Abstract
Background
A multivariate predictive model was constructed using baseline and 12-week clinical data to evaluate the rate of clearance of hepatitis B surface antigen (HBsAg) at the 48-week mark in patients diagnosed with chronic hepatitis B who are receiving treatment with pegylated interferon α (PEG-INFα).
Methods
The study cohort comprised CHB patients who received pegylated interferon treatment at Mengchao Hepatobiliary Hospital, Fujian Medical University, between January 2019 and April 2024. Predictor variables were identified (LASSO), followed by multivariate analysis and logistic regression analysis. Subsequently, predictive models were developed via logistic regression, random forest (RF), gradient boosting decision tree (GBDT), extreme gradient boosting (XGBoost), and support vector machine (SVM) algorithms. The efficacy of these models was assessed through various performance metrics, including the area under the receiver operating characteristic curve (AUC), sensitivity, specificity, and F1 score.
Results
This study included a total of 224 individuals diagnosed with chronic hepatitis B. The variables baseline log2(HBsAg), gender, age, neutrophil count at week 12, HBsAg decline rate at week 12, and HBcAb at week 12 were closely associated with functional cure and were included in the predictive model. In the validation term, the logistic regression model had an AUC of 0.858, which was better than that of the other machine learning models (AUC = 0.858,F1 = 0.753). Consequently, this model was selected for the development of the predictive tool.
Conclusions
The combined use of the baseline log2(HBsAg) value, HBsAg decline rate at week 12, gender, neutrophil count at week 12, and age can serve as a foundational predicting model for anticipating the clearance of HBsAg in individuals with chronic hepatitis B who are receiving PEG-INFα therapy.
Keywords: Chronic hepatitis B, Functional cure, Prediction model, Baseline log2(HBsAg), HBsAg decline rate at week 12
Background
Hepatitis B virus (HBV) is a DNA virus that targets the liver. It converts relaxed circular DNA into covalently closed circular DNA (cccDNA) within the nucleus of host liver cells through DNA repair processes. The formation of cccDNA is essential for the persistence of chronic infections. This condition has the potential to lead to severe hepatic disorders, including cirrhosis, hepatocellular carcinoma, and hepatic failure. Currently, HBV affects 296 million people worldwide, and each year, at least 820,000 individuals die from severe liver diseases [1].
The implementation of effective antiviral therapy for HBV is crucial for preventing the progression of liver disease. Presently, the optimal therapeutic objective centers on the eradication of HBsAg, a scenario designated a “functional cure” [2, 3].
Current antiviral drugs for hepatitis B primarily include nucleoos(t)ide analog(NA) viral DNA polymerase inhibitors and pegylated interferon alpha(PEG-INFα) [4, 5]. Nucleos(t)ide analogs (NAs) help reduce HBV replication but do not effectively target covalently closed circular DNA (cccDNA) or sufficiently restore the host antiviral immune response against HBV [6, 7]. As a result, it is difficult to achieve a significant decrease in HBsAg levels [8], and after treatment, it is common for viral activity to return, leading to liver damage [9, 10]. In contrast, individuals who respond well to pegylated interferon alpha therapy usually experience a decrease in HBsAg levels [11], after treatment ends, those receiving interferon-based therapies may show increased rates of HBsAg elimination over time [12, 13]. Pegylated interferon is effective for treatment, but its use is limited. This is due to several factors: the potential for harmful side effects, the high cost, and the need for subcutaneous injections [14]. Only a subset of patients receiving interferon therapy achieve a functional cure. Because of its drawbacks, it is important to predict early in treatment whether a patient will benefit from therapy.
Research indicates that factors as ALT fluctuation, HBsAg levels, intrahepatic cccDNA, HBV DNA, and genetic polymorphisms can help predict the likelihood of achieving a functional cure [15–18]. These studies often concentrate on individual parameters, which limits their predictive ability. They may also include markers that are challenging to use in clinical and primary care settings because of technical limitations or costs.
Machine learning is becoming a valuable tool for identifying factors that affect how CHB patients respond to treatment [19]. Researchers aim to improve the accuracy of HBsAg clearance predictions by developing predictive models that utilize large datasets from clinical populations and include various demographic and clinical parameters. This approach is especially relevant because of the multiple factors that contribute to CHB and the significant differences in how patients respond to treatment.
We developed a predictive model based on readily available clinical laboratory indicators. This model can be used as early as 12 weeks to estimate the likelihood of chronic hepatitis B (CHB) patients achieving a functional cure with pegylated interferon. We analyzed various clinical factors and their correlations with treatment response, with the goal of constructing a machine learning model that accurately predicts HBsAg clearance in CHB patients. This model is expected to provide clinicians with a more effective tool for personalizing treatment strategies and improving patient outcomes in managing chronic hepatitis B.
Study subjects
This research conducted a retrospective analysis involving 224 patients diagnosed with chronic hepatitis B, all of whom underwent PEG-INFα therapy at Mengchao Hepatobiliary Hospital, Fujian Medical University, from January 2019 to April 2024. The inclusion criteria were patients with chronic hepatitis B who had been positive for HBsAg for at least 6 months and who had undergone prior treatment with nucleoside or nucleotide analog antiviral agents, including entecavir, tenofovir, and tenofovir alafenamide. The patient’s treatment regimen was based on PEG-IFNα combination therapy. The exclusion criteria were: coinfection with hepatitis C, human immunodeficiency virus (HIV), decompensated cirrhosis, or liver tumors; and central neutrophil counts less than 1.0 × 109/L or platelet counts less than 50 × 109/L. In addition, those with concurrent psychiatric disorders, thyroid dysfunction, or autoimmune diseases were included. This study was approved by the Ethics Committee of Mengchao Hepatobiliary Hospital, Fujian Medical University, and all patients who underwent treatment provided signed informed consent.
The dataset was divided randomly at an 80:20 ratio to form a training term and a validation term.
Follow-up
Patients were administered a weekly subcutaneous injection of 180 µg of PEG-IFNα and were monitored over a period of 48 weeks. During this follow-up, assessments of liver function, as well as levels of hepatitis B surface antibody (HBsAb), hepatitis B e antigen (HBeAg), hepatitis B e antibody (HBeAb), hepatitis B core antibody (HBcAb), and complete blood count, were conducted approximately every 12 weeks. Those who showed a loss of HBsAg at any point during treatment or at week 48 were classified as responders (R), whereas those who did not show a negative conversion were classified as nonresponders (NRs).
Statistical analysis and model establishment
Statistical evaluations were executed via the SPSS software package, version 26.0 (IBM Corp., Armonk, NY, USA). For quantitative metrics, means accompanied by standard deviations were utilized to depict data adhering to a normal distribution, whereas medians alongside interquartile ranges were employed for data deviating from normality. Comparative analyses between continuous variables were performed via Student’s t test for normally distributed datasets. When the data failed to meet the assumptions of a normal distribution, nonparametric tests, including the Mann-Whitney U test and the Wilcoxon signed-rank test, were utilized. Categorical variables are expressed as frequencies and proportions, and contrasts among these variables were gauged via the chi-squared (χ²) test or Fisher’s exact test, contingent upon the fulfillment of test requirements. Univariate analysis was conducted via logistic regression, followed by stepwise regression analysis on basis of the the results of the univariate analysis. We investigated the issue of collinearity among the predictors to ensure the robustness and interpretability of our model. In the process of building the model, we first screened the predictive variables via Lasso analysis and then constructed a conventional logistic regression model grounded in the chosen predictive variables. Additionally, we built predictive models via four machine learning algorithms: random forest (RF), extreme gradient boosting (XGBoost), gradient boosting decision trees (GBDT), and support vector machines (SVMs). We evaluated the models’ performance via metrics such as the area under the curve (AUC), sensitivity, specificity, accuracy, recall, and F1 score. Calibration curves, decision curves, and receiver operating characteristic (ROC) curves were used to determine the clinical utility of the logistic regression model. Finally, the best-performing model was chosen to construct a nomogram. A p value < 0.05 was considered statistically significant in all the statistical analyses.
Results
Among the 224 patients who participated in the study, 57 cleared HBsAg by week 48 of treatment and follow-up. This resulted in a clearance rate of 25.45%. The group statistics were as follows: 132 individuals had HBsAg ≤ 1500 IU/ml, 103 had HBsAg ≤ 1000 IU/ml, 74 had HBsAg ≤ 500 IU/ml, and 36 had HBsAg ≤ 100 IU/ml. In these groups, 51, 47, 38, and 23 individuals achieved HBsAg clearance, resulting in clearance rates of 38.63%, 45.63%, 51.35%, and 63.89%, respectively.
The training term included 179 cases, including 139 male and 40 female subjects, with an average age of 36.9 years. Notably, 44 of these patients demonstrated HBsAg clearance. Statistically significant differences (p < 0.05) were observed in the baseline log2HBsAg data when the responder group was compared with the nonresponder group. The validation term consisted of 45 patients, with an average age of 39.3 years, including 32 males and 13 females. Notably, 13 patients exhibited HBsAg clearance. Detailed information can be found in Table 1.
Table 1.
Training term clinical data baseline table
| Variables | Total (n = 179) | NR (n = 135) | R (n = 44) | P | |
|---|---|---|---|---|---|
| ALT(IU/L), M (Q₁, Q₃) | 27.00 (20.00, 32.00) | 27.00 (21.50, 31.00) | 25.00 (17.00, 32.75) | Z=-0.84 | 0.401 |
| AST(IU/L), M (Q₁, Q₃) | 25.00 (21.00, 30.76) | 26.00 (21.00, 30.76) | 25.00 (19.75, 30.76) | Z=-0.84 | 0.399 |
|
log2HBsAg(log2 IU/ml) M (Q₁, Q₃) |
10.17 (8.08, 11.52) | 10.67 (9.55, 11.64) | 7.84 (4.29, 9.42) | Z=-5.79 | < 0.001 |
| HBsAb(S/CO), M (Q₁, Q₃) | 0.35 (0.00, 1.32) | 0.29 (0.00, 1.12) | 0.45 (0.07, 1.91) | Z=-1.53 | 0.127 |
| HBeAg(S/CO), M (Q₁, Q₃) | 0.06 (0.01, 0.80) | 0.15 (0.01, 0.98) | 0.03 (0.01, 0.36) | Z=-1.11 | 0.265 |
| GGT(U/L), M (Q₁, Q₃) | 22.00 (16.00, 34.00) | 23.00 (17.00, 34.00) | 19.50 (15.75, 32.50) | Z=-0.83 | 0.404 |
| ALP(U/L), M (Q₁, Q₃) | 75.00 (64.50, 91.50) | 76.00 (65.00, 89.00) | 73.00 (64.00, 99.00) | Z=-0.06 | 0.952 |
| Gender(n), M (Q₁, Q₃) | 1.00 (1.00, 1.00) | 1.00 (1.00, 1.00) | 1.00 (0.00, 1.00) | Z=-1.73 | 0.084 |
Z: Mann-Whitney test, M: Median, Q₁: 1st Quartile, Q₃: 3st Quartile, ALT: alanine aminotransferase, AST: aspartate Transaminase, GGT: gamma-glutamyl transferase, ALP: alkaline phosphatase, HBsAg: baseline Hepatitis B Surface Antigen, HBcAb: Hepatitis B Core Antibody, HBeAg: Hepatitis B e Antigen
Univariate analysis related to HBsAg clearance
In the training term, univariate analysis of baseline data and follow-up examination indicators at week 12 revealed that baseline log2 (HBsAg) (p < 0.001) and HBsAb at week 12 (p = 0.039) were associated with HBsAg clearance (Table 2).
Table 2.
Univariate analysis of clinical data from the training term regarding HBsAg clearance
| Variables | β | S.E | Z | P | OR (95%CI) |
|---|---|---|---|---|---|
| Gender | |||||
| male | |||||
| female | -0.67 | 0.39 | -1.72 | 0.086 | 0.51 (0.24 ~ 1.10) |
| Baseline HBVDNA | |||||
| ≤500 | |||||
| >500 | 0.12 | 0.61 | 0.20 | 0.845 | 1.13 (0.34 ~ 3.74) |
| HBVDNA at week 12 | |||||
| ≤500 | |||||
| >500 | 0.74 | 0.93 | 0.80 | 0.426 | 2.10 (0.34 ~ 12.96) |
| HBeAb at week 12 | |||||
| ≤25 | |||||
| >25 | 0.22 | 0.39 | 0.57 | 0.569 | 1.25 (0.59 ~ 2.65) |
| HBcAb at week 12 | |||||
| ≤25 | |||||
| >25 | -0.78 | 0.38 | -2.06 | 0.039 | 0.46 (0.22 ~ 0.96) |
| Age | -0.02 | 0.02 | -1.34 | 0.179 | 0.98 (0.94 ~ 1.01) |
| Baseline ALT | -0.01 | 0.01 | -0.67 | 0.501 | 0.99 (0.97 ~ 1.01) |
| Baseline AST | -0.03 | 0.02 | -1.22 | 0.223 | 0.97 (0.93 ~ 1.02) |
| Baseline log2(HBsAg) | -0.36 | 0.07 | -5.19 | < 0.001 | 0.70 (0.61 ~ 0.80) |
| Baseline HBeAg | 0.00 | 0.00 | 0.14 | 0.889 | 1.00 (1.00 ~ 1.00) |
| Baseline GGT | 0.00 | 0.00 | 0.35 | 0.727 | 1.00 (0.99 ~ 1.01) |
| Baseline ALP | 0.00 | 0.00 | 0.59 | 0.552 | 1.00 (0.99 ~ 1.01) |
| ALT at week 12 | 0.00 | 0.00 | 0.42 | 0.677 | 1.00 (1.00 ~ 1.01) |
| AST at week 12 | 0.00 | 0.00 | 0.27 | 0.789 | 1.00 (1.00 ~ 1.01) |
|
HBsAg decline rare at week 12 |
-0.02 | 0.05 | -0.47 | 0.640 | 0.98 (0.89 ~ 1.07) |
| N at week 12 | -0.50 | 0.26 | -1.90 | 0.057 | 0.61 (0.36 ~ 1.01) |
| PLT at week 12 | -0.01 | 0.00 | -1.34 | 0.180 | 0.99 (0.99 ~ 1.00) |
| Lym at week 12 | -0.67 | 0.45 | -1.49 | 0.137 | 0.51 (0.21 ~ 1.24) |
| GGT at week 12 | 0.00 | 0.00 | 0.13 | 0.898 | 1.00 (0.99 ~ 1.01) |
| ALP at week 12 | 0.00 | 0.00 | 0.74 | 0.459 | 1.00 (0.99 ~ 1.01) |
OR: Odds Ratio, CI: Confidence Interval; N:nertrophil; PLT: platelets; Lym: lymphocytes
Predictor selection
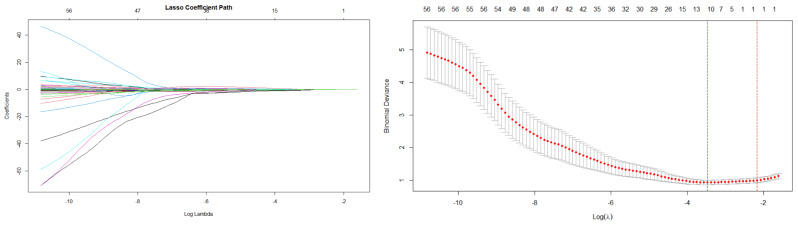
Lasso analysis was conducted using baseline and week 12 clinical indicators. The results indicated that the logarithm of the optimal lambda value was 12 (see Fig. 1). Therefore, we selected 12 variables as potential predictors for further analysis, namely gender, age, baseline ALT, baseline log2 (HBsAg), HBsAg decline rate at week 12 ((HBsAg at week 12 minus baseline HBsAg) divided by baseline HBsAg), HBsAb at week 12, HBeAg at week 12, HBcAb at week 12, HBV DNA at week 12, neutrophil count at week 12, lymphocyte count at week 12, and LMR12 (the lymphocyte-to-monocyte ratio at week 12). Binary logistic regression analysis (stepwise forward method) was subsequently with the functional cure as the target event (yes = 1, no = 0). The results showed that gender, age, baseline log2 (HBsAg), HBsAg decline rate at week 12, and HBcAb at week 12 were selected for the final model. Although the neutrophil count at week 12 had a p-value > 0.05 in the stepwise regression analysis, it was still included in the model because of its clinical significance and p value of 0.057 in the univariate analysis. The variance inflation factor (VIF) values for all the predictors were found to be below 10 (refer to Table 3), suggesting the absence of significant multicollinearity among the predictor variables.
Fig. 1.
Least absolute shrinkage and selection operator(LASSO)analysis was conducted to identify potential predictors
Table 3.
Logistic regression analysis for predictors
| Variables | β | S.E | Z | P | OR (95%CI) | VIF |
|---|---|---|---|---|---|---|
| Intercept | 9.42 | 1.97 | 4.78 | < 0.001 |
12352.87 (259.49 ~ 588055.83) |
|
| Gender | 1.078 | |||||
| male | ||||||
| female | -1.49 | 0.54 | -2.78 | 0.006 | 0.23 (0.08 ~ 0.65) | |
| Age | -0.07 | 0.03 | -2.35 | 0.019 | 0.93 (0.88 ~ 0.99) | 1.027 |
| Baseline log2(HBsAg) | -0.55 | 0.10 | -5.59 | < 0.001 | 0.58 (0.47 ~ 0.70) | 1.106 |
| HBsAg decline rate at week 12 | -0.11 | 0.06 | -1.74 | 0.082 | 0.90 (0.80 ~ 1.01) | 1.095 |
| HBcAb at week 12 | 1.046 | |||||
| ≤25 | ||||||
| >25 | -1.28 | 0.50 | -2.57 | 0.010 | 0.28 (0.11 ~ 0.74) | |
| N at week 12 | -0.62 | 0.35 | -1.78 | 0.074 | 0.54 (0.27 ~ 1.06) | 1.017 |
OR: Odds Ratio, CI: Confidence Interval
Model construction and validation
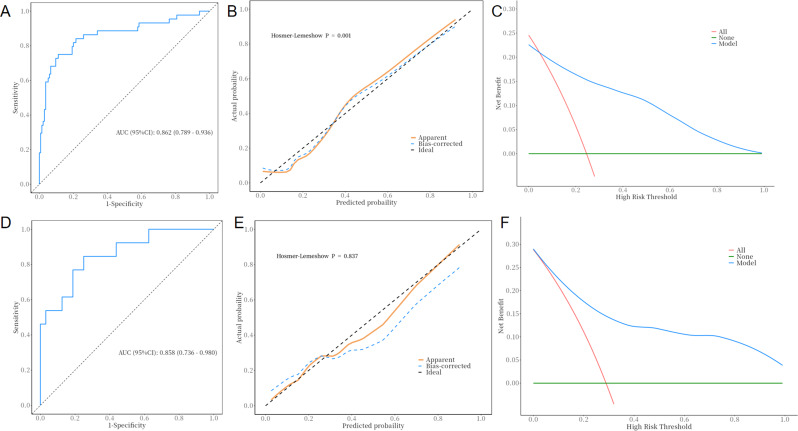
The six aforementioned variables were utilized in the construction of the machine learning models. The validation phase was employed to assess the predictive performance of the models that had been previously developed. The machine learning algorithm categorizes patients who achieve functional cure as the positive class and those who do not achieve functional cure as the negative class. In the logistic regression analysis, the predictive model demonstrated a sensitivity of 0.889 (with a 95% confidence interval of 0.836–0.942) and a specificity of 0.750 (with a 95% confidence interval of 0.622–0.878) in the training term. The model’s discriminative ability was assessed via receiver operating characteristic (ROC) curves, with an area under the curve (AUC) of 0.862 (0.789–0.936) in the training term. The analysis of the calibration curve indicated close alignment with the ideal diagonal line, whereas the decision curve analysis revealed a notable enhancement in net benefit during the training phase (see Fig. 2A-C). In the validation term, the model’s ROC curve (Fig. 2D) had an AUC of 0.858(0.736–0.980), with a sensitivity of 0.750 (0.600–0.900) and specificity of 0.769(0.540–0.998). The calibration curve (Fig. 2E) and decision curve (Fig. 2F) analysis results also indicated that the model had good predictive performance.
Fig. 2.
A-C: Training performance; D-F: Validation performance of logistic regression
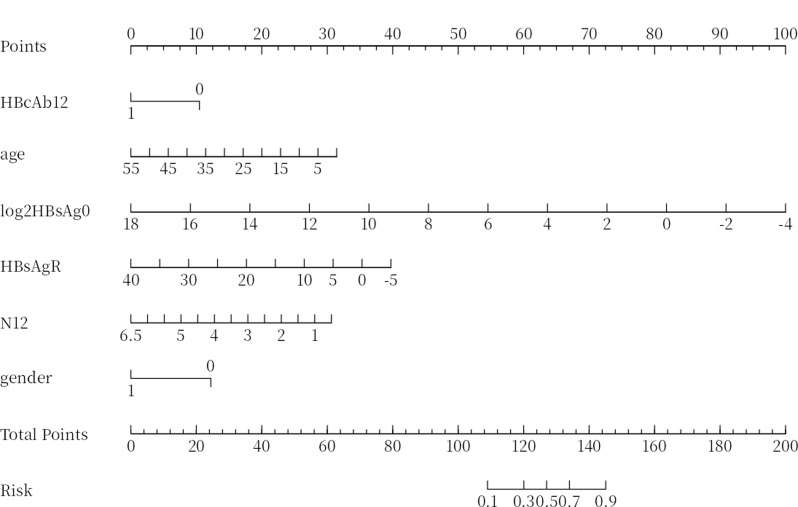
Table 4 presents the performance metrics related to the predictive capabilities of the four machine learning algorithms evaluated within the validation term. SVM had the highest specificity of 0.775, whereas logistic analysis achieved the highest accuracy of 0.756. The F1 score, which reflects the model’s positive predictive capability calculated from precision and recall, reached a maximum of 0.853 in our models. Additionally, the logistic model had the highest AUC of 0.858. On the basis of a comprehensive evaluation of predictive performance, we chose the logistic model as the predictive model. The probability of HBsAg clearance was calculated via the following equation: p = 9.42 − 1.49×gender − 0.07×age − 0.55×baseline log2(HBsAg) − 0.11×HBsAg decline rate at week 12 − 1.28×HBcAb at week 12 − 0.62×neutrophil count at week 12. This probability is then visualized via a nomogram (Fig. 3).
Table 4.
Model performance of the five algorithms in the validation term
| Models | sensitivity | specificity | precision | recall | F1score | AUC |
|---|---|---|---|---|---|---|
| XGBoost | 1 | 0.077 | 0.727 | 1 | 0.842 | 0.842 |
| RF | 0.969 | 0.077 | 0.721 | 0.969 | 0.827 | 0.523 |
| GBDT | 1 | 0.1538 | 0.744 | 1 | 0.853 | 0.762 |
| SVM | 0.800 | 0.775 | 0.308 | 0.800 | 0.444 | 0.709 |
| Logsistic regression | 0.750 | 0.769 | 0.756 | 0.750 | 0.753 | 0.858 |
Fig. 3.
Nomogram for functional cure. HBcAb12:HBcAb at week 12;log2HBsAg0: baseline log2(HBsAg); HBsAgR: HBsAg decline rate at week 12;N12: neutrophil count at week 12
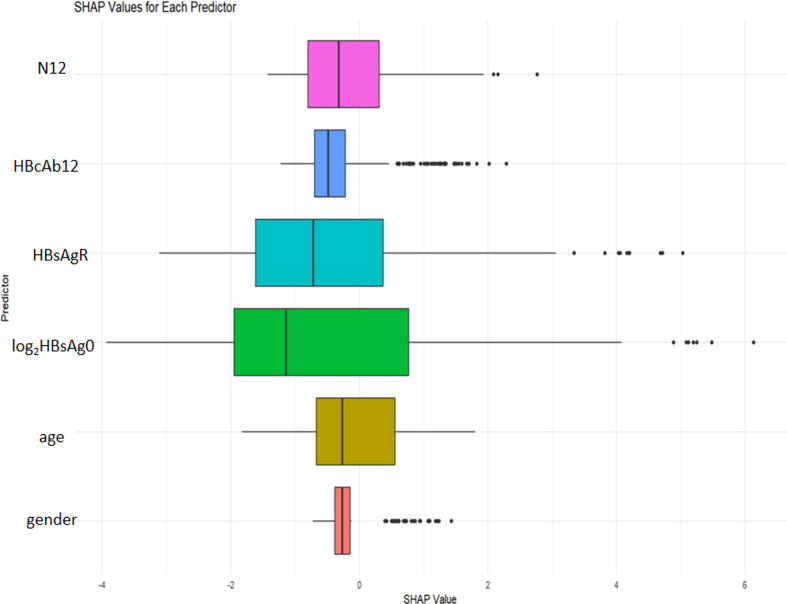
We computed the SHapley additive exPlanations (SHAP) values for every predictor, as depicted in Fig. 4. A more substantial predictor shape signifies a more significant impact on the model. Therefore, we prioritized the importance of the predictors within the model in the following order: baseline log2(HBsAg), rate of HBsAg decline at week 12, neutrophil count at week 12, HBcAb at week 12, gender, and age. The corresponding SHAP values for each predictor are as follows: baseline log2(HBsAg) (-0.456), HBsAg decline rate at week (-0.342), neutrophil count at week 12 (-0.178), HBcAb at week 12 (-0.175), gender (-0.089), and age (-0.065).
Fig. 4.
SHAP value based on predictors. HBcAb12:HBcAb at week 12;log2HBsAg0:baseline log2(HBsAg); HBsAgR: HBsAg decline rate at week 12;N12: neutrophil count at week 12
Discussion
The aim of antiviral treatment for chronic hepatitis B virus infection is to slow disease progression and reduce the risk of liver cancer and cirrhosis [20–22]. The ideal endpoint of antiviral treatment for CHB is to achieve lasting clearance of HBsAg, which improves long-term health outcomes. However, HBsAg clearance occurs in only 3% of patients treated with pegylated interferon alpha, 7% treated with pegylated interferon beta, 2% treated with entecavir, and 3% treated with tenofovir [23–27]. In patients receiving antiviral therapy with nucleoside analogs and pegylated interferon, the clearance rate of surface antigens ranges from 8.7–37.4% [28, 29]. It is crucial to find an effective antiviral strategy that involves both choosing the right treatment regimens and determining the appropriate timing for their use [30]. We evaluated HBsAg clearance rates through a 48-week follow-up of patients and identified potential predictors of treatment response via both traditional statistical methods and advanced machine learning algorithms. Our study significantly advances our understanding of predictive factors influencing the response to PEG-INFα treatment in CHB patients. This highlights that both baseline and dynamic changes in HBsAg are critical predictors of achieving a functional cure. We enhanced the robustness of our predictive model via LASSO and several machine learning algorithms. The factors in this model are easily obtained in clinical settings and can effectively predict early treatment outcomes. This enables the recognition of patients who could gain advantages from PEG-INFα, which in turn optimizes treatment strategies and resource allocation.
This study revealed that 38.63% of patients with HBsAg titers less than 1500 IU/ml achieved surface antigen clearance after receiving PEG-INFα antiviral treatment. As the surface antigen titer decreases, the occurrence rate of surface antigen seroconversion gradually increases. When the HBsAg titer falls below 100 IU/ml, the percentage at which surface antigens are cleared can reach as high as 63.89%. Most current studies indicate that baseline HBsAg significantly influences HBsAg clearance posttreatment. However, some patients with HBsAg levels less than 1500 IU/mL, regarded as a favorable group, still do not achieve HBsAg clearance after pegylated interferon therapy. To address this issue, this study develops a predictive model for HBsAg clearance on the basis of six easily obtainable parameters. These six factors are baseline log2(HBsAg), the HBsAg decline rate at week 12, the neutrophil count at week 12, age, HBcAb at week 12, and gender.
Our findings align with those of previous studies, indicating that lower baseline log2 (HBsAg) values lead to improved antiviral treatment effectiveness and HBsAg clearance [29, 31]. When the HBsAg level is less than 100, the HBsAg clearance rate can reach up to 77% [32]. A study analyzed the expression of 67 genes related to the innate immune pathway in liver tissue from 105 untreated chronic hepatitis B (CHB) patients. The findings indicated that, in comparison with those in healthy control subjects, genes associated with antiviral effects and those stimulated by interferon were notably downregulated in the livers of patients with CHB. The expression of some of these genes is negatively correlated with HBsAg levels, suggesting that in patients with high HBsAg levels, the innate immunity within the livers of patients with CHB is significantly compromised [33]. Additionally, studies have shown that HBsAg can suppress the expression of interferon-stimulating factors in NK cells, inhibiting their response to cGAMP [34]. In patients with chronic hepatitis B, HBsAg has the capacity to directly induce functional deficits in bone marrow-derived dendritic cells. This phenomenon is considered as a plausible mechanism underlying the immune evasion associated with HBV [35].These studies suggest that high titers of HBsAg are associated with persistent HBV infection.
Additionally, we found that the HBsAg decrease rate at week 12 and HBsAb level at week 12 serve as predictive markers for HBsAg clearance at week 48. These study’s results are consistent with those of previous studies, indicating that a decrease in HBsAg and HBcAb levels at week 12 is associated with HBsAg clearance [23, 36, 37]. The results of persistent hepatitis B virus (HBV) infection and the effectiveness of antiviral treatment are significantly influenced by the interactions between the virus and the host [13]. Our study revealed that neutrophil counts at week 12 are associated with HBsAg clearance. Currently, while a reduction in neutrophils during PEG-INFα treatment is known to be related to the side effects of interferon, the specific mechanisms related to immune regulation in hepatitis B virus (HBV) infection are not clear. Several studies have indicated that HBV affects mitochondrial metabolism within immune cells and that inflammatory responses in the liver during antiviral treatment may lead to a reduction in neutrophil counts [23]. Furthermore, our findings suggest that older adults and male subjects exhibit a reduced likelihood of attaining a functional cure, a conclusion that is corroborated by additional research [38, 39].
Nevertheless, this study has several limitations. The retrospective design inherently limits the ability to establish causation, and the utilization of a single-center methodology may limit the applicability of our results to diverse populations. Additionally, despite the promising predictive capabilities of our models, the sample size may not be sufficient to capture the full spectrum of patient variability, particularly in diverse populations. Future studies should aim to validate these findings in larger, multicenter cohorts and explore additional variables that may influence treatment outcomes, such as genetic factors [18]. By overcoming these constraints, future investigations can lead to a more refined comprehension of the elements that affect the effectiveness of hepatitis B treatments, ultimately paving the way for enhanced treatment protocols and improved patient care.
Conclusions
In summary, this research effectively identified multiple factors linked to the clearance of HBsAg in patients with chronic hepatitis B (CHB) receiving PEG-INFα treatment. Our findings underscore the critical role of baseline clinical parameters and early response indicators in predicting treatment outcomes when both traditional logistic regression and advanced machine learning algorithms are utilized. The logistic regression model demonstrated favorable predictive performance, as evidenced by its favorable AUC values and clinical utility metrics, thereby offering a valuable tool for tailoring individualized treatment strategies. Subsequent investigations should prioritize the confirmation of these findings in larger and more heterogeneous populations, in addition to examining other biological variables that could provide deeper insights into the intricate nature of treatment responses in CHB patients. Such efforts will ultimately enhance patient management and improve therapeutic outcomes in this challenging clinical context.
Acknowledgements
The authors acknowledge the contributions from all of the patients, and the clinical and research staf in the Mengchao Hepatobiliary Hospital, Fujian Medical University.
Abbreviations
- HBV
Hepatitis B Virus
- HCC
Hepatocellular Carcinoma
- CHB
Chronic Hepatitis B
- ALT
Alanine aminotransferase
- ULN
Upper Limit Of Normal
- HBsAg
Hepatitis B Surface Antigen
- HBeAg
Hepatitis B e Antigen
- HBeAb
Hepatitis B e Antibody
- PEG-INFα
Pegylated interferonα
- GGT
Gamma-Glutamyl Transpeptidase
- ALP
Alkaline Phosphatase
- ALT
Alanine Aminotransferase
- AST
Aspartate Aminotransferase
- PLT
Platelet
- AUC
The Area Under the Curve
- ROC
Receiver Operator Characteristic
- LASSO
Predictor Variables Were Identified
- RF
Random Forest
- GBDT
Gradient Boosting Decision Tree
- XGBoost
Extreme Gradient Boosting
- SVM
Support Vector Machine
- cccDNA
Covalently Closed Circular DNA
- NAs
Nucleos(t)ide Analogs
- OR
Odds Ratio
- CI
Confidence Interval
- N
Nertrophil
- HBeALymb
Lymphocytes
Author contributions
YMY wrote the originaldraft, wrote the reply to reviewers and modifed the manuscript. YL was involved in thedata curation and revised the manuscript. FS, WYY, LNZ, CL were involved in the data curation. CP initiated, designed the research and revised the manuscript. All authors read and approved the fnal manuscript.
Funding
This research was supported by the Fujian Province Natural Science Foundation of China(2023J11475) and the Fujian Province Natural Science Foundation of China(2024J011240).
Data availability
No datasets were generated or analysed during the current study.
Declarations
Ethics approval and consent to participate
The study was conducted in accordance with the ethical guidelines of the Declaration of Helsinki and Istanbul. The study was approved by the Human Ethics Committee of Mengchao Hepatobiliary Hospital (No. 2024-081-01).
Consent for publication
All authors read and approved the fnal manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Ya-mei Ye and Yong Lin contributed equally to this work.
Contributor Information
Chun Lin, Email: linchun200707@126.com.
Chen Pan, Email: panchencry@163.com.
References
- 1.Marjenberg Z, Wright C, Pooley N, Cheung KW, Shimakawa Y, Vargas-Zambrano JC, et al. Hepatitis B surface antigen prevalence and the rates of mother-to-child transmission of hepatitis B virus after the introduction of infant vaccination programs in South East Asia and Western Pacific regions: a systematic review. Int J Infect Dis. 2022;124:65–75. [DOI] [PubMed] [Google Scholar]
- 2.Hepatitis B. Virus infection. Nat Rev Dis Primers. 2018;4:18036. [DOI] [PubMed] [Google Scholar]
- 3.Jeng WJ, Papatheodoridis GV, Lok ASF. Hepat B Lancet. 2023;401(10381):1039–52. [DOI] [PubMed] [Google Scholar]
- 4.Farag MS, van Campenhout MJH, Pfefferkorn M, Fischer J, Deichsel D, Boonstra A, et al. Hepatitis B Virus RNA as early predictor for response to Pegylated Interferon Alpha in HBeAg-Negative chronic Hepatitis B. Clin Infect Dis. 2021;72(2):202–11. [DOI] [PubMed] [Google Scholar]
- 5.Lai CL, Yuen MF. Prevention of Hepatitis B virus-related hepatocellular carcinoma with antiviral therapy. Hepatology. 2013;57(1):399–408. [DOI] [PubMed] [Google Scholar]
- 6.Boni C, Laccabue D, Lampertico P, Giuberti T, Vigano M, Schivazappa S, et al. Restored function of HBV-specific T cells after long-term effective therapy with nucleos(t)ide analogues. Gastroenterology. 2012;143(4):963–73. e9. [DOI] [PubMed] [Google Scholar]
- 7.Rinker F, Zimmer CL, Honer Zu Siederdissen C, Manns MP, Kraft ARM, Wedemeyer H, et al. Hepatitis B virus-specific T cell responses after stopping nucleos(t)ide analogue therapy in HBeAg-negative chronic hepatitis B. J Hepatol. 2018;69(3):584–93. [DOI] [PubMed] [Google Scholar]
- 8.Hsu YC, Jun DW, Peng CY, Yeh ML, Trinh H, Wong GL, et al. Effectiveness of entecavir vs tenofovir disoproxil fumarate for functional cure of chronic hepatitis B in an international cohort. Hepatol Int. 2022;16(6):1297–307. [DOI] [PubMed] [Google Scholar]
- 9.Kim G-A, Lim Y-S, An J, Lee D, Shim JH, Kim KM, et al. HBsAg seroclearance after nucleoside analogue therapy in patients with chronic hepatitis B: clinical outcomes and durability. Gut. 2014;63(8):1325–32. [DOI] [PubMed] [Google Scholar]
- 10.Hall SAL, Vogrin S, Wawryk O, Burns GS, Visvanathan K, Sundararajan V, et al. Discontinuation of nucleot(s)ide analogue therapy in HBeAg-negative chronic hepatitis B: a meta-analysis. Gut. 2022;71(8):1629–41. [DOI] [PubMed] [Google Scholar]
- 11.Sonneveld MJ, Rijckborst V, Boucher CA, Hansen BE, Janssen HL. Prediction of sustained response to peginterferon alfa-2b for hepatitis B e antigen-positive chronic hepatitis B using on-treatment hepatitis B surface antigen decline. Hepatology. 2010;52(4):1251–7. [DOI] [PubMed] [Google Scholar]
- 12.Choi HSJ, van Campenhout MJH, van Vuuren AJ, Krassenburg LAP, Sonneveld MJ, de Knegt RJ, et al. Ultra-long-term follow-up of Interferon Alfa treatment for HBeAg-Positive chronic Hepatitis B Virus infection. Clin Gastroenterol Hepatol. 2021;19(9):1933–e401. [DOI] [PubMed] [Google Scholar]
- 13.Zhao Q, Liu H, Tang L, Wang F, Tolufashe G, Chang J, et al. Mechanism of interferon alpha therapy for chronic hepatitis B and potential approaches to improve its therapeutic efficacy. Antiviral Res. 2024;221:105782. [DOI] [PubMed] [Google Scholar]
- 14.Tang LSY, Covert E, Wilson E, Kottilil S, Chronic Hepatitis B, Infection. Rev JAMA. 2018;319(17):1802–13. [DOI] [PubMed] [Google Scholar]
- 15.Choi HSJ, Sonneveld MJ, Farag MS, Brouwer WP, Brakenhoff SM, Hirode G, et al. Effects of on-treatment ALT flares on serum HBsAg and HBV RNA in patients with chronic HBV infection. J Viral Hepatitis. 2021;28(12):1729–37. [DOI] [PubMed] [Google Scholar]
- 16.Shang H, Hu Y, Guo H, Lai R, Fu Y, Xu S et al. Using machine learning models to predict HBeAg seroconversion in CHB patients receiving pegylated interferon-α monotherapy. J Clin Lab Anal. 2022;36(11). [DOI] [PMC free article] [PubMed]
- 17.Zhong YW, Shi YM, Chu F, Liu J, Shi C, Xu JJ, et al. Prediction for HBsAg seroconversion in children with chronic hepatitis B. BMC Infect Dis. 2021;21(1):1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo M, Zhang L, Yang C, Zhou B, Hou J, Jiang DK. CXCL13 variant predicts pegylated-interferon α treatment response in HBeAg‐positive chronic hepatitis B patients. J Med Virol. 2023;95(7). [DOI] [PubMed]
- 19.Tian X, Chong Y, Huang Y, Guo P, Li M, Zhang W et al. Using Machine Learning Algorithms to Predict Hepatitis B Surface Antigen Seroclearance. Computational and Mathematical Methods in Medicine. 2019;2019:1–7.
- 20.Hosaka T, Suzuki F, Kobayashi M, Seko Y, Kawamura Y, Sezaki H, et al. Long-term entecavir treatment reduces hepatocellular carcinoma incidence in patients with hepatitis B virus infection. Hepatology (Baltimore MD). 2013;58(1):98–107. [DOI] [PubMed] [Google Scholar]
- 21.Terrault NA, Lok ASF, McMahon BJ, Chang KM, Hwang JP, Jonas MM, et al. Update on prevention, diagnosis, and treatment of chronic hepatitis B: AASLD 2018 hepatitis B guidance. Hepatology (Baltimore. Md). 2018;67(4):1560–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liaw YF, Sung JJ, Chow WC, Farrell G, Lee CZ, Yuen H, et al. Lamivudine for patients with chronic hepatitis B and advanced liver disease. N Engl J Med. 2004;351(15):1521–31. [DOI] [PubMed] [Google Scholar]
- 23.Zhou RR, Song YH, Xu CY, Zhang YY, Wu XW, Zhang L, et al. Altered counts and mitochondrial mass of peripheral blood leucocytes in patients with chronic hepatitis B virus infection. J Cell Mol Med. 2024;28(12):e18440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang TT, Gish RG, de Man R, Gadano A, Sollano J, Chao YC, et al. A comparison of entecavir and lamivudine for HBeAg-positive chronic hepatitis B. N Engl J Med. 2006;354(10):1001–10. [DOI] [PubMed] [Google Scholar]
- 25.Marcellin P, Heathcote EJ, Buti M, Gane E, de Man RA, Krastev Z, et al. Tenofovir disoproxil fumarate versus adefovir dipivoxil for chronic hepatitis B. N Engl J Med. 2008;359(23):2442–55. [DOI] [PubMed] [Google Scholar]
- 26.Lau GK, Piratvisuth T, Luo KX, Marcellin P, Thongsawat S, Cooksley G, et al. Peginterferon Alfa-2a, lamivudine, and the combination for HBeAg-positive chronic hepatitis B. N Engl J Med. 2005;352(26):2682–95. [DOI] [PubMed] [Google Scholar]
- 27.Barbui T, Vannucchi AM, De Stefano V, Masciulli A, Carobbio A, Ferrari A, et al. Ropeginterferon alfa-2b versus phlebotomy in low-risk patients with polycythaemia vera (Low-PV study): a multicentre, randomised phase 2 trial. Lancet Haematol. 2021;8(3):e175–84. [DOI] [PubMed] [Google Scholar]
- 28.Marcellin P, Bonino F, Lau GK, Farci P, Yurdaydin C, Piratvisuth T, et al. Sustained response of hepatitis B e antigen-negative patients 3 years after treatment with peginterferon alpha-2a. Gastroenterology. 2009;136(7):2169–e791. [DOI] [PubMed] [Google Scholar]
- 29.Wu FP, Yang Y, Li M, Liu YX, Li YP, Wang WJ, et al. Add-on pegylated interferon augments hepatitis B surface antigen clearance vs continuous nucleos(t)ide analog monotherapy in Chinese patients with chronic hepatitis B and hepatitis B surface antigen = 1500 IU/mL: an observational study</at. World J Gastroenterol. 2020;26(13):1525–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song A, Lin X, Chen X. Functional cure for chronic hepatitis B: accessibility, durability, and prognosis. Virol J. 2021;18(1):114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang D, Wu D, Wang P, Wang Y, Yuan W, Hu D, et al. End-of-treatment HBcrAg and HBsAb levels identify durable functional cure after Peg-IFN-based therapy in patients with CHB. J Hepatol. 2022;77(1):42–54. [DOI] [PubMed] [Google Scholar]
- 32.Wen C, Wang Y, Tian H, Lei Y, Wang Z, Cai D, et al. Clinical cure induced by pegylated interferon alpha-2b in the advantaged population of chronic hepatitis B virus infection: a retrospective cohort study. Front Cell Infect Microbiol. 2023;13:1332232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lebosse F, Testoni B, Fresquet J, Facchetti F, Galmozzi E, Fournier M, et al. Intrahepatic innate immune response pathways are downregulated in untreated chronic hepatitis B. J Hepatol. 2017;66(5):897–909. [DOI] [PubMed] [Google Scholar]
- 34.Zheng B, Yu Y, Pan Z, Feng Y, Zhao H, Han Q et al. HBsAg dampened STING Associated activation of NK cells in HBeAg-Negative CHB patients. Int J Mol Sci. 2021;22(14). [DOI] [PMC free article] [PubMed]
- 35.Op den Brouw ML, Binda RS, van Roosmalen MH, Protzer U, Janssen HL, van der Molen RG, et al. Hepatitis B virus surface antigen impairs myeloid dendritic cell function: a possible immune escape mechanism of hepatitis B virus. Immunology. 2009;126(2):280–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhong W, Yan L, Zhu Y, Shi L, He Y, Chen T, et al. A high functional cure rate was induced by pegylated interferon alpha-2b treatment in postpartum hepatitis B e antigen-negative women with chronic hepatitis B virus infection: an exploratory study. Front Cell Infect Microbiol. 2024;14:1426960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang J, Zhang Z, Zhu L, Zhang Q, Zhang S, Pan Y, et al. Association of Hepatitis B core antibody level and hepatitis B surface antigen clearance in HBeAg-negative patients with chronic hepatitis B. Virulence. 2024;15(1):2404965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yeh M-L, Huang J-F, Yu M-L, Chuang W-L. Hepatitis b infection: progress in identifying patients most likely to respond to peginterferon alfa. Expert Rev Gastroenterol Hepatol. 2020;15(4):427–35. [DOI] [PubMed] [Google Scholar]
- 39.Chemin I, Wei L, Wedemeyer H, Liaw Y-F, Chan HL-Y, Piratvisuth T et al. No association between IFNL3 (IL28B) genotype and response to peginterferon alfa-2a in HBeAg-positive or -negative chronic hepatitis B. Plos One. 2018;13(7). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No datasets were generated or analysed during the current study.