Abstract
The quality issues of ultra-high-temperature (UHT) milk, such as protein hydrolysis and aging gels throughout shelf life, are caused by proteases from psychrophilic bacteria. However, existing enzyme activity detection techniques have low sensitivity and cannot accomplish the detection of product deterioration caused by low enzyme activity. In this study, an attempt was made to analyze the relationship between enzymatically cleaved peptides and product quality using peptidomics techniques. The impact of psychrophilic bacteria proteases on the quality of UHT milk was investigated based on peptidomics by exogenously adding proteases. The results indicated that the protease activity and protein hydrolysis increased significantly in UHT milk over the storage period. Through peptidomic analysis, 2479 peptides were identified, in which 32 proteins linked to the identified peptides. 17 potential marker peptides, including αS1-casein143–156, β-lactoglobulin99–108, and β-casein39–66, were screened. The correlation of all identified peptides with protease activity and protein hydrolysis was explored by the weighted gene co-expression network analysis (WGCNA) method. The peptidomics technology helps to study the release or degradation of peptides in UHT milk during storage, which can be applied to monitor the shelf life of UHT milk and predict spoilage.
Keywords: Peptidomics, Proteases, UHT milk, Spoilage
Highlights
-
•
Peptidomics and WGCNA were applied to study peptide changes in UHT milk.
-
•
Significantly altered peptides were mainly derived from αS1-casein.
-
•
The identified peptides correlated with protease activity and protein hydrolysis.
1. Introduction
The emergence of quality problems in ultra-high-temperature (UHT) milk, including unpleasant tastes and protein hydrolysis, is attributed to the presence of residual exogenous psychrophilic protease activity. Ultra-high-temperature heat treatment can effectively kill microorganisms in milk so that milk has a relatively long shelf life (Guo et al., 2024). Nevertheless, the psychrophilic bacteria in milk can secrete thermostable extracellular enzymes before processing. In particular, proteases (such as AprX) can maintain 20 % to 40 % activity after UHT treatment (Dai et al., 2023). The alkaline metalloproteinase AprX, encoded by the aprX gene, is thermally stable and is capable of hydrolyzing κ-casein, α-casein, and β-casein, which can lead to problems of dairy quality problems (Akillioğlu et al., 2022).
It has been reported that psychrophilic bacteria such as Pseudomonas can secrete heat-resistant protease AprX (D’Incecco et al., 2022). Ultra-high-temperature treatment is effective in killing psychrophilic bacteria in milk, but their proteases secreted at low temperatures retain activity after high-temperature treatment. Age gelation induced by exogenous proteases generally involves the cleavage of κ-casein, resulting in the formation of micelles that resemble ‘para-κ-casein’ and by-products similar to ‘glycopeptides.’ This process contributes to the instability of casein micelles (Anema, 2017). When UHT milk is stored, residual proteases hydrolyze the proteins in the milk, increasing their viscosity, hydrolyzing proteins, and forming a bitter flavor, which leads to the deterioration of dairy products and a shortening of shelf life (Zhang et al., 2019). Zhang, Li, et al. (2020) added Pseudomonas and then sterilized it, storing the milk samples at 20 °C, 30 °C, and 55 °C to observe the impact on milk quality. The results showed that the decrease of sediment formation and pH, and milk protein hydrolysis took place before gelatinization.
In the early stages of storage, the protease activity of UHT milk is low so that cannot be detected directly. Current methods are not sensitive enough to probe the hydrolysis of milk protein by detecting the protease activity. Casein is enzymatically hydrolyzed to release hydrolyzed peptides, so the hydrolysis and stability of UHT milk can be indirectly determined by peptidomics to further investigate the mechanism of protein hydrolysis in milk (Zhang et al., 2023). This technology is crucial for detecting marker peptides and researching functional peptides. Also, peptidomics enables the monitoring of peptide alterations in milk, predicting the amount of protease-cleaved peptides in milk. Peptidomics can be applied to identify peptides in milk and determine the abundance of peptides, which is expected to assess dairy products for quality. (Leite et al., 2021; Ning et al., 2022).
According to Leite et al. (2021), the levels of both unspecific and targeted protease activities decreased in the casein and whey components of milk after exposure to 85 °C for 5 min, which was mainly reflected in the number of milk protease-destroying peptides. Dalabasmaz et al. (2019) analyzed the peptide profiles of 9 kinds of commercially available UHT milk until the end of the shelf life by MALDI-TOF-MS. They found that the comparative content of 22 peptides increased in the milk until reaching the expiration, surpassing the levels in the UHT milk during the shelf life. The relative quantitative examination of the MALDI-TOF peptide profiles from pasteurized and UHT sterilized milk by Ebner et al. (2016) found that all 16 peptides were affected by either heating or storage, showing very significant differences in intensity, with β-casein196–209 (m/z 1460.9 Da) being a marker peptide susceptible to heat.
In this present study, the effect of protein hydrolysis in UHT milk was investigated by adding the proteases from psychrophilic bacteria. By using LC-HRMS, the peptides and proteins in milk samples were identified. Analysis of peptides was used to identify peptides that increased significantly in relative strength during the milk storage period. The purpose of this study is to provide new perspectives for prognosticating the deterioration of UHT milk caused by proteases from psychrophilic bacteria, which could be used for quality evaluation and quality assessment of UHT milk during processing and shelf-life.
2. Materials and methods
2.1. Chemicals
Ammonium sulfate, calcium chloride (CaCl2), absolute ethyl alcohol, and sodium dodecyl sulfate (SDS) were bought from Sinopharm Chemical Reagent Co. LTD (Beijing, PRC). Azocasein was provided by Yuanye Bio-Technology Co., Ltd. (Shanghai, PRC). Trichloroacetic acid (TCA) was bought from Macklin (Shanghai, PRC). O-phthalaldehyde (OPA), dithiothreitol (DTT), trifluoroacetic acid (TFA), and β-mercaptoethanol were acquired from Aladdin (Shanghai, PRC). Acetonitrile (ACN), and formic acid were provided by Thermo Fisher Scientific (Pittsburgh, PA, USA).
2.2. Preparation of proteases
Aeromonas sp. with high protease activity was isolated from raw milk and stored in the Institute of Food Science and Technology, Chinese Academy of Agricultural Sciences. After the strain had been fully active, it was inoculated at 2 % in nutritional broth liquid medium and allowed to incubate for 25 h at 180 rpm on a shaker at 28 °C. The supernatant was extracted from the fermentation broth after it was centrifuged for 10 min at 10000 rpm. The solution was gradually supplemented with ammonium sulfate in an ice bath (0 °C) and stirred at a constant speed so that the saturation of ammonium sulfate in the enzyme solution reached 80 %. After slow stirring for 30 min, and subsequent centrifugation at 10,000 rpm for 10 min at 4 °C, the precipitate was collected. Phosphate buffered saline (PBS) was used in tiny amounts to dissolve the precipitate, and the dialysis bag with a molecular mass of 5000 Da was used for desalting the dialysate with deionized water. The dialyzed solution is freeze-dried and prepared for use.
2.3. Preparation of milk samples
UHT milk samples were purchased from supermarkets. The prepared protease powder was dissolved in PBS buffer (pH = 7.2) so that the concentration of the original protease solution was 0.5 mg/mL. 1 mL of protease solution filtered by a 0.22 μm filter was added to the UHT milk. The control was UHT milk without protease solution. Samples were stored at 37 °C and sampled every 48 h. All samples were analyzed in triplicate.
2.4. Shelf life endpoint determination
Boiling experiment: UHT milk totaling 10 mL was taken in a test tube and treated at 100 °C for 5 min. The presence of coagulation indicates a positive result.
CaCl2 experiment: add 5 mL of UHT milk to 0.3 mL of 1 % CaCl2 solution and heat it at 100 °C for 5 min. The presence of coagulation indicates a positive result.
Alcohol experiment: 5 mL of UHT milk was added in 10 mL of 70 % alcohol, and flocculation indicated a positive result.
2.5. Protease activity
The sample, PBS buffer (pH = 7.4), and 0.5 % azocasein were mixed at a ratio of 1:1:2. These were incubated at 37 °C for 2 h away from light. The reaction was terminated by adding 5 % of the TCA solution, and it was then maintained at room temperature for 30 min. The samples were centrifuged at 9500 rpm for 5 min, and then the supernatant was extracted from the samples in a 96-well plate, and the absorbance was measured at 345 nm. Suspended material in the supernatant after centrifugation could be filtered using a 0.22 μm aqueous filter. Blank control: TCA solution was added before adding the substrate.
2.6. Determination of the degree of hydrolysis
Preparation of OPA reagent: 7.620 g of sodium tetraborate, 0.2 g of SDS, 0.176 g of DTT, and 0.160 g of OPA dissolved in 4 mL of ethanol were mixed and dissolved in 200 mL of deionized water.
Preparation and determination of serine standard solution: 0.05 g of serine was fixed in 500 mL of deionized water. 400 μL of serine standard solution was taken into 3 mL of OPA reagent. The optical density (OD) of the standard was measured at 340 nm after the mixture reacted accurately for 2 min. The reaction with ionized water was used as an optical density of the blank.
Sample treatment and determination: the milk samples were centrifuged at 4500 r/min for 20 min to remove fat. The skimmed milk is added to the same amount of 12 % TCA, and centrifuged at 9500 r/min for 20 min. 400 μL of supernatant was added to 3 mL of OPA reagent, reacting accurately for 2 min, and then the OD value was measured at 340 nm.
2.7. SDS-PAGE
Appropriate modifications were made according to the method of Wang et al. (2024). 10 μL of diluted samples were added to an equal mixture of β-mercaptoethanol and 2× LaemmLi buffer, and electrophoresed after a metal bath at 100 °C for 5 min. The voltage was increased to 120 V after running for 20 min at 90 V. The staining was done with Kaumas stain for 30 min and eluted until the bands were clear.
2.8. Particle size and Zeta potential
1 mL of milk sample was diluted 1000 times with deionized water, and the average particle size of the samples was detected using the nano-particle size potential analyzer set to particle size mode. The Zeta potential of the samples was detected in the Zeta potential mode. The measurement was repeated 3 times for each sample to take the average value.
2.9. Identification of protein hydrolyzing peptides by LC-HRMS
2.9.1. Samples preparation
The samples were thawed at 4 °C and defatted by centrifugation at 3000 ×g for 10 min. The skimmed milk under the fat was taken out, and the process was repeated until no fat was observed. Skimmed milk was mixed with 20 % TCA (v/v = 1:1) at 3000 ×g and centrifuged at 4 °C for 10 min to collect the supernatant. The peptide-rich supernatant was decontaminated using a 200 mg C18 column solid-phase extraction column prepared with 99.9 % CAN-0.1 % TFA and 1 % CAN-0.1 % TFA. Carbohydrates, salts, and TCA were removed from the samples by adding 3 mL of 0.1 % CAN-0.1 % TFA, and the peptides were eluted with 80 % CAN-0.1 % TFA. The solutions obtained were lyophilized in a vacuum centrifugal concentrator and stored at −20 °C for further analysis.
2.9.2. Nano-LC-MS analysis of peptide sequences
LC-MS analysis was based on the report by Zhang et al. (2024) with some modifications. Mobile phases A (100 % water, 0.1 % formic acid) and B (80 % acetonitrile, 0.1 % formic acid) were prepared. The chromatographic gradients are as follows: 8 %–12 % B, 0 min–5 min; 12 %–30 % B, 5 min–38 min; 30 %–40 % B, 38 min–45 min; 40 %–95 % B, 45 min–46 min; 95 % B, 46 min–60 min. An Orbitrap fusion lumos mass spectrometer was used with a Nanospray Flex™ (NSI) ion source. The mass spectrum was acquired in a data-dependent acquisition mode, with a full scanning range of m/z 300–1500, a resolution of 120,000 (200 m/z) for the primary mass spectrum, an AGC of 4 × 105, and a maximum injection time of 50 ms for the C-trap. The secondary mass spectrum was detected in “Top Speed” mode, with Orbitrap detection of secondary ions, an AGC of 5 × 104. The maximum injection time is 22 ms. The collision energy for peptide fragmentation was adjusted to 35 % to generate raw data (.raw) for mass spectrometry detection.
The bovine-reviewed-6041 database was used, and the Proteome Discoverer 2.4 software was used to search the database with the following search parameter settings: no enzyme, dynamic modifications were set “Phospho/+79.966 Da (S, T), Deamidated/+0.984 Da (Q)”, precursor ion mass tolerance was ±15 ppm, fragment ion mass tolerance was ±0.02 Da, and Max Missed Cleavages was 2. The cleavage site of the protease is described in Matéos et al. (2015).
2.10. Statistical analysis
Each sample was repeated three times. Plots were made using Origin 2021 (Northampton, MA, USA.), and R (version 4.2.1) were used for data analysis (Auckland, New Zealand). Peptide plots were generated using peptigram. SPSS Statistics 26 (Chicago, IL, USA) was used for significance analysis. Peptides were analyzed using the bovine-reviewed-6041 database, and the library was searched by Proteome Discoverer 2.4 software (Pittsburgh, PA, USA). The package “WGCNA” (Langfelder et al., 2023) was used for WGCNA analysis.
3. Results and discussion
3.1. Shelf life endpoint determination
In Table 1, the boiling and CaCl2 experiments of protease-added UHT milk samples showed positive results, along with the coagulation appearing on day 2. On day 6, the alcohol-positive milk phenomenon was observed, and the samples appeared granular. The proteins in cow's milk were hydrolyzed by protease during storage to produce free amino acids, which increased the acidity of the system. The negative charge of casein was neutralized by hydrogen ions. The bound water around the casein was removed by alcohol, which reduced the stability of the casein. Then the casein aggregated and precipitated, which manifested as the alcohol-positive milk phenomenon. Samples that were positive in the boiling experiment and CaCl2 had a thicker fat upwelling phenomenon in the upper layer and a thicker protein precipitation at the bottom. This phenomenon indicated that samples had been considered unstable and κ-casein cleavage had already taken place. D’Incecco et al. (2022) assessed the stability of pasteurized milk through boiling experiments, which showed that pasteurized milk with the addition of Pseudomonas fluorescens began to show flocculation, gelation, and gel syneresis after 24 h of storage at 25 °C. This is related to the activity of protease hydrolysis.
Table 1.
Results of boiling, CaCl2, and alcohol experiments of UHT milk and UHT milk incubated with protease at day 2, day 4, and day 6 of storage.
| Samples |
0 d |
2 d |
4 d |
6 d |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | A | B | C | A | B | C | A | B | C | |
| UHT milk 1 | − | − | − | − | − | − | − | − | − | − | − | − |
| UHT milk 2 | − | − | − | − | − | − | − | − | − | − | − | − |
| UHT milk 3 | − | − | − | − | − | − | − | − | − | − | − | − |
| Protease-added UHT milk 1 | − | − | − | + | + | − | + | + | − | + | + | + |
| Protease-added UHT milk 2 | − | − | − | + | + | − | + | + | − | + | + | + |
| Protease-added UHT milk 3 | − | − | − | − | + | − | + | + | − | + | + | + |
Note: A is the boiling experiment; B is the CaCl2 experiment; C is the alcohol experiment; “−” is negative; “+” is positive; In the samples, UHT milk 1–3 are three parallel experiments of UHT milk; Protease-added UHT milk 1–3 are three parallel experiments of UHT milk with proteases.
3.2. Protease activity
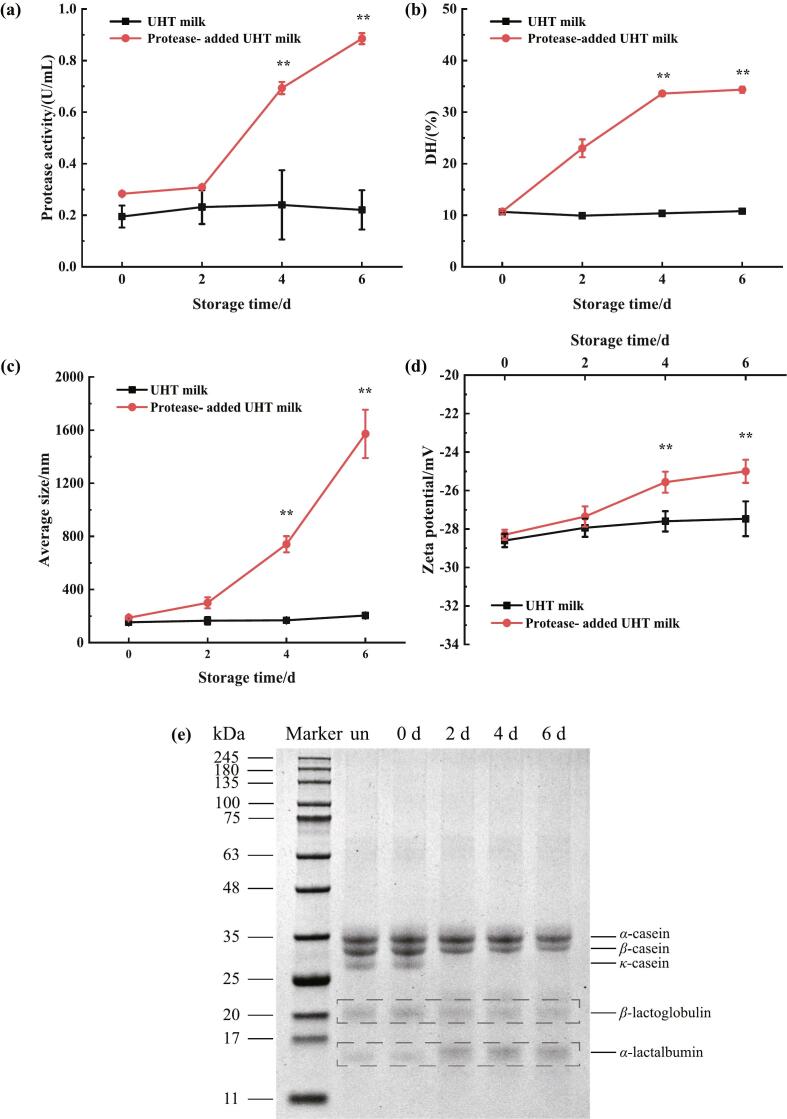
The initial protease activity of UHT with protease was 0.28 ± 0.01 U/mL. It increased significantly (P < 0.01) with time from day 4 until it peaked at 0.89 ± 0.02 U/mL on day 6, which was four times higher than that of UHT milk without protease (Fig. 1a). The control group had lower protease activity with no significant change during storage. This showed that the protease activity in UHT milk increases with longer storage time. Glück et al. (2016) isolated Pseudomonas from raw milk and investigated the thermal stability of its peptidase production, and the result showed that the retained activity of peptidase after UHT processing was 45.8 %–58.9 %. Thus, protease activity was also detected in the control group. According to Stoeckel et al. (2016), raw milk with Pseudomonas bacteria was sterilized under ultra-high temperature conditions. In that study, as the enzyme activity rises, there are subsequent changes in the quality of the milk, including the development of a bitter flavor, the formation of particles, the emulsification of fat, the production of sediment, and the gelation, which only occurs during periods of high enzyme activity.
Fig. 1.
(a) Protease activity of UHT milk and protease-added UHT milk during the storage time. (b) Degree of hydrolysis values of UHT milk and protease-added UHT milk during the storage time. (c) Particle size of UHT milk and protease-added UHT milk during storage. (d) Zeta potential of UHT milk and protease-added UHT milk during storage. (e) SDS-PAGE of UHT milk hydrolyzed by protease during the storage time. “un”: UHT milk without addition of protease; “0 d, 2 d, 4 d, 6 d”: UHT milk with protease stored for 0 days, 2 days, 4 days, and 6 days. Values are means ± SEs, n = 3. “**” represents P < 0.01.
3.3. Degree of hydrolysis
The degree of protein hydrolysis reflects the ratio of the count of cleaved peptide bonds to the overall count of peptide bonds in the protein in a protein hydrolysate (Rutherfurd, 2010). The impact of the protease on the degree of protein hydrolysis in cow's milk was investigated by the OPA method. As shown in Fig. 1b, the initial protein hydrolysis degree of UHT milk after adding protease was 10.71 ± 0.07 % on day 0, which did not differ significantly from that of UHT milk without protease addition (10.66 ± 0.16 %). The degree of protein hydrolysis in protease-added milk showed a significant increase (P < 0.01) with longer storage time. From day 4, the trend of increase leveled off. Until day 6, it increased to 34.37 ± 0.71 %, which was three times higher than that of the UHT milk on day 6. There was no significant change in the degree of protein hydrolysis in the control samples. Protein hydrolysis was accompanied by gelation and even whey separation. Under the hydrolysis effect of protease, the content of free amino acid groups rose as time went on, which is in line with the findings of Zhang et al. (2019). The proteolysis in UHT milk throughout storage is primarily related to the original heat-resistant proteases generated by psychrophilic bacteria in the milk. There was no marked rise in the degree of protein hydrolysis in UHT milk without the addition of proteases, probably due to the short storage period and low residual proteases in UHT milk. Zarei et al. (2020) found that during UHT milk's shelf life, AprX residual activity leads to an enhancement in the degree of protein hydrolysis.
3.4. SDS-PAGE
In UHT milk, proteases have different hydrolytic abilities for different milk proteins, including α-casein, β-casein, κ-casein, α-lactalbumin, β-lactoglobulin, and so on. Analysis of UHT milk protein hydrolysis by proteases was performed via SDS-PAGE. Fig. 1e shows that κ-casein was hydrolyzed initially and was almost fully hydrolyzed by the time the UHT milk underwent deterioration on day 2. β-casein bands gradually weakened after 2 days, decreasing in size over time. And α-casein was the last to be hydrolyzed. Because of their globular structure, where the cleavage site of the enzyme is buried and not easily accessible, proteases had minimal impact on the hydrolysis of α-lactalbumin and β-lactoglobulin during the storage period. (Huang et al., 2022). It can be seen that the protease will preferentially hydrolyze casein to a much greater extent than whey protein. This is consistent with prior studies (Dacres et al., 2023; Yang et al., 2021; Zhang et al., 2018). AprX preferentially cleaves κ-casein, which leads to reduced repulsion among casein micelles and ultimately to the formation of insoluble aggregates, whereas fibrinolytic enzymes specifically hydrolyze β-casein and αS-casein (Zhang, Bijl, et al., 2020). Small molecular weight bands were also observed in protease-added milk samples, which may be small-molecule peptides generated by protease hydrolysis of proteins (Paludetti et al., 2020). Matéos et al. (2015) studied the effect of purified Pseudomonas LBSA1 protease on milk proteolysis and found that β-casein was the first to be hydrolyzed, followed by αS-casein and κ-casein. The reason for this difference may be due to the amount of protease and the duration of hydrolysis, which occurs with κ-casein when samples are incubated for a prolonged period with a sufficient amount of protease (France et al., 2021).
3.5. Particle size and Zeta potential
The changes in particle size of the samples during storage are shown in Fig. 1c. The initial particle size of the UHT milk and protease-added samples were 153.1 ± 10.7 nm and 187.2 ± 14.5 nm, respectively. There was no significant change in the particle size of the UHT milk without protease during storage. From day 4, the particle size of UHT milk with protease increased significantly to 167.8 ± 7.3 nm (P < 0.01). This indicates that the samples formed a gel from day 4 and the system tended to be unstable. At 6 d, the aggregation of casein micelles, accompanied by the appearance of hydrolyzed peptides, resulted in a maximum particle size of 1572.0 ± 181.5 nm. Fan et al. (2024) observed that the particle size of UHT milk stored at 25 °C for 45 d and 60 d increased to 565.77 nm and 1070 nm, respectively. The degree of protein aggregation can be reflected by the size of casein micelles.
Fig. 1d illustrates the Zeta potential of the samples. The absolute value of the Zeta potential indicates the dispersion or aggregation of the UHT milk system. A higher absolute value indicates that the particles in the system are more dispersed and the system is more homogeneous, on the contrary, the system is unstable. There was no significant change in the Zeta potential of UHT milk when stored at 37 °C. The absolute value of UHT milk with added protease decreased significantly (P < 0.01) from 4 d. At 6 d, it decreased to −25.0 ± 0.6 mV. This shows a decrease in the stability of the system from day 4, which corresponds to the results of the particle size measurements. With the increase in storage time, the negative charge of casein micelles in protease-added UHT milk decreased, and aggregation occurred due to the weakening of electrostatic interactions between the particles, leading to a decrease in the stability of UHT milk. Qin et al. (2023) added P. fluorescens to UHT milk, stored at 25 °C, showing a decreasing trend in the absolute value of the potential of the samples, which is similar to the results of this study.
3.6. Peptidomic analysis
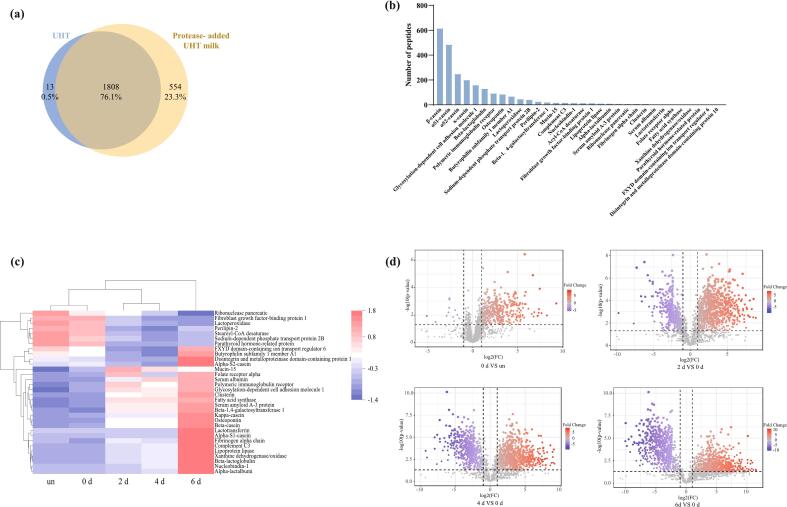
2479 peptides were detected in the samples of UHT milk and protease-added UHT milk, which were stored for 0 d, 2 d, 4 d, and 6 d. As shown in Fig. 2a, 13 and 554 peptides were identified respectively in UHT milk and protease-added UHT milk accounting for 23.8 % of the total peptides. The total peptides in the two groups of samples was 1808, accounting for 72.9 % of the total number of peptides. The quantity of peptides in UHT milk significantly increased following the protease addition. This suggests that proteases will cleave specific sites in the proteins, causing peptides released during UHT milk storage (Dalabasmaz et al., 2019; Matéos et al., 2015). This result matches the increase in protein hydrolysis, indicating that proteins in UHT milk are hydrolyzed by proteases. The 32 proteins linked to the identified peptides, which were primarily derived from caseins, are shown in Fig. 2b. Among them, peptides from β-casein, αS1-casein, αS2-casein, and κ-casein accounted for more than 8 % of the total peptides, produced 615, 485, 248, and 200 entries, respectively. This suggests that β-casein, α-casein, and κ-casein are susceptible to hydrolysis, similar to the findings of previous research analyzing endogenous peptides (Wölk et al., 2021).
Fig. 2.
(a) Venny diagram of peptides identified in UHT milk and protease-added UHT milk. The overlapped part in the figure is the peptide shared by the two groups of samples. (b) The rank of peptides produced by 32 different types of proteins linked to the peptides that were identified. (c) Heatmap analysis of proteins linked to the identified peptides. “un”: UHT milk without addition of enzyme; “0 d, 2 d, 4 d, 6 d”: UHT milk with enzyme stored for 0 days, 2 days, 4 days, and 6 days. The change of the grid from red to blue means that the peptide content goes from high to low. (d) ∼ (g) Volcano plot of identified peptides in periods. The increase in peptide content is indicated by red circular markers. The reduction in peptide content is indicated by a blue circular mark. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Heatmap analysis in Fig. 2c shows that samples stored for 2 days, 4 days, and 6 days after protease addition were significantly different compared to samples stored for 0 d and without protease addition. The distribution of protein content associated with the identified peptides was approximately the same in the samples stored for 0 d and without protease addition. The peptide content from proteins such as β-casein, osteopontin, serum albumin, and clusterin gradually increased on day 2 and day 4 of storage and peaked on day 6, when the difference in peptide content enhancement became significant. The volcano plot shows the peptide alterations during the deterioration of UHT milk. As shown in Fig. 2d, the relative content of 470 peptides was increased, and the relative content of 16 peptides was decreased in protease-added UHT milk stored for 0 d compared to UHT milk without added protease. During the storage of protease-added UHT milk, there was an increase in the relative content of 942 peptides and a decrease in the relative content of 542 peptides from 0 d to 2 d. From 0 d to 4 d, the relative content of peptides increased by 912 and decreased by 661. From 0 d to 6 d, there was an increase in the relative content of 760 peptides and a decrease in the relative content of 697 peptides. The increased number of peptides is reflected in the heatmap. The production of new peptides indicates that the released peptides and proteins are undergoing protein hydrolysis (Aksoy et al., 2024).
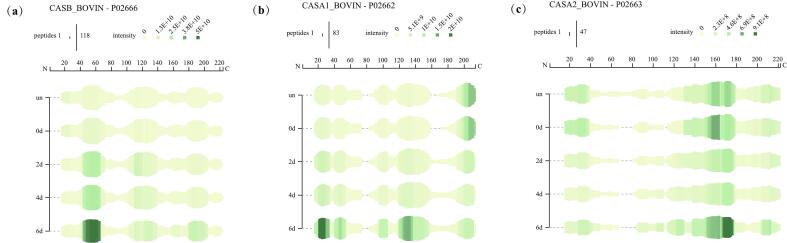
3.7. Peptide diagram analysis
Peptidomics analysis showed that significantly increased peptides (in the top 10 %) were mainly from β-casein, αS1-casein, and αS2-casein, as shown in Fig. 2b, which is similar to the findings of Aguilera-Toro et al. (2023). Fig. 3 displays the plotted peptide profile. The findings in Fig. 3a and Fig. 3b show that the N-termini of the amino acid sequences of β-casein and αS1-casein are susceptible to hydrolysis by protease. The peptide composition of the samples stored for day 2 d, day 4, and day 6 changed significantly compared to those of day 0 and samples without added protease, with an increase in peptide number starting on day 2 and peaking on day 6, indicating that the protein was hydrolyzed, which was consistent with the results of SDS-PAGE. Among the increased peptides, those related to β-casein mainly originated from positions 44–66 of the sequence, and those related to αS1-casein mainly originated from positions 21–34 and 44–54 of the sequence. The hydrolysis of αS2-casein occurs at the C-terminal end of its amino acid sequence, as seen by Fig. 3c. There is no significant difference between samples stored for 0 d and those without protease addition, and the number of peptides originating from positions 167–177 of the sequence reaches a maximum on day 6. These are different from previous studies. Dalabasmaz et al. (2019) have found that the β-casein192–206, alone or along with β-casein139–161, could monitor protein hydrolysis during UHT milk storage. The reason for this phenomenon could be differences in sample preparation (no bacterial protease was added directly to the UHT milk in this study, and the described peptides were endogenous) and enzymatic cleavage sites (the previous study was mainly based on the enzymatic cleavage site of histone B for analysis). Zhang et al. (2024) found that the C-terminus of the β-casein amino acid sequence exhibited susceptibility to hydrolysis, with the resulting peptides predominantly originating from the regions corresponding to positions 80–120 and 160–209 within the sequence. These results suggested that proteases significantly contributed to the process of age-gelatinization during the storage of UHT milk.
Fig. 3.
Peptide diagram of β-casein (a)、αS1-casein (b)、αS2-casein (c). The deepening of the green color means an increase in protein abundance. The length of the green line represents the number of peptides at a given location. The horizontal coordinate indicates the position of the peptide sequence. “un”: UHT milk without addition of protease; “0 d, 2 d, 4 d, 6 d”: UHT milk with enzyme stored for 0 days, 2 days, 4 days, 6 days. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.8. Characteristic peptide sequence analysis
Evaluation of spoilage deterioration of UHT milk can be achieved by the screened characteristic peptides. A total of 17 distinctive peptides associated with UHT milk spoilage were screened based on fold change (FC) >500 and P < 0.05, as shown in Table 2. Table 2 shows that the quantities of hydrolyzed peptides from αS1-casein, β-lactoglobulin, and β-casein grew considerably during the deterioration of UHT milk. Among them, 6 kinds of peptides were derived from αS1-casein, with FC ranging from 95.5 to 2928.4, which was greater than those contributed by β-lactoglobulin (82.9 to 2892.6) and β-casein (28.0 to 1797.7). As found by Dalabasmaz et al. (2019), marker peptides were primarily from β-casein, with a notable rise in peptide content, as the UHT milk's shelf life was coming to an end.
Table 2.
Changes in FC of the characteristic peptides during the storage period.
| serial | Peptide sequence | FC (0–2 d) |
FC (0–4 d) |
FC (0–6 d) |
Source protein | Position |
|---|---|---|---|---|---|---|
| 1 | HAQQKEPMIGVNQE | 359.5 | 590.8 | 2928.4 | αS1-casein | 143–156 |
| 2 | HAQQKEPMIG | 152.4 | 384.7 | 1909.5 | αS1-casein | 143–152 |
| 3 | IKHQGLPQE | 95.5 | 188.6 | 1189.5 | αS1-casein | 21–29 |
| 4 | FYQLDAYPSGAW | – | 235.3 | 1128.3 | αS1-casein | 168–179 |
| 5 | IVPNSVEQKHIQKEDVPSERY | 100.7 | 181.5 | 1104.7 | αS1-casein | 86–106 |
| 6 | VFGKEKVNE | 89.0 | 176.8 | 1011.7 | αS1-casein | 46–54 |
| 7 | KIDALNENKV | 202.6 | 434.8 | 2892.6 | β-lactoglobulin | 99–108 |
| 8 | AASDISLLDAQSAPLRV | 82.9 | 244.2 | 1539.8 | β-lactoglobulin | 41–57 |
| 9 | IAEKTKIPA | 114.0 | 165.2 | 894.6 | β-lactoglobulin | 88–96 |
| 10 | AMAASDISLLDAQSAP | 100.9 | 182.9 | 848.6 | β-lactoglobulin | 39–54 |
| 11 | KIIAEKTKIPA | 88.4 | 162.7 | 789.9 | β-lactoglobulin | 86–96 |
| 12 | TRINKKIEKFQSEEQQQTEDELQDKIHP | 459.3 | 547.3 | 1797.7 | β-casein | 39–66 |
| 13 | LTLTDVENLHLPLP | 92.3 | 28.0 | 1520.7 | β-casein | 140–153 |
| 14 | KVLPVPQKAVPYPQRDMP | 62.5 | 144.5 | 1289.3 | β-casein | 184–201 |
| 15 | LYQGPIVLNPWDQVKRN | 154.9 | 277.5 | 1106.1 | αS2-casein | 114–130 |
| 16 | LIKEQYEGR | 262.7 | 512.1 | 2786.3 | Polymeric immunoglobulin receptor | 403–411 |
| 17 | LNKPEDETHLE | – | 483.9 | 1661.3 | Glycosylation-dependent cell adhesion molecule 1 | 20–30 |
Note: FC is the fold change of the samples.
In Table 2, the quantity of αS1-casein143–156 (HAQQQKEPMIGVNQE) increased 359.5 times within 0–2 days, 590.8 times within 0–4 days, and 2928.4 times within 0–6 days. The peptide significantly increased in quantity during the storage period of the samples, and the FC increased with the degree of deterioration of the UHT milk, which suggested that the peptide can be used as a marker peptide in the process of UHT milk deterioration. Similarly, when a significant increase in 17 peptides, such as αS1-casein143–156 in Table 2, was detected in UHT milk, it signaled the beginning of the deterioration of UHT milk. The study of Zhang et al. (2024) differs from ours, illustrating that a total of 23 peptides were identified as characteristic markers of age-gelatinized direct-steam-infusion UHT (dUHT) milk, particularly, the peptide β-casein (L139-K176) exhibited a significant increase during the age-gelation process. In that study, the peptides ITVDDKHYQK (I71-K80) and FFVAPFPEVFGKEK (F23-K36) were recognized as hydrolysis markers for αS2-casein and αS1-casein, respectively.
3.9. Correlation analysis between peptide, protease activity, and degree of proteolysis
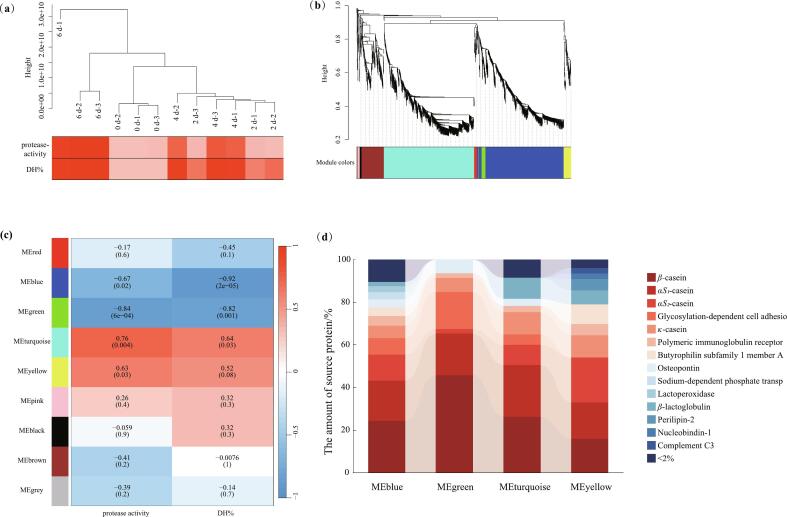
All peptides identified were analyzed for correlation with protease activity and protein hydrolysis degree by the WGCNA method. Sample dendrogram and trait heatmap (Fig. 4a) revealed the clustering of the number of peptides with protease activity and with protein hydrolysis degree. The samples' increased quantity of peptides linked to protease activity and protein hydrolysis is indicated by the light-to-dark red color. The number of peptides, protease activity, and protein hydrolysis degree of protease-added UHT milk was the lowest on day 0. The number of peptides was more similar on day 1 and day 4 of storage, and the number of peptides reached its highest on day 6 of storage. The amount of peptides increased with the rise in protease activity and protein hydrolysis degree during the storage period. To further analyze the correlation between the number of characteristic peptides, protease activity, and protein hydrolysis degree, cluster analysis was performed. The module-phenotype feature correlation mapping in Fig. 4c was performed for each module to correlate peptides with enzyme activity and protein hydrolysis degree. A positive correlation is suggested by the darker red when the P-value nears 1. Closer to −1, a negative correlation is denoted by darker blue. As can be seen from Fig. 4c, among the nine modules obtained from the analysis, four modules (MEblue, MEgreen, MEturquoise, and MEyellow) showed a strong correlation (|r| > 0.6, P < 0.05) with protease activity. The degree of protein hydrolysis was highly correlated with three modules (MEblue, MEgreen, and MEturquoise) (|r| > 0.6, P < 0.05). Among them, protease activity and protein hydrolysis showed a negative correlation with MEblue and MEgreen. Nevertheless, MEturquoise and MEyellow were positively correlated with protease activity. Proteomic analysis of dUHT milk by Zhang et al. (2024) showed an increase in proteases, such as ribonuclease A family and cathepsin D in age-gelatinised milk. These enzymes play a pivotal role in the alterations of metabolites and protein hydrolysis throughout the age gelation, leading to the instability of direct ultra-high-temperature (dUHT) milk.
Fig. 4.
(a) Sample dendrogram and trait heatmap. The height on the y-axis represents the distance between clusters, and different heights correspond to different modules, which are indicated by different colors. There are three parallels in each set of samples. (b) Cluster Dendrogram. Peptides with close clustering are classified into the same module, i.e., module colors in the figure, indicating the module colors after merging the similarities. (c) Module-trait relationships. The grid displays the P-value together with the correlation of modules on the vertical (per module) and horizontal coordinates (protease activity and proteolytic degree). (d) Percentage of source proteins of peptides in different modules. The shift in color from red to blue signifies the amount of source protein in different modules from high to low. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The percentage of proteins from which the characteristic peptides of each module originated is shown in Fig. 4d. 61.14 %, 73.91 %, 70.35 %, and 64.47 % of the characteristic peptides in the modules MEblue, MEgreen, MEturquoise, and MEyellow, respectively, originated from casein. In MEblue, MEgreen, and MEturquoise, most of them were peptides derived from β-casein, accounting for 24.23 %, 45.65 %, and 26.20 %, respectively. 21.05 % of the peptides in MEyellow were derived from αS2-casein. This suggests that β-casein is mainly associated with changes in UHT milk protease activity and protein hydrolysis, and αS2-casein is hydrolyzed with the rise in protease activity. In addition, all 17 characteristic peptides in Table 2 were detected in the MEturquoise module. This indicated that these 17 peptides were positively correlated with protein hydrolysis degree and protease activity. When the content of these peptides was reduced, the degree of protein hydrolysis and protease activity in UHT milk decreased. Class et al. (2024) monitored the shelf life of heat-treated cow's milk. The study's findings demonstrated that the a Python-based algorithm was able to identify eight potential trypsin-labeled peptides, seven of which were sourced from casein.
4. Conclusion
The peptidomics approach was used to investigate the patterns of protein hydrolysis and peptide changes during the deterioration of UHT milk under the action of psychrophilic bacteria proteases. During the deterioration of UHT milk, protease activity, and protein hydrolysis increased significantly, accompanied by the gradual degradation of casein. Under 37 °C storage conditions, the deterioration of protease-added UHT milk started on day 2, and the modified peptides primarily came from β-casein, αS1-casein, and αS2-casein. Among them, the number of peptides derived from β-casein was the highest at 25.8 %. 17 peptides, including αS1-casein143–156, β-lactoglobulin99–108, and β-casein39–66, were identified. These characteristic peptides can be used as markers to monitor and predict protein hydrolysis during UHT milk storage. Analysis of the correlations showed that β-casein was correlated with protease activity and protein hydrolysis, and αS2-casein was negatively correlated with protease activity and protein hydrolysis. This suggested that hydrolysis of UHT milk by extracellular proteases of psychrophilic bacteria altered the overall composition of UHT milk, leading to age gelation. The identified peptides correlated with changes in protease activity and protein hydrolysis, further elucidating the peptide changes in UHT milk during deterioration.
CRediT authorship contribution statement
Jinyu Xu: Writing – original draft, Software, Methodology, Formal analysis. Xiaodan Wang: Writing – review & editing, Software, Methodology, Formal analysis. Xiaoxuan Zhao: Writing – review & editing, Methodology, Formal analysis. Hongyu Cao: Writing – review & editing, Methodology, Formal analysis. Yunna Wang: Visualization, Formal analysis. Ning Xie: Visualization, Formal analysis. Xu Li: Visualization, Formal analysis. Xiaoyang Pang: Project administration, Funding acquisition, Formal analysis. Jiaping Lv: Supervision, Project administration, Funding acquisition, Conceptualization. Shuwen Zhang: Writing – review & editing, Validation, Supervision, Methodology, Funding acquisition, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Key R&D Program of Yunnan (202402AE090033), National Center of Technology Innovation for Dairy (2022-KFKT-20), Agriculture Research System of China-National Dairy Industry and Technology System (CARS-36).
Contributor Information
Xiaoyang Pang, Email: pangxiaoyang@163.com.
Jiaping Lv, Email: lvjiapingcaas@126.com.
Shuwen Zhang, Email: zswcaas@hotmail.com.
Data availability
Data will be made available on request.
References
- Aguilera-Toro M., Nielsen S.D., Kragh M.L., Xiao Y., Hansen L.T., Rauh V.…Larsen L.B. Peptidomic fingerprints of stored UHT milk inoculated with protease extracts from different Pseudomonas strains relative to AprX expression and visible spoilage. Dairy. 2023;4:83–97. doi: 10.3390/dairy4010005. [DOI] [Google Scholar]
- Akillioğlu H.G., Chatterton D.E.W., Lund M.N. Maillard reaction products and amino acid cross-links in liquid infant formula: Effects of UHT treatment and storage. Food Chemistry. 2022;396 doi: 10.1016/j.foodchem.2022.133687. [DOI] [PubMed] [Google Scholar]
- Aksoy S., Kayili H.M., Atakay M., Kirmaci H.A., Salih B. Dynamics of peptides released from cow Milk fermented by kefir microorganisms during fermentation and storage periods. International Dairy Journal. 2024;105970 doi: 10.1016/j.idairyj.2024.105970. [DOI] [Google Scholar]
- Anema S.G. Storage stability and age gelation of reconstituted ultra-high temperature skim milk. International Dairy Journal. 2017;75:56–67. doi: 10.1016/j.idairyj.2017.06.006. [DOI] [Google Scholar]
- Class L.-C., Kuhnen G., Hanisch K.L., Badekow S., Rohn S., Kuballa J. The shelf life of milk - A novel concept for the identification of marker peptides using multivariate analysis. Foods. 2024;13(6):831. doi: 10.3390/foods13060831. https://www.mdpi.com/2304-8158/13/6/831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dacres H., Weihs F., Wang J., Anderson A., Trowell S.C. Bioluminescence resonance energy transfer biosensor for measuring activity of a protease secreted by Pseudomonas fluorescens growing in milk. Analytica Chimica Acta. 2023;1270 doi: 10.1016/j.aca.2023.341401. [DOI] [PubMed] [Google Scholar]
- Dai L., Hu S., Pang X., Zhang S., Yu D., Zhang Y.…Lu G. Community diversity of psychrophilic bacteria in dairy farm raw milk and its characteristic enzyme production at different temperature. Food Bioscience. 2023;54 doi: 10.1016/j.fbio.2023.102921. [DOI] [Google Scholar]
- Dalabasmaz S., Dittrich D., Kellner I., Drewello T., Pischetsrieder M. Identification of peptides reflecting the storage of UHT milk by MALDI-TOF-MS peptide profiling. Journal of Proteomics. 2019;207 doi: 10.1016/j.jprot.2019.103444. [DOI] [PubMed] [Google Scholar]
- D’Incecco P., Rosi V., Fortina M.G., Sindaco M., Ricci G., Pellegrino L. Biochemical, microbiological, and structural evaluations to early detect age gelation of milk caused by proteolytic activity of Pseudomonas fluorescens. European Food Research and Technology. 2022;248(8):2097–2107. [Google Scholar]
- Ebner J., Baum F., Pischetsrieder M. Identification of sixteen peptides reflecting heat and/or storage induced processes by profiling of commercial milk samples. Journal of Proteomics. 2016;147:66–75. doi: 10.1016/j.jprot.2016.03.021. [DOI] [PubMed] [Google Scholar]
- Fan K., Wu P., Guo M., Wang Y., Cao Y., Wang P.…Luo J. Destabilization of ultra-instantaneous ultra-high-temperature sterilized milk stored at different temperatures. Journal of Dairy Science. 2024;107(8):5460–5472. doi: 10.3168/jds.2024-24705. [DOI] [PubMed] [Google Scholar]
- France T.C., O’Mahony J.A., Kelly A.L. Agents of Change: Enzymes in Milk and Dairy Products; 2021. The plasmin system in milk and dairy products; pp. 11–55. [Google Scholar]
- Glück C., Rentschler E., Krewinkel M., Merz M., von Neubeck M., Wenning M., Fischer L. Thermostability of peptidases secreted by microorganisms associated with raw milk. International Dairy Journal. 2016;56:186–197. doi: 10.1016/j.idairyj.2016.01.025. [DOI] [Google Scholar]
- Guo M., Wang Y., Wang P., Luo J., Qian W., Li H.…Ren F. Multiscale structure analysis reveals changes in the structure of casein micelles treated with direct-steam-infusion UHT. Food Hydrocolloids. 2024;153 doi: 10.1016/j.foodhyd.2024.110033. [DOI] [Google Scholar]
- Huang J., Zhang L., Lan H., Zhou P. How to adjust α-lactalbumin and β-casein ratio in milk protein formula to give a similar digestion pattern to human milk? Journal of Food Composition and Analysis. 2022;110 doi: 10.1016/j.jfca.2022.104536. [DOI] [Google Scholar]
- Langfelder P., Horvath S., Cai C., Dong J., Miller J., Song L., Yip A., Zhang B. 2023. Weighted correlation network analysis (WGCNA) v1.72-5 (version 1.72-5) [package "WGCNA"]http://horvath.genetics.ucla.edu/html/CoexpressionNetwork/Rpackages/WGCNA/ December, 7. [Google Scholar]
- Leite J.A.S., Montoya C.A., Loveday S.M., Maes E., Mullaney J.A., McNabb W.C., Roy N.C. Heat-treatments affect protease activities and peptide profiles of ruminants’ milk. Frontiers in Nutrition. 2021;8 doi: 10.3389/fnut.2021.626475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matéos A., Guyard-Nicodème M., Baglinière F., Jardin J., Gaucheron F., Dary A.…Gaillard J.L. Proteolysis of milk proteins by AprX, an extracellular protease identified in Pseudomonas LBSA1 isolated from bulk raw milk, and implications for the stability of UHT milk. International Dairy Journal. 2015;49:78–88. doi: 10.1016/j.idairyj.2015.04.008. [DOI] [Google Scholar]
- Ning J., Li M., Chen W., Zhao H., Chen J., Yang M.…Yue X. Peptidomics as a tool to analyze endogenous peptides in milk and milk-related peptides. Food Bioscience. 2022;50 doi: 10.1016/j.fbio.2022.102199. [DOI] [Google Scholar]
- Paludetti L.F., Kelly A.L., Gleeson D. Effect of thermoresistant protease of Pseudomonas fluorescens on rennet coagulation properties and proteolysis of milk. Journal of Dairy Science. 2020;103(5):4043–4055. doi: 10.3168/jds.2019-17771. [DOI] [PubMed] [Google Scholar]
- Qin X., Cheng J., Qi X., Guan N., Chen Q., Pei X.…Man C. Effect of thermostable enzymes produced by psychrotrophic bacteria in raw milk on the quality of ultra-high temperature sterilized milk. Foods. 2023;12(20):3752. doi: 10.3390/foods12203752. https://www.mdpi.com/2304-8158/12/20/3752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherfurd S.M. Methodology for determining degree of hydrolysis of proteins in hydrolysates: A review. Journal of AOAC International. 2010;93(5):1515–1522. [PubMed] [Google Scholar]
- Stoeckel M., Lidolt M., Achberger V., Glück C., Krewinkel M., Stressler T.…Hinrichs J. Growth of Pseudomonas weihenstephanensis, Pseudomonas proteolytica and Pseudomonas sp. in raw milk: Impact of residual heat-stable enzyme activity on stability of UHT milk during shelf-life. International Dairy Journal. 2016;59:20–28. doi: 10.1016/j.idairyj.2016.02.045. [DOI] [Google Scholar]
- Wang J., Xi Y., Sun B., Deng J., Ai N. Utilization of low-temperature heating method to improve skim milk production: Microstructure, stability, and constituents of milk fat globule membrane. Food Chemistry: X. 2024;21 doi: 10.1016/j.fochx.2024.101187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wölk M., Gebauer C., Hoffmann R., Milkovska-Stamenova S. Analysis of the endogenous peptidomes of different infant formula types and human milk. Foods. 2021;10(11) doi: 10.3390/foods10112579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Wang Z., Zhang C., Wang L., Pang L., Zhang D.…Jiang Y. Assessment of the production of Bacillus cereus protease and its effect on the quality of ultra-high temperature-sterilized whole milk. Journal of Dairy Science. 2021;104(6):6577–6587. doi: 10.3168/jds.2020-19818. [DOI] [PubMed] [Google Scholar]
- Zarei M., Mohammadpour H., Gharibi D., Pourmahdi Borujeni M. Identification of Pseudomonas jessenii and Pseudomonas gessardii as the most proteolytic Pseudomonas isolates in Iranian raw milk and their impact on stability of sterilized milk during storage. The Journal of Dairy Research. 2020;87(3):368–374. doi: 10.1017/S0022029920000709. [DOI] [PubMed] [Google Scholar]
- Zhang C., Bijl E., Hettinga K. Destabilization of UHT milk by protease AprX from Pseudomonas fluorescens and plasmin. Food Chemistry. 2018;263:127–134. doi: 10.1016/j.foodchem.2018.04.128. [DOI] [PubMed] [Google Scholar]
- Zhang C., Bijl E., Muis K.E., Hettinga K. Stability of fat globules in UHT milk during proteolysis by the AprX protease from Pseudomonas fluorescens and by plasmin. Journal of Dairy Science. 2020;103(1):179–190. doi: 10.3168/jds.2019-17150. [DOI] [PubMed] [Google Scholar]
- Zhang C., Bijl E., Svensson B., Hettinga K. The extracellular protease AprX from Pseudomonas and its spoilage potential for UHT milk: A review. Comprehensive Reviews in Food Science and Food Safety. 2019;18(4):834–852. doi: 10.1111/1541-4337.12452. [DOI] [PubMed] [Google Scholar]
- Zhang C., Boeren S., Zhao L., Bijl E., Hettinga K. The impact of low-temperature inactivation of protease AprX from Pseudomonas on its proteolytic capacity and specificity: A peptidomic study. Dairy. 2023;4(1):150–166. https://www.mdpi.com/2624-862X/4/1/11 [Google Scholar]
- Zhang D., Li S., Palmer J., Teh K.H., Leow S., Flint S. The relationship between numbers of Pseudomonas bacteria in milk used to manufacture UHT milk and the effect on product quality. International Dairy Journal. 2020;105 doi: 10.1016/j.idairyj.2020.104687. [DOI] [Google Scholar]
- Zhang T., Liu Y., Cao J., Jiang L., Lin K., Wang P.…Yi H. Milk serum peptidomics revealed the age gelation of direct UHT milk. Food Chemistry. 2024;456 doi: 10.1016/j.foodchem.2024.140012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.