Abstract
E-cadherin controls a wide array of cellular behaviors, including cell-cell adhesion, differentiation, and tissue development. We show here that E-cadherin is cleaved specifically by ADAM (a disintegrin and metalloprotease) 10 in its ectodomain. Analysis of ADAM10-deficient fibroblasts, inhibitor studies, and RNA interference-mediated down-regulation of ADAM10 demonstrated that ADAM10 is responsible not only for the constitutive shedding but also for the regulated shedding of this adhesion molecule in fibroblasts and keratinocytes. ADAM10-mediated E-cadherin shedding affects epithelial cell-cell adhesion as well as cell migration. Furthermore, the shedding of E-cadherin by ADAM10 modulates the β-catenin subcellular localization and downstream signaling. ADAM10 overexpression in epithelial cells increased the expression of the β-catenin downstream gene cyclin D1 dose-dependently and enhanced cell proliferation. In ADAM10-deficient mouse embryos, the C-terminal E-cadherin fragment is not generated, and the full-length protein accumulates, highlighting the in vivo relevance for ADAM10 in E-cadherin shedding. Our data strongly suggest that this protease constitutes a major regulatory element for the multiple functions of E-cadherin under physiological as well as pathological conditions.
Keywords: ADAM, cadherin, metalloproteinases, shedding
E-cadherin (epithelial cadherin, uvomorulin) is one of the most important molecules involved in tissue morphogenesis, wound healing, and the maintenance of tissue integrity (1, 2). The extracellular domain of this type I transmembrane glycoprotein interacts homotypically with cadherins on the surface of neighboring cells to form calcium-dependent adherens junctions. The stabilization of intercellular adhesion requires the conserved cytoplasmic domain of E-cadherin, which binds to β-catenin. β-Catenin, in turn, is linked to the cytoskeleton. Cadherin-mediated adhesion must be dynamic to accommodate epithelial growth and remodeling during development and to facilitate wound healing and turnover of epithelia in mature tissues (2, 3). Although proteolytic cleavage of E-cadherin has been suggested to cause rapid changes in cell adhesion, signaling, and apoptosis (3-5), the proteinase responsible for these processes has not been identified.
ADAMs (a disintegrin and metalloproteases), a family of zinc-dependent transmembrane proteins, have been implicated in the ectodomain shedding of various membrane-bound proteins (6, 7). ADAM17 (also known as TACE, TNF-α-converting enzyme) is required for proper epithelial tissue development in mice (8) and shares structural and functional homology with ADAM10. ADAM10 (kuzbanian) plays an essential role during neuronal development in vertebrates and Drosophila (9-11). In addition, the analysis of avian epithelial morphogenesis revealed that ADAM10 shows a very prominent expression in all epithelial tissues, especially in the epidermis, the somitic dermatome and myotome, and the epithelial tissues of the kidney, liver, and heart (12). This expression pattern suggests not only that ADAM10 might be important for neuronal development but also that it may play a significant role in the morphogenesis of epithelial tissues and in tissue remodeling. In the present study, we analyzed the potential role of different ADAMs in E-cadherin shedding and the functional relevance for keratinocyte adhesion, migration, and proliferation.
Materials and Methods
For more detailed information, see Supporting Text, which is published as supporting information on the PNAS web site.
Reagents. Reagents were obtained as follows: phorbol-12 myristate 13-acetate (PMA), staurosporine, and ionomycin were from Sigma. Hydroxamate-based inhibitors GW280264X and GI254023X are described in ref. 13. Complete EDTA-free proteinase inhibitor mixture was obtained from Roche Molecular Biochemicals. γ-Secretase inhibitor L-685,458 was obtained from Calbiochem.
Cell Culture and Transfection. Simian virus large tumor-antigen-immortalized and primary mouse embryonic fibroblast (MEF) cell lines from PS1/2-/-, ADA M10-/-, ADA M15-/-, ADAM17-/- mice and respective WT animals were generated and characterized as described in refs. 11 and 14-16. All cells were grown in DMEM (PAA, Linz, Austria) supplemented with 10% FCS and 1% penicillin/streptomycin. The human keratinocyte cell line HaCaT (17) was generously provided by N. Fusenig (Deutsches Krebsforschungszentrum, Heidelberg). Cells were transfected with FuGENE 6 (Roche Molecular Biochemicals) according to the manufacturer's instructions.
Small Interfering RNA (siRNA) Transfection. The mammalian expression vector pSUPER, kindly provided by T. R. Brummelkamp (The Netherlands Cancer Institute, Amsterdam), was used for expression of siRNA in HaCaT cells. The sequence of the human ADAM10 siRNA was as follows: 5′-GACAUUUCAACCUACGAAU-3′. The sequence was separated by a 9-nt noncomplementary spacer (tctcttgaa) from the corresponding reverse complement of the same 19-nt sequence. These sequences were inserted into the pSUPER backbone after digestion with BglII and HindIII.
Preparation of Primary Mouse Keratinocytes. Murine epidermal keratinocytes were isolated from WT mice as described in ref. 18, with the exception that 3-day-old mice were used instead of embryonic day-17.5 embryos. For each experiment, keratinocytes were freshly isolated, grown to confluence in defined keratinocyte serum-free medium (GIBCO/BRL) supplemented with epidermal growth factor (0.1 ng/ml), bovine pituitary extract (25 μg/ml), 5% streptomycin/penicillin, and 0.07 mM CaCl2.
Adhesion Assays. The adhesion assay was performed as described in refs. 13 and 19. Briefly, HaCaT cells were labeled at 2 × 106 cells per ml in PBS/0.1% BSA with 2.5 μM fluorescent dye (calcein AM, Molecular Probes) at 37°C for 30 min. After washing, cells were resuspended in growth medium and preincubated with the inhibitory E-cadherin antibody DECMA-1 (50 μg/ml), isotype control (50 μg/ml), EGTA (5 mM), PMA (200 ng/ml), and GI254023X (5 μM) plus PMA or left untreated. The labeled HaCaT cells were added to a monolayer of unstained cells at 5 × 104 cells per well in growth medium containing 1 mM CaCl2. The plate was incubated at 37°C for 20 min and then washed repeatedly. The fluorescence signal from the adherent cells was measured before and after washing by using a fluorescence plate reader (Lambda Fluoro 230, MWG Biotech, Ebersberg, Germany) at an excitation wavelength of 480 nm and an emission wavelength of 530 nm. The differences in fluorescence before and after washing were depicted as percentages of adherent cells. All analyses were performed in triplicate.
Whole-Mount Embryo Staining. Embryos were stained as described in ref. 20. Briefly, after heating and incubating in cold methanol containing 3% hydrogen peroxide for 30 min, the embryos were rehydrated with PBS containing 0.1% Triton X-100 and incubated with anti-N-terminal E-cadherin antibody (H108) 1:50 in PBS with 3% BSA. The samples were washed three times in PBS and incubated with horseradish peroxidase-conjugated secondary antibodies (1:1,000 in PBS containing 3% BSA). After washing, the embryos were incubated with diaminobenzidine (Sigma) and 0.015% hydrogen peroxide.
In Vitro Wound Healing. HaCaT cells were seeded in six-well plates (Sarstedt) and transfected with ADAM10 or empty vector and cultured until they reached confluence (48 h). To avoid a proliferative effect, cells were treated with 100 mM hydroxyurea for 24 h (Sigma-Aldrich). A cell-free area was introduced by scraping the monolayer with a pipette tip (10 μl, Sarstedt). After different periods under standard culture conditions, cells were photographed by using an inverted phase-contrast microscope (Zeiss).
Cell Proliferation Assay. HaCaT cells were seeded at an initial number of 20,000 cells into wells of microtiter plates and transfected with ADAM10 or empty vector. After 24 h of incubation under standard culture conditions, cells were pulsed with 0.25 μCi (1 Ci = 37 GBq) per well of [3H]thymidine (Amersham Pharmacia) for 16 h. After the radioactive labeling, cells were briefly frozen to detach them from the plates and harvested by a cell harvester (Inotech, Wohlen, Switzerland). The incorporated radioactivity was quantitated on a liquid scintillation counter (Wallac, Gaithersburg, MD).
Results
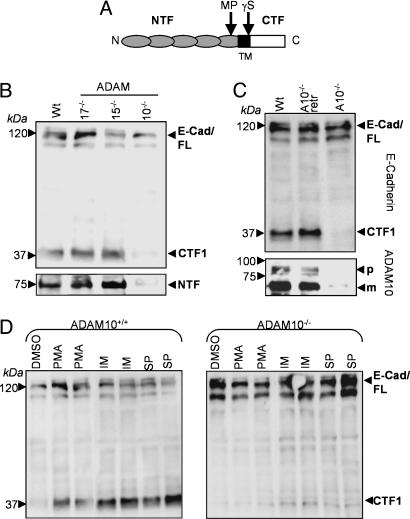
ADAM10 Mediates Shedding of E-Cadherin in MEFs. The full-length 120-kDa E-cadherin protein is cleaved in the extracellular domain by a metalloprotease, generating a 38-kDa C-terminal fragment (CTF) termed CTF1, which can be further processed by a γ-secretase-like activity into a soluble 33-kDa CTF2 (Fig. 1A) (21). To compare the influence of different ADAMs for the ectodomain cleavage of E-cadherin, we compared a panel of ADAM-deficient fibroblasts transfected with E-cadherin. Through Western blot analysis using monoclonal antibodies against the C terminus of E-cadherin, we observed a clear reduction in the generation of the E-cadherin CTF1 in ADAM10-deficient fibroblasts (Fig. 1B). Quantification considering transfection efficiencies indicated a CTF1 reduction of 90% in ADAM10-deficient cells compared with WT cells (see Fig. 6, which is published as supporting information on the PNAS web site). This reduced amount of CTF1 was independent of further processing through γ-secretase activity because the presence of the γ-secretase inhibitor L-685,458 did not change this result (see Fig. 7, which is published as supporting information on the PNAS web site). Accordingly, the released ectodomain of E-cadherin was strongly reduced in the supernatant of ADAM10-deficient cells (Fig. 1B Lower). In addition, E-cadherin shedding could be completely restored in ADAM10-deficient cells after cotransfection of WT ADAM10 (Fig. 1C). We also confirmed the ADAM10-mediated E-cadherin cleavage in a cell-free assay. Recombinant ADAM10 was able to cleave E-cadherin in vitro in a time-dependent manner, resulting in the generation of two fragments with apparent molecular masses of ≈40 and 75 kDa, as evidenced by silverstaining and immunoblotting (see Fig. 8, which is published as supporting information on the PNAS web site).
Fig. 1.
Involvement of ADAM10 in E-cadherin processing. (A) Schematic map of E-cadherin cleavage sites. Full-length E-cadherin is cleaved by a metalloprotease activity (MP) near the transmembrane domain (TM) in 80-kDa N-terminal fragments and 38-kDa CTFs (CTF1), which can be further processed by γ-secretase-like activity (γS) in soluble 33-kDa fragments (CTF2). (B) Constitutive E-cadherin cleavage is strongly reduced in ADAM10-/- fibroblasts. MEFs were transfected with E-cadherin plasmid and harvested 48 h after transfection. Immunoblots with total cell extracts from ADAM10-/-, ADAM15-/-, and ADAM17-/- fibroblasts and WT MEFs were stained with an C-terminal anti-E-cadherin antibody. Supernatants of these cells were also subjected to Western blot analysis using N-terminal anti-E-cadherin antibodies (Lower). (C) ADAM10-/- cells were retransfected with WT ADAM10 (A10-/-retr) and compared with WT and ADAM10-/- MEFs for E-cadherin expression. (D) Effect of different stimuli on E-cadherin shedding. MEFs were stimulated with PMA (200 ng/ml) or vehicle control (DMSO) for 4 h, with ionomycin (IM, 5 μM) for 30 min, or with staurosporine (SP, 1 μM) for 6 h. Cell pellets were subjected to E-cadherin (C-terminal) Western blot analysis. E-Cad/FL, full-length E-cadherin; CTF, CTF of E-cadherin; NTF, N-terminal fragment of E-cadherin; WT, WT MEFs.
For further characterization of inducible E-cadherin proteolysis, the effects of different stimuli were analyzed. Stimulation of protein kinase C using phorbol ester PMA clearly enhanced E-cadherin shedding in WT fibroblasts (Fig. 1D Left). Ionomycin, which promotes shedding of cadherins through stimulation of Ca2+ influx (21), and staurosporine, which induces apoptosis (4), also strongly increased the generation of E-cadherin CTF1. We next addressed to what extent this inducible cleavage depended on ADAM10 by analyzing the effects of these compounds in ADAM10-deficient cells. In these cells, only a very moderate stimulation of E-cadherin shedding could be observed (Fig. 1D Right), suggesting that the majority of the stimulated E-cadherin sheddase activity is due to ADAM10. The densitometric analysis verified this observation (see Fig. 9, which is published as supporting information on the PNAS web site).
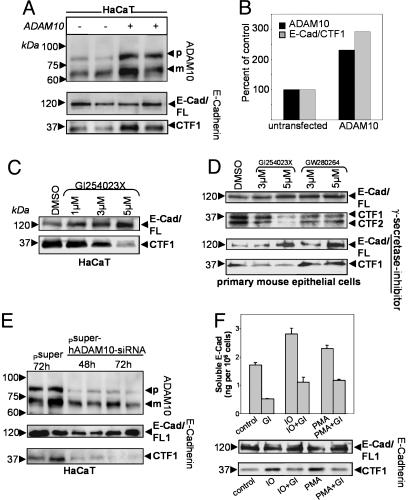
E-Cadherin Cleavage in Epithelial Cells Is ADAM10-Dependent. To analyze E-cadherin processing endogenously, we overexpressed ADAM10 in the human keratinocyte cell line HaCaT. ADAM10 transfection led to an increased level of E-cadherin CTF1 (Fig. 2 A and B). In contrast, GI254023X, an hydroxamate-based inhibitor preferentially blocking ADAM10 (13), reduced E-cadherin shedding in a dose-dependent manner in HaCaT keratinocytes (Fig. 2C; for quantification, see Fig. 10, which is published as supporting information on the PNAS web site). The ADAM10 inhibitor also reduced CTF1 generation in primary epithelial cells compared with the inhibitor GW280623X (preferentially blocking TACE and, to a lesser extent, ADAM10) (Fig. 2D), confirming the essential role of ADAM10 in this process. Comparable with our findings in MEFs, this reduction was independent of the presence of γ-secretase inhibitors in HaCaT cells (see Fig. 8) as well as in primary epithelial cells (Fig. 2D Lower). To further analyze the role of endogenously expressed ADAM10 in E-cadherin shedding in HaCaT cells, we used RNA interference (22). ADAM10 siRNA decreased E-cadherin CTF1 generation to 5% of the mock-transfected level after 72 h (Fig. 2E; see also Fig. 11, which is published as supporting information on the PNAS web site). These results clearly implicate ADAM10 in the physiological E-cadherin shedding in keratinocytes. To investigate the inducible cleavage of E-cadherin in epithelial cells, HaCaT cells were incubated with PMA or ionomycin in the presence or absence of the ADAM10 inhibitor GI254023X. Serum-free supernatants were analyzed for soluble E-cadherin by an ELISA system. As shown in Fig. 2F, the ADAM10 inhibitor abolished not only the constitutive release but also the stimulated E-cadherin release, demonstrating that inducible E-cadherin shedding in epithelial cells is also ADAM10-dependent. The intensity of decrease depended on the concentration of the applied inhibitor (data not shown). We also confirmed the important role of ADAM10 for inducible E-cadherin shedding by immunoblot analysis (Fig. 2F), demonstrating that the increase of CTF1 generation after stimulation was abrogated in the presence of the ADAM10 inhibitor.
Fig. 2.
ADAM10-mediated E-cadherin shedding in epithelial cells. (A) Overexpression of ADAM10 protein in human HaCaT cells. Cells were transiently transfected with ADAM10 or empty vector. Subsequently, cells were lysed and analyzed by Western blotting using anti-ADAM10 antibodies. The same blot was reprobed with anti-E-cadherin antibodies. (B) Quantification of A. For densitometric analysis, ADAM10 and E-cadherin CTF1s were quantificated after transfection and compared with the mock-treated cells (100%). (C and D) Effect of the inhibitor GI254023X (blocking preferentially ADAM10) and GW280623X (blocking preferentially TACE) on constitutive E-cadherin cleavage. HaCaT cells and primary mouse keratinocytes were incubated with various concentrations of GI254023X or vehicle control (DMSO) for 6 h and analyzed by Western blot. The ADAM10 inhibitor also reduced CTF1 generation in the presence of the γ-secretase inhibitor L-685,458 (1 μM). (E) HaCaT cells were transiently transfected with pSUPER-ADAM10 siRNA or empty vector. Cell pellets were harvested 48 and 72 h after transfection and analyzed for ADAM10 expression. Membranes were reprobed with anti-E-cadherin antibodies. (F) Effect of the ADAM10 inhibitor on inducible E-cadherin shedding in HaCaT keratinocytes. Cells were mock-treated (control) or treated with PMA (200 ng/ml) or ionomycin (IO, 5 μM) for 4 h or 30 min, respectively. In parallel, cells were pretreated with the ADAM10 inhibitor GI254023X (GI, 5 μM) or incubated with the ADAM10 inhibitor alone. Supernatants were harvested, and soluble E-cadherin was determined by ELISA (Upper). GI254023X treatment abolished the increased release of soluble E-cadherin but also the increase in CTF1 generation, as evidenced by E-cadherin immunoblots of comparable treated cells (Lower) (independent experiment).
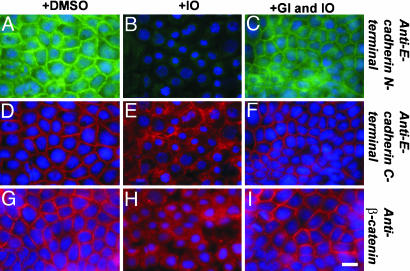
ADAM10 Affects E-Cadherin and β-Catenin Subcellular Localization. Ionomycin treatment is known to induce cleavage of E-cadherin and to disrupt cell-cell adhesion, leading to a dislocation of the C-terminal bound β-catenin (5, 21). To determine the effect of ADAM10 in this process, we analyzed the subcellular localization of E-cadherin by means of immunocytochemistry, using antibodies against the ectodomain and the cytoplasmic domain of E-cadherin in HaCaT cells (Fig. 3). Whereas E-cadherin and β-catenin immunoreactivity was very prominent at cell-cell junctions of mock-treated cells (Fig. 3 A, D, and G), stimulation with ionomycin lead to loss of the E-cadherin ectodomain from the cell surface (Fig. 3B). Accordingly, cells lost cell-cell contacts and exhibited a slightly rounded appearance. The C-terminal E-cadherin immunoreactivity as well as the β-catenin staining became more diffuse and dislocated into the cytoplasm (Fig. 3 E and H). In contrast, cotreatment with the ADAM10 inhibitor GI254023X retained E-cadherin as well as β-catenin at the cell surface, and the morphological integrity was unaffected (Fig. 3 C, F, and I). This observation confirms our biochemical data and strongly suggests that changes in E-cadherin localization, epithelial cell-cell adhesion, and β-catenin translocation are ADAM10-mediated.
Fig. 3.
The ADAM10 inhibitor GI254023X rescues E-cadherin cell surface expression and β-catenin translocation after ionomycin treatment. HaCaT cells were mock-treated (A, D, and G) or stimulated with ionomycin (5 μM) for 30 min in the presence (C, F, and I) or absence (B, E, and H) of the ADAM10 inhibitor GI254023X (GI, 5 μM). Afterward, cells were fixed with PFA and immunostained with N-terminal (green) and C-terminal (red) E-cadherin and β-catenin (red, Bottom) antibodies. Nuclei were counterstained with DAPI. (Scale bar: 10 μm.)
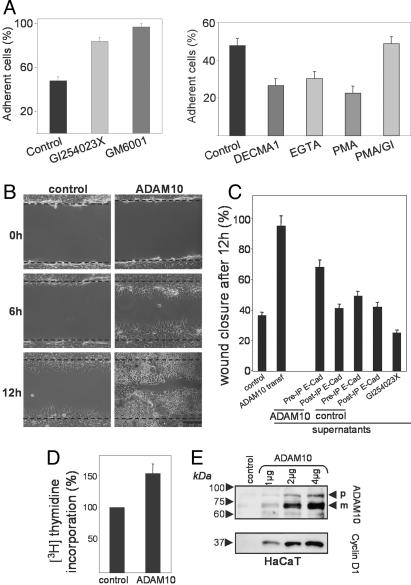
Influence of ADAM10 on Adhesion, Migration, and Proliferation of Epithelial Cells. To elucidate the functional relevance of ADAM10-mediated E-cadherin shedding for cell-cell adhesion, calcein-stained HaCaT cells were seeded on top of an unstained monolayer of corresponding cells in the presence or absence of the general metalloprotease inhibitor GM6001 or the ADAM10 inhibitor GI254023X. After 20 min of incubation, nonadherent cells were washed away, and the remaining fluorescent cells were quantified. As shown in Fig. 4A, both inhibitors strongly increased cell-cell adhesion to a nearly equal level, confirming the importance of metalloproteases in the regulation of cell-cell adhesion and demonstrating that the majority of this activity can be attributed to ADAM10. In contrast, induction of E-cadherin shedding with PMA profoundly reduced cell-cell adhesion, and this effect could be reversed with the ADAM10 inhibitor GI254023X (Fig. 4A Right). The degree of PMA-reduced cell-cell adhesion was comparable with the E-cadherin-mediated and Ca2+-dependent adhesion as evidenced by the use of the inhibitory E-cadherin antibody DECMA-1 (23) and EGTA, respectively. This experiment demonstrates that ADAM10 is critically involved in the constitutive regulation, as well as the inducible regulation, of the adhesive properties of epithelial cells.
Fig. 4.
ADAM10 regulates cell-cell adhesion and migration of epithelial cells. (A) Adhesion of calcein-labeled HaCaT cells on a monolayer of the corresponding cells after 20 min of incubation was analyzed in the presence or absence of the broad metalloprotease inhibitor GM6001 (5 μM) or the ADAM10 inhibitor GI254023X (5 μM). In a similar assay, cells were preincubated with the inhibitory anti-E-cadherin antibody DECMA-1, the corresponding IgG1 isotype (control), and EGTA (5 mM) for 30 min or with PMA (200 ng/ml) or PMA in the presence of GI254023X (GI) for 2 h. Means and SDs from three independent experiments are shown. (B) ADAM10 enhances epithelial migration. HaCaT cells were transfected with empty vector (control) or ADAM10 and grown to confluence. A cell-free area was introduced by scratching with a pipette tip, and migration was evaluated after different times. Micrographs of nonfixed cells at 0, 6, and 12 h are shown. One representative of three independent experiments is shown. (Scale bar: 100 μm.) (C) ADAM10-released soluble E-cadherin increases the migration of keratinocytes. HaCaT cells were mock-transfected (control), ADAM10-transfected, or treated with GI254023X overnight, and the supernatants of these cells were harvested. Supernatants were left untreated (PreIP E-Cad) or E-Cadherin-immunoprecipitated (PostIP E-Cad) and transferred to untreated HaCaT cells. All cells were analyzed for migration in a wound healing assay (see also Figs. 12 and 13). (D) Increased proliferation in keratinocytes overexpressing ADAM10. HaCaT cells were transfected with empty vector (control) or with ADAM10. After 24 h, cells were pulsed with 0.25 μCi per well of [3H]thymidine for an additional 16 h. Values obtained for mock-treated cells were considered 100% and compared with ADAM10-transfected cells (mean and SD; n = 6). (E) Overexpression of ADAM10 leads to a dose-dependent increase of cyclin D1 in keratinocytes. HaCaT cells were transfected with empty vector or different amounts of ADAM10 and analyzed for expression of ADAM10 and cyclin D1.
To address the role of ADAM10 in keratinocyte migration, we tested ADAM10-transfected HaCaT cells in an in vitro model for wound healing (24). In this assay, scrape wounds were generated in confluent HaCaT cultures, and cells were allowed to migrate into the denuded area for 12 h at 37°C. ADAM10-transfected HaCaT cells (40-50% transfection efficiency) started to recover the denuded area 6 h after scratching, and scratch closure was nearly completed after 12 h. In contrast, mock-transfected cells were less motile, as indicated by a lower number of cells in the denuded area after 6 and 12 h (Fig. 4B), demonstrating an ADAM10-dependent effect on epithelial cell migration.
Previously, it has been demonstrated that soluble E-cadherin can disrupt cell-cell adhesion and induce invasion into collagen type I (25, 26). To analyze the contribution of soluble E-cadherin to cell migration, we examined the influence of the supernatant of mock-transfected and ADAM10-transfected HaCaT cells on the migration of untransfected control cells. Indeed, the supernatant of ADAM10-transfected cells increased the cell migration, whereas the supernatant of mock-transfected cells showed only weak effects (Fig. 4C; see also Fig. 12, which is published as supporting information on the PNAS web site). Interestingly, immunoprecipitation of soluble E-cadherin abolished this stimulatory effect (Fig. 4C; see also Fig. 13, which is published as supporting information on the PNAS web site).
Because reepithelialization of the epidermis also requires keratinocyte proliferation, we assayed ADAM10 and mock-transfected HaCaT cells for proliferation by [3H]thymidine incorporation. ADAM10 transfection stimulated the proliferation of HaCaT cells (Fig. 4D). To explore whether this effect could be due to a modified expression of the cell cycle regulator cyclin D1, we analyzed the expression of this β-catenin downstream gene. ADAM10 transfection led to a dose-dependent increase in cyclin D1 expression (Fig. 4E), indicating that ADAM10 might influence proliferation through its effect on β-catenin translocation and β-catenin signal transduction.
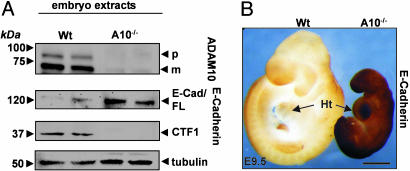
ADAM10-Mediated E-Cadherin Shedding Cannot Be Compensated in Vivo. To further elucidate the in vivo relevance of E-cadherin cleavage by ADAM10, we analyzed extracts of WT and ADAM10-deficient embryos at embryonic day 9.5 by Western blotting. The generation of the E-cadherin CTF1 was almost completely abolished in the ADAM10-deficient embryos, even though the full-length protein was expressed and equal protein was loaded (Fig. 5A). The full-length E-cadherin fragment was even increased ≈4-fold, underlining the essential role of ADAM10 in the regulation of the adhesion-competent, full-length protein. Interestingly, β-catenin downstream genes such as cyclin D1, c-myc, and c-jun were decreased in the ADAM10-deficient embryo (data not shown), supporting an effect of ADAM10 on β-catenin signaling.
Fig. 5.
Loss of E-cadherin ectodomain shedding in ADAM10-deficient embryos. (A) The same amount of proteins from WT embryo and ADAM10-/- embryo extracts (embryonic day 9.5) were analyzed by immunoblot for ADAM10 and E-cadherin cleavage products. (B) Accumulation of full-length E-cadherin in an ADAM10-/- embryo. Whole-mount staining of WT and ADAM10-/- embryos (embryonic day 9.5) with N-terminal E-cadherin (H108) antibodies is shown. Ht, heart. (Scale bar: 500 μm.)
In accordance with the E-cadherin immunoblot analysis, ADAM10-deficient embryos showed dramatically increased amounts of full-length E-cadherin on their surface, as evidenced by whole-mount staining with an N-terminal E-cadherin antibody (Fig. 5B). The specificity of the whole-mount staining was confirmed by analysis of tubulin immunoreactivity, which was comparable in both embryos (see Fig. 14, which is published as supporting information on the PNAS web site). These results demonstrate that ADAM10 is essentially involved in E-cadherin processing in vivo.
Discussion
E-cadherin plays a central role in many aspects of epithelial biogenesis and is essential for epithelial integrity during development (27-29). Regulating E-cadherin function is a dynamic process associated with cellular rearrangements, movements, and pathologic processes such as wound healing. Morphogenetic movements during development but also in postnatal life involve the continual breaking and reforming of cell-cell adhesive contacts (30). Although it has been suggested that proteolytic cleavage of E-cadherin ectodomains influences cadherin-mediated adhesion, the proteinase responsible for this process has not been identified. In this report, we have provided multiple lines of evidence that ADAM10 is critically involved in the proteolytic processing of E-cadherin in vitro and in vivo. Our experiments indicate that ADAM10-mediated cleavage of E-cadherin represents a potent mechanism for regulating cell-cell adhesion, motility, and proliferation of epithelial cells.
Recently, the cleavage of E-cadherin has been attributed to an unidentified membrane-bound metalloprotease (5). Our experiments using different ADAM-deficient fibroblasts and our retransfection studies clearly demonstrate that ADAM10 is critically involved in the constitutive cleavage of E-cadherin. To exclude that the relevance of this finding is restricted to fibroblasts, we examined the role of ADAM10 in epithelial cells. ADAM10 overexpression, inhibitor studies, and RNA interference-mediated down-regulation of endogenous ADAM10 demonstrated that in established and primary epithelial cell lines, ADAM10 represents the major E-cadherin sheddase.
Previous studies have shown that cadherin-mediated adhesion is regulated by a variety of external stimuli (4, 5). In particular, Ca2+ influx is recognized as a critical event in the stimulation of wound healing, leading to increased migration and proliferation (31, 32). Our results, which demonstrate that Ca2+ influx induces the cleavage of E-cadherin depending on ADAM10 activity, offer valuable clues to understanding these processes. Accordingly, the adhesiveness of epithelial cells was increased after inhibition of ADAM10. The important role of ADAM10 is further supported by the in vitro reepithelization assay of this study, which showed that transient transfection of ADAM10 led to increased motility of epithelial cells. In accordance with previous reports that demonstrated that soluble E-cadherin causes scattering of epithelial cells and induction of invasion (25, 26, 33), our data demonstrate that ADAM10-released soluble E-cadherin also contributes to this effect. Therefore, the increased cell migration seems to be a result of ADAM10-mediated abrogation of cell-cell contacts on the one hand and additional effects of increased amounts of soluble E-cadherin on the other hand. However, we cannot exclude that the shedding of other ADAM10 substrates like CD44 that are also important for cell migration (34, 35) might directly or indirectly contribute to the observed increase of migration. ADAM10-mediated E-cadherin shedding also affected β-catenin translocation. β-Catenin is known to bind to transcription factors of the lymphocyte enhancer-binding factor 1/T cell factor pathway to regulate expression of downstream target genes such as c-myc (36) and cyclin D1 (37), which are involved in controlling proliferation. Our data show that ADAM10 modulates β-catenin signaling through regulation of E-cadherin cell surface expression and affects the expression of β-catenin downstream genes in vitro and in vivo. Even though we cannot completely exclude that these effects are also influenced by other ADAM10 substrates, it seems reasonable that the increased cyclin D1 level due to ADAM10 overexpression contributes to the enhanced proliferation of the keratinocytes. Because transcriptional down-regulation, but also posttranscriptional down-regulation, of E-cadherin expression has been discussed as a key mechanism for the increased proliferation and migration of epithelial cancer cells, further studies will have to show whether ADAM10-mediated E-cadherin cleavage might also contribute to tumor progression.
Our data reveal that the interaction of ADAM10 and E-cadherin is also of high relevance for embryonic development in vivo. Our immunoblot analysis, as well as immunostaining of embryos, demonstrated that ADAM10-mediated E-cadherin shedding can apparently not be compensated in the knockout embryo. Because E-cadherin overexpression is known to lead to cell growth arrest and apoptosis (38), it is tempting to speculate that this E-cadherin increase might also contribute to the developmental defects of the ADAM10-deficient mice.
In conclusion, our findings demonstrate that ADAM10 is critically involved in the physiological processing of E-cadherin in vitro and in vivo. The coordinated interaction of ADAM10 and E-cadherin may be significant for an effective interplay among cell-cell adhesion, cell detachment, cell proliferation, and cell survival during development but also under pathological conditions.
Supplementary Material
Acknowledgments
We thank S. Jessen for excellent technical assistance, S. Rose-John (Biochemical Institute, Kiel, Germany) for the ADAM17-deficient MEFs, C. Blobel (Weill Medical College of Cornell University, New York) for the ADAM15-deficient MEFs, N. Fusenig for the HaCaT keratinocytes, and D. Vestweber (Max-Planck-Institut of Molecular Biomedicine, Münster, Germany) for the E-cadherin construct. This work was supported by Hensel Stiftung, Kiel, Deutsche Forschungsgemeinschaft Sonderforschungsbereich 415 (to P.S. and K.R.), Deutsche Forschungsgemeinschaft LU869/1-2 (to A.L.), Interuniversity Attraction Poles Program P5/19 of the Belgian Federal Science Policy Office, and European Union contract LSHM-CT-2003-503330 (APOPIS).
Author contributions: K.R. and P.S. designed research; T.M., K.R., A.L., J.B., F.S., D.H., and P.S. performed research; A.L., F.S., E.P., B.d.S., and D.H. contributed new reagents/analytic tools; F.S., E.P., B.d.S., and D.H. analyzed data; and K.R. and P.S. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ADAM, a disintegrin and metalloprotease; MEF, mouse embryonic fibroblast; CTF, C-terminal fragment; PMA, phorbol-12 myristate 13-acetate; siRNA, small interfering RNA.
References
- 1.Gumbiner, B. M. (1996) Cell 84, 345-357. [DOI] [PubMed] [Google Scholar]
- 2.Takeichi, M. (1991) Science 251, 1451-1455. [DOI] [PubMed] [Google Scholar]
- 3.Gumbiner, B. M. (2000) J. Cell Biol. 148, 399-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Steinhusen, U., Weiske, J., Badock, V., Tauber, R., Bommert, K. & Huber, O. (2001) J. Biol. Chem. 276, 4972-4980. [DOI] [PubMed] [Google Scholar]
- 5.Ito, K., Okamoto, I., Araki, N., Kawano, Y., Nakao, M., Fujiyama, S., Tomita, K., Mimori, T. & Saya, H. (1999) Oncogene 18, 7080-7090. [DOI] [PubMed] [Google Scholar]
- 6.Schlondorff, J. & Blobel, C. P. (1999) J. Cell Sci. 112, Part 21, 3603-3617. [DOI] [PubMed] [Google Scholar]
- 7.Seals, D. F. & Courtneidge, S. A. (2003) Genes Dev. 17, 7-30. [DOI] [PubMed] [Google Scholar]
- 8.Peschon, J. J., Slack, J. L., Reddy, P., Stocking, K. L., Sunnarborg, S. W., Lee, D. C., Russell, W. E., Castner, B. J., Johnson, R. S., Fitzner, J. N., et al. (1998) Science 282, 1281-1284. [DOI] [PubMed] [Google Scholar]
- 9.Rooke, J., Pan, D., Xu, T. & Rubin, G. M. (1996) Science 273, 1227-1231. [DOI] [PubMed] [Google Scholar]
- 10.Pan, D. & Rubin, G. M. (1997) Cell 90, 271-280. [DOI] [PubMed] [Google Scholar]
- 11.Hartmann, D., De Strooper, B., Serneels, L., Craessaerts, K., Herreman, A., Annaert, W., Umans, L., Lubke, T., Lena, I. A., von Figura, K., et al. (2002) Hum. Mol. Genet. 11, 2615-2624. [DOI] [PubMed] [Google Scholar]
- 12.Hall, R. J. & Erickson, C. A. (2003) Dev. Biol. 256, 146-159. [DOI] [PubMed] [Google Scholar]
- 13.Hundhausen, C., Misztela, D., Berkhout, T. A., Broadway, N., Saftig, P., Reiss, K., Hartmann, D., Fahrenholz, F., Postina, R., Matthews, V., et al. (2003) Blood 102, 1186-1195. [DOI] [PubMed] [Google Scholar]
- 14.Herreman, A., Hartmann, D., Annaert, W., Saftig, P., Craessaerts, K., Serneels, L., Umans, L., Schrijvers, V., Checler, F., Vanderstichele, H., et al. (1999) Proc. Natl. Acad. Sci. USA 96, 11872-11877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horiuchi, K., Weskamp, G., Lum, L., Hammes, H. P., Cai, H., Brodie, T. A., Ludwig, T., Chiusaroli, R., Baron, R., Preissner, K. T., et al. (2003) Mol. Cell. Biol. 23, 5614-5624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Black, R. A., Rauch, C. T., Kozlosky, C. J., Peschon, J. J., Slack, J. L., Wolfson, M. F., Castner, B. J., Stocking, K. L., Reddy, P., Srinivasan, S., et al. (1997) Nature 385, 729-733. [DOI] [PubMed] [Google Scholar]
- 17.Boukamp, P., Petrussevska, R. T., Breitkreutz, D., Hornung, J., Markham, A. & Fusenig, N. E. (1988) J. Cell Biol. 106, 761-771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caldelari, R., Suter, M. M., Baumann, D., De Bruin, A. & Muller, E. (2000) J. Invest. Dermatol. 114, 1064-1065. [DOI] [PubMed] [Google Scholar]
- 19.Reiss, K., Maretzky, T., Ludwig, A., Tousseyn, T., De Strooper, B., Hartmann, D. & Saftig, P. (2005) EMBO J. 24, 742-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Radice, G. L., Rayburn, H., Matsunami, H., Knudsen, K. A., Takeichi, M. & Hynes, R. O. (1997) Dev. Biol. 181, 64-78. [DOI] [PubMed] [Google Scholar]
- 21.Marambaud, P., Shioi, J., Serban, G., Georgakopoulos, A., Sarner, S., Nagy, V., Baki, L., Wen, P., Efthimiopoulos, S., Shao, Z., et al. (2002) EMBO J. 21, 1948-1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brummelkamp, T. R., Bernards, R. & Agami, R. (2002) Science 296, 550-553. [DOI] [PubMed] [Google Scholar]
- 23.Pece, S. & Gutkind, J. S. (2000) J. Biol. Chem. 275, 41227-41233. [DOI] [PubMed] [Google Scholar]
- 24.Cha, D., O'Brien, P., O'Toole, E. A., Woodley, D. T. & Hudson, L. G. (1996) J. Invest. Dermatol. 106, 590-597. [DOI] [PubMed] [Google Scholar]
- 25.Wheelock, M. J., Buck, C. A., Bechtol, K. B. & Damsky, C. H. (1987) J. Cell. Biochem. 34, 187-202. [DOI] [PubMed] [Google Scholar]
- 26.Ryniers, F., Stove, C., Goethals, M., Brackenier, L., Noe, V., Bracke, M., Vandekerckhove, J., Mareel, M. & Bruyneel, E. (2002) Biol. Chem. 383, 159-165. [DOI] [PubMed] [Google Scholar]
- 27.Levine, E., Lee, C. H., Kintner, C. & Gumbiner, B. M. (1994) Development (Cambridge, U.K.) 120, 901-909. [DOI] [PubMed] [Google Scholar]
- 28.Hermiston, M. L. & Gordon, J. I. (1995) J. Cell Biol. 129, 489-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Uemura, T., Oda, H., Kraut, R., Hayashi, S., Kotaoka, Y. & Takeichi, M. (1996) Genes Dev. 10, 659-671. [DOI] [PubMed] [Google Scholar]
- 30.Gumbiner, B. M. (1992) Cell 69, 385-387. [DOI] [PubMed] [Google Scholar]
- 31.Tran, P. O., Hinman, L. E., Unger, G. M. & Sammak, P. J. (1999) Exp. Cell Res. 246, 319-326. [DOI] [PubMed] [Google Scholar]
- 32.Coomber, B. L. & Gotlieb, A. I. (1990) Arteriosclerosis 10, 215-222. [DOI] [PubMed] [Google Scholar]
- 33.Noe, V., Willems, J., Vandekerckhove, J., Roy, F. V., Bruyneel, E. & Mareel, M. (1999) J. Cell Sci. 112, Part 1, 127-135. [DOI] [PubMed] [Google Scholar]
- 34.Nagano, O., Murakami, D., Hartmann, D., De Strooper, B., Saftig, P., Iwatsubo, T., Nakajima, M., Shinohara, M. & Saya, H. (2004) J. Cell Biol. 165, 893-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murai, T., Miyazaki, Y., Nishinakamura, H., Sugahara, K. N., Miyauchi, T., Sako, Y., Yanagida, T. & Miyasaka, M. (2004) J. Biol. Chem. 279, 4541-4550. [DOI] [PubMed] [Google Scholar]
- 36.He, T. C., Sparks, A. B., Rago, C., Hermeking, H., Zawel, L., da Costa, L. T., Morin, P. J., Vogelstein, B. & Kinzler, K. W. (1998) Science 281, 1509-1512. [DOI] [PubMed] [Google Scholar]
- 37.Shtutman, M., Zhurinsky, J., Simcha, I., Albanese, C., D'Amico, M., Pestell, R. & Ben Ze'ev, A. (1999) Proc. Natl. Acad. Sci. USA 96, 5522-5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stockinger, A., Eger, A., Wolf, J., Beug, H. & Foisner, R. (2001) J. Cell Biol. 154, 1185-1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boterberg, T., Bracke, M., Bruyneel, E. & Mareel, M. (2001) in Methods in Molecular Medicine, eds. Brooks, S. & Schumacher, U. (Humana, Totowa, NJ), pp. 33-45. [DOI] [PubMed]
- 40.Gottardi, C. J., Wong, E. & Gumbiner, B. M. (2001) J. Cell Biol. 153, 1049-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.