Abstract
The connection between the immune response and the composition of gut microbiota has been associated with an increased prevalence of atopic dermatitis in the first year of life. The study aimed to investigate gut microbiota characteristics in infants with atopic dermatitis compared to healthy infants to better understand the link between early-life microbiota composition and the development of atopic dermatitis. The study analyzed the intestinal microbiota of 121 infants with clinical signs of atopic dermatitis, divided into Group I (infants with atopic dermatitis) and Group II (healthy controls). The study showed that infants with atopic dermatitis presented increased values of proteolytic bacteria mainly represented by Enterobacter species (P = 0.041), Klebsiella species (P = 0.038), and Escherichia coli (P = 0.013), with significantly decreased levels of acidifying bacteria represented by Enterococcus species, Lactobacillus and Bifidobacterium (P < 0.05) and normal levels of Clostridium species, Candida albicans, Mould fungi and Geotrichum species. This study highlights distinct differences in the gut microbiota of infants with atopic dermatitis, providing insights into the dynamic intestinal ecosystem during early life for future personalized therapeutic strategies.
Keywords: infants, atopic dermatitis, gut microbiota, dysbiosis, proteolytic bacteria, acidifying bacteria
INTRODUCTION
Atopic dermatitis is a common skin condition frequently observed in children, caused by a combination of hereditary and environmental factors [1], substantially affecting the quality of life [2]. The incidence of atopic dermatitis in children is increasing worldwide [3]. Implications of gut microbiota in allergies and atopic dermatitis were reported by previous studies [4], emphasizing the complex relationship between gut microbiota composition and human health, which justifies the need for additional research in the field of personalized medicine [5].
Infants are highly prone to developing atopic dermatitis, often experiencing more frequent and severe flare-ups that tend to last longer [6]. They are also more likely to develop the condition at an earlier age [6]. Exposure to a food antigen can trigger a hypersensitive skin reaction, which may lead to a food allergy [7]. This highlights the significant connection between the skin and the immune response in the gut [7]. The gut microbiota plays a crucial role in autoimmune disorders [8,9] and infectious diseases [10], with long-lasting effects. Novel therapeutics targeting the gut microbiota have shown promising results [8,11].
Previous studies have demonstrated that modifying the gut microbiota can potentially impact the immune response [12,13] and have also indicated a connection between changes in gut microbiota and the development of allergy disorders [14]. Melli et al. [15] found that the gut microbiota composition differs between allergic and non-allergic children, suggesting that alterations in intestinal microbiota may be associated with the onset of atopic dermatitis symptoms. Kong et al. [16] also reported changes in microbiota composition throughout the progression of atopic dermatitis.
Gut dysbiosis in infancy has been linked to immune system development and often precedes the onset of atopic diseases, with atopic dermatitis being the initial phase of the atopic march [17]. Furthermore, microbiota patterns in the skin and gut can predict susceptibility to dietary and exterior allergens or trigger allergic reactions in the host [18].
Certain microorganisms are associated with infectious processes [19,20] due to the influence of environmental variables [21], while changes in the gut microbiota composition may contribute to the development of atopic dermatitis [22]. The existing therapeutic strategies for atopic dermatitis are limited by a lack of effective options and the heterogeneous nature of the illness [23].
Recent studies have emphasized the importance of modulating gut microbiota and its implications for allergies [14], respiratory diseases [24], and renal conditions [25], highlighting the long-term impact on overall health [26]. Modulating gut microbiota has emerged as a promising treatment strategy for infants with atopic dermatitis, demonstrating encouraging results [27]. Identifying the composition and characteristics of intestinal microbiota in infants with atopic dermatitis represents a key factor for future personalized therapy.
This study aimed to investigate gut microbiota variations in infants with atopic dermatitis compared to healthy infants. These findings reveal insights into the connection between the composition of the dynamic intestinal ecosystem and atopic dermatitis for future personalized therapeutic strategies.
MATERIAL AND METHODS
The study was conducted from April 2023 to May 2024 and included 121 infants diagnosed with atopic dermatitis. The infants were divided into two groups: Group I consisted of 91 infants with atopic dermatitis (AD), and Group II (the control group) included 30 infants without atopic dermatitis (non-AD). Demographic data were collected, and informed consent was obtained from the legal representatives of all participants for both participation and the processing of personal data. The study adhered to the ethical principles outlined in the Declaration of Helsinki.
Participants were selected based on the following criteria: age between 1 month and 1 year, a confirmed diagnosis of atopic dermatitis, and the absence of acute infectious diseases, gastrointestinal disorders, cardiovascular diseases, renal diseases, endocrine disorders, oncological conditions, autoimmune diseases, and the use of probiotics or antibiotics within four weeks before the study.
The diagnosis of AD was assessed by the dermatologist based on the presence of distinct physical characteristics and patterns of skin lesions, a persistent or recurring course, itching, and a personal or family history of atopic conditions, as outlined in the Williams criteria [28] and the Hanifin and Rajka criteria [28, 29].
Exclusion criteria included infants older than 1 year, those with gastrointestinal or genetic disorders, endocrine or metabolic diseases, blood disorders, heart, liver, or kidney conditions, recent use of antibiotics (within one month), or probiotics (within 4 weeks).
Stool samples were collected to determine the composition of gut microbiota. Fecal samples were collected during clinic visits, stored at -2°C, and promptly transported to the laboratory in sterile containers. A total of 1 g of each sample was used for bacteriological and fungal analysis. Bacteriological examination of stool samples was performed for proteolytic bacteria (Escherichia coli, Proteus species, Klebsiella species, Enterobacter species, Hafnia alvei, Serratia species, Providencia species, Morganella morganii, Kluyvera species, Citrobacter species, Pseudomonas species, Clostridium species) and acidifying bacteria (Bacteroides species, Bifidobacterium species, Lactobacillus species, Enterococcus species) and for fungal species (Candida albicans, Candida species, Geotrichum species and Mould fungi). The plates were incubated in optimal growth conditions of the target species. To facilitate understanding, we categorized the identified genera based on their abundance (high or low) in infants with atopic dermatitis.
Results were analyzed using descriptive statistics. Standard deviation (SD) was employed to measure the variability of continuous data, and a P-value of 0.05 was considered statistically significant. The reliability of differences between comparison groups was assessed using the t-test.
RESULTS
The breastfeeding rate in the atopic dermatitis (AD) group was lower compared to the control group. The analysis of the type of delivery revealed that cesarean section had a higher occurrence in the AD group. The infants were born at full-term with an average weight of 2.98 kilograms (SD = 0.24) and an average height of 50.21 centimeters (SD = 1.43). The general characteristics of the two groups are presented in Table 1.
Table 1.
Characteristics of participants
| AD group | Non-AD group | |
|---|---|---|
| Male, n (%) | 43 (47.3) | 14 (46.7) |
| Female, n (%) | 48 (52.7) | 16 (53.3) |
| Age (months) mean (SD) | 7.62 (1.7) | 6.84 (1.8) |
| Vaginal delivery, n (%) | 40 (43.9) | 17 (56.6) |
| Cesarean section, n (%) | 51 (56.1) | 13 (43.4) |
| Breastfeeding, n (%) | 40 (43.9) | 19 (63.4) |
| Formula feeding, n (%) | 35 (38.5) | 7 (23.3) |
| Mixed feeding (infant formula and breastfeeding), n (%) | 16 (17.6) | 4 (13.3) |
| Age at onset of AD, months (mean) | 5.12 | - |
| Relapse episodes of AD (mean) | 2.08 | - |
| Family history of atopy | ||
| One parent, n (%) | 68 (74.7) | 11 (36.6) |
| Both parents, n (%) | 23 (25.3) | 4 (13.3) |
The fecal microbial analysis identified significant alterations in the intestinal microbiota of infants with atopic dermatitis (Table 2). The atopic dermatitis group had a higher abundance of Proteobacteria phylum, mainly represented by gram-negative, facultatively anaerobic Enterobacteriaceae family. There were also low concentrations of Firmicutes phylum and Actinobacteria phylum. The presence of Bacteroidetes phylum was significantly higher in the AD group (P = 0.036), with concentrations of 2.31 ± 0.08 x 109 CFU/g.
Table 2.
Composition of intestinal microbiota in AD group and non-AD group
| Microorganisms (CFU/g) | AD group (n = 91) | Non-AD group (n = 30) |
|---|---|---|
| Escherichia coli (x108) | 1.8 ± 0.4 (P = 0.013) | 0.8 ± 0.32 |
| Proteus spp. (x104) | 0.83 ± 0.12 (P = 0.611) | 0.89 ± 0.04 |
| Klebsiella spp. (x104) | 1.32 ± 0.3 (P = 0.038) | 0.85 ± 0.06 |
| Enterobacter spp. (x104) | 1.12 ± 0.2 (P = 0.041) | 0.83 ± 0.12 |
| Hafnia alveii (x104) | 0.91 ± 0.02 (P = 0.311) | 0.95 ± 0.03 |
| Serratia species (x104) | 0.90 ± 0.02 (P = 0.218) | 0.92 ± 0.06 |
| Providencia species (x104) | 0.83 ± 0.03 (P = 0.183) | 0.89 ± 0.05 |
| Morganella morganii (x104) | 0.84 ± 0.02 (P = 0.164) | 0.88 ± 0.04 |
| Kluyvera species (x104) | 0.90 ± 0.02 (P = 0.913) | 0.91 ± 0.07 |
| Citrobacter species (x104) | 0.78 ± 0.04 (P = 0.875) | 0.95 ± 0.13 |
| Pseudomonas species (x104) | 0.92 ± 0.02 (P = 0.213) | 0.96 ± 0.02 |
| Clostridium species (x105) | 0.85 ± 0.05 (P = 0.098) | 0.94 ± 0.03 |
| Bacteroides species (x109) | 2.31 ± 0.08 (P = 0.036) | 0.94 ± 0.04 |
| Bifidobacterium species (x108) | 0.75 ± 0.08 (P = 0.038) | 0.93 ± 0.06 |
| Lactobacillus species (x105) | 0.82 ± 0.05 (P = 0.021) | 0.90 ± 0.07 |
| Enterococcus species (x105) | 0.71 ± 0.02 (P = 0.042) | 0.92 ± 0.03 |
| Candida albicans (x103) | 0.80 ± 0.08 (P = 0.930) | 0.96 ± 0.02 |
| Candida species (x103) | 0.91 ± 0.04 (P = 0.221) | 0.88 ± 0.08 |
| Geotrichum species (x103) | 0.89 ± 0.03 (P = 0.135) | 0.90 ± 0.07 |
The findings revealed a higher prevalence of intestinal dysbiosis in the AD group, with increased concentrations of proteolytic bacteria, including Escherichia coli (1.8. ± 0.4 x 108 CFU/g, P = 0.013), Enterobacter species (1.12 ± 0.2 x 104 CFU/g, P = 0.041), and Klebsiella species (1.32 ± 0.3 x 104 CFU/g, P = 0.038). There was also a significant increase in Bacteroides species among acidifying bacteria (2.31 ± 0.08 x 109 CFU/g, P = 0.036) in the AD group compared to the control group (Figure 1).
Figure 1.
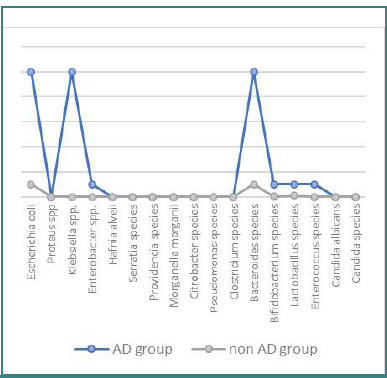
Composition of intestinal microbiota in AD and non-AD groups
Infants with atopic dermatitis had significantly lower levels of acidifying germs represented by Bifidobacterium, Lactobacillus, and Enterococcus, compared to the control group (P < 0.05). Candida albicans and Geotrichum species had normal concentrations in both groups, and no mold fungi were detected.
The quantitative analysis showed lower Bifidobacterium spp. levels in the AD group (0.75 ± 0.08 x 108 CFU/g, P = 0.038) compared to the non-AD group (0.93 ± 0.06 x 108 CFU/g). Lactobacillus spp. concentrations were also reduced in the AD group (0.82 ± 0.05 x 105 CFU/g, P = 0.021) compared to the non-AD group (0.90 ± 0.07 x 105 CFU/g). Enterococcus spp. levels were significantly lower in the AD group (0.71 ± 0.02 x 105 CFU/g, P = 0.042) compared to the non-AD group (0.92 ± 0.03 x 105 CFU/g).
Normal values were observed for Hafnia alvei, Citrobacter species, Serratia species, Providencia species, Pseudomonas species, Clostridium species, and Morganella morganii in both groups. An increase in proteolytic bacteria concentration was associated with a more alkaline fecal pH (mean value 6.5), indicating a preference for these bacteria in higher pH environments. Conversely, acidifying bacteria thrived at lower pH values. Figure 2 shows the acidifying and proteolytic bacteria concentration according to fecal pH values.
Figure 2.
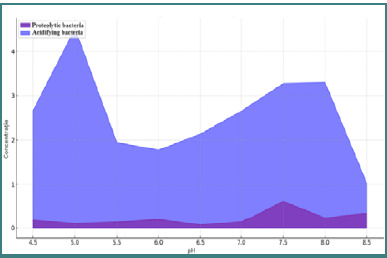
Concentration of acidifying and proteolytic bacteria according to fecal pH values
DISCUSSION
The study found significantly decreased levels of acidifying germs, such as Bifidobacterium spp., in infants with AD compared to healthy infants at the phylum level, with a high abundance of Bacteroides species. Abrahamsson et al. [30] reported reduced levels of Bacteroidetes associated with atopic dermatitis and decreased levels of Proteobacteria at one year. Additional research demonstrated a positive correlation between the incidence of AD and increased abundance of Bacteroidaceae and Bacteroides [31].
An increased abundance of Bacteroides has been linked to atopic manifestations, potentially resulting in continuous lipopolysaccharide synthesis and triggering inflammatory responses [32,33]. Bacteroides species have also been found to alter gut permeability, a characteristic observed in individuals with atopic dermatitis [32]. Additional studies showed that a high abundance of Bacteroides species and proteolytic bacteria, such as Escherichia coli and Enterobacter species, contribute to the onset of eczema during the first year of life [33,34].
Infants with atopic dermatitis presented significantly decreased levels of acidifying germs represented by Bifidobacterium, Lactobacillus, and Enterococcus, compared to the control group. Additional studies highlighted the association between the gut microbiota composition of healthy infants and AD, with a decrease in the presence of Bifidobacterium and Lactobacillus, particularly in infants with atopic dermatitis [35]. Chen et al. [36] reported an increased level of Firmicutes with a lower concentration of Bacteroidetes and increased values of Bifidobacterium, Enterococcus, Lactobacillus, Roseburia, Faecalibacterium, Ruminococcus, and Akkermansia in children with allergies.
Hong et al. [37] showed that infants with atopic dermatitis presented elevated concentrations of Klebsiella spp., the main phyla represented by Firmicutes, Proteobacteria, Actinobacteria, and Bacteroidetes. The abundance of Bifidobacterium and Enterococcus species was also linked to the occurrence of AD in infants delivered via cesarean section [37,38]. Our study findings revealed increased values of proteolytic germs represented by Escherichia coli (P = 0.013), Enterobacter species (P = 0.041), and Klebsiella species (P = 0.038).
A significant correlation between higher levels of Clostridiaceae and an increased risk of AD has been reported [34]. This association may be attributed to the release of toxins that hinder the movement of neutrophils and deactivate eosinophils, thereby exacerbating inflammation [34]. Atopic infants were shown to have elevated levels of Clostridium and reduced levels of Bifidobacterium in their stools [39]. Our study, however, showed normal concentrations of Clostridium species in infants with AD.
Parabacteroides and Klebsiella were more prevalent in infants with atopic dermatitis [40]. Our study provides additional evidence for this observation by noting a higher concentration of Klebsiella, Escherichia coli, and Enterobacter in the AD group.
Regarding fungal species, Candida albicans and Geotrichum species were found at normal concentrations in both the AD and non-AD groups. However, other studies have reported a link between high levels of Candida albicans and an increased risk of AD in the first year of life [41,42].
Enhanced therapeutic outcomes can be achieved by integrating microbiota-based therapies with conventional treatments. Recent studies focus on treatments such as multi-strain probiotics, prebiotics, and fecal microbiota transplantation [43].
Long-term intake of probiotics can modify the gut microbial habitat and maintain a healthy balance of gut microbiota and systemic immune responses [44]. Probiotics enhance the level of short-chain fatty acids (SCFA) in the intestinal lumen [44]. Specifically, short-chain fatty acids (SCFA) such as acetate, propionate, and butyrate create an intestinal environment with a low pH and compete with pathogens, suppressing excessive growth [44]. Hence, probiotics can potentially mitigate the clinical symptoms of AD by influencing the makeup of gut microbiota, metabolic processes, and immunological reactions [44]. The preventive impact of Lactobacillus rhamnosus and mixed probiotics is beneficial due to the decreased abundance of Bifidobacterium and Lactobacillus species in infants with atopic dermatitis [45]. Some studies have suggested administering probiotics, specifically Lactobacillus or Bifidobacterium strains, to pregnant women in the last trimester to prevent AD in infants [46]. Additionally, administering probiotics and vitamin D supplements during pregnancy and infancy while minimizing needless antibiotic use may decrease the risk of atopic dermatitis [46].
Novel technologies such as CRISPR-based genome editing and high-throughput screening could improve therapeutic capabilities on probiotic strains [47]. Microencapsulation techniques improve the stability, viability, and targeted distribution of probiotics, enhancing their effectiveness [47].
Fecal microbiota transplantation (FMT) is a new therapeutic approach used to restore the gut microbiota. The therapeutic potential of FMT was examined through the alteration of gut microbiota, modulation of the immune system, and measurement of fecal metabolites in an AD mice model [48], emphasizing the necessity for additional research in this area.
The current study is subject to significant limitations. Firstly, it did not examine stool samples before the onset of AD, hence impeding the establishment of a temporal relationship. It is important to note that intestinal microbiota composition might differ significantly across individuals. In this particular age group, the intestinal flora is still undergoing active and ongoing development. Furthermore, the description of potential confounding factors was inadequate due to the omission of some influential factors that impact the gut microbiota, such as maternal antibiotic exposure.
CONCLUSION
The study reveals characteristics in the composition of gut microbiota in infants with atopic dermatitis, with specific variation in acidifying and proteolytic germs, with higher diversity of proteolytic bacteria represented by Escherichia coli, and increased colonization of Enterobacter spp. and Klebsiella spp., with a lower diversity of acidifying germs represented by Lactobacillus, Enterococcus and significantly decreased values of Bifidobacterium spp. The study shows distinct variations of gut microbiota in the genus characteristics in infants with atopic dermatitis compared to healthy infants. This research provides valuable insights into the relationship between intestinal bacterial composition and atopic dermatitis, paving the way for future therapeutic strategies in personalized medicine.
Conflict of interest
The authors declare no conflict of interest.
Ethical approval
The study received ethical approval from the Ethics Committee of the Clinical Emergency Hospital of Constanta, Romania, according to the Faculty of Medicine, Ovidius University of Constanta, Romania (no. 19856/11.04.2024).
Consent to participate
Informed consent was obtained from legal representatives, and personal data processing consent was obtained.
Authorship
ACP contributed to conceptualizing the study. ACP, ALB, and CMM contributed to the methodology. ACP contributed to writing the original draft. ACP, WN, MAKK, TC, LM, AU, CEF, SIC, and SCC contributed to editing the manuscript. ACP, ALB, and AU contributed to data collection.
References
- 1.Chong AC, Visitsunthorn K, Ong PY. Genetic/Environmental Contributions and Immune Dysregulation in Children with Atopic Dermatitis. J Asthma Allergy. 2022 Nov 23;15:1681–1700. doi: 10.2147/JAA.S293900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang EJ, Beck KM, Sekhon S, Bhutani T, Koo J. The impact of pediatric atopic dermatitis on families: A review. Pediatr Dermatol. 2019 Jan;36(1):66–71. doi: 10.1111/pde.13727. [DOI] [PubMed] [Google Scholar]
- 3.Ilic I, Stojkovic A, Velickovic V, Zivanovic Macuzic I, Ilic M. Atopic Dermatitis in Children Under 5: Prevalence Trends in Central, Eastern, and Western Europe. Children (Basel) 2023 Jul 25;10(8):1275. doi: 10.3390/children10081275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Graham F, Eigenmann PA. Atopic dermatitis and its relation to food allergy. Curr Opin Allergy Clin Immunol. 2020 Jun;20(3):305–310. doi: 10.1097/ACI.0000000000000638. [DOI] [PubMed] [Google Scholar]
- 5.Nekrasova AI, Kalashnikova IG, Bobrova MM, Korobeinikova AV, Bakoev SY, Ashniev GA, et al. Characteristics of the Gut Microbiota in regard to Atopic Dermatitis and Food Allergies of Children. Biomedicines. 2024;12(3):553. doi: 10.3390/biomedicines12030553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Łoś-Rycharska E, Gołębiewski M, Grzybowski T, Rogalla-Ładniak U, Krogulska A. The microbiome and its impact on food allergy and atopic dermatitis in children. Postepy Dermatol Alergol. 2020 Oct;37(5):641–650. doi: 10.5114/ada.2019.90120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Florsheim EB, Sullivan ZA, Khoury-Hanold W, Medzhitov R. Food allergy as a biological food quality control system. Cell. 2021 Mar 18;184(6):1440–1454. doi: 10.1016/j.cell.2020.12.007. [DOI] [PubMed] [Google Scholar]
- 8.Lupu VV, Butnariu LI, Fotea S, Morariu ID, Badescu MC, Starcea IM, et al. The Disease with a Thousand Faces and the Human Microbiome—A Physiopathogenic Intercorrelation in Pediatric Practice. Nutrients. 2023;15(15):3359. doi: 10.3390/nu15153359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lupu VV, Jechel E, Mihai CM, Mitrofan EC, Lupu A, Starcea IM, et al. Connection between Celiac Disease and Systemic Lupus Erythematosus in Children—A Development Model of Autoimmune Diseases Starting from What We Inherit to What We Eat. Nutrients. 2023;15(11):2535. doi: 10.3390/nu15112535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mihai CM, Chisnoiu T, Cambrea CS, Frecus CE, Mihai L, Balasa AL, et al. Neurological manifestations found in children with multisystem inflammatory syndrome. Exp Ther Med. 2022;23:261. doi: 10.3892/etm.2022.11187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kassim MAK, Pantazi AC, Nori W, Tuta LA, Balasa AL, Mihai CM, et al. Non-Pharmacological Interventions for Pain Management in Hemodialysis: A Narrative Review. J Clin Med. 2023;12(16):5390. doi: 10.3390/jcm12165390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nori W, Akram NN, Mueen Al-kaabi M. Probiotics in women and pediatrics health: A narrative review. Al-Anbar Med J. 2023 Jun 1;19(1):10–6. doi: 10.33091/amj.2023.138442.1021. [DOI] [Google Scholar]
- 13.Penders J, Stobberingh EE, Thijs C, Adams H, Vink C, van Ree R, et al. Molecular fingerprinting of the intestinal microbiota of infants in whom atopic eczema was or was not developing. Clin Exp Allergy. 2006 Dec;36(12):1602–8. doi: 10.1111/j.1365-2222.2006.02599.x. [DOI] [PubMed] [Google Scholar]
- 14.Pantazi AC, Mihai CM, Balasa AL, Chisnoiu T, Lupu A, Frecus CE, et al. Relationship between Gut Microbiota and Allergies in Children: A Literature Review. Nutrients. 2023 May 29;15(11):2529. doi: 10.3390/nu15112529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Melli LCFL, Carmo-Rodrigues MSD, Araújo-Filho HB, Mello CS, Tahan S, Pignatari ACC, et al. Gut microbiota of children with atopic dermatitis: Controlled study in the metropolitan region of São Paulo, Brazil. Allergol Immunopathol (Madr) 2020 Mar-Apr;48(2):107–115. doi: 10.1016/j.aller.2019.08.004. [DOI] [PubMed] [Google Scholar]
- 16.Kong HH, Oh J, Deming C, Conlan S, Grice EA, Beatson MA, et al. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res. 2012 May;22(5):850–9. doi: 10.1101/gr.131029.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim JE, Kim HS. Microbiome of the Skin and Gut in Atopic Dermatitis (AD): Understanding the Pathophysiology and Finding Novel Management Strategies. J Clin Med. 2019;8(4):444. doi: 10.3390/jcm8040444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park HJ, Lee SW, Hong S. Regulation of Allergic Immune Responses by Microbial Metabolites. Immune Netw. 2018 Feb 26;18(1):e15. doi: 10.4110/in.2018.18.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Halichidis S, Balasa AL, Ionescu EV, Iliescu MG, Cambrea SC, Petcu LC, et al. Evolution of salmonellosis in Constanta area in correlation with environmental factors. J Environ Prot Ecol. 2019;20(3):1496–1504. Available from: https://www.cabidigitallibrary.org/doi/full/10.5555/20203158441. [Google Scholar]
- 20.Cambrea SC, Petcu LC, Mihai CM, Hangan TL, Iliescu DM. Influence of environmental factors on the evolution of shigellosis in Constanta County, Romania. J Environ Prot Ecol. 2019;20:986–994. Available from: https://scibulcom.net/en/article/VjIJ8TiiFbDccEnL2q1Q. [Google Scholar]
- 21.Stoicescu RM, Mihai CM, Arghir O, Cambrea C, Halichidis S, Lilios G. Soil ingestion among children from Constanta County during 2002-2012. J Environ Prot Ecol. 2014;15(1):321–325. Available from: https://scibulcom.net/en/article/odny9DttlGKoAMflLyF0. [Google Scholar]
- 22.Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell. 2014 Mar 27;157(1):121–41. doi: 10.1016/j.cell.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chovatiya R, Silverberg JI. The Heterogeneity of Atopic Dermatitis. J Drugs Dermatol. 2022 Feb 1;21(2):172–176. doi: 10.36849/JDD.6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lupu A, Jechel E, Mihai CM, Mitrofan EC, Fotea S, Starcea IM, et al. The Footprint of Microbiome in Pediatric Asthma—A Complex Puzzle for a Balanced Development. Nutrients. 2023;15(14):3278. doi: 10.3390/nu15143278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pantazi AC, Kassim MAK, Nori W, Tuta LA, Mihai CM, Chisnoiu T, et al. Clinical Perspectives of Gut Microbiota in Patients with Chronic Kidney Disease and End-Stage Kidney Disease: Where Do We Stand? Biomedicines. 2023;11(9):2480. doi: 10.3390/biomedicines11092480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pantazi AC, Balasa AL, Mihai CM, Chisnoiu T, Lupu VV, Kassim MAK, et al. Development of Gut Microbiota in the First 1000 Days after Birth and Potential Interventions. Nutrients. 2023;15(16):3647. doi: 10.3390/nu15163647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alam MJ, Xie L, Yap YA, Marques FZ, Robert R. Manipulating Microbiota to Treat Atopic Dermatitis: Functions and Therapies. Pathogens. 2022 Jun 2;11(6):642. doi: 10.3390/pathogens11060642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yao X, Song ZQ, Li W, Liang YS, Zhao Y, Cao H, et al. Guidelines for diagnosis and treatment of atopic dermatitis in China 2020. Int J Dermatol Venereol. 2021;4(1):1–9. doi: 10.1097/JD9.0000000000000143. [DOI] [Google Scholar]
- 29.Hanifin JM, Rajka G. Diagnostic features of atopic dermatitis. Acta Derm Venereol. 1980;(Suppl 92):44–47. [Google Scholar]
- 30.Abrahamsson TR, Jakobsson HE, Andersson AF, Björkstén B, Engstrand L, Jenmalm MC. Low diversity of the gut microbiota in infants with atopic eczema. J Allergy Clin Immunol. 2012 Feb;129(2):434–40. doi: 10.1016/j.jaci.2011.10.025. 440.e1-2. [DOI] [PubMed] [Google Scholar]
- 31.Xue Y, Zhang L, Chen Y, Wang H, Xie J. Gut microbiota and atopic dermatitis: a two-sample Mendelian randomization study. Front Med. 2023;10:1174331. doi: 10.3389/fmed.2023.1174331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reddel S, Del Chierico F, Quagliariello A, Giancristoforo S, Vernocchi P, Russo A, et al. Gut microbiota profile in children affected by atopic dermatitis and evaluation of intestinal persistence of a probiotic mixture. Sci Rep. 2019 Mar 21;9(1):4996. doi: 10.1038/s41598-019-41149-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leo S, Cetiner OF, Pittet LF, Messina NL, Jakob W, Falquet L, et al. The association between the composition of the early-life intestinal microbiome and eczema in the first year of life. Front Microbiomes. 2023;2:1147082. doi: 10.3389/frmbi.2023.1147082. [DOI] [Google Scholar]
- 34.Wrześniewska M, Wołoszczak J, Świrkosz G, Szyller H, Gomułka K. The role of the microbiota in the pathogenesis and treatment of atopic dermatitis—a literature review. Int J Mol Sci. 2024;25(12):6539. doi: 10.3390/ijms25126539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Björkstén B, Sepp E, Julge K, Voor T, Mikelsaar M. Allergy development and the intestinal microflora during the first year of life. J Allergy Clin Immunol. 2001 Oct;108(4):516–20. doi: 10.1067/mai.2001.118130. [DOI] [PubMed] [Google Scholar]
- 36.Chen C-C, Chen K-J, Kong M-S, Chang H-J, Huang J-L. Alterations in the gut microbiotas of children with food sensitization in early life. Pediatr Allergy Immunol. 2016;27:254–262. doi: 10.1111/pai.12522. [DOI] [PubMed] [Google Scholar]
- 37.Hong PY, Lee BW, Aw M, Shek LP, Yap GC, Chua KY, et al. Comparative analysis of fecal microbiota in infants with and without eczema. PLoS One. 2010 Apr 1;5(4):e9964. doi: 10.1371/journal.pone.0009964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yap GC, Loo EX, Aw M, Lu Q, Shek LP, Lee BW. Molecular analysis of infant fecal microbiota in an Asian at-risk cohort-correlates with infant and childhood eczema. BMC Res Notes. 2014 Mar 20;7:166. doi: 10.1186/1756-0500-7-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kalliomäki M, Kirjavainen P, Eerola E, Kero P, Salminen S, Isolauri E. Distinct patterns of neonatal gut microflora in infants in whom atopy was and was not developing. J Allergy Clin Immunol. 2001 Jan;107(1):129–34. doi: 10.1067/mai.2001.111237. [DOI] [PubMed] [Google Scholar]
- 40.Ye S, Yan F, Wang H, Mo X, Liu J, Zhang Y, et al. Diversity analysis of gut microbiota between healthy controls and those with atopic dermatitis in a Chinese population. J Dermatol. 2021;48(2):158–67. doi: 10.1111/1346-8138.15530. [DOI] [PubMed] [Google Scholar]
- 41.Leo S, Cetiner OF, Pittet LF, Messina NL, Jakob W, Falquet L, et al. The association between the composition of the early-life intestinal microbiome and eczema in the first year of life. Front Microbiomes. 2023;2:1147082. doi: 10.3389/frmbi.2023.1147082. [DOI] [Google Scholar]
- 42.Mok K, Suratanon N, Roytrakul S, Charoenlappanit S, Patumcharoenpol P, Chatchatee P, et al. ITS2 Sequencing and Targeted Meta-Proteomics of Infant Gut Mycobiome Reveal the Functional Role of Rhodotorula sp. during Atopic Dermatitis Manifestation. J Fungi (Basel) 2021 Sep 13;7(9):748. doi: 10.3390/jof7090748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sebastian VJ, Sanjana J. Targeting the gut microbiome for atopic dermatitis: a comprehensive review of mechanisms and therapeutic approaches. Med Res Arch. 2024 May;12(5) doi: 10.18103/mra.v12i5.5290. Available from: https://esmed.org/MRA/mra/article/view/5290. [DOI] [Google Scholar]
- 44.Fang Z, Li L, Zhang H, Zhao J, Lu W Chen W. Gut Microbiota, Probiotics, and Their Interactions in Prevention and Treatment of Atopic Dermatitis: A Review. Front Immunol. 2021;12:720393. doi: 10.3389/fimmu.2021.720393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang F, Wu F, Chen H, Tang B. The effect of probiotics in the prevention of atopic dermatitis in children: a systematic review and meta-analysis. Transl Pediatr. 2023 Apr 29;12(4):731–748. doi: 10.21037/tp-23-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chu DK, Koplin JJ, Ahmed T, Islam N, Chang CL, Lowe AJ. How to Prevent Atopic Dermatitis (Eczema) in 2024: Theory and Evidence. J Allergy Clin Immunol Pract. 2024 Jul;12(7):1695–1704. doi: 10.1016/j.jaip.2024.04.048. [DOI] [PubMed] [Google Scholar]
- 47.Pires L, González-Paramás AM, Heleno SA, Calhelha RC. Exploring Therapeutic Advances: A Comprehensive Review of Intestinal Microbiota Modulators. Antibiotics (Basel) 2024 Aug 1;13(8):720. doi: 10.3390/antibiotics13080720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim JH, Kim K, Kim W. Gut microbiota restoration through fecal microbiota transplantation: a new atopic dermatitis therapy. Exp Mol Med. 2021 May 20;53:907–916. doi: 10.1038/s12276-021-00593-4. [DOI] [PMC free article] [PubMed] [Google Scholar]


