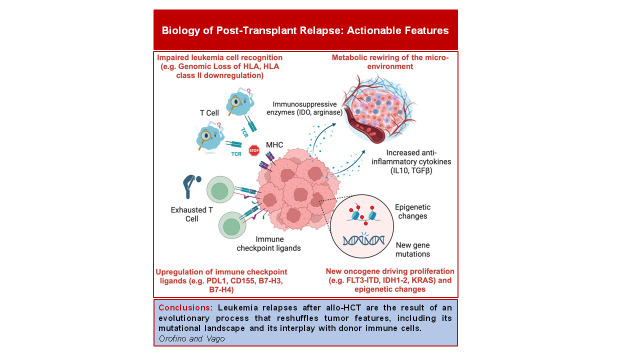
Visual Abstract
Abstract
In patients receiving allogeneic hematopoietic cell transplantation to cure acute myeloid leukemia (AML), recurrence of the underlying disease, or relapse, represents a crucial unanswered issue and prominent cause of mortality. Still, over recent years, advancements in omic technologies have allowed us to gain new insights into the dynamic changes occurring in cancer and the host over the course of treatments, providing a novel evolutionary perspective on the issue of disease relapse. In this review, we summarize current knowledge on the molecular features of relapsing AML, with a specific focus on changes in the mutational asset of the disease and in the interplay between the tumor and the donor-derived immune system. In particular, we discuss how this information can be translated into relevant indications for monitoring transplanted patients and selecting the most appropriate therapeutic options to prevent and treat relapse.
Learning Objectives
Review the mutational landscape of relapsing leukemia, with a focus on mutations relevant for molecular monitoring and targeted intervention
Summarize knowledge on post-transplant immune escape modalities and on specific strategies to circumvent them
CLINICAL CASE
A 25-year-old man with no relevant previous medical history is diagnosed with acute myeloid leukemia (AML). Disease characterization at presentation evidences monosomal karyotype and an oncogenic mutation in ASXL1, thus the patient is stratified as high risk according to European Leukemia Net (ELN) 2022 classification.1 After 2 induction cycles of intensive chemotherapy, the patient achieves complete remission with incomplete hematologic recovery and proceeds to allogeneic hematopoietic cell transplantation (alloHCT). Due to the lack of human leukocyte antigens (HLA)-identical siblings and of rapidly available and adequately matched unrelated donors, the HLA-haploidentical 22-year-old brother is selected as donor, and a myeloablative peripheral blood stem cell transplant is performed, followed by high-dose cyclophosphamide, tacrolimus, and mycophenolate mofetil as graft-versus-host disease (GvHD) prophylaxis. No major clinical complication occurs over the early post-transplant phase, and the patient is discharged from the hospital at day 32 after alloHCT.
Introduction
Disease relapse after transplantation represents a crucial unsolved clinical issue, jeopardizing the results obtained in a complex and delicate procedure such as alloHCT, with major medical, psychological, and socioeconomic implications.2
A traditional and rather superficial perception regarding relapse is that treatment “was not enough” to eradicate residual malignant cells, often leading to considerations about the feasibility of increasing doses and the associated toxicities. Whereas in some cases this concept may lead to remarkable results (as for instance demonstrated by the effectiveness in some patients of infusion of fresh donor lymphocytes to boost the immunotherapeutic effects of alloHCT), frequently this is not feasible or effective.
Recently, the perception of the biological causes of treatment failure has changed, thanks to studies that investigated intrapatient cancer heterogeneity and the genetic and nongenetic changes that accumulate during cancer progression. This perspective embraces the Darwinian evolutionary theory and identifies relapse as the selection of variants of the original disease that are resistant to the mechanism of action of the employed therapy; for this reason, caution in reusing treatments that have the same targets is warranted. In particular, in the fine balance between leukemia clones (and epiclones), alloHCT represents a dramatic evolutionary bottleneck of importance: to be able to persist, subclones must carry the genetic mutations that grant them resistance to chemotherapy and be able to evade immune recognition mediated by donor cells transferred as part of the graft (graft-versus-tumor [GvT] effect).3
In this review, we provide an overview on current knowledge about how the mutational repertoire of AML is sculpted upon transplant and on known mechanisms employed by malignant cells to thwart immune elimination. In particular, we provide practical indications on how this biological evidence can inform the design of post-transplantation monitoring strategies and therapy selection.
Changes in the mutational landscape over the course of treatments
One of the major achievements of modern oncology is the possibility, thanks to next-generation sequencing (NGS) technologies, to easily and rapidly obtain the mutational asset of a tumor. At the time of disease presentation, a detailed disease mutational profile serves mainly 3 purposes: (1) determining the prognostic category to which the patient belongs; (2) identifying mutations that can confer sensitivity or resistance to specific drugs (summarized in Table 1); and (3) picking 1 or more alterations to employ as disease-specific markers during the subsequent follow-up.
Table 1.
AML mutations selectively targeted by drugs, and available studies in post-transplant maintenance and relapse
| Mutated gene | Drug | Trials for maintenance after alloHCT | Trials in relapsed AML |
|---|---|---|---|
| FLT3 | Midostaurin | 120 pts randomized to midostaurin vs standard of care. OS 85% vs 76% at 24 months (P = 0.34). NCT018833624 | 99 pts treated. ORR 71% in FLT3mut pts and 42% in FLT3wt pts. NCT000459429 |
| Sorafenib (active only in FLT3-ITD) | 83 pts randomized to sorafenib vs placebo. OS 90.5% vs 66.2% at 24 months (P = 0.007). DRKS000005915 202 pts randomized to sorafenib vs placebo. 2-year OS 82% vs 68% (P = 0.012). NCT024742906 |
29 pts treated. ORR 37, 9%, median OS 7.1 months.10 | |
| Crenolanib | A phase 2 trial is currently ongoing. NCT02400255 | 38 pts treated; ORR 47% at 14 weeks.11 | |
| Gilteritinib | 356 pts randomized to gilteritinib vs placebo. No difference in OS, but in pts with MRD higher RFS with gilteritinib (P = 0.0065). NCT029972027 | 371 pts randomized to gilteritinib vs chemotherapy. ORR 67.6% vs 25.8%, OS 9.3 months vs 5.6 months (P < 0.001). NCT0242193912 | |
| Quizartinib (active only in FLT3-ITD) | 13 pts treated, acceptable toxicities, OS 13-142 weeks; no difference in OS vs historical cohort. NCT014684678 | 367 pts randomized to quizartinib vs chemotherapy. OS 6.2 months vs 4.7 months (P = 0.02). NCT02039726 (Cortes et al.)13 | |
| IDH1 | Ivosidenib | 16 pts treated, PFS 81% at 2 years, OS 88% at 2 years. NCT0356482115 | 258 pts treated. ORR 41.6%. NCT0207483918 |
| Olutasidenib | 31 pts treated. ORR 22%. NCT0271957416 | ||
| IDH2 | Enasidenib | 19 pts treated, PFS 69% at 2 years, OS 74% at 2 years. NCT0351551214 | 229 pts treated. ORR 40.3%. NCT0191549817 |
| KMT2A fusion NPM1 NUP98 fusion |
Revumenib | 9 pts treated, CR maintained in 6/9 (66%). NCT0406539919 | 68 pts treated. ORR 53%, CR/CRh 30%. NCT0406539920 |
| JNJ-72576617 | A phase 1 trial is currently ongoing. NCT05453903 | ||
| Ziftomenib | A phase 1 trial is currently ongoing. NCT06440135 | A phase 1 trial is currently ongoing. NCT04067336 |
CR, complete remission; CRh, complete remission with partial hematological recovery; mut, mutated; ORR, overall response rate; OS, overall survival; PFS, progression-free survival; pts, patients; RFS, relapse-free survival; wt, wild type.
In detail, FLT3 mutations represent a relevant therapeutic target, with at least 5 drugs (midostaurin, sorafenib, crenolanib, gilteritinib, quizartinib) tested in both post HCT maintenance4-8 and therapy for relapsed disease9-13; among them, midostaurin and quizartinib are active only in the presence of FLT3-ITD mutations, while the others are effective in both ITD and TKD mutations.
Mutations in IDH1 and IDH2 also can be targeted with specific compounds (ivosidenib, enasidenib, and olutasidenib), and the new class of drugs termed menin inhibitors (revumenib, JNJ-72576617, ziftomenib) showed promising efficacy in patients carrying NPM1 mutations.14-20
Repeating a similar profiling at the time of relapse, after either chemotherapy or alloHCT, has shown that often the mutational asset changes, with some mutations that alter their quantitative representation in the tumor bulk (quantified through the variant allele frequency) and others that are gained or lost.21-25
Table 2 summarizes available literature on genetic changes occurring in AML upon alloHCT. In particular, one of the most relevant findings emerging from such studies is that the spectrum of gained and lost mutations after transplantation is similar to its counterpart after chemotherapy, with no specific recurrent mutation or recurrently altered pathway (with the remarkable exception of HLA genes, discussed in the next section). However, as shown by Hong and colleagues in a more recent study, relapses after alloHCT are often more different from the original disease presentation than their counterparts after chemotherapy and present a wider repertoire of possible novel alterations. For instance, in the authors' analysis, new IDH1 mutations (potentially targetable by small molecules under development as discussed previously and detailed in Table 1) were detected only in posttransplant relapses.21
Table 2.
Studies on the genetic changes occurring in AML at post-transplantation relapse
| Reference | Disease | N | Analysis of macroalterations | Mutational profiling |
|---|---|---|---|---|
| Bacher et al68 | AML | 26 | Yes (by standard cytogenetics and FISH): new genomic macroalterations in 20/26 relapses | No |
| Waterhouse et al25 | AML | 21 | Yes (by SNP arrays): new genomic macroalterations in 16/21 relapses, FLT3-ITD CN-LOH in 1/21 | No |
| Quek et al22 | AML | 29 | Yes (by standard cytogenetics and FISH): new genomic macroalterations in 16/29 relapses | Yes (by targeted NGS panel): changes in mutational profile in 13/29 relapses, new mutations in TET2, NRAS, WT1, ETV6, RUNX1, DNMT3A, TP53, NPM1, IDH1, FLT3 ITD, PHF6 |
| Christopher et al24 | AML | 15 | No | Yes (by WES): changes in mutational profile in 13/15 relapses, new mutations in NRAS, FLT3, WT1, STAG2 |
| Vosberg et al27 | AML | 12 | No | Yes (by WES): new WT1 mutations in 6/12 relapses |
| Toffalori et al41 | AML | 12 | Yes (by SNP arrays): new genomic macroalterations in 7/12 relapses, FLT3-ITD CN-LOH in 2/12 | No |
| Hong et al21 | AML/MDS | 49 | No | Yes (by targeted NGS panel): changes in mutational profile in 46/49 relapses, new IDH1 mutations |
| Pagliuca et al69 | AML/MDS | 55 | No | Yes (by targeted NGS panel, including HLA genes): HLA mutations in 9/40 diagnoses and 17/44 relapses, variable changes in non–HLA genes |
| Wienecke et al23 | AML | 59 | No | Yes (by WES): changes in mutational profile in 28/59 relapses, mutations in spliceosome and epigenetic modifiers stable, in signal transduction genes unstable |
FISH, fluorescence in situ hybridization; NGS, next-generation sequencing; SNP, single nucleotide polymorphism; WES, whole-exome sequencing.
Mutations in FLT3 and other genes that boost leukemia cell proliferation, including KRAS, NRAS, PTPN11, and KIT, are often found in subclonal fractions of the disease, indicating they are often late events in disease evolution.21-24 As such they are frequently gained (but also sometimes lost) in the massive reshaping that accompanies relapse. In particular, Waterhouse and collaborators were the first to show frequent copy neutral loss of heterozygosity (CN-LOH) of chromosome 13 at relapse in cases that originally carried the FLT3-ITD mutation, leading to double the allelic burden of the alteration.25 It should also be mentioned that targeting FLT3-ITD by specific inhibitors has the added effect of inducing the release from leukemic cells of IL-15, which after alloHCT might promote the GvT effect and synergize with donor lymphocyte infusion.26
Other mutations that have been described to appear frequently at relapse, or to increase their variant allele frequency if preexistent, include TP53 and WT1.21,22,24,27 Regarding the latter, loss-of-function mutations are reportedly positively selected upon alloHCT, suggesting that they might represent a modality of immune escape.27
Of particular relevance are those mutations that are present and clonal at disease diagnosis, and that are in most cases detected at all subsequent disease presentations, since they represent the best options to track eventual disease persistence. Belonging to this category are core binding factor gene fusions and mutations in epigenetic modifiers (ie, DNMT3A, TET2, ASLX1, IDH1-IDH2), spliceosome genes, and NPM1. In an interesting recent work, Wienecke and collaborators profiled the mutational asset of diagnoses and post-transplantation relapses and employed this information to analyze peripheral blood remission samples for presence of the mutations. They showed that this would allow detection of up to 38% and 64% of relapses, respectively, if monitoring was performed every 3 months or monthly. Also in this study, 27% of relapse mutations were newly acquired.23
Immune-related changes at relapse after alloHCT
AlloHCT represents a paradigmatic form of adoptive immunotherapy, transferring from the donor to the patient an entire immune system, composed of cells with a different specificity, mechanism of action, and maturation state. This allows for targeting simultaneously a plethora of targets on malignant cells, attacking residual malignant cells in multiple, possibly synergistic ways. However, clinical and experimental data have clearly shown over the years that some elements of the immune orchestra are dominant over others upon alloHCT, and in particular primary alloreactivity of T cells against incompatible HLA molecules supersedes and displaces responses against minor histocompatibility antigens and tumor-specific antigens.
Disease recurrence often originates when leukemic cells gain features that allow them to evade this dominant response, and it is becoming increasingly clear that different alternative modalities of immune escape exist, with different frequency in relation to time after transplant, donor-recipient matching, and other transplant-related variables, including graft composition and occurrence of GvHD, suggesting that all these factors shape the dominant immune response and thus the countermeasure necessary for leukemia to evade it.28-30 In this perspective, treating relapse should aim to shift the immune response to new targets, or even to equilibrate a deranged immunodominant GvT effect. In the next sections, we briefly summarize current knowledge on modalities of post-transplantation leukemia immune escape and present strategies to detect and counteract them.
Genomic loss of 1 HLA haplotype (HLA loss)
As mentioned in the previous paragraph, HLA molecules that are present in the patient but not in the donor represent the most potent targets of donor T-cell-mediated primary alloreactivity, being naturally recognized by an extremely high number of individual donor T cells without need of prior priming. Fifteen years ago, our group described for the first time that in the setting of alloHCT from haploidentical family donors a consistent proportion of relapses (up to one-third) presented genomic loss of 1 HLA haplotype through CN-LOH.31 Losing mismatched HLAs by duplicating the compatible haplotype dramatically reduces the immunogenicity of leukemic cells, rendering them “invisible” to the donor immune system. Thus, HLA loss represented the first evidence of a recurrent modality of immune escape responsible for leukemia relapse after alloHCT.
A number of subsequent studies confirmed the very high incidence of HLA loss in relapses after haploidentical HCT, independently from the type of post-transplant GvHD prophylaxis employed, although this modality of relapse is less frequent after HCT from partially mismatched unrelated donors (accounting for approximately 10% of relapses) and very rare after 10/10 matched unrelated donor transplants and, unexpectedly, umbilical cord blood transplants.32-34
The initial description of HLA loss was performed in the setting of T-cell-depleted haploidentical transplant combined with prophylactic donor lymphocyte infusion (DLI), evidencing this genetic rearrangement in 30% of the relapses.31 A similar frequency was documented in T-cell-replete platforms, including those with the use of post-transplant cyclophosphamide.32-35 No specific analysis is available regarding T-cell-depleted haploidentical transplants without subsequent use of DLIs.
Several methods are currently available to detect genomic loss of HLA haplotype, originally performed through HLA typing of purified leukemia blasts and single nucleotide polymorphism (SNP) arrays,36 and now possible through quantitative polymerase chain reaction (using the HLA-KMR assay)37 and NGS.38
Since their original description, HLA loss relapses have prompted considerations of salvage therapeutic approaches. The first clinically relevant observation is that use of DLIs should be avoided in this setting, since leukemia has lost their main targets while other tissues continue to present them, configuring a situation in which the risk of GvHD surpasses by far the expected therapeutic benefit. In fit patients, a viable option is second transplantation from a different donor, selected not only based on matching with the patient but specifically for being mismatched against the HLA-rearranged leukemia. While this could be achieved with cord blood units and adult unrelated donors, the best example comes from choosing a second haploidentical donor, matched with the patient for the other haplotype. Moreover, therapeutic strategies not based on conventional interactions between the donor T-cell receptors and target HLAs gain a specific rationale in this setting. In particular, we presented preclinical evidence on the rationale of using bispecific antibodies to bridge back donor T cells toward HLA loss blasts,39 and Wu and colleagues coherently administered the anti-CD19/CD3 bispecific T-cell engager blinatumomab to 4 patients with HLA loss relapses, all of them achieving complete remission, with minimal residual disease (MRD) negativity reached in 3.40
Based on current evidence, HLA loss testing should absolutely be performed for relapses after haploidentical HCT but would be preferable also in those after unrelated donor transplants, especially if relapse occurred late after transplant, since it has been shown that this type of relapse tends to occur later than its “classical” counterpart.33 However, regardless of the method employed, HLA loss detection must be performed in samples containing a significant proportion of malignant cells (preferably over 5%), thus necessitating either enrichment of these cells based on their immunophenotype or performance only at time of hematological relapse.
Downregulation of HLA class II molecules
Two independent studies focused on relapses without genomic HLA loss–identified downregulation of HLA class II genes (HLA-DR, -DQ, -DP) and of their master regulator (the class II major histocompatibility transactivator, CIITA) as an alternative modality employed by leukemic cells to hide from donor T cells and result in relapse after transplantation.41,42 Different from the mechanism presented in the previous section, this modality is primarily epigenetic and occurs with similar frequency (approximately 40% of relapses) after all type of transplants, including HLA-identical ones. Given the epigenetic basis of this alteration, it is possible to revert it. One possibility is to expose leukemic cells to interferon y (IFN-γ), which through an alternative CIITA promoter can drive the re-expression of HLA class II molecules on the cell surface. A clinical trial testing the use of recombinant human IFN-γ is ongoing, with promising preliminary results.43 Alternatively, IFN-γ can be released by activated T cells, either upon activation against other tissues (such as when GvHD occurs41) or when redirected against non–HLA targets on leukemic cells, as recently shown by an elegant study from Rimando and colleagues.44
A more ambitious possibility is to rewire the epigenetic alterations underpinning HLA class downregulation. Our group has identified polycomb repressive complex 2 (PRC2) as the key epigenetic driver of this immune escape modality, showing that the inhibition of EZH2, the catalytic subunit of PRC2 (in particular using tazemetostat, already clinically approved for hematological and solid tumors), was able to rescue HLA class II expression both in vitro and in vivo, reinstating leukemic recognition by T cells.45 A phase 1clinical trial testing the combination of tazemetostat with chemotherapy in refractory/relapsed AML is currently ongoing, permitting the enrollment of patients relapsed after alloHCT (NCT05627232).
Upregulation of ligands for inhibitory T-cell checkpoints
Another frequent immune-related alteration observed at relapse in leukemic cells is the upregulation of ligands for T-cell inhibitory receptors, in particular involving PD-L1, CD276/B7-H3, and CD155/PVRL2 in variable and composite combinations. This is accompanied by upregulation of multiple inhibitory receptors on donor T cells, including PD-1, TIM-3, and LAG-3, often mirroring the changes occurring in leukemia and more evident in central memory and memory stem T cells infiltrating the bone marrow of relapsed patients.41,46-48
The most rationale countermeasure to these changes is to block the inhibitory checkpoints using monoclonal antibodies. Ipilimumab, an anti-CTLA4 antibody, conveyed promising responses in initial studies, shown to be associated with upregulation of PD-1, HLA-DR, and ICOS on CD8+ cells.49,50 Recently, Garcia et al tested the combined administration of ipilimumab with decitabine in a multicenter phase 1 trial (ETCTN/CTEP 10026 study, NCT02890329).51 In transplanted patients, the overall response rate was only 20%, with rather short-lived remissions. Immune-related adverse events occurred in 44% of patients and did not appear to be associated with differential response; strong association of response with a high baseline ratio of T to AML cells was detected with single-cell RNA sequencing. Immune activation was only evident after ipilimumab exposure, which drove CD4+ T-cell differentiation and increased the frequency of marrow-infiltrating regulatory T cells. Of note, immune changes were more evident in extramedullary leukemia sites as compared with the bone marrow, suggesting a relevant role of microenvironmental niches in shaping the GvL effect. Similarly, PD-1 inhibitors, such as nivolumab and pembrolizumab,52,53 have been tested in the post-transplant relapse setting, with rare responses reported and significant toxicities (especially GvHD). Also the combination of nivolumab with hypomethylating agents tested in the recent phase 2 NIFAR study yielded only a 25% overall response rate, with another 25% of the patients achieving stable disease.54
Overall it appears that blockade of a single immune checkpoint, even in combination with other drugs, is insufficient to revert this escape modality, and new preclinical and clinical studies are urgently needed to identify alternative approaches.
In addition, TP53-mutated AML can generate around itself a cold tumor microenvironment, characterized by low infiltration of cytotoxic T cells and by the accumulation of immunosuppressive cells such as regulatory T cells and myeloid-derived suppressor cells.55 Similarly, FLT3-mutated AML are also frequently associated with reduced infiltration by effector T cells in the bone marrow, dampening their immune recognition.56
Microenvironmental changes
It is increasingly recognized that in hematological malignancies the tumor microenvironment (TME) is also significantly rewired and plays a key role in disease progression. This aspect is particularly interesting in a setting such as alloHCT, in which the immune and hematopoietic components of the bone marrow are replaced, but all the stromal components remain of host origin. One of the main modalities by which the TME acts on all its components in a concerted way is metabolic rewiring of T-cell activity: recently, Uhl and colleagues described indeed that upon alloHCT AML blasts enhance their production of lactic acid and that this in turn impairs T-Cell activity57; in this setting, an ongoing clinical trial is testing the administration of sodium bicarbonate following DLIs (NCT04321161) in relapsed AML. Also, drugs can interact with multiple components of the TME: in an interesting study, Vallet and colleagues showed that azithromycin alters the proportion of immune subsets circulating in patients and inhibits T-cell cytotoxicity against tumor cells by altering their metabolism, ultimately explaining the excess of relapse incidence observed in the ALLOZITHRO trial in patients receiving this antibiotic. In addition, they identified a Bacteroides taxon enriched in the enterobiome of relapsing patients, showing an association to a specific plasm a metabolite signature that ultimately favored the accumulation of exhausted T cells,58 in line with the findings from Van de Brink's group and suggesting a key role of microbiome for prediction and treatment of leukemia relapse.59 Moreover, the tissue damage associated with conditioning regimen and immune complications, such as GvHD, leads to oxidative stress and increases reactive oxygen species in the cells, which in turn have been shown to cause oxidative DNA damage and to dramatically hamper T-cell activation,60 supporting the rationale for implementing antioxidant therapies.
Although mostly investigated outside the setting of alloHCT, the production of cytokines from leukemic cells has also been convincingly shown to dampen immune recognition. For instance, chronic myeloid leukemia cells produce transforming growth factor (TGF-β),59 which antagonizes the CIITA/MHC-II axis and thereby renders leukemia cells less immunogenic.60 In addition, AML blasts can produce both cytokines (IL-4 and IL-10)61,62 and enzymes (indoleamine 2,3-dioxygenase-1 and arginase)63,64 that rewire the surrounding immune microenvironment toward a tolerant/immunosuppressive program, enriched in regulatory T cells and M2-like monocytes.65,66
CLINICAL CASE (continued)
Since the NGS panel employed at the time of diagnosis to characterize the mutational asset of the patient is not designed or validated for quantification of MRD, alloHCT monitoring is performed employing multiparametric flow cytometry, cytogenetics, and bone marrow chimerism. After 1 year of remission, a small cluster of CD34+ cells (0.57% of bone marrow mononucleated cells) displaying the original pathological phenotype is detected. CD34+ cells are enriched by magnetic bead selection, and complete mutational profiling by NGS is performed together with HLA loss analysis by HLA-KMR. Mutational profile results are largely superimposable with the one at disease presentation, and in particular no new targetable mutation is present. HLA-KMR provides evidence of HLA haplotype loss. Based on this result, DLIs are withheld, and the patient receives 2 cycles of preemptive azacytidine + venetoclax while undergoing fitness evaluation for a second transplant. The patient reobtains multiparametric flow cytometry complete remission and is currently in remission 3 years after a second haploidentical HCT from the mother, sharing the other HLA haplotype.
Conclusions
The flow chart provided in Figure 1 provides some practical indications on the diagnostic and therapeutic management of AML patients after alloHCT.
Figure 1.
Suggested workflow for post-transplantation disease monitoring and selection of treatment options. CBF, Core binding factor; HSCT, hematopoietic stem cell transplanation; qPCR, quantitative polymerase chain reaction.
The multitude of variables that determine the overall risk of relapse and that are increasingly recognized as determinants of the relapse modality are becoming too difficult to compute by standard means, and new artificial intelligence technologies are needed to calculate patient-specific hazards and devise the most appropriate monitoring and intervention plan. In solid tumors, mathematical modeling has even allowed tracing of the temporal trajectory of clonal evolution, thereby developing algorithms able to predict the most probable mutational asset at relapse.67
In this rapidly evolving context, the design and enrollment of patients into clinical trials is highly warranted: not only would they permit the collection of punctual high-quality information needed for advancing relapse knowledge and for the development of AI-powered predictors, but they might represent the only way to test personalized approaches to relapse in a reasonable time frame and with a business volume of potential interest for drug developers.
Acknowledgments
Work in the Vago lab is supported by the Associazione Italiana per la Ricerca sul Cancro (Investigator Grants #22197 and #28953 to LV), by the European Commission and Fondazione Regionale Ricerca Biomedica (ERA-NET-JTC2021 TRANSCAN-3 “PIXEL” to LV), by the Italian Ministry of Health (GR-2016-02364847 to Cristina Toffalori, GR-2018-12367860 to LV), by the Cariplo Foundation (Cariplo Giovani Ricercatori 2019-1708 to Cristina Toffalori), by the DKMS Stiftung Leben Spenden (John Hansen Grant 2022 to Cristina Toffalori), and by the Leukemia & Lymphoma Society (LLS SCOR to Robert Soiffer). The authors apologize to all authors who were not quoted in this review because of space constraints.
Conflict-of-interest disclosure
Giorgio Orofino has no competing financial interests to declare.
Luca Vago receives royalties from GenDx, B.V. (Utrecht, The Netherlands).
Off-label drug use
Giorgio Orofino: nothing to disclose.
Luca Vago: nothing to disclose.
References
- 1.Dohner H, Wei AH, Appelbaum FR, et al.. Diagnosis and management of AML in adults: 2022 recommendations from an international expert panel on behalf of the ELN. Blood. 2022;140(12):1345-1377. [DOI] [PubMed] [Google Scholar]
- 2.Horowitz M, Schreiber H, Elder A, et al.. Epidemiology and biology of relapse after stem cell transplantation. Bone Marrow Transplant. 2018;53(11):1379-1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vago L. Clonal evolution and immune evasion in posttransplantation relapses. Hematology Am Soc Hematol Educ Program. 2019;2019(1):610-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maziarz RT, Levis M, Patnaik MM, et al.. Midostaurin after allogeneic stem cell transplant in patients with FLT3-internal tandem duplication-positive acute myeloid leukemia. Bone Marrow Transplant. 2021;56(5):1180-1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burchert A, Bug G, Fritz LV, et al.. Sorafenib maintenance after allogeneic hematopoietic stem cell transplantation for acute myeloid leukemia with FLT3-internal tandem duplication mutation (SORMAIN). J Clin Oncol. 2020;38(26):2993-3002. [DOI] [PubMed] [Google Scholar]
- 6.Xuan L, Wang Y, Huang F, et al.. Sorafenib maintenance in patients with FLT3-ITD acute myeloid leukaemia undergoing allogeneic haematopoietic stem-cell transplantation: an open-label, multicentre, randomised phase 3 trial. Lancet Oncol. 2020;21(9):1201-1212. [DOI] [PubMed] [Google Scholar]
- 7.Levis MJ, Hamadani M, Logan B, et al; BMT-CTN 1506/MORPHO Study Investigators. Gilteritinib as post-transplant maintenance for AML with internal tandem duplication mutation of FLT3. J Clin Oncol. 2024;42(15):1766-1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sandmaier BM, Khaled S, Oran B, Gammon G, Trone D, Frankfurt O.. Results of a phase 1 study of quizartinib as maintenance therapy in subjects with acute myeloid leukemia in remission following allogeneic hematopoietic stem cell transplant. Am J Hematol. 2018;93(2):222-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fischer T, Stone RM, Deangelo DJ, et al.. Phase IIB trial of oral midostaurin (PKC412), the FMS-like tyrosine kinase 3 receptor (FLT3) and multi-targeted kinase inhibitor, in patients with acute myeloid leukemia and high-risk myelodysplastic syndrome with either wild-type or mutated FLT3. J Clin Oncol. 2010;28(28):4339-4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Metzelder SK, Schroeder T, Lübbert M, et al.. Long-term survival of sorafenib-treated FLT3-ITD-positive acute myeloid leukaemia patients relapsing after allogeneic stem cell transplantation. Eur J Cancer. 2017;86:233-239. [DOI] [PubMed] [Google Scholar]
- 11.Randhawa JK, Kantarjian HM, Borthakur G, et al.. Results of a phase II study of crenolanib in relapsed/refractory acute myeloid leukemia patients (pts) with activating FLT3 mutations. Blood. 2014;124(21):389. [Google Scholar]
- 12.Perl AE, Martinelli G, Cortes JE, et al.. Gilteritinib or chemotherapy for relapsed or refractory FLT3-mutated AML. N Engl J Med. 2019;381(18):1728-1740. [DOI] [PubMed] [Google Scholar]
- 13.Cortes JE, Khaled S, Martinelli G, et al.. Quizartinib versus salvage chemotherapy in relapsed or refractory FLT3-ITD acute myeloid leukaemia (QuANTUM-R): a multicentre, randomised, controlled, open-label, phase 3 trial. Lancet Oncol. 2019;20(7):984-997. [DOI] [PubMed] [Google Scholar]
- 14.Fathi AT, Kim HT, Soiffer RJ, et al.. Enasidenib as maintenance following allogeneic hematopoietic cell transplantation for IDH2-mutated myeloid malignancies. Blood Adv. 2022;6(22):5857-5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fathi AT, Kim HT, Soiffer RJ, et al.. Multicenter phase I trial of ivosidenib as maintenance treatment following allogeneic hematopoietic cell transplantation for IDH1-mutated acute myeloid leukemia. Clin Cancer Res. 2023;29(11):2034-2042. [DOI] [PubMed] [Google Scholar]
- 16.de Botton S, Fenaux P, Yee K, et al.. Olutasidenib (FT-2102) induces durable complete remissions in patients with relapsed or refractory IDH1-mutated AML. Blood Adv. 2023;7(13):3117-3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stein EM, DiNardo CD, Pollyea DA, et al.. Enasidenib in mutant IDH2 relapsed or refractory acute myeloid leukemia. Blood. 2017;130(6):722-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DiNardo CD, Stein EM, de Botton S, et al.. Durable remissions with ivosidenib in IDH1-mutated relapsed or refractory AML. N Engl J Med. 2018;378(25): 2386-2398. [DOI] [PubMed] [Google Scholar]
- 19.Zucenka A, Issa GC, Arellano M, et al.. Revumenib maintenance therapy following revumenib-induced remission and transplant. Blood. 2023;142(suppl 1):4950. [Google Scholar]
- 20.Issa GC, Aldoss I, DiPersio J, et al.. The menin inhibitor revumenib in KMT2A- rearranged or NPM1-mutant leukaemia. Nature. 2023;615(7954):920-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hong S, Rybicki L, Gurnari C, et al.. Pattern of somatic mutation changes after allogeneic hematopoietic cell transplantation for acute myeloid leukemia and myelodysplastic syndromes. Bone Marrow Transplant. 2022;57(10):1615-1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Quek L, Ferguson P, Metzner M, et al.. Mutational analysis of disease relapse in patients allografted for acute myeloid leukemia. Blood Adv. 2016;1(3): 193-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wienecke CP, Heida B, Venturini L, et al.. Clonal relapse dynamics in acute myeloid leukemia following allogeneic hematopoietic cell transplantation. Blood. 2024;144(3):296-307. [DOI] [PubMed] [Google Scholar]
- 24.Christopher M, Petti AA, Miller C, et al.. Clonal evolution of acute myeloid leukemia following allogeneic stem cell transplantation. Blood. 2016; 128(22):1528.27495140 [Google Scholar]
- 25.Waterhouse M, Pfeifer D, Pantic M, Emmerich F, Bertz H, Finke J.. Genome-wide profiling in AML patients relapsing after allogeneic hematopoietic cell transplantation. Biol Blood Marrow Transplant. 2011;17(10):1450-1459.e11459e1. [DOI] [PubMed] [Google Scholar]
- 26.Mathew NR, Baumgartner F, Braun L, et al.. Sorafenib promotes graft- versus-leukemia activity in mice and humans through IL-15 production in FLT3-ITD-mutant leukemia cells. Nat Med. 2018;24(3):282-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vosberg S, Hartmann L, Metzeler KH, et al.. Relapse of acute myeloid leukemia after allogeneic stem cell transplantation is associated with gain of WT1 alterations and high mutation load. Haematologica. 2018;103(12):e581-e584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tameni A, Toffalori C, Vago L.. Tricking the trickster: precision medicine approaches to counteract leukemia immune escape after transplant. Blood. 2024;143(26):2710-2721. [DOI] [PubMed] [Google Scholar]
- 29.Zeiser R, Vago L.. Mechanisms of immune escape after allogeneic hematopoietic cell transplantation. Blood. 2019;133(12):1290-1297. [DOI] [PubMed] [Google Scholar]
- 30.Rovatti PE, Gambacorta V, Lorentino F, Ciceri F, Vago L.. Mechanisms of leukemia immune evasion and their role in relapse after haploidentical hematopoietic cell transplantation. Front Immunol. 2020;11:147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vago L, Perna SK, Zanussi M, et al.. Loss of mismatched HLA in leukemia after stem-cell transplantation. N Engl J Med. 2009;361(5):478-488. [DOI] [PubMed] [Google Scholar]
- 32.Toffalori C, Cavattoni I, Deola S, et al.. Genomic loss of patient-specific HLA in acute myeloid leukemia relapse after well-matched unrelated donor HSCT. Blood. 2012;119(20):4813-4815. [DOI] [PubMed] [Google Scholar]
- 33.Crucitti L, Crocchiolo R, Toffalori C, et al.. Incidence, risk factors and clinical outcome of leukemia relapses with loss of the mismatched HLA after partially incompatible hematopoietic stem cell transplantation. Leukemia. 2015;29(5):1143-1152. [DOI] [PubMed] [Google Scholar]
- 34.Vago L, Toffalori C, Ahci M, et al.. Incidence of HLA loss in a global multicentric cohort of post-transplantation relapses: results from the Hlaloss Collaborative Study. Blood. 2018;132:818. [Google Scholar]
- 35.Muñiz P, Kwon M, Carbonell D, et al.. Clinical utility of the detection of the loss of the mismatched HLA in relapsed hematological patients after haploidentical stem cell transplantation with high-dose cyclophosphamide. Front Immunol. 2021;12:642087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mazzi B, Clerici TD, Zanussi M, et al.. Genomic typing for patient-specific human leukocyte antigen-alleles is an efficient tool for relapse detection of high-risk hematopoietic malignancies after stem cell transplantation from alternative donors. Leukemia. 2008;22(11):2119-2122. [DOI] [PubMed] [Google Scholar]
- 37.Ahci M, Toffalori C, Bouwmans E, et al.. A new tool for rapid and reliable diagnosis of HLA loss relapses after HSCT. Blood. 2017;130(10):1270-1273. [DOI] [PubMed] [Google Scholar]
- 38.Wu H, Shi J, Luo Y, et al.. Assessment of patient-specific human leukocyte antigen genomic loss at relapse after antithymocyte globulin-based T-cell-replete haploidentical hematopoietic stem cell transplant. JAMA Netw Open. 2022;5(4):e226114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rovatti PE, Zito L, Draghi E, et al.. Exploiting an anti-CD3/CD33 bispecific antibody to redirect donor T cells against HLA loss leukemia relapses. Blood. 2019;134(suppl 1):513. [Google Scholar]
- 40.Wu H, Cai Z, Shi J, Luo Y, Huang H, Zhao Y.. Blinatumomab for HLA loss relapse after haploidentical hematopoietic stem cell transplantation. Am J Cancer Res. 2021;11(6):3111-3122. [PMC free article] [PubMed] [Google Scholar]
- 41.Toffalori C, Zito L, Gambacorta V, et al.. Immune signature drives leukemia escape and relapse after hematopoietic cell transplantation. Nat Med. 2019;25(4):603-611. [DOI] [PubMed] [Google Scholar]
- 42.Christopher MJ, Petti AA, Rettig MP, et al.. Immune escape of relapsed AML cells after allogeneic transplantation. N Engl J Med. 2018;379(24):2330-2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Geramita E, Ventura K, Bhise S, et al.. Pilot trial of interferon-γ and donor lymphocyte infusion to treat relapsed myeloblastic malignancies after allogeneic hematopoietic stem cell transplantation. Transplant Cell Ther. 2023;29:S45-S46. [Google Scholar]
- 44.Rimando JC, Chendamarai E, Rettig MP, et al.. Flotetuzumab and other T-cell immunotherapies upregulate MHC class II expression on acute myeloid leukemia cells. Blood. 2023;141(14):1718-1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gambacorta V, Beretta S, Ciccimarra M, et al.. Integrated multiomic profiling identifies the epigenetic regulator PRC2 as a therapeutic target to counteract leukemia immune escape and relapse. Cancer Discov. 2022;12(6):1449-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Noviello M, Manfredi F, Ruggiero E, et al.. Bone marrow central memory and memory stem T-cell exhaustion in AML patients relapsing after HSCT. Nat Commun. 2019;10(1):1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Norde WJ, Maas F, Hobo W, et al.. PD-1/PD-L1 interactions contribute to functional T-cell impairment in patients who relapse with cancer after allogeneic stem cell transplantation. Cancer Res. 2011;71(15):5111-5122. [DOI] [PubMed] [Google Scholar]
- 48.Zhou M, Sacirbegovic F, Zhao K, Rosenberger S, Shlomchik WD. T cell exhaustion and a failure in antigen presentation drive resistance to the graft-versus-leukemia effect. Nat Commun. 2020;11(1):4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Penter L, Zhang Y, Savell A, et al.. Molecular and cellular features of CTLA-4 blockade for relapsed myeloid malignancies after transplantation. Blood. 2021;137(23):3212-3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Davids MS, Kim HT, Bachireddy P, et al; Leukemia and Lymphoma Society Blood Cancer Research Partnership. Ipilimumab for patients with relapse after allogeneic transplantation. N Engl J Med. 2016;375(2):143-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Garcia JS, Flamand Y, Penter L, et al.. Ipilimumab plus decitabine for patients with MDS or AML in posttransplant or transplant-naïve settings. Blood. 2023;141(15):1884-1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Davids MS, Kim HT, Costello C, et al.. A multicenter phase 1 study of nivolumab for relapsed hematologic malignancies after allogeneic transplantation. Blood. 2020;135(24):2182-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Godfrey J, Liu H, Yu J, et al.. Pembrolizumab for the treatment of disease relapse after allogeneic hematopoietic stem cell transplantation. Blood Adv. 2023;7(6):963-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Apostolova P, Kreutmair S, Toffalori C, et al.. Phase II trial of hypomethylating agent combined with nivolumab for acute myeloid leukaemia relapse after allogeneic haematopoietic cell transplantation—Immune signature correlates with response. Br J Haematol. 2023;203(2):264-281. [DOI] [PubMed] [Google Scholar]
- 55.Sallman DA, McLemore AF, Aldrich AL, et al.. TP53 mutations in myelodysplastic syndromes and secondary AML confer an immunosuppressive phenotype. Blood. 2020;136(24):2812-2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lau CM, Nish SA, Yogev N, Waisman A, Reiner SL, Reizis B.. Leukemia-associated activating mutation of Flt3 expands dendritic cells and alters T cell responses. J Exp Med. 2016;213(3):415-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Uhl FM, Chen S, O'Sullivan D, et al.. Metabolic reprogramming of donor T cells enhances graft-versus-leukemia effects in mice and humans. Sci Transl Med. 2020;12(567):eabb8969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vallet N, Le Grand S, Bondeelle L, et al.. Azithromycin promotes relapse by disrupting immune and metabolic networks after allogeneic stem cell transplantation. Blood. 2022;140(23):2500-2513. [DOI] [PubMed] [Google Scholar]
- 59.Peled JU, Devlin SM, Staffas A, et al.. Intestinal microbiota and relapse after hematopoietic-cell transplantation. J Clin Oncol. 2017;35(15):1650-1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Karl F, Liang C, Böttcher-Loschinski R, et al.. Oxidative DNA damage in reconstituting T cells is associated with relapse and inferior survival after allo-SCT. Blood. 2023;141(13):1626-1639. [DOI] [PubMed] [Google Scholar]
- 61.Naka K, Hoshii T, Muraguchi T, et al.. TGF-beta-FOXO signalling maintains leukaemia-initiating cells in chronic myeloid leukaemia. Nature. 2010; 463(7281):676-680. [DOI] [PubMed] [Google Scholar]
- 62.Lee YJ, Han Y, Lu HT, et al.. TGF-beta suppresses IFN-gamma induction of class II MHC gene expression by inhibiting class II transactivator messenger RNA expression. J Immunol. 1997;158(5):2065-2075. [PubMed] [Google Scholar]
- 63.Park HH, Kim M, Lee B-H, et al.. Intracellular IL-4, IL-10, and IFN-gamma levels of leukemic cells and bone marrow T cells in acute leukemia. Ann Clin Lab Sci. 2006;36(1):7-15. [PubMed] [Google Scholar]
- 64.DiLillo DJ, Weinberg JB, Yoshizaki A, et al.. Chronic lymphocytic leukemia and regulatory B cells share IL-10 competence and immunosuppressive function. Leukemia. 2013;27(1):170-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Munn DH, Sharma MD, Baban B, et al.. GCN2 kinase in T cells mediates proliferative arrest and anergy induction in response to indoleamine 2,3-dioxygenase. Immunity. 2005;22(5):633-642. [DOI] [PubMed] [Google Scholar]
- 66.Mussai F, De Santo C, Abu-Dayyeh I, et al.. Acute myeloid leukemia creates an arginase-dependent immunosuppressive microenvironment. Blood. 2013;122(5):749-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Caravagna G, Heide T, Williams MJ, et al.. Subclonal reconstruction of tumors by using machine learning and population genetics. Nat Genet. 2020;52(9):898-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bacher U, Haferlach T, Alpermann T, et al.. Comparison of cytogenetic clonal evolution patterns following allogeneic hematopoietic transplantation versus conventional treatment in patients at relapse of AML. Biol Blood Marrow Transplant. 2010;16(12):1649-1657. [DOI] [PubMed] [Google Scholar]
- 69.Pagliuca S, Gurnari C, Hercus C, et al.. Leukemia relapse via genetic immune escape after allogeneic hematopoietic cell transplantation. Nat Commun. 2023;14(1):3153. [DOI] [PMC free article] [PubMed] [Google Scholar]