Abstract
Background
Cardiac macrophages are a heterogeneous population with high plasticity and adaptability, and their mechanisms in heart failure (HF) remain poorly elucidated.
Methods
We used single-cell and bulk RNA sequencing data to reveal the heterogeneity of non-cardiomyocytes and assess the immunoreactivity of each subpopulation. Additionally, we employed four integrated machine learning algorithms to identify macrophage-related genes with diagnostic value, and in vivo validation was performed. To assess the immune infiltration characteristics in HF, we utilized the CIBERSORT and single sample gene set enrichment analysis (ssGSEA). An unsupervised consensus clustering algorithm was applied to identify the macrophage-related HF subtypes. Furthermore, the scMetabolism was employed to explore the specific metabolic patterns of the macrophage subtypes. Finally, CellChat was used to investigate cell-cell interactions among the identified subtypes.
Results
The immunoreactivity score of macrophages in the HF was higher than that in the other cell types. GSEA of macrophage clusters indicated a significant enrichment of leukocyte-mediated immune processes, antigen processing, and presentation. The intersection of the results from machine learning revealed that SERPINA3, GPAT3, ANPEP, and FCER1G can serve as feature genes and form a diagnostic model with a good predictive capability. Unsupervised consensus clustering algorithms reveal the immune and metabolic subtypes of macrophages. The metabolic heterogeneity of macrophage subpopulations can lead to macrophage polarization into different types, which may be related to the metabolic reprogramming between glycolysis and mitochondrial oxidative phosphorylation. Cellular communication revealed that macrophages form a network of interactions with neutrophils to support each other’s functions and maintenance. The complex efferent and afferent signals are closely associated with myocardial fibrosis.
Conclusion
SERPINA3, GPAT3, ANPEP, and FCER1G can potentially serve as immune therapeutic targets and central biomarkers. The immunological and metabolic heterogeneity of macrophages may offer a more precise direction to explore the mechanisms underlying HF and novel immunotherapies.
Keywords: heart failure, single cell sequencing, machine learning, macrophage, diagnostic models, immune infiltration, metabolic reprogramming
Introduction
Heart failure (HF) is a complex clinical syndrome that represents the end-stage manifestation of cardiovascular diseases.1,2 Despite significant improvements in diagnostic and therapeutic approaches, including the application of minimally invasive artificial heart technologies, leading to enhanced diagnosis and survival rates in patients with HF, the incidence of HF remains high.3 This underscores the need to further explore the molecular and cellular mechanisms underlying HF. Emerging evidence indicates a close association between inflammatory activation, immune infiltration, and the occurrence, progression, and prognosis of HF.4,5 Infiltrating immune cells release cytokines such as TGF-β1 and TNF-α, promoting cardiac remodeling.6 Therefore, investigating changes in the immune microenvironment and key regulatory factors during HF development is of great significance for early diagnosis and prognosis prediction in HF.
Macrophages play a significant role as integral components of the innate and adaptive immune responses and contribute to electrical conduction, angiogenesis, vascular development, and mitochondrial homeostasis.7–10 Studies have shown that IL-10 derived from macrophages is upregulated during the development of hypertension or HF and promotes cardiac fibrosis.11 In hypertensive patients with HF and TREM2 deficiency, macrophages enhance the expression of proinflammatory cytokines and suppress the expression of vascular growth genes.12 Furthermore, immune cell activation is closely associated with metabolic alterations.13 Changes in macrophage metabolic patterns are associated with several signaling pathways, such as mechanistic targets of rapamycin and AMP-activated protein kinase pathways.14,15 Previous studies have demonstrated that metabolic pathways regulate macrophage polarization towards different phenotypes during cardiac injury.16 The pro-inflammatory M1 macrophages exhibit glycolytic activity, and the upregulation of glycolysis activates the pentose phosphate pathway, increasing NADPH-oxidase and producing reactive oxygen species like hydrogen peroxide and superoxide.17 Glycolytic metabolism also promotes cytoskeletal remodeling and the migration of macrophages to injury sites.18 In vitro studies indicate that macrophages lacking glucose transporter 1 or pyruvate dehydrogenase kinase 1 show reduced glycolysis and transition to a reparative M2 macrophage phenotype, which relies on oxidative metabolism and fatty acid oxidation.19,20 Previous research on cardiac innate immune cells has primarily focused on acute injury, with significantly less emphasis on the role of macrophages in chronic remodeling and HF. Pressure overload induced by transverse aortic constriction (TAC) leads to the expansion of CCR2- macrophages, peaking at 1 week.21 Following increased pressure overload and pathological remodeling, there is a depletion of resident cardiac macrophages, reduced angiogenesis, and exacerbated fibrosis.22 These observations highlight the adaptive role of cardiac macrophages during chronic stress. Unfortunately, the metabolic profile of macrophages in chronic HF has yet to be determined, and research on the role of macrophage metabolism in chronic cardiac remodeling is lacking. Therefore, gaining a comprehensive understanding of the metabolic landscape of macrophages in HF could provide further insights into targeted immunotherapies that focus on metabolism.
Although significant progress has been made in the study of macrophages, detailed mechanisms underlying HF remain unclear. Bulk RNA sequencing (RNA-seq) has provided important insights into the pathogenesis of HF, but lacks cell-specific information. The development of single-cell technologies is considered indispensable for investigating immune heterogeneity and providing additional detailed information to uncover the immune landscape in HF. Therefore, this study utilized a combination of bulk RNA-seq and single-cell RNA sequencing (scRNA-seq) to further elucidate macrophage-associated gene features, the metabolic landscape, and intercellular communication in HF.
Materials and Methods
Data Collection
The datasets used in this study were sourced from Gene Expression Omnibus (GEO), a comprehensive gene expression database. GSE222144 is an scRNA-seq dataset consisting of a normal and a HF samples.23 GSE57338 is a bulk RNA-seq dataset comprising 313 individuals with or without HF.24 The GSE26887 dataset contained 24 samples with or without HF and served as an external validation set to assess the accuracy of the diagnostic model further.25
scRNA-Seq Data Processing
Quality control of the scRNA-seq data was performed using the Seurat package.26 After excluding low-quality cells (gene expression <300 or mitochondrial gene expression >25%), we utilized the harmony package to integrate and remove batch effects across different treatment datasets.27 Dimensionality reduction using PCA was conducted to explore cell heterogeneity. Specific cluster-defining genes were identified using the FindAllMarker function. Cell subpopulations were annotated as known cell types using CellMarker 2.0, and marker genes from the literature.28
Selection of Differentially Expressed Genes and Immune Infiltration Analysis in Bulk RNA-Seq
The raw data of GSE57338 were downloaded from the GEO database using the GEO query package and subsequently normalized. Differential expression analysis using the limma package resulted in the identification of differentially expressed genes with an adjusted p-value < 0.05 and |logFC| > 0.5 as differentially expressed genes. Moreover, based on the CIBERSORT algorithm, we deduced the relative abundance of distinct immune cell types in both the normal and HF patient samples. Further analyses were performed to assess the heterogeneity of the immune-infiltrating cells in patients with HF.
Scoring Immune-Related Gene Sets Based on Single-Cell Data
Using the AUCell R package, we computed the area under the curve (AUC) for immune-related gene sets and generated gene expression rankings for each cell, thereby estimating the proportion of highly expressed immune genes within individual cells.29 The threshold for identifying genomically active cells was determined using the AUCell_exploreThresholds function. Following the acquisition of gene set scores, the internal functions of Seurat were used to map the AUC scores of each cell-to-cell subgroup, enabling the evaluation of immune activity within each cell subgroup.
Differential Gene Enrichment Analysis
Employing the clusterProfiler R package, KEGG pathway analysis and GO analysis were conducted on the differentially expressed genes to elucidate their functional roles. Additionally, gene set enrichment analysis (GSEA) was performed on the genes obtained from the Bulk RNA-seq differential analysis, revealing the key functions associated with the differentially expressed genes.
Establishment and Validation of a Diagnostic Model for Macrophage-Related Gene Signatures
The key to predicting the onset and progression risk of HF is the identification of stable and significant features. To select macrophage-related genes with diagnostic significance, we constructed four machine learning models: light gradient boosting machine (LightGBM), extreme gradient boosting (XGBoost), random forest (RF), and support vector machine (SVM). In these models, disease classification was the response variable and differentially expressed genes were used as predictor variables. Finally, from the aforementioned machine learning algorithms, we selected overlapping genes as macrophage-related hub genes. These hub genes were visualized using a Venn diagram to illustrate their common presence across the different models.
The construction of a forest plot using the R software package “forestplot” allowed us to assess the impact of diagnostic gene expression changes on HF. Moreover, utilizing the “rms” package, we developed a nomogram to evaluate the probability of predicting HF occurrence based on diagnostic genes. Finally, the discriminative performance of the model was evaluated by analyzing receiver operating characteristic (ROC) curves.
Identification of Macrophage-Related Gene Expression Patterns
Consistency cluster analysis of differentially expressed macrophage-related genes was performed using the R software package “ConsensusClusterPlus.” The model underwent 1000 iterations to classify patients with HF into distinct molecular subtypes. Moreover, we performed gene set variation analysis (GSVA) with the “h.all.v2023.1. The Hs.symbols.gmt” gene set was used to elucidate biological functional disparities among distinct subtypes. Using a single-sample gene set enrichment analysis (ssGSEA) algorithm, we assessed discrepancies in immune cell infiltration between the two subtypes. Additionally, we investigated immune activity at the single-cell level to gain a comprehensive understanding of the differences between the two subtypes.
Identification of Metabolism-Related Hub Genes in Heart Failure Based on WGCNA
Weighted gene co-expression network analysis (WGCNA) allows the association of modules with phenotypes, thereby identifying modules containing characteristic genes. First, 177 samples from patients with HF were systematically clustered using the “hclust” function, and clearly aberrant samples were removed. Next, low-expression genes were filtered using the absolute deviation median method. Subsequently, the “pickSoftThreshold” function from the WGCNA package was employed to select an appropriate soft-thresholding parameter β based on the scale-free network criterion. Subsequently, WGCNA was performed to identify the gene modules associated with fatty acid metabolism and oxidative phosphorylation.
Observation of Macrophage Subtype-Specific Metabolic Patterns
We performed clustering analysis at a higher resolution to observe the metabolic heterogeneity of macrophages. Using the KEGG metabolic gene set and the “VISION” method, we visualized and quantified the metabolic activity of individual cells within each macrophage subpopulation using scMetabolism.30
Intercellular Communication Analysis
We utilized CellChat to infer cell-cell communication interactions between subpopulations of cells.31 We predicted potential interaction strengths and inferred cell state-specific communication based on the expression of receptors and ligands. Furthermore, we screened for differentially expressed signaling pathways and associated genes across all cell groups.
Construction and Evaluation of Mouse Models of Heart Failure
Male C57BL/6 mice were adaptively housed for one week at 25°C, 50% relative humidity, and a daily 12-hour light cycle. The experimental group (n=3) underwent a moderate incision using a 27G needle, whereas the sham-operated group (n=3) underwent only an incision without further manipulation. In the TAC model, a portable small animal anesthesia machine (Shanghai Yuyan, ACS) was used for inhalation anesthesia with isoflurane (Shanghai Yuyan), followed by the fixation of the mouse’s head and limbs. The skin of the neck and chest was incised, and an incision was made between the second intercostal space at the left edge of the sternum. The incision was expanded, the thymus was separated, and the aortic arch and its branches were exposed. A 6–0 suture was passed through the aortic arch, and a 27G needle was placed alongside the aorta. The suture was tightened to create aortic constriction. The chest cavity and skin were then sutured, and measures were taken to prevent infection and alleviate pain. After modeling, the mice were housed for four weeks, and their cardiac function was assessed using echocardiography. The hearts were rapidly excised and immersed in 10% KCl solution to arrest the heart in diastole. After blotting with filter paper, heart weight was measured to calculate the heart weight-to-body weight ratio (HW/BW). Subsequently, the hearts were embedded in paraffin, sectioned, and subjected to hematoxylin and eosin (HE) and Masson staining.
Experimental Validation of Macrophage-Related Characterized Genes
Considering the high expression levels and increased differential fold-change of SERPINA3 in the heart, we further investigated its expression levels in an animal model. After isolating the heart tissue from the HF mouse model, total RNA was extracted for qPCR analysis of the SERPINA3 transcript levels. The primer-F of SERPINA3 was TGCCATGTTCATCCTCCCTG, and the primer-R was GCTGTAGTCGGTGGAGATGG. Primer-F of the reference gene Actin was TTCTACAATGAGCTGCGTG and primer-R was CTCAAACATGATCTGGGTC. Total protein was extracted for Western blot analysis of SERPINA3 (Bioss, bs-0094R) protein expression, using a pre-stained protein marker (Thermo Scientific, 26616).
Results
Single-Cell Landscape in Heart Failure Tissues
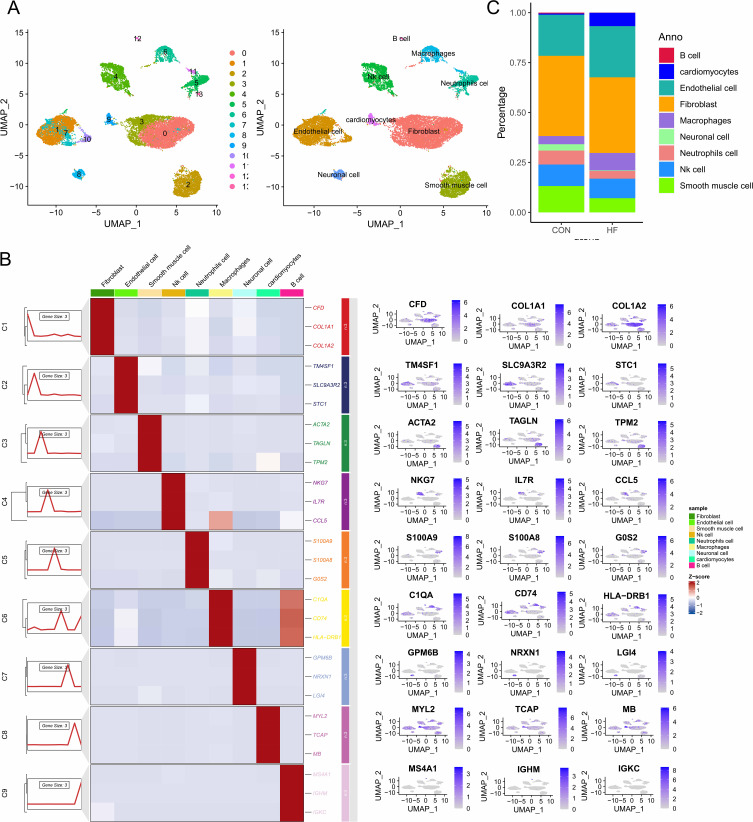
After quality control, a dataset containing 18,104 cells and 24,781 genes was obtained from single-cell samples of normal and HF tissues (Supplementary Figure 1). Based on the marker genes of different cells, the cell clusters were annotated into nine distinct cell lineages: fibroblasts, endothelial cells, B cells, neutrophils, natural killer cells, macrophages, smooth muscle cells, neural cells, and cardiomyocytes (Figure 1A and B). Interestingly, compared to normal patients, we found an increased proportion of macrophages and a significantly decreased proportion of neutrophils in HF patients (Figure 1C). Therefore, we hypothesize that dysregulation of the immune microenvironment mediated by macrophages or neutrophils may be associated with adverse cardiac remodeling.
Figure 1.
Single-cell landscape in heart failure tissues. (A) UMAP mapping of 18104 cells. (B) Expression levels of marker genes in different cell subpopulations.(C) Proportion of cells in heart failure and normal samples.
Immune-Related Gene Set Scoring and Cell-Cell Communication Analysis
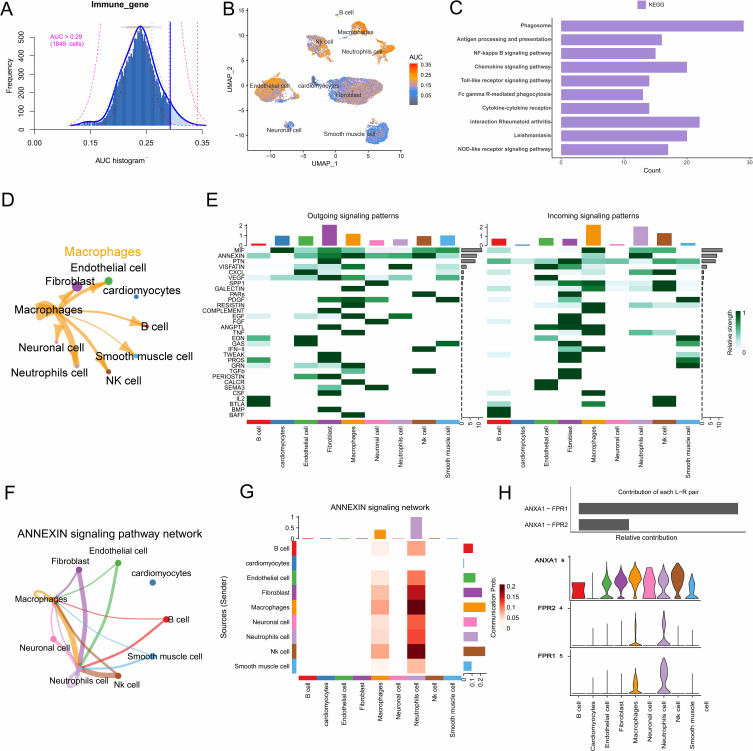
To investigate the immune characteristics of HF, we utilized the AUCell R package to evaluate the immune activity of each cell type (Figure 2A). As shown in Figure 2B, we observed that macrophages and neutrophils expressed a higher number of immune-related genes. Given the significant dysregulation of these cells in HF, we conducted differential gene enrichment analysis within the macrophage cluster. Phagosome, antigen processing and presentation, cytokine-cytokine receptors, along with several classical signaling pathways, were significantly enriched (Figure 2C). Additionally, CIBERSORT results indicated significant differences in immune cell infiltration between HF and healthy samples (Supplementary Figure 2A–C). In summary, macrophages play a promotive role in the progression of HF, where their proliferation enhances immune and inflammatory processes in HF patients.
Figure 2.
Immune-related gene set scoring and cell communication analysis. (A) Immune gene set scoring with a threshold set at 0.29. (B) UMAP plot of immune gene set scores in clusters. Macrophages and neutrophils expressed more immune genes and exhibited higher AUC values. (C) Differential gene enrichment analysis of macrophage clusters. (D) Interactions between macrophages and other cell types. (E) The contribution of cells to the outgoing and incoming signals in different signaling pathways. (F) Cell-cell communication mediated by the ANNEXIN signaling pathway. (G) The bar chart shows the contribution of each ligand-receptor pair to the ANNEXIN signaling pathway. (H) The violin plot shows the expression levels of ANNEXIN signaling ligand-receptor genes across all subclusters.
Given the significant enrichment of cytokine and receptor-related pathways, we performed an analysis of the nine identified cell clusters using CellChat (Supplementary Figure 2D). CellChat results revealed that the communication strength between macrophages and neutrophils was the highest (Figure 2D), with numerous receptor-ligand pairs being sent from the macrophage cluster to the neutrophil cluster (Figure 2E and Supplementary Figure 2E). This may be related to the activation of the ANNEXIN signaling pathway (Figure 2F and G). In addition, we identified specific ligand-receptor pairs within the ANNEXIN signaling pathway, finding that ANXA1 can specifically bind to FPR1 or FPR2 receptors to activate intrinsic pathways in these cells. This may induce macrophage polarization towards a pro-inflammatory phenotype, thereby promoting the immune response in HF (Figure 2H).
Construction and Validation of the Macrophage-Associated Diagnostic Model
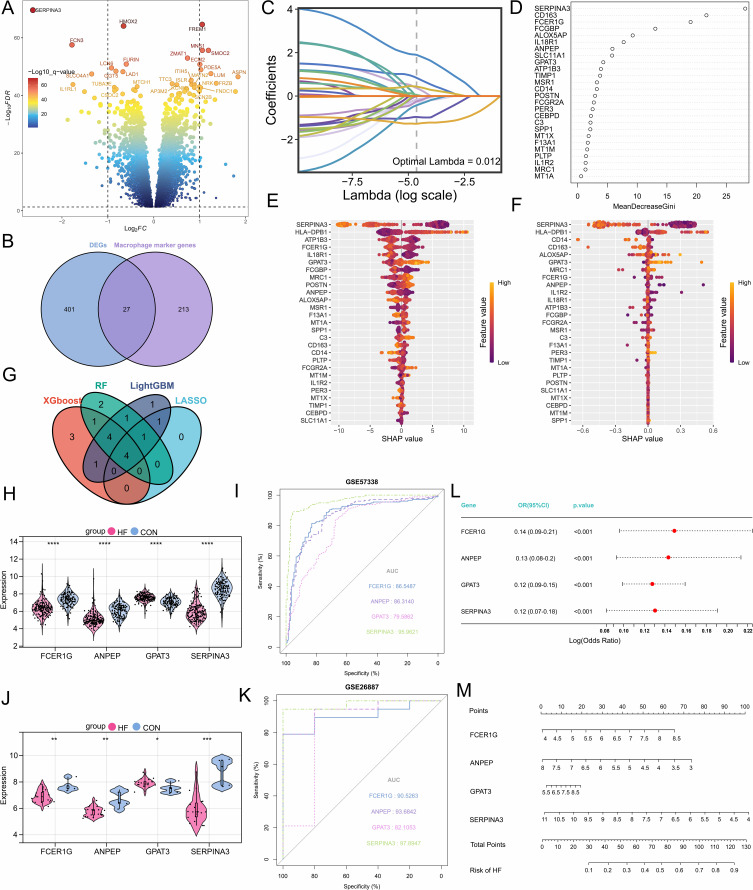
Differential expression analysis was performed on the expression data of HF patients and normal samples from the GSE57338 dataset, identifying 428 differentially expressed genes (Figure 3A). GSEA showed that the differentially expressed genes were significantly enriched in phagocytosis, immune, and inflammatory responses (Supplementary Figure 3A). These findings indicate that macrophages play a key role in the immune processes of HF patients. After intersecting with differentially expressed genes associated with macrophage subpopulations, 27 macrophage-related differentially expressed genes were identified (Figure 3B). Subsequently, four machine learning algorithms were employed to select feature variables, and the intersection of the results indicated that SERPINA3, GPAT3, ANPEP, and FCER1G may play a role in regulating the progression of HF and are closely associated with macrophages (Figure 3C–G). Additionally, the expression levels of these four genes showed significant differences in two external datasets (Figure 3H and J). ROC analysis performed on the training and external validation cohorts revealed that these four diagnostic genes have high predictive value, particularly SERPINA3 (Figure 3I and K). Using the ssGSEA algorithm, we calculated the infiltration levels of immune cells in each sample and explored the correlation between hub genes and immune cells. The ssGSEA results showed that SERPINA3, ANPEP, and FCER1G were significantly positively correlated with macrophage infiltration, while GPAT3 exhibited a significant negative correlation with macrophage infiltration (Supplementary Figure 3B–I). It is noteworthy that the ssGSEA results indicated that SERPINA3, GPAT3, and ANPEP not only participate in immune-related signaling pathways but also affect oxidative phosphorylation and fatty acid metabolism. Therefore, changes in the expression of SERPINA3, GPAT3, and ANPEP may mediate macrophage phenotype and immune activity by influencing macrophage metabolism. Multivariate logistic regression analysis showed that the expression of these four marker genes is independently associated with HF (Figure 3L). We also created a nomogram to score each feature variable and summed the scores of all feature variables to obtain a total risk score for the occurrence of HF (Figure 3M). In summary, the four macrophage-related genes exhibit high diagnostic value and can be utilized for the development of clinical targets.
Figure 3.
Selection, validation of signature genes, and construction of the diagnostic model. (A) Volcano plot of differentially expressed genes. (B) The Venn diagram shows the differentially expressed macrophage-related genes. (C) Feature genes were selected using the LASSO algorithm with λ = 0.012. (D) The RF algorithm ranks the importance of 27 differentially expressed genes. (E) SHAP dependency analysis based on the XGBoost model describes the importance of individual feature variables in the predictive model. (F) SHAP dependency analysis based on the LightGBM model describes the importance of individual feature variables in the predictive model. (G) Hub genes with diagnostic value were identified through the intersection of multiple machine learning algorithms. (H and I) Expression levels of hub genes and evaluation of the diagnostic model in the GSE57338 dataset. (J and K) Expression levels of hub genes and evaluation of the diagnostic model in the GSE26887 dataset. (L) Multivariable logistic regression analysis. (M) The nomogram evaluates the total risk score for developing heart failure. *, P<0.05; **, P<0.01; ***, P<0.001; ****, P<0.0001.
Establishment of the Mouse Heart Failure Model and Validation of Indicators
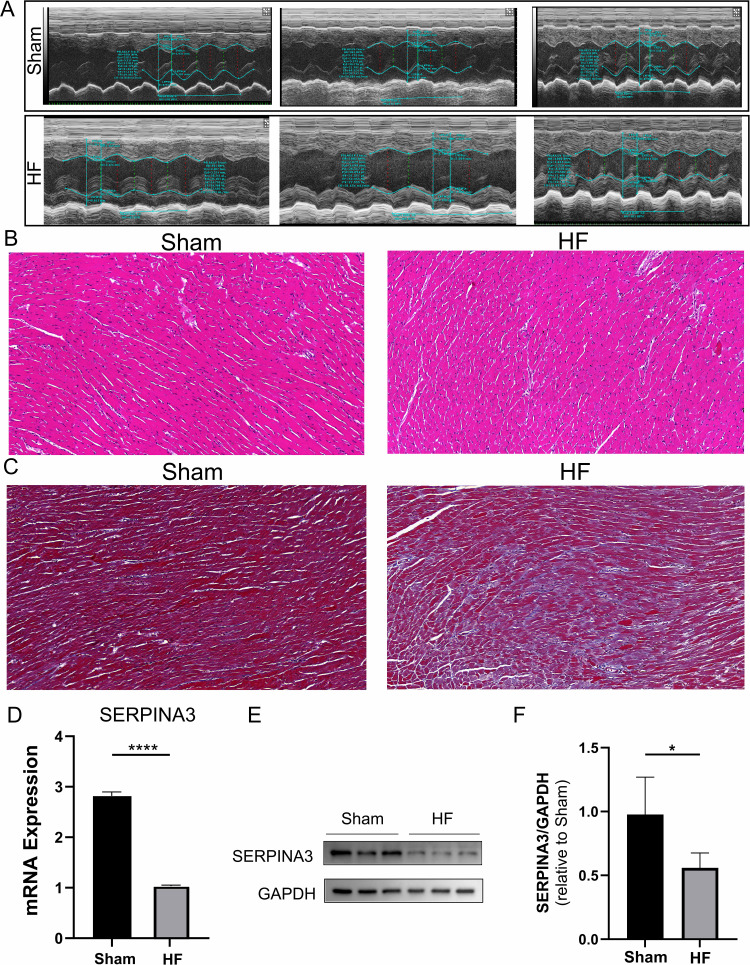
After TAC modeling, mice underwent echocardiographic assessment of cardiac function. Cardiac function was measured using M-mode for left ventricular ejection fraction (LVEF), left ventricular fraction shortening (LVFS), left ventricular internal diameter at end-diastole (LVIDd), and left ventricular internal diameter at end-systolic (LVIDs). The results indicated a decrease in LVEF and LVFS, while LVIDd and LVIDs increased (Figure 4A and Supplementary Figure 4). HE staining showed that myocardial hypertrophy and fibrosis were more pronounced in the HF group compared to the sham-operated group (Figure 4B). Masson staining revealed that the degree of fibrosis in the HF group was higher than that in the sham-operated group (Figure 4C), indicating the successful establishment of the HF model. Subsequently, cardiac tissues from TAC-induced HF mice were collected, and qPCR results showed that SERPINA3 expression was downregulated in the HF group compared to the sham group (Figure 4D). Western blotting results also demonstrated that SERPINA3 expression was downregulated in the HF group compared to the sham group (Figure 4E and F).
Figure 4.
Construction of TAC-induced heart failure mouse model and experiments. (A) Echocardiography of the mouse model. (B) HE staining of rat myocardial tissue. Scale bar: 20 μm; original magnification, ×20. (C) Masson staining shows higher fibrosis levels in the HF group compared to the sham group. Scale bar: 20 μm; original magnification, ×40. (D) q-PCR validation of SERPINA3 expression at the mRNA level. (E and F) Western blot bands and histograms of SERPINA3 protein expression in myocardial tissue. *, P < 0.05; ****, P < 0.0001.
Characterization of Macrophage-Associated Gene Expression Patterns in Heart Failure
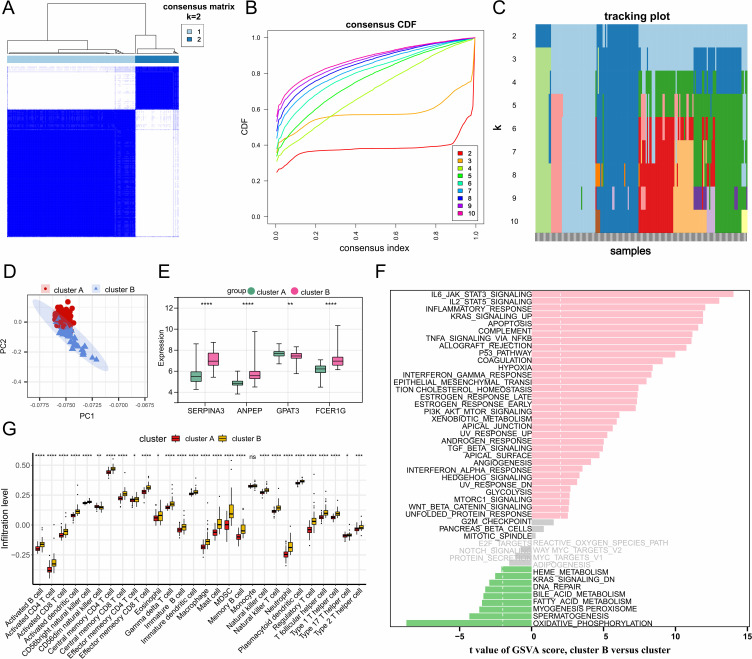
In HF, macrophages exhibit distinct immune and metabolic activities. Therefore, unsupervised clustering analysis based on the expression profiles of 27 macrophage-associated genes clearly divided the samples into two clusters (Cluster A, 132 samples; Cluster B, 45 samples) (Figure 5A–C). PCA revealed significant differences in expression patterns between the two clusters (Figure 5D). Box plots and heatmaps demonstrated significant heterogeneity in the expression of hub genes between the two clusters (Figure 5E). To investigate the differences in pathway expression between the two HF clusters, GSVA enrichment analysis was performed based on the MSigDB Hallmark gene set. Notably, immune and inflammation-related pathways, such as IL6_JAK_STAT3_SIGNALING, IL2_STAT5_SIGNALING, and INFLAMMATORY_RESPONSE, were significantly enriched in Cluster B, while metabolic pathways, including oxidative phosphorylation and fatty acid metabolism, were significantly enriched in Cluster A (Figure 5F). Additionally, we used the ssGSEA algorithm to quantify the abundance differences of 28 immune cell types between the two clusters. As shown in Figure 5G, immune cell infiltration levels were markedly higher in Cluster B compared to Cluster A. In conclusion, the analysis results suggest that HF patients exhibit distinct immune and metabolic characteristics. Patients in Cluster B may have heightened immune and inflammatory responses, while those in Cluster A show dysregulation of metabolism-related pathways.
Figure 5.
Identification of macrophage-related gene expression patterns in heart failure. (A–C) Unsupervised clustering analysis based on the expression profile data of 27 macrophage-related genes. When K=2, heart failure samples can be clearly clustered into two subtypes. (D) PCA reveals the clustering trend of the two subgroups in HF samples. (E) Bar chart showing the expression changes of hub genes in the two subgroups. (F) GSVA enrichment analysis results based on the Hallmark gene sets from the MSigDB database. (G) Differences in immune cell infiltration between the two subgroups. *, P<0.05; **, P<0.01; ***, P<0.001; ****, P<0.0001.
Identification of Heart Failure Metabolism-Related Hub Genes Based on Weighted Gene Co-Expression Network Analysis
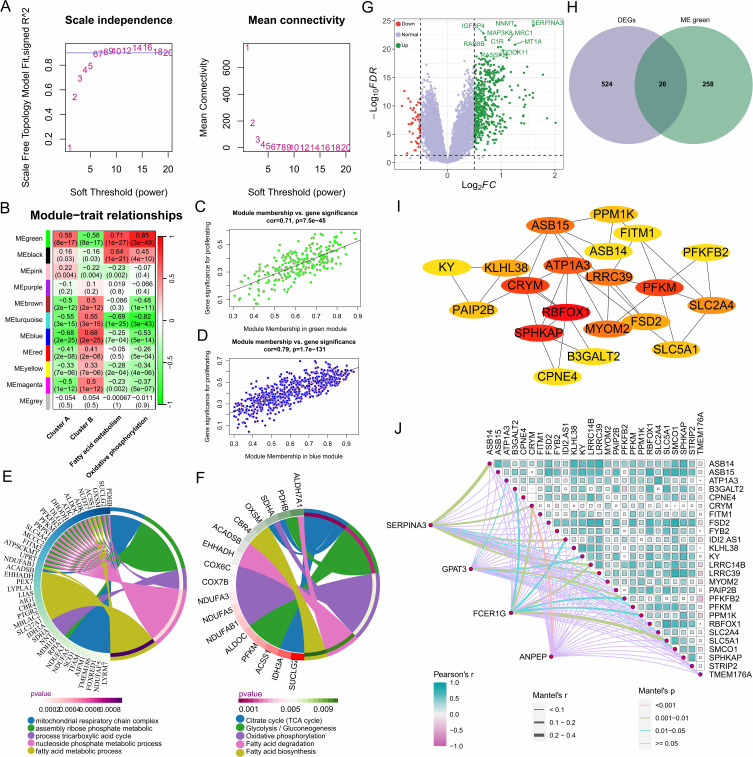
The consistency between GSVA enrichment analysis and ssGSEA results underscores the importance of metabolic pathways in HF. Therefore, we performed WGCNA on 177 hF samples. After excluding two outlier samples (GSM1379815 and GSM1380018), the “pickSoftThreshold” function was used to determine the optimal soft-threshold parameter β = 6, where the scale-free R² reached 0.9 with high average connectivity, making it suitable for further analysis (Figure 6A). A total of 11 modules were identified through gene clustering. The module-trait correlation heatmap revealed that genes in the MEgreen module were significantly positively correlated with fatty acid metabolism and oxidative phosphorylation, while genes in the MEblue module were significantly positively correlated with Cluster B (Figure 6B–D). Further enrichment analysis of MEgreen module genes showed significant enrichment in metabolic pathways such as the tricarboxylic acid cycle, oxidative phosphorylation, glycolysis, and fatty acid metabolism (Figure 6E and F). Next, we intersected the 550 differentially expressed genes between Clusters A and B, obtained using the Limma R package, with the 284 genes in the MEgreen module, identifying 26 differentially expressed metabolism-related genes (Figure 6G and H). A protein-protein interaction (PPI) network was constructed using these 26 genes, and hub genes were identified using the cytoHubba plugin in Cytoscape (Figure 6I). Mantel test results showed a significant and strong correlation between differentially expressed metabolism-related genes and hub genes (Figure 6J). Among these, CRYM, ATP1A3, LRRC39, RBFOX1, PFKM, ASB15, and MYOM2 may be associated with changes in cardiac function in HF patients (Supplementary Figure 5).
Figure 6.
Identification of metabolic hub genes in heart failure based on WGCNA analysis. (A) Selection of 6 as the appropriate soft threshold based on the scale-free topology fitting index, which provides optimal network connectivity. (B) Correlation between module and trait. The redder the color, the higher the positive correlation between the module and the trait; the greener the color, the higher the negative correlation. (C and D) Correlation between the green and blue modules and the trait. (E) Biological processes involved by genes in the green module. (F) Signaling pathways involved by genes in the green module. (G) Volcano plot showing differentially expressed genes between the two clusters. (H) Differentially expressed genes related to fatty acid metabolism and oxidative phosphorylation. (I) Protein-protein interaction network of differentially expressed metabolic-related genes between the two clusters. (J) Correlation between diagnostic markers in cluster A and metabolic-related genes.
High-Resolution Macrophage Metabolic Landscape
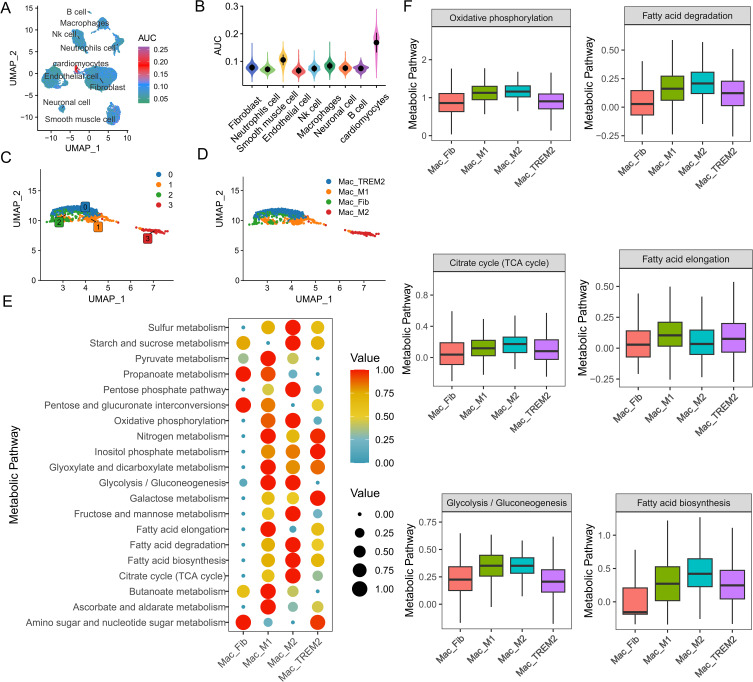
However, the relationship between immunometabolism and HF phenotypes remains unclear. Therefore, we investigated the metabolic heterogeneity among different macrophage subtypes. As shown in Figure 7A and B, macrophages exhibited the highest metabolic activity among all immune cells. Here, we performed more detailed clustering analysis of macrophages and classified them into four main clusters based on gene set scores of marker genes, including M1-like macrophages (Mac_M1), M2-like macrophages (Mac_M2), fibroblast-like macrophages (Mac_Fib), and TREM2-high macrophages (Mac_TREM2) (Figure 7C and D). We selected the top 20 pathways with the most significant changes across all subgroups (Figure 7E). Notably, Mac_M1 and Mac_M2 exhibited the highest metabolic activity among all macrophage subtypes. As shown in Figure 7F, glycolysis and the pentose phosphate pathway were highly expressed in Mac_M1, indicating their crucial role in macrophage polarization toward a pro-inflammatory phenotype. Previous studies have shown that aerobic glycolysis is mainly associated with M1 macrophages. In this study, we found that Mac_M2 not only enhanced mitochondrial oxidative phosphorylation but also upregulated glycolytic rates. The role of glycolysis in M2 macrophages should not be overlooked and warrants further investigation. Additionally, fatty acid biosynthesis and metabolism may contribute to the polarization of tissue macrophages toward the M2 phenotype. In summary, a deeper understanding of the diverse immunometabolic phenotypes of immune cells may help elucidate the mechanisms of HF and guide the development of personalized therapeutic strategies.
Figure 7.
Macrophage metabolic landscape at high resolution. (A) Gene set scoring for fatty acid metabolism and oxidative phosphorylation. (B) Metabolic activity scoring of cell subpopulations, with macrophages exhibiting the highest metabolic activity among immune cells. (C and D) Identification of macrophage subpopulations. (E) Activity of the top 20 variable metabolic pathways in macrophage subpopulations. (F) Metabolic activity of fatty acid metabolism, oxidative phosphorylation, the tricarboxylic acid cycle, and glycolysis in macrophage subpopulations.
Discussion
HF is a cardiovascular clinical syndrome characterized by a high incidence of complications and a poor prognosis.32 Cardiac remodeling in patients with HF is accompanied by a more severe inflammatory state, cardiac fibrosis, cellular apoptosis and changes in cardiac function.33 With the advancement of single-cell technologies, new tools have been developed to dissect cell types present in both healthy and diseased tissues. Recently, these methods have been used to gain deeper insights into how diseases affect the cellular and transcriptional landscapes in the heart.34 In this study, we used a combination of scRNA-seq and bulk RNA-seq data to reveal the relationship between gene expression levels, immune cell infiltration, and immune-related functions in patients with HF. First, using the AUCell package to score immune gene sets in each cell, we found that macrophages scored the highest, indicating that macrophages are the cell type most closely associated with immune function in patients with HF. Second, by employing four machine learning algorithms, we constructed a diagnostic nomogram model with excellent diagnostic performance for HF, as demonstrated by the ROC curves, validating the diagnostic efficacy of the model. Based on the differentially expressed macrophage-related genes, we clustered patients with HF into two distinct subtypes (immune-related and metabolism-related). These two subtypes exhibit distinct biological functional differences. Based on these findings, we further evaluated the immune metabolic landscape within the macrophage-related subtypes. Finally, intercellular communication revealed crosstalk between fibroblasts, macrophages, and neutrophils during myocardial remodeling.
An increasing number of studies indicate that inflammatory responses play a crucial role in the development of HF.35 Cells regulate the chemotactic behavior of immune cells in HF by secreting certain chemokines and cytokines, which are of paramount importance in myocardial remodeling. Cardiac macrophages represent a heterogeneous population with high plasticity and adaptability and play a key role in fibrotic tissue responses.36 Resident macrophages respond to the immune microenvironment by modulating the synthesis of cytokines and growth factors, thus generating a substantial amount of pro-fibrotic growth factors that regulate fibrotic processes.37 In our study, the percentage of macrophages in the single-cell lineage significantly increased during HF progression. Therefore, macrophage-related biomarkers may offer a more effective approach to HF diagnosis and treatment. Based on the aforementioned findings, we further employed machine learning algorithms to identify macrophage-related gene diagnostic features in HF and validated them through experimental research. Feature selection algorithms based on machine learning are employed to eliminate redundancies and unrelated features, thereby improving the accuracy of the model.38 Additionally, in our research, we integrated four machine learning algorithms: light gradient boosting machine (LightGBM), extreme gradient boosting (XGBoost), random forest (RF), and support vector machine (SVM). Cross-referencing of the features from these four algorithms revealed that SERPINA3, GPAT3, ANPEP, and FCER1G are potential diagnostic markers for HF, which was validated through quantitative PCR, Western blotting, and immunofluorescence staining. This study utilized the ssGSEA algorithm to analyze the relationship between hub genes and immune cell infiltration. In addition to identifying a correlation between hub genes and macrophage infiltration, T cells also appeared to be involved to some extent. Considering the bidirectional regulatory relationship between T cells and macrophages, as well as their joint role in shaping the inflammatory microenvironment, it is possible that hub genes may indirectly influence the different phenotypes of macrophages by regulating T cell activation and cytokine secretion.39 This warrants further exploration in future studies. Furthermore, multivariate logistic regression analysis demonstrated that the diagnostic features of these four genes exhibited a high discriminatory ability in predicting HF. The AUC values of the ROC curve for the external dataset confirmed the reliability of the results.
Existing research indicates a close association between SERPINA3 and the immune response.40 SERPINA3 inhibits cytokine activity in macrophages by suppressing IKK complex formation and NF-κB activation.41 High SERPINA3 expression adversely affects cardiac function and increases the risk of mortality or adverse cardiac events.42 SERPINA3 shows significant potential as a predictive biomarker for HF. In our study, the downregulation of protective SERPINA3 expression in macrophages may reduce the inhibition of protease activity, thereby diminishing the limitation on protease-mediated tissue degradation, contributing to the progression of HF and being detrimental to tissue repair, anti-inflammatory responses, and immune regulation. GPAT3 is a gene associated with lipid metabolism, and growing evidence suggests that activated macrophages alter lipid composition and that targeted regulation of fatty acid synthesis may affect macrophage inflammatory responses.43,44 Dysregulation of GPAT3 expression may affect lipid metabolites, thereby regulating the pro-inflammatory or anti-inflammatory status of macrophages. Additionally, lipid accumulation resulting from disturbed lipid metabolism in macrophages may lead to further tissue damage.45 Thus, elevated GPAT3 levels may contribute to dysregulation of macrophage lipid metabolism and drive inflammatory responses in HF. ANPEP is a widely expressed ectoenzyme that plays crucial roles in various inflammatory diseases.46 As an enzyme, ANPEP regulates cytokine activity by cleaving its N-terminus and modulates immune cell development and activity by reducing peptides involved in antigen processing that bind to MHC II.47 In our study, antigen processing and presentation were significantly enriched, consistent with our findings. FCER1G is an important inflammation-related molecule involved in antigen-antibody binding reactions that activates cellular effector functions.48,49 Extensive research has shown that FCER1G is widely expressed in different types of immune cells, including macrophages, mast cells, and eosinophils.50,51 Immune infiltration analysis revealed a positive correlation between FCER1G expression and macrophage infiltration. Additionally, FCER1G participates in various immune responses such as phagocytosis and cytokine release.52 Previous studies have shown that FCER1G is expressed in M1 pro-inflammatory cells in the heart and is primarily involved in transmitting pro-inflammatory signals from immune cells.53 However, in patients with HF, FCER1G expression was lower than that in the normal samples. Therefore, our study maintains that downregulation of FCER1G in patients with HF may weaken the inflammatory response by favoring M1 polarization of macrophages. In addition to single-cell sequencing, genome-wide association study (GWAS) is also a valuable approach for investigating the impact of genetic changes on disease development. Single-cell sequencing and GWAS have different research focuses and technical limitations, which can sometimes result in key genes identified by single-cell sequencing not being captured in GWAS studies. GWAS focuses on identifying common genetic variants in large populations, while single-cell sequencing focuses on dynamic gene expression at the cellular level. Therefore, by integrating the results of both methods and conducting further functional validation and in-depth research, a more comprehensive understanding of the genetic mechanisms and gene regulation underlying HF can be achieved.
Based on the expression of macrophage-related genes. Cluster A was characterized by dominant metabolic processes, whereas cluster B was primarily associated with immunity. Numerous studies have indicated that macrophages exhibit distinct immune functions and metabolic phenotypes that participate in different stages of the inflammatory responses.54 In our study, M1 and M2 macrophages from patients demonstrated the highest metabolic activity, which may be associated with glycolysis, oxidative phosphorylation, and fatty acid metabolism. Metabolic reprogramming of macrophages leads to polarization of different phenotypes. M1 macrophages typically show enhanced glycolysis to meet energy demands.55,56 Glycolysis can rapidly provide the ATP required for macrophage immune responses and meet the high-energy demands of phagocytosis.57–59 Additionally, glycolysis can enhance the activity of the pentose phosphate pathway, leading to increased levels of reactive oxygen species in M1 macrophages.17,60 In vitro studies have shown that macrophages lacking glucose transporters exhibit reduced glycolysis and undergo phenotypic transition to anti-inflammatory M2 macrophages.20 M2 macrophages are typically associated with oxidative phosphorylation and fatty acid metabolism.61,62 Previous studies indicated that M2 macrophages do not exhibit increased glycolysis. However, our study revealed a significant increase in glycolysis in the M2 macrophages. The role of glycolysis in M2 macrophages may be more important than previously thought, necessitating further investigation.
To decipher the ligand-receptor interactions between different cells, we employed CellChat analysis to investigate the communication between the nine cell clusters. Fibroblast-macrophage and macrophage-neutrophil interactions exhibited stronger interplay in HF, indicating that cell crosstalk fundamentally impacts the progression of HF. Interestingly, fibroblasts showed elevated expression of macrophage migration inhibitory factor (MIF). MIF-related ligand-receptor interactions are highly activated in HF. MIF exerts a protective effect against lipotoxicity, leading to a reduction in macrophage numbers.63 Furthermore, the co-expression of CXCR4, CD74, and MIF enhances cell survival and migratory capabilities. Individually or in combination, these three factors promote cell death and eliminate MIF-driven migratory responses.64 ANNEXIN signaling is primarily mediated by ANXA1-FPR1 and ANXA1-FPR2 ligand-receptor interactions. Studies suggest that ANXA1 and its receptor FPRs promote neutrophil and macrophage maturation, directing their migration to injured lung tissues.65 During myocardial ischemia-reperfusion, downregulation of ANXA1 exacerbates the inflammatory response and weakens the cardiac contractile function.66 In conclusion, this study postulates that the binding of these ligands and receptors transmits pro-inflammatory or anti-inflammatory signals, thereby mediating the immune responses involved in HF.
However, our study has some limitations. First, the datasets used in this study were obtained from public databases and lacked relevant clinical information about the included patients with HF, which might lead to individual differences in the severity of the disease. Second, the single-cell dataset used in this study had a relatively small sample size, necessitating further evaluation of the accuracy of the results using larger sample sets. Third, this study utilized a smaller sample size for preliminary exploration and validation, providing a solid foundation for subsequent larger-scale research. In future studies, we plan to increase the sample size to ensure greater robustness. Fourth, TAC-induced HF in mice can simulate the pathological process of human HF to a certain extent, but due to the physiological differences between species, the translational applicability of the research results still needs to be evaluated. Fifth, in future studies, techniques such as flow cytometry sorting and single-cell sequencing can be employed to validate the expression levels of the identified key genes at the cellular level and explore their mechanisms. This will help to better elucidate the role of gene regulation in macrophages and their further involvement in HF. Sixth, the classification of macrophages and deeper exploration will help to better understand their roles in HF. In future studies, we plan to extend our analysis by further distinguishing between resident and infiltrating macrophages, as well as examining the specific roles and subtypes of resident macrophages in HF pathophysiology. Seventh, while we identified four key genes (SERPINA3, GPAT3, ANPEP, and FCER1G) associated with macrophage infiltration in HF, we did not fully elucidate the specific mechanisms through which these genes regulate macrophage involvement, particularly their metabolic interactions and potential crosstalk with fibroblasts. Future explorations will focus on the pathways linking these genes to macrophage polarization or metabolic regulation and the extent to which these genes influence fibroblast interactions.
Conclusions
In conclusion, the four key genes selected in this study, SERPINA3, GPAT3, ANPEP, and FCER1G, are promising biomarkers with significant diagnostic value and offer valuable insights into the diagnosis and treatment of HF. Moreover, we identified two distinct biological phenotypes in patients with HF that could aid in precision therapy for HF. Additionally, we deciphered the crucial role of ligand-receptor interactions between different cells in the immune microenvironment of HF. These findings provide novel insights and potential targets for a comprehensive understanding of the impact of immune responses and metabolic reprogramming on HF.
Acknowledgments
We thank the Gene Expression Omnibus for providing the data. We thank Dr. Jin Rao, Pengchao Cheng, Yue Yu, and Xiangyu Chen for the model construction and experiments in this study.
Data Sharing Statement
Publicly available datasets (ID: GSE222144, GSE57338, and GSE26887) were obtained from https://www.ncbi.nlm.nih.gov/geo/.
Ethics Statement
All methods and animal experimental procedures in this study were approved by the Medical Ethics Committee and the Animal Ethics Committee of Shanghai Changzheng Hospital. Animal welfare was in compliance with the Guide for the Care and Use of Laboratory Animals published by the NIH.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors declare no conflicts of interest in this work.
References
- 1.Ponikowski P, Voors AA, Anker SD, et al. 2016 ESC guidelines for the diagnosis and treatment of acute and chronic heart failure: the Task Force for the diagnosis and treatment of acute and chronic heart failure of the European Society of Cardiology (ESC) developed with the special contribution of the Heart Failure Association (HFA) of the ESC. Eur Heart J. 2016;37(27):2129–2200. doi: 10.1093/eurheartj/ehw128 [DOI] [PubMed] [Google Scholar]
- 2.Yancy CW, Jessup M, Bozkurt B, et al. 2017 ACC/AHA/HFSA focused update of the 2013 ACCF/AHA guideline for the management of heart failure: a report of the American College of Cardiology/American Heart Association Task Force on clinical practice guidelines and the Heart Failure Society of America. Circulation. 2017;136(6):e137–e161. doi: 10.1161/cir.0000000000000509 [DOI] [PubMed] [Google Scholar]
- 3.Grinstein J, Belkin MN, Kalantari S, Bourque K, Salerno C, Pinney S. Adverse hemodynamic consequences of continuous left ventricular mechanical support: JACC review topic of the week. J Am Coll Cardiol. 2023;82(1):70–81. doi: 10.1016/j.jacc.2023.04.045 [DOI] [PubMed] [Google Scholar]
- 4.Murphy SP, Kakkar R, McCarthy CP, Januzzi JL Jr. Inflammation in heart failure: JACC state-of-the-art review. J Am Coll Cardiol. 2020;75(11):1324–1340. doi: 10.1016/j.jacc.2020.01.014 [DOI] [PubMed] [Google Scholar]
- 5.Swirski FK, Nahrendorf M. Cardioimmunology: the immune system in cardiac homeostasis and disease. Nat Rev Immunol. 2018;18(12):733–744. doi: 10.1038/s41577-018-0065-8 [DOI] [PubMed] [Google Scholar]
- 6.Schultheiss HP, Fairweather D, Caforio ALP, et al. Dilated cardiomyopathy. Nat Rev Dis Primers. 2019;5(1):32. doi: 10.1038/s41572-019-0084-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arts RJW, Netea MG. Adaptive characteristics of innate immune responses in macrophages. Microbiol Spectr. 2016;4(4). doi: 10.1128/microbiolspec.MCHD-0023-2015 [DOI] [PubMed] [Google Scholar]
- 8.Hulsmans M, Clauss S, Xiao L, et al. Macrophages facilitate electrical conduction in the heart. Cell. 2017;169(3):510–522.e20. doi: 10.1016/j.cell.2017.03.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gula G, Rumiński S, Niderla-Bielińska J, et al. Potential functions of embryonic cardiac macrophages in angiogenesis, lymphangiogenesis and extracellular matrix remodeling. Histochem Cell Biol. 2021;155(1):117–132. doi: 10.1007/s00418-020-01934-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nicolás-ávila JA, Lechuga-Vieco AV, Esteban-Martínez L, et al. A network of macrophages supports mitochondrial homeostasis in the heart. Cell. 2020;183(1):94–109.e23. doi: 10.1016/j.cell.2020.08.031 [DOI] [PubMed] [Google Scholar]
- 11.Hulsmans M, Sager HB, Roh JD, et al. Cardiac macrophages promote diastolic dysfunction. J Exp Med. 2018;215(2):423–440. doi: 10.1084/jem.20171274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smart CD, Fehrenbach DJ, Wassenaar JW, et al. Immune profiling of murine cardiac leukocytes identifies Trem2 as a novel mediator of hypertensive heart failure. Cardiovasc Res. 2023;119:2312–2328. doi: 10.1093/cvr/cvad093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sack MN. Mitochondrial fidelity and metabolic agility control immune cell fate and function. J Clin Invest. 2018;128(9):3651–3661. doi: 10.1172/jci120845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mounier R, Théret M, Arnold L, et al. AMPKα1 regulates macrophage skewing at the time of resolution of inflammation during skeletal muscle regeneration. Cell Metab. 2013;18(2):251–264. doi: 10.1016/j.cmet.2013.06.017 [DOI] [PubMed] [Google Scholar]
- 15.Palmieri EM, Menga A, Martín-Pérez R, et al. Pharmacologic or genetic targeting of glutamine synthetase skews macrophages toward an M1-like phenotype and inhibits tumor metastasis. Cell Rep. 2017;20(7):1654–1666. doi: 10.1016/j.celrep.2017.07.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Banerjee D, Tian R, Cai S. The role of innate immune cells in cardiac injury and repair: a metabolic perspective. Curr Cardiol Rep. 2023;25(7):631–640. doi: 10.1007/s11886-023-01897-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Erlich JR, To EE, Luong R, et al. Glycolysis and the pentose phosphate pathway promote LPS-induced NOX2 oxidase- and IFN-β-dependent inflammation in macrophages. Antioxidants. 2022;11(8):1488. doi: 10.3390/antiox11081488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Semba H, Takeda N, Isagawa T, et al. HIF-1α-PDK1 axis-induced active glycolysis plays an essential role in macrophage migratory capacity. Nat Commun. 2016;7:11635. doi: 10.1038/ncomms11635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Freemerman AJ, Zhao L, Pingili AK, et al. Myeloid Slc2a1-deficient murine model revealed macrophage activation and metabolic phenotype are fueled by GLUT1. J Immunol. 2019;202(4):1265–1286. doi: 10.4049/jimmunol.1800002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tan Z, Xie N, Cui H, et al. Pyruvate dehydrogenase kinase 1 participates in macrophage polarization via regulating glucose metabolism. J Immunol. 2015;194(12):6082–6089. doi: 10.4049/jimmunol.1402469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weisheit C, Zhang Y, Faron A, et al. Ly6C(low) and not Ly6C(high) macrophages accumulate first in the heart in a model of murine pressure-overload. PLoS One. 2014;9(11):e112710. doi: 10.1371/journal.pone.0112710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liao X, Shen Y, Zhang R, et al. Distinct roles of resident and nonresident macrophages in nonischemic cardiomyopathy. Proc Natl Acad Sci USA. 2018;115(20):E4661–e4669. doi: 10.1073/pnas.1720065115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Du L, Sun X, Gong H, et al. Single cell and lineage tracing studies reveal the impact of CD34(+) cells on myocardial fibrosis during heart failure. Stem Cell Res Ther. 2023;14(1):33. doi: 10.1186/s13287-023-03256-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Y, Morley M, Brandimarto J, et al. RNA-Seq identifies novel myocardial gene expression signatures of heart failure. Genomics. 2015;105(2):83–89. doi: 10.1016/j.ygeno.2014.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Greco S, Fasanaro P, Castelvecchio S, et al. MicroRNA dysregulation in diabetic ischemic heart failure patients. Diabetes. 2012;61(6):1633–1641. doi: 10.2337/db11-0952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hao Y, Hao S, Andersen-Nissen E, et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184(13):3573–3587.e29. doi: 10.1016/j.cell.2021.04.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Korsunsky I, Millard N, Fan J, et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods. 2019;16(12):1289–1296. doi: 10.1038/s41592-019-0619-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu C, Li T, Xu Y, et al. CellMarker 2.0: an updated database of manually curated cell markers in human/mouse and web tools based on scRNA-seq data. Nucleic Acids Res. 2023;51(D1):D870–d876. doi: 10.1093/nar/gkac947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aibar S, González-Blas CB, Moerman T, et al. SCENIC: single-cell regulatory network inference and clustering. Nat Methods. 2017;14(11):1083–1086. doi: 10.1038/nmeth.4463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu Y, Yang S, Ma J, et al. Spatiotemporal immune landscape of colorectal cancer liver metastasis at single-cell level. Cancer Discov. 2022;12(1):134–153. doi: 10.1158/2159-8290.Cd-21-0316 [DOI] [PubMed] [Google Scholar]
- 31.Jin S, Guerrero-Juarez CF, Zhang L, et al. Inference and analysis of cell-cell communication using CellChat. Nat Commun. 2021;12(1):1088. doi: 10.1038/s41467-021-21246-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mosterd A, Hoes AW. Clinical epidemiology of heart failure. Heart. 2007;93(9):1137–1146. doi: 10.1136/hrt.2003.025270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tanai E, Frantz S. Pathophysiology of heart failure. Compr Physiol. 2015;6(1):187–214. doi: 10.1002/cphy.c140055 [DOI] [PubMed] [Google Scholar]
- 34.Litviňuková M, Talavera-López C, Maatz H, et al. Cells of the adult human heart. Nature. 2020;588(7838):466–472. doi: 10.1038/s41586-020-2797-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martini E, Kunderfranco P, Peano C, et al. Single-cell sequencing of mouse heart immune infiltrate in pressure overload-driven heart failure reveals extent of immune activation. Circulation. 2019;140(25):2089–2107. doi: 10.1161/circulationaha.119.041694 [DOI] [PubMed] [Google Scholar]
- 36.Ma Y, Mouton AJ, Lindsey ML. Cardiac macrophage biology in the steady-state heart, the aging heart, and following myocardial infarction. Transl Res. 2018;191:15–28. doi: 10.1016/j.trsl.2017.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Frangogiannis NG. Cardiac fibrosis: cell biological mechanisms, molecular pathways and therapeutic opportunities. Mol Aspect Med. 2019;65:70–99. doi: 10.1016/j.mam.2018.07.001 [DOI] [PubMed] [Google Scholar]
- 38.Liang S, Ma A, Yang S, Wang Y, Ma Q. A review of matched-pairs feature selection methods for gene expression data analysis. Comput Struct Biotechnol J. 2018;16:88–97. doi: 10.1016/j.csbj.2018.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Song M, Xu S, Zhong A, Zhang J. Crosstalk between macrophage and T cell in atherosclerosis: potential therapeutic targets for cardiovascular diseases. Clin Immunol. 2019;202:11–17. doi: 10.1016/j.clim.2019.03.001 [DOI] [PubMed] [Google Scholar]
- 40.Sánchez-Navarro A, González-Soria I, Caldiño-Bohn R, Bobadilla NA. An integrative view of serpins in health and disease: the contribution of SerpinA3. Am J Physiol Cell Physiol. 2021;320(1):C106–c118. doi: 10.1152/ajpcell.00366.2020 [DOI] [PubMed] [Google Scholar]
- 41.Wang X, Ding Y, Li R, et al. N(6)-methyladenosine of Spi2a attenuates inflammation and sepsis-associated myocardial dysfunction in mice. Nat Commun. 2023;14(1):1185. doi: 10.1038/s41467-023-36865-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Delrue L, Vanderheyden M, Beles M, et al. Circulating SERPINA3 improves prognostic stratification in patients with a de novo or worsened heart failure. ESC Heart Fail. 2021;8(6):4780–4790. doi: 10.1002/ehf2.13659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hsieh WY, Zhou QD, York AG, et al. Toll-like receptors induce signal-specific reprogramming of the macrophage lipidome. Cell Metab. 2020;32(1):128–143.e5. doi: 10.1016/j.cmet.2020.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yan J, Horng T. Lipid metabolism in regulation of macrophage functions. Trends Cell Biol. 2020;30(12):979–989. doi: 10.1016/j.tcb.2020.09.006 [DOI] [PubMed] [Google Scholar]
- 45.Fan G, Li Y, Zong Y, et al. GPAT3 regulates the synthesis of lipid intermediate LPA and exacerbates Kupffer cell inflammation mediated by the ERK signaling pathway. Cell Death Dis. 2023;14(3):208. doi: 10.1038/s41419-023-05741-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Glyn-Jones S, Palmer AJ, Agricola R, et al. Osteoarthritis. Lancet. 2015;386:9991):376–87. doi: 10.1016/s0140-6736(14)60802-3 [DOI] [PubMed] [Google Scholar]
- 47.Lu C, Amin MA, Fox DA. CD13/Aminopeptidase N is a potential therapeutic target for inflammatory disorders. J Immunol. 2020;204(1):3–11. doi: 10.4049/jimmunol.1900868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Küster H, Thompson H, Kinet JP. Characterization and expression of the gene for the human Fc receptor gamma subunit. Definition of a new gene family. J Biol Chem. 1990;265(11):6448–6452. [PubMed] [Google Scholar]
- 49.Brandsma AM, Hogarth PM, Nimmerjahn F, Leusen JH. Clarifying the confusion between cytokine and Fc Receptor ”Common Gamma Chain”. Immunity. 2016;45(2):225–226. doi: 10.1016/j.immuni.2016.07.006 [DOI] [PubMed] [Google Scholar]
- 50.Yu M, Eckart MR, Morgan AA, et al. Identification of an IFN-γ/mast cell axis in a mouse model of chronic asthma. J Clin Invest. 2011;121(8):3133–3143. doi: 10.1172/jci43598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Podgórska D, Cieśla M, Kolarz B. FCER1G gene hypomethylation in patients with rheumatoid arthritis. J Clin Med. 2022;11(16):4664. doi: 10.3390/jcm11164664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fu L, Cheng Z, Dong F, et al. Enhanced expression of FCER1G predicts positive prognosis in multiple myeloma. J Cancer. 2020;11(5):1182–1194. doi: 10.7150/jca.37313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhao W, Wu T, Zhan J, Dong Z. Identification of the immune status of hypertrophic cardiomyopathy by integrated analysis of bulk- and single-cell RNA sequencing data. Comput Math Methods Med. 2022;2022:7153491. doi: 10.1155/2022/7153491 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 54.Povo-Retana A, Landauro-Vera R, Fariñas M, et al. Defining the metabolic signatures associated with human macrophage polarisation. Biochem Soc Trans. 2023;51:1429–1436. doi: 10.1042/bst20220504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Boscá L, González-Ramos S, Prieto P, et al. Metabolic signatures linked to macrophage polarization: from glucose metabolism to oxidative phosphorylation. Biochem Soc Trans. 2015;43(4):740–744. doi: 10.1042/bst20150107 [DOI] [PubMed] [Google Scholar]
- 56.Ganeshan K, Chawla A. Metabolic regulation of immune responses. Ann Rev Immunol. 2014;32:609–634. doi: 10.1146/annurev-immunol-032713-120236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ómaoldomhnaigh C, Cox DJ, Phelan JJ, Malone FD, Keane J, Basdeo SA. The Warburg effect occurs rapidly in stimulated human adult but not umbilical cord blood derived macrophages. Front Immunol. 2021;12:657261. doi: 10.3389/fimmu.2021.657261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.O’Neill LA, Kishton RJ, Rathmell J. A guide to immunometabolism for immunologists. Nat Rev Immunol. 2016;16(9):553–565. doi: 10.1038/nri.2016.70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu Y, Xu R, Gu H, et al. Metabolic reprogramming in macrophage responses. Biomarker Res. 2021;9(1):1. doi: 10.1186/s40364-020-00251-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mills EL, Kelly B, O’Neill LAJ. Mitochondria are the powerhouses of immunity. Nat Immunol. 2017;18(5):488–498. doi: 10.1038/ni.3704 [DOI] [PubMed] [Google Scholar]
- 61.Zhang S, Bories G, Lantz C, et al. Immunometabolism of phagocytes and relationships to cardiac repair. Front Cardiovasc Med. 2019;6:42. doi: 10.3389/fcvm.2019.00042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pearce EL, Pearce EJ. Metabolic pathways in immune cell activation and quiescence. Immunity. 2013;38(4):633–643. doi: 10.1016/j.immuni.2013.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Heinrichs D, Berres ML, Coeuru M, et al. Protective role of macrophage migration inhibitory factor in nonalcoholic steatohepatitis. FASEB J. 2014;28(12):5136–5147. doi: 10.1096/fj.14-256776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thavayogarajah T, Sinitski D, El Bounkari O, et al. CXCR4 and CD74 together enhance cell survival in response to macrophage migration-inhibitory factor in chronic lymphocytic leukemia. Exp Hematol. 2022;115:30–43. doi: 10.1016/j.exphem.2022.08.005 [DOI] [PubMed] [Google Scholar]
- 65.Leslie J, Millar BJ, Del Carpio Pons A, et al. FPR-1 is an important regulator of neutrophil recruitment and a tissue-specific driver of pulmonary fibrosis. JCI Insight. 2020;5(4). doi: 10.1172/jci.insight.125937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Qin C, Buxton KD, Pepe S, et al. Reperfusion-induced myocardial dysfunction is prevented by endogenous annexin-A1 and its N-terminal-derived peptide Ac-ANX-A1(2-26). Br J Pharmacol. 2013;168(1):238–252. doi: 10.1111/j.1476-5381.2012.02176.x [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Publicly available datasets (ID: GSE222144, GSE57338, and GSE26887) were obtained from https://www.ncbi.nlm.nih.gov/geo/.