Abstract
αβ-Tubulin heterodimers, from which the microtubules of the cytoskeleton are built, have a complex chaperone-dependent folding pathway. They are thought to be unique to eukaryotes, whereas the homologue FtsZ can be found in bacteria. The exceptions are BtubA and BtubB from Prosthecobacter, which have higher sequence homology to eukaryotic tubulin than to FtsZ. Here we show that some of their properties are different from tubulin, such as weak dimerization and chaperone-independent folding. However, their structure is strikingly similar to tubulin including surface loops, and BtubA/B form tubulin-like protofilaments. Presumably, BtubA/B were transferred from a eukaryotic cell by horizontal gene transfer because their high degree of similarity to eukaryotic genes is unique within the Prosthecobacter genome.
Keywords: cytoskeleton, tubulin family, Prosthecobacter dejongeii, Verrucomicrobia, polymerization
Genomic sequencing of the bacterium Prosthecobacter dejongeii has revealed the genes btubA and btubB that share higher sequence similarity with eukaryotic αβ-tubulin than with the bacterial tubulin-homologue FtsZ, raising questions about the evolutionary origins of these genes (1). Historically, bacteria were thought not to contain a cytoskeleton, a view that has only recently been overturned (2, 3). Tubulin and the bacterial tubulin-homologue FtsZ are thought to have evolved from a common ancestor as is indicated by a similar structure of the monomer and protofilament. In addition, they share a polymerization-dependent GTPase activation mechanism (4). It has been argued, however, that the evolutionary distance between these proteins (and also actin and the bacterial actin homologue MreB) seems extraordinarily large considering that tubulin is amongst the most conserved proteins in eukaryotes (5, 6). αβ-Tubulin differ from the single subunit bacterial protein FtsZ in that they form stable heterodimers with nonexchangeable GTP being trapped in the interface. αβ-Tubulin contain a C-terminal domain, absent in FtsZ, that forms the outside of microtubules (7), structures that have never been reported for FtsZ.
There have been several reports showing microtubule-like structures in bacteria (8). The best examples are so-called “epixenosomes,” bacteria growing as ectosymbionts on Euplotidium ciliates (9). These organisms belong to the Verrucomicrobia, a phylum of bacteria of uncertain lineage (10). Recently, two genes, called btubA and btubB, sharing ≈35% sequence identity with α- and β-tubulin have been found in the free-living species Prosthecobacter dejongeii (1), which are also part of Verrucomicrobia (11). These genes are cotranscribed from one operon, but on the basis of theoretical modeling, Prothecobacter tubulin BtubA and BtubB were predicted not to form heterodimers (1). However, a recent study pelleting N-terminally His-tagged BtubA and -B showed that equimolar amounts of the two proteins assembled into protofilaments in a cooperative manner (12).
Here we show that, contrary to eukaryotic tubulin, soluble and functional BtubA and BtubB proteins can be expressed in Escherichia coli and can be unfolded reversibly. No tag is necessary. The purified proteins form heterodimers and assemble into protofilaments, like αβ-tubulin and FtsZ. Their crystal structures show such far-reaching similarities to αβ-tubulin as to indicate that BtubA/B were transferred from a eukaryotic cell by horizontal gene transfer.
Materials and Methods
Protein Expression and Purification. BtubA from P. dejongeii DSM 12251 was expressed with an N-terminal thioredoxin fusion protein partner and a C-terminal hexahistidine tag in E. coli C41(DE3) cells, induced with 1 mM isopropyl β-d-thiogalactoside for 3 h at 37°C. BtubA-trx (predicted molecular weight, 64,356) was purified by using Ni-NTA agarose (Qiagen, Valencia, CA) resin (buffer A: 50 mM Tris·HCl/300 mM NaCl, pH 7.0; buffer B: buffer A plus 1 M imidazole) and further purified using a S200 Sephacryl (Amersham Pharmacia) size-exclusion column (20 mM Tris·HCl/1 mM azide/1 mM EDTA, pH 7.5). Typically, 34 mg was obtained from 12 liters of culture.
For bicistronic coexpression, genes btubA and btubB were cloned by leaving the intergenic region intact and not adding any extra residues. Expression conditions were as above. The proteins were purified by using Q Sepharose (buffer A: 20 mM Tris·HCl/1 mM azide/5 mM magnesium acetate, pH 8.5; buffer B: buffer A plus 1 M NaCl) and size-exclusion chromatography (S300 Sephacryl, Amersham Pharmacia; 20 mM Tris·HCl/1 mM azide/1 mM EDTA, pH 7.5). The separated BtubA (predicted pI, 5.2; molecular weight, 51,265) and BtubB (predicted pI, 6.1; molecular weight, 46,417) proteins were kept apart or pooled. Twelve liters of culture typically yielded 100 mg of protein.
Selenomethionine-substituted proteins were produced by the feed-back inhibition method (13), and proteins were purified as described above with 5 mM DTT or β-mercaptoethanol.
Nucleotide Analysis. The guanine nucleotide of Btub proteins was extracted with perchloric acid, measured spectrophotometrically, and analyzed by HPLC (14). The protein concentration was determined from absorption spectra in 6 M guanidinium hydrochloride, after subtraction of the nucleotide contribution, employing the extinction coefficients BtubA, 47,770 M-1·cm-1; BtubB, 38,780 M-1·cm-1; and BtubA-trx, 61,360 M-1·cm-1 at 280 nm.
Pelleting Assays and Light Scattering. Each reaction contained 10 μM BtubA/B (i.e., 10 μM BtubA and 10 μM BtubB protein in a state of association that depends on the solution conditions), 100 mM Pipes·NaOH (pH 7.0), 0 or 5 mM MgCl2, and 0 or 1 mM nucleotide (50 μl). After 30 min at room temperature, samples were centrifuged at 80,000 rpm for 20 min at 20°C in a TLA100 rotor (Beckman). The supernatants were removed and mixed with an equal volume of gel-loading buffer. The pellets were washed with 50 μl of identical solution without protein and then solubilized in 100 μl of loading buffer. For electrophoresis, pellets and supernatants were loaded with a delay on the same SDS/polyacrylamide gel. The presence or absence of filaments was verified by using electron microscopy as described below.
For static 90° light scattering, 400 μl of solution containing 10 μM BtubA/B, 100 mM Pipes·NaOH (pH 6.8), 5 mM MgCl2, 200 mM KCl, and 0.5 mM GTP was measured in a PerkinElmer LS 50B luminescence spectrometer at 350 nm and 37°C (10-mm path length). Measurements were taken every 2 s, and fresh GTP was added after the signal approached baseline.
Analytical Ultracentrifugation. Sedimentation velocity experiments were conducted in a Beckman Optima XL-I analytical ultracentrifuge with interference optics, using an An50Ti rotor at 50,000 rpm at 20°C (buffer: 100 mM Pipes·NaOH/200 mM KCl, pH 6.8). Differential sedimentation coefficient distributions, c(s), were calculated with sedfit (15). Sedimentation equilibrium measurements were made as described in ref. 16.
Electron Microscopy. Fifty-microliter reactions were performed at room temperature for 30 min, containing 5 or 10 μM BtubA/B, 100 mM Pipes·NaOH (pH 6.8), 5 mM MgCl2, 0 or 200 mM KCl, and 0.5-1 mM GTP. Ten microliters was then transferred to glow-discharged carbon-coated copper electron microscopic grids and negatively stained with 2% (wt/vol) uranyl acetate. Images were taken in an 80 keV Phillips 208 electron microscope on film and digitized at 7 μm with a Zeiss SCAI film scanner. Diffraction analysis was performed by using the mrc program suite (17).
Crystallization and Data Collection. For initial screening, 1,440 crystallization conditions were tested by using the MRC Laboratory of Molecular Biology nanoliter crystallization robotics (18). For BtubA-trx, crystals were grown in hanging drops of 1 μl of protein at a concentration of 17 mg/ml plus 1 μl of reservoir solution (1.6 M sodium potassium phosphate, pH 6.0) at 19°C. Crystals were soaked in cryo-buffer [2 M Na K phosphate, pH 6.0/15% (vol/vol) ethylene glycol] and cryo-cooled. BtubA/B was crystallized in sitting drops using 1 μl of protein (10 mg/ml in total) and 1.4 μl of reservoir (1.5 M Li2SO4/0.4 M Tris·Cl, pH 7.5). Cryoprotection was obtained by adding 15% glycerol. Data sets were collected at 100 K.
Structure Determination. The structure of BtubA-trx was solved by multiple anomalous dispersion using solve (19) and sharp (20). Manual model building was performed with main (21) and refinement with refmac5 (22) with TLS refinement for separate chains. The structure of the BtubA/B heterodimer was solved by molecular replacement using the BtubA-trx structure and refined using cns (23). Crystallographic data are summarized in Table 1; the final parameters of the structures are summarized in Table 2.
Table 1. Crystallographic data.
| Crystal | λ, Å | res., Å | I/σI* | Rm,† % | Multipl.‡ | Compl.,§ % |
|---|---|---|---|---|---|---|
| P. dejongeii BtubA-trx | ||||||
| space group P321, a = b = 180.5 Å, c = 84.2 Å | ||||||
| PEAK (Se) | 0.9793 | 3.1 | 19.7 (5.1) | 0.076 (0.332) | 8.3 | 99.9 (100.0) |
| INFL (Se) | 0.9796 | 3.1 | 20.3 (5.0) | 0.072 (0.347) | 8.4 | 99.9 (100.0) |
| HREM (Se) | 0.9184 | 3.1 | 19.0 (4.4) | 0.078 (0.392) | 8.2 | 99.9 (100.0) |
| NATI | 0.9340 | 2.5 | 12.5 (3.0) | 0.083 (0.452) | 4.9 | 99.9 (100.0) |
| P. dejongeii BtubA/B heterodimer | ||||||
| space group P6522, a = b = 154.4 Å, c = 256.3 Å | ||||||
| PEAK (Se) | 0.9790 | 4.0 | 11.4 (4.0) | 0.157 (0.429) | 8.1 | 99.7 (100.0) |
| NATI | 0.9340 | 3.2 | 12.8 (2.6) | 0.090 (0.379) | 3.6 | 99.0 (99.5) |
Signal-to-noise ratio of merged intensities; values in parentheses are for the highest resolution shell.
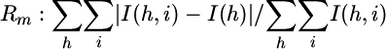 were I(h, i) are symmetry-related in tensities and I(h) is the mean intensity of the reflection with unique index h. The highest resolution bin is in parentheses.
were I(h, i) are symmetry-related in tensities and I(h) is the mean intensity of the reflection with unique index h. The highest resolution bin is in parentheses.
Multiplicity for unique (anomalously unique where appropriate) reflections.
Completeness for unique reflections; anomalous completeness is identical because inverse beam geometry was used. Values in parentheses are for the highest resolution bin. Correlation coefficients of anomalous differences at different wavelengths for the MAD experiment (cut at 4 Å): PEAK vs. INFL, 0.51; PEAK vs. HREM, 0.64; and INFL vs. HREM, 0.40.
Table 2. Refinement statistics.
| P. dejongeii BtubA-trx | |
| Model | Two NCS-related BtubA molecules, chains A, B |
| One E. coli thioredoxin chain T | |
| A: resid. 3-58, 61-246, 253-283, 289-326, 331-432, 1 GTP | |
| B: residues 3-177, 183-347, 350-432, 1 GTP | |
| T: residues 23-125 | |
| 216 water molecules | |
| Diffraction data | 2.5 Å, all data, data set BtubA-trx NATI |
| R factor, Rfree* | 0.202 (0.270), 0.235 (0.275), REFMAC5 with TLS refinement |
| B average/bonded† | 55.1 Å2, 0.721 Å2 |
| Geometry bonds/angles‡ | 0.017 Å, 1.369° |
| NCS rmsd | 0.5 Å |
| Ramachandran§, % | 91.1/0.0 |
| PDB ID code | 2BTO |
| P. dejongeii BtubA/B heterodimer | |
| Model | One BtubA/B heterodimer, chains A: BtubA, B: BtubB |
| A: residues 2-283, 289-442, 1 GDP | |
| B: residues 2-38, 56-71, 84-273, 285-320, 325-426, 4 SO4 | |
| No water molecules | |
| Diffraction data | 3.2 Å, all data, data set BtubAB NATI |
| R factor, Rfree* | 0.215 (0.354), 0.247 (0.334), CNS 1.1 |
| B average/bonded† | 73.7 Å2, 3.246 Å2 |
| Geometry bonds/angles‡ | 0.008 Å, 1.376° |
| Ramachandran,§ % | 83.4/0.0 |
| PDB ID code | 2BTQ |
5% of reflections were randomly selected for determination of the free R factor, prior to any refinement. High-resolution bin is in parentheses.
Temperature factors averaged for all atoms and rmsd of temperature factors between bonded atoms.
rmsd from ideal geometry for bond lengths and restraint angles.
Percentage of residues in the “most favored region” of the Ramachandran plot and percentage of outliers.
Refolding. Purified BtubA/B protein (100 μM) was denatured in 5.5 or 8 M guanidinium hydrochloride for 1.5 h at 26°C. The denatured protein then was injected into refolding buffer (20 mM Tris·HCl/1 mM azide/5 mM magnesium acetate, pH 8.5/1 mM GDP/1 mM DTT) while stirring fast. Precipitation was removed by centrifugation, and the protein was run on the S300 size exclusion column as described above. Circular dichroism (CD) spectra were recorded on a Jasco J-720 (Great Dunmow, U.K.) with 16 measurements averaged per data point at 2.7 μM in 2 mM Tris·HCl, 0.1 mM azide, 0.1 mM EDTA (pH 7.5), or 5.5 M guanidinium hydrochloride, respectively. To test for polymerization formation, the refolded protein was analyzed as described for electron microscopy.
Results and Discussion
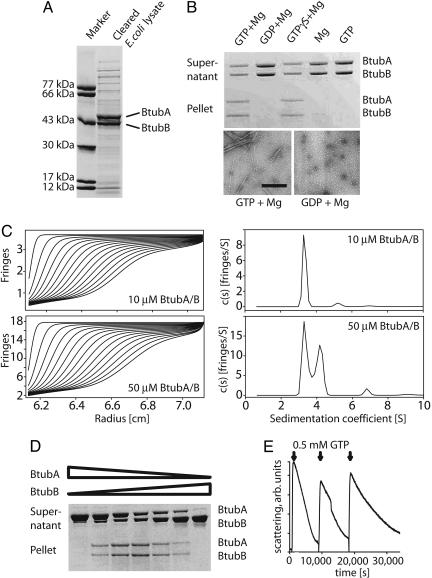
We have cloned P. dejongeii btubAB as one cistron into an expression plasmid under the control of a T7 promoter, maintaining the intergenic region between btubA and btubB. This procedure allowed high expression levels in E. coli (Fig. 1A). The proteins were purified by anion-exchange and size-exclusion chromatography. During both steps it became apparent that the two proteins do not form a tight complex, and pure BtubA and BtubB could be produced separately that way.
Fig. 1.
Heterodimerization and GTP-dependent dynamic polymerization of BtubA/B. (A) Bicistronic expression of P. dejongeii BtubA/B in E. coli. After 3 h of induction, soluble BtubA/B protein makes up the majority of proteins in the lysate (Coomassie-stained polyacrylamide SDS gel). (B) Nucleotide-dependent pelleting and polymerization of BtubA/B. BtubA/B was incubated at 10 μM in 100 mM Pipes·NaOH (pH 7.0) with and without nucleotides and magnesium and centrifuged at 80,000 rpm. Polymerization depends on nucleotide and magnesium binding. The electron micrographs are taken of samples from the pelleting reactions just before centrifugation to demonstrate fiber formation in these reactions. (Negative stain, scale bar: 200 nm.) (C) BtubA/B dimers form only at high concentrations as shown by analytical ultracentrifugation. Shown are sedimentation velocity profiles (Left) and the corresponding sedimentation coefficient distributions (Right). BtubA/B at low concentration (10 μM) gave one main peak with a sedimentation coefficient s20,w = 3.5S (92%) (Upper), whereas BtubA/B at higher concentration (50 μM) gave two main peaks of 3.5S (48%, monomers) and 4.4S (48%, dimers), respectively (Lower), plus a minor peak of 6.8S (4%; 2% in other experiments, which could correspond to incipient association to larger oligomers or to protein aggregation). Buffer contained 100 mM Pipes·NaOH and 200 mM KCl (pH 6.8). (D) BtubA/B polymerizes as a mixed polymer. Different ratios of purified BtubA and BtubB were pelleted under the same conditions as in B. Maximum pelleting occurs at a ratio of 1:1, indicating that polymer formation depends on the BtubA/B interaction. A/B concentrations were 11.2/0.01, 9.2/2, 7.4/3.6, 5.4/5.4, 3.6/7.4, 2/9.2, and 0.01/11.2 μM, respectively. (E) BtubA/B polymers are dynamic. A 90° light-scattering assay at 350 nm with 10 μM BtubA/B and 0.5 mM GTP showed rapid polymerization and slow depolymerization. More rounds of polymerization can be induced by adding fresh GTP.
BtubA/B polymerizes in the presence of GTP and magnesium as demonstrated by a pelleting assay (Fig. 1B), but not with GDP and magnesium. Interestingly, the slowly hydrolyzable GTP analogue GTPγS polymerizes BtubA/B as well as GTP. When samples from the pelleting assay were checked by electron microscopy (Fig. 1B Lower), it appeared that other than the GDP condition, the GTP-containing reaction produced filaments, confirming that the experiment assays filament formation.
Sedimentation velocity runs in an analytical ultracentrifuge indicated that BtubA and BtubB are monomeric at low concentrations (Fig. 1C Upper) and associate into dimers at high concentrations (Fig. 1C Lower). The weak dimerization was confirmed by sedimentation equilibrium analysis at different protein concentrations, which indicated monomers (approximate molecular weight, 50,000) in equilibrium with dimers (see Supporting Materials and Methods and Fig. 5, which are published as supporting information on the PNAS web site).
To see whether BtubA and BtubB polymerize as homo- or hetero-oligomers, we tested GTP-dependent polymerization with different ratios of BtubA and BtubB in a pelleting assay (Fig. 1D). It can clearly be seen that polymerization only occurs when both BtubA and BtubB are present and is best at a ratio of 1:1, indicating heterodimer formation.
GTP-dependent polymerization of BtubA/B is reversible and is induced by GTP (Fig. 1E) as shown by a 90° light scattering assay at 350 nm. During these reactions it became apparent that a slight increase in salt concentration (0.2 M KCl) strongly enhances polymerization. The protein polymerizes rapidly and then slowly depolymerizes over ≈10,000 s because of GTP turnover. Fresh addition of GTP initiates another round of polymerization with a slower depolymerization rate because the GDP from the first reaction acts as an inhibitor of the GTP consumption.
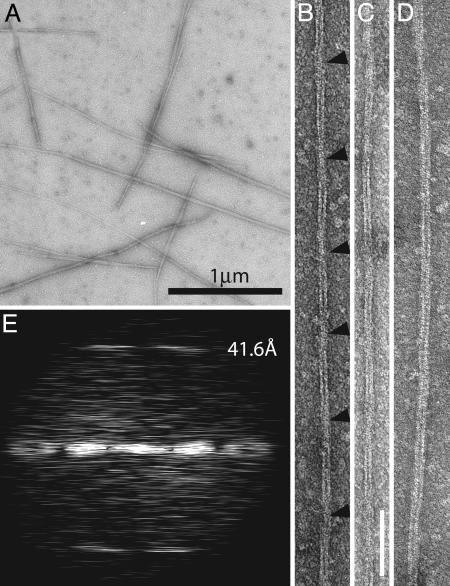
The polymers of BtubA/B we have produced are not microtubules (Fig. 2) but are made up of protofilaments similar to those formed by tubulin and FtsZ. By using our conditions (0.1 M Pipes·NaOH, pH 6.8/200 mM KCl/1 mM GTP/5 mM MgCl2), a large proportion of the protein forms filaments that stain well with uranyl acetate negative staining (Fig. 2A). When observed at higher magnification (Fig. 2 B-D), these filaments appear to be either double protofilaments or bundles of double protofilaments. Most of the double protofilament fibers are straight and have a twist (Fig. 2 B and C). Whether these double filaments are composed of parallel or antiparallel protofilaments remains to be investigated. They most likely have the conserved structure of bona fide tubulin/FtsZ-like protofilaments as demonstrated by the 41.6-Å longitudinal repeat, the length of which is between that of the tubulin repeat (40 Å) and the FtsZ repeat (41-43 Å) (Fig. 2E). The conditions for polymerization of BtubA/B appear also different from eukaryotic tubulin. In typical microtubule assembly buffers (such as 80 mM Pipes·NaOH/1 mM EGTA/2 mM MgCl2, pH 6.8, with 1 mM GTP or 0.2 mM guanylyl α,β-methylene diphosphonate), BtubA/B only polymerized in the presence of additives, such as 200 mg/ml Ficoll 70 or 1 M glutamate, forming protofilament bundles with the same longitudinal repeat (data not shown).
Fig. 2.
BtubA/B polymers have a longitudinal repeat similar to α, β-tubulin indicating protofilament formation. (A) Low-magnification micrograph showing BtubA/B filaments after polymerization in the presence of GTP. Protein at 10 μM was incubated for 30 min at ambient temperature with 100 mM Pipes·NaOH (pH 6.8), 5 mM MgCl2, 200 mM KCl, and 1 mM GTP and was negatively stained with 2% uranyl acetate. (B-D) BtubA/B double filaments. These are the most commonly formed filaments, presumably consisting of two BtubA/B protofilaments. Most filaments twist (B and C), indicated by arrowheads at the crossover points. Filament C has an average width of ≈109 Å. (Scale bar: 100 nM.) (E) Computed diffraction pattern of filament B. Layer lines are clearly visible at ≈42 Å, representing the subunit repeat along the protofilament axis. This repeat matches the repeat seen in the BtubA/B crystal structure and is close to that of tubulin (40 Å).
A recent report pelleting N-terminally His-tagged BtubA and BtubB showed that equimolar amounts of the two proteins polymerize into protofilament bundles and hydrolyze GTP (12). However, the His-tags are not necessary but interfere with the polymerization.
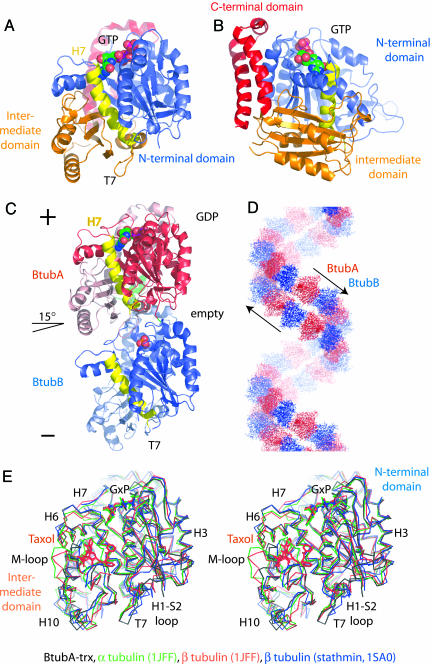
We solved the crystal structure of BtubA alone, fused to thioredoxin at 2.5-Å resolution by selenomethionine multiple anomalous dispersion. It very closely resembles the structure of tubulin with the typical three-domain architecture (Fig. 3 A and B). The N-terminal domain (blue) provides loops T1-T6 for nucleotide binding and is separated by helix H7 (yellow) from the intermediate domain (orange). The latter provides loop T7 that activates nucleotide hydrolysis in the protofilament when inserted into the active site of the next subunit (4). Importantly, BtubA also contains the C-terminal domain of tubulin (red), mostly consisting of two large helices that form the outside in microtubules and that are absent from FtsZ (7). Although there was no nucleotide added during crystallization, the structure of BtubA contains GTP and is matched by a nucleotide content of 0.68 GTP (0.02 GDP) per BtubA in the protein preparation.
Fig. 3.
Crystal structures of BtubA and BtubA/B. (A) Crystal structure of BtubA at 2.5-Å resolution. BtubA's structure is closely related to tubulin. The fold is divided into the N-terminal nucleotide-binding domain (blue), separated by helix H7 (yellow) from the intermediate domain (orange) and two large helices forming the C-terminal domain (red) (4). (B) BtubA contains the C-terminal tubulin domain. Shown is a view rotated 90° around the y axis from A. The two large helices (red) at the C terminus of tubulin form the outside of microtubules (7) and make the biggest difference between tubulin and FtsZ. (C) Crystal structure of BtubA/B heterodimer (asymmetric unit of the crystals) at 3.2-Å resolution. BtubA/B form the same heterodimer as tubulin (24, 25). The protofilament axis is vertical. BtubA is situated at the plus (+) end (red), and BtubB is at the minus (-) end (blue). In the crystals, BtubA contains GDP, whereas BtubB has a sulfate ion in the nucleotide-binding site. The heterodimer is not completely straight; the two subunits are rotated by ≈15° around the z axis [same direction as in the tubulin-stathmin complex (26)], tangential to the microtubule wall. (D) The BtubA/B crystals contain a continuous double filament. The 6522 space group symmetry produces an antiparallel double filament with repeating BtubA/B units in the crystal packing. The bend per heterodimer is 60°, divided into 15° between BtubA and -B (intradimer; see C) and 45° between B and A (interdimer). (E) BtubA/B are very closely related to tubulin. Shown is the superposition of BtubA (black) (rmsd to BtubB, 1.34 Å; 36% sequence identity; 82% aligned) with α-tubulin (25) (green; rmsd 1.5 Å; 37% sequence identity; 85% aligned; Protein Data Bank ID code 1JFF), β-tubulin (25) (red; rmsd 1.71 Å; 35% sequence identity; 85% aligned; Protein Data Bank ID code 1JFF), and subunit B from the tubulin-stathmin complex (26) (blue; rmsd 1.3 Å; 35% sequence identity; 85% aligned; Protein Data Bank ID code 1SA0). Differences are small and mainly located in the T7-loop, the M-loop, which is involved in microtubule formation for tubulin (7), helix H6, and loop H1-S2, which are part of the protofilament contact. BtubA/B have a short S9-S10 loop that in α-tubulin covers the Taxol-binding pocket completely. (Figure was generated with pymol.)
The crystal structure of the BtubA/B dimer at 3.2-Å resolution (Fig. 3C) shows essentially a tubulin-like heterodimer (24, 25) with BtubA (bound to GDP) in the β-tubulin position and BtubB (no nucleotide) in the α-tubulin position. We have confirmed experimentally the correct placement of BtubA and -B by the positions of selenium in an anomalous Fourier map calculated from selenomethionine-substituted BtubA/B (data not shown). We prefer not to assign BtubA or BtubB to β- or α-tubulin because both BtubA and BtubB have an activating T7 loop sequence (1) and a short S9-S10 loop in the Taxol binding pocket (Fig. 3E), mixing characteristics of both α- and β-tubulin, respectively. In the crystal structure of the BtubA/B heterodimer, the nucleotide-binding pocket of BtubA contains GDP, deriving from the protein preparation, and BtubB has no nucleotide but a sulfate ion bound. This finding is confirmed by a nucleotide content of 1.27 GDP (0.12 GTP) per BtubA/B heterodimer in the protein preparation (no nucleotide was added to the crystallization conditions), although preparations differed in their nucleotide content. In noncrystalline polymers as shown in Fig. 2, both BtubA and B most likely have nucleotide bound, as have tubulin and FtsZ in protofilaments.
The BtubA/B heterodimer is bent by ≈15° (Fig. 3C) in the same direction as seen in a tubulin-stathmin complex (26) and similar to the bend seen in a medium-resolution reconstruction of tubulin tubes containing bent protofilaments (27). In all three cases, the bend is mostly tangential to the corresponding microtubule wall, and deviations from this orientation are most likely related to the fact that the protofilaments actually form helices (27). In our case, the bend occurs because BtubA/B forms a continuous double filament in the crystals (Fig. 3D). The crystal geometry (space group P6522) produces a helix made of an antiparallel double filament with a 60° bend, 15° intradimer, and 45° interdimer.
To find the same direction of bending in our structure, the tubulin-stathmin complex, and curved tubulin protofilaments might indicate that tubulin has a built-in hinge between the monomers that allows bending only in one direction and that the nucleotide state of that particular interface controls the probability of bending.
When superimposed on available tubulin and FtsZ structures it becomes apparent that BtubA [and also BtubB, because BtubB is very similar to BtubA with 1.3-Å root-mean-square deviation (rmsd) and 36% sequence identity] is very closely related to tubulin (see also the Fig. 3E legend). The rmsd of Cα backbone atoms is 1.3 Å compared with chain B in the tubulin-stathmin complex (26), 1.5 Å to α-tubulin (25), and 2.7 Å to FtsZ from Methanococcus jannaschii (28). A structural alignment is provided in Table 3, which is published as supporting information on the PNAS web site. Differences to tubulin are mostly conformational, whereas FtsZ is missing the C-terminal domain and parts of several large loops (such as the M-loop involved in microtubule formation, loop H1-S2) (4). Also the sequence similarity to FtsZ is much lower with 17% identity. We consider the structural similarity between αβ-tubulins and BtubA/B as surprisingly high, even for proteins with 36-37% sequence identity.
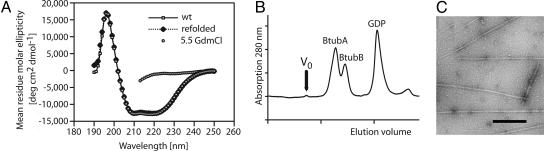
Given the high degree of similarity to tubulin and the high levels of BtubA/B when expressed in E. coli, we investigated whether BtubA/B can fold without any of the chaperones needed for tubulin folding (29) as FtsZ does (30). Pure BtubA/B was denatured in 5.5 M guanidinium hydrochloride and then refolded by injecting it slowly into a large volume of buffer containing GDP and magnesium. Refolding of the protein was then assayed by taking CD spectra (Fig. 4A). GTP-dependent polymerization of the refolded protein was confirmed by using electron microscopy (Fig. 4C) after concentration and size-exclusion chromatography (Fig. 4D). Identical results were obtained when 8 M guanidinium hydrochloride was used (data not shown). Surprisingly, both proteins refold reasonably in vitro (typical efficiency 16%), and the proteins are soluble and active. This result indicates that the chaperone dependence of eukaryotic tubulin is not inherent in the overall fold or domain architecture but may be needed for the building of the tight heterodimer or depend on the amino acid sequence.
Fig. 4.
Chaperone independent folding of BtubA/B in vitro.(A) BtubA/B refold in vitro in the presence of GDP. CD spectra showing the folding state of BtubA/B, in 5.5 M guanidinium hydrochloride (GdmCl) and after refolding by dilution and subsequent size exclusion chromatography. The unfolded spectrum was cut at 213 nm because of light absorption by guanidinium hydrochloride. (B) Refolded BtubA/B are monomeric. Size exclusion chromatography of refolded BtubA/B on a Sephacryl S300 column (Amersham Pharmacia) in 20 mM Tris·HCl, 1 mM azide, and 1 mM EDTA (pH 7.5). Refolded protein shows no aggregation. Never-unfolded protein shows the same chromatogram with BtubA and BtubB running separately (data not shown). (C) Refolded BtubA/B polymerizes. An electron micrograph shows filaments made from refolded BtubA/B protein. The filaments are indistinguishable from wild-type filaments. Conditions are as for Figs. 1B and 2. (Scale bar: 200 nm.)
How did BtubA/B proteins with a rmsd of ≈1.5Åto αβ-tubulin, forming a similar heterodimer, having the characteristic C-terminal tubulin helices and polymerizing into the same protofilaments, end up in bacterial organisms? We can envisage two main possibilities explaining the short evolutionary distance between BtubA/B and tubulin. BtubA/B and tubulin might have a common ancestor on the same lineage, implying that Prosthecobacter as an organism or at least a subset of its genome is closer to eukaryotes or parts of their genome than other bacteria. This possibility is clearly not the case: Although the P. dejongeii genome is only completed to ≈90% (see www.integratedgenomics.com), other genes clearly group very closely with their bacterial counterparts (for example MreB, the bacterial actin homologue, and 16S RNA), and almost no eukaryotic signature proteins have been found in Prosthecobacter (31), certainly not more than in any other bacterium.
We prefer horizontal gene transfer as a more likely explanation for the uniquely close relationship between BtubA/B and eukaryotic tubulin. FtsZ (in bacteria) and tubulin (in eukaryotes) share a common ancestor and were separated a very long evolutionary distance ago, as indicated by their weak sequence similarity and modest structural similarity (5). Besides the weak similarities, the fact that they polymerize into the same protofilaments makes them true homologues (32). However, as shown here, the structure of BtubA/B is much closer to tubulin than FtsZ, and that includes all surface loops that are unique to tubulin. This structure also includes the C-terminal domain, which is on the outside of eukaryotic microtubules where its function is to interact with motor proteins that so far have not been found in prokaryotes. We think it is therefore unlikely that BtubA/B are an intermediate between FtsZ and tubulin because the C-terminal domain and many surface loops involved in microtubule formation only are required in the eukaryotic context. We propose that at some point one or possibly two tubulin genes were transferred to Prosthecobacter where they were modified not to form a tight heterodimer and to fold without chaperones, whose function in eukaryotic tubulin folding could be regulatory.
Supplementary Material
Acknowledgments
We thank Carlos Alfonso (Servicio de Ultracentrifugación Analítica Centro de Investigaciones Biológicas, Consejo Superior de Investigaciones Científicas) for analytical ultracentrifugation, Christopher Johnson (Medical Research Council Centre for Protein Engineering) for help with the CD measurements, and Linda Amos for helpful discussion. We thank everyone at the following beamlines for excellent support: Beamline 10.1 at the SRS (Daresbury, U.K.) and ID14-1, ID14-4, ID29 and BM14, all at the ESRF (Grenoble, France). This work was supported by Ministerio de Ciencia y Tecnología Grants BIO 2002-03665 (to J.M.A.) and a Phillip Leverhulme Prize (to J.L.).
Author contributions: D.S., J.M.A., and J.L. designed research; D.S., M.A.O., J.M.A., and J.L. performed research; D.S., M.A.O., J.M.A., and J.L. analyzed data; and D.S. and J.L. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviation: rmsd, rms deviation.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 2BTO (BtubA-trx) and 2BTQ (BtubAB)].
References
- 1.Jenkins, C., Samudrala, R., Anderson, I., Hedlund, B. P., Petroni, G., Michailova, N., Pinel, N., Overbeek, R., Rosati, G. & Staley J. T. (2002) Proc. Natl. Acad. Sci. USA 99, 17049-17054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carballido-Lopez, R. & Errington, J. A. (2003) Trends Cell Biol. 13, 577-583. [DOI] [PubMed] [Google Scholar]
- 3.Van den Ent, F., Amos, L. & Lowe, J. (2001) Curr. Opin. Microbiol. 4, 634-638. [DOI] [PubMed] [Google Scholar]
- 4.Nogales, E., Downing, K. H., Amos, L. A. & Lowe, J. (1998) Nat. Struct. Biol. 5, 451-458. [DOI] [PubMed] [Google Scholar]
- 5.Doolittle, R. F. & York, A. L. (2002) BioEssays 24, 293-296. [DOI] [PubMed] [Google Scholar]
- 6.Faguy, D. M. & Doolittle, W. F. (1998) Curr. Biol. 8, R338-R341. [DOI] [PubMed] [Google Scholar]
- 7.Nogales, E., Whittaker, M., Milligan, R. A. & Downing, K. H. (1999) Cell 96, 79-88. [DOI] [PubMed] [Google Scholar]
- 8.Bermudes, D., Hinkle, G. & Margulis, L. (1994) Microbiol. Rev. 58, 387-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosati, G., Lenzi, P. & Verni, F. (1993) Micron 24, 465-471. [Google Scholar]
- 10.Petroni, G., Spring, S., Schleifer, K.-H., Verni, F. & Rosati, G. (2000) Proc. Natl. Acad. Sci. USA 97, 1813-1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hedlund, B. P., Gosink, J. J. & Staley, J. T. (1997) Antonie Leeuwenhoek 72, 29-38. [DOI] [PubMed] [Google Scholar]
- 12.Sontag, C. A., Staley J. T. & Erickson, H. P. (2005) J. Cell Biol. 169, 233-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van Duyne, G. D., Standaert, R. F., Karplus, P. A., Schreiber, S. L. & Clardy, J. (1993) J. Mol. Biol. 229, 105-124. [DOI] [PubMed] [Google Scholar]
- 14.Huecas, S. & Andreu, J. M. (2003) J. Biol. Chem. 278, 46146-46154. [DOI] [PubMed] [Google Scholar]
- 15.Schuck, P. (2000) Biophys. J. 78, 1606-16019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rivas, G., Lopez, A., Mingorance, J., Ferrandiz, M. J., Zorrilla, S., Minton, A. P., Vicente, M. & Andreu, J. M. (2000) J. Biol. Chem. 275, 11740-11749. [DOI] [PubMed] [Google Scholar]
- 17.Crowther, R. A., Henderson, R. & Smith, J. M. (1996) J. Struct. Biol. 116, 9-16. [DOI] [PubMed] [Google Scholar]
- 18.Stock, D., Perisic, O. & Lowe, J. (2005) Prog. Biophys. Mol. Biol. 88, 311-327. [DOI] [PubMed] [Google Scholar]
- 19.Terwilliger, T. C. & Berendzen, J. (1999) Acta Crystallogr. D 55, 849-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De La Fortelle, E. & Bricogne, G. (1997) Methods Enzymol. 276, 472-494. [DOI] [PubMed] [Google Scholar]
- 21.Turk, D. (1992) Ph.D. thesis (Technische Univ., Munich).
- 22.Murshudov, G. N. (1997) Acta Crystallogr. D 53, 240-255. [DOI] [PubMed] [Google Scholar]
- 23.Brunger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, 905-921. [DOI] [PubMed] [Google Scholar]
- 24.Nogales, E., Wolf, S. G. & Downing, K. H. (1998) Nature 391, 199-203. [DOI] [PubMed] [Google Scholar]
- 25.Lowe, J., Li, H., Downing, K. H. & Nogales, E. (2001) J. Mol. Biol. 313, 1045-1057. [DOI] [PubMed] [Google Scholar]
- 26.Ravelli, R. B., Gigant, B., Curmi, P. A., Jourdain, I., Lachkar, S., Sobel, A. & Knossow, M. (2004) Nature 428, 198-202. [DOI] [PubMed] [Google Scholar]
- 27.Wang, H.-W. & Nogales, E. (2005) Nature, in press.
- 28.Lowe, J. & Amos, L. A. (1998) Nature 391, 203-206. [DOI] [PubMed] [Google Scholar]
- 29.Lewis, A. A., Tian, G. & Cowan, N. J. (1997) Trends Cell Biol. 7, 479-484. [DOI] [PubMed] [Google Scholar]
- 30.Andreu, J. M., Oliva, M. A. & Monasterio, O. (2002) J. Biol. Chem. 277, 43262-43270. [DOI] [PubMed] [Google Scholar]
- 31.Staley, J. T., Bouzek, H. & Jenkins, C. (2005) FEMS Microbiol. Lett. 243, 9-14. [DOI] [PubMed] [Google Scholar]
- 32.Oliva, M. A., Cordell, S. C. & Lowe, J. (2004) Nat. Struct. Mol. Biol. 11, 1243-1250. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.