Abstract
Background
Embryogenic callus (EC) has strong regenerative potential, useful for propagation and genetic transformation. miRNAs have been confirmed to play key regulatory roles in EC regeneration across various plants. However, challenges in EC induction have hindered the breeding of drumstick (Moringa oleifera Lam.), a tree with significant commercial potential. Understanding the regulatory networks of miRNAs-lncRNAs during EC formation in drumstick is crucial for overcoming these barriers.
Results
In this study, three drumstick EC small RNA libraries were sequenced using an Illumina Nova 6000 system. We identified 50 known miRNAs and 233 novel miRNAs. Target prediction and functional analysis showed that these miRNAs are involved in plant hormone signal transduction. Notably, miR319a and miR319b were upregulated throughout the entire process, while miR171 and miR160 were downregulated in the earlier stage but upregulated in the later stage. The expression patterns of 6 miRNAs detected by qRT-PCR were consistent with those observed in RNA-seq. The regulatory relationships between 6 selected highly expressed miRNAs and their target genes generally conformed to a negative regulatory pattern. Furthermore, miR156 and MolncRNA2275 were identified as key regulators in miRNA-mRNA-lncRNA network.
Conclusions
In summary, our study provides valuable insights into the molecular mechanisms underlying EC formation and enhances the understanding of the miRNA networks involved in this process.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12870-024-05983-9.
Keywords: Moringa oleifera Lam, miRNAs, Shoot regeneration, Plant hormone signal, lncRNAs
Introduction
De novo shoot organogenesis is one of the primary methods of in vitro plant regeneration, widely used in both breeding and basic research [1]. Various regeneration systems have been developed for different plant species, including Arabidopsis thaliana, Nicotiana benthamiana and Toona ciliate [2–4]. High regeneration efficiency is crucial for genetic transformation, which is a key tool in gene function analysis. During in vitro plant culture, explants are capable of perceiving external plant hormones, particularly cytokinin and auxin, to gain organogenic competence [5]. However, shoot regeneration is a complex and dynamic process, with highly intricate intracellular molecular regulatory mechanisms.
MicroRNAs (miRNAs) play a crucial regulatory role in various developmental processes by modulating gene expression at the post-transcriptional level. There is evidence suggesting that miRNAs are involved in maintaining pluripotency and have also emerged as key regulators of callus induction and regeneration [6–8]. Notably, miR160 has been identified as a repressor of organogenesis by mediating interactions between auxin and cytokinin [9]. miR160 negatively regulates the shoot regeneration capacity in Arabidopsis by targeting ARF10, which in turn modulates the expression of shoot meristem-specific genes. Additionally, it is demonstrated that miR319 inhibits the transcript levels of TEOSINTE BRANCHED 1/CYCLOIDEA/PCF 3 (TCP3) and TEOSINTE BRANCHED 1/CYCLOIDEA/PCF 4 (TCP4), with the suppression of shoot regeneration being alleviated by modulating cytokinin responses [10]. During the early stage of in vitro plant regeneration, the accumulation of most miRNAs coincides with the establishment of the shoot apical meristem, especially miR156 and miR166 [1]. miR156 is a highly conserved microRNA in plants. During shoot regeneration, miR156 promotes the formation of adventitious shoots by suppressing the expression of SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) transcription factors. This suppression indirectly regulates downstream genes associated with plant hormones, such as auxin and cytokinin, thereby influencing the balance and distribution of hormones during the regeneration process [11]. Multiple miRNAs are involved in the process of shoot regeneration in plants, and investigating their regulatory roles across different species is of great significance. This is particularly true for economically important tree species such as drumstick, as it can contribute to advancing breeding efforts and facilitating large-scale propagation.
Moringa oleifera Lam., known for its rapid growth and high economic value, belongs to the Moringaceae family and is primarily cultivated in subtropical regions [12]. Commonly referred to as the drumstick tree, it has been consumed for thousands of years due to its remarkable nutritional value, earning it the widely publicized title of the “miracle tree”. Numerous studies have shown that drumstick leaves are a rich source of amino acids, proteins, vitamins, and minerals [13–15]. Additionally, their high content of functional components, such as flavonoids and polysaccharides, endows them with various biological activities, including antioxidant, hypotensive, and anti-inflammatory effects [16, 17]. As a result, the drumstick tree holds immense commercial value, with promising applications in health-related fields such as food and medicine. Despite its industrial potential, limited progress has been made in breeding drumstick resources. Although tissue culture systems have been developed using different plant tissues, regeneration efficiency remains largely constrained by genotype [18, 19]. In previous studies, we found that the MoIAA13 gene likely influences adventitious shoot induction efficiency in the drumstick leaf regeneration system [20]. However, the role of miRNAs in regulating adventitious shoot induction in drumstick tree leaves remains understudied. In this research, we identified a total of 230 differentially expressed miRNAs and their target genes involved in shoot regeneration from in vitro drumstick leaves, comprising 41 known miRNAs and 189 novel miRNAs. We constructed a core regulatory miRNA-mRNA-lncRNA network for drumstick shoot regeneration. Our results preliminarily reveal that miRNAs play an important role in the process of embryogenic callus formation. Moreover, miR156 and MolncRNA2275 interact to play a central regulatory role. In summary, the findings of this study provide valuable insights into the comprehensive miRNA–mRNA regulatory network involved in drumstick shoot organogenesis, offering important information for enhancing the propagation efficiency of drumstick and contribute to molecular breeding efforts.
Materials and methods
Induction of shoot formation in in vitro drumstick leaves
Drumstick seeds from PKM-1 cultivar trees, collected at the germplasm conservation farm of South China Agricultural University, were disinfected and germinated in sterilized MS (Murashige & Skoog) medium. Young leaves from the sterile seedlings were then used as explants. The leaves were trimmed to remove the petioles and cut into pieces approximately 0.5 cm² in size. A few incisions were gently made on each leaf using a sterile scalpel before inoculating them onto MS medium supplemented with 0.8 mg/L 6-BA (6-Benzylamino purine), 0.2 mg/L KT (Kinetin), and 0.05 mg/L NAA (1-naphthaleneacetic acid). All media were autoclaved at 121 °C for 20 min. The cultures were maintained at a room temperature of approximately 26 °C under a 12-hour photoperiod.
RNA extraction and small RNA sequencing
Drumstick leaf explants cultured for 0, 10 and 20 days were collected for small RNA sequencing. Total RNA was extracted from samples collected at each developmental stage, with three biological replicates per stage, using the Simple Total RNA Kit (OMEGA, Guangzhou, China). The quality of the extracted RNA was rigorously evaluated to ensure its suitability for small RNA library construction. Specifically, RNA concentration and purity were quantified using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, USA), while RNA integrity was analyzed with the RNA Nano 6000 Assay Kit on the Agilent Bioanalyzer 2100 platform (Agilent Technologies, CA, USA). Small RNA was ligated with adaptors, reverse transcribed into cDNA, and the resulting fragments were amplified by PCR and purified using the AMPure XP system [21]. Finally, the qualified PCR products were sequenced by Biomarker Technologies (Beijing, China). RNA-seq data was submitted to National Center for Biotechnology Information (NCBI, https://www.ncbi.nlm.nih.gov/) under accession number PRJNA771463. Small RNA-seq data was submitted to NCBI under accession number PRJNA814844. miRNA sequences were submitted to GenBank under accession number PQ469266 - PQ469548.
Identification of miRNAs and target genes prediction
Raw reads from small RNA sequencing underwent initial quality assessment using FastQC (v0.11.9). Adapter sequences were then removed using Cutadapt (v1.9.1), followed by base quality control with Trimmomatic (v0.36). Small RNAs (sRNAs) ranging from 18 to 30 nucleotides in length were aligned to the Rfam and Ensembl databases using Bowtie software to filter out ribosomal RNA (rRNA), transfer RNA (tRNA), small nuclear RNA (snRNA), small nucleolar RNA (snoRNA), other non-coding RNAs (ncRNAs), and repetitive sequences. The remaining reads were mapped to the drumstick reference sequence [22] using Bowtie to analyze their distribution [23]. Known miRNAs were identified through the miRBase database [24], while novel miRNAs were predicted using mirDeep2 [25]. The secondary structures of the predicted novel miRNAs were further analyzed using Randfold software. The potential target genes of the identified miRNAs were predicted using previously described methods [26]. Based on the identified known miRNAs and newly predicted miRNAs, and using the drumstick gene sequence information, target gene prediction was performed using the TargetFinder software [27]. Differentially expressed miRNAs were identified using DESeq2 with a fold change (FC) ≥ 1.5 and a false discovery rate (FDR) ≤ 0.05. Additionally, the target genes were mapped to the KEGG database to predict their potential functions.
Identification of lncRNA and target genes prediction
The RNA-seq data in our previous study [28] were used to identify lncRNA. Raw data were filtered with fastp software [29]. Clean reads were mapped to the reference genome [22] using HISAT2 (version 2.1.0). StringTie (version 2.1.3) was used for transcript assembly and expression quantification. Transcripts shorter than 200 nt and those significantly overlapping with known protein-coding genes were filtered out. Subsequently, transcripts with FPKM values less than 0.5 were removed. The remaining transcripts were further analyzed for coding potential using LGC [30], CPC2 [31], and CNCI [32]. Transcripts predicted as non-coding by all three tools were subjected to Pfam database for further filtering. Transcripts encoding proteins were excluded, and the final retained transcripts were classified as lncRNAs. Differential expression analysis was performed using DESeq2, applying thresholds of |log2FC| ≥ 1 and p-value ≤ 0.05. RIblast was used to predict the target genes of lncRNAs [33].
qRT-PCR validation of miRNAs
qRT-PCR was performed using the stem-loop RT-PCR method to verify miRNA expression levels, with U6 selected as the internal control [34]. Briefly, cDNA was synthesized from the RNA used for sRNA-seq, employing the PrimeScript™ RT Master Mix (Takara, Japan). All primers, including those for stem-loop RT-PCR and miRNAs, are listed in Table S1. The method for estimating the relative expression level of each miRNA can be found in Yang et al. [20].
Results
Differential expression miRNAs during shoot induction
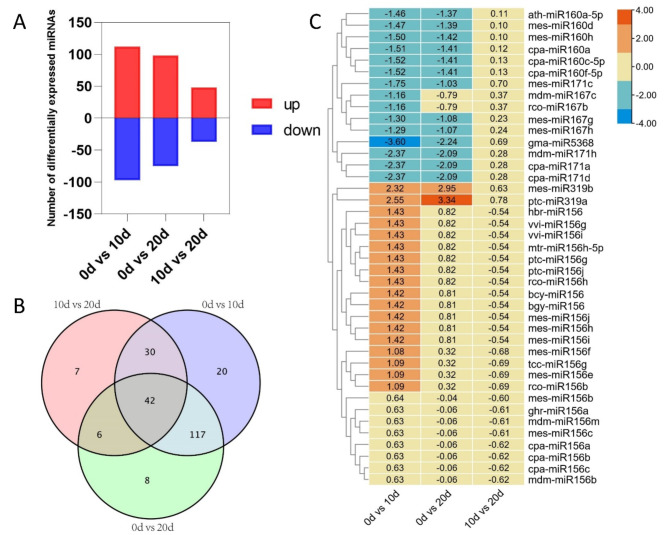
Leaf explants produced abundant callus after 10 days of culture, which gradually developed into adventitious shoots after 20 days (Fig. S1). To investigate the role of miRNAs in regulating adventitious shoot induction in the drumstick tree, nine miRNA libraries were constructed from three explants at three stages (0 d, 10 d, 20 d). A total of 205.45 million clean reads were obtained, with each sample exhibiting a Q30 (base quality > 30) of not less than 96.34% (Table S2). In total, 283 miRNAs were identified, including 50 known miRNAs and 233 novel miRNAs, with 41 known and 189 novel miRNAs differentially expressed during shoot induction (FDR = 0.05, FC = 1.5). Of these differentially expressed miRNAs (DEMs), 209 were identified in the 0 d vs. 10 d group, with 112 upregulated and 97 downregulated. As the induction process progressed, the number of DEMs decreased: only 85 DEMs were identified between the 10 d and 20 d stages, of which 48 were upregulated and 37 downregulated (Fig. 1A). Additionally, several DEMs were found to be differentially expressed only in single comparison groups. Notably, 42 miRNAs exhibited differential expression across all stages (Fig. 1B). Among the 41 known DEMs, ptc-miR319a showed the highest expression level in explants cultured for 20 days, while gma-miR5368 was significantly downregulated in explants at 10 days (Fig. 1C). Moreover, several novel miRNAs were significantly upregulated after 10 days of explant induction culture and maintained consistently high expression levels throughout the entire shoot induction process, including novel_miR_110, novel_miR_133 and novel_miR_115 (Table S3). These results suggest that miRNA expression varies significantly across different stages of shoot regeneration, and some DEMs are likely required for regulation throughout the entire process.
Fig. 1.
Identification of differentially expressed miRNAs (DEMs) at different stages of shoot induction. (A) Bar graph of up-regulated and down-regulated miRNAs from pairwise comparisons. (B) Venn diagram showing the DEMs. (C) Heatmap of 41 known DEMs, the scale represents the log2FC value
Analysis of target genes of DEMs
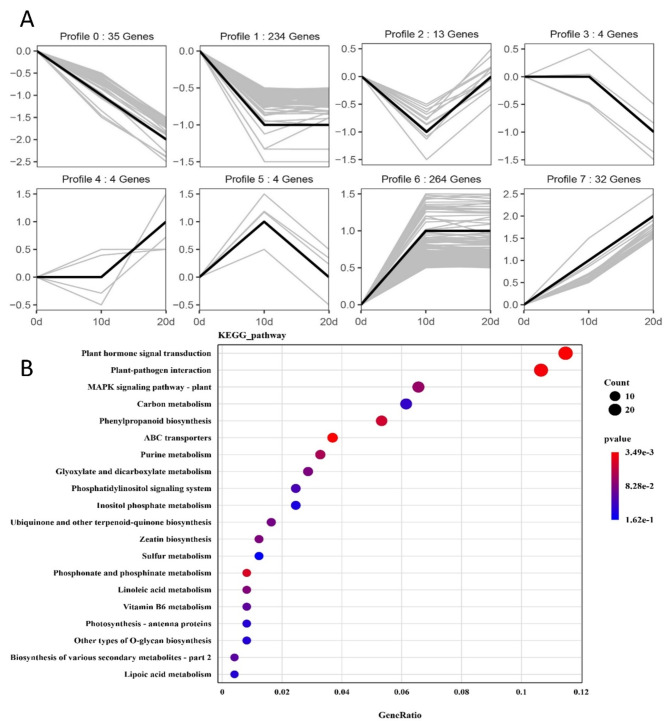
miRNAs exert their biological functions by negatively regulating their target genes [35]. To clarify the expression of target genes of DEMs, we analyzed RNA-seq data from drumstick tree callus and adventitious shoot induction, which had been previously submitted to NCBI under accession number PRJNA771463. In total, 230 DEMs identified through sRNA-seq were found to collectively target 2271 protein-coding genes (Table S4). Enrichment analysis of target genes for both known and novel miRNAs consistently revealed significant enrichment in the plant hormone signal transduction pathway, highlighting the critical role of this pathway (Fig. S2). Furthermore, a total of 671 target genes of DEMs were differentially expressed during shoot induction (FDR = 0.01, FC = 2). These 671 differentially expressed genes (DEGs) were grouped into eight clusters based on their expression patterns using STEM software (Fig. 2A). Cluster 6 contained the most DEGs (264), with gene expression increasing at 10 days and remaining stable between 10 and 20 days. Cluster 1 exhibited the opposite trend and had the second most DEGs (234). This was consistent with the observation that the number of DEMs decreased in the later stages of shoot induction, and most target genes were not differentially expressed. KEGG pathway analysis revealed that many DEGs were significantly enriched in plant hormone signal transduction pathways, highlighting the critical role of plant hormones and their signaling in the adventitious shoot induction of Moringa oleifera.
Fig. 2.
The expression trend and KEGG pathway analysis of target genes of DEMs. (A) The trend clusters of 671 target DEGs for 230 DEMs were obtained using STEM software with P-value ≤ 0.05, the y-axis of each cluster indicates the log2FC value. (B) KEGG pathway analysis of all target DEGs
Analysis of the top 10 DEMs at different stages
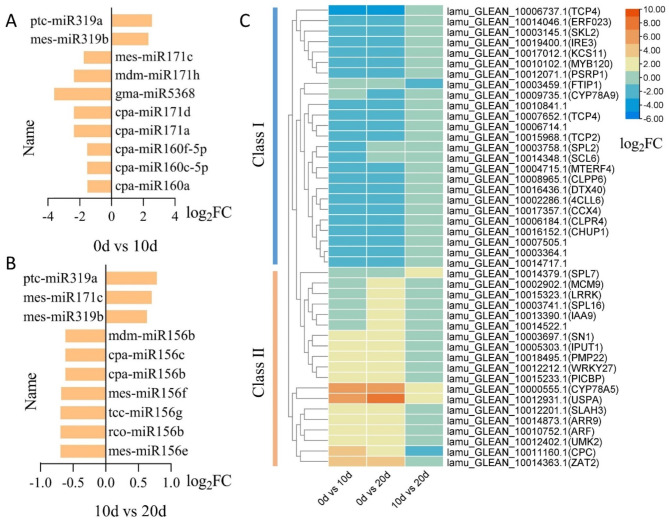
The top 10 DEMs from 0 to 10 days and 10 to 20 days of explant development were further analyzed. DEMs expression levels in explants from 0 to 10 days were generally higher than those from 10 to 20 days (Fig. 3A, B). Most DEMs were downregulated at both stages, while ptc-miR319a and mes-miR319b were continuously upregulated throughout explant development. It is also worth noting that all DEMs downregulated in the 10 to 20-day stage belonged to the miR156 family (Fig. 3B). Totally, 44 miRNA target genes were differentially expressed between these two stages. These 44 DEGs were classified into two groups based on their expression patterns (Fig. 3C). Generally, genes whose expression decreased from 0 to 10 days were grouped into Class I, while those with the opposite trend were classified as Class II. Four of these DEGs were annotated in the KEGG pathway of plant hormone signal transduction, including lamu_GLEAN_10013390.1 (IAA9) and lamu_GLEAN_10014873.1 (ARR9).
Fig. 3.
The top 10 DEMs at two stages (0 d-10 d, 10 d-20 d) and their 44 DEGs. (A) Map of the top 10 DEMs by their log2FC value at 0–10 d. (B) Map of the top 10 DEMs by their log2FC value at 10–20 d. (C) Heatmap of 44 target DEGs, the scale represents the log2FC value
qRT-PCR verification of miRNAs
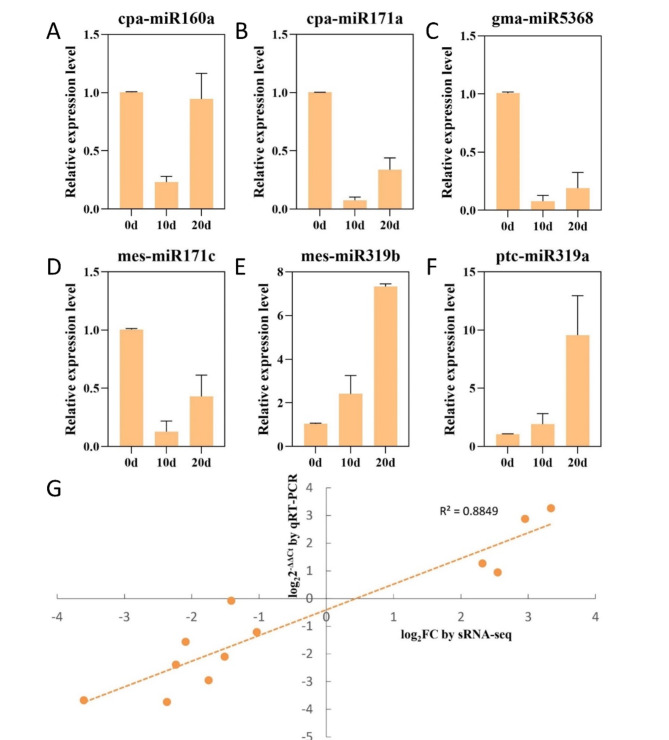
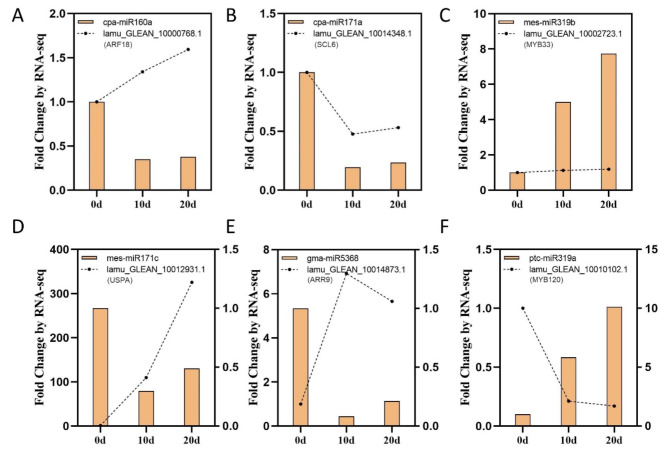
To verify the expression of six miRNAs with higher differential expression levels and confirm the reliability of the sequencing data, qRT-PCR was performed at three developmental stages of explants (Fig. 4A–F). The expression level of miR319 gradually increased throughout the cultivation process, reaching approximately 8 times the initial level after 20 days of culture. The other four DEMs followed a similar expression pattern, with an initial decrease followed by a slight increase at 20 days, though still lower than their initial levels. The qRT-PCR results showed a high correlation with the sequencing data, with an R² value of 0.88 (Fig. 4G). These findings suggest that the six DEMs may play important regulatory roles in shoot induction in the drumstick tree and further support the reliability of the sequencing analysis.
Fig. 4.
Verification of the expression patterns of 6 selected miRNAs in three stages. (A-F) qPCR analysis of 6 selected miRNAs expression levels. (G) Fitting curve of sequencing data and qPCR analysis
Analysis of 6 selected miRNAs target genes
Furthermore, six target genes of the six selected DEMs with high expression across the three stages were chosen for expression level analysis. These target genes generally adhered to the expected negative regulatory relationship with their corresponding miRNAs, except for lamu_GLEAN_10002723.1 (MYB33). The expression of lamu_GLEAN_10012931.1 (USPA) increased dramatically as explant development progressed, while mes-miR171c was only slightly downregulated. Conversely, although ptc-miR319a was significantly upregulated, its target gene lamu_GLEAN_10010102.1 (MYB120) showed only a slight downregulation. These results confirmed that miRNAs closely and precisely regulate their target genes, although further validation is still required.
Fig. 5.
miRNA and their target gene expression levels in three stages
Discussion
Many miRNAs are involved in SAM regeneration
miRNAs are endogenous negative regulatory factors that modulate the expression of target genes primarily at the post-transcriptional level [36]. The miRNA regulatory pathway is crucial for plant growth and development. With the rapid development of bioinformatics and transcriptomics, the function of miRNAs has been studied more and more deeply [37, 38]. At present, it has been confirmed that miRNA plays an important regulatory role in the process of somatic cell regeneration in many plants, which lays a foundation for further research on the mechanism of miRNA regulation of somatic cell regeneration [39–41]. In the current study, we also identified 50 conserved miRNAs and 233 novel miRNAs via small RNA sequencing of the shoot apical meristem (SAM) regeneration process derived from drumstick explant. A total of 230 miRNAs were differentially expressed during this process, with their 671 target genes also showing differential expression levels. Analysis of the expression trends and functional enrichment of these differentially expressed target genes revealed that genes involved in the plant hormone signal transduction pathway were significantly regulated by miRNAs and played a role in the formation of adventitious shoots.
Among the top 10 highly expressed miRNAs, miR319, miR171, miR156 and miR160 were shared by the dedifferentiated tissues. A recent study on miRNA responses in Arabidopsis during a wide range of abiotic stress identified miR319b as a multi-stress-responsive miRNA, with higher expression levels during metal stress and lower levels during drought, heat, and salinity [42]. In our study, the expression levels of miR319a and miR319b gradually increased during the process of SAM regeneration. In Arabidopsis, miR319b is also one of the miRNAs identified as highly expressed in early and advanced stages of somatic embryogenesis [43]. miR156, one of the largest miRNA families in plants, plays pivotal regulatory roles in plant regeneration. Its expression was significantly higher in embryogenic callus than in non-embryogenic callus in citrus, and higher in differentiated callus compared to undifferentiated tissues [44]. Elevated levels of miR156 enhance the capacity for somatic embryogenesis by prolonging the juvenile phase and suppressing premature differentiation. During the shoot induction of drumstick tree, miR156 expression increased significantly, peaking at the stage of extensive callus formation after 10 days of culture. This suggests that miR156 plays a crucial role in promoting cellular dedifferentiation and maintaining totipotency during organogenesis. In the present study, the expression level of several miR156 members decreased at callus differentiation stage, likely reflecting the need to reduce totipotency to facilitate shoot formation. The conserved miR160 and miR171 were downregulated during the process of callus induction and upregulated in the stage of callus differentiation. A similar pattern was observed in Arabidopsis, where miR160a is also expressed highly during the advanced stages of somatic embryogenesis induction and plays a role in regulating embryonic development [43]. As drumstick callus formation and adventitious shoot development progress, the downregulation of miR160 appears to relieve the repression of ARF genes, thereby enhancing auxin signal transduction and facilitating organ regeneration. miRNAs play a crucial role in organogenesis by finely regulating the synthesis and distribution of endogenous hormones through the modulation of genes involved in plant hormone signal transduction pathways.
Potential regulatory networks of miRNAs during SAM regeneration
Auxin and cytokinin establish molecular links between hormone signaling and embryogenic regeneration by activating or inhibiting multiple genes within their respective pathways [45–47]. There is increasing evidence that miRNAs are involved callus formation and adventitious bud regeneration by targeting genes in hormone signaling pathways [48]. In our study, KEGG pathway analysis of the predicted targets of DEMs was performed to explore the function of these differential genes. Most DEGs were significantly enriched in plant hormone signal transduction pathways. miR160a was barely detectable in the early stages of embryogenic callus induction, but was highly expressed in the middle and late stages of induction [49]. miR160 regulates embryogenesis by targeting auxin response factors ARF10, ARF16 and ARF17 [50]. miR156 was significantly expressed mainly in embryogenic callus, but hardly expressed in non-embryogenic callus, suggesting that miR156 plays an important role in regulating embryogenic callus maintenance in citrus [44]. The SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) transcription factor is the target gene of miR156. SPL can bind to type-B ARABIDOPSIS RESPONSE REGULATOR (B-ARR), a key transcription factor in the cytokinin signaling pathway, resulting in inhibition of the transcriptional activation activity of B-ARR. It showed insensitivity to cytokinin [51]. Overexpression of miR156 in weakly embryogenic wild kumquat led to downregulation of SPL expression, and the embryogenic ability of transgenic callus was significantly higher than that of wild type [52].
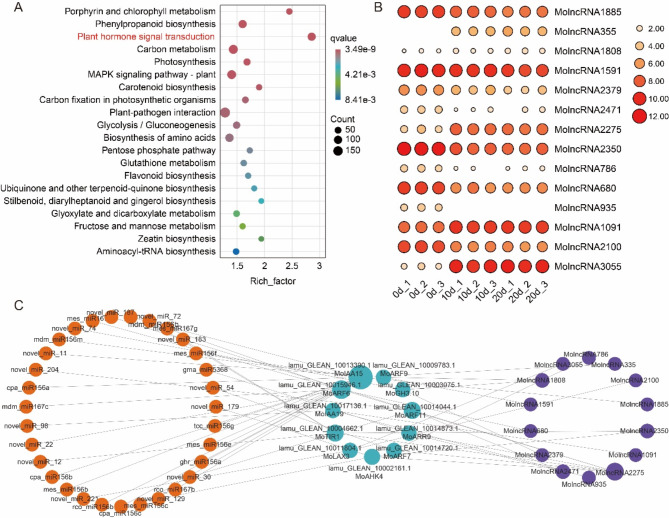
lncRNAs function as competitive endogenous RNAs (ceRNAs) by sequestering miRNAs, thereby alleviating their repression on target genes and indirectly enhancing the expression of these genes [53]. A total of 819 lncRNAs were identified as differentially expressed during the process of SAM regeneration (Fig. S3 and S4). KEGG pathway analysis of their target genes revealed significant enrichment in the plant hormone signal transduction pathway (Fig. 6A). This pathway includes 11 genes also identified as target genes of DEMs, such as MoARR9, several ARFs, and IAAs. Previous studies have demonstrated that MoIAAs play critical roles in the induction of adventitious shoots in drumstick [20]. Consistent with these findings, we constructed a correlation matrix between mRNAs and lncRNAs (Table S5) and identified 14 lncRNAs significantly associated with the expression of these 11 genes (Fig. 6B). Additionally, 31 DEMs, including 17 known miRNAs and 14 novel miRNAs, were identified as directly targeting these 11 genes. A regulatory network of miRNA-mRNA-lncRNA interactions was constructed (Fig. 6C), highlighting MoIAA15 and MoARF6 as central nodes, suggesting their pivotal roles in the SAM regeneration. novel_miR_22 was identified as a miRNA targeting MoIAA15. Moreover, miR156 and MolncRNA2275 were identified as key regulators in this network (Fig. 6C). These findings are in line with previous studies reporting the critical role of miRNA-lncRNA interactions in developmental and regenerative processes in plants [26].
Fig. 6.
Regulatory network of miRNA-mRNA-lncRNA for DEGs in the plant hormone signal transduction pathway. (A) KEGG enrichment pathways of target genes for differentially expressed lncRNAs. (B) Heatmap of expression levels of lncRNAs targeting DEGs in the plant hormone signal transduction pathway. (C) miRNA-mRNA-lncRNA network diagram
Conclusions
In this study, miRNAs and transcriptomic data were analyzed to gain further insights into the embryogenic callus induction of the drumstick tree. miRNAs were found to play a key regulatory role in the SAM regeneration process. Target prediction and functional analysis revealed their involvement in plant hormone signal transduction. Specifically, miR319a and miR319b were upregulated throughout the entire process, inhibiting their target genes, including MYB transcription factors in the auxin signaling pathway, which are closely related to embryogenic callus formation. The miRNA-mRNA-lncRNA regulatory network was constructed during shoot regeneration, MoIAA15 and MoARF6 are central genes in the network, with novel_miR_22 targeting MoIAA15. Additionally, miR156 and MolncRNA2275 have been identified as key regulatory factors within the network. These findings provide valuable insights into the molecular mechanisms of miRNA involvement in the shoot regeneration process.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Material 1: Table S1: RT and qPCR primers list of miRNAs.
Supplementary Material 2: Table S2: Statistics of data quality.
Supplementary Material 3: Table S3: The FPKM of novel miRNA.
Supplementary Material 4: Table S4: mRNA target genes prediction of DEM.
Supplementary Material 5: Table S5: Correlation between differentially expressed lncRNAs and DEGs
Supplementary Material 6: Fig. S1: Induction process of drumstick shoot regeneration.
Supplementary Material 7: Fig. S2: Functional enrichment analysis of target genes of DEMs. (A) KEGG pathway enrichment of target genes of 41 differentially expressed known miRNA. (B) KEGG pathway enrichment of target genes of 189 differentially expressed novel miRNA.
Supplementary Material 8: Fig. S3: Heatmap of lncRNA expression levels.
Supplementary Material 9: Fig. S4: Venn analysis of DElncRNAs of different comparison groups.
Author contributions
Conceptualization, J.Z. and X.C, methodology, E.Y, validation, J.Z., X.C. and E.Y, formal analysis, M.Z, investigation, L.Z, resources, E.Y. and J.Z, data curation, M.Z. and L.Z, writing—original draft preparation, E.Y, writing—review and editing, E.Y, visualization, M.Z. and L.Z, supervision, J.Z, project administration, X.C, funding acquisition, J.Z. and X.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China, grant number 32301607.
Data availability
The raw sequence data were deposited in the National Center for Biotechnology Information (NCBI, https://www.ncbi.nlm.nih.gov/) under accession number PRJNA771463. Small RNA-seq data was submitted to NCBI under accession number PRJNA814844. miRNA sequences were submitted to GenBank under accession number PQ469266 - PQ469548.
Declarations
Ethics approval and consent to participate
Clinical trial number: Not applicable. All experimental research on plants, no specific permits were required for the collection of specimens for this study.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Xiaoyang Chen, Email: xychen@scau.edu.cn.
Junjie Zhang, Email: zhangjunjie3168@163.com.
References
- 1.Lopez-Ruiz BA, Juarez-Gonzalez VT, Sandoval-Zapotitla E, Dinkova TD. Development-Related miRNA Expression and Target Regulation during Staggered In Vitro Plant Regeneration of Tuxpeno VS-535 Maize Cultivar. Int. J. Mol. Sci. 2019; 20(9): 2079. 10.3390/ijms20092079 [DOI] [PMC free article] [PubMed]
- 2.Yan A, Borg M, Berger F, Chen Z. The atypical histone variant H3.15 promotes callus formation in in Arabidopsis thaliana. Development. 2020;147(11):dev184895. 10.1242/dev.184895 [DOI] [PubMed] [Google Scholar]
- 3.Pathi KM, Tula S, Tuteja N. High frequency regeneration via direct somatic embryogenesis and efficient Agrobacterium-mediated genetic transformation of tobacco. Plant Signal Behav. 2013;8:e24354–24354. 10.4161/psb.24354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mao W, Song H, Li Y, Wang Y, Lin H, Yao C, Zhou W, Yang B, Chen X, Li P. Efficient plant regeneration and genetic transformation system of the precious fast-growing tree Toona ciliata. IND CROP PROD. 2021;172:114015. 10.1016/j.indcrop.2021.114015 [Google Scholar]
- 5.Passamani LZ, Reis RS, Vale EM, Sousa KR, Aragao VPM, Santa-Catarina C, Silveira V. Long-term culture with 2,4-dichlorophenoxyacetic acid affects embryogenic competence in sugarcane callus via changes in starch, polyamine and protein profiles. PLANT CELL TISS ORG. 2020;140:415–29. 10.1007/s11240-019-01737-w [Google Scholar]
- 6.Bravo-Vázquez LA, Angulo-Bejarano PI, Bandyopadhyay A, Sharma A, Paul S. Regulatory roles of noncoding RNAs in callus induction and plant cell dedifferentiation. Plant Cell Rep. 2023;42:689–705. 10.1007/s00299-023-02992-0 [DOI] [PubMed] [Google Scholar]
- 7.Chen Y, Zhang M, Jin X, Tao H, Wang Y, Peng B, Fu C, Yu L. Transcriptional reprogramming strategies and miRNA-mediated regulation networks of Taxus media induced into callus cells from tissues. BMC Genomics. 2020;21:168. 10.1186/s12864-020-6576-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Z, Li J, Wang L, Li Q, Lu Q, Yu Y, Li S, Bai MY, Hu Y, Xiang F. Repression of callus initiation by the miRNA-directed interaction of auxin-cytokinin in Arabidopsis thaliana. Plant J. 2016;87:391–402. 10.1111/tpj.13211 [DOI] [PubMed] [Google Scholar]
- 9.Turner M, Nizampatnam NR, Baron M, Coppin S, Damodaran S, Adhikari S, Arunachalam SP, Yu O, Subramanian S. Ectopic expression of miR160 results in auxin hypersensitivity, cytokinin hyposensitivity, and inhibition of symbiotic nodule development in soybean. Plant Physiol. 2013;162(4):2042–55. 10.1104/pp.113.220699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang W, Choi MH, Noh B, Noh YS. Deshoot regenerationrcontrolledrolled by HEN1 and TCP3/4 in Arabidopsis. Plant Cell Physiol. 2020;61:1600–13. 10.1093/pcp/pcaa083 [DOI] [PubMed] [Google Scholar]
- 11.Zhang TQ, Lian H, Tang H, Dolezal K, Zhou CM, Yu S, Chen JH, Chen Q, Liu H, Ljung K, Wang JW. An intrinsic microRNA timer regulates progressive decline in shoot regenerative capacity in plants. Plant Cell. 2015;27(2):349–60. 10.1105/tpc.114.135186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anzano A, Ammar M, Papaianni M, Grauso L, Sabbah M, Capparelli R, Lanzotti V. Moringa oleifera aaphyto Phyto-chemicapharmacologicalooverviewerview. Horticulturae. 2021;7:409. 10.3390/horticulturae7100409 [Google Scholar]
- 13.Zarina, Wani AW, Rawat M, Kaur H, Das S, Kaur T, Akram N, Faisal Z, Jan SS, Oyshe NN. Medicinal utilization and nutritional properties of drumstick (Moringa oleifera)-A comprehensive review. Food SCI NUTR. 2024;12:4546–68. 10.1002/fsn3.4139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drisya-Ravi RS, Nair BR, Siril EA. Morphological diversity, phenotypic and genotypic variance and heritability esti-mates in Moringa oleifera Lam.: a less used vegetable with substantial nutritional value. GENET RESOUR CROP EV. 2021;68:3241–56. 10.1007/s10722-021-01183-8 [Google Scholar]
- 15.Lyons G, Gondwe C, Banuelos G, Mendoza C, Haug A, Christophersen O, Ebert AW. Drumstick tree (Moringa oleifera) leaves as a source of dietary selenium, sulphur and pro-vitamin A. Acta Hortic. 2017;1158:287–91. 10.17660/ActaHortic.2017.1158.32 [Google Scholar]
- 16.Rodriguez GM, Sibaja JC, Espitia PJP, Otoni CG. Antioxidant active packaging based on papaya edible films incorpo-rated with Moringa oleifera and ascorbic acid for food preservation. Food Hydrocolloids. 2020;103:105630. 10.1016/j.foodhyd.2019.105630 [Google Scholar]
- 17.de-Siqueira-Patriota LL, Marques-Ramos DB, Lima-Amorim-dos-Santos AC, Silva YA, Gama-e-Silva M, Lira-Torres DJ, Procopio TF, de -Oliveira AM, Breitenbach-Barroso-Coelho LC, Pontual EV. Antitumor activity of Moringa oleifera (drumstick tree) flower trypsin inhibitor (MoFTI) in sarcoma 180-bearing mice. Food Chem Toxicol. 2020;145:111691. 10.1016/j.fct.2020.111691 [DOI] [PubMed] [Google Scholar]
- 18.Zhang J, Pian R, Yang E, Zhou W, He Q, Chen X. In vitro induction and characterisation of tetraploid drumstick tree (Moringa oleifera Lam). Open Life Sci. 2020;15:840–7. 10.1515/biol-2020-0087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng M, Yang H, Yang E, Zou X, Chen X, Zhang J. Efficient in vitro shoot bud proliferation from cotyledonary nodes and apical buds of Moringa oleifera Lam. IND CROP PROD. 2022;187:115394. 10.1016/j.indcrop.2022.115394 [Google Scholar]
- 20.Yang E, Yang H, Li C, Zheng M, Song H, Zou X, Chen X, Zhang J. Genome-wide identification and expression analysis of the Aux/IAA Gene Family of the Drumstick Tree (Moringa oleifera Lam.) Reveals Regulatory effects on shoot regeneration. Int J Mol Sci. 2022;23:15729. 10.3390/ijms232415729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hess JF, Kohl TA, Kotrová M, Rönsch K, Paprotka T, Mohr V, Hutzenlaub T, Brüggemann M, Zengerle R. Nie-Mann S. Library preparation for next generation sequencing: a review of automation strategies. Biotechnol Adv. 2020;41:107537. 10.1016/j.biotechadv.2020.107537 [DOI] [PubMed] [Google Scholar]
- 22.Shyamli PS, Pradhan S, Panda M, Parida A. De novo Whole-Genome Assembly of Moringa olehelps identifyegenes regulatinglating DrstressStoleranceerance. Front Plant Sci. 2021;12:766999. 10.3389/fpls.2021.766999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kozomara A, Birgaoanu M, Griffiths-Jones S. miRBase: from microRNA sequences to function. Nucleic Acids Res. 2019;47:d155–62. 10.1093/nar/gky1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Friedländer MR, Mackowiak SD, Li N, Chen W, Rajewsky N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012;40(1):37–52. 10.1093/nar/gkr688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao Y, Cui Y, Zhao R, Chen X, Zhang J, Zhao J, Kong L. Cryo-Treatment enhances the embryogenicity of mature somatic embryos via the lncRNA–miRNA–mRNA network in White Spruce. Int J Mol Sci. 2022;23:1111. 10.3390/ijms23031111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fahlgren N, Carrington JC. miRNA target prediction in plants. Methods Mol Biol. 2010;592:51–7. 10.1007/978-1-60327-005-2_4 [DOI] [PubMed] [Google Scholar]
- 28.Yang E, Zheng M, Zou X, Huang X, Yang H, Chen X, Zhang J. Global transcriptomic analysis reveals differentially expressed genes involved in Embryogenic Callus induction in Drumstick (Moringa oleifera Lam). Int J Mol Sci. 2021;22(22):12130. 10.3390/ijms222212130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen S, Zhou Y, Chen Y, Gu J. Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34(17):i884–90. 10.1093/bioinformatics/bty560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang G, Yin H, Li B, Yu C, Wang F, Xu X, Cao J, Bao Y, Wang L, Abbasi AA, Bajic VB, Ma L, Zhang Z. Characterization and identification of long non-coding RNAs based on feature relationship. Bioinformatics. 2019;35(17):2949–56. 10.1093/bioinformatics/btz008 [DOI] [PubMed] [Google Scholar]
- 31.Kang YJ, Yang DC, Kong L, Hou M, Meng YQ, Wei L, Gao G. CPC2: a fast and accurate coding potential calculator based on sequence intrinsic features. Nucleic Acids Res. 2017;45(W1):W12–6. 10.1093/nar/gkx428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun L, Luo H, Bu D, Zhao G, Yu K, Zhang C, Liu Y, Chen R, Zhao Y. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res. 2013;41(17):e166. 10.1093/nar/gkt646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fukunaga T, Hamada M. RIblast: an ultrafast RNA-RNA interaction prediction system based on a seed-and-extension approach. Bioinformatics. 2017;33(17):2666–74. 10.1093/bioinformatics/btx287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ge W, Zhang Y, Cheng Z, Hou D, Li X, Gao J. Main regulatory pathways, key genes and microRNAs involved in flower formation and development of moso bamboo (Phyllostachys edulis). Plant Biotechnol J. 2017;15:82–96. 10.1111/pbi.12593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang X, Wang L, Yuan D, Lindsey K, Zhang X. Small RNA and degradome sequencing reveal complex miRNA regulation during cotton somatic embryogenesis. J Exp Bot. 2013;64:1521–36. 10.1093/jxb/ert013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Siddiqui ZH, Abbas ZK, Ansari MW, Khan MN. The role of miRNA in somatic embryogenesis. Genomics. 2019;111:1026–33. 10.1016/j.ygeno.2018.11.022 [DOI] [PubMed] [Google Scholar]
- 37.Song X, Li Y, Cao X, Qi Y. MicroRNAs and their Regulatory roles in Plant-Environment interactions. Annu Rev Plant Biol. 2019;70:489–525. 10.1146/annurev-arplant-050718-100334 [DOI] [PubMed] [Google Scholar]
- 38.Wu CC, Hsieh KT, Yeh SY, Lu YT, Chen LJ, Ku MSB, Li WH. Simultaneous detection of miRNA and mRNA at the single-cell level in plant tissues. Plant Biotechnol J. 2023;21:136–49. 10.1111/pbi.13931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wojcik AM. Research Tools for the Functional Genomics of Plant miRNAs during zygotic and somatic embryogenesis. Int J Mol Sci. 2020;21:4969. 10.3390/ijms21144969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ramakrishnan M, Zhou M, Ceasar SA, Ali DJ, Maharajan T, Vinod KK, Sharma A, Ahmad Z, Wei Q. Epigenetic modifications and miRNAs determine the transition of somatic cells into somatic embryos. Plant Cell Rep. 2023;42:1845–73. 10.1007/s00299-023-03071-0 [DOI] [PubMed] [Google Scholar]
- 41.Xu XP, Cao QY, Guan QX, Mohammadi MA, Di CR, Chen XH, Zhang ZH, Chen YK, Xu HX, Lin YL. Genome-wide identification of miRNAs and targets associated with cell wall biosynthesis: Differential roles of dlo-miR397a and dlo-miR408-3p during early somatic embryogenesis in longan. Plant Sci. 2022;323:111372. 10.1016/j.plantsci.2022.111372 [DOI] [PubMed] [Google Scholar]
- 42.Yasar S, Pulat E, Cakir O. Effects of nitrogen deficiency and drought stresses on miRNA expressions in Arabidopsis thaliana. PLANT CELL TISS ORG. 2024;157:11240. 10.1007/s11240-024-02754-0 [Google Scholar]
- 43.Szyrajew K, Bielewicz D, Dolata J, Wojcik AM, Nowak K, Szczygiel-Sommer A, Szweykowska-Kulinska Z, Jar-molowski A, Gaj MD. MicroRNAs are intensively regulated during induction of somatic embryogenesis in Arabidopsis. Front Plant Sci. 2017;8:18. 10.3389/fpls.2017.00018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu XM, Kou SJ, Liu YL, Fang YN, Xu Q, Guo WW. Genomewide analysis of small RNAs in nonembryogenic and embryogenic tissues of citrus: microRNA-and siRNA-mediated transcript cleavage involved in somatic embryogenesis. Plant Biotechnol J. 2015;13:383–94. 10.1111/pbi.12317 [DOI] [PubMed] [Google Scholar]
- 45.Qi Y, Wang S, Shen C, Zhang S, Chen Y, Xu Y, Liu Y, Wu Y, Jiang D. OsARF12, a transcription activator on auxin response gene, regulates root elongation and affects iron accumulation in rice (Oryza sativa). New Phytol. 2012;193:109–20. 10.1111/j.1469-8137.2011.03910.x [DOI] [PubMed] [Google Scholar]
- 46.Wang L, Liu N, Wang T, Li J, Wen T, Yang X, Lindsey K, Zhang X. The GhmiR157a-GhSPL10 regulatory module controls initial cellular dedifferentiation and callus proliferation in cotton by modulating ethylene-mediated flavonoid biosynthesis. J Exp Bot. 2018;69:1081–93. 10.1093/jxb/erx475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cai H, Yang C, Liu S, Qi H, Wu L, Xu LA, Xu M. MiRNA-target pairs regulate adventitious rooting in Populus: a functional role for miR167a and its target Auxin response factor 8. Tree Physiol. 2019;39:1922–36. 10.1093/treephys/tpz085 [DOI] [PubMed] [Google Scholar]
- 48.Dexheimer PJ, Cochella L, MicroRNAs. From mechanism to Organism. Front Cell Dev Biol. 2020;8:409. 10.3389/fcell.2020.00409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin Y, Lai Z, Tian Q, Lin L, Lai R, Yang M, Zhang D, Chen Y, Zhang Z. Endogenous target mimics down-regulate miR160 mediation of ARF10, -16, and – 17 cleavage during somatic embryogenesis in Dimocarpus longan lour. Front Plant Sci. 2015;6:956. 10.3389/fpls.2015.00956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dai X, Lu Q, Wang J, Wang L, Xiang F, Liu Z. MiR160 and its target genes ARF10, ARF16 and ARF17 modulate hypo-cotyl elongation in a light, BRZ, or PAC-dependent manner in Arabidopsis miR160 promotes hypocotyl elongation. Plant Sci. 2021;303:110686. 10.1016/j.plantsci.2020.110686 [DOI] [PubMed] [Google Scholar]
- 51.Xu Z, Zhang X, Su Y, Hu Y, Xu L, Wang J. Plant cell totipotency and regeneration. Scientia Sinica Vitae. 2019;49:1282–300. 10.1360/SSV-2019-0199 [Google Scholar]
- 52.Long JM, Liu CY, Feng MQ, Liu Y, Wu XM, Guo WW. miR156-SPL modules regulate induction of somatic embryogenesis in citrus callus. J Exp Bot. 2018;69:4141–4141. 10.1093/jxb/ery197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang J, Ariel F, Wang D. Plant long non-coding RNAs: biologically relevant and mechanistically intriguing. J Exp Bot. 2023;74(7):2364–73. 10.1093/jxb/erac482 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material 1: Table S1: RT and qPCR primers list of miRNAs.
Supplementary Material 2: Table S2: Statistics of data quality.
Supplementary Material 3: Table S3: The FPKM of novel miRNA.
Supplementary Material 4: Table S4: mRNA target genes prediction of DEM.
Supplementary Material 5: Table S5: Correlation between differentially expressed lncRNAs and DEGs
Supplementary Material 6: Fig. S1: Induction process of drumstick shoot regeneration.
Supplementary Material 7: Fig. S2: Functional enrichment analysis of target genes of DEMs. (A) KEGG pathway enrichment of target genes of 41 differentially expressed known miRNA. (B) KEGG pathway enrichment of target genes of 189 differentially expressed novel miRNA.
Supplementary Material 8: Fig. S3: Heatmap of lncRNA expression levels.
Supplementary Material 9: Fig. S4: Venn analysis of DElncRNAs of different comparison groups.
Data Availability Statement
The raw sequence data were deposited in the National Center for Biotechnology Information (NCBI, https://www.ncbi.nlm.nih.gov/) under accession number PRJNA771463. Small RNA-seq data was submitted to NCBI under accession number PRJNA814844. miRNA sequences were submitted to GenBank under accession number PQ469266 - PQ469548.