Abstract
Background
Different species of sea cucumbers in various regions have diverse preferred habitats and feeding habits. However, detailed research on the correlation between food selection and habitat preference of sea cucumbers, as well as their adaptive adjustments to specific habitat types, is still lacking.
Methods
A field study was carried out to explore the relationship between food selection and habitat preference, as well as the adaptation process, of the tropical sea cucumber Stichopus chloronotus, which has specific food preferences. This was achieved using an in situ mesocosm method with three single habitat types: sandy, broken coral branches, and reef. Changes is the prokaryotic community structure of gut contents, revealed by high-throughput sequencing analysis of the 16S rRNA gene, were used as the biomarker. Tax4Fun assessed the metabolic pathways of samples, and FAPROTAX evaluated the biogeochemical cycling processes.
Results
Alpha diversity, PCoA, and UPGMA tree analyses consistently revealed that bacterial community structures in the gut contents of S. chloronotus in reef bottom cages (GRee) are closely related to those of wild S. chloronotus collected in September (GWS) and October (GWO) than those from the sandy bottom (GSan) and broken coral branches bottom (GBra) cages. The relative abundance of Ruegeria is one of the dominant genera in the control groups (GWS and GWO), while Synechococcus CC9902 is among the predominant genera in the treatment groups (GSan, GBra and GRee). Functional prediction outcomes from Tax4Fun and FAPROTAX also indicate that the metabolic pathways in the gut contents of the treatment groups are distinct from those of the control groups.
Conclusions
Compared with S. chloronotus in single habitat types, wild S. chloronotus showed stronger feeding selectivity and ingested actively larger proportion of Ruegeria sp. For this picky species, hard-substrate habitats that can keep it away from strong waves seem to be more important to than those with good sedimentary food. Inappropriate habitats without stable substrate for attachment may cause an unusual change in food preference of S. chloronotus. Tax4Fun and FAPROTAX functional annotation also confirmed that the adaptive adjustment of S. chloronotus can be completed within a month.
Keywords: Stichopus chloronotus, Feeding adaptability, Habitat type, 16S rRNA, Gut contents
Background
Tropical sea cucumbers have high commercial value and huge market demand, but long-term overfishing has led to a serious decline in their populations [1–3]. Sea cucumbers also have important ecological functions and play an important role in the benthic ecosystem [4]. Sedimentary sea cucumbers can purify the benthic environment and disturb the sediment by feeding, which accelerate the exchange of inorganic nutrients in sediments with seawater [5–9]. Moreover, their fecal deposits can enhance coral calcification and provide a buffer against ocean acidification [10–13]. At present, research on tropical sea cucumbers mainly focuses on resource surveys, classification and diversity, and physiological ecology [14–16]. In comparison, research on the correlation between their food selection and habitat preference is relatively scarce.
The habitat selection of animals is often closely related to their feeding habits, covering their food search and ingestion. Whether or not food is easy to obtain is the first question that animals need to consider, followed by factors such as avoiding enemies [17]. Is there a similar phenomenon with Stichopus chloronotus? A few studies have revealed the distinct species-specific substrate and food preference of several tropical coral reef sea cucumbers. Holothuria edulis and Holothuria atra prefer distributing in sandy bottom areas and have no clear selectivity to the sediment [18], whereas S. chloronotus and Stichopus monotuberculatus prefer areas with hard substrates (e.g., rocks and coral reef blocks) and are picky about food with fine grain size and high organic content [2, 19, 20]. In particular, S. chloronotus is more inclined to eat sediments with high microalgae content, and S. monotuberculatus actively eats sediments with higher nutrient content [21]. However, which is taken into account first between food quality and physical substrate during the habitat selection of sea cucumbers remain unclear.
The preferred habitats of S. chloronotus vary in different regions and life history stages, such as reefs shoals, lagoons [22], soft substrates [23], and other hard substrates [18]. The reason may be that S. chloronotus have made adaptive adjustments to the habitat characteristics and food conditions of specific sea areas, though detailed studies are still lacking. The present research aims to explore whether the strategic adjustment of sea cucumber response to habitat changes is a positive rapid process or a passive long-term adaptation. In what aspects have sea cucumbers made adaptive adjustments, and what do these adjustments mean? To sum up, studying the natural ecology of sea cucumbers will help us to understand their ecological habits, grasp the ecological adaptation rules, and provide a basis for the protection and sustainable development of sea cucumber resources.
DNA barcoding requires only an accurate DNA sequence to detect many biological species from a small number of samples, offering the advantages of sensitivity, accuracy, and speed [24]. 16S rRNA gene high-throughput sequencing technology has been widely used to analyze microflora in various biomes, such as water, sediments, and gut content of animals [25–28]. This study aims to explore the feeding adjustment ability of sea cucumbers on a small spatial scale when their food sources are consistent. Prokaryotes in their gut contents can be used as indicator microorganisms to determine their feeding adjustment in a single habitat type.
The sea area of Wuzhizhou Island is a typical example of tropical marine ranching, with effective protection of the coral reef ecosystem in the South China Sea [29]. S. chloronotus is the dominant species here [3]. Our previous study found that this species has a strong preference for hard substrates such as reefs and rocks [2]. Therefore, we selected S. chloronotus as the experimental species and conducted an in situ mesocosm experiment for one month. 16S rRNA gene high-throughput sequencing was used to reveal the changes in microbial community structure of gut contents of S. chloronotus. The goals of this study are to explore the feeding selectivity of S. chloronotus and the correlation between its habitat preferences and feeding choices. Moreover, it aims to investigate the adaptability adjustment of S. chloronotus in net cages with a single habitat type, namely sandy, broken coral branches, and reef, over the short term.
Materials and Methods
Study area and period
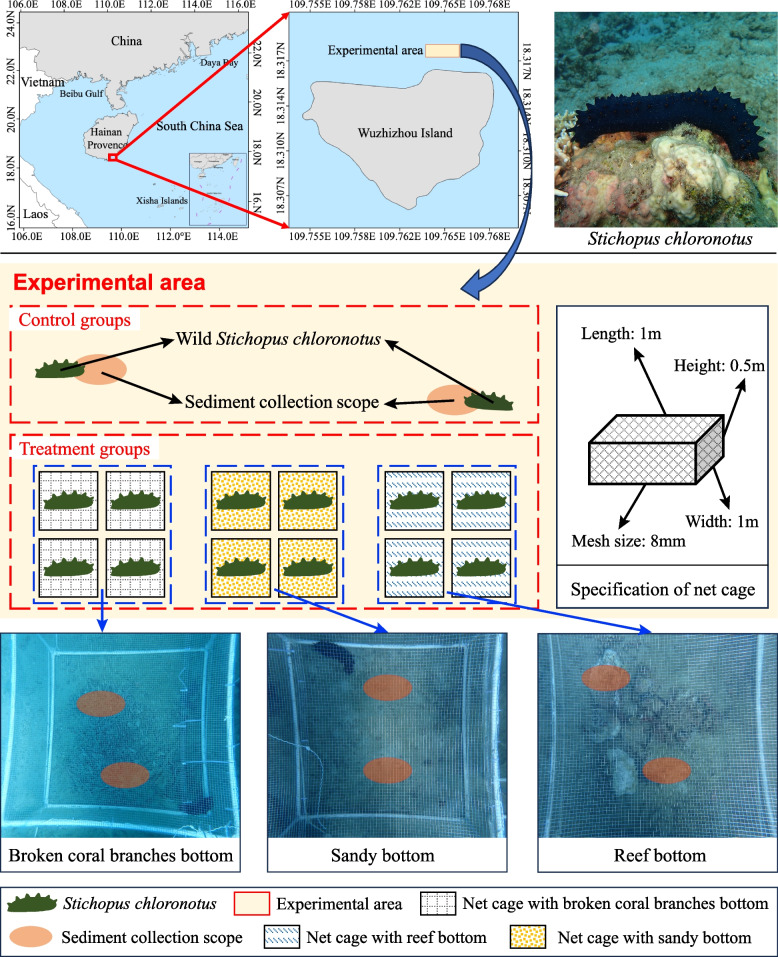
The northeast of Wuzhizhou Island is a compound distribution area of living coral reefs and sandy bottom, with well-developed coral reefs and diverse habitat types [30, 31]. Numerous artificial reefs are deployed here to restore fishery resources and provide reef diving tourism. The experimental area (18°322′N, 109°775′E) is between two sunken vessels (length, 30 m; width, 6 m; and height, 8 m) that are parallel to the coastline to effectively weaken the intrusion of typhoons and wind waves (Fig. 1). Net cages were placed in the sandy bottom for easy deployment. A previous study found that the population of S. chloronotus increases during the rainy season and that changes in preferred habitats tend to stabilize in September [2]. Consequently, the experiment commenced on September 20, 2021, and lasted for a full month.
Fig. 1.
Study area and experimental design
Design of experiment
Twelve custom net cages (length, 1 m; width, 1 m; height, 0.5 m; mesh size, 8 mm) were fixed by divers between two sunken vessels and divided into three treatment groups (n = 4). Each group had a different substrate type: sand, broken coral branches, and reef blocks (Fig. 1). Dead broken coral branches and small reef blocks (< 45 cm in diameter) were collected nearby and laid into the net cages, covering 60% of the bottom on average (0.6 m2).
At the beginning of the study, 16 wild S. chloronotus with similar sizes (306.97 ± 38.65 g) were collected via SCUBA diving. Of this number, 12 were placed into 12 cages, which were sealed to prevent escape, and the remaining 4 individuals (serving as the control group in their natural habitat) were brought back to the laboratory for gut content sampling. At the end of the 30-day experiment, divers sampled all the sea cucumbers within the cages and additionally sampled 4 wild ones of similar weight (373.34 ± 80.37 g) outside the net cages, which served as the control group in October.
Sample collection of gut contents
The collected S. chloronotus were transported to the Wuzhizhou Island Laboratory within half an hour under oxygenation and low temperature conditions. Subsequently, the samples were put on ice, and 5 ml potassium chloride solution (0.35 mol/L) was administered with a pinhole syringe to facilitate the expulsion of gut tissue. The gut contents were then collected and stored in a 2 ml sterile cryopreservation tube. After each sample collection, the tray and tweezers were disinfected with 75% ethanol and sterilized seawater to minimize bacterial cross-contamination between samples. According to the experimental design, the gut content samples from wild S. chloronotus in September and October were labeled as GWS1–GWS4 and GWO1–GWO4, respectively. The gut content samples of S. chloronotus housed in net cages with sandy bottom, broken coral branches bottom, and reef bottom (with one invalid sample) were labeled as GSan1–GSan4, GBra1–GBra4, and GRee2–GRee4, respectively.
Sample collection of environmental sediments
The collection of environmental sediment samples corresponded to that of S. chloronotus. Sediment samples collected near wild S. chloronotus in September and October were labeled as SWS1–SWS4 and SWO1–SWO4, respectively. Sediment samples in the four net cages with sandy bottom were directly collected by divers using 2 ml sterile cryopreservation tubes and labeled as SSan1–SSan4. Sediment samples collected in the net cages with broken coral branches bottom and reef bottom were scraped from dead coral branches and reef surfaces and labeled as SBra1–SBra4 and SRee1–SRee4, respectively. The samples of gut contents and sediments were stored at −80° C until required for analysis.
DNA extraction, PCR amplification, and high-throughput sequencing
The genomic DNA of the gut contents and sediment samples was extracted by CTAB (hexadecyl trimethyl ammonium Bromide) method, and the purity and concentration of DNA were detected by agarose gel electrophoresis. Appropriate DNA was placed in a centrifuge tube and diluted to 1 ng/μl with sterile water. Using the diluted genomic DNA as the template, based on the selection of sequenced regions (V4) [24, 32], the universal primer set, 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) [33, 34], was used for amplification of the 16S rRNA gene from the gut contents and sediments. A total of 30-μL PCR mixture contained 15 μL of Phusion® High-Fidelity PCR Master Mix (New England Biolabs), 3 μL of each primer (2 μM), 10 μL genomic DNA (1 ng/μL), and 2 μL sterile water. Thermal cycling was as follows: initial denaturation at 98 °C for 1 min, followed by 30 cycles of denaturation at 98 °C for 10 s, annealing at 50 °C for 30 s, and elongation at 72 °C for 30 s. The process concluded with a final extension at 72 °C for 5 min. PCR products were mixed at equal concentration and purified by GeneJETTM Gel Extraction Kit (Thermo Scientific) [28].
Sequencing libraries were generated using the TruSeq® DNA PCR-Free Library Preparation Kit (Illumina, USA) following the manufacturer’s instructions. The constructed library quality was assessed on the Qubit@ 2.0 Fluorometer (Thermo Scientific, USA) system. After qualification, the constructed library was sequenced by NovaSeq6000 platform (Novogene, China).
Statistical analysis
FLASH (V1.2.7, http://ccb.jhu.edu/software/FLASH/) was used to splice the reads of each sample and obtain raw tags [35]. Uparse algorithm (Uparse v7.0.1001, http://www.drive5.com/uparse/) was used to cluster all the effective tags of all samples [36]. The sequences are clustered into operational taxonomic units (OTUs) with 97% identity, and representative sequence of the OTUs were selected. The Mothur method and the SSUrRNA database of SILVA138 (http://www.arb-silva.de/) were used for species annotation analysis, with the threshold set at 0.8 ~ 1 [37, 38]. Next, the taxonomic information was obtained at each classification level. MUSCLE software (version 3.8.31, http://www.drive5.com/muscle/) was used to perform fast multi-sequence alignment to obtain the phylogenetic relationships of all representative sequences of the OTUs. Finally, the data of all samples were homogenized, and the subsequent alpha diversity and beta diversity analyses were based on the homogenized data.
R (version 2.15.3) was used to analyze the inter-group differences of alpha diversity indices, including abundance-based coverage estimator (ACE), Chao1 richness estimator, Shannon diversity index, and Simpson diversity index. R software was also used for principal coordinates analysis (PCoA) and species analysis, with significant differences between groups (T-test). Analysis of similarities (Anosim) used the Anosim function of the R vegan package. Qiime (version 1.9.1) was used to calculate the unweighted-unifrac distance and construct the unweighted pair-group method with arithmetic means (UPGMA) sample cluster tree. Tax4Fun analysis (R Version 3.0.3) and the FAPROTAX software were used for functional prediction of samples.
Results
At the end of the experiment, all 12 net cages were intact and there was no instance of S. chloronotus escaping. The S. chloronotus in the treatment groups lost weight, but none were reported of injured or deceased. Specifically, the weight of S. chloronotus in GSan, GBra, and GRee decreased by 21.25 ± 10.36 g, 18.39 ± 7.20 g, and 11.53 ± 3.58 g, respectively.
A total of 2 492 373 effective reads, with an average length of 253 base pairs (bp), were obtained from 19 gut content samples and 20 sediment samples through 16S rRNA gene sequencing. The number of effective reads for each gut content samples ranged from 61 680 to 69 316, with an average of 65 270 ± 2 585. For the sediment samples, the number of effective reads ranged from 40 484 to 70 232, with an average of 62 612 ± 7 805. The Good's coverage estimate for all samples was between 91.80% and 97.85%, indicating that the sequencing depth was sufficient to capture the majority of microorganisms present.
Richness and diversity analysis of gut and sediment samples
A total of 25 650 OTUs were identified from 39 samples based on a 97% similarity threshold. GWS and GWO harbored 4 777 and 4 540 OTUs, respectively, while GSan, GBra, and GRee contained 6 182, 6 937, and 4 147 OTUs, respectively (Fig. 2). Among these OTUs, 860, 652, 1 661, 2 082 and 559 were uniquely detected in GWS, GWO, GSan, GBra, and GRee, respectively (Fig. 2). A total of 1 849 OTUs (15.53%) were shared across all gut content samples; 2 982 (47.07%) were shared between GWS and GWO; and 2 454 (24.20%) were shared among GSan, GBra, and GRee (Fig. 2). Correspondingly, the sediment samples (SWS, SWO, SSan, SBra, and SRee) contained 10 406, 12 599, 11 179, 9 925, and 11 782 OTUs, respectively.
Fig. 2.
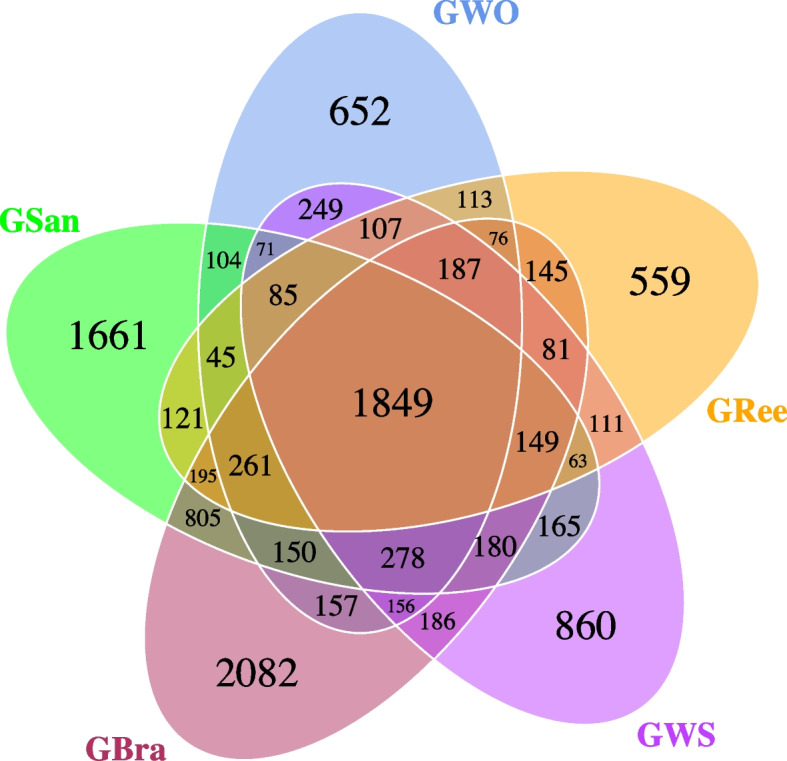
Venn diagram of core OTUs among the gut contents of five group of S. chloronotus
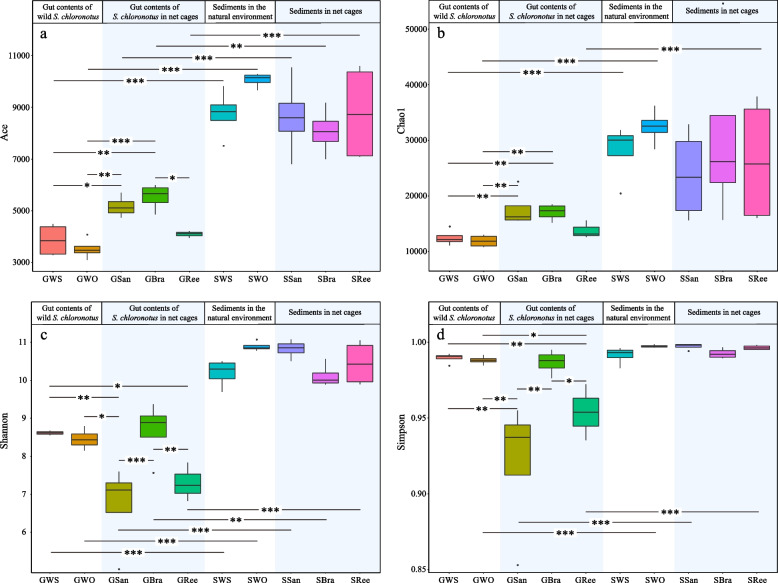
The richness indices (ACE, Chao1) and diversity indices (Shannon, Simpson) for each sample group are shown in Fig. 3. The ACE and Shannon diversity indices for the sediment groups (SWS, SWO, SSan, SBra, SRee) were extremely significantly higher than those for the corresponding gut content groups (GWS, GWO, GSan, GBra, GRee) (P < 0.01) (Fig. 3a, c), indicating that the bacterial communities in the sediments exhibited greater richness and diversity compared to those in the gut contents. Upon comparison of the gut content samples, the ACE and Chao1 indices for GSan and GBra were found to be significantly higher than those for GWS and GWO (P < 0.05), whereas no clear differences were observed among GRee, GWS, and GWO (Fig. 3a, b). The Shannon and Simpson diversity indices for GWS, GWO, and GBra were significantly higher than those for GSan and GRee (expect for GWO and GRee in the Shannon index) (P < 0.05) (Fig. 3c, d). Furthermore, no significant difference was observed between GWS and GWO, which were characterized by high diversity indices and low richness indices (Fig. 3).
Fig. 3.
Alpha-diversity of the gut content samples and sediment samples. a ACE, b Chao1, c Shannon index, d Simpson index. *: P < 0.05; **: P < 0.01; ***: P < 0.001. Dark point represents the abnormal value
Bacterial community structure in gut contents and sediments
The gut content groups (GWS: 36.25 ± 3.30, GWO: 37.25 ± 2.87, GSan: 51.50 ± 3.51, GBra: 51.75 ± 5.62, GRee: 41.33 ± 1.53) and sediment groups (SWS: 67.50 ± 2.65, SWO: 69.50 ± 2.89, SSan: 64.25 ± 2.99, SBra: 63.50 ± 1.29, SRee: 61.50 ± 8.27) exhibited varying numbers of phyla. Significantly, the sediment groups had a higher number of phyla compared to the gut content groups (P < 0.05), with GSan and GBra showing a significant increase over GWS, GWO, and GRee (P < 0.05).
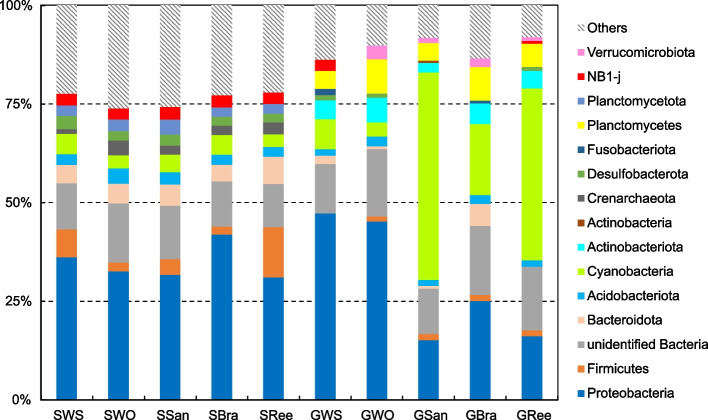
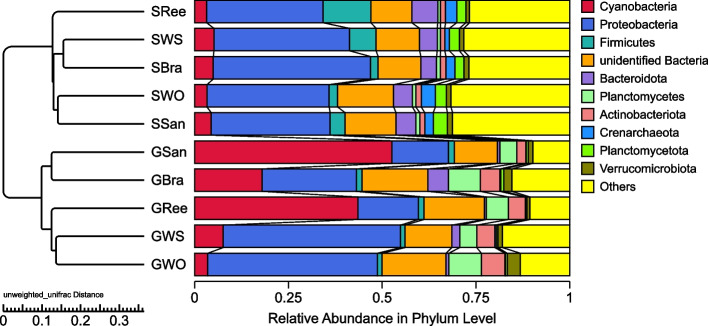
The 10 most abundant phyla across different groups accounted for 73.89%–91.86% of the total reads (Fig. 4). For the sediment groups, the top 10 phyla were consistent, with variations in their relative abundances. Proteobacteria (34.68 ± 4.50%) dominated the sediment groups, followed by Firmicutes (5.62 ± 4.47%), Bacteroidota (5.22 ± 1.04%), and Cyanobacteria (4.22 ± 0.91%), which also constituted relatively significant proportions. However, the top 10 phyla in the five gut content groups varied, with Proteobacteria, Acidobacteriota, Cyanobacteria, Actinobacteriota and Planctomycetes appearing in every group and comprising 59.06%–76.10% of the total.
Fig. 4.
Relative abundance of the 10 most abundant phyla
Proteobacteria was the predominant phylum in GWS (47.29 ± 4.43%) and GWO (45.23 ± 4.74%), whereas Cyanobacteria was the dominant phylum in GSan (52.59 ± 16.07%) and GRee (43.51 ± 10.47%). In the case of GBra, both Proteobacteria (25.06 ± 7.91%) and Cyanobacteria (18.01 ± 9.93%) constituted substantial fractions (Fig. 4). The relative abundances of Proteobacteria, Planctomycetes, and Actinobacteriota in GWS and GWO were also significantly higher than those in SWS and SWO, whereas the relative abundances of Cyanobacteria and Planctomycetes in GSan and GRee were significantly higher than those in SSan and SRee (T-test, P < 0.05).
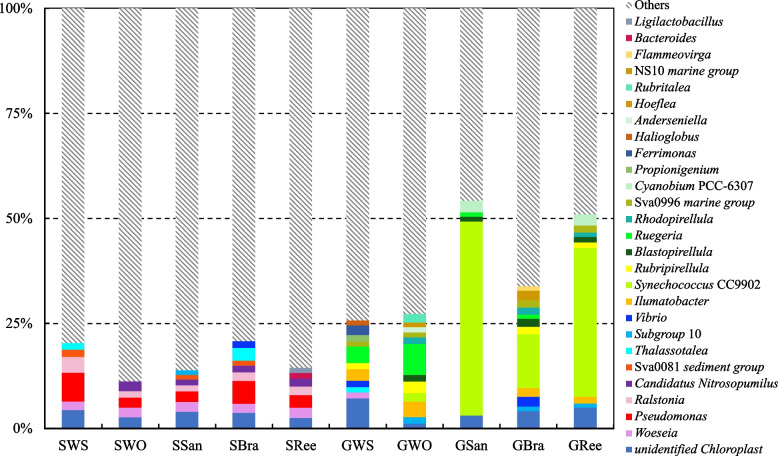
Prokaryotes with a relative abundance of over 1% at the genus level in each group are shown in Fig. 5. In five sediment groups, 10 genera of prokaryotes had relative abundances over 1%, with Pseudomonas (4.05 ± 2.01%), Woeseia (2.23 ± 0.15%), Ralstonia (2.15 ± 0.93%), and Candidatus Nitrosopumilus (1.82 ± 0.42%) present in at least four groups (Fig. 5). In five gut content groups, 21 genera of prokaryotes had relative abundance over 1%, with Synechococcus CC9902 (24.12 ± 20.24%), Ruegeria (3.35 ± 3.03%), Ilumatobacter (2.53 ± 0.93%), Rubripirellula (1.82 ± 0.63%), Blastopirellula (1.48 ± 0.35%), and Sva0996 marine group (1.43 ± 0.33%) present in at least four groups (Fig. 5). Dominant prokaryotes in the sediment and gut content samples were markedly different at the genus level. A comparison of the relative abundances of dominant genera within each gut content group showed that Synechococcus CC9902 had the highest relative abundance in GSan (46.16%), GBra (12.83%), and GRee (35.45%), whereas it was less than 2.03% in GWS and GWO (Fig. 5). The relative abundance of Sva0996 marine group exceeded 1% in GWS (1.14%), GWO (1.15%), GBra (1.77%), and GRee (1.65%), but only 0.75% in GSan. In addition, the relative abundance of Ruegeria in GWO (7.39 ± 1.38%) was extremely significantly higher than that in GSan (1.06 ± 1.11%), GBra (1.01 ± 0.60%), and GRee (0.53 ± 0.10%) (P < 0.001) (Fig. 5).
Fig. 5.
Prokaryotes with relative abundance of more than 1% at the genus level in each group
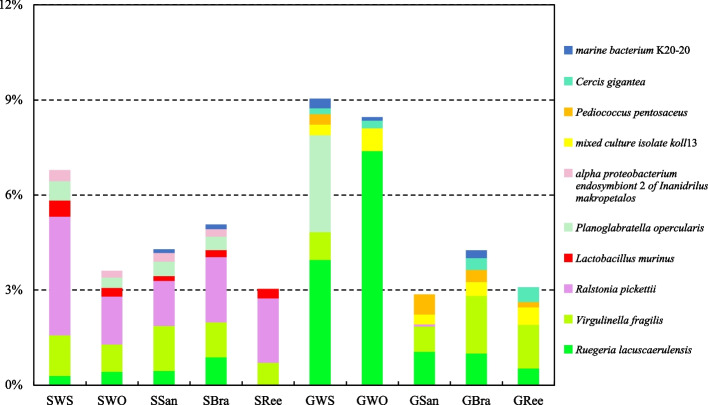
The top 10 prokaryotes in relative abundance at the species level and present in at least four sample groups are shown in Fig. 6. Among them, Virgulinella fragilis (1.08 ± 0.29%), Ralstonia pickettii (2.15 ± 0.93%), and Lactobacillus murinus (0.29 ± 0.14%) were present in all sediment groups. Ruegeria lacuscaerulensis (0.51 ± 0.25%), Planoglabratella opercularis (0.45 ± 0.12%), and alpha proteobacterium endosymbiont 2 of Inanidrilus makropetalos (0.27 ± 0.06%) were present in SWS, SWO, SSan, and SBra (Fig. 6). By contrast, dominant prokaryotes in the gut content samples were markedly different from those in the sediment samples at the species level. They were characterized by the presence of R. lacuscaerulensis (2.79 ± 2.91%) and mixed culture isolate koll13 (0.47 ± 0.17%) in all five gut content groups. V. fragilis (1.22 ± 0.48%) and Pediococcus pentosaceus (0.38 ± 0.19%) were present in GWS, GSan, GBra, and GRee (Fig. 6). Note that the relative abundance of R. lacuscaerulensis in the control groups (GWS: 3.96%, GWO: 7.39%) was higher than in the treatment groups (GSan: 1.06%; GBra: 1.01%, GR: 0.53%), with GWO showing statistically significant differences (T-test, P < 0.001) (Fig. 6). Moreover, the relative abundance of P. opercularis in GWS reached 3.05% but was less than 0.12% in GWO, GSan, GBra, and GRee (T-test, P < 0.001) (Fig. 6).
Fig. 6.
Prokaryotes with relative abundances of the top 10 at the species level and present in at least four sample groups
Relationships among the bacterial communities from gut content and sediment samples
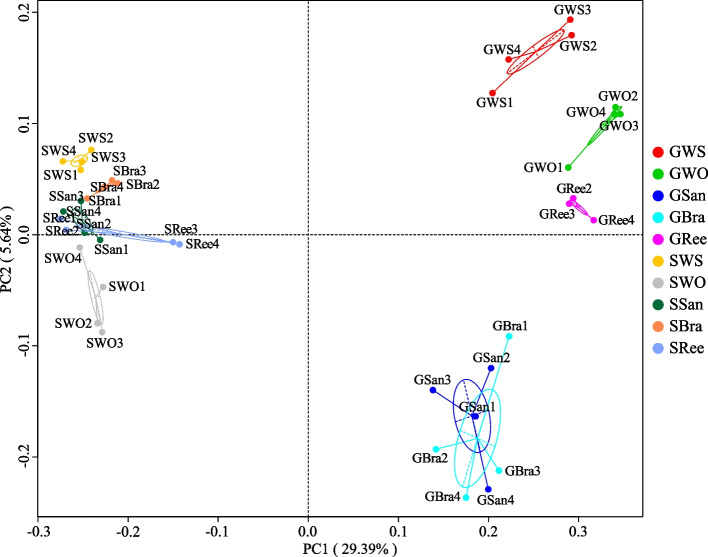
PCoA analysis and UPGMA tree were conducted to evaluate the bacterial compositional similarities among different samples (Figs. 7 and 8). The PCoA analysis showed the samples segregated into three groups (Fig. 7). Among them, all sediment samples (SWS, SWO, SSan, SBra, SRee) were clustered on the left side of the figure along the first principal component axis (PC1), accounting for 29.39% of the total variation (Fig. 7). The gut content samples of GWS, GWO, and GRee were clustered along the second principal component axis (PC2) on the upper right side of the figure, whereas the GSan and GBra samples were grouped on the lower right side, accounting for 5.64% of the total variation (Fig. 7). UPGMA tree at the phylum level (Fig. 8) corroborated the results of the PCoA analysis. These results indicate that the gut content and sediment samples harbor distinct bacterial community characteristics, with two single habitat types (sandy bottom and broken coral branches bottom) altering the gut content bacterial community structure of S. chloronotus. On the contrary, the bacterial community structure of GRee bore the closest resemblance to that of GWO and GWS.
Fig. 7.
Two-dimensional PCoA of each sample based on Unweighted unifrac distance at the OTU level
Fig. 8.
UPGMA tree at the phylum level based on Unweighted unifrac distance of each group
Anosim revealed that the R values between the control groups (GWS, GWO) and the experimental groups (GSan, GBra, GRee) were greater than 0, and the differences were statistically significant (Table 1, P < 0.05). This finding suggests that both monthly variations and habitat changes can significantly affect the gut bacterial community structure of S. chloronotus.
Table 1.
Analysis of similarities (ANOSIM) between control groups (GWS, GWO) and experimental groups (GSan, GBra, GRee)
| GWS-GWO | GWS-GSan | GWS-GBra | GWS-GRee | GWO-GSan | GWO-GBra | GWO-GRee | ||
|---|---|---|---|---|---|---|---|---|
| R value | 0.96 | 1 | 0.81 | 1 | 1 | 0.59 | 1 | |
| P value | 0.04* | 0.03* | 0.03* | 0.02* | 0.03* | 0.03* | 0.03* | |
R value is between (- 1, 1), and R-value greater than 0 indicates that the difference between groups is greater than the difference within groups
*P < 0.05, indicates statistical significance
Functional prediction of microbiota in gut contents and sediments
Tax4Fun is adept at functional prediction for both gut and sediment samples. FAPROTAX demonstrates robust predictive capabilities for the biochemical cycling of sediment samples.
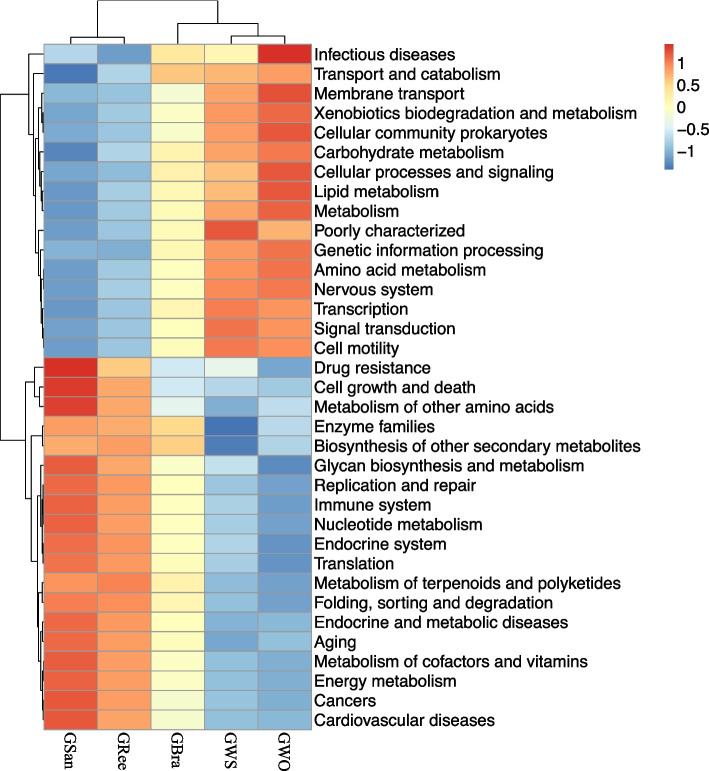
Utilizing the KEGG pathway database, Tax4Fun analysis delineated 7 level-1 pathways and 44 level-2 pathways across five groups of gut contents. The relative abundance of microbial flora functions within the level-2 pathways was normalized, revealing a bifurcation where five intestinal content groups were categorized into two primary branches: GSan and GRee clustered on one branch, and GWS, GWO, and GBra formed another (Fig. 9). The main pathways for GSan and GRee encompassed Drug resistance, Immune system, Cancers, Cardiovascular diseases, Metabolism of other amino acids, Enzyme families, Biosynthesis of other secondary metabolites, Glycan biosynthesis and metabolism, Nucleotide metabolism, Metabolism of terpenoids and polyketides, Metabolism of cofactors and vitamins, Energy metabolism, among many others. By contrast, the principal pathways for GWS and GWO focused on Transport and catabolism, Membrane transport, Xenobiotics biodegradation and metabolism, Carbohydrate metabolism, Lipid metabolism, Metabolism, Amino acid metabolism, Genetic information processing, Transcription. Noteworthy differences in metabolic pathways were observed between GSan and GBra, as well as in GWO and GWS (Fig. 9). The T-test also indicated no significant differences in level-3 pathways between GSan and GRee (P > 0.05).
Fig. 9.
Level-2 cluster heatmap (Top 35) for predicting Gene function of gut contents based on Tax4Fun
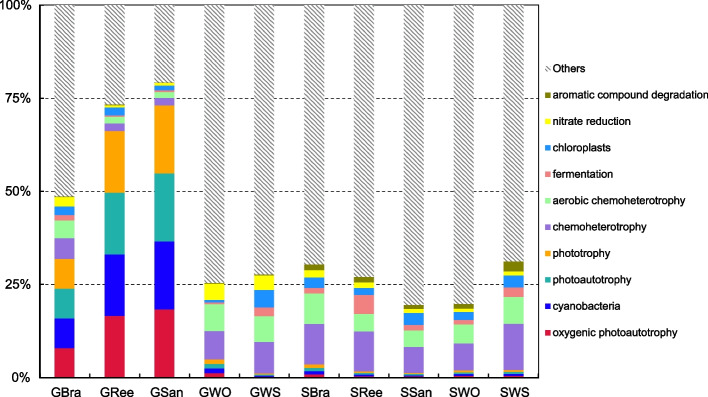
The top 10 biochemical cycling processes of gut content groups and sediment groups predicted by FAPROTAX are shown in Fig. 10. The relative abundances of oxygenic photoautotrophy, cyanobacteria, photoautotrophy, and phototrophy in GBra (7.95%), GRee (16.55%), and GSan (18.28%) were significantly higher than those observed in GWO (1.24%), GWS (0.29%), and five sediments groups (0.53% ± 0.21%) (P < 0.05) (Fig. 10). Conversely, the relative abundances of chemoheterotrophy and aerobic chemoheterotrophy in GBra (5.62% and 4.72%), GSan (2.10% and 1.75%), and GRee (2.02% and 1.54%) were significantly lower compared to those of GWO (7.55% and 7.16%), GWS (8.51% and 6.81%), and five sediments groups (9.64% ± 2.40% and 5.91% ± 1.71%) (P < 0.05) (Fig. 10). The predicted functional structure of S. chloronotus within complex multi-habitat types (GWS and GWO) was similar to those of the sediment groups (SWS, SWO, SSan, SBra, and SRee), whereas that of S. chloronotus in single habitat types (GSan, GBra, and GRee) exhibited significant alterations.
Fig. 10.
Relative abundance of the 10 most abundant functional predicted by FAPROTAX
Discussion
Feeding selectivity of S. chloronotus
S. chloronotus has feeding preferences and exhibits selective enrichment of microbiomes in their gut contents. The number of microbial phyla in the gut contents (80) was remarkably lower than those in ambient sediments (100), with the microbial community structure in the gut contents characterized by reduced richness and diversity. Dominant genera within the gut contents were also markedly distinct from those in the ambient sediment. Alpha-diversity, PCoA analysis, and UPGMA tree consistently revealed that the microbial communities of sediments were distinctly different from those of gut contents [38–42]. S. chloronotus is known to have a propensity for ingesting sediments with finer grain size [18, 21], actively consuming sediments with higher organic content [19, 20, 43], and selectively feeding on eukaryotes [32]. Moriarty et al. (1985) found that the assimilation efficiency of S. chloronotus for heterotrophic bacteria ranged from 32 to 44%, surpassing that of organic matter in sediments [44]. The tentacles of sea cucumbers, particularly the sensory receptors at the tips, may facilitate the selection of specific sediment patches by sea cucumbers [45, 46].
In comparison to S. chloronotus in three single habitat types (GSan, GBra, and GRee), wild S. chloronotus in complex multi-habitat types (GWS and GWO) display enhanced feeding selectivity. The reduced richness (ACE, Chao1) and increased diversity (Shannon, Simpson) of the microbial community in GWS and GWO suggest that wild S. chloronotus possesses stable food sources and actively searches for specific sediment patches. This is corroborated by its conspicuous patchy clustering behavior [2, 21, 22]. The weight loss of S. chloronotus in net cages also indicates a stronger feeding selectivity and a broader feeding range for wild S. chloronotus. The relative abundance of Ruegeria (R. lacuscaerulensis) was notably high in GWS and GWO, with GWO showing a significant increase compared to GSan, GBra, and GRee (P < 0.001). Ruegeria is known to secrete a variety of bioactive molecules, such as cyclic dipeptides, which can inhibit Vibrio [47], and is a potential probiotic for corals [48, 49]. R. lacuscaerulensis possesses significant ecological functions and plays a crucial role in the marine ecosystem by participating in the metabolic process of dimethylsulfoniopropionate through specific metabolic pathways [50]. The elevated abundance of Ruegeria has been correlated with an enhanced capacity for cultured aquatic organisms to fend off pathogenic invasions [51, 52]. Ruegeria also constitutes a dominant genus in the gut microbiota of pound-cultured S. monotuberculatus and Holothuria scabra [53]. Consequently, it is hypothesized that wild S. chloronotus actively seeks out and ingests higher quantities of Ruegeria (R. lacuscaerulensis) across a larger territory to maintain the stability of its gut microbiota and overall health.
Key factor influencing the niche differentiation of S. chloronotus
In comparison to food selectivity, our findings indicate that habitat preference is the key factor influencing the niche differentiation of S. chloronotus. This study was conducted in in situ habitats with consistent food sources. However, the microbial community structure within the gut contents of S. chloronotus exhibited differentiation across the three single habitat types (GSan, GBra, and GRee). S. chloronotus prefers to inhabit hard substates such as reefs and rocks [2], and its microbial community structure in the hard habitat (GRee) closely resembles that of wild S. chloronotus sampled in October (GWO) and September (GWS). By contrast, GSan and GBra are closer to each other and distinct from the aforementioned groups (based on PCoA analysis and UPGMA clustering). Furthermore, wild S. chloronotus is known to feed during the day and perch on rocks at night [3, 23]. For S. chloronotus in single habitat types characterized by sandy bottoms (GSan) and broken coral branches (GBra), they were often observed attaching to the sides of net cages during the daytime, with their tentacles in a foraging stance. However, the sides and top of the cages are clearly not suitable foraging locations and cannot provide adequate sustenance. Surprisingly, when confronted with unsuitable bottom types, S. chloronotus dramatically altered their original feeding habits. Securing a stable substrate for attachment became their primary objective. This shift contradicts conventional cognitive and animal ecological principles, which posit that the procurement of food is the paramount consideration for animals [17]. Our previous laboratory microcosm research likewise found that S. chloronotus consistently prefers the reef bottom, while H. edulis, which prefers sandy bottoms in the wild, also opts to inhabit the reef bottom. These studies and observations collectively substantiate that habitat selection occupies a greater weight when food sources and habitat environments undergo changes. The inference that sea cucumbers exhibit reduced dependency on food sources may provide a compelling explanation for why the majority of tropical sea cucumbers, particularly within coral reef ecosystems, opt to feed on sediments.
The reasons for this phenomenon may be multifaceted and complex. Previous study identified that the S. chloronotus population is extensively distributed across shallow coral reef areas at approximately 5 m depth. Such areas are prone to wind and wave impacts [3], making the discovery of a stable attachment substrate especially critical for sea cucumbers. This may also be associated with the physiological structure of S. chloronotus, whose flattened ventral surface and powerful adhesive foot enable it to adhere firmly to reefs and effectively withstand severe marine conditions [2]. It is evident that loose sands and unstable broken coral branches are unsuitable for the attachment of S. chloronotus. Consequently, in the study, only sea cucumbers in the reef bottom (GRee) maintained consistent feeding habits with their wild counterparts, albeit with a slight decrease in the micro-diversity index due to the restricted foraging area within the net cages.
Adaptive modulation of S. chloronotus to food in single habitat types
The R values from Anosim indicated significant shifts in the microbial community structure within the gut contents of S. chloronotus in single habitat types. At the phylum level, Proteobacteria predominated in GWS, GWO, and five sediment groups but was significantly decreased in GSan, GBra, and GRee, with a corresponding significant increase of Cyanobacteria (52.59%, 18.01%, and 43.51%). As the largest bacterial phylum, Proteobacteria is the dominant phylum detected in the gut contents of numerous marine invertebrates, such as Litopenaeus vannamei, Fenneropenaeus chinensis and Tedania sp. [54–56]. Proteobacteria also exhibits absolute dominance among the symbiotic and epiphytic microorganisms of Holothuroidea. Gao et al. (2022) and Zhang (2021) identified Proteobacteria as the predominant phylum in the gut contents of H. atra, Holothuria leucospilota, S. monotuberculatus and ambient sediments in Wuzhizhou Island [24, 28], consistent with our findings for wild S. chloronotu and sediments. The increase of Cyanobacteria in GSan, GBra, and GRee was also primarily reflected by the increase of Synechococcus CC9902 (GSan: 46.16%, GBra: 12.83%, GRee: 35.45%) at the genus level, whereas the relative abundance of Synechococcus CC9902 in wild S. chloronotus and sediments were less than 2.03%. This suggests that S. chloronotus in single habitats (with restricted feeding areas) adjusts their diet by altering their feeding selectivity. Wu (2018) found that Synechococcophycideae, which is symbiotic with sponges, can synthesize polyphosphates (polyP) as energy reserves and degrade them to maintain the normal metabolic activities of the host under harsh conditions [55]. Additionally, symbiotic algae of invertebrates (corals) and symbiotic bacteria of bivalves (Lucinoma aequizonata) have also been reported to accumulate polyP under normal conditions and degrade it to supply energy to the host during extreme conditions such as hypoxia [57, 58]. Currently, research is limited on the symbiotic and epiphytic microorganisms of sea cucumber [59]. Therefore, we speculate that Synechococcus CC9902 may be a potential probiotic of S. chloronotus, and its increase ensures the energy supply and normal metabolic activities of S. chloronotus when habitat types and feeding areas are restricted.
The prokaryotic composition and species-level proportions in the gut contents of S. chloronotus from the treatment groups (GSan, GBra, GRee) were more similar to each other and differed from those in the wild (GWO) during the same period. This difference was characterized by a significant increase in V. fragilis and P. pentosaceus. Notably, V. fragilis adapts to grow in anoxic environments with sulfides and contains symbionts of diatom origin [60, 61], making it the predominant species in the liquid of Chaetoceros muelleri (accounting for 68.8%) [62]. We hypothesize that the observed alterations are a consequence of the dietary and foraging behavior adjustments of S. chloronotus in response to specific substrate conditions, and according to the aforementioned findings, adaptive adjustment is likely to be completed within the span of a month.
Microbial function variation of S. chloronotus in single habitat types
Results from microbial functional analysis revealed the high abundance of carbohydrate metabolism, amino acid metabolism, lipid metabolism, and membrane transport in GWS and GWO, indicating that the gut content microbiota of wild S. chloronotus is stable and actively engaged in its daily metabolic processes. Carbohydrates, proteins, and lipids are essential nutrients required for the growth of sea cucumbers [63]. Membrane transport can enhance the stability of the gut microbiota, thereby improving its biological resistance [64]. However, the functional metabolic pathways of the gut microbiota of S. chloronotus in single habitat types undergo changes, with GSan and GRee showing remarkable differences compared to those of wild S. chloronotus. Glycine serves as an energy source for the colon microbiota during metabolic processes, wherein various vitamins play a crucial regulatory role.
Therefore, we speculate that the high abundance of glycan biosynthesis and metabolism, metabolism of cofactors and vitamins, and energy metabolism, along with changes in level-1 metabolic pathways (e.g., genetic information processing and organismal systems), may be the way for adaptive adjustment of S. chloronotus in unsuitable habitats with restricted feeding areas.
Functional annotation of microbial communities can provide insights into the overall metabolism of the host [65]. Regarding the FAPROTAX outcomes, the significant increase in cyanobacteria, oxygenic photoautotrophy, and photoautotrophy in GSan, GBra, and GRee corresponds to the substantial intake of Cyanobacteria by S. chloronotus in single habitat types. The FAPROTAX predictions also suggest that the biochemical cycling of the gut microbiota of S. chloronotus in single habitat types has changed, whereas the functional relative abundance of GWS and GWO is similar to that of sediments. This result further confirms that S. chloronotus in single habitat types can actively modify their diet. Furthermore, whether changes in the biochemical cycling of gut contents can alter the organic properties of their feces and the possible impact on coral reef warrants further research.
Conclusion
Through in situ mesocosm experiments and 16S rRNA gene high-throughput sequencing technology, we found that S. chloronotus possesses a strong capacity for habitat adaptation and can actively adjust its diet. We observed changes in the feeding and habitat behavior of caged S. chloronotus, as well as shifts in the functionality of the microbial community in their gut contents. This adaptive adjustment can be completed within one month. Moreover, compared to the acquisition of food resources, the availability of a suitable habitat is a more critical factor influencing the ecological niche differentiation of S. chloronotus. Securing a stable substrate for attachment has become the primary concern.
Acknowledgements
The authors are grateful to Fengguo WANG, Sunming Li and Yupeng Cai from Wuzhizhou Island Tourism Company for their help in fieldwork.
Financial interests
The authors have no relevant financial or non-financial interests to disclose.
Abbreviations
- GWS
The gut contents from wild S.chloronotus in September
- GWO
The gut contents from wild S.chloronotus in October
- GSan
The gut contents of S.chloronotus housed in net cages with sandy bottom
- GBra
The gut contents of S.chloronotus housed in net cages with broken coral branches bottom
- GRee
The gut contents of S.chloronotus housed in net cages with reef bottom
- SWS
The sediments collected near wild S.chloronotus in September
- SWO
The sediments collected near wild S.chloronotus in October
- SSan
The sediments collected in net cages with sandy bottom
- SBra
The sediments collected in net cages with broken coral branches bottom
- SRee
The sediments collected in net cages with reef bottom
- OTUs
Operational taxonomic units
- PCoA
Principal coordinates analysis
- Anosim
Analysis of similarities
- ACE
Abundance-based coverage estimator
- UPGMA
Unweighted Pair-group Method with Arithmetic Mean
- VPE1
Vibrioparahaemolyticus E1
- polyp
Polyphosphates
Authors’ contributions
C.Y. Sun contributed to the conception and design of the experiment, the acquisition, processing, and analysis of the data, as well as the writing and revision of the manuscript. Y.N. Wang, C.H. Jia, Y. Rong, B.X. Feng and K.Z. Yao contributed to the execution of the experiment and the analysis of the data. F. Gao provided technical support and participated in the revision of the manuscript. Q. Xu was responsible for the overall planning, design, and implementation of the research, providing financial and technical support. The final manuscript was read and approved by all authors.
Funding
This study was financially supported by the National Natural Science Fund of China (42076097, 42166005), and the National Key R and d Project of China (2019YFD0901304).
Data availability
The datasets generated and analysed during the current study are available in the NCBI repository, (No. PRJNA1113006).
Declarations
Ethics approval and consent to participate
Not applicable. Our research did not involve human participants or samples. No specific permits were required for the described field studies.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Reichenbach N, Holloway S. Potential for asexual propagation of several commercially important species of tropical sea cucumber (Echinodermata). J World Aquacult Soc. 1995;26(3):272–8. 10.1111/j.1749-7345.1995.tb00255.x. [Google Scholar]
- 2.Sun CY, Huang DJ, Xu Q, Gao F, Li XB, Wang AM. Diverse habitat preferences of two sea cucumber species and the seasonal change in a coral reef area. J Oceanol Limnol. 2022;40(4):1578–91. 10.1007/S00343-021-1254-Z. [Google Scholar]
- 3.Huang DJ. Community structure of echinoderms and population ecology of sea cucumber in Wuzhizhou Island, Sanya, China. Hainan University. 10.27073/d.cnki.ghadu.2020.001378. Accessed May 2020. (in Chinese)
- 4.Liao YL, Xiao N. Species composition and faunal characteristics of echinoderms in China seas. Biodiversity Science. 2011;19(06):729–36. 10.3724/SP.J.1003.2011.08155. (in Chinese with English abstract). [Google Scholar]
- 5.Mercier A, Battaglene SC, Hamel JF. Daily burrowing cycle and feeding activity of juvenile sea cucumbers Holothuria scabra in response to environmental factors. J Exp Mar Biol Ecol. 1999;239:125–56. 10.1016/S0022-0981(99)00034-9. [Google Scholar]
- 6.Uthicke S. Sediment bioturbation and impact of feeding activity of Holothuria (Halodeima) atra and Stichopus chloronotus, two sediment feeding holothurians, at Lizard Island, Great Barrier Reef. Bull Mar Sci. 1999;64(1):129–41. [Google Scholar]
- 7.Reise K. Sediment mediated species interactions in coastal waters. J Sea Res. 2002;48(2):127–41. 10.1016/S1385-1101(02)00150-8. [Google Scholar]
- 8.Lohrer AM, Thrush SF, Gibbs MM. Bioturbators enhance ecosystem function through complex biogeochemical interactions. Nature. 2004;431(7012):1092–5. [DOI] [PubMed] [Google Scholar]
- 9.Tian SY, Zhang WL, Zhang R. Role of macrobenthos in marine ecosystem. Journal of Salt and Chemical Industry. 2009;38(02):50–4. 10.16570/j.cnki.issn1673-6850.2009.02.003. (in Chinese with English abstract). [Google Scholar]
- 10.Schneider K, Silverman J, Woolsey E, Eriksson H, Byrne M, Caldeira K. Potential influence of sea cucumbers on coral reef CaCO3 budget: a case study at One Tree Reef. J Geophys Res. 2011;116(G4):G04032. 10.1029/2011JG001755.
- 11.Schneider K, Silverman J, Kravitz B, Rivlin T, Schneider-Mor A, Barbosa S, Byrne M, Caldeira K. Inorganic carbon turnover caused by digestion of carbonate sands and metabolic activity of holothurians. Estuar Coast Shelf Sci. 2013;133:217–23. 10.1016/j.ecss.2013.08.029. [Google Scholar]
- 12.Wheeling RJ, Verde EA, Nestler JR. Diel cycles of activity, metabolism, and ammonium concentration in tropical holothurians. Mar Biol. 2007;152(2):297–305. 10.1007/s00227-007-0683-3. [Google Scholar]
- 13.Zaneveld JR, Burkepile DE, Shantz AA, Pritchard CE, McMinds R, Payet JP, Welsh R, Correa AMS, Lemoine NP, et al. Overfishing and nutrient pollution interact with temperature to disrupt coral reefs down to microbial scales. Nat Commun. 2016;7(1):11833. 10.1038/ncomms11833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao MX, Yu KF, Zhang QM. Review on coral reefs biodiversity and ecological function. Acta Ecol Sin. 2006;26(1):186–94 (in Chinese with English abstract). [Google Scholar]
- 15.Fan SG, Hu CQ, Zhang LP, Wen J. Morphological character of ossicles in eight tropical holothurians in the Xisha Islands of China. J Trop Oceanogr. 2010;29(04):148–53 (in Chinese with English abstract). [Google Scholar]
- 16.Xue YL, Gao F, Xu Q, Huang DJ, Wang AM, Sun T. Study on feeding selection of environmental sediments and digestive function adaptability of Holothuria atra. Oceanologia Et Limnologia Sinica. 2019;50(5):1070–9. 10.11693/hyhz20190200033. (in Chinese with English abstract). [Google Scholar]
- 17.Sun RY, Wang DH, Niu CJ, Liu DZ, Zhang LI. Principles of animal ecology. 4th ed. Beijing: Beijing Normal University Press; 2019. (in Chinese).
- 18.Liao YL. Fauna sinica: Echinodermata. Beijing: Science Press; 1997. (in Chinese). [Google Scholar]
- 19.Xue YL. Study on the feeding selection and the adaptability of digestive system structure and function of three common sea cucumbers in tropical coral reefs. Hainan University. 10.27073/d.cnki.ghadu.2019.001065. Accessed May 2019. (in Chinese)
- 20.Wu PL, Huang DJ, Ma WG, Gao F, Wang AM, Xu Q. Determination of the food sources of two tropical coral reef sea cucumbers Holothuria edulis and Stichopus chloronotus using a fatty acid biomarker analysis. Marine Sci. 2021;45(09):58–68. 10.11759/hykx20210318002.
- 21.Uthicke S, Karez R. Sediment patch selectivity in tropical sea cucumbers (Holothurioidea: Aspidochirotida) analysed with multiple choice experiments. J Exp Mar Biol Ecol. 1999;236(1):69–87. 10.1016/S0022-0981(98)00190-7. [Google Scholar]
- 22.Bellchambers LM, Meeuwig JJ, Evans SN. Modelling habitat associations of 14 species of holothurians from an unfished coral atoll: implications for fisheries management. Aquat Biol. 2011;14(1):57–66. 10.3354/ab00381. [Google Scholar]
- 23.Eriksson H, Jamon A, Wickel J. Observations on habitat utilization by the sea cucumber Stichopus chloronotus. SPC Beche-de-Mer Information Bulletin. 2012;32:39–42. [Google Scholar]
- 24.Zhang Y. Study on gut morphological structure and food sources of juvenile sea cucumber Stichopus monotuberculatus. Hainan University. 10.27073/d.cnki.ghadu.2021.001522. Accessed May 2021. (in Chinese)
- 25.Budarf AC, Burfeind DD, Loh WKW, Tibbetts IR. Identification of seagrasses in the gut of a marine herbivorous fish using DNA barcoding and visual inspection techniques. J Fish Biol. 2011;79(1):112–21. 10.1111/j.1095-8649.2011.02999.x. [DOI] [PubMed] [Google Scholar]
- 26.Hatmanti A, Purwati P. Bacteria associated holothurians: the key of habitat preference, diet, and functions. Jurnal Ilmu Dan Teknologi Kelautan Tropis. 2011;3(1):73–81. 10.29244/jitkt.v3i1.7836. [Google Scholar]
- 27.Bade LM, Balakrishnan CN, Pilgrim EM, Mcrae SB, Luczkovich JJ. A genetic technique to identify the diet of cownose rays, Rhinoptera bonasus: analysis of shellfish prey items from North Carolina and Virginia. Environ Biol Fishes. 2014;97(9):999–1012. 10.1007/s10641-014-0290-3. [Google Scholar]
- 28.Gao F, Zhang Y, Wu PL, Chen ML, He LW, Xu Q, Wang AM. Bacterial community composition in gut content and ambient sediment of two tropical wild sea cucumbers (Holothuria atra and H.leucospilota). Journal of Oceanology and Limnology. 2022;40(01):360–72. 10.1007/S00343-021-1001-5. [Google Scholar]
- 29.Yan HH (颜慧慧). Study on the evolution and evaluation of ecological environment in the marine ranching tourism area of Wuzhizhou Island, Sanya (三亚蜈支洲岛海洋牧场旅游区生态环境演变与评价研究[D]). Hainan University (海南大学). Accessed May 2017. (in Chinese). https://kns.cnki.net/kcms2/article/abstract?v=NK8hpUzgeRV_lD9HmxzwTF9hcBTAiBEd4whuMJkdMouz0Nw7tiMZXH3fbQHzHHhD1Be1vZPE8voKL3fwuvNYfDuCPoFccb_yd_MMItcxlgSSWWR3EDJqAbxMUC9BhZPyQy749fvICr_Gt2Cif7jzlbilDfRaU1cMtzaSwhczVs=&uniplatform=NZKPT.
- 30.Li XB, Li YC, Xu Q. Current situation, ecological restoration and protection countermeasures of coral reefs in Wuzhizhou Island, Sanya. Beijing: Science Press; 2019. p. 32–9 (in Chinese). [Google Scholar]
- 31.Huang DJ, Xu Q, Li XB, Xue YL, Wu PL, Gao F. The community structure of echinoderms in sandy coral reef area in Wuzhizhou Island, Sanya, China. Oceanologia et Limnologia Sinica. 2020;51(1):103–13. 10.11693/hyhz20190900174. (in Chinese with English abstract). [Google Scholar]
- 32.Jia CH, Zhang Y, Xu Q, Sun CY, Wang YN, Gao F. Comparative analysis of in situ eukaryotic food sources in three tropical sea cucumber species by metabarcoding. Animals. 2022;12:17. 10.3390/ani12172303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tao XY, Feng JJ, Yang YF, Wang GS, Tian RM, Fan FL, Ning DL, Bates CT, Hale L, Yuan MT, et al. Winter warming in Alaska accelerates lignin decomposition contributed by Proteobacteria. Microbiome. 2020;8(1):84. 10.1186/s40168-020-00838-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang KP, Delgado-Baquerizo M, Zhu YG, Chu HY. Space is more important than season when shaping soil microbial communities at a large spatial scale. mSystems. 2020;5:3. 10.1128/mSystems.00783-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Magoč T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics (Oxford, England). 2011;27(21):2957–63. 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10(10):996–8. 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 37.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic acids research. 2013;41(Database issue):D590-596. 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73(16):5261–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gao F, Tan J, Sun HL, Yan JP. Bacterial diversity of gut content in sea cucumber (Apostichopus japonicus) and its habitat surface sediment. Journal of Ocean University of China. 2014;13(02):303–10. 10.1007/s11802-014-2078-7. [Google Scholar]
- 40.Gao F, Li FH, Tan J, Yan JP, Sun HL. Bacterial community composition in the gut content and ambient sediment of sea cucumber Apostichopus japonicus revealed by 16S rRNA gene pyrosequencing. PLoS One. 2014;9(6):e100092. 10.1371/journal.pone.0100092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang L, Wei C, Chang YQ, Ding J. Response of bacterial community in sea cucumber Apostichopus japonicus intestine, surrounding water and sediment subjected to high-temperature stress. Aquaculture. 2021;535. 10.1016/J.AQUACULTURE.2021.736353.
- 42.Yamazaki Y, Sakai Y, Mino S, Suda W, Hattori M, Meirelles PM, Thompson F, Sawabe T. Repeated selective enrichment process of sediment microbiota occurred in sea cucumber gut. Environmental microbiology reports. 2019;11(6):797–807. 10.1111/1758-2229.12791. [DOI] [PubMed] [Google Scholar]
- 43.Moriarty DJW. Feeding of Holothuria atra and Stichopus chloronotus on bacteria, organic carbon and organic nitrogen in sediments of the Great Barrier Reef. Mar Freshw Res. 1982;33(2):255–63. 10.1071/mf9820255. [Google Scholar]
- 44.Moriarty DJW, Pollard PC, Hunt WG, Moriarty CM, Wassenberg TJ. Productivity of bacteria and microalgae and the effect of grazing by holothurians in sediments on a coral reef flat. Mar Biol. 1985;85(3):293–300. 10.1007/BF00393250. [Google Scholar]
- 45.Smith TB. Tentacular ultrastructure and feeding behaviour of Neopentadactyla mixta (Holothuroidea: Dendrochirota). J Mar Biol Assoc UK. 1983;63:301–11. 10.1017/S0025315400070697. [Google Scholar]
- 46.Foster GG, Hodgson AN. Feeding, tentacle and gut morphology in five species of southern African intertidal holothuroids (Echinodermata). African Zoology. 1996;31(2):70–9. 10.1080/02541858.1996.11448396. [Google Scholar]
- 47.Miura N, Motone K, Takagi T, Aburaya S, Watanabe S, Aoki W, Ueda M. Ruegeria sp. strains isolated from the reef-building coral Galaxea fascicularis inhibit growth of the temperature-dependent pathogen Vibrio coralliilyticus. Marine biotechnology (NY). 2019;21(1):1–8. 10.1007/s10126-018-9853-1. [DOI] [PubMed] [Google Scholar]
- 48.Kitamura R, Miura N, Okada K, Motone K, Kataoka M. Design of novel primer sets for easy detection of Ruegeria species from seawater. Biosci Biotechnol Biochem. 2020;84(4):854–64. 10.1080/09168451.2019.1700776. [DOI] [PubMed] [Google Scholar]
- 49.Kitamura R, Miura N, Ito M, Takagi T, Yamashiro H, Kataoka M. Specific detection of coral-associated Ruegeria, a potential probiotic bacterium, in corals and subtropical seawater. Mar Biotechnology. 2021;23(4):576–89. 10.1007/s10126-021-10047-2. [DOI] [PubMed] [Google Scholar]
- 50.Wirth JS, Wang T, Huang Q, White RH, Whitman WB. Dimethylsulfoniopropionate sulfur and methyl carbon assimilation in Ruegeria species. mBio. 2020;11:e00329-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dittmann KK. Interaction between fish probiotic Roseobacters and the natural microbiota in aquaculture settings. Accessed: Technical University of Denmark; 2019. https://findit.dtu.dk/en/catalog/5db71eabd9001d013530afe3?single_revert=%2Fen%2Fcatalog%3Fq%3DInteraction%2Bbetween%2Bfish%2Bprobiotic%2Band%2Bthe%2Bnatural%2Bmicrobiota%2Bin%2Baquaculture%2Bsettings%26show_sing.
- 52.Pintado J, Ruiz P, Olmo DG, Makridis P. Co-Culturing microalgae with Roseobacter clade bacteria as a strategy for Vibrionaceae control in microalgae-enriched Artemia. Microorganisms. 2023;11(11):2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jia CH, Wang YH, Zheng BJ, Wang YN, He LW, Xu Q, Gao F. Comparative analysis of gut bacterial community composition in two tropical economic sea cucumbers under different seasons of artificial environment. Int J Mol Sci. 2024;25(8):4573. 10.3390/ijms25084573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu HD, Wang L, Liu M, Wang BJ, Jiang KY, Ma SS, Li QF. The intestinal microbial diversity in Chinese shrimp (Fenneropenaeus chinensis) as determined by PCR–DGGE and clone library analyses. Aquaculture. 2011;317(1):32–6. 10.1016/j.aquaculture.2011.04.008. [Google Scholar]
- 55.Wu SF (吴淑妃). Structure of sponge symbiont microbial communities during different life stages and preliminary study of functions of stage-specific microorganisms (海绵生长阶段共生微生物群落结构及特征性微生物功能初步研究[D]). Xiamen University (厦门大学). Accessed May 2018. (in Chinese). https://kns.cnki.net/kcms2/article/abstract?v=NK8hpUzgeRWHZVMyW3_svny6dWDM0Uj-NgyFljMEkDdKZcjig16GU8bQIedwUHxoXhsJgBio2rGF73wsQG2F8k5-WjXluM72mVIQ7JqbWaswruiCKwz-fH6ZCWKoorxQOCddtL-Sp4E2i_mvn6VQ822PRL1-gjUIyZl0bIKfcfU=&uniplatform=NZKPT.
- 56.Gao S, Pan LQ, Huang F, Song MS, Tian CC, Zhang MY. Metagenomic insights into the structure and function of intestinal microbiota of the farmed Pacific white shrimp (Litopenaeus vannamei). Aquaculture. 2019;499:109–18. 10.1016/j.aquaculture.2018.09.026. [Google Scholar]
- 57.Arndt-Sullivan C, Lechaire JP, Felbeck H. Extreme tolerance to anoxia in the lucinoma aequizonata symbiosis. J Shellfish Res. 2008;27(1):119–27. 10.2983/0730-8000(2008)27[119:ETTAIT]2.0.CO;2. [Google Scholar]
- 58.Yellowlees D, Rees TAV, Leggat W. Metabolic interactions between algal symbionts and invertebrate hosts. Plant, Cell Environ. 2008;31(5):679–94. 10.1111/j.1365-3040.2008.01802.x. [DOI] [PubMed] [Google Scholar]
- 59.Xia XK, Liu X, Zhang YG, Yuan WP, Zhang MS, Wang XJ, Meng XM, Liu CH. Study on the second metabolisms from fungus HS-1 Epicoccum spp. from the sea cucumber in Yellow Sea. J Chin Med Mater. 2010; 33:10:1577–9. 10.13863/j.issn1001-4454.2010.10.048. (in Chinese with English abstract). [PubMed]
- 60.Tsuchiya M, Toyofuku T, Uematsu K, Brüchert V, Collen J, Yamamoto H, Kitazato H. Cytologic and genetic characteristics of endobiotic bacteria and kleptoplasts of Virgulinella fragilis(Foraminifera). J Eukaryot Microbiol. 2015;62(4):454–69. [DOI] [PubMed] [Google Scholar]
- 61.Tsuchiya M, Grimm GW, Heinz P, Stögerer K, Ertan KT, Collen J, Brüchert V, Hemleben C, Hemleben V, Kitazato H. Ribosomal DNA shows extremely low genetic divergence in a world-wide distributed, but disjunct and highly adapted marine protozoan (Virgulinella fragilis, Foraminiferida). Mar Micropaleontol. 2009;70(1–2):8–19. [Google Scholar]
- 62.Tang YP (唐亚鹏). Effects of two kinds of microalgae on microbial community structure and water environmental factors of larval rearing of Penaeus monodon (两种微藻对斑节对虾育苗微生物群落结构和水环境因子的影响[D]). Tianjin Agricultural University (天津农学院). Accessed June 2019. (in Chinese). https://kns.cnki.net/kcms2/article/abstract?v=NK8hpUzgeRWwFd-NR_QL9t0AMWT2psPDjnqLJd2MGSkBhF4cgl0ZAyaIR2FzfokjEEj6o23bOTaaklOMpbPSII8FaNizKkI_d8FHNFvOeNTVbZBBl1VTgVm_QMPSPwIN3DrQmjj1YEYescCaVZXookzTSiCKcb7JcjX7GgZCs=&uniplatform=NZKPT, 10.27717/d.cnki.gtjnx.2019.000015.
- 63.Xia B, Gao QF, Wang JY, Li PY, Zhang LM, Zhang ZD. Effects of dietary carbohydrate level on growth, biochemical composition and glucose metabolism of juvenile sea cucumber Apostichopus japonicus (Selenka). Aquaculture. 2015;448:63–70. 10.1016/j.aquaculture.2015.05.038. [Google Scholar]
- 64.Mamphogoro TP, Maboko MM, Babalola OO, Aiyegoro OA. Bacterial communities associated with the surface of fresh sweet pepper (Capsicum annuum) and their potential as biocontrol. Sci Rep. 2020;10(1):8560. 10.1038/s41598-020-65587-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zeng CX, Lin M, Li ZQ, Ma Y, Wang SH. The structural and functional characteristics of the gut microbiota of Marsupenaeus japonicus as revealed by 16S rRNA gene amplicon sequencing. Microbiology China. 2020;47(06):1857–66. 10.13344/j.microbiol.china.190749. (in Chinese with English abstract). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and analysed during the current study are available in the NCBI repository, (No. PRJNA1113006).