Abstract
It has been proposed that the anti-double-stranded DNA (dsDNA) response in patients with systemic lupus erythematosus (SLE) is antigen driven and that DNA or nucleosomes select anti-DNA reactive, somatically mutated B cells. We have used site-directed mutagenesis to systematically revert the somatic mutations of two human anti-dsDNA antibodies from SLE patients to analyze the resulting changes in DNA binding as well as binding to other autoantigens. Our data demonstrate that high-affinity binding to dsDNA and nucleosomes is acquired by somatic replacement mutations in a stepwise manner. Reactivity to surface structures of apoptotic cells is acquired by the same somatic mutations that generate high-affinity dsDNA binding. Importantly, revertant antibodies with germ-line V regions did not show any measurable DNA reactivity. We propose that anti-DNA autoantibodies are generated from nonautoreactive B cells during a normal immune response. B cells may acquire autoreactivity de novo during the process of somatic hypermutation. Nucleosomes, if available in lupus patients because of defects in clearing of apoptotic debris, might subsequently positively select high affinity anti-DNA B cells.
Keywords: somatic hypermutation, systemic lupus erythematosus
Systemic lupus erythematosus (SLE) is an autoimmune disease characterized by the production of autoantibodies binding to nuclear antigens. Antibodies to double-stranded DNA (anti-dsDNA) are particularly important as serum anti-dsDNA levels correlate with disease activity (1), and anti-dsDNA antibodies have been found deposited in the glomeruli of lupus patients (2). Pathogenic anti-DNA antibodies typically have an IgG isotype and demonstrate high avidity for double-stranded DNA (dsDNA) (2). Sequence analysis of monoclonal anti-DNA antibodies from various murine models for SLE proved that the variable region genes of the H and L chains are somatically mutated. A high ratio of replacement to silent mutations in the complementary determining regions (CDRs) suggested antigen-driven expansion (3-5). Basic amino acids like arginines that are introduced by somatic mutations frequently increase the affinity to the negatively charged DNA (6). This result suggested that DNA or nucleosomes might act as selecting antigens in the process of affinity maturation of anti-dsDNA autoantibodies.
This pattern, consistent with positive selection, has also been described for anti-DNA antibodies from SLE patients (7, 8). Less evidence for DNA-driven selection is available for human anti-dsDNA antibodies as clonally related autoantibodies have not been isolated from individual patients, however. Therefore the affinity maturation toward DNA by somatic mutations remains speculative for human autoantibodies. In addition, the specificity of the primary B cell that gives rise to the high-affinity autoantibody in SLE patients is completely unknown. For the clonally related antibodies from murine models, a low affinity anti-DNA B cell has been suggested as the origin (6).
Whereas it has been shown in mice that anti-dsDNA B cells are deleted in the bone marrow, low-affinity anti-ssDNA B cells are not subjected to central tolerance mechanisms (9, 10). Low-affinity autoreactive B cells normally are anergic but can be activated in an autoimmune background, which leads to the generation of pathological autoantibodies (11). Alternatively, autoreactive B cells with very low affinity to self that are not tolerized might be activated by dual engagement of the antigen-receptor and Toll-like receptors (12). Whether the failure of any of these regulatory checkpoints for autoantibody production is responsible for the generation of anti-dsDNA autoantibodies in SLE remains elusive.
We wanted to directly test whether SLE derived anti-dsDNA antibodies were affinity maturated toward DNA. Furthermore, we intended to characterize the anti-DNA reactivity of the primary B cell that initiated the autoimmune response in the patient. For this purpose, human monoclonal anti-DNA antibodies were systematically subjected to side-directed mutagenesis reverting amino acid substitutions to the germ-line sequence, and the antibody reactivity against DNA and other autoantigens was tested.
Experimental Procedures
Site Directed Mutagenesis and Expression of Human Anti-DNA Antibodies. The Ig variable gene segments from genomic DNA from 33.C9 and 33.F12 hybridomas (8) were cloned into the pCR2.1 vector (Invitrogen). The resulting plasmids were used as a template for the QuikChange Mutagenesis Kit (Stratagene). All plasmids were sequenced to identify clones containing the reversions.
An expression vector was constructed for the expression of IgH genes in myeloma cells. In brief, an EcoRI fragment containing the SP6 genomic VH-D-JH fragment, including the VH promotor and IgH enhancer from the plasmid pRSP6 (13) and a 3-kb XmnI fragment containing the constant region exons of mouse γ2a excised from pCD4-mγ2a-C (obtained from A. Traunecker, formerly of Basel Institute for Immunology, Basel), were cloned into pKO SelectPuro (Stratagene). The VH-D-JH cassette was then replaced by a VH-D-JH fragment from 33.C9 or 33.F12 genomic DNA obtained by PCR with unique NcoI and HindIII sites. VH-D-JH fragments obtained from site-directed mutagenesis were cloned in the same way into the expression vector. For the expression of Igκ chains, the Vκ-Jκ region of 33.C9 or 33.F12 were amplified from genomic DNA and inserted into the SacI and HindIII restriction sites of the plasmid pkm-kp (obtained from A. Traunecker). The vector pkm-kp contains the mouse Igκ constant region, Igκ promotor and enhancer elements, and a neomycin resistance gene. Vκ-Jκ fragments obtained from site-directed mutagenesis were cloned in the same way into the expression vector.
Sp2/0 myeloma cells were cotransfected with linearized H and L chain expression plasmids (30 μg). G418 and puromycin double-resistant clones were screened for IgG production in ELISA as well as heavy- and light-chain protein by intracellular flow cytometry analysis, and the positive clones were cultured in miniPERM cell culture reactors (Vivascience, Hannover, Germany). The culture supernatants were loaded onto a HiTrap protein A column (Amersham Pharmacia) and bound antibodies eluted with ImmunoPure Gentle Elution Buffer (Pierce).
ELISA. Anti-dsDNA and anti-ssDNA ELISA was performed exactly as described in ref. 14 by using native and heat-denatured calf thymic DNA. Antibody binding to dioleoyl phosphatidylserine was measured as described in ref. 15. A commercially available enzyme immunoassay was used for the detection of anti-nucleosome reactivity (Orgentec, Mainz, Germany). Antibodies binding to phospholipids were measured by using the AESKULISA Phospholipid-8Pro ELISA (AESKU Diagnostics, Wendelsheim, Germany).
Surface Plasmon Resonance. The kinetics of Abs binding to immobilized oligonucleotides was determined by using a Biacore 2000 instrument. 3′-biotinylated 32 bp single-stranded and double-stranded oligonucleotides (5′-GTCTGTCTACTTTA CTTGCCTAATCTAGCTAG-3′) were coupled to streptavidin sensor chip surfaces (Sensor Chip SA, Biacore). The binding experiments were performed at a flow rate of 30 μl/min in HBS buffer (10 mM Hepes/0.15 M NaCl/3 mM EDTA/0.05% surfactant P20). Serial dilutions of analytes were injected for 6 min, followed by a 10 min dissociation phase. The surface was regenerated with a 0.15 M MgCl2 pulse. Nonspecific and bulk responses were substracted from an in-line blank reference surface. To calculate kinetic parameters for the Aboligonucleotide interaction, a global analysis of the sensograms was performed with a bivalent model by using biaevaluation 3.0 (Biacore).
Flow Cytometry. To induce apoptosis, Jurkat cells irradiated with UV-B (180 mJ/cm2) were incubated at 37°C for 16 h in RPMI 1640 medium (Gibco), supplemented with 10% FCS. The staining of the apoptotic cells with the 33.C9 and variant antibodies was performed as follows: 2 × 105 cells were incubated for 30 min at 4°C with 10 μg/ml antibody in Ringer's solution. After washing, bound antibodies were detected with FITC-conjugated goat anti-mouse IgG (BD Pharmingen). The samples were measured employing an EPICS flow cytometer (Coulter). Apoptotic and viable cells were gated according to their forward and side scatter characteristics, and membrane integrity was controlled by exclusion of PI-positive cells.
Results
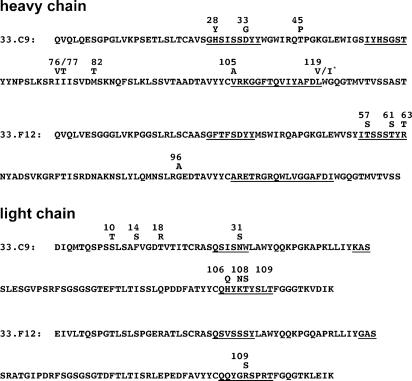
Somatic Mutations Are Essential for dsDNA Binding of the Antibody 33.C9. To evaluate the role of somatic mutations for DNA binding, we choose the human IgG anti-dsDNA antibody 33.C9 because it shares several features of anti-dsDNA antibodies characteristic for SLE (16), and it has been shown to be pathogenic in mice (17). By searching the international ImMuno-GeneTics information system (http://imgt.cines.fr; ref. 18) with the variable region genes (V genes) of 33.C9 (8), we determined the VH-4 gene IGHV4-b (allele 01) and the Vκ gene IGKV1-5 (allele 03) as the corresponding germ-line genes (Fig. 1). Sequencing of the germ-line IGHV4-b gene amplified from genomic DNA of the patient revealed 100% identity with the IGHV4-b allele 01 (GenBank accession no. Z12367). Several somatic mutations in the H and L chain V genes were identified (Fig. 1), which were systematically reverted to germ-line sequences.
Fig. 1.
Amino acid sequences of the anti-DNA antibodies 33.C9 and 33.F12. Positions of mutated amino acid residues are indicated with the germ-line-encoded amino acids shown above the sequence. Underlined amino acids represent the CDRs. *, at position 119 of the VH of 33.C9, the germ-line alleles IGHJ3*01 or IGHJ3*02 encode valine or isoleucin.
The first set of 33.C9 variant antibodies combined H chains encoded by reverted V genes with the original L chain (Table 1). In the VH gene, the replacement mutation Gly to Asp at position 33 in the CDR1 is essential for anti-dsDNA binding, because all mutant antibodies containing either a single D33G reversion or the D33G reversion combined with additional back mutations completely lost detectable dsDNA binding (H chain revertants A, F, and H, Table 1). All of the other revertants of the H chain showed dsDNA binding in ELISA indistinguishable to 33.C9, even the antibody HC-G with all somatic mutations in the VH gene except Asp-33 reverted to germ-line sequences (Table 1). All of the revertants of the L chain initially designed and expressed together with the 33.C9 H chain lost dsDNA binding activity (L chain revertants A-G, Table 1). The only possible explanation for this observation was that the somatic mutations S31N in CDR1 and N108K in CDR3 are both essential for anti-dsDNA binding when paired with the 33.C9 H chain. An additionally created antibody LC-H with the H106Q and the T109S reversions in the CDR3 of the L chain bound dsDNA similar to 33.C9. Because nucleosomes have been suggested to be the selecting antigen for anti-dsDNA B cells (19), we tested all revertants for reactivity to nucleosomes by ELISA. The reactivity to nucleosomes perfectly correlated with dsDNA reactivity (Table 1).
Table 1. Binding of 33.C9 and variants to dsDNA and nucleosomes.
| Reversions to germ-line sequence*
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| H chain revertants
|
L chain revertants
|
||||||||||||||
| Antibody | H28Y | D33G | T45P | I76V | I77T | M82T | S10T | F14S | T18R | N31S | H106Q | K108N | T109S | dsDNA† | Nucleosomes† |
| 33.C9 | 100 | 100 | |||||||||||||
| HC-A | • | <1 | <1 | ||||||||||||
| HC-B | • | 102 | 108 | ||||||||||||
| HC-C | • | • | • | 98 | 102 | ||||||||||
| HC-D | • | • | • | • | 105 | 114 | |||||||||
| HC-E | • | 98 | 91 | ||||||||||||
| HC-F | • | • | • | <1 | <1 | ||||||||||
| HC-G | • | • | • | • | • | 90 | 101 | ||||||||
| HC-H | • | • | • | • | • | • | <1 | <1 | |||||||
| LC-A | • | <1 | <1 | ||||||||||||
| LC-B | • | <1 | nd | ||||||||||||
| LC-C | • | • | • | <1 | <1 | ||||||||||
| LC-D | • | • | • | • | <1 | <1 | |||||||||
| LC-E | • | • | • | • | • | <1 | <1 | ||||||||
| LC-F | • | • | • | • | • | • | <1 | <1 | |||||||
| LC-G | • | • | • | • | • | • | • | <1 | <1 | ||||||
| LC-H | • | • | 80 | 110 | |||||||||||
nd, not determined.
Somatic mutations were reverted to germ-line sequence at the positions indicated by black circles. H chain revertants were combined with the original 33.C9 L chain and L chain revertants with the original 33.C9 H chain.
Binding of 33.C9 and variants in solid-phase ELISA. Results are expressed in relative units using 33.C9 as standard, which was set to 100 relative units.
These results provided direct evidence that somatic replacement mutations are essential for dsDNA binding of a human anti-dsDNA antibody. Each of three somatic mutations, one in the heavy chain and two in the light chain, appeared indispensable for dsDNA binding.
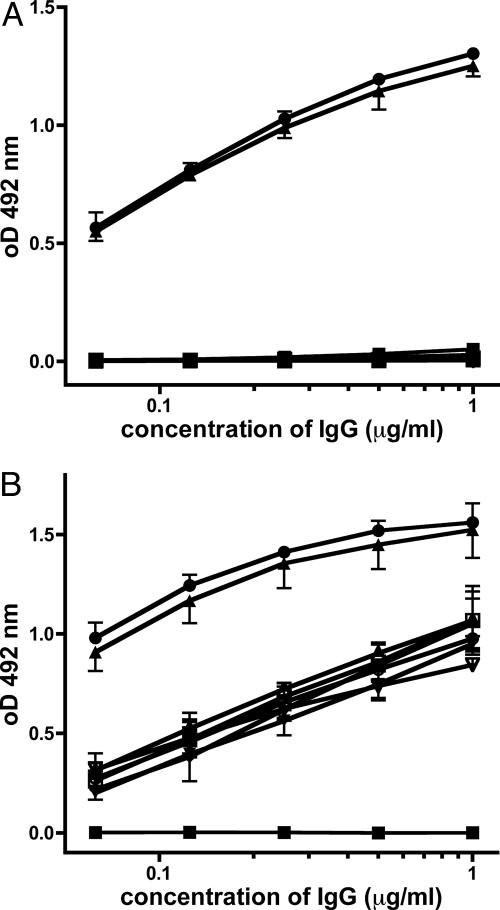
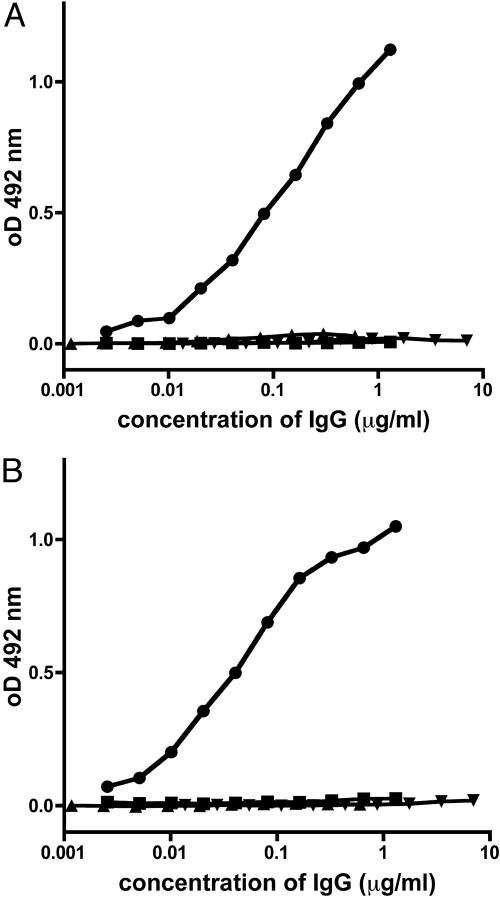
Stepwise Acquisition of ssDNA and dsDNA Reactivity Through Somatic Mutation. We next combined the H and L chains, which have been fully reverted to germ-line sequence to analyze DNA reactivity. In ELISA, the germ-line revertant did not bind significantly to dsDNA, ssDNA, or nucleosomes (Fig. 2 and Table 2). Even with surface plasmon resonance, which is able to detect low-affinity interactions (20), no binding to single- or double-stranded oligonucleotides was detectable (Table 2). We then introduced the three somatic mutations, which were identified before as being essential for dsDNA binding individually and in all possible combinations into the scaffold of the germ-line revertant. Interestingly, ssDNA reactivity was already gained by each of the three individual mutations, the G33D mutation of the H chain and the S31N and N108K mutations of the L chain (Fig. 2 and Table 2). Combinations of each of two mutations did not significantly alter ssDNA binding, whereas the triple-mutant and 33.C9 showed significantly stronger binding. Importantly, only when all three somatic mutations were introduced, dsDNA reactivity of the antibody and reactivity to nucleosomes could be detected (Fig. 2 A and Table 2). The triple mutant and the 33.C9 antibody showed identical dsDNA binding. Therefore, the three somatic mutations identified were not only essential for dsDNA binding, but also sufficient to generate an anti-dsDNA antibody with binding characteristics indistinguishable from the original 33.C9 antibody. Taken together, our systematic remutation analysis of the antibody 33.C9 demonstrates that high-affinity binding to dsDNA and nucleosomes is acquired by somatic replacement mutations. Importantly, the germ-line-encoded revertant antibody did not show any measurable DNA specificity.
Fig. 2.
Anti-DNA reactivity of 33.C9 and variants. Binding of purified IgG to dsDNA (A) and binding to ssDNA in solid phase ELISA (B). Absorbance readings corresponding to the indicated concentrations of IgG were taken at 492 nm. Representative results for 33.C9 (•), its complete reversion to germ-line sequence (▪) and the mutants 33D/31N/108K (▴), 33D (▾), 31N (⋄), 108K (○), 31N/108K (□), 33D/31N (▵), and 33D/108K (▿).
Table 2. Binding of 33.C9 and variants to autoantigens.
| Presence of mutation*
|
Specificity
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| H chain
|
L chain
|
dsDNA ELISA†
|
dsOligo SPR,‡ M
|
Nucleosomes ELISA†
|
ssDNA ELISA†
|
ssOligo SPR,‡ M
|
Apoptotic cells FACS§
|
||
| Antibody | 33D | 31N | 108K | ||||||
| 33.C9 | • | • | • | 100 | 1 × 10−8 | 100 | 100 | 3 × 10−9 | + |
| Germ line | <1 | nb | <1 | <1 | nb | ns | |||
| 33D/31N/108K | • | • | • | 109 | 1 × 10−8 | 89 | 91 | 3 × 10−9 | + |
| 33D/31N | • | • | <1 | nb | <1 | 7 | 2 × 10−8 | ns | |
| 33D/108K | • | • | <1 | nb | <1 | 6 | 2 × 10−8 | nd | |
| 31N/108K | • | • | <1 | nb | <1 | 7 | 7 × 10−8 | ns | |
| 33D | • | <1 | nb | <1 | 8 | 3 × 10−8 | ns | ||
| 31N | • | <1 | nb | <1 | 7 | 6 × 10−8 | ns | ||
| 108K | • | <1 | nb | <1 | 6 | 3 × 10−8 | nd | ||
ns, not significant; nb, no binding detectable at 20 μg/ml IgG; nd, not determined.
Somatic mutations present at the positions indicated by black circles.
Binding of 33.C9 and variants in solid-phase ELISA. Results are expressed in relative units by using 33.C9 as a standard, which was set to 100 relative units.
Binding kinetics were analyzed by SPR with oligonucleotides as ligands and antibodies as analytes and calculated Kd values are presented.
Flow cytometric analysis of binding to apoptotic Jurkat cells, +, significant binding (P < 0.05, t test).
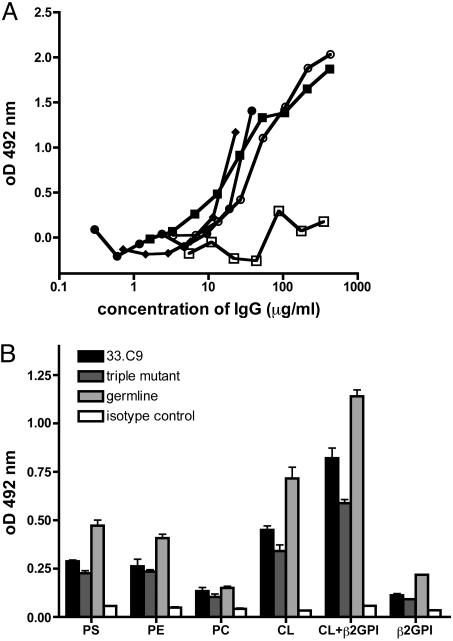
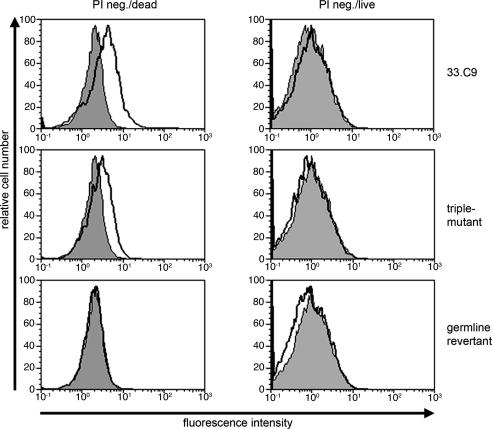
Antibody Reactivity to Phospholipids and Apoptotic Cells. Many anti-DNA antibodies show crossreactions with phospholipids (21), and murine anti-DNA antibodies have been shown to bind to apoptotic cells (10, 15), most likely due to binding of phosphatidylserine exposed on the surfaces of apoptotic cells (22). We therefore tested the antibody 33.C9 and antibody revertants for binding to phosphatidylserine by ELISA. As shown in Fig. 3A, the antibody 33.C9 as well as the germ-line revertant and the triple mutant exhibited detectable, albeit low, binding to dioleoyl phosphatidylserine when compared with control monoclonal antibodies. Further analyses of binding to various phospolipids in ELISA revealed a broad reactivity of 33.C9 and the germ-line revertant and the triple mutant to phospholipids (Fig. 3B). In addition, 33.C9, the triple mutant, and the germ-line revertant all bind to cardiolipin (Fig. 3B). β2-glycoprotein 1 enhances the binding to cardiolipin but is not recognized by 33.C9 and the antibody variants (Fig. 3B). When the antibodies were tested for binding to apoptotic cells generated by UV irradiation, only the antibody 33.C9 and the triple-mutant antibody bound significantly and reproducibly to PI-negative cells with forward and side scatter characteristics of apoptotic cells (Fig. 4 and Table 2). This population of cells entirely stained positive for annexin V and, therefore, represented apoptotic cells. No binding to viable cells above background levels was observed for any of the antibodies tested (Fig. 4). Late apoptotic particles (PI positive) were recognized by the 33.C9 and the triple-mutant antibody but not by the germ-line revertant and control antibodies (data not shown). These results suggest that reactivity to surface structures of apoptotic cells is acquired by the same somatic mutations that generate high-affinity dsDNA binding.
Fig. 3.
Antibody reactivity to phospholipids. (A) Phosphatidylserine ELISA of 33.C9 and variants. Dioleoyl phosphatidylserine binding is given as OD at 492 nm minus the average value of the blank wells. In contrast to the mouse IgG2a isotype control (□), the 33.C9 mAb (•), its complete reversion to germ-line sequence (▪), and the triple mutant 33D/31N/108K (♦) all bind to dioleoyl phosphatidylserine in solid-phase ELISA. A serum pool from lupus-prone MRL/lpr mice with equilibrated IgG concentration (○) was used as a positive control; representative binding curves are from two independent experiments (B) Phospholipid ELISA of 33.C9 and variants. Binding of purified IgG (2 μg/ml) to phosphatidylserine (PS), phosphatidylethanolamine (PE), phosphatidylcholine (PC), and cardiolipin (CL), with or without β2-glycoprotein I (β2GPI) are given as absorbance readings at OD at 492 nm (mean ± SD).
Fig. 4.
Antibody reactivity to apoptotic cells. Flow cytometric analysis of 33.C9 and variants binding to apoptotic Jurkat cells. UV-B treated Jurkat cells stained with 33.C9 or antibody variants were gated according light scatter and PI uptake into viable and apoptotic PI-negative cells. Stainings with an isotype control antibody and secondary reagent are overlaid as shaded histograms.
Specificity of the 33.C9 Germ-Line Revertant. To further determine whether the revertant antibody with germ-line V genes is self-reactive, we tested for binding to nuclear and cytoplasmic antigens on HEp-2 cells and a variety of organ sections used in standard clinical assays for autoantibodies. No binding above background was observed on HEp-2 cells or on kidney, lung, liver, and stomach sections (data not shown). Because it has been shown that DNA reactive B cells might arise as a consequence of somatic hypermutation from antibacterial antibodies (23), we also tested binding of the germ-line revertant to Salmonella typhimurium. No specific binding was observed (data not shown). Interestingly, however, binding of the germ-line-encoded antibody to the flagellum and the basal body of Crithidia luciliae was observed (data not shown). No binding to the kinetoplast, which is used as a diagnostic substrate for anti-dsDNA antibodies, was detectable. Because natural autoantibodies frequently exhibit binding to tubulin (24), which is a major constituent of the protozoan flagellum, we suspected reactivity of the germ-line revertant to tubulin. This possibility is unlikely, because a monoclonal anti-tubulin antibody showed a different staining pattern with the C. luciliae substrate (data not shown) and because the antibody did not bind to ciliae in lung tissue containing tubulin (data not shown). The antibody 33.C9 and the triple-mutant antibody showed staining of the kinetoplast but no binding to the flagellum (data not shown). Taken together the germ-line V sequence-encoded antibody, from which the 33.C9 anti-dsDNA antibody had originated, has no detectable autoreactivity.
Loss of DNA Reactivity in a Second Anti-dsDNA Antibody by Reversion of Somatic Mutations. Because the antibody 33.C9 might be exceptional for the findings described above, we systematically reverted the somatic mutations in a second anti-dsDNA antibody 33.F12 (Fig. 1; refs. 8 and 16). As shown in Fig. 5, reversion of the somatic mutations in the CDR2 of the H chain and the R109S reversion independently resulted in complete loss of detectable dsDNA and ssDNA binding. In addition, the complete germ-line revertant antibody did not show any detectable binding to DNA (Fig. 5). Thus, also for a second SLE-derived anti-dsDNA antibody, high-affinity binding to dsDNA is acquired by somatic replacement mutations.
Fig. 5.
Anti-DNA reactivity of 33.F12 and variants. Binding of purified IgG to dsDNA (A) and binding to ssDNA (B) in solid-phase ELISA. Absorbance readings corresponding to the indicated concentrations of IgG were taken at 492 nm. Representative results for 33.F12 (•), its complete reversion to germ-line sequence (▪), H chain revertant (T57S, T61S and R63T; ▾), and L chain revertant (R109S; ▴).
Discussion
In this report, we show that anti-dsDNA autoantibodies from SLE patients can develop from non-DNA reactive B cells through somatic hypermutation. A stepwise maturation from a non-anti-DNA reactive B cell to an anti-dsDNA autoreactive B cell is suggested by our data. These findings are very similar to anti-DNA reactive B cells developing in murine models of SLE and suggest that positive selection by DNA, nucleosomes, or a crossreactive antigen operated on these autoantibodies (3, 5, 25). In contrast to the best characterized and prototypic murine anti-dsDNA antibody 3H9 derived from MRL/lpr mice (6), the complete germ-line revertants of the V regions of two SLE-derived autoantibodies 33.C9 and 33.F12 exhibited no detectable ssDNA or dsDNA binding. For a third SLE derived anti-dsDNA antibody 32.B9 (8, 16), a complete revertant of the light chain also did not bind to ssDNA or dsDNA (26). Given the limited number of SLE derived human anti-dsDNA antibodies analyzed here, other mechanisms for the development of anti-dsDNA autoantibodies in SLE cannot be excluded, however.
Arginines have been shown to frequently enhance antibody affinity for DNA (27-29). Here, we show that this situation is clearly also the case for the human antibody 33.F12. For the antibody 33.C9, however, none of the three amino acid exchanges that are essential and sufficient to create a high-affinity anti-dsDNA antibody are arginines. Whereas it is compatible with our current understanding of protein-DNA interactions that the asparagine and lysine residues derived from somatic mutation in the light chain of 33.C9 are involved in the contact to DNA, the role of the aspartic acid in the CDR1 of the 33.C9 heavy chain remains difficult to explain. One possible explanation would be that the aspartic acid is involved in the interaction with histones in nucleosomes most likely being the selecting antigen. However, our finding that the aspartic acid at position 33 of the H chain is likewise crucial for the binding capacity of 33.C9 to synthetic dsDNA oligonucleotides argues against this explanation. Without a crystal structure of the variable region of the antibody bound to DNA, the exact role of the aspartic acid residue in mediating DNA binding remains elusive.
Our results with the 33.C9 antibody show that reactivity to surface structures of apoptotic cells is acquired by the same somatic mutations that generate high affinity dsDNA and nucleosome binding. The work of Casciola-Rosen et al. (30) demonstrated that several autoantigens targeted in SLE are present in the blebs at the surfaces of apoptotic cells. However, for being a selecting antigen for B lymphocytes, the autoantigens need to be accessible, which is easily conceivable in the situation when the apoptotic cells undergo secondary necrosis caused by delayed clearance. Furthermore, our findings here are compatible with recent findings that nucleosomes are accessible for anti-DNA autoantibodies on apoptotic cells (31).
For the initial activation of the B cell in the case of 33.C9, phospholipids might have contributed. The strong binding of the antibody with germ-line-reverted V genes to cardiolipin and recent findings that exposure of cardiolipin molecules on the cell plasma membrane is an early event of the apoptotic cellular program (32) support this view. Subsequent mutations gave rise to DNA binding, creating an antibody with specificity to phospholipids and DNA. Antibodies with shared specificity for phospholipids and DNA are frequently found in SLE (33).
It has been shown earlier that DNA reactive B cells may arise as a consequence of somatic hypermutation of phosphorycholine and phenylarsonate reactive B cells (23, 34). The generation of autoreactive B cell receptors is an inherent hazard of the B cell mutator, even with a tendency to generate anti-DNA antibodies because of the frequent replacement of serine residues with arginine in the CDRs (27) as part of the mutationally active RGYW motif. Because somatic mutations can create autoreactive B cells from harmless, xenoreactive B cells, mechanisms that censor autoreactive mutants are needed in the germinal centers. These mechanisms might be defective in genetically predisposed individuals.
Germinal center B cells are clonally deleted by high concentrations of soluble antigen (35, 36), and similar mechanisms might control autoreactive B cells arising in the germinal center reaction. This censoring mechanism for germinal center B lymphocytes might not be operating if complement-binding nuclear autoantigens are displayed on the surfaces of follicular dendritic cells (FDCs). Rather positive selection of B cells expressing high-affinity BCRs could be mediated through the interaction of the BCR with immune complexes deposited on FDCs (37). Positively selecting nuclear autoantigens might derive from defective clearance of apoptotic material in the germinal centers of SLE patients. Apoptotic cells and nuclear debris were found to be bound to FDCs, most likely through CR2/CD21 (38). Chromatin is well known to activate complement (39) and may thereby represent the antigen for positive selection of anti-DNA reactive germinal center B cells. In the same direction, mice with altered clearance of apoptotic material develop high titers of anti-DNA autoantibodies and a lupus-like disease (40-43). Notably, in autoimmune MFG-E8-deficient mice, the tingible body macrophages have been shown to exhibit an engulfment defect for apoptotic cells (43).
Taken together, at least some human anti-DNA autoantibodies are generated from non-DNA reactive B cells during the process of somatic hypermutation. Defective clearance of apoptotic cells in the germinal center might favor the positive selection of high-affinity anti-DNA antibodies.
Acknowledgments
We thank Drs. Reinhard Voll, Ole-Petter Rekvig, and Ari Waisman for critical reading of our manuscript. This work was supported by the Deutsche Forschungsgemeinschaft through SFB 423.
Author contributions: U.W., J.R.K., and T.H.W. designed research; U.W., M.L., M.H., and S.A. performed research; U.W., M.L., M.H., and T.H.W. analyzed data; and T.H.W. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: dsDNA, double-stranded DNA; anti-dsDNA, antibodies to dsDNA; CDR, complementary determining region; SLE, systemic lupus erythematosus.
References
- 1.ter Borg, E., Horst, G., Hummel, E., Limburg, P. & Kallenberg, C. (1990) Arthritis Rheum. 33, 634-643. [DOI] [PubMed] [Google Scholar]
- 2.Winfield, J. B., Faiferman, I. & Koffler, D. (1977) J. Clin. Invest. 59, 90-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shlomchik, M. J., Aucoin, A. H., Pisetsky, D. S. & Weigert, M. G. (1987) Proc. Natl. Acad. Sci. USA 84, 9150-9154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eilat, D., Hochberg, M., Tron, F., Jacob, L. & Bach, J.-F. (1989) Eur. J. Immunol. 19, 1241-1246. [DOI] [PubMed] [Google Scholar]
- 5.Marion, T. N., Tillman, D. M. & Jou, N. T. (1990) J. Immunol. 145, 2322-2332. [PubMed] [Google Scholar]
- 6.Radic, M. Z., Mackle, J., Erikson, J., Mol, C., Anderson, W. F. & Weigert, M. (1993) J. Immunol. 150, 4966-4977. [PubMed] [Google Scholar]
- 7.van Es, J. H., Gmelig-Meyling, F. H., van-de-Akker, W. R., Aanstoot, H., Derksen, R. H. & Logtenberg, T. (1991) J. Exp. Med. 173, 461-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Winkler, T. H., Fehr, H. & Kalden, J. R. (1992) Eur. J. Immunol. 22, 1719-1728. [DOI] [PubMed] [Google Scholar]
- 9.Chen, C., Nagy, Z., Radic, M. Z., Hardy, R. R., Huszar, D., Camper, S. A. & Weigert, M. (1995) Nature 373, 252-255. [DOI] [PubMed] [Google Scholar]
- 10.Xu, H., Li, H., Suri-Payer, E., Hardy, R. R. & Weigert, M. (1998) J. Exp. Med. 188, 1247-1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brard, F., Shannon, M., Prak, E. L., Litwin, S. & Weigert, M. (1999) J. Exp. Med. 190, 691-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leadbetter, E. A., Rifkin, I. R., Hohlbaum, A. M., Beaudette, B. C., Shlomchik, M. J. & Marshak-Rothstein, A. (2002) Nature 416, 603-607. [DOI] [PubMed] [Google Scholar]
- 13.Rusconi, S. & Kohler, G. (1985) Nature 314, 330-338. [DOI] [PubMed] [Google Scholar]
- 14.Wellmann, U., Werner, A. & Winkler, T. H. (2001) Eur. J. Immunol. 31, 2800-2810. [DOI] [PubMed] [Google Scholar]
- 15.Cocca, B. A., Seal, S. N., D'Agnillo, P., Mueller, Y. M., Katsikis, P. D., Rauch, J., Weigert, M. & Radic, M. Z. (2001) Proc. Natl. Acad. Sci. USA 98, 13826-13831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Winkler, T. H., Jahn, S. & Kalden, J. R. (1991) Clin. Exp. Immunol. 85, 379-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ehrenstein, M. R., Katz, D. R., Griffiths, M. H., Papadaki, L., Winkler, T. H., Kalden, J. R. & Isenberg, D. A. (1995) Kidney Int. 48, 705-711. [DOI] [PubMed] [Google Scholar]
- 18.Lefranc, M. P. (2003) Nucleic Acids Res. 31, 307-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burlingame, R. W., Boey, M. L., Starkebaum, G. & Rubin, R. L. (1994) J. Clin. Invest. 94, 184-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jonsson, U., Fagerstam, L., Ivarsson, B., Johnsson, B., Karlsson, R., Lundh, K., Lofas, S., Persson, B., Roos, H., Ronnberg, I., et al. (1991) BioTechniques 11, 620-627. [PubMed] [Google Scholar]
- 21.Lafer, E. M., Rauch, J., Andrzejewski, C., Jr., Mudd, D., Furie, B., Schwartz, R. S. & Stollar, B. D. (1981) J. Exp. Med. 153, 897-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fadok, V. A., Voelker, D. R., Campbell, P. A., Cohen, J. J., Bratton, D. L. & Henson, P. M. (1992) J. Immunol. 148, 2207-2216. [PubMed] [Google Scholar]
- 23.Diamond, B. & Scharff, M. D. (1984) Proc. Natl. Acad. Sci. USA 81, 5841-5844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dighiero, G., Lymberi, P., Mazie, J. C., Rouyre, S., Butler-Browne, G. S., Whalen, R. G. & Avrameas, S. (1983) J. Immunol. 131, 2267-2272. [PubMed] [Google Scholar]
- 25.Pewzner-Jung, Y., Simon, T. & Eilat, D. (1996) J. Immunol. 156, 3065-3073. [PubMed] [Google Scholar]
- 26.Fehr, H. (1997) Dissertation (Naturwissenschafliche Fakultät, Friedrich-Alexander-Universität Erlangen-Nürnberg, Erlangen, Germany).
- 27.Radic, M. Z. & Weigert, M. (1994) Annu. Rev. Immunol. 12, 487-520. [DOI] [PubMed] [Google Scholar]
- 28.Roben, P., Barbas, S. M., Sandoval, L., Lecerf, J. M., Stollar, B. D., Solomon, A. & Silverman, G. J. (1996) J. Clin. Invest. 98, 2827-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rahman, A., Haley, J., Radway-Bright, E., Nagl, S., Low, D. G., Latchman, D. S. & Isenberg, D. A. (2001) J. Mol. Biol. 307, 149-160. [DOI] [PubMed] [Google Scholar]
- 30.Casciola-Rosen, L. A., Anhalt, G. & Rosen, A. (1994) J. Exp. Med. 179, 1317-1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Radic, M., Marion, T. & Monestier, M. (2004) J. Immunol. 172, 6692-6700. [DOI] [PubMed] [Google Scholar]
- 32.Sorice, M., Circella, A., Misasi, R., Pittoni, V., Garofalo, T., Cirelli, A., Pavan, A., Pontieri, G. M. & Valesini, G. (2000) Clin. Exp. Immunol. 122, 277-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rahman, A., Menon, S., Latchman, D. S. & Isenberg, D. A. (1996) Semin. Arthritis Rheum. 26, 515-525. [DOI] [PubMed] [Google Scholar]
- 34.Casson, L. P. & Manser, T. (1995) J. Exp. Med. 182, 743-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pulendran, B., Kannourakis, G., Nouri, S., Smith, K. G. & Nossal, G. J. (1995) Nature 375, 331-334. [DOI] [PubMed] [Google Scholar]
- 36.Shokat, K. M. & Goodnow, C. C. (1995) Nature 375, 334-338. [DOI] [PubMed] [Google Scholar]
- 37.MacLennan, I. C. (1994) Annu. Rev. Immunol. 12, 117-139. [DOI] [PubMed] [Google Scholar]
- 38.Baumann, I., Kolowos, W., Voll, R. E., Manger, B., Gaipl, U., Neuhuber, W. L., Kirchner, T., Kalden, J. R. & Herrmann, M. (2002) Arthritis Rheum. 46, 191-201. [DOI] [PubMed] [Google Scholar]
- 39.Hicks, P. S., Saunero-Nava, L., Du Clos, T. W. & Mold, C. (1992) J. Immunol. 149, 3689-3694. [PubMed] [Google Scholar]
- 40.Botto, M., Dell'Agnola, C., Bygrave, A. E., Thompson, E. M., Cook, H. T., Petry, F., Loos, M., Pandolfi, P. P. & Walport, M. J. (1998) Nat. Genet. 19, 56-59. [DOI] [PubMed] [Google Scholar]
- 41.Bickerstaff, M. C., Botto, M., Hutchinson, W. L., Herbert, J., Tennent, G. A., Bybee, A., Mitchell, D. A., Cook, H. T., Butler, P. J., Walport, M. J. & Pepys, M. B. (1999) Nat. Med. 5, 694-697. [DOI] [PubMed] [Google Scholar]
- 42.Napirei, M., Karsunky, H., Zevnik, B., Stephan, H., Mannherz, H. G. & Moroy, T. (2000) Nat. Genet. 25, 177-181. [DOI] [PubMed] [Google Scholar]
- 43.Hanayama, R., Tanaka, M., Miyasaka, K., Aozasa, K., Koike, M., Uchiyama, Y. & Nagata, S. (2004) Science 304, 1147-1150. [DOI] [PubMed] [Google Scholar]