-way repeated-measures ANOVA and pairwise comparisons (*p < 0.05). The white part represented the light phase and the gray part represented the dark phase (ZT0, 7:00 am). CONGFP, the control group with empty vector; CONBMAL1, the control group with AAV-genetic overexpression of BMAL1; SDGFP, the chronic SD group with empty vector; SD.BMAL1, the chronic SD with AAV-genetic overexpression of BMAL1
Role of BMAL1 in modulating feeding behavior
As elucidated above, BMAL1 might impact the transduction of the JAK2/STAT3 signal pathway; therefore, we next verified the role of BMAL1 in the modulation of cell signals. Representative protein blots are shown in Fig. 6Q and Additional file 4: Fig. S6. We found that the phosphorylation JAK2 and STAT3 in the hypothalamus were significantly reduced in the SDGFP group (p < 0.05); subsequently, AgRP and NPY were upregulated and POMC was downregulated in the SDGFP group, compared to the CONGFP group. However, the compromised JAK2/STAT3 signals and their downstream target genes, AgRP, NPY, and POMC, could be reversed in the SDBMAL1 group (Fig. 6R–Y, f–I, and Additional file 5: Fig. S7). The changes in mRNA levels of POMC, AgRP, and NPY in the hypothalamus were consistent with protein levels (Fig. 6j–l). The in situ expression of p-STAT3, AgRP, NPY, and POMC proteins in ARC are shown in Fig. 7A–H, and immunostaining of the ARC showed the same trends of change as protein expressions derived from the entire hypothalamus. Aberrant expressions of AgRP, NPY, and POMC proteins in ARC were rescued in the SDBMAL1 group. Accordingly, the contents of serum α-MSH, a product of POMC, were significantly decreased in the SDGFP group as compared to the CONGFP group (p < 0.05, shown in Fig. 6e) and rescued in the SDBMAL1 group as expected. Consistent with the expression of NPY in protein and mRNA, the serum NPY level was also upregulated in the SDGFP group (p < 0.05, shown in Fig. 6d), and reversed to the baseline level in the SDBMAL1 group, compared to the CONGFP group.
Fig. 7.
Compensation of BMAL1 rescued in situ expressions of p-STAT3 and appetite-related peptides in the ARC. A–B Representative immunofluorescence (IF) staining for p-STAT3, DAPI, and merged images in the ARC (scale bars = 200 μm). C–D Representative IF staining for AgRP, DAPI, and merged images in the ARC (scale bars = 200 μm). E–F Representative IF staining for NPY, DAPI, and merged images in the ARC (scale bars = 200 μm). G–H Representative IF staining for POMC, DAPI, and merged images in the ARC (scale bars = 200 μm). The average IF intensities were determined by Image J software and presented as bar graphs (n = 3) for A–H. Mean intensity = integrated density/area. I Representative autofluorescence (green), DAPI, and merged images in the SCN (scale bars, left, 200 μm; right, 50 μm). J Representative autofluorescence (green), DAPI, and merged images in the ARC (scale bars, left, 500 μm; right 50 μm). Data were presented as the means ± standard deviations. Significant difference (single asterisk) was defined as p < 0.05 by one-way ANOVA. CONGFP, the control group with empty vector; CONBMAL1, the control group with AAV-genetic overexpression of BMAL1; SDGFP, the chronic SD group with empty vector; SD.BMAL1, the chronic SD with AAV-genetic overexpression of BMAL1. Samples were taken at ZT6 (1:00 pm)
The suppressor of cytokine signaling 3 (SOCS3) contains the SH2 domain, which also binds to phosphor-Tyr985 and in turn suppresses the activation of JAK2/STAT3 signals [49]. The intracellular level of SOCS3 provides a negative feedback mechanism to downregulate JAK2 signal action [50]. Our present result also showed that the expressions of SOCS3 in the hypothalamus at both the protein and mRNA levels were significantly upregulated in the SDGFP group, compared to the CONGFP group (p < 0.05, shown in Fig. 6b and c). It was also notable for the reversed expressions of SOCS3 in the SDBMAL1 group, indicating that overexpression of BMAL1 might work against SOCS3 to release its inhibition for the STAT3 signals. One previous study revealed that the mRNA level of SOCS3 in the SCN exhibited a day/night variation in mice [51]. Another study also reported the role of CLOCK in modulating the expression of SOCS3 in the ARC of the hypothalamus [52]. These data indicated that the expression of SOCS3 might be regulated by the circadian clock genes, but the specific target site needs further exploration in the future.
PTP1B and TCPTP are non-receptor protein tyrosine phosphatases (PTP), which can dephosphorylate JAK2 and STAT3, respectively [53, 54]. Evidence showed that higher expressions of PTP1B and TCPTP contribute to leptin resistance and promote obesity development [55–57]. However, our results did not show any change of PTP1B and TCPTP expressions in the hypothalamus of mice between the CONGFP group and SDGFP group in the present sleep deprivation model; therefore, these two tyrosine phosphatases may not be the reason that contributed to the impediment of JAK2/STAT3 signals.
Potential neural circuit mechanism on the dysregulation of feeding behavior induced by chronic sleep deprivation
The molecular oscillation of the TTFL in SCN synchronizes postsynaptic cell autonomous through interlinking electrical activity and neurotransmitters [58, 59]. In the intact circuit, TTFL orchestrates the spontaneous firing rate of neurons via driving Na+ and K+ conductance and Ca2+ oscillation in both diurnal and nocturnal active species [60]. The circadian profile of intracellular Ca2+ is modulated by voltage-gated calcium channels (VGCC) that can be activated by electrical firing [61]. Subsequently, SCN output neurons release several output factors including GABA, AVP, VIP, and prokineticin 2 (Prok2) onto target neurons of extra-SCN [62–65]. The neural firing modulated by ion channels and neurotransmitters further drives TTFL in the downstream region of SCN [58, 59]. Therefore, we subsequently observed projection neurons indicated by GFP green fluoresce through the confocal images. AAV exhibits anterograde trans-neuronal transport after infection with AAV for 6 weeks in our animal model, indicating a synaptic projection via GFP-tagged neurons. Our data here showed that fewer GFP-tagged neurons were observed in the ARC region of mice in the SDGFP group, whereas the green fluoresce seemed to be recovered in the SDBMAL1 group, compared to the CONBMAL1 group (shown in Fig. 7I and J). This finding indicated that a neuronal circuit projection from SCN to ARC disrupted by chronic sleep deprivation was restored through compensation BMAL1 in SCN since BMAL1 also regulates the expression of AVP, GABAAR, and Prok2 in the dorsal SCN [61].
Chronic sleep deprivation reduces energy expenditure
Obesity development is the outcome of disruption of energy homeostasis resulting from food intake excessing and reduced energy expenditure. As we described above, chronic sleep deprivation could cause aberrant feeding behavior in adolescent mice. Next, we further investigated the metabolic significance of chronic sleep deprivation. The gas exchange, whole-body energy expenditure, and locomotor activity of mice were measured after 4 weeks of sleep deprivation in metabolic cages. We found that sleep-deprived mice showed decreased energy expenditure as compared to the CONGFP group in both light and dark phases (p < 0.05, shown in Figure S1A and B). The diurnal rhythm of oxygen consumption and carbon dioxide generation of mice in the SDGFP group were also reduced, compared to the CONGFP group, especially in the light phase (p < 0.05, shown in Figure S1G-J). Notably, average daily feeding was also increased in these mice of the SDGFP group in the dark phase (p < 0.05, shown in Figure S1C and D). In addition, chronic sleep deprivation did not lead to any change in the diurnal rhythmicity of locomotion (shown in Figure S1E).
SCN efferent in modulating energy homeostasis
On the other hand, the total energy expenditure in mice of the SDBMAL1 group was dramatically upregulated, compared to the SDGFP group (p < 0.05, Figure S1A and B). The diurnal rhythm of oxygen consumption and carbon dioxide generation of mice in the SDBMAL1 group were also reversed (Figure S1C and D). These results meant that AAV-genetic compensation of BMAL1 in SCN could rescue the disrupted diurnal patterns in the metabolism of mice after 4 weeks of sleep deprivation. Meanwhile, this finding set us thinking about how SCN impacts the metabolism under sleep deprivation.
A wealth of evidence shows the important role of PVN in the control of feeding and metabolism; bilateral lesions of PVN lead to obesity development through dampening this nucleus which tonically controls energy balance [66–68]. PVN contains key neuroendocrine neurons controlling systemic metabolism; its neuron activity was demonstrated to be an essential component of PVN projections to autonomic control beta cell function and glucose metabolism in pancreas postsynaptic circuits [69]. There are also pre-autonomic neurons in PVN that control the sympathetic and parasympathetic branches of the autonomic nervous system (ANS), whose pivotal role in controlling energy balance was widely acknowledged [70, 71]. The dorsal and periventricular parvocellular divisions, and magnocellular neurons in PVN receive dense fiber projections from efferent connections of the SCN [36, 72]. Moreover, a previous study has demonstrated that BMAL1 in PVN neurons has shown its modulation of diurnal rhythmicity in metabolism, indicating the key role of the circadian clock [30]. In our present study, confocal images of PVN slices showed that green fluoresces, pointing to the projection neurons from SCN, were obviously reduced in the SDGFP group compared to the CONGFP group indicating a compromised projection from SCN to PVN under chronic sleep deprivation. But in the SDBMAL1 group, the projection neurons with BMAL1 overexpression were almost recovered to the level of the CONBMAL1 group observed in PVN (shown in Fig. 8A).
Neuroendocrine parvocellular neurons in PVN produce a corticotropin-releasing hormone (CRH), which targets the anterior pituitary gland to modulate the secretion of adrenocorticotrophic hormone (ACTH), in turn, releases into the bloodstream and acts on the adrenals to impetus glucocorticoid secretion; this loop is so-called hypothalamic–pituitary–adrenal (HPA) axis [73]. The cortisol/corticosterone (CORT) is widely known for its function in modulating carbohydrate ingestion and metabolism during the solar circadian cycle, whose oscillation amplitude is strongly coordinated by SCN [74–76]. AVP cells in SCN may directly innervate and/or act via inhibitory interneurons (GABAergic neurons) in DMH (dorsomedial hypothalamus) and/or SPZ (subparaventricular zone) to inhibit CRH neurons in PVN [77, 78], thereby inhibiting CORT secretion. Elevated CORT interferes with energy conservation and inhibits energy storage [79]. High CORT levels in serum may result in significant enlargement in white adipose tissue, plasma insulin, and triglyceride levels [80, 81]. The circulation CORT of mice in the SDGFP group was slightly upregulated, compared to the CONGFP group (shown in Fig. 8B) in the present chronic sleep deprivation model, which could be a fallback similar to the control in the SDBMAL1 group. Moreover, it was slightly and significantly upregulated at ZT6 and ZT12, respectively, in the wild sleep-deprived mice (p < 0.05, shown in Fig. 8C). Therefore, the elevated level of serum CORT is a well-reasoned illustration of the disrupted neuroendocrine regulation of the SCN after chronic sleep deprivation. Collectively, these findings might forward a potential mechanism underlying the disruption of SCN-PVN neural circuit leading to energy homeostasis disorder in mice caused by chronic sleep deprivation.
On the other hand, a variety of peripheral tissues might receive timing information from the SCN via neural connections in order to help regulate fundamental physiological functions such as energy metabolism [82, 83]. Specifically, the sympathetic nervous system (SNS) is responsible for SCN communication with peripheral tissues in addition to ANS, and for some tissues, this communication also involves the parasympathetic nervous system (PSNS) [84]. Accordingly, we also measured BMAL1 expression in the liver, connected with SCN by PSNS; our data showed that protein levels of BMAL1 in the liver of mice in the SDGFP group were significantly decreased as compared to the CONGFP group and surprisingly reversed in the SDBMAL1 group (p < 0.05, shown in Fig. 8E and Additional file 6: Fig. S8).
SIRT1 is regarded as a master metabolic regulator in the recent decade due to its deacetylation of several transcription factors and co-factors to the cleavage of NAD+. SIRT1 in the liver plays a vital role in gluconeogenesis and fatty acid metabolism [85, 86]. In our sleep deprivation model, compensation of BMAL1 in the SCN rescued the aberrant expressions of NAMPT and SIRT1 in the liver (shown in Fig. 8E). Additionally, the liver tissue of postmortem assumed a fatty liver change in mice of the SDGFP group, but not seen in the SDBMAL1 group, compared to the CONGFP group.
NAMPT has two different forms in mammals: intracellular NAMPT (iNAMPT) and extracellular NAMPT (eNAMPT); the latter is secreted by iNAMPT to the bloodstream in the form of extracellular vesicles [87]. SIRT1 interacts directly with its deacetylation target, K53 at iNAMPT to regulate eNAMPT secretion in adipocytes [88]. The circadian oscillation of NAD+ levels in the hypothalamus is thought to be driven by blood eNAMPT [87]. Moreover, genetic ablation of SIRT1 in adipose tissues leads to increased adiposity and insulin resistance [89]. Next, we detected SIRT1 and NAMPT expressions in the adipose and hypothalamus. The SIRT1 and NAMPT expressions in the hypothalamus were significantly decreased at ZT6, ZT12, and ZT18 in the chronic SD group, compared to controls (p < 0.05, shown in Fig. 8G–I and Additional file 7: Fig. S9), and similar expression patterns were observed in the brown adipose tissues (BAT), although there was only a significance difference at the light phase. However, the expression patterns of SIRT1 and NAMPT in BAT were different from their BMAL1 (Fig. 8K–O and Additional file 8: Fig. S10). The expression patterns of SIRT1 and NAMPT in the white adipose tissue (WAT) exhibited a different oscillation profile from BAT and the hypothalamus (shown in Fig. 8P-T and Additional file 9: Fig. S11), which were consistent with BMAL1, in contrast to BAT, probably connected with SCN by SNS. On the other hand, the circadian oscillation of serum NAMPT was altered in the chronic SD group; it was significantly upregulated at the dark phase (p < 0.05, shown in Fig. 8J), which seemed to show an association with iNAMPT in BAT, rather than the WAT.
Discussion
SCN is important for rhythmic metabolism and feeding; its outputs modulate the hypothalamic balance of life [90, 91]. In mammals, SCN is the central pacemaker and oscillator, orchestrating circadian biological rhythms in the organism. It contains ~ 20,000, mostly GABAergic neurons simultaneously co-expressing neuropeptides, generating autonomous circadian oscillations. Individual SCN neuron can be a molecular clock, driven by TTFL in cooperation with cytosolic signaling molecules, which will be activated by light through intrinsically photosensitive retinal ganglion cells (ipRGCs). IpRGCs project to retinohypothalamic tract (RHT) to release glutamate onto retinorecipient SCN neurons, a changeover into inhibitory outputs including synaptic neuronal connections and various neuropeptides [92, 93].
The two-process model of sleep regulation has been widely considered as the conceptual framework in the sleep process. The model indicates a homeostatic process (Process S) depending on sleep and wakefulness, which interacts with a process controlled by the circadian pacemaker in SCN (Process C). Many neurobehavioral functions were modulated by homeostatic sleep pressure during the circadian cycle. High sleep pressure is generally associated with lower circadian amplitude and decreased light responsiveness of the circadian clock, indicating sleep pressure alters how the clock affects behavior and physiology [94–96]. Our present results first demonstrated that sleep deprivation for 4 weeks with 6 h of opportunity sleep per day dampened the expressions of core circadian, BMAL1, in the SCN of mice. Moreover, the downregulations occurred at each time point, further verifying that high sleep pressure caused by chronic sleep deprivation led to a lower circadian “set point” (amplitude). AVP and VIP are robustly expressed in the dorsomedial “shell” or ventrolateral “core” sub-region of the SCN, respectively. AVP has been implicated in feeding, energy homeostasis, and locomotor activity [33, 97, 98] and is considered to serve as an SCN output [99]. Our confocal images showed that the reductions of BMAL1 in situ expressions were mainly manifested in the “shell” co-stained with AVP at each time point. BMAL1 is one of the essential transcription factors of the TTFL, and its absence from different types of SCN neurons is sufficient for aberrant behaviors in mice [33, 62]. Especially, the lack of BMAL1 in AVP neurons of SCN causes extinct disruption of circadian behavior rhythm in mice; besides, AVP is also a downstream target of BMAL1 [33]. On this ground, we speculated that chronic sleep deprivation might mainly weaken the BMAL1 expressions in AVP neurons, located in the shell of SCN. Additionally, BMAL1 regulates the expression of Prok2 and Rgs16 in the dorsal SCN. Prok2 signaling pathways may be crucial for the interneurons of the SCN and neuronal connection with the extra-SCN region via the Prok2 receptor, such as ARC [63]. Rgs16 increases cAMP production by inactivating Gi signaling through GTP hydrolysis. A noteworthy characteristic of VIP receptor 2 is that it is Gs-coupled, raising the possibility that the expression of Rgs16 may enhance VIP signaling to coordinate synchronization between pacemaker neurons within SCN [100, 101]. Given that activation of VIP cells in the core, for the most part, depends on light exposure, chronic sleep deprivation might compromise cellular synchrony via disrupting the VIP receptor function, although the light/dark cycle was not changed in our animal model.
Next, we determined the mechanisms underlying BMAL1 modulation in the SCN by injection of AAV1-hSyn-Bmal1 into the shell region. The results implicated that compensation BMAL1 could rescue the decreased expressions of core clock genes in the hypothalamus. BMAL1 and CLOCK form heterodimers and active transcription of Per, Cry, and Rev-Erbα genes through E-box enhancers. For the positive feedback loop, increasing REV-ERBα levels act through Rev-Erb/ROR response elements in the Bmal1 promoter to repress its transcription [102]. In our results, REV-ERBα was upregulated at both the transcriptional and translational levels after chronic sleep deprivation, inhibiting the mRNA levels of BMAL1. Interestingly, compensation of BMAL1 could also downregulate the REV-ERBα expressions both in protein and mRNA levels, which means the positive feedback of this loop could be rescued, maybe through upregulating the expressions of the CCGs, including RORα and NAMPT that also be reversed as compared to chronic SD group. In addition to TTFL, circadian oscillates also in kinase pathways, energy metabolism (AMP/ATP ratio), and redox state (NAD+/NADH ratio) linking the molecular clock to cellular physiology and metabolism [103]. SIRT1, a sirtuin of the HDAC class III family, acts as a rheostat of the circadian clock, exerting its control predominantly on the amplitude of CCG expression in the SCN [104]. SIRT1 could participate in circadian control by regulating the HAT function of CLOCK; its reaction requires the coenzyme NAD+ [46]. It also directly activates the transcription of Bmal1 via its co-activator PGC-1α to increase its expression amplitude in the SCN [48]. Our results revealed that the protein levels of SIRT1 in the hypothalamus were also decreased at each time point, indicating its role in downregulated Bmal1 induced by chronic sleep deprivation. NAD+ acts as a coenzyme of SIRT1, poly (ADP-ribose) polymerase-1 (PARP-1), and cADP-ribose synthases (CD38) to deplete its intracellular stores. NAM can be catalyzed into NAM mononucleotide (NMN) by NAMPT. Nampt is one of the metabolic output targets of CLOCK:BMAL1, and this feedback loop depends on the oscillation of NAD+ level in a circadian pattern in different cell types. Furthermore, the NAD+-activated SIRT1 feeds back into the NAD+ salvage pathway by directly regulating Nampt [34]. The NAMPT expression in the hypothalamus was reasonably well-reversed by the compensation of BMAL1 under our chronic sleep deprivation model and might, in turn, increase the production of NMN, the key ingredient of NAD+. Furthermore, Wang et al. conducted a convincible study that redox state regulated excitability in SCN neurons through non-transcriptional regulation of multiple K+ channels [105]. Hence, their results implied that changes in cellular metabolic state could be the “cause,” rather than the “result,” of neuronal activity. On the other hand, the sensitivity of clock gene transcription to the redox state will modulate circadian and energetic cycles at the transcriptional level [106, 107]. The circadian redox oscillations in rodent SCN require functional molecular clockwork involving the clock gene, Bmal1 [105]. Collectively, chronic sleep deprivation may firstly impact the redox state of the SCN, in turn weakening SIRT1 activity, and then the core gene Bmal1 was downregulated at the transcriptional level. Hence, the decreased expression of BMAL1 further induced dysfunction of SCN neurons including an imbalance of redox state directly concerning decreased Nampt expression and reduced neurotransmitter levels and neuronal activity, which dampened the neuronal projection to extra-SCN in the brain. Further studies need to be done in the future to confirm our hypothesis, whether redox state is the primary target of chronic sleep deprivation.
The present examinations were performed at an earlier age window since we recorded EEG for the 5–9 weeks age of mice, which is equivalent to the adolescence to young adult stage for humans. Our data indicated that the AAV-genetic compensation of BMAL1 did not change the power spectra during the NREM and WK stages at baseline. One reason for this phenomenon could be due to the TTFL regulation under physiological conditions, since the expressions of clock genes and downstream signals were not altered in the CONBMAL1, compared to the CONGFP. Additionally, a previous study has demonstrated that the SCNVIP neurons, but not SCNAVP neurons, are responsible for the amplitude of the sleep–wake circadian rhythm [62]. Moreover, as deciphered above, the AVP neurons located in the shell of the SCN, rather than the core where the VIP neurons are mainly expressed, were involved in our chronic sleep deprivation model. It has been well established that there is a significant negative correlation was obtained between SCN neuronal activity and SWA in the EEG [108, 109]. The neuronal activity in the SCN was significantly higher during the NREM deprivation and SWA declined below the baseline, but once the mice recovered to sleep, then SWA increased [108, 109]. Mice in the SDGFP and SDBMAL1 groups exhibit decreased SWA relative to the baseline, and the SWA of the SDGFP group was only elevated by 30% in the opportunity sleep state, compared to the sleep deprivation state. That meant the sleep state of mice could not be compensated during 6-h opportunity sleep each day after sleep deprivation for 4 weeks. Meanwhile, there was no obvious difference in the SWA between the SDGFP and SDBMAL1 groups. This result indicated that exogenous BMAL1 in the SCN did not alter the sleep–wake profile. Interestingly, after sleep deprivation, mice in the SDBMAL1 group showed less sleep debt to adapting sleep deprivation and recovery.
There is mounting epidemiological evidence that sleep loss is linked to obesity in children and adults worldwide [110]. Lack of sleep alters appetite-regulating hormones and increases calorie consumption [111]. The mechanisms by which insufficient sleep leads to the disorder of appetite regulation are uncertain. Our findings revealed that chronic sleep deprivation increased food consumption by upregulating the expressions of AgRP and NPY and reducing the expression of POMC in the ARC of the hypothalamus. This disorder could be attributed to compromised JAK2/STAT3 signal pathways in AgRP/NPY and POMC neurons. The phosphorylated STAT3 actives satiety signaling POMC neurons and inhibits hunger-signaling AgRP/NPY neurons [32, 112, 113]. POMC neurons exhibit the anorexigenic function and augment energy expenditure through the post-transcriptional product of the POMC gene, including α-MSH, that binds to the MC3/4R in PVN [112]. NPY/AgRP neurons increase food intake and decrease energy expenditure through the release of NPY, GABA, and AgRP. AgRP exerts GABAergic-mediated inhibition on POMC neurons and antagonizes α-MSH action [114]. For answers to whether the SCN comes into play intermediate mechanisms underlying chronic sleep deprivation changed expressions for appetite regulation peptides in the ARC, since it was a target of Prok2 afferent fibers from the SCN [63, 115], we further observed the neuronal projection from SCN to ARC, which was impaired after chronic sleep deprivation, in contrast, the impaired projections could be rescued by injection with AAV1 carrying Bmal1 into the SCN. AAV1 can also be expressed across anterograde trans-synapses after infection for 6 weeks in our chronic sleep deprivation animal model. Although its transmission efficiency is low and without cell specificity, it can still partially interpret the disruption of the neural circuit between the SCN and its doubtless downstream area of the brain, ARC. The molecular oscillation of the TTFL in SCN synchronizes clocks in the extra-SCN region through interlinking electrical activity and neurotransmitters [58, 59]. TTFL orchestrates the spontaneous firing rate of neurons via driving Na+ and K+ conductance and Ca2+ oscillation, thus synchronizing postsynaptic neurons [60]. SCN output neurons release several output factors including GABA, AVP, VIP, and Prok2 onto target neurons of extra-SCN [62–65]. Furthermore, BMAL1 was found to modulate the GABAA γ2 receptor in the PVN [30]. Therefore, with all the opinions above, the BMAL1-mediated molecular clock in the neurons of the SCN may act on the TTFL of the downstream areas of the brain through neuronal firing and neurotransmitters including GABA, AVP, and Prok2. Accordingly, compensation of BMAL1 in the SCN could rescue the depletion of BMAL1 in the ARC through the restored innervation to the ARC and then rescue the disruption of appetite regulation through reversing STAT3 signals and the disorder of expressions for orexigenic and anorexigenic peptides in the ARC of the hypothalamus. SOCS3, a target gene of STAT3, can dephosphorylate JAK2 to terminate this signal transduction [116], which was increased in our chronic sleep deprivation model. The upregulated SOCS3 was also reversed by overexpression of BMAL1, which is probably due to the direct action of BMAL1: CLOCK since the role of CLOCK in the modulation of the expression of SOCS3 in the ARC of the hypothalamus has been reported [52]. It should be noted that feeding behavior can also be regulated by SIRT1 in the ARC by deacetylation of FoxO1 in POMC and AgRP/NPY neurons [117–119]. FoxO1 activates POMC and suppresses AgRP transcription [118]. The deacetylation by SIRT1 in POMC neurons is necessary for homeostasis in defenses against diet-induced obesity. Diet-induced obesity is hypersensitive to SIRT1 deficiency due to decreased energy expenditure in POMC neurons [117]. SIRT1 in POMC neurons is required for the brown adipose tissue-like remodeling of the perigonadal white adipose tissue through sympathetic activation [117].
The present study further revealed a reduction in whole-body energy expenditure of adolescent mice after chronic sleep deprivation without changing locomotion activity, indicating abnormal neuroendocrine mechanisms related to metabolism. The important role of PVN in regulating energy balance is well established; it can integrate inputs from other sub-regions of the hypothalamus [120, 121]. The dorsal and periventricular parvocellular divisions of PVN receive dense fiber projections from the SCN [72]. The disrupted projections from SCN to PVN were observed under chronic sleep deprivation in the present study, which could be rescued with overexpression of BMAL1. There are direct SCN inhibition projections to CRH neurons in the PVN to regulate daily CORT rhythmicity through AVP and VIP [78, 92]. The CORT plays an important role for its function in modulating carbohydrate ingestion and metabolism [74–76]. In our sleep deprivation animal model, the CORT levels in the serum of chronic sleep-deprived mice were slightly upregulated at ZT6 as compared to the CONGFP group, which was reversed in the SDBMAL1 group. Interestingly, the CORT level in the serum of chronic sleep deprivation mice was first slightly increased at ZT6, consistent with our compensation intervention experiments, and significantly increased at ZT12 after recovery to opportunity sleep for 6 h. The elevated circulating CORT level reflected a disorder of SCN efferent since SCN neurons are multi-synaptically connected to the adrenal via SNS or pre-autonomic PVN neurons involved in the HPA axis [84]. On the other hand, AgRP neurons in the ARC release GABAergic signaling to the MC4R neurons in the PVN to promote feeding [122]. MC4Rs are densely expressed in the PVN and action in the PVN is significant for restraining feeding [123]. Reciprocally, there are abundant glucocorticoid receptors in AgRP neurons of the ARC, providing feedback to the autonomic PVN neurons [124]. Therefore, higher circulating CORT during the light phase caused by chronic sleep deprivation may activate the AgRP neurons to magnify the appetite-promoting effect, verified by the increased expressions of AgRP also occurring in the light phase.
As illustrated above, SIRT1 plays an important role in maintaining oscillation machinery in the SCN and acts as a master metabolic regulator [46]. The protein levels of SIRT1 and NAMPT in the hypothalamus were decreased at each time point, and those in WAT and BAT were mainly reduced during the light phase in the wild sleep-deprived mice, compared to the control. A previous study has demonstrated that genetic ablation of SIRT1 in adipose tissues leads to increased adiposity and insulin resistance [89]. SIRT1 promotes fat mobilization in WAT and drives lipid utilization in the liver and muscle via PPARγ [125] and also enhances energy expenditure by deacetylating PPARγ to facilitate Prdm16 binding [126]. Our results indicated that chronic sleep deprivation might compromise the SIRT1 function in WAT and BAT, predisposing adipose accumulation and decreased thermogenesis. Besides, it has been reported that in both brown and white adipose tissues, Nampt ablation impairs many core clock genes and disrupts the circadian rhythm of many metabolic pathways, especially those related to lipid metabolism [127]. Our results showed that the oscillation amplitude of NAMPT was dampened with reductions of protein levels in both WAT and BAT after chronic sleep deprivation. SIRT1 deacetylates the iNAMPT to regulate the eNAMPT secretion from adipose tissues [88]. The circulating eNAMPT can be derived from adipocytes, hepatocytes, and leukocytes [87, 88, 128, 129]. A prior investigation has indicated that the release of eNAMPT from adipocytes rises during fasting periods [88]. This study argues against adipose tissue-derived eNAMPT being a contributor to eNAMPT elevations during the physiological feeding period [87]. Our result indicated that chronic sleep deprivation obviously disrupted the circulating eNAMPT rhythmicity. The most recent study suggested that the circadian rhythm of circulating eNAMPT can be altered in individuals with obesity [130], though the relationship between circulating eNAMPT and obesity is still uncertain. Furthermore, this study also demonstrated that systemic eNAMPT activity failed to significantly affect the SCN clock activity but could regulate Pomc and Agrp transcription in the ARC [130]. A finely conducted work has revealed that Nampt knockout in the liver dampens circadian oscillations [131]. SIRT1 in the liver supports gluconeogenesis via PGC-1α and FOXO1. SIRT1 also promotes fatty acid oxidation by activating PPARα and inhibits fatty acid synthesis by targeting SREBP1c for degradation in the liver [85, 86]. Our results revealed that chronic sleep deprivation could reduce the expressions of BMAL1, PER1, SIRT1, and NAMPT in the liver that were rescued with compensation of BMAL1 in the SCN, which could be deciphered as the communications of the SCN with the liver through PSNS [84].
Conclusions
Given the steep rise in obesity in industrialized nations, coupled with the continuous decline in sleep duration, it is crucial to understand the association between sleep loss and obesity. The SCN emits temporal afferents to regulate energy balance by firing rate and neurotransmitters to fine-tune its charge of different endocrine functions, but how the codes changed under chronic sleep deprivation is unknown. Our study revealed that chronic sleep deprivation for 4 weeks impaired appetite regulation and decreased energy expenditure through dampening BMAL1 expression, probably in the transcription level, in the SCN, which in turn affected the neuron projections to ARC and PVN. SIRT1 may be a potential intervention target since it participates in the disruption of the BMAL1 regulation loop and metabolic disorder induced by chronic sleep deprivation. Lacking specific targeting of the overexpression of BMAL1 in the AVP neurons using AVP-Cre mice is one scientific limitation of the present manuscript, and we will further explore the causality between sleep deprivation and BMAL1 in AVP neurons in AVP-Cre mice in the future and artificially activate the AVP neurons using optogenetic and chemogenetic technologies to demonstrate its role in regulating food intake, especially under chronic sleep deprivation conditions.
Methods
Animals
Mice were bred and housed in the animal facilities in the Animal Department in China Medical University, in accordance with National Institutes of Health Guide for the Care and Use of Laboratory Animals. All experimental protocols were approved by the China Medical University Animal Care and Use Committee, following the National Institutes of Health Guide for the Care and Use of Laboratory Animals. The experimental animals in this study were Kunming mice (SPF grade, a wild nocturnal active type) purchased from Huafukang Biotechnology (China). All mice were fed standard chow ad libitum under a 12:12 h light:dark cycle (lights on 7:00 am–7:00 pm, ZT0-ZT12; lights off 7:00 pm–7:00 am of next day, ZT12-ZT24) and housed at a constant temperature (22 ± 2℃) and humidity (40–60%). Adaptive feeding was performed for 1 week before the experiment started. EEG and EMG electrode implantation surgery and virus injection were performed at the age of 3 weeks (shown in Fig. 1A).
Reagents
TRIzol reagent was obtained from Takara (Japan). RIPA lysis buffer and bicinchoninic acid (BCA) protein assay kit were purchased from Beyotime Biotechnology (China). The enhanced chemiluminescence (ECL) plus kit was obtained from MilliporeSigma (USA). The primary antibodies against phosphorylated-JAK2 (p-JAK2, Tyr1007/Tyr1008, #3771S, RRID: AB_330403), JAK2 (#3230S, RRID: AB_2128522), SOCS3 (#52113S, RRID: AB_2799408), phosphorylated-STAT3 (p-STAT3, #9145S, RRID: AB_2491009), STAT3 (#4904 T, RRID: AB_331269), BMAL1 (#14020S, RRID: AB_2728705), REV-ERBα (#13418S, RRID: AB_2630359), NPY (#11976S, RRID: AB_2716286), GAPDH (#2118S, RRID: AB_561053), and β-actin (#4970, RRID: AB_2223172) were procured from Cell Signaling Technology (USA). The primary antibodies against LepRb (#ab5593, RRID: AB_304969), CLOCK (#ab3517, RRID: AB_303866), and POMC (#ab210605, RRID: AB_3073977) were purchased from Abcam (USA). The primary antibody against AgRP (#Sc-518077, RRID: AB_3099749) was purchased from Santa Cruz Biotechnology (USA). The primary antibodies against PER2 (#AF4601, RRID: AB_2844559) and PER1 (#DF9080, RRID: AB_2842276) were purchased from Affinity Biosciences (China). The primary antibodies against SIRT1 (#A11267, RRID: AB_2861537), RORα (#A6971, RRID: AB_2767528), CRY1 (#A13662, RRID: AB_2760523), and CRY2 (#A6891, RRID: AB_2767450) were purchased from ABclonal (China). The primary antibodies against TCPTP (#YN1458, RRID: AB_3099750), PTP1B (#YT3900, RRID: AB_3099752), and NAMPT (#YT6086, RRID: AB_3099751) were purchased from ImmunoWay (USA).
All other chemicals were of analytical grade and obtained from local chemical suppliers. These chemical reagents were prepared as stock solutions with sterile water and then diluted to the final concentrations before application. The water used in this study was double distilled.
Chronic sleep deprivation procedure
The paradigm of chronic sleep deprivation (SD) is shown in Fig. 1A. Mice were deprived of sleep with an automated sleep deprivation (ViewPoint Life Sciences) device. The animals were flexible in a shaker apparatus (cylindrical, which is transparent). There is a shaker device at the bottom of the deprivation chamber, which forms a connection between vibration and the computer via an adaptor. The computer is installed with an EEG/EMG signal collection program. Mice were settled into the chamber before sleep deprivation started, where they could obtain water and food ad libitum. Then a 4-week SD (total 18 h per day for SD, ZT6-ZT12 for opportunity sleep of each day) was produced by activating the device. The shaking of the chamber bottom platform is launched by programmed electromagnetic pulses to keep the mice awake during sleep deprivation. The parameters of the electromagnetic pulses are as follows: SD, a sequence of random 2 to 4 pulses (20 ms duration) sent at 2 Hz every random 0.15–0.33 min. For mice with simultaneous EEG/EMG recording, four mice were sleep-deprived/recorded at the same time. The parameters of the SD protocol have shown that the SD process may not induce acute stress since the circulating corticosterone levels did not change after acute sleep deprivations for 6, 12, 18, or 24 h (data not shown).
Animal experiments timeline
In the first part of our study, mice were randomly divided into two groups: the control group (CON) and the chronic sleep deprivation group (SD); the brain tissues and serum were taken immediately at ZT6, ZT12, ZT18, and ZT24 on the last sleep deprivation day and stored at − 80℃ until analysis. EEG/EMG electrodes were implanted 1 week before the sleep deprivation process (as shown in Fig. 1A).
In the intervention study, mice were randomly divided into four groups: the control group with empty vector (CON + GFP, CONGFP), the control group with AAV-genetic overexpression of BMAL1 (CON + AAV1-hSyn-Bmal1, CONBMAL1), the chronic SD group with empty vector (SD + GFP, SDGFP), and the chronic SD with AAV-genetic overexpression of BMAL1 group (SD + AAV1-hSyn-Bmal1, SDBMAL1). When mice were under anesthetization, EEG/EMG electrodes were implanted directly followed by the viral injection (as shown in Fig. 4A and B); 5 weeks were allowed for viral expression in mice after viral injection surgery, and 1 week was allowed for mice to recover from electrode implantation. Mice could acclimatize to the deprivation chamber for 1 h per day, at least 5 days before chronic sleep deprivation started. After baseline recording, mice were sleep-deprived for 18 h per day starting at night onset (7:00 p.m. to 1:00 p.m. the next day) for up to 4 weeks. EEG/EMG signal was collected on the last day of 4 weeks of chronic sleep deprivation day from 7:00 am to 7:00 am the next day (total for 24 h, ZT6-ZT12 for opportunity sleep), then the animals were immediately sacrificed by decapitation at the end of chronic sleep deprivation at 1:00 p.m. (ZT6). Samples were taken immediately and stored at − 80℃ until analysis.
Viral injection
AAV1-hSyn-Bmal1 was purchased from Genechem (Shanghai, China). Three-week-old mice were anesthetized using isoflurane inhalation, placed on a brain stereotactic holder, and drilled with bilateral holes into the skull (AP = − 0.47 mm; ML = ± 0.45 mm; DV = − 5.65 mm). A glass pipette was attached to a microsyringe (Hamilton), which led to a bilateral infection of the hypothalamus; 1 μL of AAV1-hSyn-Bmal1 (1.725 × 1012 viral genomes per mL) viral infusions were injected at a rate of 200 nL min-1. The AAV vector expressing GFP alone served as the control vector. The recombinant AAV virus was injected at the SCN bilaterally in the mice. After each injection, the needle or glass pipette was kept still for 5 min before slowly withdrawing. Mice were recovered for 6 weeks after surgery to achieve the viral induction process.
EEG/EMG electrode implantation
Three weeks of age mice were fixed on the brain stereotaxic apparatus after anesthetization in an isoflurane-inducing chamber. The body temperature of mice was maintained using a heating pad and a gas mask was used to maintain isoflurane anesthesia throughout the surgery procedure. Before surgery, an erythromycin eye ointment was applied to protect mice’s eyes. The mice were placed in a cage on the heating pad when EEG and EMG electrodes were implanted until fully awake. The mouse head was shaved and the skull surface was exposed with a midline scalp incision, then wipe off the connective tissue on the skull surface. Three small skull holes (0.5–1 mm in diameter) were drilled with the skull drill, then stainless steel mini-screws (China) for EEG were implanted into the skull above the frontal electrode (stereotaxic coordinate bregma as a reference: AP = + 2 mm, LAT = + 1 mm), parietal electrode (AP = − 2 mm, LAT = − 1.5 mm), and reference electrode (AP = − 6 mm, LAT = + 2 mm). Miniscrews were soldered to Teflon® coated semi-rigid silver wires (0.003 inch in diameter). Two semi-rigid silver wires served as EMG electrodes and were inserted into the trapezius muscle ~ 5 mm apart and sutured in place for EMG recording. The electrodes were previously soldered to a connector, which was mounted on the skull using dental cement.
EEG/EMG recording
Implanted surgery was described above, mice were allowed to recover for 1 week in their cage and fed standard chow ad libitum. EEG and EMG recording across a complete 24-h light–dark cycle was performed on the day before chronic sleep deprivation and recorded as the baseline.
EEG/EMG signals were collected by a new device to quantify sleep (ONEIROS), the system can be used either as a data logger, by using embedded media storage, or as a telemetry device for real-time monitoring and analysis of the signals [132]. Wakefulness (WK), NREM, and REM episodes longer than 5 s were quantified based on previously described criteria [108]. In brief, WK is characterized by desynchronized small-amplitude EEG and extensive EMG activity. NREM is characterized by synchronized, large-amplitude delta (0.25–4 Hz)-dominant EEG and reduced EMG activity compared to WK. REM is featured by an EEG of high frequency (4–9 Hz-dominant) and relatively uniform amplitude smaller than that in NREM and often associated with a flat EMG. The relative power of delta (0.5–4 Hz), theta (4–9 Hz), and alpha (9–20 Hz) bands are calculated as a fraction of total power (0.5–20 Hz) for each animal. NREM sleep pressure is indicated as the calculation of the average power density of NREM, also known as slow-wave activity (SWA, power in the 0.5–4 Hz range) is determined for 30 s epochs [133, 134]. Polygraphic signals are amplified and bandpass filtered (0.3–30 and 30–300 Hz for EEG and EMG, respectively) by a polysomnograph (Grass Model 9, Grass Instruments Co.). Along with signals from the motion detectors, these polygraphic signals are digitized (sampling rate of 250 Hz) and stored on a computer using data collection software (SleepSign, Kissei Comptec Co.) for subsequent offline analysis.
Body weights and food intake
The body weights of the mice were measured twice a week for 4 weeks. Each measurement was taken at 1:00 p.m. (ZT6). Food intake was measured daily (ZT0 and ZT12) by calculating the amounts of food consumed in each cage and dividing them by the number of mice in the cage. Food intake was presented as g/day/mouse.
Western blot analysis
Mice were sacrificed at the end of chronic sleep deprivation. Samples for Western blot experiments were taken immediately into tubes then were put in liquid nitrogen and stored at − 80℃ until analysis. Total protein samples derived from the tissues were lysed with RIPA lysis buffer. Aliquots of each sample (20 µg per lane) were loaded onto 8–12% SDS–polyacrylamide gel electrophoresis (PAGE). The gels after electrophoresis were transferred onto polyvinylidene difluoride (PVDF, Millipore, USA) membranes in trans-buffer electronically. The membranes were incubated with primary antibodies against BMAL1 (1:1000), CLOCK (1:500), PER1 (1:1000), PER2 (1:1000), CRY1 (1:1000), CRY2 (1:1000), LepRb (1:1000); p-JAK2 and JAK2 (1:500); p-STAT3 (1:1000); STAT3 (1:1000); SOCS3 (1:1000); POMC (1:1000), NPY (1:500), AgRP (1:1000), SIRT1 (1:1000), NAMPT (1:2000), REV-ERBα (1:2000), RORα (1:2000), TCPTP (1:2000), PTP1B (1:2000), β-actin (1:1000), and GAPDH (1:1000) overnight at 4℃. On the following day, membranes were incubated with corresponding secondary antibodies for 1 h at 22 ± 2℃. The BCA protein assay kit was used to determine protein concentrations in lysates. Analysis was performed with the multifunctional imaging system (Tanon5200, China). β-actin or GAPDH was used as the internal standard. Semi-quantifications of the bands were performed using Image J software.
Realtime RT-qPCR
Quantitative real-time PCR (RT-qPCR) was conducted according to the minimum information for publication of quantitative real-time PCR experiments (MIQE) guidelines; 1 µg of total RNA was extracted from the tissues with Trizol reagent. The cDNA was synthesized from total RNA using a PrimeScript RT reagent kit (Takara, Japan) and served as a template for real-time PCR amplification. TB Green (Takara, Japan) was used in QuantStudio 6 Flex fluorescence quantitative PCR instrument (Thermo Fisher, USA) for real-time PCR progress. Primer sequences for target genes are in the list of oligos in Table 1. The comparative CT method (ΔΔCT) was used to quantify the target genes [135]. RNA abundance was expressed as 2−ΔΔCT for the target gene normalized against β-actin (as the internal control) and presented as fold change vs. control sample.
Energy expenditure, basal locomotor activity, and body composition
Energy expenditure of mice was measured after sleep deprivation for 4 weeks; briefly, mice were housed for 5 days in the eight-channel Promethion system (Sable Systems International) cages. Mice were settled into the cages and were acclimatized for 24 h. Individual cages include a ceiling-mounted food hopper and water spigot, where they could obtain water and food ad libitum, and simultaneously, carbon dioxide production (Vco2), oxygen consumption (Vo2), food intake, and locomotor activity were measured. Locomotor activity was recorded by the photoelectric beam motion detectors. Airflow through the chamber was negative (2000 mL/min), and gas analyses were recorded once per minute. The cages are mounted inside thermally controlled cabinets that are maintained at 24℃ to diminish heat loss. Energy expenditure was calculated as 3.815 × Vo2 + 1.232 × Vco2. Data collected from days 2–5 were used for analyses.
At the end of chronic sleep deprivation, mice underwent body composition analyses after multiplexed phenotyping. Body composition was assessed using time-domain nuclear magnetic resonance (TD-NMR, Bruker Minisp LF50, Germany). These in vivo experiments were performed once, with numbers of animals (n values) indicated in figure legends.
Leptin, NPY, α-MSH, NAMPT, and corticosterone levels in serum
Blood was taken immediately under anesthetization with inhaled isoflurane from the retrobulbar intraorbital capillary plexus then centrifuged and obtained serum was frozen at − 80℃. The serum leptin level was assessed using an ELISA kit (Quantikine ELISA Kit R&D Systems, United States) according to the manufacturer’s instructions. The level of α-MSH was measured using commercially available ELISA kits (Enzyme-Linked Biotechnology Co., Shanghai, China). The serum NPY, NAMPT, and corticosterone levels were measured using ELISA kits purchased from Elabscience (China). The ELISA kits use the double-antibody sandwich method, the absorbance of samples is read at 450 nm. Data were calculated using an appropriate standard curve generated by the ELX800 ELISA plate reader (BioTek Instruments, USA).
Immunofluorescence staining
The brains were collected from cardiac-perfused mice with 50 mL saline followed by 50 mL 4% paraformaldehyde via the left ventricle of the heart under anesthesia and quickly placed in a deep freezer at − 80℃. The brain slices (20 μm thickness) were blocked with 5% BSA and 0.5% Triton X-100 in phosphate-buffered saline (PBS). Bovine serum albumin (BSA) was added and blocked for 30 min, followed by incubation with primary antibodies, respectively: rabbit anti-p-STAT3, anti-AgRP, anti-NPY, and anti-POMC antibody at 4℃ overnight. The primary antibody dilution was 1:50. After washing with PBS, slices were incubated with Alexa Fluor 594-conjugated anti-rabbit secondary antibody or Alexa Fluor 488-conjugated anti-mouse secondary antibody at room temperature for 1 h, with a dilution of 1:500. Slices were washed, mounted, and imaged using a multi-channel laser scanning confocal microscope (Nikon AXR). Immunofluorescence staining for detecting the protein expressions in the SCN and ARC for multiple time points was performed using the Quadruple-Fluorescence immunohistochemical mouse/rabbit kit (RS0037, ImmunoWay Biotechnology), following the manufacturer’s protocol. Two slices of three brains for each group were stained. For negative controls, the primary antibodies were omitted.
Statistical analysis
Results were represented as means ± standard deviations unless otherwise specified in the legends. The samples “n” represented biological replicates (outliers were omitted), and data analysis was performed with SPSS v22.0 (SPSS Inc., USA). The band of Western blot intensities and average immunofluorescence intensities were quantified using Image J and presented as bar graphs or line charts (for multiple time points) to show relative quantification. Normality was tested by the Shapiro–Wilk test. The comparisons between two groups with multiple time points were analyzed with a two-way repeated-measures ANOVA and pairwise comparisons. Multi-group data for different time points, such as food intake and weight gain of the mice were analyzed with two-way repeated-measures ANOVA with Tukey’s post hoc test. One-way ANOVA with LSD post hoc test was used to determine the differences in the intervention experiments, and p < 0.05 was considered statistically significant.
Supplementary Information
Supplementary Material 1: Figure S1 Chronic sleep deprivation-induced disruption of energy expenditure and food consumption. A and B. Representative energy expenditurerecorded by metabolic cages in light-dark conditions in the mice under continuous monitoringor total energy expenditure. C and D. Representative food consumedrecorded by metabolic cages in light-dark conditions in the mice under continuous monitoringor total food consumed. E and F. Representative locomotor activityrecorded by metabolic cages in light-dark conditions in the mice under continuous monitoringor total locomotor activity. G and H. Representative oxygen consumptionrecorded by metabolic cages in light-dark conditions in the mice under continuous monitoringor total oxygen consumption. I and J. Representative carbon dioxide productionrecorded by metabolic cages in light-dark conditions in the mice under continuous monitoringor total carbon dioxide production. Data were presented as the means ± standard deviations and presented as bar graphs. Significant difference * was defined as p<0.05 by one-way ANOVA. CONGFP, the control group with empty vector; CONBMAL1, the control group with AAV-genetic overexpression of BMAL1; SDGFP, the chronic SD group with empty vector; SDBMAL1, the chronic SD with AAV-genetic overexpression of BMAL1. ZT, Zeitgeber time. Data were presented as the means ± standard deviations. Significant difference * was defined as p<0.05 by one-way ANOVA. CONGFP, the control group with empty vector; CONBMAL1, the control group with AAV-genetic overexpression of BMAL1; SDGFP, the chronic SD group with empty vector; SDBMAL1, the chronic SD with AAV-genetic overexpression of BMAL1. Figure S3 The original blots for Figure 2A. Figure S4 The original blots for Figure 2H. Figure S5 The original blots for Figure 6A. Figure S6 The original blots for Figure 6Q. Figure S7 The original blots for Figure 6T. Figure S8 The original blots for Figure 8E. Figure S9 The original blots for Figure 8G. Figure S10 The original blots for Figure 8K.Figure S11 The original blots for Figure 8P. Figure S2 Compensation of BMAL1 in the SCN rescued the in situ expressions of BMAL1 in the ARC A. Representative IF staining for BMAL1, DAPI and merged imagesin the ARC. B. The average IF intensities of BMAL1 among groups were determined by Image J software and presented as bar graph. Mean intensity = Integrated Density/Area.
Acknowledgements
Appreciation for the National Natural Science Foundation of China (NSFC, 81903342) and the High-Quality Development of China Medical University supported by the Science and Technology Department of Liaoning Province (2022 JH2/20200013).
Data sharing statement
All the authors listed approve to publish the data of this study, and dataset generated during and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Abbreviations
- AAV
Adeno-associated virus
- ACTH
Adrenocorticotrophic hormone
- AgRP
Agouti-related peptide
- ANS
Autonomic nervous system
- AP
Anteroposterior
- ARC
Arcuate nucleus
- AVP
Arginine vasopressin
- BCA
Bicinchoninic acid
- BL
Baseline
- BMAL1
Brain and muscle arnt-like 1
- BSA
Bovine serum albumin
- cAMP
Cyclic adenosine monophosphate
- CART
Cocaine- and amphetamine-regulated transcript
- CCGs
Clock-controlled genes
- CLOCK
Circadian locomotor output cycles kaput
- CON
Control
- CORT
Corticosterone
- CRH
Corticotropin-releasing hormone
- CRY
Cryptochrome
- DV
Dorsoventral
- ECL
Enhanced chemiluminescence
- EEG
Electroencephalogram
- ELISA
Enzyme-linked immunosorbent assay
- EMG
Electromyogram
- eNAMPT
Extracellular NAMPT
- GAPDH
Glyceraldehyde-3-phosphate dehydrogenase
- GFP
Green fluorescent protein
- HPA
Hypothalamo–pituitary–adrenal
- IF
Immunofluorescence
- iNAMPT
Intracellular NAMPT
- ipRGCs
Intrinsically photosensitive retinal ganglion cells
- JAK2
Janus kinase 2
- LAT
Lateral
- LepRb
Leptin receptor
- NAMPT
Nicotinamide phosphoribosyltransferase
- NAD
Nicotinamide adenine dinucleotide
- NPY
Neuropeptide Y
- PAGE
Polyacrylamide gel electrophoresis
- PER
Period
- POMC
Proopiomelanocortin
- PTP1B
Protein tyrosine phosphatase 1B
- PTP
Protein tyrosine phosphatases
- MC3R
Melanocortin 3 receptor
- MC4R
Melanocortin 4 receptor
- ML
Mediolateral
- NMN
Nicotinamide mononucleotide
- NREM
Non-rapid eye movement
- PBS
Phosphate-buffered saline
- ProK2
Prokineticin 2
- Prdm16
PR domain-containing 16
- PSNS
Parasympathetic nervous system
- PVDF
Polyvinylidene difluoride
- PVN
Paraventricular nucleus
- REM
Rapid eye movement
- REV-ERBα
Nuclear receptor subfamily1
- Rgs16
Regulator of G protein signaling 16
- RHT
Retinohypothalamic tract
- RORα
Retinoid acid receptor-related orphan receptor alpha
- RRE
REV response element
- RT-qPCR
Quantitative real-time PCR
- SCN
Suprachiasmatic nucleus
- SD
Sleep deprivation
- SH2
Src homology 2
- SIRT1
Sirtuin1
- SOCS3
Suppressor of cytokine signaling 3
- SNS
Sympathetic nervous system
- STAT3
Signal transducer and activator of transcript 3
- SWA
Slow-wave activity
- TCPTP
T cell protein tyrosine phosphatase
- TD-NMR
Time-domain nuclear magnetic resonance
- TTFL
Transcriptional translation feedback loop
- VGCC
Voltage-gated calcium channels
- VIP
Vasoactive intestinal polypeptide
- WK
Wakefulness
- ZT
Zeitgeber time
- α-MSH
α-Melanocyte-stimulating hormone
Authors’ contributions
TD and SL performed and interpreted the experiments. TD and QS wrote the manuscript. LH and JH performed metabolic cages parts of the experiments. TH performed EEG recordings and analysis. HY and LG performed fluorescence image acquisition. XS performed parts of the experiments. LJ, YY, and QS edited the manuscript. QS conceived the study, and designed and revised the manuscript. All authors read and approved the final manuscript.
Funding
This study was funded by NSFC (81903342) and High-Quality Development of China Medical University supported by the Science and Technology Department of Liaoning Province (2022 JH2/20200013).
Data availability
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Declarations
Ethics approval and consent to participate
This study was approved by the Ethics Committee of China Medical University (KT2022384), following the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Consent for publication
All authors consent for this manuscript to be published.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Tingting Du and Shuailing Liu are co-first authors.
References
- 1.Robbins R, Beebe DW, Byars KC, Grandner M, Hale L, Tapia IE, Wolfson AR, Owens JA. Adolescent sleep myths: identifying false beliefs that impact adolescent sleep and well-being. Sleep Health. 2022;8(6):632–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Owens JA, Weiss MR. Insufficient sleep in adolescents: causes and consequences. Minerva Pediatr. 2017;69(4):326–36. [DOI] [PubMed] [Google Scholar]
- 3.Suglia SF, Kara S, Robinson WR. Sleep duration and obesity among adolescents transitioning to adulthood: do results differ by sex? J Pediatr. 2014;165(4):750–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun Q, Bai Y, Zhai L, Wei W, Jia L. Association between Sleep Duration and Overweight/Obesity at Age 7–18 in Shenyang, China in 2010 and 2014. Int J Environ Res Public Health. 2018;15(5):854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun Q, Liu Y, Wei W, Wu D, Lin R, Wen D, Jia L. Chronic timed sleep restriction attenuates LepRb-mediated signaling pathways and circadian clock gene expression in the rat hypothalamus. Front Neurosci. 2020;14:909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu S, Liu X, Wang Y, Zhang R, Wei S. Melatonin protects against body weight gain induced by sleep deprivation in mice. Physiol Behav. 2022;257:113975. [DOI] [PubMed] [Google Scholar]
- 7.Al Dweik R, Sheble Y, Ramadan H, Issa H, Sheble A. The association between sleeping behavior, obesity, psychological depression, and eating habits among adolescents in the emirate of Abu Dhabi-United Arab Emirates. PLoS ONE. 2022;17(8):e0269837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tomaso CC, Yaroch AL, Hill JL, Jackson T, Nelson JM, James T, Mason WA, Espy KA, Nelson TD. The roles of sleep and executive function in adolescent nighttime eating. Eat Behav. 2022;46:101657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Manasse SM, Haedt-Matt AA, Smith KE, Egbert AH, O’Sullivan K, Koren D, Engel S, Goldschmidt AB. The moderating role of sleep duration on momentary relations between negative affect and loss-of-control eating in children and adolescents. Eur Eat Disord Rev. 2022;30(6):815–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcez MR, de Castro MA, César CLG, Goldbaum M, Fisberg RM. A chrononutrition perspective of diet quality and eating behaviors of Brazilian adolescents in associated with sleep duration. Chronobiol Int. 2021;38(3):387–99. [DOI] [PubMed] [Google Scholar]
- 11.DiFrancesco MW, Alsameen M, St-Onge M-P, Duraccio KM, Beebe DW. Altered neuronal response to visual food stimuli in adolescents undergoing chronic sleep restriction. Sleep. 2024; 47(4):zsad036. [DOI] [PMC free article] [PubMed]
- 12.Akhlaghi M, Kohanmoo A. Sleep deprivation in development of obesity, effects on appetite regulation, energy metabolism, and dietary choices. Nutr Res Rev. 2023;31:1–21. [DOI] [PubMed] [Google Scholar]
- 13.Carmo-Silva S, Cavadas C. Hypothalamic dysfunction in obesity and metabolic disorders. Adv Neurobiol. 2017;19:73–116. [DOI] [PubMed] [Google Scholar]
- 14.Marraudino M, Bonaldo B, Farinetti A, Panzica G, Ponti G, Gotti S. Metabolism disrupting chemicals and alteration of neuroendocrine circuits controlling food intake and energy metabolism. Front Endocrinol (Lausanne). 2018;9:766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seong J, Kang JY, Sun JS, Kim KW. Hypothalamic inflammation and obesity: a mechanistic review. Arch Pharm Res. 2019;42(5):383–92. [DOI] [PubMed] [Google Scholar]
- 16.Alqaderi H, Abdullah A, Finkelman M, Abufarha M, Devarajan S, Abubaker J, Ramesh N, Tavares M, Al-Mulla F, Bin-Hasan S. The relationship between sleep and salivary and serum inflammatory biomarkers in adolescents. Front Med (Lausanne). 2023;10:1175483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stenvers DJ, Jongejan A, Atiqi S, Vreijling JP, Limonard EJ, Endert E, Baas F, Moerland PD, Fliers E, Kalsbeek A, et al. Diurnal rhythms in the white adipose tissue transcriptome are disturbed in obese individuals with type 2 diabetes compared with lean control individuals. Diabetologia. 2019;62(4):704–16. [DOI] [PubMed] [Google Scholar]
- 18.Tsang AH, Koch CE, Kiehn J-T, Schmidt CX, Oster H. An adipokine feedback regulating diurnal food intake rhythms in mice. Elife. 2020;9:e55388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scheer FAJL, Morris CJ, Shea SA. The internal circadian clock increases hunger and appetite in the evening independent of food intake and other behaviors. Obesity (Silver Spring). 2013;21(3):421–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chao OY, Nikolaus S, Yang Y-M, Huston JP. Neuronal circuitry for recognition memory of object and place in rodent models. Neurosci Biobehav Rev. 2022;141:104855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang W, Ramsey KM, Marcheva B, Bass J. Circadian rhythms, sleep, and metabolism. J Clin Invest. 2011;121(6):2133–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stoynev AG, Ikonomov OC, Usunoff KG. Feeding pattern and light-dark variations in water intake and renal excretion after suprachiasmatic nuclei lesions in rats. Physiol Behav. 1982;29(1):35–40. [DOI] [PubMed] [Google Scholar]
- 23.Li A-J, Wiater MF, Oostrom MT, Smith BR, Wang Q, Dinh TT, Roberts BL, Jansen HT, Ritter S. Leptin-sensitive neurons in the arcuate nuclei contribute to endogenous feeding rhythms. Am J Physiol Regul Integr Comp Physiol. 2012;302(11):R1313–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cone RD, Cowley MA, Butler AA, Fan W, Marks DL, Low MJ. The arcuate nucleus as a conduit for diverse signals relevant to energy homeostasis. Int J Obes Relat Metab Disord. 2001;25(Suppl 5):S63–7. [DOI] [PubMed] [Google Scholar]
- 25.Schwartz MW, Woods SC, Porte D, Seeley RJ, Baskin DG. Central nervous system control of food intake. Nature. 2000;404(6778):661–71. [DOI] [PubMed] [Google Scholar]
- 26.Tong Q, Ye C-P, Jones JE, Elmquist JK, Lowell BB. Synaptic release of GABA by AgRP neurons is required for normal regulation of energy balance. Nat Neurosci. 2008;11(9):998–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buijs RM, Wortel J, Van Heerikhuize JJ, Feenstra MG, Ter Horst GJ, Romijn HJ, Kalsbeek A. Anatomical and functional demonstration of a multisynaptic suprachiasmatic nucleus adrenal (cortex) pathway. Eur J Neurosci. 1999;11(5):1535–44. [DOI] [PubMed] [Google Scholar]
- 28.O’Hare JD, Zsombok A. Brain-liver connections: role of the preautonomic PVN neurons. Am J Physiol Endocrinol Metab. 2016;310(3):E183–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clemenzi MN, Martchenko A, Loganathan N, Tse EK, Brubaker PL, Belsham DD. Analysis of Western diet, palmitate and BMAL1 regulation of neuropeptide Y expression in the murine hypothalamus and BMAL1 knockout cell models. Mol Cell Endocrinol. 2020;507:110773. [DOI] [PubMed] [Google Scholar]
- 30.Kim ER, Xu Y, Cassidy RM, Lu Y, Yang Y, Tian J, Li D-P, Van Drunen R, Ribas-Latre A, Cai Z-L, et al. Paraventricular hypothalamus mediates diurnal rhythm of metabolism. Nat Commun. 2020;11(1):3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leng Y, Musiek ES, Hu K, Cappuccio FP, Yaffe K. Association between circadian rhythms and neurodegenerative diseases. Lancet Neurol. 2019;18(3):307–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bravo Santos R, Delgado J, Cubero J, Franco L, Ruiz-Moyano S, Mesa M, Rodríguez AB, Uguz C, Barriga C. Activity/inactivity circadian rhythm shows high similarities between young obesity-induced rats and old rats. Physiol Int. 2016;103(1):65–74. [DOI] [PubMed] [Google Scholar]
- 33.Mieda M, Ono D, Hasegawa E, Okamoto H, Honma K-I, Honma S, Sakurai T. Cellular clocks in AVP neurons of the SCN are critical for interneuronal coupling regulating circadian behavior rhythm. Neuron. 2015;85(5):1103–16. [DOI] [PubMed] [Google Scholar]
- 34.Sassone-Corsi P. Minireview: NAD+, a circadian metabolite with an epigenetic twist. Endocrinology. 2012;153(1):1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pei H, Sutton AK, Burnett KH, Fuller PM, Olson DP. AVP neurons in the paraventricular nucleus of the hypothalamus regulate feeding. Mol Metab. 2014;3(2):209–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saeb-Parsy K, Lombardelli S, Khan FZ, McDowall K, Au-Yong IT, Dyball RE. Neural connections of hypothalamic neuroendocrine nuclei in the rat. J Neuroendocrinol. 2000;12(7):635–48. [DOI] [PubMed] [Google Scholar]
- 37.Zhan C, Zhou J, Feng Q, Zhang J-E, Lin S, Bao J, Wu P, Luo M. Acute and long-term suppression of feeding behavior by POMC neurons in the brainstem and hypothalamus, respectively. J Neurosci. 2013;33(8):3624–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bouffet E, Doumi N, Thiesse P, Mottolese C, Jouvet A, Lacroze M, Carrie C, Frappaz D, Brunat-Mentigny M. Brain metastases in children with solid tumors. Cancer. 1997;79(2):403–10. [DOI] [PubMed] [Google Scholar]
- 39.Vaisse C, Halaas JL, Horvath CM, Darnell JE, Stoffel M, Friedman JM. Leptin activation of Stat3 in the hypothalamus of wild-type and ob/ob mice but not db/db mice. Nat Genet. 1996;14(1):95–7. [DOI] [PubMed] [Google Scholar]
- 40.Gropp E, Shanabrough M, Borok E, Xu AW, Janoschek R, Buch T, Plum L, Balthasar N, Hampel B, Waisman A, et al. Agouti-related peptide-expressing neurons are mandatory for feeding. Nat Neurosci. 2005;8(10):1289–91. [DOI] [PubMed] [Google Scholar]
- 41.Chaput J-P, McHill AW, Cox RC, Broussard JL, Dutil C, da Costa BGG, Sampasa-Kanyinga H, Wright KP. The role of insufficient sleep and circadian misalignment in obesity. Nat Rev Endocrinol. 2023;19(2):82–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu S, Wang X, Zheng Q, Gao L, Sun Q. Sleep deprivation and central appetite regulation. Nutrients. 2022;14(24):5196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim SJ, Hotta-Hirashima N, Asano F, Kitazono T, Iwasaki K, Nakata S, Komiya H, Asama N, Matsuoka T, Fujiyama T, et al. Kinase signalling in excitatory neurons regulates sleep quantity and depth. Nature. 2022;612(7940):512–8. [DOI] [PubMed] [Google Scholar]
- 44.Wilckens KA, Ferrarelli F, Walker MP, Buysse DJ. Slow-wave activity enhancement to improve cognition. Trends Neurosci. 2018;41(7):470–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, Kreppel F, Mostoslavsky R, Alt FW, Schibler U. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell. 2008;134(2):317–28. [DOI] [PubMed] [Google Scholar]
- 46.Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, Guarente LP, Sassone-Corsi P. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134(2):329–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science. 2009;324(5927):654–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chang H-C, Guarente L. SIRT1 mediates central circadian control in the SCN by a mechanism that decays with aging. Cell. 2013;153(7):1448–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bjorbak C, Lavery HJ, Bates SH, Olson RK, Davis SM, Flier JS, Myers MG. SOCS3 mediates feedback inhibition of the leptin receptor via Tyr985. J Biol Chem. 2000;275(51):40649–57. [DOI] [PubMed] [Google Scholar]
- 50.Bjørbaek C, El-Haschimi K, Frantz JD, Flier JS. The role of SOCS-3 in leptin signaling and leptin resistance. J Biol Chem. 1999;274(42):30059–65. [DOI] [PubMed] [Google Scholar]
- 51.Sadki A, Bentivoglio M, Kristensson K, Nygård M. Suppressors, receptors and effects of cytokines on the aging mouse biological clock. Neurobiol Aging. 2007;28(2):296–305. [DOI] [PubMed] [Google Scholar]
- 52.Xie X, Yang S, Zou Y, Cheng S, Wang Y, Jiang Z, Xiao J, Wang Z, Liu Y. Influence of the core circadian gene “Clock” on obesity and leptin resistance in mice. Brain Res. 2013;1491:147–55. [DOI] [PubMed] [Google Scholar]
- 53.Krajewska M, Banares S, Zhang EE, Huang X, Scadeng M, Jhala US, Feng G-S, Krajewski S. Development of diabesity in mice with neuronal deletion of Shp2 tyrosine phosphatase. Am J Pathol. 2008;172(5):1312–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kaszubska W, Falls HD, Schaefer VG, Haasch D, Frost L, Hessler P, Kroeger PE, White DW, Jirousek MR, Trevillyan JM. Protein tyrosine phosphatase 1B negatively regulates leptin signaling in a hypothalamic cell line. Mol Cell Endocrinol. 2002;195(1–2):109–18. [DOI] [PubMed] [Google Scholar]
- 55.Loh K, Fukushima A, Zhang X, Galic S, Briggs D, Enriori PJ, Simonds S, Wiede F, Reichenbach A, Hauser C, et al. Elevated hypothalamic TCPTP in obesity contributes to cellular leptin resistance. Cell Metab. 2011;14(5):684–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Morrison CD, White CL, Wang Z, Lee S-Y, Lawrence DS, Cefalu WT, Zhang Z-Y, Gettys TW. Increased hypothalamic protein tyrosine phosphatase 1B contributes to leptin resistance with age. Endocrinology. 2007;148(1):433–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.White CL, Whittington A, Barnes MJ, Wang Z, Bray GA, Morrison CD. HF diets increase hypothalamic PTP1B and induce leptin resistance through both leptin-dependent and -independent mechanisms. Am J Physiol Endocrinol Metab. 2009;296(2):E291–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu AC, Welsh DK, Ko CH, Tran HG, Zhang EE, Priest AA, Buhr ED, Singer O, Meeker K, Verma IM, et al. Intercellular coupling confers robustness against mutations in the SCN circadian clock network. Cell. 2007;129(3):605–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Patton AP, Chesham JE, Hastings MH. Combined pharmacological and genetic manipulations unlock unprecedented temporal elasticity and reveal phase-specific modulation of the molecular circadian clock of the mouse suprachiasmatic nucleus. J Neurosci. 2016;36(36):9326–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Whitt JP, Montgomery JR, Meredith AL. BK channel inactivation gates daytime excitability in the circadian clock. Nat Commun. 2016;7:10837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Irwin RP, Allen CN. Calcium response to retinohypothalamic tract synaptic transmission in suprachiasmatic nucleus neurons. J Neurosci. 2007;27(43):11748–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Todd WD, Venner A, Anaclet C, Broadhurst RY, De Luca R, Bandaru SS, Issokson L, Hablitz LM, Cravetchi O, Arrigoni E, et al. Suprachiasmatic VIP neurons are required for normal circadian rhythmicity and comprised of molecularly distinct subpopulations. Nat Commun. 2020;11(1):4410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cheng MY, Bullock CM, Li C, Lee AG, Bermak JC, Belluzzi J, Weaver DR, Leslie FM, Zhou Q-Y. Prokineticin 2 transmits the behavioural circadian rhythm of the suprachiasmatic nucleus. Nature. 2002;417(6887):405–10. [DOI] [PubMed] [Google Scholar]
- 64.Hermes MLHJ, Kolaj M, Doroshenko P, Coderre E, Renaud LP. Effects of VPAC2 receptor activation on membrane excitability and GABAergic transmission in subparaventricular zone neurons targeted by suprachiasmatic nucleus. J Neurophysiol. 2009;102(3):1834–42. [DOI] [PubMed] [Google Scholar]
- 65.Tousson E, Meissl H. Suprachiasmatic nuclei grafts restore the circadian rhythm in the paraventricular nucleus of the hypothalamus. J Neurosci. 2004;24(12):2983–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang J, Ling S, Usami T, Murata T, Narita K, Higuchi T. Effects of ghrelin, corticotrophin-releasing hormone, and melanotan-II on food intake in rats with paraventricular nucleus lesions. Exp Clin Endocrinol Diabetes. 2007;115(10):669–73. [DOI] [PubMed] [Google Scholar]
- 67.Wang J, Yuan Z, Dong J, Zhang D, Usami T, Murata T, Narita K, Higuchi T. Neuropeptide Y loses its orexigenic effect in rats with lesions of the hypothalamic paraventricular nucleus. Endocr Res. 2013;38(1):8–14. [DOI] [PubMed] [Google Scholar]
- 68.Herrera Moro Chao D, Kirchner MK, Pham C, Foppen E, Denis RGP, Castel J, Morel C, Montalban E, Hassouna R, Bui L-C et al. Hypothalamic astrocytes control systemic glucose metabolism and energy balance. Cell Metab. 2022; 34(10):1532–47. [DOI] [PMC free article] [PubMed]
- 69.Papazoglou I, Lee J-H, Cui Z, Li C, Fulgenzi G, Bahn YJ, Staniszewska-Goraczniak HM, Piñol RA, Hogue IB, Enquist LW, et al. A distinct hypothalamus-to-β cell circuit modulates insulin secretion. Cell Metab. 2022;34(2):285–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Licht CMM, de Geus EJC, Penninx BWJH. Dysregulation of the autonomic nervous system predicts the development of the metabolic syndrome. J Clin Endocrinol Metab. 2013;98(6):2484–93. [DOI] [PubMed] [Google Scholar]
- 71.Thayer JF, Yamamoto SS, Brosschot JF. The relationship of autonomic imbalance, heart rate variability and cardiovascular disease risk factors. Int J Cardiol. 2010;141(2):122–31. [DOI] [PubMed] [Google Scholar]
- 72.Berk ML, Finkelstein JA. An autoradiographic determination of the efferent projections of the suprachiasmatic nucleus of the hypothalamus. Brain Res. 1981;226(1–2):1–13. [DOI] [PubMed] [Google Scholar]
- 73.Gjerstad JK, Lightman SL, Spiga F. Role of glucocorticoid negative feedback in the regulation of HPA axis pulsatility. Stress. 2018;21(5):403–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tempel DL, Leibowitz SF. Adrenal steroid receptors: interactions with brain neuropeptide systems in relation to nutrient intake and metabolism. J Neuroendocrinol. 1994;6(5):479–501. [DOI] [PubMed] [Google Scholar]
- 75.Moore RY, Eichler VB. Loss of a circadian adrenal corticosterone rhythm following suprachiasmatic lesions in the rat. Brain Res. 1972;42(1):201–6. [DOI] [PubMed] [Google Scholar]
- 76.Waite EJ, McKenna M, Kershaw Y, Walker JJ, Cho K, Piggins HD, Lightman SL. Ultradian corticosterone secretion is maintained in the absence of circadian cues. Eur J Neurosci. 2012;36(8):3142–50. [DOI] [PubMed] [Google Scholar]
- 77.Kalsbeek A, van der Spek R, Lei J, Endert E, Buijs RM, Fliers E. Circadian rhythms in the hypothalamo-pituitary-adrenal (HPA) axis. Mol Cell Endocrinol. 2012;349(1):20–9. [DOI] [PubMed] [Google Scholar]
- 78.Vrang N, Larsen PJ, Mikkelsen JD. Direct projection from the suprachiasmatic nucleus to hypophysiotrophic corticotropin-releasing factor immunoreactive cells in the paraventricular nucleus of the hypothalamus demonstrated by means of Phaseolus vulgaris-leucoagglutinin tract tracing. Brain Res. 1995;684(1):61–9. [DOI] [PubMed] [Google Scholar]
- 79.Rich EL, Romero LM. Exposure to chronic stress downregulates corticosterone responses to acute stressors. Am J Physiol Regul Integr Comp Physiol. 2005;288(6):R1628–36. [DOI] [PubMed] [Google Scholar]
- 80.Shahanoor Z, Sultana R, Savenkova M, Karatsoreos IN, Romeo RD. Metabolic dysfunctions following chronic oral corticosterone are modified by adolescence and sex in mice. Physiol Behav. 2023;269:114289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Burke SJ, Batdorf HM, Huang T-Y, Jackson JW, Jones KA, Martin TM, Rohli KE, Karlstad MD, Sparer TE, Burk DH, et al. One week of continuous corticosterone exposure impairs hepatic metabolic flexibility, promotes islet β-cell proliferation, and reduces physical activity in male C57BL/6 J mice. J Steroid Biochem Mol Biol. 2019;195:105468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.van Esseveldt KE, Lehman MN, Boer GJ. The suprachiasmatic nucleus and the circadian time-keeping system revisited. Brain Res Brain Res Rev. 2000;33(1):34–77. [DOI] [PubMed] [Google Scholar]
- 83.Watts PD, Ferguson KL, Draper BA. Energetic output of subadult polar bears (Ursus maritimus): resting, disturbance and locomotion. Comp Biochem Physiol A Comp Physiol. 1991;98(2):191–3. [DOI] [PubMed] [Google Scholar]
- 84.Bartness TJ, Song CK, Demas GE. SCN efferents to peripheral tissues: implications for biological rhythms. J Biol Rhythms. 2001;16(3):196–204. [DOI] [PubMed] [Google Scholar]
- 85.Liu Y, Dentin R, Chen D, Hedrick S, Ravnskjaer K, Schenk S, Milne J, Meyers DJ, Cole P, Yates J, et al. A fasting inducible switch modulates gluconeogenesis via activator/coactivator exchange. Nature. 2008;456(7219):269–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rodgers JT, Puigserver P. Fasting-dependent glucose and lipid metabolic response through hepatic sirtuin 1. Proc Natl Acad Sci U S A. 2007;104(31):12861–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Friebe D, Neef M, Kratzsch J, Erbs S, Dittrich K, Garten A, Petzold-Quinque S, Blüher S, Reinehr T, Stumvoll M, et al. Leucocytes are a major source of circulating nicotinamide phosphoribosyltransferase (NAMPT)/pre-B cell colony (PBEF)/visfatin linking obesity and inflammation in humans. Diabetologia. 2011;54(5):1200–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yoon MJ, Yoshida M, Johnson S, Takikawa A, Usui I, Tobe K, Nakagawa T, Yoshino J, Imai S-i. SIRT1-Mediated eNAMPT secretion from adipose tissue regulates hypothalamic NAD+ and function in mice. Cell Metab. 2015; 21(5):706–17. [DOI] [PMC free article] [PubMed]
- 89.Chalkiadaki A, Guarente L. High-fat diet triggers inflammation-induced cleavage of SIRT1 in adipose tissue to promote metabolic dysfunction. Cell Metab. 2012;16(2):180–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kalsbeek A, Palm IF, La Fleur SE, Scheer FAJL, Perreau-Lenz S, Ruiter M, Kreier F, Cailotto C, Buijs RM. SCN outputs and the hypothalamic balance of life. J Biol Rhythms. 2006;21(6):458–69. [DOI] [PubMed] [Google Scholar]
- 91.Guzmán-Ruiz MA, Ramirez-Corona A, Guerrero-Vargas NN, Sabath E, Ramirez-Plascencia OD, Fuentes-Romero R, León-Mercado LA, Basualdo Sigales M, Escobar C, Buijs RM. Role of the suprachiasmatic and arcuate nuclei in diurnal temperature regulation in the rat. J Neurosci. 2015;35(46):15419–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Paul S, Brown T. Direct effects of the light environment on daily neuroendocrine control. J Endocrinol. 2019; 1:JOE-19–0302.R1. [DOI] [PubMed]
- 93.Starnes AN, Jones JR. Inputs and outputs of the mammalian circadian clock. Biology (Basel). 2023;12(4):508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dijk DJ, Beersma DG. Effects of SWS deprivation on subsequent EEG power density and spontaneous sleep duration. Electroencephalogr Clin Neurophysiol. 1989;72(4):312–20. [DOI] [PubMed] [Google Scholar]
- 95.Boivin DB, Czeisler CA, Dijk DJ, Duffy JF, Folkard S, Minors DS, Totterdell P, Waterhouse JM. Complex interaction of the sleep-wake cycle and circadian phase modulates mood in healthy subjects. Arch Gen Psychiatry. 1997;54(2):145–52. [DOI] [PubMed] [Google Scholar]
- 96.Wyatt JK, Ritz-De Cecco A, Czeisler CA, Dijk DJ. Circadian temperature and melatonin rhythms, sleep, and neurobehavioral function in humans living on a 20-h day. Am J Physiol. 1999;277(4 Pt 2):R1152–63. [DOI] [PubMed] [Google Scholar]
- 97.Meyer AH, Langhans W, Scharrer E. Vasopressin reduces food intake in goats. Q J Exp Physiol. 1989;74(4):465–73. [DOI] [PubMed] [Google Scholar]
- 98.Spruce BA, McCulloch AJ, Burd J, Orskov H, Heaton A, Baylis PH, Alberti KG. The effect of vasopressin infusion on glucose metabolism in man. Clin Endocrinol (Oxf). 1985;22(4):463–8. [DOI] [PubMed] [Google Scholar]
- 99.Kalsbeek A, Yi C-X, La Fleur SE, Fliers E. The hypothalamic clock and its control of glucose homeostasis. Trends Endocrinol Metab. 2010;21(7):402–10. [DOI] [PubMed] [Google Scholar]
- 100.Aton SJ, Colwell CS, Harmar AJ, Waschek J, Herzog ED. Vasoactive intestinal polypeptide mediates circadian rhythmicity and synchrony in mammalian clock neurons. Nat Neurosci. 2005;8(4):476–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Doi M, Ishida A, Miyake A, Sato M, Komatsu R, Yamazaki F, Kimura I, Tsuchiya S, Kori H, Seo K, et al. Circadian regulation of intracellular G-protein signalling mediates intercellular synchrony and rhythmicity in the suprachiasmatic nucleus. Nat Commun. 2011;2:327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Saper CB, Scammell TE, Lu J. Hypothalamic regulation of sleep and circadian rhythms. Nature. 2005;437(7063):1257–63. [DOI] [PubMed] [Google Scholar]
- 103.Scammell TE, Arrigoni E, Lipton JO. Neural circuitry of wakefulness and sleep. Neuron. 2017;93(4):747–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zocchi L, Sassone-Corsi P. Joining the dots: from chromatin remodeling to neuronal plasticity. Curr Opin Neurobiol. 2010;20(4):432–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wang TA, Yu YV, Govindaiah G, Ye X, Artinian L, Coleman TP, Sweedler JV, Cox CL, Gillette MU. Circadian rhythm of redox state regulates excitability in suprachiasmatic nucleus neurons. Science. 2012;337(6096):839–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rutter J, Reick M, Wu LC, McKnight SL. Regulation of clock and NPAS2 DNA binding by the redox state of NAD cofactors. Science. 2001;293(5529):510–4. [DOI] [PubMed] [Google Scholar]
- 107.Dioum EM, Rutter J, Tuckerman JR, Gonzalez G, Gilles-Gonzalez M-A, McKnight SL. NPAS2: a gas-responsive transcription factor. Science. 2002;298(5602):2385–7. [DOI] [PubMed] [Google Scholar]
- 108.Deboer T, Vansteensel MJ, Détári L, Meijer JH. Sleep states alter activity of suprachiasmatic nucleus neurons. Nat Neurosci. 2003;6(10):1086–90. [DOI] [PubMed] [Google Scholar]
- 109.Colwell CS, Michel S. Sleep and circadian rhythms: do sleep centers talk back to the clock? Nat Neurosci. 2003;6(10):1005–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Dashti HS, Scheer FA, Jacques PF, Lamon-Fava S, Ordovás JM. Short sleep duration and dietary intake: epidemiologic evidence, mechanisms, and health implications. Adv Nutr. 2015;6(6):648–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ward AL, Galland BC, Haszard JJ, Meredith-Jones K, Morrison S, McIntosh DR, Jackson R, Beebe DW, Fangupo L, Richards R, et al. The effect of mild sleep deprivation on diet and eating behaviour in children: protocol for the Daily Rest, Eating, and Activity Monitoring (DREAM) randomized cross-over trial. BMC Public Health. 2019;19(1):1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Cowley MA, Smart JL, Rubinstein M, Cerdán MG, Diano S, Horvath TL, Cone RD, Low MJ. Leptin activates anorexigenic POMC neurons through a neural network in the arcuate nucleus. Nature. 2001;411(6836):480–4. [DOI] [PubMed] [Google Scholar]
- 113.Fioramonti X, Contié S, Song Z, Routh VH, Lorsignol A, Pénicaud L. Characterization of glucosensing neuron subpopulations in the arcuate nucleus: integration in neuropeptide Y and pro-opio melanocortin networks? Diabetes. 2007;56(5):1219–27. [DOI] [PubMed] [Google Scholar]
- 114.Zagmutt S, Mera P, Soler-Vázquez MC, Herrero L, Serra D. Targeting AgRP neurons to maintain energy balance: lessons from animal models. Biochem Pharmacol. 2018;155:224–32. [DOI] [PubMed] [Google Scholar]
- 115.Zhang C, Truong KK, Zhou Q-Y. Efferent projections of prokineticin 2 expressing neurons in the mouse suprachiasmatic nucleus. PLoS ONE. 2009;4(9):e7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Engin A. Diet-induced obesity and the mechanism of leptin resistance. Adv Exp Med Biol. 2017;960:381–97. [DOI] [PubMed] [Google Scholar]
- 117.Ramadori G, Fujikawa T, Fukuda M, Anderson J, Morgan DA, Mostoslavsky R, Stuart RC, Perello M, Vianna CR, Nillni EA, et al. SIRT1 deacetylase in POMC neurons is required for homeostatic defenses against diet-induced obesity. Cell Metab. 2010;12(1):78–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Cakir I, Perello M, Lansari O, Messier NJ, Vaslet CA, Nillni EA. Hypothalamic Sirt1 regulates food intake in a rodent model system. PLoS ONE. 2009;4(12):e8322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sasaki T, Kim H-J, Kobayashi M, Kitamura Y-I, Yokota-Hashimoto H, Shiuchi T, Minokoshi Y, Kitamura T. Induction of hypothalamic Sirt1 leads to cessation of feeding via agouti-related peptide. Endocrinology. 2010;151(6):2556–66. [DOI] [PubMed] [Google Scholar]
- 120.Sandoval D, Cota D, Seeley RJ. The integrative role of CNS fuel-sensing mechanisms in energy balance and glucose regulation. Annu Rev Physiol. 2008;70:513–35. [DOI] [PubMed] [Google Scholar]
- 121.Sutton AK, Myers MG, Olson DP. The role of PVH circuits in leptin action and energy balance. Annu Rev Physiol. 2016;78:207–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Mangieri LR, Lu Y, Xu Y, Cassidy RM, Xu Y, Arenkiel BR, Tong Q. A neural basis for antagonistic control of feeding and compulsive behaviors. Nat Commun. 2018;9(1):52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Balthasar N, Dalgaard LT, Lee CE, Yu J, Funahashi H, Williams T, Ferreira M, Tang V, McGovern RA, Kenny CD, et al. Divergence of melanocortin pathways in the control of food intake and energy expenditure. Cell. 2005;123(3):493–505. [DOI] [PubMed] [Google Scholar]
- 124.Perry RJ, Resch JM, Douglass AM, Madara JC, Rabin-Court A, Kucukdereli H, Wu C, Song JD, Lowell BB, Shulman GI. Leptin’s hunger-suppressing effects are mediated by the hypothalamic-pituitary-adrenocortical axis in rodents. Proc Natl Acad Sci U S A. 2019;116(27):13670–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Picard F, Kurtev M, Chung N, Topark-Ngarm A, Senawong T, Machado De Oliveira R, Leid M, McBurney MW, Guarente L. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature. 2004; 429(6993):771–6. [DOI] [PMC free article] [PubMed]
- 126.Qiang L, Wang L, Kon N, Zhao W, Lee S, Zhang Y, Rosenbaum M, Zhao Y, Gu W, Farmer SR, et al. Brown remodeling of white adipose tissue by SirT1-dependent deacetylation of Pparγ. Cell. 2012;150(3):620–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Basse AL, Nielsen KN, Karavaeva I, Ingerslev LR, Ma T, Havelund JF, Nielsen TS, Frost M, Peics J, Dalbram E, et al. NAMPT-dependent NAD+ biosynthesis controls circadian metabolism in a tissue-specific manner. Proc Natl Acad Sci U S A. 2023;120(14):e2220102120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Yoshida M, Satoh A, Lin JB, Mills KF, Sasaki Y, Rensing N, Wong M, Apte RS, Imai S-I. Extracellular vesicle-contained eNAMPT delays aging and extends lifespan in mice. Cell Metab. 2019;30(2):329–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Garten A, Petzold S, Barnikol-Oettler A, Körner A, Thasler WE, Kratzsch J, Kiess W, Gebhardt R. Nicotinamide phosphoribosyltransferase (NAMPT/PBEF/visfatin) is constitutively released from human hepatocytes. Biochem Biophys Res Commun. 2010;391(1):376–81. [DOI] [PubMed] [Google Scholar]
- 130.Park JW, Roh E, Kang GM, Gil SY, Kim HK, Lee CH, Jang WH, Park SE, Moon SY, Kim SJ, et al. Circulating blood eNAMPT drives the circadian rhythms in locomotor activity and energy expenditure. Nat Commun. 2023;14(1):1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Levine DC, Hong H, Weidemann BJ, Ramsey KM, Affinati AH, Schmidt MS, Cedernaes J, Omura C, Braun R, Lee C, et al. NAD+ controls circadian reprogramming through PER2 nuclear translocation to counter aging. Mol Cell. 2020;78(5):835–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Cetin A, Komai S, Eliava M, Seeburg PH, Osten P. Stereotaxic gene delivery in the rodent brain. Nat Protoc. 2006;1(6):3166–73. [DOI] [PubMed] [Google Scholar]
- 133.Brunner DP, Dijk DJ, Borbély AA. Repeated partial sleep deprivation progressively changes in EEG during sleep and wakefulness. Sleep. 1993;16(2):100–13. [DOI] [PubMed] [Google Scholar]
- 134.Akerstedt T, Kecklund G, Ingre M, Lekander M, Axelsson J. Sleep homeostasis during repeated sleep restriction and recovery: support from EEG dynamics. Sleep. 2009;32(2):217–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-stime quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material 1: Figure S1 Chronic sleep deprivation-induced disruption of energy expenditure and food consumption. A and B. Representative energy expenditurerecorded by metabolic cages in light-dark conditions in the mice under continuous monitoringor total energy expenditure. C and D. Representative food consumedrecorded by metabolic cages in light-dark conditions in the mice under continuous monitoringor total food consumed. E and F. Representative locomotor activityrecorded by metabolic cages in light-dark conditions in the mice under continuous monitoringor total locomotor activity. G and H. Representative oxygen consumptionrecorded by metabolic cages in light-dark conditions in the mice under continuous monitoringor total oxygen consumption. I and J. Representative carbon dioxide productionrecorded by metabolic cages in light-dark conditions in the mice under continuous monitoringor total carbon dioxide production. Data were presented as the means ± standard deviations and presented as bar graphs. Significant difference * was defined as p<0.05 by one-way ANOVA. CONGFP, the control group with empty vector; CONBMAL1, the control group with AAV-genetic overexpression of BMAL1; SDGFP, the chronic SD group with empty vector; SDBMAL1, the chronic SD with AAV-genetic overexpression of BMAL1. ZT, Zeitgeber time. Data were presented as the means ± standard deviations. Significant difference * was defined as p<0.05 by one-way ANOVA. CONGFP, the control group with empty vector; CONBMAL1, the control group with AAV-genetic overexpression of BMAL1; SDGFP, the chronic SD group with empty vector; SDBMAL1, the chronic SD with AAV-genetic overexpression of BMAL1. Figure S3 The original blots for Figure 2A. Figure S4 The original blots for Figure 2H. Figure S5 The original blots for Figure 6A. Figure S6 The original blots for Figure 6Q. Figure S7 The original blots for Figure 6T. Figure S8 The original blots for Figure 8E. Figure S9 The original blots for Figure 8G. Figure S10 The original blots for Figure 8K.Figure S11 The original blots for Figure 8P. Figure S2 Compensation of BMAL1 in the SCN rescued the in situ expressions of BMAL1 in the ARC A. Representative IF staining for BMAL1, DAPI and merged imagesin the ARC. B. The average IF intensities of BMAL1 among groups were determined by Image J software and presented as bar graph. Mean intensity = Integrated Density/Area.
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files.