Abstract
In eukaryotes, misfolded proteins must be distinguished from correctly folded proteins during folding and transport processes by quality control systems. Yeast peptide:N-glycanase (yPNGase) specifically deglycosylates the denatured form of N-linked glycoproteins in the cytoplasm and assists proteasome-mediated glycoprotein degradation by forming a complex with 26S proteasome through DNA repair protein, yRad23. Here, we describe the crystal structures of a yPNGase and XPC-binding domain of yRad23 (yRad23XBD, residues 238-309) complex and of a yPNGase-yRad23XBD complex bound to a caspase inhibitor, Z-VAD-fmk. yPNGase is formed with three domains, a core domain containing a Cys-His-Asp triad, a Zn-binding domain, and a Rad23-binding domain. Both N- and C-terminal helices of yPNGase interact with yRad23 through extensive hydrophobic interactions. The active site of yPNGase is located in a deep cleft that is formed with residues conserved in all PNGase members, and three sugar molecules are bound to this cleft. Complex structures in conjunction with mutational analyses revealed that the walls of the cleft block access to the active site of yPNGase by native glycoprotein, whereas the cleft is sufficiently wide to accommodate denatured glycoprotein, thus explaining the specificity of PNGase for denatured substrates.
Keywords: deep cleft, PNGase, zinc-metalloenzyme
The correct folding of newly synthesized glycoproteins is an essential part of the protein manufacturing process and is important for many cellular functions such as trafficking and cell-cell interactions. In eukaryotes, newly synthesized proteins are transported to the endoplasmic reticulum (ER) where they are correctly folded and glycosylated on the Asn residue of the Asn-Xaa-Ser/Thr consensus moiety to form N-linked glycoproteins, which then enter the secretory pathway (1). However, misfolded glycoproteins are initially retained in the ER and are then dislocated to the cytosol where they undergo ubiquitination, deglycosylation, and degradation by proteasomes (2, 3). Failures in the protein quality control process may cause several diseases, including autosomal dominant neurohypophyseal diabetes insipidus or α1-antitrypsin disease (4, 5).
Peptide:N-glycanase (PNGase) is responsible for the deglycosylation of unfolded N-linked glycoproteins dislocated from the ER to the cytosol (6-9). This highly conserved enzyme in eukaryotes cleaves the β-aspartyl-glucosamine bond between an Asn residue and the N-acetylglucosamine (GlcNAc) residue at the reducing end of the glycan moiety and releases a free carbohydrate. Although PNGase has a broad range of substrate specificity, it specifically functions on misfolded glycoproteins (9-12).
Recent studies have shown that PNGase is physically associated with 26S proteasome through Rad23 in yeast and mouse, which suggests that PNGase is an important component of the ER-associated degradation pathway (13-15). PNGase is believed to facilitate the efficient proteasomal proteolysis of N-linked glycoproteins by removing glycan (8). Rad23, an essential DNA repair protein, participates in both nucleotide excision repair and ubiquitin-mediated protein degradation and thus is involved in the coordination of these two biologically important pathways (16, 17).
Recently, a pro-caspase inhibitor, carbobenzyloxy-Val-Ala-Asp-α-fluoromethylketone (Z-VAD-fmk) was shown to irreversibly inhibit yeast PNGase (yPNGase) (18). To understand the substrate specificity of PNGase for denatured glycoproteins, the interaction between PNGase-Rad23, and the structural basis of PNGase inhibition, we determined the crystal structures of a yPNGase-yRad23 complex and of a Z-VAD-fmk-bound yPNGase-yRad23 complex (see Table 1, which is published as supporting information on the PNAS web site). We identified the XPC-binding domain (yRad23XBD; residues 238-309) of yRad23 as a primary yPNGase binding site and found that both the N- and C-terminal helices of yPNGase bind to the hydrophobic patch of yRad23XBD. Structural and mutational analyses revealed that the deep cleft containing the active site plays a crucial role in the differentiation of denatured glycoprotein substrates from glycoproteins in their native conformations.
Materials and Methods
Protein Expression and Purification. See Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
Crystallization and Data Collection. Crystals were grown by hanging drop vapor diffusion method against crystallization buffer containing 0.1 M Tris·HCl, 3.5 M sodium chloride, and 10 mM DTT (pH 7.8) at 4°C. For inhibitor binding, a crystal was soaked in a solution containing crystallization buffer and 3 mM Z-VAD-fmk for 2 days before collecting data. The crystals formed in space group P3121 with a = 129.2 Å, b = 129.2 Å, and c = 128.5 Å and contained one complex molecule in an asymmetric unit. Diffraction data were collected at -170°C with crystals flash-frozen in crystallization buffer containing 30% sucrose. Assuming one complex per asymmetric unit, the solvent content was estimated to be 81%.
Structure Determination and Refinement. A single-wavelength data set was collected by using a Se-Met crystal on beamline 5A at the Photon Factory (Tsukuba, Japan). Data from an inhibitor-bound crystal were collected on 4A at Pohang Advanced Light Source (Pohang, South Korea). Integration, scaling, and merging of the diffraction data were performed by using the hkl2000 program suites (19). Five Se sites and initial phases were determined by using the cns program (20). After density modification, the electron density map calculated to 3.5-Å resolution was of excellent quality, which allowed us to trace most of the chains. Successive rounds of model building using o (21), refinement using cns and refmac programs (20, 22), and phase combination allowed the complete structure to be built (Table 1). The final model contained residues 8-327 of yPNGase and residues 253-309 of yRad23XBD.
Atomic Absorption Spectroscopy. A typical atomic absorption spectroscopy analysis used 10 ml of a solution of 13 μM PNGase in buffer containing 20 mM Tris·HCl and 5 mM DTT (pH 7.5) and yielded 0.3 parts per million of Zn. The stoichiometry corresponded to 0.38 mol of Zn per mol of yPNGase. An analysis of the buffer used showed no contaminating metal.
Mutagenesis, Deglycosylation Assay, and Circular Dichroism Analysis. See Supporting Materials and Methods for details. See also Figs. 7-12, which are published as supporting information on the PNAS web site.
Results and Discussion
yPNGase Binds to the XPC-Binding Domain of yRad23. Full-length recombinant yPNGase has poor solubility, and the amounts of soluble proteins produced in Escherichia coli were insufficient for structural studies. However, the solubility of yPNGase significantly increased in the presence of full-length yRad23, and gel filtration analysis showed that yRad23 and yPNGase form a 1:1 complex (15).
The limited proteolysis of the yPNGase-yRad23 complex revealed that XBD of yRad23 is critically required and sufficient to form a complex with yPNGase (see Fig. 7). Several lines of evidence support the importance of yRad23XBD for recognition of yPNGase. First, in the elucidated complex structure, the N-terminal region of yPNGase extensively interacts with yRad23XBD (Fig. 1). Second, biochemical analyses showed that deletion of the N-terminal region of yPNGase, which binds yRad23XBD, abrogates the complex formation between yPNGase and yRad23 (15). Thus, it is likely that the XBD is the primary yPNGase-binding region in yRad23. Based on this biochemical analysis, we determined the structure of the yPNGase (residues 8-342) and yRad23XBD complex.
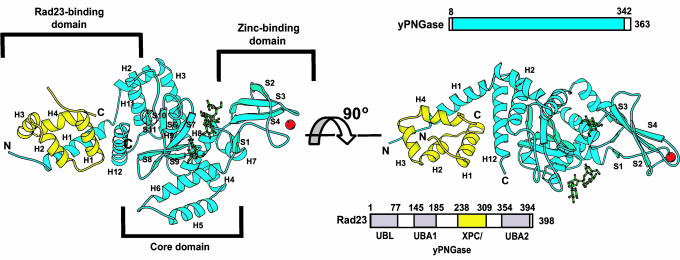
Fig. 1.
Schematic representation of the yPNGase-yRad23 complex, providing two different views of the yPNGase-yRad23 complex structure. yPNGase is shown in blue and yRad23 is in yellow. yPNGase comprises three domains, an N-terminal Rad23-binding, a core, and a Zn-binding domain. Three sucrose molecules (green) are located in the deep cleft. A Zn atom (red) is coordinated by Cys-129, -132, -165, and -168 in yPNGase.
yPNGase Is a Zinc Metalloenzyme. In the course of model building, we observed a strong density coordinated to four Cys residues (Cys-129, -132, -165, and -168) in two CxxC motifs (Figs. 2 and 8). This finding raised the possibility that yPNGase binds a metal ion. To address this possibility, we examined the effects of metal chelating agent on deglycosylation activity. The addition of metal chelating agent, EDTA, abolished deglycosylation activity of yPNGase (Fig. 3A). The metal content of yPNGase was further investigated by atomic absorption spectroscopy, and the metal was identified as Zn. Moreover, this Zn content consistently corresponds to a ratio of ≈0.4 mol of Zn per mol of yPNGase. We presume that this low stoichiometry is due to the dissociation of Zn from the protein during purification.
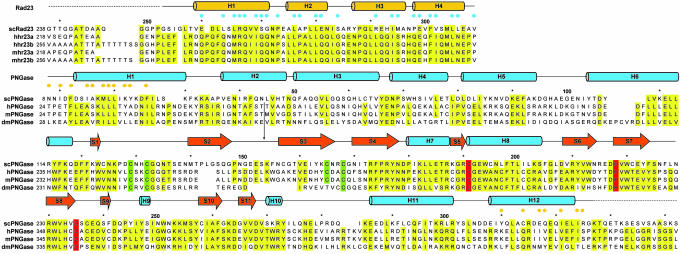
Fig. 2.
Structural alignment and conservation in yRad23XBD and yPNGase. Shown at the top is the sequence identity between Saccharomyces cerevisiae Rad23XBD (yRad23XBD) and its orthologues from human (Hs) and mouse (Mm). Blue dots indicate yRad23XBD residues that contact yPNGase. Every 10th residue is marked with an asterisk. The secondary structural elements are indicated above the alignment. At the bottom, sequence identity between S. cerevisiae PNGase (yPNGase) and its orthologues from human (Hs), mouse (Mm), and fly (Dm, bottom) is shown. Conserved residues are shaded in yellow, and the catalytic triad is highlighted in red. The four Cys residues that coordinate the Zn atom are shown in green. Orange dots indicate yPNGase residues that contact yRad23XBD. Omitted regions of HsPNGase (residues 64-174), MmPNGase (residues 64-171), and DmPNGase (residues 68-165) are indicated by arrows.
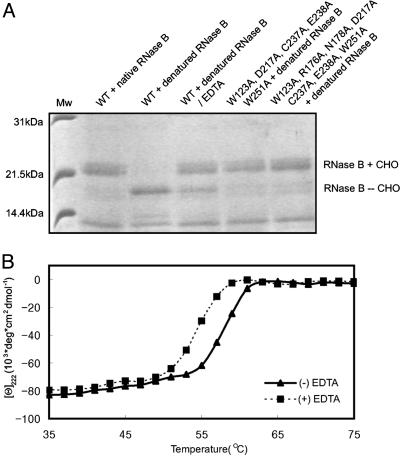
Fig. 3.
Effects of the metal chelating agent EDTA on the deglycosylation activity and stability of yPNGase. (A) Deglycosylation activities of the wild-type (WT) and mutant yPNGase proteins. Lane 1, standard molecular mass; lane 2, WT yPNGase and native RNase B; lane 3, WT yPNGase and the denatured RNase B; lane 4, WT yPNGase and the denatured RNase B in the presence of EDTA; lane 5, five residues were mutated simultaneous in yPNGasemis5; lane 6, seven residues were mutated simultaneous in yPNGasemis7. RNaseB + CHO and RNaseB - CHO represent glycosylated and deglycosylated RNase B, respectively. (B) Thermal melting curves of yPNGase without and with EDTA are determined by circular dichroism.
The above results together with our findings that metal-chelating agent abolishes deglycosylation activity suggest that yPNGase is a Zn metalloenzyme. Although the Zn ion is somewhat distantly located from the active site, it plays an important role in yPNGase stability because mutation of any of four Cys residues to Ala completely abolished deglycosylation activity (ref. 23; Fig. 1); moreover, the Tm of yPNGase value was reduced by 4°C in the presence of EDTA (Fig. 3B).
Overall Structure of the yPNGase-yRad23 Complex. yPNGase folds into α/β structure with a dimension of 92 × 55 × 37 Å (Fig. 1). The overall structure is formed with three domains, a Rad23 binding, a core, and a Zn-binding domain. The Rad23-binding domain consists of four helices (H1, H2, H11, and H12), and the N-terminal helix adopts an extended conformation. The core domain contains six central β-strands (S6 to S11), which are buttressed by three α-helices (H3, H5, and H6) and several surface loops. The Zn-binding domain is comprised of five strands (S1, S4, S3, S2, and S5) and two helices (H7 and H8). Two loops spanning S1 and S2, and S3 and S4, and a strand S3 located at the opposite end of a Rad23-binding domain binds to the Zn ion. An antiparallel β-sheet, formed by four (S6, S7, S8, and S10) of the six β-strands and helix H3 from the core, are packed against a helix H8 and the strands S1, S4, and S5, forming a deep interdomain cleft. This cleft can be largely divided into two parts: a carbohydrate-binding region in which sucrose molecules are bound and a protein-binding region where a peptide inhibitor is located (Figs. 4 and 9).
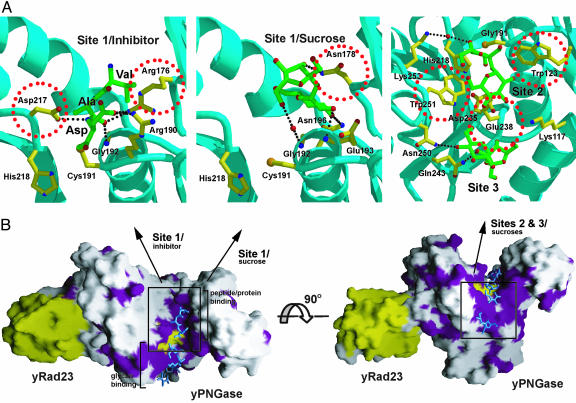
Fig. 4.
Active site of yPNGase and the surface representation of the yPNGase-yRad23XBD complex. (A) Interactions between yPNGase and the inhibitor (Left) or sucrose molecules (Center and Right). A sucrose molecule in site 1 of yPNGase (Center) is replaced by an inhibitor, Z-VAD-fmk, but the other two sucrose molecules in sites 2 and 3 remained in the same position upon inhibitor binding (Right). H-bonds are represented by dashed lines. O, N, and S atoms are shown in red, blue, and orange, respectively. Residues that replaced in mutational analyses are marked with red circles. (B) The molecular surfaces of yPNGase and yRad23XBD are colored in white and yellow, respectively. The surface of the yPNGase residues that is >80% conserved in four yPNGase orthologues (Fig. 2) is colored in purple. A catalytic triad is shown in yellow. Axes indicate the close-up views for the inhibitor and sugar-binding sites.
The N-terminal helix, H1, is extended against the C-terminal helix, H12, at a 45° angle, and four helices from yRad23XBD bind to these two helices. This region of the yPNGase surface is enriched with hydrophobic amino acids.
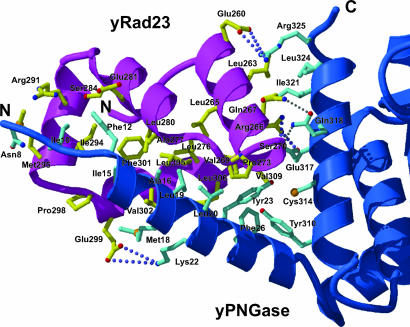
Overall Structure of the XPC Binding Domain of yRad23. The structure of yRad23XBD contains four helices and is similar to its human Rad23 homologues, hHR23A and hHR23B, with rms deviation values of 2.0 and 2.8 Å for 54 Cα atoms, respectively (24, 25). A conserved hydrophobic patch containing residues Leu-265, Val-269, Pro-273, Leu-276, Ala-277, Leu-280, Ile-283, Leu-290, Ile-294, Pro-298, Phe-301, Val-302, Leu-305, Leu-306, Leu-305, and Val-309 is located at the center of these four helices and forms a groove where the N-terminal helix of yPNGase binds (Fig. 5).
Fig. 5.
Close-up view of the yPNGase-yRad23 interface. Residues from the hydrophobic patch of yRad23XBD bind to the residues from both N- and C-terminal helices of yPNGase. The secondary structures of yPNGase (blue) and yRad23XBD (magenta) and the side chains of yPNGase (cyan) and yRad23XBD (yellow) are shown. O and N atoms are shown in red and blue, respectively. The dotted lines indicate intermolecular H-bonds and ion pairs between yPNGase and yRad23XBD.
Overall Structure of yPNGase and Its Structural Homologues. Sequence alignment and mutational analysis suggested that yPNGase contains a transglutaminase-like fold with a catalytic triad, Cys-191, His-218, and Asp-235 (23). A database search revealed that the core domain of yPNGase most closely resembles that of coagulation factor XIII transglutaminase, arylamine N-acetyltransferase, and avrpphB hydrolase (see Fig. 9) (26-28). The rms deviation values of these proteins are between 2.6 and 3.4 Å for 77-121 Cα atoms. yPNGase and these structural homologues share a common core of two α-helices (H6 and H8) and five strands of an antiparallel β-sheet.
However, yPNGase and these structural homologues are markedly different in terms of other structural features. The major difference between yPNGase and these three homologues is its deep cleft (width 8 Å, length ≈30 Å), which is formed at the interface between the core and the Zn-binding domain of yPNGase. This large cleft runs halfway around the interdomain interface and contributes to the substrate-binding site, in which a catalytic triad and three sucrose molecules are located. Figs. 1 and 4 show the structural framework of the front face of the cleft, which is formed by a helix H9 and strands S8 and S9 forming one side and strand S1 forming the other. The bottom of the cleft is lined with residues from the H6 helix. The back face of the cleft is formed from H3 on one side and from S3 and S4 on the other. However, in the structural homologues of yPNGase described above, the two domains are much more tightly packed, creating a shallow substrates-binding cavity.
The residues forming the large cleft in yPNGase are highly conserved across the species. These residues include Trp-123, Arg-176, Asn-178, Asp-217, Cys-237, Glu-238, Ile-246, and Trp-251 (Figs. 2 and 4). However, these residues are not conserved in other structural homologues, suggesting that the large cleft observed in yPNGase is a highly specific feature in the PNGase family.
Active Site of yPNGase. Although substrates of PNGase have high contents of mannose molecules, they also contain other monosaccharides (9, 23). We observed strong densities for the three sucrose molecules in the deep cleft. We presume that the unusually high content of solvent in the crystal assisted the transportation of these sucrose molecules from the cryosolution to the deep cleft. Overall, sucrose molecules interact with yPNGase residues through multiple H-bonds and van der Waals interactions (Figs. 4A and 10). In site 1, the glucose ring is sandwiched between the aromatic ring of Tyr-211 and aliphatic part of the side chain of Asn-178. The O2 atom of glucose interacts with the carbonyl oxygen of Tyr-177, and the O3 and O6 atoms of fructose make H-bonds with the side chain of Asn-178 and the backbone amide of Gly-192, respectively. In site 2, the glucose ring is packed against the Trp-251 ring, while the fructose ring is packed against Trp-123. The O3 and O4 atoms of the glucose moiety make H-bond contact to the side chain of Glu-238 and Trp-251, respectively.
Overlap of the yPNGase catalytic triad with those of the three structural homologues described above yields rms deviation values on the three Cα atoms of between 1.2 and 1.4 Å. In the superimposed structures of yPNGase and these homologues, the central helix and the catalytic Cys and His residues aligned well. To define the active site more clearly and to understand the catalytic mechanism of PNGase, we soaked the crystals for yPNGase-yRad23 with the irreversible inhibitor, Z-VAD-fmk (Fig. 8). Upon binding of the Z-VAD-fmk, the thiol group of Cys-191 that was H-bonded to His-218 moved away and bound to the methyl group adjacent to the inhibitor's carbonyl group (Figs. 4A and 10). A sucrose molecule in site 1 was displaced by the inhibitor, whereas those in sites 2 and 3 remained unchanged. Within the active site, the peptide chain, Val-Ala-Asp, is well positioned to make a number of H-bonds and van der Waals interactions with residues from strands S4 and S5, whereas the carbobenzyloxy group was not clearly defined. The side chain of Asp at the P1 position of the Z-VAD-fmk forms ion pairs with side chains of Arg-176 and -190. The backbone carbonyl groups of Val and Ala residues of an inhibitor also form H-bonds with the side chains of Arg-176 and -190. In addition, the side chain of Ala of an inhibitor makes van der Waals contact with the side chain of Val-219.
Interaction Between yPNGase and yRad23. The yPNGase-yRad23 interactions result in the burial of 4,514 Å2 exposed surface area (Fig. 5). Biochemical studies using mouse PNGase and HR23B showed that both N-terminal and middle (equivalent to the C-terminal region of yPNGase) regions of mouse PNGase are required for interaction with mouse HR23B (14). Consistent with this analysis, the complex structure revealed that yRad23 makes an extensive interaction with both the N- and C-terminal helices of yPNGase. The yPNGase-yRad23 interface is best described in two interface regions from the perspective of yPNGase. In the first interface, the N-terminal helix of yPNGase binds to the groove formed by all four helices of yRad23 through multiple van der Waals contacts and a H-bond, with 3,222 Å2 of buried surface area. The side chains of Ile-10 and Phe-12 of yPNGase fit into the hydrophobic groove created by the side chains of Met-295, Pro-298, Ile-294, Phe-301, and Leu-280 of yRad23. In addition, the side chains of Ile-15 and Ala-16 of yPNGase interact with the aliphatic parts of the side chains of Glu-281 and Arg-291 of yRad23. Most of the residues in this interface are highly conserved in both PNGase and Rad23 orthologues (Figs. 2 and 5). Several lines of evidences support the importance of hydrophobic interactions in this yPNGase-yRad23 complex interface. First, the deletion of the N-terminal helix of yPNGase abolishes complex formation between yPNGase and yRad23 (15). Second, all four helices of yRad23XBD participate in the interaction. Third, the yPNGase-yRad23 complex dissociates in the presence of the nonionic detergent Triton X-100 (data not shown).
In the second part, the C-terminal helix of yPNGase, H12, exclusively interacts with residues from the N-terminal helix of yRad23 through two H-bonds and two ion pairs. Because the N-terminal helix of yPNGase alone is insufficient to interact with yRad23, this interface also plays an important role in formation of a yPNGase-yRad23 complex (14).
Recent studies suggest that mouse or human PNGase is not only present as a free form in the cytosol but that it is also associated with the ER membrane (14). It is unclear whether yPNGase also binds to the ER surface. In the primary structure, no hydrophobic region was predicted to facilitate anchoring to the ER membrane surface. We propose from our complex structure that if yPNGase binds to the ER membrane, it is likely that the N-terminal helix that associates with yRad23 by hydrophobic interactions would be involved in this binding. No other regions in yPNGase contain high contents of exposed hydrophobic surface area. Residues spanning the N-terminal helix H1 of yPNGase are highly conserved from Drosophila to human and are known to interact with other proteins during proteasome-mediated glycoprotein degradation (ref. 8; Fig. 2).
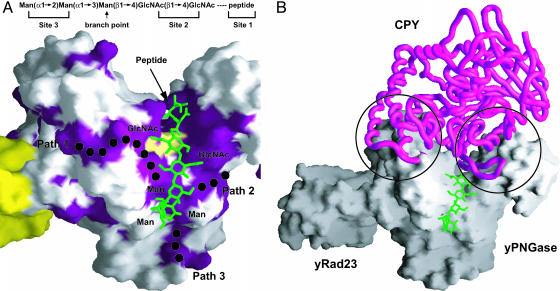
Insights into the Specificity of yPNGase for Denatured Glycoproteins. Although yPNGase specifically deglycosylates glycopeptides or denatured glycoproteins, all three structural homologues of yPNGase described above function on substrates in a native conformation (12, 26-28). Then, how can PNGase distinguish the glycopeptide or unfolded glycoproteins from glycoproteins in their native conformation? The structures presented in this work contain both carbohydrate molecules and a tripeptide inhibitor and thus may mimic the binding form of the glycopeptide (Fig. 6A). To understand the conformational selectivity of PNGase, we performed modeling analysis using several glycoprotein structures including yeast carboxypeptidase Y, RNase B, and ovalbumin (29-32). Previous studies have revealed that yPNGase deglycosylates only unfolded forms of glycoproteins (11, 12). We superimposed the GlcNAc moiety, glycosylated Asn, and neighboring residues of substrates listed above, respectively, onto the sucrose moiety and the Cα position of peptide inhibitor on the active site of yPNGase (Fig. 6B). In yPNGase, the active site is located deep inside the large cleft, and to access this active site, substrates must have dimensions that properly fit the cleft. However, in our model, glycosylated Asn residues in substrates could not be positioned correctly into the active site of yPNGase without being obstructed by both sides of the cleft. The strands S2 and S3 in the Zn-binding domain and two loops between H10 and H11 helices and between H9 and S10 of the core domain located at each side of the cleft would block access to the active site by the native form of glycoproteins as substrates enter the active site. In contrast to native glycoproteins, denatured glycoproteins are believed to be highly flexible and to form extended random conformations, as we observed for our tripeptide inhibitor bound structure, and thus these denatured forms can access the active site without constraint by the cleft walls. In homologous structures described above, the active site cavity is located on the shallow groove, and substrates with native conformation can dock properly with this shallow active site cavity (refs. 26-28; Fig. 9).
Fig. 6.
Molecular model of the yPNGase-substrate complex. (A) A glycan moiety containing two GlcNAc residues and three mannose molecules was modeled on the active site of the cleft. The dotted lines indicate the three conserved regions where the additional carbohydrate molecules could bind. The common structure of a glycan motif is shown at the top. The close-up view on these conserved regions is shown in Fig. 11. (B) Three Cα atoms from Val-366, Arg-367, and Asn-368 and the GlcNAc residue of yeast carboxypeptidase Y (CPY; Protein Data Bank ID code IYSC) were superimposed onto those from three residues of the inhibitor, Z-VAD-fmk, and a fructose molecule in the active site of yPNGase, respectively. The glycosylated Asn residue of CPY in native form is prevented from accessing the active site by both sides of the deep cleft, which are formed by the strands S2 and S3 in the Zn-binding domain and two loops between H10 and H11 helices and between H9 and S10 of the core domain.
The substrates of yPNGase commonly contain two GlcNAc and six to eight mannose molecules or other monosaccharides that are connected by α-1,6 and/or α-1,3 linkages, and these carbohydrates would be expected to bind broadly over the large cleft of yPNGase (Figs. 6A and 11). The observation in our structure that three sucrose molecules continuously bind to the conserved residues in the cleft may reflect the binding mode of the carbohydrates of glycoproteins to yPNGase. Because the carbohydrate-binding site is likely to be broad and continuous, a single-residue perturbation at the binding site would have a weak effect on binding affinity. Previous studies have shown that a single mutant on the conserved surface of this cleft, such as Trp-123 or -251, did not affect the deglycosylase activity of yPNGase (23). To corroborate our structural analysis findings, we constructed two cleft mutants. In these mutants, we replaced five or seven highly conserved residues on the surface of the cleft simultaneously (Figs. 4A and 12). All these mutants lost deglycosylation activity, which supports our structure-based model for carbohydrate binding (Fig. 3).
It is unclear from the elucidated structures how yPNGase recognizes extended or branched carbohydrates moieties that bind to yPNGase precisely. Our structure exhibits three conserved regions that are connected to the deep cleft (Figs. 6A and 11). Although a glycan motif may bind to only one of these regions, it is also possible that branched chains of a glycan motif participate in interacting with all three conserved regions. How a glycan entity would interact with PNGase requires further structural analysis.
In summary, the structure of yPNGase-yRad23XBD complex provides the structural insight into the folding-dependent deglycosylation mechanism and the interaction between yPNGase and yRad23. It reveals (i) the general mechanism for the deglycosylation reaction in which a catalytic triad is involved; (ii) a deep interdomain cleft that provides the basis for high stringency for misfolded proteins over native form of glycoproteins; and (iii) the predominant hydrophobic interface in the yPNGase-yRad23XBD complex.
Supplementary Material
Acknowledgments
We thank Yugene Kim for help in initial protein purification and Kwang Yeon Hwang, Heung Soo Lee, and Kyung Hwa Kim for help with data collection. This work was supported by the National Creative Research Initiatives (Ministry of Science and Technology).
Author contributions: J.-H.L. and Y.C. designed research; J.-H.L., J.M.C., C.L., and K.J.Y. performed research; J.-H.L., J.M.C., and Y.C. analyzed data; and J.-H.L. and Y.C. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ER, endoplasmic reticulum; GlcNAc, N-acetylglucosamine; PNGase, peptide:N-glycanase; yPNGase, yeast PNGase; Z-VAD-fmk, carbobenzyloxy-Val-Ala-Asp-α-fluoromethylketone.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 1X3W (yPNGase-yRad23) and 1X3Z (yPNGase-yRad23 inhibitor)].
References
- 1.Helenius, A. & Aebi, M. (2001) Science 291, 2364-2369. [DOI] [PubMed] [Google Scholar]
- 2.Goldberg, A. L. (2003) Nature 426, 895-899. [DOI] [PubMed] [Google Scholar]
- 3.McCracken, A. A. & Brodsky, J. L. (1996) J. Cell Biol. 132, 291-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rutishauser, J. & Spiess, M. (2002) Swiss Med. Wkly. 132, 211-222. [DOI] [PubMed] [Google Scholar]
- 5.Bailey, C. K., Andriola, I. F., Kampinga, H. H. & Merry, D. E. (2002) Hum. Mol. Genet. 11, 515-523. [DOI] [PubMed] [Google Scholar]
- 6.Wiertz, E. J., Jones, T. R., Sun, L., Bogyo, M., Geuze, H. J. & Ploegh, H. L. (1996) Cell 84, 769-779. [DOI] [PubMed] [Google Scholar]
- 7.Suzuki, T., Yan, Q. & Lennarz, W. J. (1998) J. Biol. Chem. 273, 10083-10086. [DOI] [PubMed] [Google Scholar]
- 8.Suzuki, T., Park, H. & Lennarz, W. J. (2002) FASEB J. 16, 635-641. [DOI] [PubMed] [Google Scholar]
- 9.Suzuki, T. & Lennarz, W. J. (2003) Biochem. Biophys. Res. Commun. 302, 1-5. [DOI] [PubMed] [Google Scholar]
- 10.Hirsch, C., Blom, D. & Ploegh, H. L. (2003) EMBO J. 22, 1036-1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hirsch, C., Misaghi, S., Blom, D., Pacold, M. E. & Ploegh, H. L. (2004) EMBO Rep. 5, 201-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Joshi, S., Katiyar, S. & Lennarz, W. J. (2005) FEBS Lett. 579, 823-826. [DOI] [PubMed] [Google Scholar]
- 13.Suzuki, T., Park, H., Kwofie, M. A. & Lennarz, W. J. (2001) J. Biol. Chem. 276, 21601-21607. [DOI] [PubMed] [Google Scholar]
- 14.Katiyar, S., Li, G. & Lennarz, W. J. (2004) Proc. Natl. Acad. Sci. USA 101, 13774-13779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Biswas, S., Katiyar, S., Li, G., Zhou, X., Lennarz, W. J. & Schindelin, H. (2004) Biochem. Biophys. Res. Commun. 323, 149-155. [DOI] [PubMed] [Google Scholar]
- 16.Schauber, C., Chen, L., Tongaonkar, P., Vega, I., Lambertson, D., Potts, W. & Madura, K. (1998) Nature 391, 715-718. [DOI] [PubMed] [Google Scholar]
- 17.Xie, Z., Liu, S., Zhang, Y. & Wang, Z. (2004) Nucleic Acids Res. 32, 5981-5990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Misaghi, S., Pacold, M. E., Blom, D., Ploegh, H. L. & Korbel, G. A. (2004) Chem. Biol. 11, 1677-1687. [DOI] [PubMed] [Google Scholar]
- 19.Otwinowski, Z. & Minor, W. (1997) Methods Enzymol. 276, 307-326. [DOI] [PubMed] [Google Scholar]
- 20.Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crstallogr. D 54, 905-921. [DOI] [PubMed] [Google Scholar]
- 21.Kleywegt, G. J. & Jones, T. A. (1996) Acta Crystallogr. D 50, 829-832. [DOI] [PubMed] [Google Scholar]
- 22.Collaborative Computational Project B (1994) Acta Crystallogr. D 50, 760-763.15299374 [Google Scholar]
- 23.Katiyar, S., Suzuki, T., Balgobin, B. J. & Lennarz, W. J. (2002) J. Biol. Chem. 277, 12953-12959. [DOI] [PubMed] [Google Scholar]
- 24.Walters, K. J., Lech, P. J., Goh, A. M., Wang, Q. & Howley, P. M. (2003) Proc. Natl. Acad. Sci. USA 100, 12694-12699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kamionka, M. & Feigon, J. (2004) Protein Sci. 13, 2370-2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yee, V. C., Pedersen, L. C., Trong, I. L., Bishop, P. D., Stenkamp, R. E. & Teller, D. C. (1994) Proc. Natl. Acad. Sci. USA 91, 7296-7300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sinclair, J. C., Sandy, J., Delgoda, R., Sim, E. & Noble, M. E. M. (2000) Nat. Struct. Biol. 7, 560-564. [DOI] [PubMed] [Google Scholar]
- 28.Zhu, M., Shao, F., Innes, R. W., Dixon, J. E. & Xu, Z. (2004) Proc. Natl. Acad. Sci. USA 101, 302-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suzuki, T., Park, H., Kitajima, K. & Lennarz, W. J. (1998) J. Biol. Chem. 273, 21526-21530. [DOI] [PubMed] [Google Scholar]
- 30.Endrizzi, J. A., Breddam, K. & Remington, S. J. (1994) Biochemistry 33, 11106-11120. [DOI] [PubMed] [Google Scholar]
- 31.Stein, P. E., Leslie, A. G., Finch, J. T. & Carrell, R. W. (1991) J. Mol. Biol. 221, 941-959. [DOI] [PubMed] [Google Scholar]
- 32.Williams, R. L., Greene, S. M. & McPherson, A. (1987) J. Biol. Chem. 262, 16020-16031. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.