Abstract
Objective
We aimed to characterize integrative and conjugative elements (ICEs) in antimicrobial resistant Streptococcs uberis isolates from bovine milk in Chiba, Japan, based on whole-genome sequence (WGS) data.
Results
Of the 101 isolates, we found the 36 isolates harboring erm(B)–tet(O), showing resistance to macrolides–lincosamides–tetracyclines. The 22 isolates were randomly selected and subject to WGS determination. The genomes measured 1.991–2.517 Mbp, with G + C contents of 35.8–36.9%. We used ResFinder–ICEfinder (web-based applications) to search for the antimicrobial resistant genes and ICEs. ResFinder detected combined erm(B)–tet(O)–ant(6)-Ia at the identical contig in each WGS. ICEfinder detected ICEs belonging to the same contigs, which contained erm(B)–tet(O)–ant(6)-Ia complete or partial sequences. Detection of putative ICEs using comparative genomic analysis was performed with identification of other streptococcal ICE resembling S. uberis ICEs. Using comparative genomic analysis (a reference WGS in NZ01 strain), putative ICE base size in UB37 isolate was 77,386-bp that was identical in other 13 isolates. Another similar streptococcal ICE was S. suis ICEnsui78–tet(O)–erm(B) mobile element. For ICE characterization in S. uberis with WGSs, a comparative genomic analysis is required with use of ICEfinder and other annotation tools.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13104-024-07065-3.
Keywords: Integrative and conjugative element, Erm(B), Tet(O), Streptococcus uberis, CompArative GEne cluster analysis Toolbox, ICEfinder
Introduction
Mobile genetic elements (MGEs) play a major role in horizontal gene transfer among microorganisms. Integrative and conjugative elements (ICEs) of MGEs are self-transmissible in bacteria and are characterized by both integrative and conjugative features [1]. ICEs are mosaic elements with both bacteriophage- and plasmid-like characteristics that integrate into and replicate with host cell chromosomes [2]. Additionally, ICEs transfer their antimicrobial resistance (AMR) determinants as well as the genes involved in their mobility/regulation/maintenance. Many ICEs are demonstrated among the genus Streptococcus [3]. ICEs belonging to Tn916-Tn1545 family transfer the widely disseminated tetracycline-resistance determinant tet(M), along with additional determinant erm(B) [4]. erm(B) gene encodes methylase that generates a posttranscriptional methylation of 23 S rRNA, leading to cross-resistance to macrolides, lincosamides, and streptogramin B antimicrobials [5].
Bovine clinical/subclinical mastitis in dairy cattle affect udder health, milk quality, and milk production, resulting in economic losses in dairy farms. Major causative microorganisms of the mastitis contain Escherichia coli, Staphylococcus aureus [6], coagulase-negative staphylococci, and contagious/environmental streptococci [7]. Contagious Streptococcus is S. agalactiae, whereas environmental streptococci contain S. dysgalactiae, S. canis, and S. uberis [8, 9].
S. uberis is isolated from environmental sources (soil, pasture, bedding materials, and bovine feces) and is present on the skin surface of dairy cows [8]. This microorganism develops bovine (sub)clinical mastitis during lactating/non-lactating periods after direct contact with the teat apex [10]. Molecular epidemiological investigation regarding S. uberis-associated clinical mastitis in dairy herds described that either predominant or a limited number of isolates may produce intramammary infections or transmission among cows (including potential transmission of the isolates via milking machine/environment) [11].
Macrolides, lincosamides, and beta-lactams are administered for treatment of S. uberis-associated mastitis [12]. Excessive usage of antimicrobials in dairy herds can result in elevated AMR among mastitis bacteria [13]. Zhang et al. [12] reported AMR phenotypes/genotypes among S. uberis isolates from bovine mastitis in Thailand. The isolates were resistant to tetracycline (82.0%), ceftiofur (19.3%), and erythromycin (8.3%). Prevalent AMR genes were tet(M) (87.3%), erm(B) (66.2%), and blaZ (6.6%). We have showed AMR patterns of S. uberis from bovine milk in Japan [14]. AMR monitoring of bovine mastitis-associated S. uberis may support an antimicrobial stewardship program for dairy farms.
Based on the findings by polymerase chain reaction (PCR) method, Haenni et al. [15] documented diverse and mobile features of ICEs among bovine S. uberis isolates. We searched for related articles by entering the keywords “streptococcus uberis, integrative and conjugative element, whole-genome” in PubMed database (https://pubmed.ncbi.nlm.nih.gov/) [16]. There is limited literature available on this topic, although sequence characterization and novel insights into bovine mastitis-associated S. uberis in dairy herds have been reported [17]. The aim of our work is to characterize ICE characterization in S. uberis AMR isolates from bovine milk based on whole-genome sequence (WGS) data.
Methods
AMR phenotyping/genotyping
All bovine milk-origin isolates (n = 106) identified based on mass spectrometry results were provided by Sanritsu Zelkova Veterinary Laboratory during Mar–Oct 2022. This laboratory determined minimum inhibitory concentrations of 14 antimicrobials (penicillin-G/ampicillin/minocycline/erythromycin/azithromycin/clindamycin/levofloxacin/chloramphenicol/cefotaxime/ceftriaxone/cefepime/cefozopran/meropenem/vancomycin) using broth microdilution method (MICroFAST Panel Types 7 J for Streptococcus spp.; Beckman Coulter Inc., Tokyo, Japan) recommended in Clinical and Laboratory Standards Institute (CLSI) M100-S26 document for alpha-hemolytic streptococci [18]. Quality control was performed using Enterococcus faecalis American Type Culture Collection (ATCC) 29,212, Streptococcus pneumoniae ATCC 49,619, and S. uberis ATCC 700,407 strains with the antimicrobial susceptibilities. To determine susceptibility/resistance to minocycline, we used tetracycline breakpoints in accordance with the CLSI guidelines.
One isolate per host was stored at − 70 °C to − 80 °C in Kitasato laboratory. We performed PCR methods to differentiate S. uberis and S. parauberis according to PCR fragments of 16 S and 23 S rRNA genes [19]. One hundred one isolates were analyzed. AMR genotyping [blaZ–erm(A)–erm(B)–mef(A)–lnu(B)–lnu(D)–tet(M)–tet(O)–tet(K)–tet(L)–tet(S)] was conducted using PCR assay [20, 21].
Randomized selection of S. uberis isolates with erm(B)–tet(O)
We found the isolates harboring both erm(B) and tet(O) (n = 36): all isolates indicated resistance to macrolides, lincosamides, and tetracyclines (the AMR rates of 33.7%, 54.5%, and 48.5%) [14]. Combined erm(B)–tet(O) genotype seems to be a representative marker to find ICEs in corresponding isolates, because both genes are located within MGEs [15]. The twenty-two isolates with erm(B)–tet(O) were selected based on random sampling numbers using Excel application. S. uberis ATCC 700,407 having WGS (https://genomes.atcc.org/genomes/26250c65fb774abc) at ATCC site was used as a reference strain. The enrolled strains’ list is shown in Supplementary Table 1.
WGS determination
DNAs from the twenty-two isolates were extracted as previously reported [22]. Whole-genome sequencing was carried out on a DNBSEQ-G400RS platform (MGI-Tech, Tokyo, Japan) using DNA Nanoball technology [23]. Sequencing library was generated using MGIEasy FS DNA Library Prep Set. Paired-end runs were performed with a read length of 2 × 150-bp.
We assessed read lengths, ambiguous bases, and quality scores for quality control. The reads were trimmed using quality trimming tool in CLC Genomics Workbench (v.8.0.2) with default parameters (quality limit and maximum number of ambiguous bases). De novo assembly was conducted using CLC Genomics Workbench with modified parameters, wherein the minimum contig length was set to 800-bp. Draft genome sequences were annotated using DDBJ Fast Annotation and Submission Tool (DFAST; https://dfast.nig.ac.jp) [24]. We determined assembly metrics and annotated features including genome size, number of contigs, average coverage, N50, numbers of coding DNA sequences (CDSs)/tRNAs/rRNAs/clustered regularly interspaced short palindromic repeats (CRISPR), G + C content, and coding ratio.
Additionally, the sequence type/clonal complex were determined using S. uberis PubMLST Isolate database (https://pubmlst.org/bigsdb?db=pubmlst_suberis_isolates). We also constructed the phylogenetic tree of 22 isolates and ATCC 700,407 as previously reported [25].
Web-based applications
We used ResFinder v.4.1 (https://cge.food.dtu.dk/services/ResFinder/) and ICEfinder v.1.0 (https://bioinfo-mml.sjtu.edu.cn/ICEfinder/index.php) with WGSs, to comprehensively search for related AMR genes and ICEs [25, 26]. Among the ICEs found, we selected the ICE containing erm(B)–tet(O) genes as a candidate from each isolate for further characterization.
Detection of putative ICEs using comparative genomic analysis
Flanking products of each candidate ICE were confirmed based on DFAST annotations. We selected a complete S. uberis WGS sequence without combined erm(B)–tet(O) as a reference. This WGS needed to possess flanking products identical to those of selected ICE based on the ICEfinder results. When we subtracted flanking products in reference WGS from corresponding products in candidate WGSs (comparative genomic analysis), we could detect putative ICEs containing erm(B)–tet(O). The subtraction procedure was based on concept “integration” resulting from ICE definition.
The putative ICE base size and product organization were determined for each WGS. To conduct homologous gene cluster searches and to generate publication-quality figures of gene cluster comparison between reference and candidate WGSs, we applied CompArative GEne Cluster Analysis Toolbox (CAGECAT, https://cagecat.bioinformatics.nl) consisting of two components of cblaster (for searches) and clinker (for figures) [27–29]. A Prokaryotic Genomes Annotation Pipeline (PGAP) in National Center for Biotechnology Information (NCBI) was applied for re-annotation. To specify ICE components, ICEscreen software was also applied [30]. To speculate protein function similarities, we used basic local alignment search tool x (blastx, translated nucleotide to protein homology search) on NCBI.
We found out where ICEUB37 was inserted into the genome selected.
Searching for other streptococcal ICE resembling S. uberis ICEs
Product organization of each determined ICE was inserted into homology search box on NCBI site. Other streptococcal ICEs with high-percent identity with the inserted query were extracted. One ICE showing the high-percent identity with inserted ICEs in our 22 isolates was specified. Using clinker, we constructed a visualized and interactive figure among the specified ICE and our ICEs.
Results
Assembly metrics and annotated features
WGS assembly metrics and annotated features (genome size, number of contigs, average coverage, N50, numbers of CDSs/tRNAs/rRNAs/CRISPR, G + C content, and coding ratio) are shown in Supplementary Table 2.
Additionally, the sequence type/clonal complex are shown in Supplementary Table 2. Of 22 strains, twenty belonged to CC996, suggesting that this population is closely related. The constructed phylogenetic tree of 22 isolates and ATCC 700,407 is shown in Supplementary Fig. 1.
Web-based application results
Web-based application results with WGSs (AMR genes and ICEs) are shown in Supplementary Table 3. There were combined genes erm(B)–tet(O)–ant(6)-Ia located at identical contig for each WGS. We observed other AMR genes [lnu(C), lsa(E)–lnu(B), or lnu(C)–fosB] at different contigs of WGSs in three isolates (UB25, UB78, or UB97). ICEfinder detected ICEs belonging to the same contigs containing combined erm(B)–tet(O)–ant(6)-Ia complete or partial sequences. All ICEs included erm(B)–ant(6)-Ia complete sequences, whereas some possessed tet(O) complete sequence: others had tet(O) partial sequence (Supplementary Table 3). Sometimes ICEfinder does not delimit the ICEs correctly and misses some part of the ICE.
Detection of putative ICEs using comparative genomic analysis
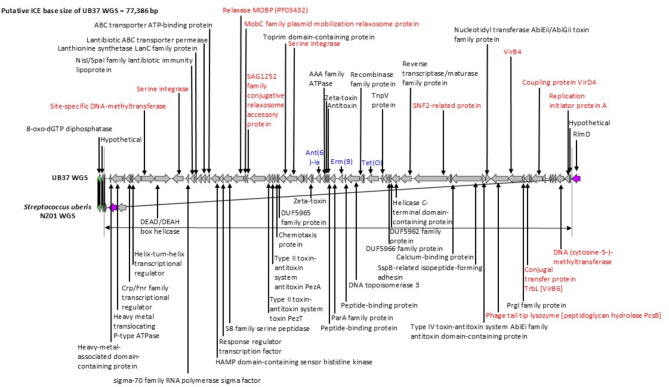
Detection approach of putative ICEs using comparative genomic analysis is shown in Fig. 1. As a reference WGS, we selected a complete S. uberis sequence (nucleotide accession no. CP022435.1) in bovine mastitis-associated NZ01 strain from New Zealand in 2014. UB37 isolate was chosen as an analyzed WGS. NZ01 and UB37 WGSs shared identical flanking products of 8-oxo-dGTP diphosphatase and RlmD [23 S rRNA (uracil(1939)-C(5))-methyltransferase].
Fig. 1.
Detection approach of putative Integrative and conjugative elements (ICEs) using comparative genomic analysis. As a reference, we applied a complete genome sequence (nucleotide accession no. CP022435.1) in bovine NZ01 strain from New Zealand in 2014: UB37 isolate was chosen. Similar/identical product comparisons between reference and candidate WGSs are shown in the same colors. Black gradations in interspace among the corresponding genes indicate percent identity. NZ01 and UB37 WGSs possessed identical flanking products of 8-oxo-dGTP diphosphatase and RlmD [23 S rRNA (uracil(1939)-C(5))-methyltransferase]. Putative ICE base size was 77,386-bp. Product organization in ICE was determined. The core products are shown in red letters; the antimicrobial resistance products are shown in blue letters. We performed the re-annotation for ICEUB37 using a Prokaryotic Genomes Annotation Pipeline. To specify ICE components, ICEscreen was applied. To speculate protein function similarities, we used basic local alignment search tool x; the protein functions are shown in brackets
Using comparative genomic analysis, the putative ICE base size of UB37 WGS was considered 77,386-bp, and the product organization of ICEUB37 was determined (Fig. 1). This ICE contained site-specific DNA-methyltransferase, serine integrase, relaxase MOBP (PF03432), MobC family plasmid mobilization relaxosome protein, SAG1252 family conjugative relaxosome accessory protein, SNF2-related protein, phage tail tip lysozyme, conjugal transfer protein TrbL, DNA (cytosine-5-)-methyltransferase, and replication initiator protein A. Using ICEscreen, we confirmed that ICEUB37 included a component set consisting of VirB4 and coupling protein VirD4. Its virB4 and virD4 sequences (sizes 2,352-bp and 1,818-bp) were similar with those of S. suis D9 complete genome (nucleotide accession no. CP002641.1) belonging to the Tn5252 (Supplementary Fig. 3). Furthermore, using blastx, we found that conjugal transfer protein TrbL has VirB6 function and that phage tail tip lysozyme has in part peptidoglycan hydrolase PcsB in Streptococcus.
Supplementary Fig. 2 indicates where ICEUB37 was inserted into the NZ01 genome.
Similarities/differences in ICE results (base start to end and size) between comparative genomic analysis and ICEfinder are shown in Table 1. There were two prevalent ICE base sizes by comparative genomic analysis consisting of 77,386-bp (UB7/UB15/UB25/UB36/UB37/UB41/UB56/UB58/UB63/UB77/UB81/UB82/UB89/UB97) and 76,735-bp (UB68/UB85/UB92). In contrast, we found three prevalent ICE base sizes by ICEfinder containing 48,105-bp (UB15/UB25/UB37/UB38/UB56/UB63/UB82/UB89/UB97), 49,736-bp (UB7/UB36/UB41/UB58/UB81), and 85,330-bp (UB68/UB85).
Table 1.
Similarities/differences in ICE results between ICEfinder and comparative genomic analysis
| Isolate | ICE base start to end [contig no.] by ICEfinder | ICE base start to end [contig no.] by CAGECAT | ICE base size by ICEfinder (bp) | ICE base size by CAGECAT (bp) |
|---|---|---|---|---|
| UB2 | 244,514 to 294,228 [1] | 224,660 to 302,024 [1] | 49,715 | 77,365 |
| UB7 | 244,883 to 294,618 [5] | 225,029 to 302,414 [5] | 49,736 | 77,386 |
| UB15 | 854,622 to 902,726 [3] | 858,346 to 935,731 [3] | 48,105 | 77,386 |
| UB23 | 577,061 to 640,616 [3] | 572,095 to 643,092 [3] | 63,556 | 70,998 |
| UB25 | 820,660 to 868,764 [1] | 824,384 to 901,769 [1] | 48,105 | 77,386 |
| UB36 | 244,515 to 294,250 [1] | 224,661 to 302,046 [1] | 49,736 | 77,386 |
| UB37 | 818,668 to 866,772 [3] | 822,392 to 899,777 [3] | 48,105 | 77,386 |
| UB38 | 818,651 to 866,755 [5] | 822,375 to 899,766 [5] | 48,105 | 77,392 |
| UB41 | 244,729 to 294,464 [3] | 224,875 to 302,260 [3] | 49,736 | 77,386 |
| UB56 | 819,593 to 867,697 [2] | 823,317 to 900,702 [2] | 48,105 | 77,386 |
| UB58 | 244,515 to 294,250 [3] | 224,661 to 302,046 [3] | 49,736 | 77,386 |
| UB63 | 781,903 to 830,007 [3] | 785,627 to 863,012 [3] | 48,105 | 77,386 |
| UB68 | 789,597 to 874,926 [2] | 818,820 to 895,554 [2] | 85,330 | 76,735 |
| UB77 | 576,689 to 657,436 [1] | 582,527 to 659,912 [1] | 80,748 | 77,386 |
| UB78 | 771,598 to 857,048 [1] | 800,942 to 878,348 [1] | 85,451 | 77,407 |
| UB81 | 244,514 to 294,249 [1] | 224,660 to 302,045 [1] | 49,736 | 77,386 |
| UB82 | 819,632 to 867,736 [1] | 823,356 to 900,741 [1] | 48,105 | 77,386 |
| UB85 | 789,395 to 874,724 [1] | 818,618 to 895,352 [1] | 85,330 | 76,735 |
| UB89 | 815,907 to 864,011 [2] | 819,631 to 897,016 [2] | 48,105 | 77,386 |
| UB92 | 188,482 to 268,578 [2] | 194,320 to 271,054 [2] | 80,097 | 76,735 |
| UB97 | 819,497 to 867,601 [3] | 823,221 to 900,606 [3] | 48,105 | 77,386 |
| UB99 | 817,430 to 870,565 [2] | 823,268 to 872,342 [2]* | 53,136 | > 49,075 |
*Accurate estimation of UB99 ICE was impossible because of the analyzed position at contig terminal
Most prevalent ICE base size by comparative genomic analysis is shown in bold letters
However, we obtained partial ICE base size (> 49,075-bp) in UB99. The accurate estimation was impossible because of the analyzed position at contig terminal (Table 1/Fig. 2) (see the Limitations).
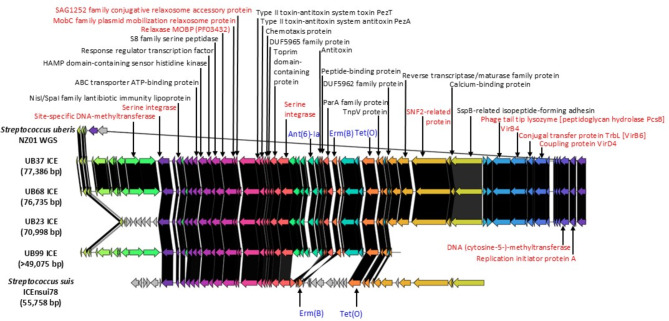
Fig. 2.
Putative identification of other streptococcal Integrative and conjugative element (ICE) resembling S. uberis ICEs (UB37/UB68/UB23/UB99), with S. uberis NZ01 WGS. Similar/identical product comparisons between reference and candidate WGSs are shown in the same colors. Black gradations in interspace among the corresponding genes indicate percent identity. Another similar streptococcal ICE was Streptococcus suis strain STC78 ICEnsui78–tet(O)–erm(B) mobile element (nucleotide accession no. ON944185.1), based on the similar product arrangements between UB37/UB68/UB23/UB99 ICEs and ICEnsui78. The core products are shown in red letters; the antimicrobial resistance products are shown in blue letters. We performed the re-annotation for ICEUB37 using a Prokaryotic Genomes Annotation Pipeline. To specify ICE components, ICEscreen was applied. To speculate protein function similarities, we used basic local alignment search tool x; the protein functions are shown in brackets
Putative identification of other streptococcal ICE resembling S. uberis ICEs
Putative identification of other streptococcal ICE resembling S. uberis ICEs (UB37/UB68/UB23/UB99) is shown in Fig. 2. Another similar streptococcal ICE was Streptococcus suis strain STC78 ICEnsui78–tet(O)–erm(B) mobile element (nucleotide accession no. ON944185.1; 55,758-bp), based on the similar product organization between UB37/UB68/UB23/UB99 ICEs and ICEnsui78. In contrast, this element did not possess arrangement of Ant(6)-Ia.
Discussion
Firstly, ICEfinder was applied to comprehensively search for ICEs. Some ICEs possessed tet(O) complete sequence, whereas others had tet(O) partial sequence (Supplementary Table 3), although phenotypic resistance to minocycline was confirmed in all isolates (Supplementary Table 1). This suggests that ICEfinder did not consider the entire ICE. Secondary, we performed putative ICE detection using comparative genomic analysis (a reference WGS in NZ01 strain). Twenty-one ICEs were determined (Table 1/Fig. 2). We could not obtain the accurate results of UB99 ICE because of the analyzed position at contig terminal. Therefore, we should construct complete WGSs using short- and long-read sequencing as we previously reported complete WGSs of Streptococcus canis strains from dogs [31].
The facts that ICEUB37 can be found in other streptococci is significant topics for discussion. Huang et al. [2] reported genomic organization of ICESa2603 (nucleotide accession no. AE009948.1; 54,349-bp) in S. agalactiae strain 2603 V/R. ICESa2603 comprised three proteins of SAG1250 (similar with prokaryotic DNA relaxase), SAG1251 (similar with MobC of prokaryotic plasmid), and SAG1252: ICEUB37 contained SAG1250 family conjugative relaxase [relaxase MOBP (PF03432)], MobC family plasmid mobilization relaxosome protein, and SAG1252 family conjugative relaxosome accessory protein. We found the similarity in ICE component involved with mobilization between ICESa2603 and ICEUB37. A structure, which was very similar with ICEUB37 [66,036/66,071 (99.95%)], was also found in S. ruminantium WGS (nucleotide accession no. AP025333.1; 67,641-bp spanning nucleotides from 1,168,946 to 1,236,586), using homology search in NCBI nucleotide database. S. ruminantium is the causative agent of several bovine and ovine diseases and S. ruminantium-related sheep mastitis outbreak has recently been reported from Italy [32]. Another structure similar with ICEUB37 [33,405/34,281 (97.44%)] was observed in S. pluranimalium WGS (nucleotide accession no. CP120510.1; 34,273-bp spanning nucleotides from 26,385 to 60,657). The other structure similar with ICEUB37 [27,974/28,781 (97.20%)] was also confirmed in S. macedonicus WGS (nucleotide accession no. CP113440.1; 28,775-bp spanning nucleotides from 1,544,153 to 1,572,927).
Another streptococcal ICE resembling S. uberis ICEs was S. suis ICEnsui78–tet(O)–erm(B) [33]. However, S. suis ICEnsui78 seems incomplete, because of lacking VirB4, coupling protein VirD4, and replication initiator protein A in the ICE. This could be possibly resulting from shotgun WGS using Illumina that did not fully resolve the ICE. Additionally, we analyzed the conjugation family [34] to which ICEUB37 belongs. This ICE fits into the Tn5252 (conjugation family) (Supplementary Fig. 3).
In conclusion, for full ICE characterization in S. uberis with WGSs, a comparative genomic analysis is required with use of ICEfinder and other annotation tools.
Limitations
Characterizing ICEs of selected S. uberis strains (n = 22) only by short-read WGS technology may be a limitation as repetitive sequences may be dismissed. Given the significant clinical implications of this research, it is recommended that whole-genome sequencing needs to be conducted using long-reads to enable more meaningful linear comparisons. We may consider requesting an extension for further modifications to solve the limitations of this study. Creations of hybrid genomes on two platforms making long- and short-read WGSs should be performed to completely identify ICEs. To the best of our knowledge, conjugation experiments using S. uberis have not been reported. We need to perform in vitro conjugation experiments to confirm ICE transfer between selected donor and recipient strains. Detailed information (antimicrobial dosing and duration) should be clarified for the relationships between ICEs and their clinical implications.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors wish to thank Mr. Goro Kurita DVM (Laboratory of Infectious Diseases, Graduate School of Infection Control Sciences and Ōmura Satoshi Memorial Institute, Kitasato University, Tokyo, Japan) for his helpful suggestions.
Abbreviations
- AMR
Antimicrobial resistance
- ATCC
American Type Culture Collection
- Blastx
Basic Local Alignment Search Tool X
- CAGECAT
CompArative GEne Cluster Analysis Toolbox
- CDS
Coding DNA sequence
- CLSI
Clinical and Laboratory Standards Institute
- CRISPR
Clustered regularly interspaced short palindromic repeats
- DFAST
DDBJ Fast Annotation and Submission Tool
- ICE
Integrative and conjugative element
- MGE
Mobile genetic element
- NCBI
National Center for Biotechnology Information
- PCR
Polymerase chain reaction
- PGAP
Prokaryotic Genomes Annotation Pipeline
- WGS
Whole-genome sequence
Author contributions
Each author is expected to have made substantial contributions. The study was conceived and designed by TT, TM, and YT. The data were collected by TM and TT. The data were analyzed by TM, HY, MG, and YT. The manuscript was drafted by TT. The manuscript was critically revised by TT and TM. All authors read and approved the final manuscript.
Funding
This study was supported in part by the Basic Research Grant (“Establishment of molecular bases in bovine mastitis-associated Streptococcus uberis to develop its vaccines”) from the Matsuoka Research Institute for Science (to TT).
Data availability
Sequence data that support the findings of this study have been deposited in NCBI, US. Supplementary Table 2 lists the corresponding GenBank nucleotide accession numbers of WGSs.
Declarations
Ethics approval and consent to participate
The ethics committee of the Sanritsu Zelkova Veterinary Laboratory reviewed and approved our study design to maintain the anonymity and privacy of companion animals (approval no. SZ20220324). Background information (host species, collection year, geographic location, and isolation source) for the WGSs is available in the NCBI database. A total of twenty-two isolate-related background information was enrolled in the study. The consent to participate is not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Burrus V, Pavlovic G, Decaris B, Guédon G. Conjugative transposons: the tip of the iceberg. Mol Microbiol. 2002;46:601–10. 10.1046/j.1365-2958.2002.03191.x. [DOI] [PubMed] [Google Scholar]
- 2.Huang J, Liang Y, Guo D, Shang K, Ge L, Kashif J, et al. Comparative genomic analysis of the ICESa2603 family ICEs and spread of erm(B)- and tet(O)-carrying transferable 89K-subtype ICEs in swine and bovine isolates in China. Front Microbiol. 2016;7:55. 10.3389/fmicb.2016.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beres SB, Musser JM. Contribution of exogenous genetic elements to the group a Streptococcus metagenome. PLoS ONE. 2007;2:e800. 10.1371/journal.pone.0000800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Varaldo PE, Montanari MP, Giovanetti E. Genetic elements responsible for erythromycin resistance in Streptococci. Antimicrob Agents Chemother. 2009;53:343–53. 10.1128/AAC.00781-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leclercq R. Mechanisms of resistance to macrolides and lincosamides: nature of the resistance elements and their clinical implications. Clin Infect Dis. 2002;34:482–92. 10.1086/324626. [DOI] [PubMed] [Google Scholar]
- 6.Zadoks RN, van Leeuwen WB, Kreft D, Fox LK, Barkema HW, Schukken YH, et al. Comparison of Staphylococcus aureus isolates from bovine and human skin, milking equipment, and bovine milk by phage typing, pulsed-field gel electrophoresis, and binary typing. J Clin Microbiol. 2002;40:3894–902. 10.1128/JCM.40.11.3894-3902.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng WN, Han SG. Bovine mastitis: risk factors, therapeutic strategies, and alternative treatments - a review. Asian-Australas J Anim Sci. 2020;33:1699–713. 10.5713/ajas.20.0156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zadoks RN, Tikofsky LL, Boor KJ. Ribotyping of Streptococcus uberis from a dairy’s environment, bovine feces and milk. Vet Microbiol. 2005;109:257–65. 10.1016/j.vetmic.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 9.Kabelitz T, Aubry E, van Vorst K, Amon T, Fulde M. The role of Streptococcus spp. Bovine Mastitis Microorganisms. 2021;9:1497. 10.3390/microorganisms9071497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leigh JA. Streptococcus uberis: a permanent barrier to the control of bovine mastitis? Vet J. 1999;157:225–38. 10.1053/tvjl.1998.0298. [DOI] [PubMed] [Google Scholar]
- 11.Davies PL, Leigh JA, Bradley AJ, Archer SC, Emes RD, Green MJ. Molecular epidemiology of Streptococcus uberis clinical mastitis in dairy herds: strain heterogeneity and transmission. J Clin Microbiol. 2016;54:68–74. 10.1128/JCM.01583-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang T, Niu G, Boonyayatra S, Pichpol D. Antimicrobial resistance profiles and genes in Streptococcus uberis associated with bovine mastitis in Thailand. Front Vet Sci. 2021;8:705338. 10.3389/fvets.2021.705338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saini V, McClure JT, Léger D, Dufour S, Sheldon AG, Scholl DT, et al. Antimicrobial use on Canadian dairy farms. J Dairy Sci. 2012;95:1209–21. 10.3168/jds.2011-4527. [DOI] [PubMed] [Google Scholar]
- 14.Tsuyuki Y, Maeda T, Torii K, Yoshida H, Ikeda N, Yoshida S, et al. Antimicrobial resistance patterns of Streptococcus uberis isolates from bovine milk in Chiba prefecture, Japan: association between multidrug resistance and clonal complex 996. J Vet Med Sci. 2024;86:468–73. 10.1292/jvms.23-0526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haenni M, Saras E, Bertin S, Leblond P, Madec JY, Payot S. Diversity and mobility of integrative and conjugative elements in bovine isolates of Streptococcus agalactiae, S. Dysgalactiae subsp. dysgalactiae, and S. Uberis. Appl Environ Microbiol. 2010;76:7957–65. 10.1128/AEM.00805-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.The National Library of Medicine. PubMed. https://pubmed.ncbi.nlm.nih.gov/. Accessed 24 June 2024.
- 17.Vezina B, Al-Harbi H, Ramay HR, Soust M, Moore RJ, Olchowy TWJ, et al. Sequence characterisation and novel insights into bovine mastitis-associated Streptococcus uberis in dairy herds. Sci Rep. 2021;11:3046. 10.1038/s41598-021-82357-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clinical and Laboratory Standards Institute (CLSI). Performance standards for antimicrobial susceptibility testing; 26th informational supplement. M100-S26. Wayne, PA: CLSI; 2016. [Google Scholar]
- 19.Hassan AA, Khan IU, Abdulmawjood A, Lämmler C. Evaluation of PCR methods for rapid identification and differentiation of Streptococcus uberis and Streptococcus parauberis. J Clin Microbiol. 2001;39:1618–21. 10.1128/JCM.39.4.1618-1621.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Malhotra-Kumar S, Lammens C, Piessens J, Goossens H. Multiplex PCR for simultaneous detection of macrolide and tetracycline resistance determinants in Streptococci. Antimicrob Agents Chemother. 2005;49:4798–800. 10.1128/AAC.49.11.4798-4800.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takayama Y, Tanaka T, Oikawa K, Fukano N, Goto M, Takahashi T. Prevalence of blaZ gene and performance of phenotypic tests to detect penicillinase in Staphylococcus aureus isolates from Japan. Ann Lab Med. 2018;38:155–9. 10.3343/alm.2018.38.2.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maeda T, Yoshida H, Abe N, Murakami K, Goto M, Takahashi T. Draft genome sequence of emm103/ST1363 Streptococcus pyogenes strain AB1, isolated from the blood of a woman with peritonitis and toxic shock syndrome. Microbiol Resour Announc. 2024;13:e0102723. 10.1128/mra.01027-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anslan S, Mikryukov V, Armolaitis K, Ankuda J, Lazdina D, Makovskis K, et al. Highly comparable metabarcoding results from MGI-Tech and Illumina sequencing platforms. PeerJ. 2021;9:e12254. 10.7717/peerj.12254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tanizawa Y, Fujisawa T, Nakamura Y. DFAST: a flexible prokaryotic genome annotation pipeline for faster genome publication. Bioinformatics. 2018;34:1037–9. 10.1093/bioinformatics/btx713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koide S, Nagano Y, Takizawa S, Sakaguchi K, Soga E, Hayashi W, et al. Genomic traits associated with virulence and antimicrobial resistance of invasive group B Streptococcus isolates with reduced penicillin susceptibility from elderly adults. Microbiol Spectr. 2022;10:e0056822. 10.1128/spectrum.00568-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. 2012;67:2640–4. 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gilchrist CLM, Booth TJ, van Wersch B, van Grieken L, Medema MH, Chooi YH. Cblaster: a remote search tool for rapid identification and visualization of homologous gene clusters. Bioinform Adv. 2021;1:vbab016. 10.1093/bioadv/vbab016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gilchrist CLM, Chooi YH. Clinker & clustermap.js: automatic generation of gene cluster comparison figures. Bioinformatics. 2021;37:2473–5. 10.1093/bioinformatics/btab007. [DOI] [PubMed] [Google Scholar]
- 29.van den Belt M, Gilchrist C, Booth TJ, Chooi YH, Medema MH, Alanjary M. The CompArative GEne Cluster Analysis Toolbox for rapid search and visualisation of homologous gene clusters. BMC Bioinformatics. 2023;24:181. 10.1186/s12859-023-05311-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lao J, Lacroix T, Guédon G, Coluzzi C, Payot S, Leblond-Bourget N, et al. ICEscreen: a tool to detect Firmicute ICEs and IMEs, isolated or enclosed in composite structures. NAR Genom Bioinform. 2022;4:lqac079. 10.1093/nargab/lqac079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim JS, Sakaguchi S, Fukushima Y, Yoshida H, Nakano T, Takahashi T. Complete genome sequences of four Streptococcus canis strains isolated from dogs in South Korea. Microbiol Resour Announc. 2020;9:e00818–20. 10.1128/MRA.00818-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosa MN, Vezina B, Marogna G, Canu A, Molotzu MR, Tola S. Streptococcus ruminantium-associated sheep mastitis outbreak detected in Italy is distinct from bovine isolates. Vet Res. 2023;54:118. 10.1186/s13567-023-01248-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brizuela J, Kajeekul R, Roodsant TJ, Riwload A, Boueroy P, Pattanapongpaibool A, et al. Streptococcus suis outbreak caused by an emerging zoonotic strain with acquired multi-drug resistance in Thailand. Microb Genom. 2023;9:mgen000952. 10.1099/mgen.0.000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ambroset C, Coluzzi C, Guédon G, Devignes MD, Loux V, Lacroix T, et al. New insights into the classification and integration specificity of Streptococcus integrative conjugative elements through extensive genome exploration. Front Microbiol. 2016;6:1483. 10.3389/fmicb.2015.01483. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence data that support the findings of this study have been deposited in NCBI, US. Supplementary Table 2 lists the corresponding GenBank nucleotide accession numbers of WGSs.