Abstract
Peptide generation by the proteasome is rate-limiting in MHC class I-restricted antigen presentation in response to IFN-γ. IFN-γ-induced de novo formation of immunoproteasomes, therefore, essentially supports the rapid adjustment of the mammalian immune system. Here, we report that the molecular interplay between the proteasome maturation protein (POMP) and the proteasomal β5i subunit low molecular weight protein 7 (LMP7) has a key position in this immune adaptive program. IFN-γ-induced coincident biosynthesis of POMP and LMP7 and their direct interaction essentially accelerate immunoproteasome biogenesis compared with constitutive 20S proteasome assembly. The dynamics of this process is determined by rapid LMP7 activation and the immediate LMP7-dependent degradation of POMP. Silencing of POMP expression impairs recruitment of both β5 subunits into the proteasome complex, resulting in decreased proteasome activity, reduced MHC class I surface expression, and induction of apoptosis. Furthermore, our data reveal that immunoproteasomes exhibit a considerably shortened half-life, compared with constitutive proteasomes. In consequence, our studies demonstrate that the cytokine-induced rapid immune adaptation of the proteasome system is a tightly regulated and transient response allowing cells to return rapidly to a normal situation once immunoproteasome function is no longer required.
Keywords: antigen presentation, immunoproteasome, MHC class I
The proteasome is the key enzyme in the proteolytic cascade required for the generation of peptides presented to cytotoxic T lymphocytes by MHC class I molecules. Within this cascade, the 26S proteasome is responsible for the initial selective degradation of polyubiquitinated cellular protein substrates. This multisubunit enzyme is formed by the catalytic 20S core complex and two 19S regulator complexes that are responsible for the binding and unfolding of ubiquitinated substrates (1, 2). The 20S proteasome is composed of 14 nonidentical subunits building four stacked rings of seven subunits each. Seven different but related α subunits form the two outer rings, whereas the two inner rings contain seven different β subunits. The hydrolyzing activities of the 20S core are conferred by three of the seven β subunits, i.e., subunits β1, β2, and β5 (3), located in both of the two inner heptameric β rings.
In vertebrates, specific catalytically active proteasome subunits, collectively referred to as immunosubunits (4), have evolved that improve proteasome-dependent antigen processing (5-7). IFN-γ induces the synthesis of the immunosubunits β1i [also named low molecular weight protein 2 (LMP2)], β2i [multicatalytic endopeptidase complex-like 1 (MECL1)], and β5i (LMP7). These subunits are cooperatively incorporated into nascent proteasomes, thereby replacing their constitutive homologues β1, β2, and β5 (8-11). Thus, there exist two types of proteasomes, i.e., constitutive proteasomes (c20S) that are constitutively expressed in all cells and immunoproteasomes (i20S) that are formed upon exposure of cells to IFN-γ.
Both c20S and i20S are exclusively formed de novo following a sophisticated and not yet fully understood biogenesis program. We previously showed that assembly of mammalian 20S proteasomes is a multistep process that occurs via the formation of distinct proteasome precursor complexes with different β subunit compositions. Active-site β subunits are synthesized and incorporated as proproteins that essentially mature by a two-step procedure within the precursor complexes (12, 13). Final activation of the β subunits requires the formation of the preholoproteasome assembly intermediate. Concomitantly, the cis- and trans-autocatalytic removal of the β subunit propeptides liberates the active-site threonines of the now fully active 20S core proteasome (8, 10, 12-16).
Eukaryotic proteasome biogenesis requires accessory proteins to promote its assembly and final maturation steps. A protein that is directly associated with proteasome precursor complexes is proteasome maturation protein (POMP), also named proteassemblin or human/mouseUmp1 according to their yeast homologue Ump1 (17-20). These factors are proposed to be involved in the coordinate processing of the β subunits, thereby becoming the first substrate of the fully assembled and activated 20S proteasome. Thus, degradation of Ump1p/POMP signals the successful completion of the proteasome biogenesis program (17, 20). Importantly, in mammalian cells, IFN-γ was found to enhance POMP mRNA levels, suggesting that POMP may play an important role in i20S biogenesis (17, 19).
Highlighting the functional importance of i20S, it was shown that deficiency in the immunosubunits β1i or β5i reduces the cytotoxic T lymphocyte repertoire and thus the efficiency of the immune response (5, 6). Impairment of i20S formation has been observed as consequences of oncogenesis (21-23) and virus-induced immune evasion strategies (24).
The availability of peptides for loading of MHC class I molecules limits the assembly of MHC I complexes in the endoplasmic reticulum (25). Moreover, priming of CD8+ T cells upon IFN-γ treatment or infection is a surprisingly rapid process and was shown to be a transient response (26). Thus, all known data indicate that the timely formation and availability of i20S are decisive steps in the rapid adaptation of the antigen processing machinery to the immunological requirements of a challenged organism. The molecular mechanisms of this proteasomal immune adaptation, however, remain purely defined.
The present studies were therefore undertaken to investigate the molecular basis and kinetics of i20S biogenesis and to analyze the role of POMP, β5i/LMP7, and proteasome turnover in this process. Our experiments demonstrate that both POMP and β5i/LMP7 are essential for the accelerated up-regulation of i20S. In combination with the observed drastically reduced half-life of i20S, our experiments explain that proteasomal immune adaptation is designed to be a highly dynamic and transient response that permits the rapid return to the constitutive situation once i20S function is no longer required.
Materials and Methods
Cell Culture and Transfection. Human cell lines were cultivated under standard conditions in RPMI medium 1640 (DLD-1, HeLa, and T2) or Basal Iscove's medium (SW-480), each containing 10% FCS, 2 mM l-glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin. Stable transfectants of T2 cells have been described (11, 27). For induction of i20S, cells were incubated with 150 units/ml human IFN-γ up to 24 h.
Northern and Western Blotting. For Northern blots, 5 μg of total RNA per lane were processed and hybridized with digoxygenin-labeled riboprobes of LMP7/β5i or POMP, as described (28). Equal RNA loading and RNA quality were monitored by ethidium bromide-stained 28S rRNA.
Equal amounts of protein extracts were separated on SDS-Laemmli gels, transferred by electroblotting onto poly(vinylidene difluoride) membranes, and immunodetected for 20S proteasomes (MP3), LMP7, LMP2, β5 (all laboratory stock), POMP (17), or GAPDH (Santa Cruz Biotechnology), as indicated. The protein amount in each lane was equalized by amidoblack staining before immunodetection and served as a loading control.
Cloning Procedures. DNA manipulations and transformation of Escherichia coli were performed according to standard protocols (29). All plasmids were verified by sequencing.
Cloning of both untagged β5 subunits in pIVEX2.3_MCS plasmid (30) and expression of His-6-POMP have been described (17). For expression of untagged POMP, the cDNA was subcloned into pRSETA by NdeI/BamHI. The plasmid encoding the Archaeoglobus fulgidus β subunit was kindly provided (31). His-6-tagged β subunit full-length and Δpropeptide constructs were generated by PCR amplification of the inserts and subcloning in the pIVEX2.4a plasmid (Roche Molecular Biochemicals) by NotI/XhoI. The chimeric β subunits were constructed by PCR amplification of the coding sequence of the A. fulgidus proteasome β subunit and the fragments encoding the propeptide of the human β5or β5i subunit. Both fragments were subcloned in pIVEX2.4a. For yeast two-hybrid studies, the cDNAs of POMP and both β5 subunits were subcloned into pAS2-1 for binding domain fusion and into pACT2 for activation domain fusion (BD Biosciences).
Gene Silencing. For silencing of POMP expression with siRNAs, we used the RNA-oligonucleotide duplex technique (32). Two duplexes were designed (1.1 GGACAGUAUUCCAGUUACUdTdT; 1.2 GGACAGUAUUCCAGUUACUdTdT) and used in combination in 100 nM concentration. HeLa cells were transfected with duplexes following the JetSI-ENDO protocol (Eurogentech, Brussels). A universal control oligonucleotide duplex (Eurogentech) or mock-transfected cells served as controls. Four hours after transfection, cells were stimulated either immediately with IFN-γ for 24 h or after 24 h, as indicated. Immunoprecipitation was performed with a monoclonal α2 antibody and immunostained for β5, LMP7, and 20S proteasomes. Chymotryptic activity of the proteasome was assessed by using the peptide substrate Suc-Leu-Leu-Val-Tyr-aminomethylcoumarin. The onset of apoptosis was measured by assaying caspase 3 and 7 activity (Apo-ONE Homogeneous Caspase-3/7 assay, Promega). MHC class I surface expression was determined by FACSCalibur flow cytometry (BD Biosciences) by using anti-class I antibody (One Lambda, Los Angeles) staining after 16-h IFN-γ stimulation (23).
Protein Interaction. Yeast two-hybrid interaction trap experiments were performed by using the Saccharomyces cerevisiae strain HF7c with the auxotrophic markers leu2-3, trp1-901, his3-200, and the interaction reporters HIS3 and lacZ. Protein interactions were assessed by His-prototrophy and by a colony-lift filter assay for β-galactosidase expression (MATCHMAKER Yeast Two-Hybrid assay, BD Biosciences).
For in vitro interaction, His-tagged β subunits and POMP or untagged β subunits and His-tagged POMP were coexpressed in the presence of TRAN35S-Label (ICN) in reticulocyte lysate (Promega). Pull-down assays were done under stringent washing conditions (0.5% Tween 20/500 mM NaCl) by using magnetic nickel beads (Qiagen, Valencia, CA). For interaction of His-tagged β subunits with POMP, the interacting proteins were cleaved off the beads by factor Xa proteolysis (Roche Molecular Biochemicals). Interacting β subunits of the His-6-POMP pull-down were eluted by 300 mM imidazole.
Metabolic [35S] Labeling and Immunoprecipitation. For standard pulse-chase experiments, cells were cultured for 24 h with or without IFN-γ, pulsed with TRAN35S-Label (ICN) for 1 h, washed 3-fold, and chased for the times indicated. To determine the stability of c20S or i20S, T2 or T2 LMP2 + 7 cells were labeled for 8 h, washed intensively, and grown for 24 h in the presence or absence of IFN-γ in cold medium and further chased up to 4 days in the absence of cytokines. Radioactivity was determined by liquid scintillation counting. Equal counts were supplied to immunoprecipitation and processed as described (17). The following polyclonal antisera were used: POMP (17), β5 and LMP7 (all laboratory stock), or anti-C8, specifically recognizing precursor complexes (10). The radioactive protein pattern was detected by phosphoimaging (Fuji FLA3000) and evaluated by aida software (Raytest, Straubenhardt, Germany). The turnover of single proteasome precursor proteins or complete proteasome complexes was calculated by adjusting the evaluated pixel densities to the background and integrating to obtain intensity values per area. Logarithmizing the intensities and bringing them into a function of chase time resulted in an approximated linear function. The slope of the line of the calculated linear equation served as a value for protein turnover, and their half-life values were estimated. Alterations of turnover rates were calculated from ratios of the slopes. For proteasome stability, intensity values per area were calculated, and time-point zero was set to 100%.
Results
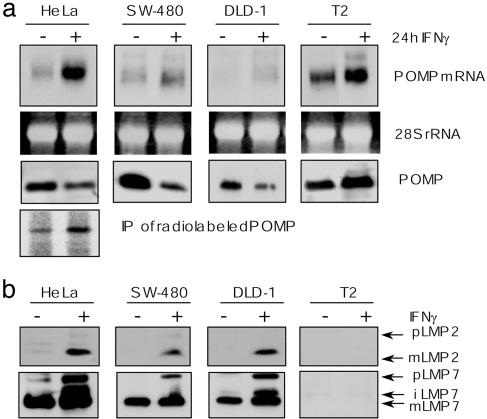
POMP Is Up-Regulated by IFN-γ, and Its Levels Reciprocally Correlate with the Presence of Immunosubunits. To study the role of POMP in i20S formation, POMP expression was analyzed with respect to its induction by IFN-γ. In all cell lines tested, POMP mRNA levels increased significantly after stimulation with the cytokine (Fig. 1a). However, despite increased POMP mRNA levels in HeLa, DLD-1 and SW-480 cells immunoblotting revealed no increase in the amount of POMP (Fig. 1a). In fact, when analyzed under steady-state conditions, cytokine treatment even resulted in a decrease of cellular POMP levels (Fig. 1a). In contrast, analysis of the human lymphoblastoma cell line T2, which lack the immunosubunits LMP2 and LMP7 (33, 34), gave opposite results and revealed an up-regulation of POMP upon IFN-γ induction (Fig. 1a).
Fig. 1.
Increased POMP levels after IFN-γ stimulation correlate with the absence of immunosubunits. (a) POMP mRNA levels are increased by 24-h IFN-γ stimulation (+) in different human cell lines analyzed by Northern blotting. Ethidium bromide-stained 28S rRNA bands are shown as an internal control (Top). Cellular POMP levels did not reflect mRNA levels, as shown by Western blot analysis of total lysates by using a POMP-specific antibody (Middle). Induction of POMP synthesis in response to IFN-γ as visualized by immunprecipitation of radio-labeled POMP from protein extracts of pulsed HeLa cells (Bottom). (b) Expression of immunosubunits reciprocally correlates with the cellular amount of POMP. Western blot analysis of the proforms (p) and the matured forms (m) of LMP2 (Upper) and LMP7 (Lower) in total cell lysates of different human cell lines in the presence (+) or absence (-) of IFN-γ.
To resolve this apparent contradiction, POMP biosynthesis was analyzed by pulse labeling and immunoprecipitation before and after IFN-γ stimulation of HeLa cells. In contrast to the steady-state situation reflected by immunoblotting on total cell extracts, we now detected a significant up-regulation of POMP also in HeLa cells (Fig. 1a). We therefore hypothesized that the POMP induction observed by immunoblotting in T2 cells may be due to an abolished or reduced expression of immunosubunits. To test this, HeLa, DLD-1, and SW-480 cells were analyzed with regard to their expression of LMP2 and LMP7. HeLa, DLD-1, and SW-480 cells revealed normal IFN-γ induction of the two immunosubunits (Fig. 1b). As expected, no expression of LMP2 and LMP7 was detectable in T2 cells. These experiments therefore showed that POMP levels reciprocally correlate with the presence of the immunosubunits and led us to hypothesize that INF-γ treatment of HeLa cells and the concomitant up-regulation of immunosubunits result in an enhanced turnover of POMP.
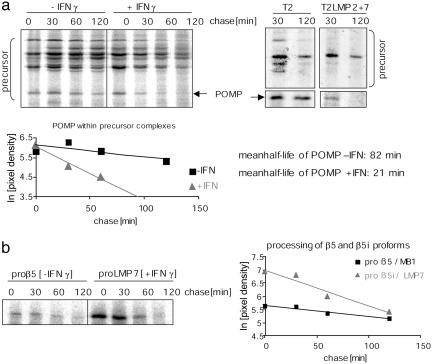
Immunoproteasome Formation Is Accelerated and Independent of IFN-γ Signaling. The above data led us to investigate whether the reciprocal relationship between POMP turnover and up-regulation of immunosubunits also influences the dynamics of i20S formation. We therefore compared the maturation kinetics of c20S with that of i20S. Because degradation of POMP signals completion of proteasome maturation (17, 19, 20), the turnover of POMP and the processing of β5 proproteins served as markers for maturation progress. Metabolically labeled precursor complexes from untreated and IFN-γ-treated HeLa cells were specifically immunoprecipitated (10). Comparison of the POMP half-life in untreated vs. IFN-γ-stimulated cells revealed an ≈4-fold acceleration of POMP turnover upon IFN-γ exposure (Fig. 2a). This accelerated degradation of POMP was accompanied by a faster turnover of i20S precursor complexes (Fig. 2a). Calculation of the turnover rates of POMP revealed a mean half-life of 82 min in untreated vs. 21 min in IFN-γ-treated HeLa cells. Importantly, comparison of the turnover of proteasome precursors in T2 LMP2 + 7 and T2 cells revealed that the rapid turnover of i20S precursors is independent of any cytokine signal and considerably faster than that of precursors of constitutive proteasomes (Fig. 2a). The difference in maturation kinetics was also reflected at the level of individual β5 subunit processing. Comparison of the maturation kinetics of the constitutive subunit β5 and the immunosubunit β5i also revealed an accelerated maturation of β5i/LMP7 (Fig. 2b).
Fig. 2.
i20S formation is accelerated and independent of IFN-γ signaling. (a) Turnover of precursor complexes and incorporated POMP is faster in the presence (+) than in the absence (-) of IFN-γ in metabolically labeled HeLa cells (Left). Turnover of i20S precursor complexes of labeled T2 LMP2 + 7 cells is accelerated in comparison to constitutive precursors of T2 independent of the cytokine signal (Right). Precursor complexes were specifically immunoprecipitated from total cell lysates at the different chase times indicated. Quantification of autoradiograms and calculated half-lives are shown (Lower). (b) For visualization of faster individual LMP7 subunit maturation compared with β5, subunits were immunoprecipitated from radiolabeled HeLa cell lysates (-/+ IFN-γ) at different chase times. The quantifications of POMP, proteasome precursors, or β5 proprotein turnover by phosphoimaging represent the mean of at least two independent experiments.
Thus, taking the turnover kinetics of POMP and POMP-containing precursor complexes and the processing of both β5 individual subunits as indicative for completion of proteasome assembly and activation, we conclude that the generation of i20S occurs ≈4-fold faster than that of c20S and is independent of other IFN-γ-induced proteins.
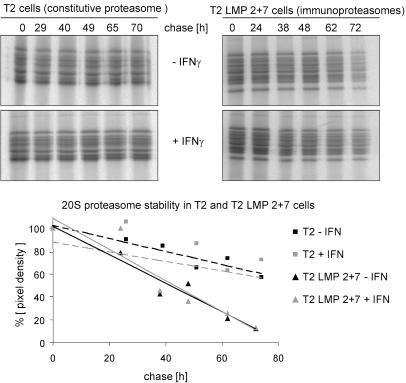
Immunoproteasome Formation Is a Transient Response. The accelerated de novo formation of i20S raised questions regarding the influence of INF-γ on the fate of c20S as well as i20S. To study this, we used human T2 cells expressing c20S and T2 LMP2 + 7 cells expressing i20S independent of INF-γ stimulation and pulsed the cells in the absence of cytokines to label proteasomes. Subsequently, the cells were either exposed for 24 h to IFN-γ or not and chased in the absence of IFN-γ. For a period of 5 days, no effect of IFN-γ on T2 or T2 LMP2 + 7 cell viability was observed on the basis of a proliferation assay (not shown).
In agreement with the reported half-life of 5 days of c20S (35), IFN-γ showed no effect on the turnover of c20S (mean half-life 133 h) or of i20S (Fig. 3). However, with a calculated mean half-life of 27 h, i20S displayed a much shorter half-life than c20S and thus were, independent of IFN-γ, strikingly less stable than c20S (Fig. 3). Thus, the up-regulation of i20S in response to IFN-γ is a transient response.
Fig. 3.
i20S are less stable than c20S independent of IFN-γ. c20S (T2 cells) or i20S (T2 LMP2 + 7 cells) were specifically immunoprecipitated at different time points during the chase period in the absence (Upper) or presence of 24-h IFN-γ (Lower). The diagram shows percent pixel density of time-point zero with trend lines. The calculated half-lives of 133 h for c20S and 27 h for i20S are the mean of five independent experiments. A representative experiment is shown.
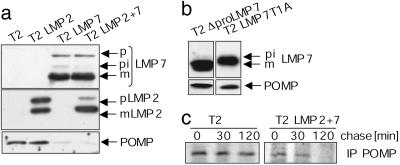
Rapid Turnover of POMP Requires the Active β5i/LMP7 Subunit. The presence or absence of immunosubunits seemed to affect the stability of POMP. Therefore, we analyzed whether POMP levels were affected in T2 cells stably expressing either LMP2 or LMP7 (Fig. 4a). Nontransfected T2 cells exhibited high levels of POMP. However, as soon as wild-type LMP7 was expressed, the amount of POMP dramatically decreased (T2 LMP7 and T2 LMP2 + 7). In contrast, expression of LMP2 alone had no effect on the amount of POMP (T2 LMP2; Fig. 4a). Independent of the type of immunosubunit expressed, POMP mRNA levels were not changed in the different transfected cell lines (not shown). To examine whether POMP destabilization requires the presence of an active LMP7 subunit or whether the LMP7 structure itself is sufficient to signal POMP degradation, we used T2 cells expressing mutated LMP7 derivatives. These mutated and proteolytically inactive forms of LMP7 were previously reported to be incorporated into the nascent proteasome complex (17). Inactivation of LMP7 by deletion of the prosequence (T2 ΔproLMP7) or by substitution of the active-site threonine to alanine (T2 LMP7T1A) led to POMP amounts similar to that observed in untransfected T2 cells (Fig. 4b). Thus, the amount of POMP depends on the efficient maturation and activity of LMP7.
Fig. 4.
Rapid turnover of POMP requires the presence of active LMP7. (a) The presence of LMP7 affected POMP stability as analyzed by Western blots of POMP, LMP2, and LMP7 in cell lysates of different T2 cell lines, stably expressing LMP7 and/or LMP2. The proforms (pi, p) and the matured forms (m) of the subunits LMP7 and LMP2 are indicated. (b) POMP stability is restored by expression of inactive variants of LMP7. Western blot analyses of POMP and LMP7 in cell lysates of T2 cell lines stably expressing LMP7 without the propeptide (ΔpLMP7) and the active-site mutation LMP7T1A. (c) POMP turnover in T2 is lower than in T2 LMP2 + 7 cells independent of the IFN-γ signal. Cells were metabolically labeled, and POMP was immunoprecipitated from total cell lysates at the different chase times indicated.
The interdependence of POMP stability and LMP7 was further investigated by immunoprecipitation experiments with protein extracts of metabolically labeled T2 and T2 LMP2 + 7 cells (Fig. 4c). During a 2-hr chase, the amount of precipitated POMP in T2 cells was only moderately reduced, whereas the amount of POMP in T2 LMP2 + 7 cells dramatically decreased after 30 min (Fig. 4c). Together, these experiments demonstrate that the varying stability of POMP is essentially controlled by the activity of the β5i subunit LMP7 and is independent of other cytokine-induced proteins.
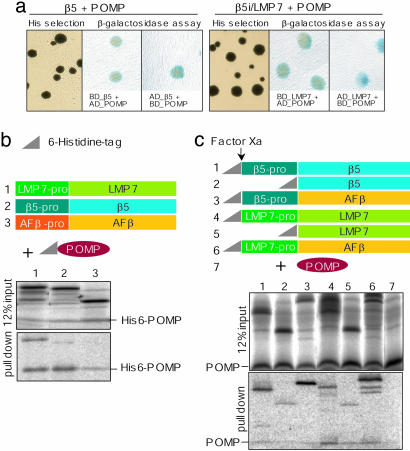
POMP Directly Interacts with the β5i Subunit LMP7. Supported by several lines of evidence, it was previously discussed that POMP-like factors interact with the constitutive β5 subunit (20, 36, 37). Based on our results, we tested whether POMP can also bind to the β5i subunit. This appeared to be of particular importance, because it had been reported that POMP and β5i did not interact (37). Our yeast two-hybrid screens, however, demonstrated that POMP interacts not only with the constitutive β5 subunit but also with LMP7 (Fig. 5a). To verify this interaction, we performed pull-down assays by using either His-tagged POMP (His-6-POMP) and untagged β5 and β5i subunits or His-tagged β subunits and untagged POMP (Fig. 5 b and c). Indeed, His-6-POMP was found to interact with both untagged proforms of the β5 and β5i subunits (Fig. 5b, lanes 1 and 2). Reversely, the proforms of both β5 subtypes pulled down untagged POMP (Fig. 5c, lanes 1 and 4). In contrast, the wild-type form of the A. fulgidus β subunit, serving as control, did not interact with His-6-POMP, indicating the specificity of the observed interaction of POMP with the human β5 subunits (Fig. 5b, lane 3). To reveal a potential interaction of POMP with the subunit propeptides, we tested constructs encoding β5 and β5i without propeptides as well as chimeric β subunit constructs expressing either the β5 or the β5i propeptide fused to the archaebacterial β subunit of A. fulgidus. These experiments (Fig. 5c, lanes 3 and 6) showed that both chimeric β5 subunits interacted with POMP, and that the binding of the chimeric β subunits to POMP was mediated by the propeptides of the β5 and β5i subunits, respectively. Strikingly, in all experimental subsets, the interaction of POMP with the proform of LMP7 appeared to be stronger than that with the β5 proform, indicating a higher affinity of POMP to LMP7. Surprisingly, even those forms of β5 and β5i that lack the propeptides interacted with POMP (Fig. 5c, lanes 2 and 5), suggesting that there exist at least two interaction sites for POMP, one within the propeptide and another within the sequence of the matured β5 subunits.
Fig. 5.
POMP interacts directly with LMP7. (a) Yeast two-hybrid assay for interaction of POMP with the proforms of β5 (Left) or LMP7 (Right). Interaction is shown by selection for His-prototrophy and β-galactosidase expression as detected by using activation-domain POMP fusion (AD-POMP) and the binding domain fused to the β5 subunit (BD-β5 or BD-LMP7) or vice versa. (b) Schematic representation of untagged β subunits and His-6-tagged POMP (His-tag = triangle). Both proforms of the human β5 subunits (1, β5; or 2, LMP7) bind to His-6-POMP, whereas the A. fulgidus β subunit did not bind (3, AFβ; specificity control). (c) The N-terminal His-6-fusions of human β5 subunits or their chimeras and the factor Xa site are schematically illustrated. (Upper) Input controls (12% input). (Lower) His-tagged proforms (1 and 4), β5 subunits without propeptides (2 and 5), as well as chimeric β subunits containing a human propeptide and the β subunit of A. fulgidus (3, β5-pro AFβ; 6, LMP7-pro AFβ) pulled down untagged POMP. Untagged POMP did not bind to the nickel beads (7, negative control)
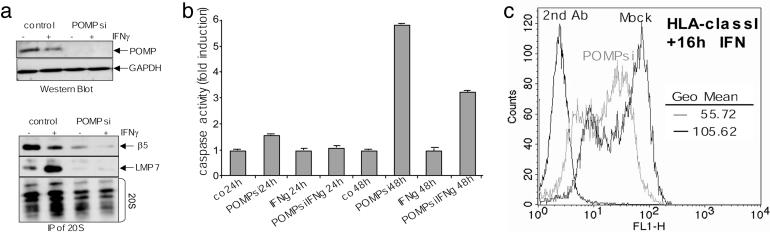
POMP Expression Is Essential for Proteasome Formation. So far, our experiments revealed an interaction between POMP and LMP7 and a LMP7-mediated rapid degradation of POMP during i20S formation. To analyze whether, in a reverse relationship, incorporation and maturation of the LMP7 subunit depend on POMP, the expression of POMP in HeLa cells was silenced by using short interfering RNAs (siRNAs). The siRNA targeting POMP mRNA led to silencing of POMP expression in the absence or presence of IFN-γ, compared with mock-transfected cells (Fig. 6a). As checked by RT-PCR, mRNA expression of LMP7 and actin was affected neither by transfection of POMPsi nor by an unspecific control siRNA (not shown). Silencing of POMP expression abolished incorporation of β5 into c20S complexes as well as severely impaired incorporation of the β5i subunit LMP7 into i20S complexes (Fig. 6a). As a result, the absence of POMP caused a strongly decreased total amount of 20S complexes (Fig. 6a).
Fig. 6.
Silencing of POMP gene expression by RNA interference results in a decrease of proteasomes. (a) Transfection of siRNA targeting POMP (POMPsi) silenced POMP expression in the presence or absence of IFN-γ (+/-) but not mock transfection of cells (control). GAPDH levels served as loading control (Upper). Silencing of POMP led to a strong reduction of incorporated β5 and β5i/LMP7 and immunoprecipitated 20S proteasome complexes (IP of 20S; Lower). GAPDH levels represent 15% input. (b) Prolonged knockdown of POMP (POMPsi) up to 48 h caused the induction of apoptosis as measured by caspase 3/7 activity in the presence (IFNg) or absence (co) of IFN-γ.(c) POMP depletion caused a decrease in MHC class I surface expression as measured by HLA class I fluorescence staining of mock (black line) or POMPsi RNA-transfected cells (gray line) after IFN-γ stimulation. Background staining with the second antibody only (2nd ab; IgG1) is shown (Left). Mean fluorescence levels are indicated as geometric (Geo) mean.
Thus, knockdown of POMP led to a considerable reduction of the proteasomal hydrolyzing activity and thereby to an accumulation of ubiquitin conjugates. In fact, after 24 h, proteolytic activity was reduced to ≈60% of the respective controls with or without IFN-γ (not shown). A prolonged depletion of POMP for 48 h resulted in a dramatic decrease of the proteasomal hydrolyzing activity to ≈40% of the untransfected control (not shown). Consequently, we found that the cells induced apoptosis under prolonged silencing of POMP (Fig. 6b). Moreover, MHC class I surface expression was reduced by ≈50% in POMP-depleted cells after IFN-γ (Fig. 6c). Therefore, these experiments reveal not only that POMP is required for both the incorporation of the LMP7 subunit and its timely maturation but also that it is essential for proteasome formation in general and consequently for cell survival.
Discussion
Our study demonstrates that immune adaptation of the proteasome system caused by infection and concomitant IFN-γ production (38, 39) is an extremely rapid and transient response. The dynamics of this process is controlled by the molecular interplay between the LMP7 subunit and POMP and the consequent accelerated formation of i20S.
Challenging the postulated importance of POMP in i20S biogenesis, we observed that, despite an up-regulation of POMP mRNA in IFN-γ-stimulated cells (17, 19), POMP levels were reduced even under steady-state conditions. Up-regulation of POMP was detected only after a short radioactive pulse or in the absence of a fully processed functional LMP7 subunit. Our data therefore extend the present knowledge by revealing a strong functional interdependence of POMP and the β5i subunit LMP7 in i20S biogenesis. In fact, POMP stabilization in the presence of an inactive LPM7 subunit demonstrates that LMP7 essentially drives the rapid degradation of POMP.
The accelerated degradation of POMP is directly connected with the faster maturation of LMP7 (compared with β5) and, in consequence, with the ≈4-fold faster formation of i20S. This rapid maturation of β5i may also reflect the preferential incorporation of LMP7 into nascent proteasomes (40). Thus, the kinetics of proteasome biogenesis seems to be differentially determined by the two β5 subunits and their molecular interplay with POMP, allowing a rapid switch from c20S to i20S function.
Importantly, the rapid degradation of POMP and the strongly accelerated maturation of i20S do not, as might have been expected, require any additional IFN-γ-induced factors. Consequently, the dynamics of i20S formation appears to be essentially self-controlled and the result of a functional interdependence of LMP7 and POMP. Moreover, the up-regulation of i20S is a transient response because, independent of IFN-γ signaling, i20S exhibit a much shorter half-life than c20S. Thus, both i20S-specific characteristics may be due to intrinsic properties of the enzyme complex. As shown by immunological experiments (27), the incorporation of LMP7 can affect the structural properties of the proteasome. This also becomes apparent by the altered chromatographic properties of immunoproteasomes (41).
The key position of POMP and LMP7 in the accelerated formation of i20S is supported by their direct interaction. In contrast with our experiments, an interaction of POMP with the β5 subunit but not with LMP7 was recently reported based on a yeast two-hybrid interaction screen (37). This negative result was interpreted to reflect the observed biased incorporation of β5 and LMP7 into the respective proteasome complexes due to their different prosequences (40). As shown here, interaction of POMP with both mammalian β5 homologues is independent of the prosequences, suggesting a second interaction site within domains of the matured β subunits. This is in agreement with our previous observation that the β5i prosequence is not absolutely essential for its incorporation into 20S proteasomes, and that POMP incorporation into precursor complexes is independent of the β5i prosequence (17). Although the β5i prosequence is not essential for β5i subunit incorporation, the presence of the correct prosequence strongly supports the subunit's incorporation efficiency (17).
In the presence of sufficient amounts of POMP, the availability of LMP7 seems to be the rate-limiting factor of this process and vice versa. Our POMP knockdown experiments demonstrate that POMP determines the recruitment of the LMP7 subunit (and that of β5) into the proteasome complex. In contrast to the role of the yeast Ump1p homologue (20), POMP function turns out to be essential for proteasome biogenesis and consequently also for mammalian cell viability. As an immunological consequence, the limited generation of antigenic peptides also is reflected by the reduction of MHC class I surface expression in POMP-depleted cells (25). These data therefore demonstrate that POMP possesses an essential coordinative function in proteasome assembly that is independent of the different prosequences and not directly correlated with the differential incorporation of the two β5 subunits. IFN-γ-induced amounts of POMP therefore will permit an efficient recruitment of the LMP7 subunit, which concomitantly facilitates the accelerated formation of i20S.
The effectiveness of the MHC class I immune response of an attacked organism is largely determined by the rapid and coordinated generation of antigenic peptides and their presentation on the cell surface. In conclusion, the accelerated formation of i20S forced by the interplay between POMP and LMP7 meets the demands of an efficient and rapid answer to an immunological challenge. Subsequently, the observed strongly reduced half-life of i20S permits cells to return more rapidly to a normal situation once i20S functions is no longer needed, also supporting the finding of a transient nature of CD8+ T cell priming (26). Thus, our model is in good agreement with the current knowledge of MHC class I antigen presentation, following the hypothesis that most antigenic peptides derived from defective ribosomal products, allowing cells to cope immediately with rapidly replicating viruses (42). Therefore, we present an immune-adaptation mechanism by the proteasome system, which is essentially self-controlled and rapid enough to contribute to an efficient immune response.
Acknowledgments
We are grateful to J. Monaco (University of Cincinnati, College of Medicine, Cincinnati) for kindly providing the C8 antiserum, to C. Beier for excellent technical assistance, and to the members of the Kloetzel laboratory for support and helpful discussions and for critically reading the manuscript. This study was supported by the Deutsche Forschungsgemeinschaft (Kl 427/8-5/SFB 421).
Author contributions: S.H., P.-M.K., and E.K. designed research; S.H., D.L., and E.K. performed research; S.H., D.L., and E.K. analyzed data; and P.-M.K. and E.K. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: POMP, proteasome maturation protein; LMPn, low molecular weight protein n; siRNA, short interfering RNA.
See Commentary on page 9089.
References
- 1.Kloetzel, P. M. (2001) Nat. Rev. Mol. Cell. Biol. 2, 179-187. [DOI] [PubMed] [Google Scholar]
- 2.Rock, K. L., York, I. A., Saric, T. & Goldberg, A. L. (2002) Adv. Immunol. 80, 1-70. [DOI] [PubMed] [Google Scholar]
- 3.Groll, M., Ditzel, L., Lowe, J., Stock, D., Bochtler, M., Bartunik, H. D. & Huber, R. (1997) Nature 386, 463-471. [DOI] [PubMed] [Google Scholar]
- 4.Aki, M., Shimbara, N., Takashina, M., Akiyama, K., Kagawa, S., Tamura, T., Tanahashi, N., Yoshimura, T., Tanaka, K. & Ichihara, A. (1994) J. Biochem. (Tokyo) 115, 257-269. [DOI] [PubMed] [Google Scholar]
- 5.Chen, W., Norbury, C. C., Cho, Y., Yewdell, J. W. & Bennink, J. R. (2001) J. Exp. Med. 193, 1319-1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Toes, R. E., Nussbaum, A. K., Degermann, S., Schirle, M., Emmerich, N. P., Kraft, M., Laplace, C., Zwinderman, A., Dick, T. P., Muller, J., et al. (2001) J. Exp. Med. 194, 1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kloetzel, P. M. (2004) Nat. Immunol. 5, 661-669. [DOI] [PubMed] [Google Scholar]
- 8.Frentzel, S., Pesold-Hurt, B., Seelig, A. & Kloetzel, P. M. (1994) J. Mol. Biol. 236, 975-981. [DOI] [PubMed] [Google Scholar]
- 9.Groettrup, M., Standera, S., Stohwasser, R. & Kloetzel, P. M. (1997) Proc. Natl. Acad. Sci. USA 94, 8970-8975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nandi, D., Woodward, E., Ginsburg, D. B. & Monaco, J. J. (1997) EMBO J. 16, 5363-5375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmidt, M., Zantopf, D., Kraft, R., Kostka, S., Preissner, R. & Kloetzel, P. M. (1999) J. Mol. Biol. 288, 117-128. [DOI] [PubMed] [Google Scholar]
- 12.Schmidtke, G., Kraft, R., Kostka, S., Henklein, P., Frommel, C., Lowe, J., Huber, R., Kloetzel, P. M. & Schmidt, M. (1996) EMBO J. 15, 6887-6898. [PMC free article] [PubMed] [Google Scholar]
- 13.Schmidtke, G., Schmidt, M. & Kloetzel, P. M. (1997) J. Mol. Biol. 268, 95-106. [DOI] [PubMed] [Google Scholar]
- 14.Chen, P. & Hochstrasser, M. (1996) Cell 86, 961-972. [DOI] [PubMed] [Google Scholar]
- 15.Heinemeyer, W., Fischer, M., Krimmer, T., Stachon, U. & Wolf, D. H. (1997) J. Biol. Chem. 272, 25200-25209. [DOI] [PubMed] [Google Scholar]
- 16.Jager, S., Groll, M., Huber, R., Wolf, D. H. & Heinemeyer, W. (1999) J. Mol. Biol. 291, 997-1013. [DOI] [PubMed] [Google Scholar]
- 17.Witt, E., Zantopf, D., Schmidt, M., Kraft, R., Kloetzel, P. M. & Krüger, E. (2000) J. Mol. Biol. 301, 1-9. [DOI] [PubMed] [Google Scholar]
- 18.Griffin, T. A., Slack, J. P., McCluskey, T. S., Monaco, J. J. & Colbert, R. A. (2000) Mol. Cell. Biol. Res. Commun. 3, 212-217. [DOI] [PubMed] [Google Scholar]
- 19.Burri, L., Hockendorff, J., Boehm, U., Klamp, T., Dohmen, R. J. & Levy, F. (2000) Proc. Natl. Acad. Sci. USA 97, 10348-10353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ramos, P. C., Hockendorff, J., Johnson, E. S., Varshavsky, A. & Dohmen, R. J. (1998) Cell 92, 489-499. [DOI] [PubMed] [Google Scholar]
- 21.Cerundolo, V., Kelly, A., Elliott, T., Trowsdale, J. & Townsend, A. (1995) Eur. J. Immunol. 25, 554-562. [DOI] [PubMed] [Google Scholar]
- 22.Seliger, B., Wollscheid, U., Momburg, F., Blankenstein, T. & Huber, C. (2000) Tissue Antigens 56, 327-336. [DOI] [PubMed] [Google Scholar]
- 23.Sun, Y., Sijts, A. J., Song, M., Janek, K., Nussbaum, A. K., Kral, S., Schirle, M., Stevanovic, S., Paschen, A., Schild, H., Kloetzel, P. M. & Schadendorf, D. (2002) Cancer Res. 62, 2875-2882. [PubMed] [Google Scholar]
- 24.Ehrlich, R. (1997) Hum. Immunol. 54, 104-116. [DOI] [PubMed] [Google Scholar]
- 25.Benham, A. & Neefjes, J. (1997) J. Immunol. 159, 5896-5904. [PubMed] [Google Scholar]
- 26.Wong, P. & Pamer, E. (2003) Immunity 18, 499-511. [DOI] [PubMed] [Google Scholar]
- 27.Sijts, A. J., Ruppert, T., Rehermann, B., Schmidt, M., Koszinowski, U. & Kloetzel, P. M. (2000) J. Exp. Med. 191, 503-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krüger, E., Msadek, T. & Hecker, M. (1996) Mol. Microbiol. 20, 713-723. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook, J., Fritsch, E. F. & Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Woodbury, NY).
- 30.Apcher, G. S., Heink, S., Zantopf, D., Kloetzel, P. M., Schmid, H. P., Mayer, R. J. & Krüger, E. (2003) FEBS Lett. 553, 200-204. [DOI] [PubMed] [Google Scholar]
- 31.Groll, M., Brandstetter, H., Bartunik, H., Bourenkow, G. & Huber, R. (2003) J. Mol. Biol. 327, 75-83. [DOI] [PubMed] [Google Scholar]
- 32.Elbashir S. M., H. J., Lendeckel W., Yalcin A., Weber K., Tuschl T. (2001) Nature 411, 494-498. [DOI] [PubMed] [Google Scholar]
- 33.Salter, R. D. & Cresswell, P. (1986) EMBO J. 5, 943-949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ortiz-Navarrete, V., Seelig, A., Gernold, M., Frentzel, S., Kloetzel, P. M. & Hammerling, G. J. (1991) Nature 353, 662-664. [DOI] [PubMed] [Google Scholar]
- 35.Hendil, K. B. (1988) Biochem. Int. 17, 471-477. [PubMed] [Google Scholar]
- 36.Cagney, G., Uetz, P. & Fields, S. (2001) Physiol. Genom. 7, 27-34. [DOI] [PubMed] [Google Scholar]
- 37.Jayarapu, K. & Griffin, T. A. (2004) Biochem. Biophys. Res. Commun. 314, 523-528. [DOI] [PubMed] [Google Scholar]
- 38.Khan, S., van den Broek, M., Schwarz, K., de Giuli, R., Diener, P. A. & Groettrup, M. (2001) J. Immunol. 167, 6859-6868. [DOI] [PubMed] [Google Scholar]
- 39.Kuckelkorn, U., Ferreira, E. A., Drung, I., Liewer, U., Kloetzel, P. M. & Theobald, M. (2002) Eur. J. Immunol. 32, 1368-1375. [DOI] [PubMed] [Google Scholar]
- 40.Kingsbury, D. J., Griffin, T. A. & Colbert, R. A. (2000) J. Biol. Chem. 275, 24156-24162. [DOI] [PubMed] [Google Scholar]
- 41.Dahlmann, B., Ruppert, T., Kuehn, L., Merforth, S. & Kloetzel, P. M. (2000) J. Mol. Biol. 303, 643-653. [DOI] [PubMed] [Google Scholar]
- 42.Yewdell, J.W., Reits, E. & Neefjes, J. (2003) Nat. Rev. Immunol. 3, 952-961. [DOI] [PubMed] [Google Scholar]