Abstract
Background
Undetected atrial fibrillation (AF) poses a significant risk of stroke and cardiovascular mortality. However, diagnosing AF in real-time can be challenging as the arrhythmia is often not captured instantly. To address this issue, a deep-learning model was developed to diagnose AF even during periods of arrhythmia-free windows.
Methods
The proposed method introduces a novel approach that integrates clinical data and electrocardiograms (ECGs) using a colorization technique. This technique recolors ECG images based on patients' demographic information while preserving their original characteristics and incorporating color correlations from statistical data features. Our primary objective is to enhance atrial fibrillation (AF) detection by fusing ECG images with demographic data for colorization. To ensure the reliability of our dataset for training, validation, and testing, we rigorously maintained separation to prevent cross-contamination among these sets. We designed a Dual-input Mixed Neural Network (DMNN) that effectively handles different types of inputs, including demographic and image data, leveraging their mixed characteristics to optimize prediction performance. Unlike previous approaches, this method introduces demographic data through color transformation within ECG images, enriching the diversity of features for improved learning outcomes.
Results
The proposed approach yielded promising results on the independent test set, achieving an impressive AUC of 83.4%. This outperformed the AUC of 75.8% obtained when using only the original signal values as input for the CNN. The evaluation of performance improvement revealed significant enhancements, including a 7.6% increase in AUC, an 11.3% boost in accuracy, a 9.4% improvement in sensitivity, an 11.6% enhancement in specificity, and a substantial 25.1% increase in the F1 score. Notably, AI diagnosis of AF was associated with future cardiovascular mortality. For clinical application, over a median follow-up of 71.6 ± 29.1 months, high-risk AI-predicted AF patients exhibited significantly higher cardiovascular mortality (AF vs. non-AF; 47 [18.7%] vs. 34 [4.8%]) and all-cause mortality (176 [52.9%] vs. 216 [26.3%]) compared to non-AF patients. In the low-risk group, AI-predicted AF patients showed slightly elevated cardiovascular (7 [0.7%] vs. 1 [0.3%]) and all-cause mortality (103 [9.0%] vs. 26 [6.4%]) than AI-predicted non-AF patients during six-year follow-up. These findings underscore the potential clinical utility of the AI model in predicting AF-related outcomes.
Conclusions
This study introduces an ECG colorization approach to enhance atrial fibrillation (AF) detection using deep learning and demographic data, improving performance compared to ECG-only methods. This method is effective in identifying high-risk and low-risk populations, providing valuable features for future AF research and clinical applications, as well as benefiting ECG-based classification studies.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12874-024-02421-0.
Keywords: Atrial fibrillation, Demographic information, Deep learning, Dual-input mixed neural network, ECG colorization, Sinus rhythm
Background
Atrial fibrillation (AF) is a common cardiac arrhythmia that increases the risk of stroke, heart failure, and even death. Its diagnosis relies on the recording of the diagnostic rhythm on electrocardiograms (ECGs). However, the early use of ECG in diagnosing AF disease is subject to several limitations. One major limitation is the reliance on capturing AF episodes during medical evaluation, which can be challenging due to AF’s intermittent and self-terminating nature. Patients may not exhibit arrhythmia during the assessment, leading to underdiagnosis. This constraint could lead to missed chances for early AF intervention, raising risks like stroke and mortality. AF is a complex disease often caused by underlying cardiac pathologies, such as structural heart disease, hypertension, or valve abnormalities. These underlying conditions can precipitate the onset and progression of AF and potentially be detected during sinus rhythm (SR) [1, 2]. Early approaches by machine learning may only partially consider subtle or hidden ECG features that may indicate an AF attack [3, 4]. With the advancement of artificial intelligence (AI) technology [5–7], these hidden ECG features can be analyzed and used for AF diagnosis even without overt arrhythmia. However, the diagnostic performance of these deep learning algorithms still needs improvement. Although AF detection by AI algorithm is probably superior to the usual practice (hazard ratio: 2.85), more than 92.4% of patients diagnosed as high risk did not have AF [5]. Furthermore, AI risk stratification is not linked to clinically relevant outcomes, including stroke or cardiovascular death.
Enhancing diagnostic performance is essential for implementing AI-assisted AF diagnosis at SR in real-world clinical settings [1, 2, 5–7]. This can be achieved by integrating comprehensive patient data like medical history, risk factors, and imaging findings into the AI algorithm [6]. Employing image processing techniques to convert ECG signals into images enhances feature analysis and might allow AF detection even during non-arrhythmic periods. Unlike the traditional focus on signal variations, this approach embraces color, shape, and texture for nuanced feature learning. Including richer patient data through color in ECG images facilitates intricate feature learning while maintaining the connection between medical conditions and clinical information. This innovation might foster a more robust and precise AF diagnosis model.
In the present work, we transfer the graphical representation of electrocardiograms through color transformation accompanied by clinical information. By embedding age details within the color-coded ECG images, we enhance the capability to discern AF, leveraging a dual-input mixed neural network architecture. Incorporating AI-assisted AF diagnosis into the traditional risk assessment enhances the categorization of patients based on their susceptibility to cardiovascular and overall mortality. Incorporating both patients’ clinical information and ECG features into the image processing significantly improves the diagnostic accuracy of AF, which holds the potential to transform AF screening and advance patient outcomes by enabling early interventions.
Methods
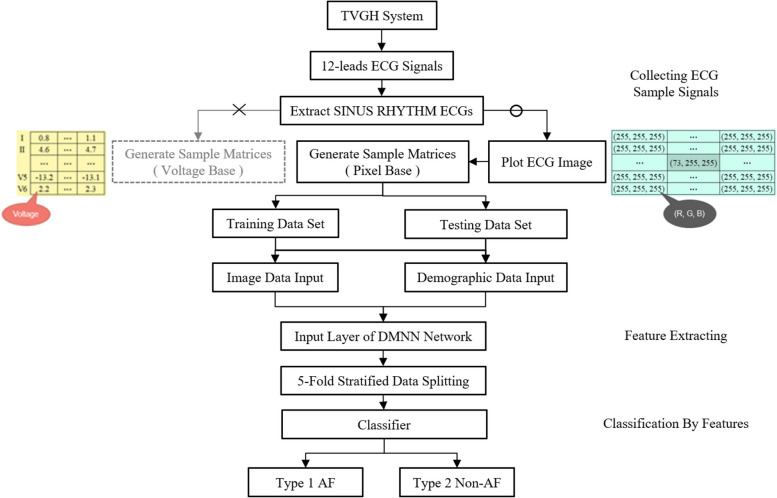
This paper develops an AI model for recognizing potential AF patients by sinus rhythm ECG. The general plan structure is displayed in Fig. 1. We propose a novel idea in order to permit the model to learn all the more successfully the ECG distinctions between patients. We transform demographic information into a coloring palette for more efficient processing to recolor the patient’s ECG to expand the advantage of image informational learning.
Fig. 1.
Program flow diagram
The more significant part of the past studies zeroed in on the learning of the original ECG signal or the ensemble learning of the clinical helper information and ECG, and no study has been done to add color information to the images by directly infusing the clinical helper information into the images in a color-coded way. Accordingly, we should utilize this review to look at the presentation distinction between the two. We propose a Dual-input Mixed Neural Network (DMNN) framework that combines patients’ ECG images and demographic information. The framework uses two training input data: the first being the image. We filter the sinus rhythm ECG from the original XML file and obtain 12 leads numerical data. Our developed program reproduces images similar to the Philips machines’. After completing the ECG image, we can quickly obtain the pixel values of the images and observe the spatial correlation between the leads more efficiently. Besides the location relationship between objects in an image being an influential feature in image classification, another significant feature is the image’s color. This part is often neglected in ECG studies. Because of this, we hope to enhance the feature of ECG image learning. We inject demographic information into ECG images by coloring them with rainbow colormap mapping in Matplotlib as one of the inputs for deep learning.
The second part of the dataset is numerical demographic data as another input for training. That data is put into the backbone network in two forms for deep learning. This chapter first describes the various datasets used in this study. It then explains signal processing, the network architecture design, implementation, and training, and finally explains the metrics used for evaluation.
Materials
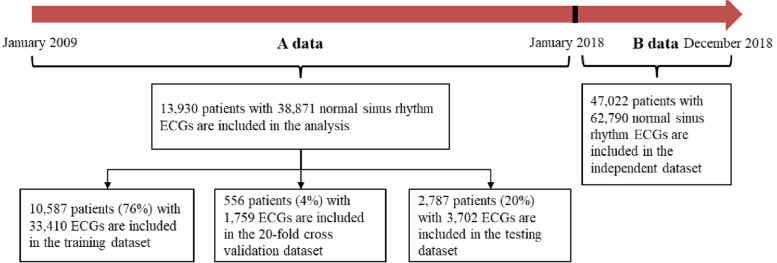
We included all patients aged 20 years or older who had at least one digital, normal sinus rhythm, standard 10-s, 12-lead ECG acquired from the Taipei Veterans General Hospital (TVGH) database between January 2009 and December 2017. We excluded patients with AF if their ECGs were obtained AF ECG through all the ECG recordings, those with poor ECG image quality, and those with a diagnosis code for atrial fibrillation but no ECG documentation to confirm the condition to eliminate diagnostic ambiguity. ECGs indicating paced rhythms were also excluded from the study. Ultimately, the cohort comprised 13,930 patients with 38,871 standard 12-lead sinus rhythm ECGs, all of whom had well-documented clinical and imaging data at the study's outset. The ECGs were recorded using the Philips M4994 device, with a sample rate of 500 Hz. All experimental protocols were approved by the Institutional Review Board (IRB) of the Taipei Veterans General Hospital, where data collection was performed. Based on medical diagnostic reports, patients were classified into two categories: atrial fibrillation (AF) and non-atrial fibrillation (NAF). AF patients were defined as those who had their first recorded ECG as the index ECG during the study period annotated as atrial fibrillation (including atrial flutter). Any subsequent sinus rhythm ECGs recorded after the index ECG were categorized as AF. Conversely, NAF patients had no detected records of atrial fibrillation throughout the study period additionally without reference to AF in the diagnostic codes in their electronic medical record, and all their sinus rhythm ECGs were classified as NAF. According to the patient identity numbers, 80% were in the training set (11,143 patients), and 20% were in the test set (2,787 patients), with a 1:3 ratio between AF and NAF. A statistical analysis of the training and test data is displayed in Fig. 2. A patient cannot be classified into both the training and test sets at the same time to prevent cross-contamination, as shown in Fig. 3. However, the same patient could have multiple ECG recordings at different time points and be only grouped into a single side dataset.
Fig. 2.
Patient flow diagram. A separate dataset was taken out of 2018 (B data) for extra validation, and data before 2018 (A data) was split into training, validation, and test sets. ECG = electrocardiograph
Fig. 3.
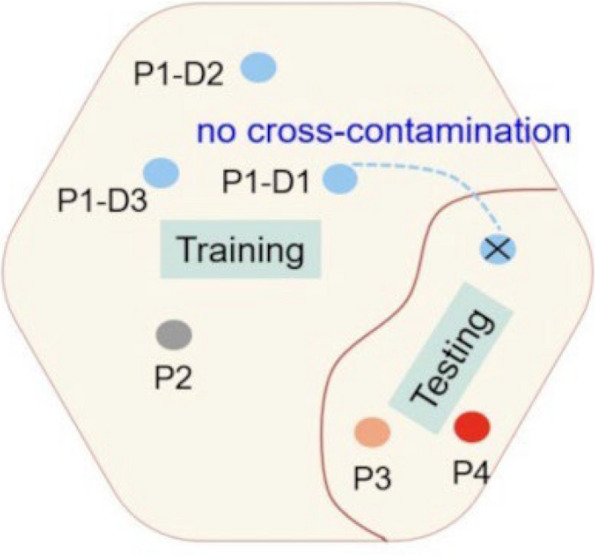
Ensure that training, validation, and test data sets are kept free of cross-contamination. The goal is to ensure a robust and reliable data set for the network’s training, validation, and testing. Once a patient’s data is placed in one of the datasets, that patient’s data is only used in that set, thus avoiding “cross-contamination” between training, validation, and testing datasets. A list of four patients (P1, P2, P3, and P4) and P1-D1, P1-D2, and P1-D3 represent three different ECGs taken by patient1 at different dates and times
Signal processing
The sinus rhythm ECG was used as the module training data source. Philip’s machine produces ECG signals in XML format, and the patient’s personal information, previous medical records, and ECG signal value are extracted for input characteristics of the module development. Analyze the XML text file using an SPxml parser to obtain the patient number, medical record number, gender, age, ECG acquisition date, and sinus rhythm tag. The AI module will utilize these demographic values as one of its input data. We use a specially developed ECG generator to standardize and denoise the data. The first step is to filter out noise from all ECGs. Low-frequency noise from breathing or patient movement, which typically causes baseline drift in ECGs, is removed using a high-pass filter to remove frequencies below 1 Hz. A low-pass filter removes excessive frequencies to ensure the ECG signals are free from high-frequency noise. The second step is to transpose the input of the 12-lead ECG. We conducted experiments to confirm the robustness and accuracy of the model. The input ECG datasets consist of 12 leads, each containing 5000 time points, sampled at a frequency of 500 Hz. After transposition, the input ECG is set to have a time step of 5000, with the input data for each time step having a size of 12. This configuration allows the DMNN model to process the information from each time point for all leads sequentially. The generator then extracts signal values from these 12 leads and converts them into 10-s 12-lead ECG images.
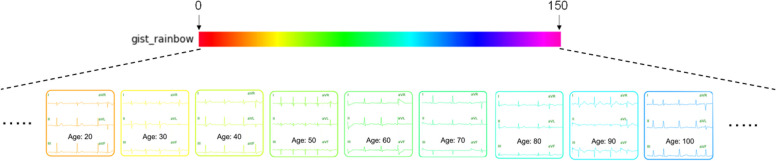
Next, we will colorize the ECG images, and the coloring rules are shown in Fig. 4 and Eqs. (1)-(3). Each patient’s ECG is assigned a color regarding the patient’s age which is the most distinctive feature. The following briefly describes how the ECG is colored: First, a palette color range is defined, here set to 150, meaning that all patient ages are included. Equation (1) maps age to color space location:
| 1 |
where age is the patient’s age in years, cage is the corresponding color space location, and ⌊·⌋ denotes the floor function. In (2), SCPhex denotes the hexadecimal color space representation of the rainbow color palette. From rainbow color palette, a color space location is selected in the #x1y1x2y2x3y3 representation to correspond to the age space location.
| 2 |
where SCPhex is a function that returns the hexadecimal color code from the rainbow color palette at the given position, and color is the selected color. The color is then converted to the RGB space representation by (3).
| 3 |
where x1, y1, x2, y2, x3, and y3 are the hexadecimal digits of the color code in #x1y1x2y2x3y3 format, and (R, G, B) is the corresponding RGB color value. The ECG will be re-colored with this color, and patients of different ages will have different colors of ECG, as shown in Fig. 4. Using the above method, the characteristic information of age can be enlarged to the whole image. In addition, the original correlation between age and disease can be maintained.
Fig. 4.
The ECG coloring mechanism converts to rainbow color distribution according to age-feature values. A maximum age range is set as the color distribution range, and each patient’s age is mapped to a color (HTML color) in #RRGGBB format. The patient’s ECG was recolored with updated R, G, and B values converted from HTML color to (R, G, B) tuples. By age-coding ECG coloring, a simple single feature value is amplified and strengthened while maintaining a correlation between age and disease
In contrast with the traditional approach, they do not consider color but only numerical values or waves of leads in space. For image classification, color is also a key feature. Performance can be improved by making efficient use of the correspondence between numerical values and color. The colorized image is used as the second input to the model. Lastly, the deep learning model is trained based on dual input. As a side note, we are focusing on a change in wave patterns. To avoid the influence of grid features, we remove the grid and patient record data from the image. The final generated image is used as training data and test data. We present a dual input model that creatively integrates ECG images with demographic data. We describe our approach of mapping demographic features onto a color space and applying this innovative color-coding directly to the ECG images. This technique transforms demographic data from simple numerical values into a visually colorized distinct format and enriches the waveform data. By converting demographic information into appropriate colors for different groups, we enhance the model's ability to identify complex patterns. This increased variety and spatial representation of image features enriches the interpretative depth of our model and exploits the inherent correlations between clinical and demographic data. These advances significantly improve the performance and classification capabilities of our model.
Network design and training
To predict atrial fibrillation, we designed a Dual-input Mixed Neural Network. Figure 5 shows the framework diagram for this model. As mentioned earlier, we combine two types of training data (colored ECG images and demographic data) and input them into the training model. Our development is based on the Xception backbone model due to its efficiency and suitability for our limited hardware resources, which made it the most practical choice for our needs. To address the issue of data imbalance, we employed a technique that automatically assigns balanced class weights, which are inversely proportional to their respective frequencies. This approach ensures that the model is not biased towards more frequent classes. Utilizing a pre-trained model from ImageNet [8] for transfer learning, we selected binary cross-entropy as our loss function due to its suitability for binary classification problems. For hyperparameter optimization, we harnessed the power of the Hyperband search algorithm. This efficient algorithm thoroughly explores the hyperparameter space, enabling us to identify the optimal settings. To counter the risk of overfitting, we introduced L2 regularization [9]. This technique, by adding a penalty proportional to the square of the magnitude of the model coefficients to the loss function, effectively reduces the model's complexity and prevents it from fitting noise in the training data. ImageNet [8], a significant visual database designed for visual object recognition software research, played a pivotal role in our transfer learning process. It provided a robust starting point, allowing us to leverage pre-trained weights and make substantial improvements to our model's performance. We also implemented L2 regularization [9], a widely used technique in machine learning, to prevent overfitting by penalizing large coefficients and promoting simpler models. Following the 20-fold validation evaluation, we evaluated the AUC, accuracy, sensitivity, specificity, and F1 scores. We selected the patient as the target in the data selection process and adopted measures to prevent cross-contamination. If a patient has multiple ECGs, the patient’s data may be included in training and the test sets. These ECGs may be related to each other, resulting in cross-contamination problems. To avoid this issue, in this study, all ECGs from the same patient will only appear in either one training set or one testing set when selecting data collection.
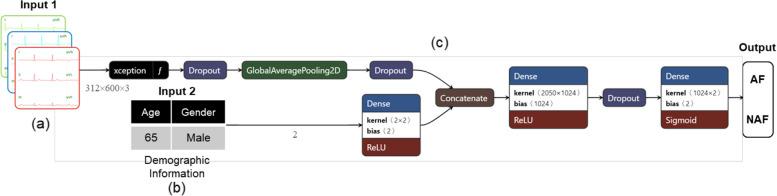
Fig. 5.
The architecture of a Dual-input Mixed Neural Network with ECG coloring technology (a) generates colored ECG based on the patient’s demographic information. The conversion mechanism is illustrated in Fig. 4. The demographic information (b) in the original numerical format is treated as another input data and put into (c) together with (a). Xception is used as the backbone for model training. The final prediction of AF and Non-AF prediction models are obtained
The DMNN model combines numerical classification data and image data to handle different types of inputs in a more beneficial way for prediction performance and incorporates the mixed characteristics of the data into the training model. Figure 5(a) shows the ECGs with coloring. Ages that are close to each other are configured with similar colors. When a particular age group has a high incidence rate, the weight for that color will also increase. Figure 5(b) is the demographic data. The label for each patient corresponds to two input forms. The first input form is pixel-format image data, and the second is primary patient data with numerical classification. Next, combine the two inputs and train the model. Prediction models are developed by combining information from various data sources. We propose a framework that is significantly more flexible than the neural network trained only on ECG signals. Since hardware resources are limited, we chose Xception, which increases computational efficiency without increasing network complexity (model parameters), as the backbone network. The trained models were validated with three data sets: the validation set (20-fold), the pre-2018 test set, and the 2018 independent test data. The proposed network architecture is evaluated by assuming the class containing AF signals as the positive class and using various metrics to measure its performance. These metrics include accuracy, sensitivity, specificity, F1 score, and AUC (Area Under the Curve).
| 4 |
| 5 |
| 6 |
| 7 |
| 8 |
True Positive (TP) is the number of AF signals that are correctly classified as such.
False Positive (FP) is the number of signals without AF incorrectly classified as AF.
True Negative (TN) is the number of signals without AF that are correctly classified as such.
False Negative (FN) is the number of AF signals incorrectly classified without AF.
Results
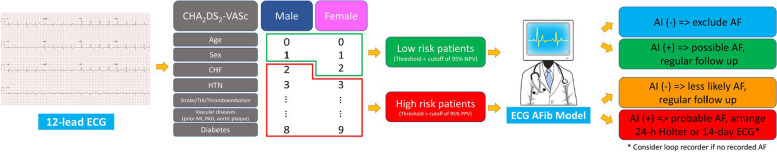
Among 13,930 patients, the distribution between AF and NAF groups was at a 1:3 ratio, with 3,480 patients in the AF group and 10,450 patients in the NAF group. The performance of the proposed network architecture is evaluated using various metrics, including accuracy, sensitivity, specificity, F1 score, and AUC. The evaluation compares the input data as signal values (Scenario I) with colored ECG images (Scenario II) in Sect. 3.1. Section 3.2 proposes a risk-scoring algorithm for clinical application (Fig. 6), which classifies patients into high-risk and low-risk populations based on CHA2DS2-VASc risk scores and AI prediction.
Fig. 6.
Integrating Clinical Risk Scoring with AI Prediction in the Clinical Workflow. An ECG from a specific patient, captured using a 12-lead system, is initially categorized based on the CHA2DS2-VASc score: men with a score ≥ 2 and women with a score ≥ 3 are classified as high-risk. Conversely, men with a score of 0 or 1 and women with scores of 0, 1, or 2 are deemed low-risk. This classification is based on the presence of associated comorbidities. Upon classification, the ECG is processed through the AI model, which predicts the risk as either AF or non-AF for both the high-risk and low-risk groups. The resultant cardiovascular and all-cause mortality risks are represented in blue, green, orange, and red, corresponding to the survival curves illustrated in Fig. 8. Of paramount concern is the high-risk group where AI predicts AF. Individuals within this category require intensive monitoring due to the significantly elevated cardiovascular and all-cause mortality rates. Conversely, the remaining populations necessitate regular follow-ups, which are deemed adequate
Evaluation of performance measures
We compare the input data as signal values (Scenario I) with our colored ECG images (Scenario II). The results of the experiment between the two are recorded in Table 1. The data results in the table are calculated using the threshold defined by the optimal sensitivity and specificity that maximizes sensitivity + specificity—1 (based upon the Youden index [10]) for all metric performance evaluations.
Table 1.
Overall classification results for AF
| Models | Inputs | Coloring | Dataset | Accuracy | Sensitivity | Specificity | F1 score | AUC |
|---|---|---|---|---|---|---|---|---|
| CNN (Xception) | Signal (Voltage) | No | Validation (Best) | 0.860 | 0.842 | 0.863 | 0.645 | 0.919 |
| Validation (Avg.) | 0.842 | 0.800 | 0.850 | 0.607 | 0.892 | |||
| Test (< 2018) | 0.867 | 0.803 | 0.882 | 0.695 | 0.903 | |||
| Test (2018) | 0.698 | 0.720 | 0.696 | 0.314 | 0.758 | |||
| DMNN (Xception) | Colored Image (Pixel) + Demographic | Yes | Validation (Best) | 0.951 | 0.906 | 0.958 | 0.848 | 0.972 |
| Validation (Avg.) | 0.955 | 0.894 | 0.966 | 0.858 | 0.964 | |||
| Test (< 2018) | 0.939 | 0.847 | 0.960 | 0.839 | 0.945 | |||
| Test (2018) | 0.782 | 0.761 | 0.784 | 0.400 | 0.834 |
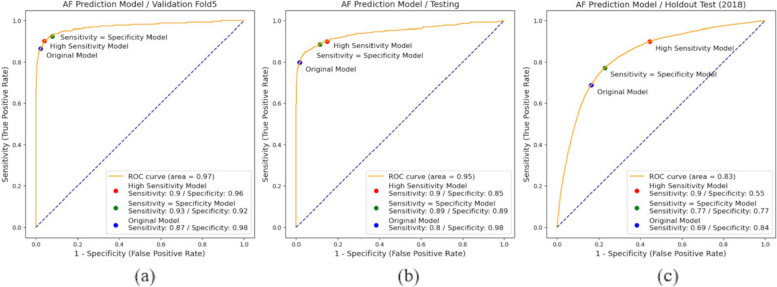
The DMNN network demonstrated compelling performance on the test set, showcasing an AUC of 94.5%, ac-curacy at 93.9%, sensitivity reaching 84.7%, specificity of 96%, and an F1 score of 83.9%. When tested on an independent dataset from beyond the classified 2018 dataset, the AUC was 83.4%. For the evaluation of AUC performance improvement, the DMNN architecture exhibited a 4.2% enhancement over the signal-based CNN model in the pre-2018 test set. Moreover, in the 2018 independent test data, the DMNN architecture achieved a performance boost of 7.6% compared to the signal-based CNN model. These outcomes notably outperformed the use of solely original signal values as input for the convolutional network. The ROC curves of DMNN for AF in validation, pre-2018, and 2018 independent test data are shown in Fig. 7. Figure 7(a) shows the fifth fold of the 20-fold cross-validation (with the highest AUC). Figure 7(b) contains the test set before 2018. Figure 7(c) displays the ROC curve of the independent test dataset for 2018. We adjusted the diagnostic thresholds of AUC for AF detection including high sensitivity (sensitivity score greater than or equal to 90%), sensitivity = specificity, and original (threshold = 0.5). These thresholds have been applied in the clinical diagnostic tools [11]. For example, in Fig. 7(b), the threshold difference is shown for high sensitivity (threshold = 0.12), sensitivity = specificity (threshold = 0.15), and original cases (threshold = 0.5).
Fig. 7.
Dual-input Mixed Neural Network with varying threshold models for (a) optimized AUC split validation, (b) testing data ranging from 2009 to 2018, and (c) individual testing data in 2018
Clinical risk scoring combined with AI prediction
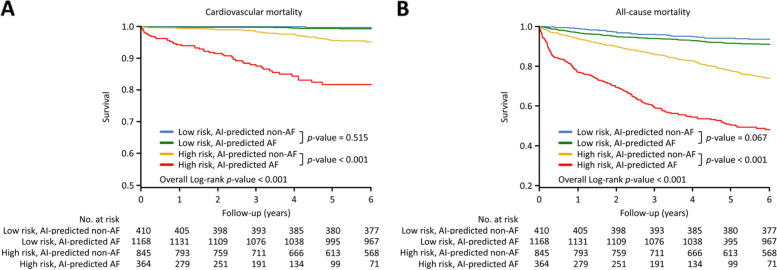
A risk-scoring algorithm was proposed for clinical application (Fig. 6). A total of 2,787 patients in the test set (Pre-2018) were applied according to the guideline-based risk scores (Supplementary Table s1) [12–14]. Patients were divided into high-risk (Table 2, male with CHA2DS2-VASc scoring ≥ 2; female with CHA2DS2-VASc scoring ≥ 3) and low-risk (male with CHA2DS2-VASc scoring 0 and 1; female with CHA2DS2-VASc scoring 0, 1 and 2) populations. The patients were followed for a period of 71.6 ± 29.1 months since the ECG test. In the high-risk population during six-year follow-up, AI-predicted AF patients had higher cardiovascular mortality (AF vs. non-AF; 47 [18.7%] vs. 34 [4.8%] patients, p < 0.001) and all-cause mortality (176 [52.9%] vs. 216 [26.3%] patients, p < 0.001) than AI-predicted non-AF patients by the Kaplan–Meier survival analysis (Fig. 8). In the low-risk population, AI-predicted AF patients had marginally higher cardiovascular (7 [0.7%] vs. 1 [0.3%] patients, p = 0.515), and all-cause mortality (103 [9.0%] vs. 26 [6.4%] patients, p = 0.067) than AI-predicted non-AF patients during six-year follow-up.
Table 2.
Basic characteristics of the study population
| Baseline | Group 1 | Group 2 | Group 3 | Group 4 | p value |
|---|---|---|---|---|---|
| characteristics | (N = 410) | (N = 1168) | (N = 845) | (N = 364) | |
| Age | 47.5 ± 14.1 | 49.4 ± 14.7 | 73.4 ± 11.5 | 77.1 ± 10.8 | < 0.001 |
| Male, n (%) | 172 (42) | 577 (49.4) | 508 (60.1) | 236 (64.8) | < 0.001 |
| HTN, n (%) | 53 (12.9) | 204 (17.5) | 624 (73.9) | 276 (75.8) | < 0.001 |
| DM, n (%) | 17 (4.2) | 40 (3.4) | 330 (39.1) | 129 (35.4) | < 0.001 |
| CHF, n (%) | 2 (0.5) | 21 (1.8) | 115 (13.6) | 164 (45.1) | < 0.001 |
| Prior stroke/ TIA/ TE, n (%) | 0 (0) | 0 (0) | 142 (16.8) | 101 (27.8) | < 0.001 |
| Prior vascular diseases, n(%)* | 4 (1) | 3 (0.3) | 67 (7.9) | 57 (15.7) | < 0.001 |
Values are numbers and percentages (%) of the variables or the mean ± standard deviation.
Abbreviations: CHF chronic heart failure, DM diabetes mellitus, HTN hypertension, TE thromboembolism, TIA transient ischemic attack
*Prior vascular diseases included prior myocardial infarction, peripheral artery disease or aortic plaque
Fig. 8.
Kaplan–Meier survival curves for AI-classified AF or non-AF patients. AI-predicted AF: patients classified by the AI model as having AF. AI-predicted non-AF: patients classified by the AI model as having non-AF. a Cardiovascular mortality. b All-cause mortality. AF, atrial fibrillation; AI, artificial intelligence
Discussion
There are several challenges to be addressed in this research. Firstly, the prediction of concealed atrial fibrillation relies on identifying its diagnostic features from normal electrocardiograms (ECGs) that exhibit a sinus rhythm. Clinicians cannot diagnose atrial fibrillation solely based on these ECGs. Secondly, various comorbidities have made training more challenging when conducting binary classification problems for atrial fibrillation (AF) and non-AF cases. In previous studies [15–20], machine learning techniques were used to classify 12-lead ECG signals. There have been several studies using deep learning for classification [1, 2, 21–28] and studies using single-lead classification [29–32]. Other studies have employed deep learning convolutional training and machine learning classifiers for AF classification [33, 34]. Moreover, some studies have incorporated demographic information into ensemble learning [35] and analyzed AF prediction according to different demographic groups [6]. This article introduces a novel approach incorporating demographic information into the image transfer process using 12-lead signal values. The demographic features are mapped to different colors within the rainbow to recolor the original ECG image. For example, this method ensures that ECGs of patients aged 30 and 70 are visually differentiated based on their demographic characteristics [36]. The study makes significant contributions in two main areas: (1) Using chromatics and images combined with the dual input model containing demographic information enhances feature learning efficiency, enables comprehensive sample learning, reduces training time, and improves prediction accuracy. (2) The precise screening of AF patients is essential to reduce clinical waste. By combining the CHA2DS2-VASc clinical risk scoring with the predicted results from the DMNN atrial fibrillation AI model, patients can be categorized into high-risk, moderate-risk, and low-risk groups. This twofold check component enables more accurate identification of patients requiring clinical attention. The study’s results demonstrate that AI can effectively utilize ECGs recorded during normal sinus rhythm to detect atrial fibrillation, which is typically easily accessible and cost-effective. The ability to identify undetected atrial fibrillation through these tests has significant practical implications, particularly in screening and managing atrial fibrillation patients. Furthermore, it provides valuable decision-making support for cardiology specialists in outpatient diagnoses.
DMNN framework in ECG analysis
Image classification is a mature technology in deep learning that can effectively distinguish differences between images by learning graphics’ shape, color, and texture features. Image classification has been widely used in many research areas, such as facial recognition and traffic violation enforcement. However, image classification has yet to be extensively studied in the ECG classification of medical field. In early ECG classification problems, machine learning [16–19] or deep learning techniques [1, 23, 24] were developed directly by reading ECG signal patterns. There needs to be more research that re-visualizes ECG signal patterns into images and then trains deep learning models. In addition, the coloring clustering approach of ECG images is first proposed in this study. It decisively overcomes the problems of not being able to color ECG signal patterns directly (because it is not pixel-based) and ignoring the critical use of color features in image classification while only considering the waveform features (shape) of ECG images. This new method learns the variations of ECG waveforms and, importantly, injects demographic feature clustering (represented by different colors). For patients of the same age group, the relationship between waveform (shape), age (color), and disease is extended, rather than just the relationship between waveform (shape) and disease. The use of the ECG coloring method with dual input and image transformation in experimental results has shown significant improvements in performance metrics compared to traditional models that only utilize machine signal voltage values. In the pre-2018 test set, there was a 4.2% increase in AUC performance, a 7.2% increase in accuracy, a 4.1% increase in sensitivity, and a 7.8% increase in specificity. In the 2018 independent test data, there was a 7.6% increase in AUC performance, an 11.3% increase in accuracy, a 9.4% increase in sensitivity, and an 11.6% increase in specificity. Additionally, the F1 score showed an 8.6% improvement in the pre-2018 test set and a 25.1% improvement in the 2018 independent test data. In addition, our model showed significant performance in clinical trials, and we will next discuss the experimental analysis of clinical applications.
An Innovative approach using hierarchical AI-Assisted ECG screening
Diagnosing AF can be challenging, primarily due to its intermittent and often asymptomatic nature. The under-diagnosis of AF is tied to increased adverse cardiovascular outcomes including stroke, heart failure, and mortality. Therefore, various risk prediction models, such as the FHS (Framingham Heart Study) [37], ARIC (Atherosclerosis Risk in Communities Study) [38], and CHARGE-AF (Cohorts for Heart and Aging Research in Genomic Epidemiology–Atrial Fibrillation) score [39], have been proposed. However, these models rely heavily on extensive clinical variables, some of which are not easily accessible, particularly for general practitioners. These include parameters like heart murmurs and echocardiographic readings. Moreover, their predictive accuracy remains unsatisfactory, with AUCs hovering around 0.77 to 0.78.
Several AI-assisted ECG algorithms are capable of detecting AF during sinus rhythm with AUCs of 0.79 to 0.87 [1, 5–7]. One of these algorithms has been tested in the multicenter clinical trial [5]. The AI-guided targeted screening approach increased the yield for atrial fibrillation detection and could improve the effectiveness of atrial fibrillation screening. However, it’s concerning that overdiagnosis risk might be significant, given that more than 92.4% of patients initially classified as high-risk did not manifest AF. Furthermore, a critical question looms over whether the AI algorithm designed for AF detection during sinus rhythm can effectively distinguish adverse cardiovascular outcomes. The determination of the AI algorithm’s clinical relevance in the context of stroke or mortality could potentially reshape clinical decision-making and prompt early interventions, including the consideration of anticoagulants [12]. Our model has demonstrated a diagnostic performance that is comparable to previous models. Moreover, when incorporated into AF risk scoring for stroke, the current model exhibits the potential to enhance risk differentiation for cardiovascular and overall mortality. Within the subset of patients with high stroke-risk scores, noteworthy cardiovascular and all-cause mortalities were observed during long-term follow-up among those with AI-predicted AF, in contrast to those without such predictions. In this context, it is imperative to arrange comprehensive ECG monitoring to validate AF diagnoses, aligning closely with existing diagnostic and treatment protocols [10]. Conversely, within low-risk AF populations, survival rates remained consistently modest both among AI-predicted AF patients and non-AF individuals. This finding suggests that due to the limited morbidity risks associated with AF in these cases, the survival advantages of screening this group are marginal.
In the clinical scenario and practice, while the risk of overdiagnosis cannot be overlooked, the use of AI significantly enhances the ability to detect undetected AF, which is crucial for preventing serious outcomes such as stroke and cardiovascular mortality. To mitigate the issue of false positives, the AI model should be integrated into a structured clinical workflow where its predictions serve as an initial screening tool, followed by confirmatory tests, such as 24-h Holter, 7-day, 14-day or 30-day ECG patches or longer ECG monitoring, before any invasive procedures are undertaken. This approach not only preserves the clinician's judgment but also reduces unnecessary patient anxiety and treatment. Furthermore, the early detection and management of AF facilitated by AI can lead to significant healthcare savings by avoiding the costs associated with treating advanced-stage cardiovascular conditions. Therefore, the clinical and economic advantages of using AI for AF detection strongly support its adoption in medical practice, provided it is used judiciously within a robust diagnostic framework.
Limitations
One limitation of our study is the potential geographic and demographic bias, as our initial dataset was derived from a single institution. Additionally, while our innovative approach of color-coding demographic information into ECG images has shown promising results, its effectiveness needs further validation across diverse populations, institutions, and different types of equipment. We achieved an AUC of 83.4% in our initial validation using an independent dataset from 2018, but ongoing evaluations are necessary to ensure the model's robustness and generalizability in varied clinical settings in the medical community. A pragmatic randomized controlled trial is essential to validate our AI model across a diverse patient population.
In this study, we represent an initial approach to utilizing a color-coding mechanism with demographic data for feature enhancement. Collecting more physiological parameters may further enrich the color mapping process across different leads, capturing a broader range of features. This expansion could enhance the model's ability to discern nuanced variations and improve classification performance. Therefore, future research exploring these methods and integrating more advanced machine learning models may enhance prediction accuracy and overall robustness.
Conclusions
This study introduces a novel approach to enhance atrial fibrillation (AF) detection using deep learning and demographic data, improving performance compared to ECG-only methods. This method effectively aids in identifying high and low-risk populations, offering valuable features for future AF research and clinical applications, while also benefiting ECG-based classification studies.
Supplementary Information
Abbreviations
- AF
Atrial Fibrillation
- AUC
Area Under the Curve
- CNN
Convolutional Neural Network
- DMNN
Dual-input Mixed Neural Network
- ECG
Electrocardiogram
- SR
Sinus Rhythm
Authors’ contributions
WWC and CML wrote the main manuscript text. YFH, SAC, and HHSL coordinated the project and approved the final version for submission. CCT, CCH, ICW, PFC, SLC, YJL, LWL, FPC, TFC, TCT, JNL, CYL, TYC, LK, CIW, SHL, and JCHW collected and verified the data. All authors reviewed the manuscript.
Funding
This work received funding from various sources, including the National Science and Technology Council (Grants:110–2811-M-A49-550-MY2, 110–2118-M-A49-002-MY3, 111–2634-F-A49-014-, 112–2321-B-075–002-, 112–2634-F-A49-003-, 113–2118-M-A49-007-MY2, 113–2314-B-075–034-MY3, 113–2923-M-A49-004-MY3, 113–2321-B-075A-002-, 113–2622-E-A49-010-, 113–2628-B-A49-016-), the Higher Education Sprout Project of the National Yang Ming Chiao Tung University from the Ministry of Education, and the Yushan Scholar Program of the Ministry of Education, Taiwan. Additional support was provided by Taipei Veterans General Hospital (Grants: VGH108C-019, VN108-12, VN109-03, V109C-070, V110C-039, V110B-043, V113B-008), the Ministry of Science and Technology (Grants: 108–2628-B-075–003, 109–2628-B-075–017, 109–2321-B-009–007, 109–2314-B-075–077, 110–2628-B-075–015, 110–2314-B-075–063-MY3, 110–2321-B-075–002, 110–2321-B-A49-003, 110–2634-F-A49-005-SP3, 110–2118-M-A49-002-MY3, 110–2634-F-A49-005-, 111–2634-F-A49-014-), the National Yang Ming Chiao Tung University (Grants: 113W090271, 113W080409), the National Health Research Institutes (Grants: EX113-11314SI), and Academia Sinica (Grant: AS-TM-112–01-01). We also thank Wan-Yi Tai for her valuable assistance and acknowledge the National Center for High-performance Computing for providing computing resources. The funding bodies played no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Data availability
The data that support the findings of this study are available from the corresponding author on request.
Declarations
Ethics approval and consent to participate
This study was approved by the Institutional Review Board (2017–10-009BC) at Taipei Veterans General Hospital, Taipei, Taiwan. All methods were carried out following the regulations of the Institutional Review Board. The Internal Review Board of Taipei Veterans General Hospital granted an exemption from the need to secure informed consent due to the thorough de-identification of patient data.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Wei-Wen Chen and Chih-Min Liu contributed equally to this work.
Contributor Information
Yu-Feng Hu, Email: yfhu0609@nycu.edu.tw.
Shih-Ann Chen, Email: epsachen@ms41.hinet.net.
Henry Horng-Shing Lu, Email: henryhslu@kmu.edu.tw.
References
- 1.Attia ZI, Noseworthy PA, Lopez-Jimenez F, Asirvatham SJ, Deshmukh AJ, Gersh BJ, et al. An artificial intelligence-enabled ECG algorithm for the identification of patients with atrial fibrillation during sinus rhythm: a retrospective analysis of outcome prediction. Lancet. 2019;394(10201):861–7. 10.1016/S0140-6736(19)31721-0. [DOI] [PubMed] [Google Scholar]
- 2.Baek YS, Lee SC, Choi W, Kim DH. A new deep learning algorithm of 12-lead electrocardiogram for identifying atrial fibrillation during sinus rhythm. Sci Rep. 2021;11(1):12818. 10.1038/s41598-021-92172-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liaqat S, Dashtipour K, Zahid A, Assaleh K, Arshad K, Ramzan N. Detection of atrial fibrillation using a machine learning approach. Information. 2020;11(12):549. 10.3390/info11120549. [Google Scholar]
- 4.Tseng AS, Noseworthy PA. Prediction of atrial fibrillation using machine learning: a review. Front Physiol. 2021;12:1873. 10.3389/fphys.2021.752317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Noseworthy PA, Attia ZI, Behnken EM, Giblon RE, Bews KA, Liu S, et al. Artificial intelligence-guided screening for atrial fibrillation using electrocardiogram during sinus rhythm: a prospective non-randomised interventional trial. Lancet. 2022;400(10359):1206–12. 10.1016/S0140-6736(22)01637-3. [DOI] [PubMed] [Google Scholar]
- 6.Melzi P, Tolosana R, Cecconi A, Sanz-Garcia A, Ortega GJ, Jimenez-Borreguero LJ, Vera-Rodriguez R. Analyzing artificial intelligence systems for the prediction of atrial fibrillation from sinus-rhythm ECGs including demographics and feature visualization. Sci Rep. 2021;11(1):22786. 10.1038/s41598-021-03535-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hygrell T, Mant J. An artificial intelligence-based model for prediction of atrial fibrillation from single-lead sinus rhythm ECGs facilitating screening. Europace. 2023. 10.1093/europace/euad036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deng, J., Dong, W., Socher, R., Li, L.J., Li, K, Fei-Fei, L. ImageNet: A large-scale hierarchical image database. IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Miami, 2009:248–255. 10.1109/CVPR.2009.5206848
- 9.Ng, A.Y. Feature Selection, L1 vs. L2 Regularization, and Rotational Invariance. Proceedings of the 21st International Conference on Machine Learning (ICML), Banff, 2004;78. 10.1145/1015330.1015435
- 10.Schisterman EF, Perkins NJ, Liu A, Bondell H. Optimal cut-point and its corresponding Youden Index to discriminate individuals using pooled blood samples. Epidemiology. 2005;16:73–81. 10.1097/01.ede.0000147512.81966.ba. [DOI] [PubMed] [Google Scholar]
- 11.Galloway CD, Valys AV, Shreibati JB, Treiman DL, Petterson FL, Gundotra VP, Albert DE, Attia ZI, Carter RE, Asirvatham SJ, Ackerman MJ, Noseworthy PA, Dillon JJ, Friedman PA. Development and validation of a deep-learning model to screen for hyperkalemia from the electrocardiogram. JAMA cardiology. 2019;4(5):428–36. 10.1001/jamacardio.2019.0640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.January CT, Wann LS, Calkins H, Chen LY, Cigarroa JE, Cleveland JC Jr, Ellinor PT, Ezekowitz MD, Field ME, Furie KL, Heidenreich PA, Murray KT, Shea JB, Tracy CM, Yancy CW. 2019 AHA/ACC/HRS Focused Update of the 2014 AHA/ACC/HRS Guideline for the Management of Patients With Atrial Fibrillation: A Report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines and the Heart Rhythm Society in Collaboration With the Society of Thoracic Surgeons. Circulation. 2019;140(2):e125–51. 10.1161/CIR.0000000000000665. [DOI] [PubMed] [Google Scholar]
- 13.Hindricks G, Potpara T, Dagres N, Arbelo E, Bax JJ, Blomström-Lundqvist C, Boriani G, Castella M, Dan GA, Dilaveris PE, Fauchier L, Filippatos G, Kalman JM, La Meir M, Lane DA, Lebeau JP, Lettino M, Lip GYH, Pinto FJ, Thomas GN, et al. 2020 ESC Guidelines for the diagnosis and management of atrial fibrillation developed in collaboration with the European Association for Cardio-Thoracic Surgery (EACTS): The Task Force for the diagnosis and management of atrial fibrillation of the European Society of Cardiology (ESC) Developed with the special contribution of the European Heart Rhythm Association (EHRA) of the ESC. European heart journal. 2021;42(5):373–498. 10.1093/eurheartj/ehaa612. [DOI] [PubMed] [Google Scholar]
- 14.Chao TF, Joung B, Takahashi Y, Lim TW, Choi EK, Chan YH, Guo Y, Sriratanasathavorn C, Oh S, Okumura K, Lip GYH. 2021 focused update consensus guidelines of the asia pacific heart rhythm society on stroke prevention in atrial fibrillation: executive summary. Thromb Haemost. 2022;122(1):20–47. 10.1055/s-0041-1739411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Monedero I. A novel ECG diagnostic system for the detection of 13 different diseases. Eng Appl Artif Intell. 2022;107: 104536. 10.1016/j.engappai.2021.104536. [Google Scholar]
- 16.Aziz S, Ahmed S, Alouini MS. ECG-based machine-learning algorithms for heartbeat classification. Sci Rep. 2021;11(1):18738. 10.1038/s41598-021-97118-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alfaras M, Soriano MC, Ortín S. A fast machine learning model for ECG-based heartbeat classification and arrhythmia detection. Front Physics. 2019;7:103. 10.3389/fphy.2019.00103. [Google Scholar]
- 18.Wen X, Huang Y, Wu X, Zhang B. A feasible feature extraction method for atrial fibrillation detection from BCG. IEEE J Biomed Health Inform. 2019;24(4):1093–103. 10.1109/JBHI.2019.2927165. [DOI] [PubMed] [Google Scholar]
- 19.Zheng J, Fu G, Abudayyeh I, Yacoub M, Chang A, Feaster WW, et al. A high-precision machine learning algorithm to classify left and right outflow tract ventricular tachycardia. Front Physiol. 2021;12:641066. 10.3389/fphys.2021.641066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu X, Zheng Y, Chu CH, He Z. Extracting deep features from short ECG signals for early atrial fibrillation detection. Artif Intell Med. 2020;109: 101896. 10.1016/j.artmed.2020.101896. [DOI] [PubMed] [Google Scholar]
- 21.Kwon S, Hong J, Choi EK, Lee E, Hostallero DE, Kang WJ, et al. Deep learning approaches to detect atrial fibrillation using photoplethysmographic signals: algorithms development study. JMIR mHealth and uHealth. 2019;7(6):e12770. 10.2196/12770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ribeiro AH, Ribeiro MH, Paixão GM, Oliveira DM, Gomes PR, Canazart JA, et al. Automatic diagnosis of the 12-lead ECG using a deep neural network. Nat Commun. 2020;11(1):1760. 10.1038/s41467-020-16172-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Darmawahyuni A, Nurmaini S, Rachmatullah MN, Tutuko B, Sapitri AI, Firdaus F, et al. Deep learning-based electrocardiogram rhythm and beat features for heart abnormality classification. PeerJ Comput Sci. 2022;8:e825. 10.7717/peerj-cs.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang H, Dai H, Zhou Y, Zhou B, Lu P, Zhang H, Wang Z. An effective feature extraction method based on GDS for atrial fibrillation detection. J Biomed Inform. 2021;119: 103819. 10.1016/j.jbi.2021.103819. [DOI] [PubMed] [Google Scholar]
- 25.Attia ZI, Kapa S, Lopez-Jimenez F, McKie PM, Ladewig DJ, Satam G, et al. Screening for cardiac contractile dysfunction using an artificial intelligence–enabled electrocardiogram. Nat Med. 2019;25(1):70–4. 10.1038/s41591-018-0240-2. [DOI] [PubMed] [Google Scholar]
- 26.Lih OS, Jahmunah V, San TR, Ciaccio EJ, Yamakawa T, Tanabe M, et al. Comprehensive electrocardiographic diagnosis based on deep learning. Artificial Intell Med. 2020;103:101789. 10.1016/j.artmed.2019.101789. [DOI] [PubMed] [Google Scholar]
- 27.Lai D, Bu Y, Su Y, Zhang X, Ma CS. Non-standardized patch-based ECG lead together with deep learning based algorithm for automatic screening of atrial fibrillation. IEEE J Biomed Health Inform. 2020;24(6):1569–78. 10.1109/JBHI.2020.2980454. [DOI] [PubMed] [Google Scholar]
- 28.Katsushika S, Kodera S, Nakamoto M, Ninomiya K, Inoue S, Sawano S, et al. The effectiveness of a deep learning model to detect left ventricular systolic dysfunction from electrocardiograms. International Heart Journal. 2021;62(6):1332–41. 10.1536/ihj.21-407. [DOI] [PubMed] [Google Scholar]
- 29.Mathews SM, Kambhamettu C, Barner KE. A novel application of deep learning for single-lead ECG classification. Comput Biol Med. 2018;99:53–62. 10.1016/j.compbiomed.2018.05.013. [DOI] [PubMed] [Google Scholar]
- 30.Van Zaen, J., Chételat, O., Lemay, M., Calvo, E. M., & Delgado-Gonzalo, R. (2019). Classification of cardiac arrhythmias from single lead ECG with a convolutional recurrent neural network. arXiv preprint arXiv:1907.01513. 10.5220/0007347900330041
- 31.Hatamian, F. N., Ravikumar, N., Vesal, S., Kemeth, F. P., Struck, M., & Maier, A. (2020, May). The effect of data augmentation on classification of atrial fibrillation in short single-lead ECG signals using deep neural networks. In ICASSP 2020–2020 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP) (pp. 1264–1268). IEEE. 10.1109/ICASSP40776.2020.9053800
- 32.Pitman, B. M., Chew, S. H., Wong, C. X., Jaghoori, A., Iwai, S., Thomas, G., ... & Lau, D. H. (2021). Performance of a mobile single-lead electrocardiogram technology for atrial fibrillation screening in a semirural African population: insights from “The Heart of Ethiopia: Focus on Atrial Fibrillation”(TEFF-AF) Study. JMIR mHealth and uHealth, 9(5), e24470. 10.2196/24470 [DOI] [PMC free article] [PubMed]
- 33.Li Z, Feng X, Wu Z, Yang C, Bai B, Yang Q. Classification of atrial fibrillation recurrence based on a convolution neural network with SVM architecture. IEEE Access. 2019;7:77849–56. 10.1109/ACCESS.2019.2920900. [Google Scholar]
- 34.Dang H, Sun M, Zhang G, Qi X, Zhou X, Chang Q. A novel deep arrhythmia-diagnosis network for atrial fibrillation classification using electrocardiogram signals. IEEE Access. 2019;7:75577–90. 10.1109/ACCESS.2019.2918792. [Google Scholar]
- 35.Kwon JM, Jeon KH, Kim HM, Kim MJ, Lim SM, Kim KH, et al. Comparing the performance of artificial intelligence and conventional diagnosis criteria for detecting left ventricular hypertrophy using electrocardiography. EP Europace. 2020;22(3):412–9. 10.1093/europace/euz324. [DOI] [PubMed] [Google Scholar]
- 36.Choosing colormaps in matplotlib (2022) - matplotlib 3.6.2 documentation. https://matplotlib.org/stable/ tutorials/colors/colormaps.html
- 37.Schnabel RB, Sullivan LM, Levy D, Pencina MJ, Massaro JM, D’Agostino RB Sr, Newton-Cheh C, Yamamoto JF, Magnani JW, Tadros TM, Kannel WB, Wang TJ, Ellinor PT, Wolf PA, Vasan RS, Benjamin EJ. Development of a risk score for atrial fibrillation (Framingham Heart Study): a community-based cohort study. Lancet (London, England). 2009;373(9665):739–45. 10.1016/S0140-6736(09)60443-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chamberlain AM, Agarwal SK, Folsom AR, Soliman EZ, Chambless LE, Crow R, Ambrose M, Alonso A. A clinical risk score for atrial fibrillation in a biracial prospective cohort (from the Atherosclerosis Risk in Communities [ARIC] study). Am J Cardiol. 2011;107(1):85–91. 10.1016/j.amjcard.2010.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alonso A, Krijthe BP, Aspelund T, Stepas KA, Pencina MJ, Moser CB, Sinner MF, Sotoodehnia N, Fontes JD, Janssens AC, Kronmal RA, Magnani JW, Witteman JC, Chamberlain AM, Lubitz SA, Schnabel RB, Agarwal SK, McManus DD, Ellinor PT, Larson MG, et al. Simple risk model predicts incidence of atrial fibrillation in a racially and geographically diverse population: the CHARGE-AF consortium. J American Heart Assoc. 2013;2(2):e000102. 10.1161/JAHA.112.000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author on request.