Abstract
Populations of jellyfish, Mastigias sp., landlocked in tropical marine lakes during the Holocene, show extreme genetic isolation (0.74 ≤ φST ≤ 1.00), founder effects (genetic diversity: 0.000 ≤ π ≤ 0.001), rapid morphological evolution, and behavioral adaptation. These results demonstrate incipient speciation in what we propose may be modern analogues of Plio-Pleistocene populations isolated in ocean basins by glacially lowered sea level and counterparts to modern marine populations isolated on archipelagos and other distant shores. Geographic isolation in novel environments, even if geologically brief, may contribute much to marine biodiversity because evolutionary rates in marine plankton can rival the most rapid speciation seen for limnetic species, such as cichlids and sticklebacks. Marine lakes present situations rare in their clarity for studying evolution in marine taxa.
Keywords: founder effect, island, rate of evolution
Marine species often are expected to have large geographic ranges, large population sizes, and high gene flow because of weekslong planktonic stages that may be dispersed in currents traveling many kilometers per day (1-3). Such widespread panmictic populations result in relatively few (4), and relatively slowly evolving, marine species (2, 5-7). However, marine molecular genetics and physical oceanography are increasingly revealing biotic and physical discontinuities in an ecologically heterogeneous environment (3, 8-10), raising questions about the relative frequencies of different modes and rates of evolution.
Tempo and mode of evolution can be inferred from species distributions, the fossil record, population differentiation, and genetic variation set against a backdrop of geologic and climatic history (2, 6, 11-14). Areas with high endemism or high species diversity, particularly, are enigmatic, and their study may elucidate processes predominant in speciation (11). Speciation in small isolated populations, for example, may be quicker than average but, in peripheral locations, contribute little to overall patterns of diversity (11). Large, centrally situated, populations, in contrast, may speciate less often but may be more successful when they do, thus dominating patterns of biodiversity on oceanic scales (11). However, isolation is not necessarily unique to peripheral populations. The Indo-West Pacific center of marine biodiversity in the “East Indies Triangle” (11) is a particularly complex region of the seas composed of many basins and island chains with a long history of tectonic activity and, like other parts of the world, fluctuating sea level in response to climate change. Although many modern Indo-West Pacific species were present during the late Pliocene, most molecular studies suggest that modern patterns of intraspecific genetic variation in the Indo-West Pacific originated during the Pleistocene (12) as a result of greater isolation of ocean basins during glacial low stands (12-14). Similar patterns are seen elsewhere. For example, 65-70% of modern south Australian marine species were present in the late Pliocene, by the middle Pleistocene marine molluscan faunas were composed entirely of modern species (15), and patterns of intraspecific genetic variation in southeast Australia taxa are consistent with subdivision of species ranges during Pleistocene low stands (16). Marine fossil faunas of North and Central America show pulse extinction and speciation events ≈2 million years ago (17). Thus, much modern marine diversity appears to have originated during the relatively short periods of glacially lowered sea levels that predominated during the late Pliocene and Pleistocene “icehouse” climate (2, 18, 19).
The details of such events are difficult to study in many marine taxa; they are impossible to study in gelatinous marine zooplankton, an ecologically important and phyletically diverse group including ctenophores, salps, pteropods, cubozoan, hydrozoan, and scyphozoan jellyfishes, because exceptionally few ever leave a fossil or subfossil record. Populations of jellyfishes landlocked in “marine lakes” during the Holocene therefore may provide a valuable modern analogue of Plio-Pleistocene seas and a window into the evolutionary processes that predominated millions of years ago.
Marine lakes are small bodies of seawater entirely surrounded by land. A cluster of ≈70 marine lakes formed in Palau as a topographically complex karst landscape (uplifted Miocene reef) permeated with caverns and fissures was progressively flooded by rising sea level after the last glacial maximum (≈18,000 years ago). Lakes formed in chronological series, deeper depressions flooded first (≈12,000 years ago) and shallower depressions later (≈5,000 years ago); the floodwaters carried propagules from the surrounding lagoon varying distances inland, some of which became the progenitors of modern-day lake populations. Contemporary connectivity between marine lakes and the lagoon is limited and varied, as indicated by damped and delayed tides (20). Modern marine lakes therefore differ from the lagoon and from lake to lake in attributes such as depth, age, size, isolation, physical and chemical structure, and biota, even though all lakes within Palau have similar geological history and are subject to similar climate, weather, and potential propagules from the surrounding sea (ref. 20; Table 1), providing an unprecedented opportunity to examine the evolution of multiple peripatric marine populations.
Table 1. Sample locations, their physical characteristics, and estimates of genetic diversity in populations of Mastigias medusae.
| Habitat type
|
Shortest distance overland, m†
|
Nucleotide diversity, π·10-3
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Site* | Physical structure | Coral in lake? | Depth, m | Area, m2·103 | Sample size, n | No. of haplotypes | Haplotype diversity, h | ||
| Lake | |||||||||
| NLK | Meromictic | N | 38 | 43 | 210 | 14 | 3 | 0.47 ± 0.14 | 1.03 ± 1.04 |
| GLK | Meromictic | N | 15 | 21 | 110 | 15 | 1 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| OLO | Holomictic | N | 4 | 9 | 150 | 15 | 1 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| CLM | Meromictic | N | 30 | 39 | 260 | 12 | 2 | 0.53 ± 0.08 | 1.08 ± 1.08 |
| OTM | Meromictic | N | 30 | 50 | 150 | 14 | 1 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| TLM‡ | Stratified | Y | 60 | 470 | 75 | 8 | 2 | 0.57 ± 0.10 | 1.16 ± 1.19 |
| Coves and lagoon | |||||||||
| NCK | Holomictic | Y | 9 | ≫1 | 0 | 12 | 4 | 0.71 ± 0.11 | 2.27 ± 1.79 |
| RCA | Holomictic | Y | 20 | ≫2 | 0 | 14 | 5 | 0.70 ± 0.10 | 3.55 ± 2.47 |
| Lagoon§ | Holomictic | Y | <4 to >60 | ≫103 | 0 | 18 | 4 | 0.60 ± 0.11 | 2.19 ± 1.69 |
The island or region of Palau in which each site lies is given in parentheses: NLK (Uet era Ngermeuangel or “Big Jellyfish Lake” Koror), GLK (Goby Lake, Koror), OLO (Uet era Ongael, Ongael), CLM (Clearwater Lake, Mecherchar), OTM (Ongeim'l Tketau, Mecherchar), TLM (Tketau Lake, Mecherchar), NCK (Ngermeuangel or “Big Jellyfish” Cove, Koror), RCA (Risong Cove, Auluptagel).
Present-day distance from “mainland” lagoon to “island” marine lake. Present-day distance is correlated (Pearson's r = 0.91, P = 0.032) with shortest distance from the center of each lake basin to the edge of the nearest reef face, which approximates the shortest overland distance 10,000 years ago when sea level was -25 to -30 m (n = 5, excluding OLO, which is only 4 m deep).
TLM's physical structure is intermediate between “holomictic” and “meromictic”; vertical stratification varies in strength, resulting in the formation of highly turbid, presumably dysoxic, waters as shallow as 30 m (unpublished data) or limited mixing and oxygenation to >50 m (20).
Malakal (n = 7), Ngerchaol Cove (n = 4), Tab Kukau Cove (n = 4), and Mecherchar (n = 3).
In Palau, putative ancestral populations of golden jellyfish, Mastigias sp. occur in semienclosed coves and in the reef-lagoon waters that surround the archipelago; at least six marine lakes have been colonized by Mastigias (Fig. 1A). In contrast to lagoon populations, which all have similar morphologies and behaviors (e.g., daily migration from west to east), each lake population has a distinct morphology characterized by different degrees of vestigialization and has different behavior (21-23). For example, in Goby Lake (GLK), one of the youngest lakes, Mastigias migrate similarly to their recent ancestors in the lagoon, aggregating densely in the east during late afternoon. In Uet era Ngermeuangel (NLK), one of the oldest lakes, Mastigias migrate weakly both eastward and westward during both morning and afternoon and, comparatively, are quite evenly distributed. In contrast, in Ongeim'l Tketau (OTM), which is of intermediate age, up to 24 million medusae migrate 0.5 km eastward each morning, accumulating at the east end of the lake around noon at average densities of ≈600 m-3 before they turn to migrate westward in the afternoon (21-22). The distinct behaviors and morphologies of lake medusae indicate isolation, in situ evolution, and local adaptation during the Holocene. Here, we use molecular analyses to assess population structure and to provide a framework for interpreting evolutionary history in Mastigias in Palau.
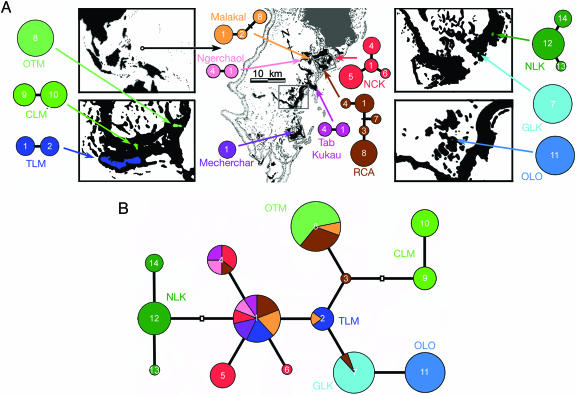
Fig. 1.
The geography of Palau and genetic diversity in Mastigias. (A Upper Left Inset) Palau lies between 6° 53′ N 134° 08′ E and 8° 12′ N 134° 44′ E in the western equatorial North Pacific. (Center) Karst islands with high relief (black) surrounded by the lagoon and harboring semienclosed coves and land-locked marine lakes (boxes). Modern barrier and fringing reefs and the large igneous island in the northeast are shown in gray. Mastigias COI haplotype networks are shown for six cove or lagoon sites (center) and six marine lake populations (Left Lower, Right Upper, and Right Lower); the area of each circle is proportional to haplotype frequency; small crossbars show haplotypes that were not sampled in Malakal or Risong Cove (RCA) but were present in other populations in Palau (B). Table 1 describes each location and sample sizes. (B) COI haplotype network for 122 Mastigias medusae. Each circle represents a distinct haplotype; small empty squares are unsampled inferred haplotypes. Haplotypes are color coded as in A. In addition, marine lake haplotypes are labeled, whereas cove or lagoon haplotypes are unlabeled. In both A and B, each branch indicates one nucleotide difference, each haplotype is identified by a number, and the colors of circles and segments indicate the geographic origins of haplotypes. The area of each circle, or portion thereof, is proportional to the frequency of the haplotype (largest to smallest: 23, 21, 16, 15, 10, 8, 7, 6, 5, 3, and 1).
Materials and Methods
Molecular Analyses. A total of 122 medusae were collected from 12 locations (Table 1), each location being sampled usually on two occasions between 1996 and 2001. Cytochrome c oxidase subunit I (COI) was amplified with primers LCOjf (5′-ggtcaacaaatcataaagatattggaac) and MpHCO (5′-caaaatagatgctgatacaaaatagg) and sequenced at the University of New South Wales' Ramaciotti Centre. All electropherograms and sequences were checked and aligned by eye, ambiguous positions removed (leaving ≈500 nucleotides per sequence), then analyzed by using unweighted maximum parsimony in paup*4.0b10 (24) and pairwise sequence difference in arlequin 2.0 (25). The proportion of unique haplotypes (number of unique haplotypes/sample size) was chosen as the measure of diversity because it best represents the number of successful colonizations and makes some accommodation for small variations in sample sizes among sites. The proportion of unique haplotypes was correlated with haplotype diversity and nucleotide diversity (Pearson's r = 0.877 to 0.939, P ≤ 0.002, uncorrected for three tests), so this choice did not influence the overall interpretation.
To provide some theoretical context for our study, we compare patterns of genetic diversity in Mastigias with the predicted genetic consequences of evolution on islands (26). Although heterodox, the comparison is valuable because the physical and biological setting of marine lakes (20, 27) fits the conceptual model of terrestrial islands (26) in four key respects. Dispersal occurs from the ocean (mainland) to the lakes (islands). Dispersal rate is approximately constant over time, being a function of sea level rise from ≈12,000-5,000 years ago and tidal exchange. Mutation rate is constant, in this case zero, over the period considered. The molecular marker COI is neutral or nearly neutral, as indicated by 10 of 13 parsimony informative positions being third position synonymous changes. Finally, there is no evidence that marine lakes deviate from other assumptions of terrestrial island theory, for example, that extinction rate is constant through time (26), except where noted in Results and Discussion. Small deviations from the assumptions are inconsequential (26).
Morphological Analyses. Forty morphological features were measured on 64 medusae 100 mm in bell diameter: 10 per lake [except Tketau Lake (TLM)] and 14 from several lagoon locations (see ref. 23 for detailed methods). Features were normalized on a 0-1 scale and downweighted if correlated during multidimensional scaling and plotting in two dimensions (x and y) by spss 10 for Mac (23). Morphological distance (d) between a pair of medusae was calculated as 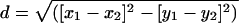 . Morphological disparity was calculated for each lake (and lagoon) medusa as the mean of all of its pairwise morphological distances from every (other) lagoon medusa.
. Morphological disparity was calculated for each lake (and lagoon) medusa as the mean of all of its pairwise morphological distances from every (other) lagoon medusa.
Results and Discussion
Geographic variation in COI shows greater diversity in lagoon populations than lake populations, consistent with an ancestral lagoonal range of Mastigias and founder effects at, or bottlenecks after, colonization of marine lake habitats (Fig. 1 A and Table 1). Lagoon populations are genetically more similar to each other than are either (i) lagoon and lake populations excepting TLM or (ii) lake populations to each other (Fig. 1B and Table 2), a pattern that is consistent with independent colonization of each marine lake from the ancestral lagoonal population. Population differentiation between the lagoon populations and TLM (φST = 0.2320, SD 0.1243, range 0.0981-0.3436) was comparable with differentiation among lagoon populations (mean φST 0.2962, SD 0.1319, range 0.1761-0.4374), mirroring their similar morphologies (23) and consistent with shared ecosystem characteristics (Table 1). Thus, TLM is apparently well connected with the lagoon (or only very recently disconnected) in contrast to all other lake populations, which are isolated from TLM (mean φST 0.8762, SD 0.0458, range 0.8097-0.9214), from the lagoon (mean φST 0.7439, SD 0.1279, range 0.3577-0.8693), and from each other (mean φST 0.9610, SD 0.0287, range 0.9247-1.000; Table 2). No marine lake shares a single COI haplotype with any other lake, consistent with independent, chance colonization of lakes by haplotypes from the lagoon. As a rule of thumb, if 14 haplotypes occurred with a similar frequency in the Palau lagoon during the Holocene (Fig. 1B), there is an ≈1-in-13 chance two lakes would contain the same haplotype; the probability that common haplotypes (e.g., those sampled by OTM and TLM) are shared is higher, but this effect is offset by the fact that haplotype compliments and frequencies have changed with time. Consequently, genetic dissimilarity was not significantly related to the three metrics used to distinguish habitat type: circumscription by land (i.e., the definition of a lake), physical structure of the water column, or the presence vs. absence of a coral reef community [analysis of molecular variance (AMOVA) P ≥ 0.38, Table 3). Genetic similarity was also unrelated to the geographic proximity of populations to each other (Mantel's r = 0.061, P = 0.283; AMOVA P = 0.31, Table 3), refuting a hypothesis of isolation by distance. However, genetic distance of marine lake populations from lagoon populations was highly correlated with the degree of geographic isolation as measured by modern [or estimated early Holocene (Table 1)] shortest overland distance from lagoon to lake (R2 = 0.858, F1,4 = 24.072, P = 0.008; Fig. 2A), indicating that colonization routes into lakes constitute more formidable dispersal filters than pathways between lagoon locations and that the intensity of the filter scales with distance. Genetic distance was not correlated with depth, i.e., time since isolation (linear regression: F1,4 = 2.061, P ≥ 0.224, including or excluding TLM), consistent with the low (but nonzero) probability of observing a mutation in COI during the ≈10,000-year time scale considered here, assuming the benchmark 1% divergence per 106 years (16). Thus, the patterns of geographic variation in COI reported here most parsimoniously result from the random redistribution (i.e., dispersal) of genetic diversity that existed in the lagoon during the formation of each lake. Not all lake haplotypes were observed in the lagoon, indicating that some are rare in the lagoon, or they are no longer in Palau, having been advected away or extirpated. Genetic diversity was positively correlated with habitat area (Spearman's rank correlation r = 0.975, P = 0.005), although neither a power-curve nor linear relationship fitted well with the quantitative data (P ≥ 0.094, Fig. 2B). Thus, patterns of genetic variation among these peripatric populations of Mastigias in Palau are generally consistent with predictions for taxa evolving in isolation on peripheral islands (Fig. 2; ref. 26).
Table 2. Estimates of population isolation (φST) in Palau Mastigias.
| Lagoon and cove populations
|
Lake populations
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Body of water type | Site | Lagoon | NCK | RCA | TLM | OLO | GLK | OTM | CLM |
| Lagoon and coves | NCK | 0.176 | |||||||
| RCA | 0.275 | 0.437 | |||||||
| Lakes | TLM | 0.098ns | 0.344 | 0.255 | |||||
| OLO | 0.822 | 0.869 | 0.735 | 0.921 | |||||
| GLK | 0.746 | 0.822 | 0.620 | 0.866 | 1.000 | ||||
| OTM | 0.789 | 0.865 | 0.358 | 0.918 | 1.000 | 1.000 | |||
| CLM | 0.816 | 0.846 | 0.692 | 0.866 | 0.958 | 0.948 | 0.930 | ||
| NLK | 0.717 | 0.746 | 0.717 | 0.810 | 0.954 | 0.943 | 0.952 | 0.925 | |
All values except one (ns) were statistically different from zero [based on 5,040 permutations in ARLEQUIN 2.0 (25)] after Bonferroni adjustment for 36 tests. NCK, Ngermeuangel Cove; RCA, Risong Cove; TLM, Tketau Lake; OLO, Uet era Ongael; GLK, Goby Lake; OTM, Ongeim'l Tketau; CLM, Clearwater Lake; NLK, Uet era Ngermeuangel.
Table 3. Mitochondrial genetic variation (COI) in Mastigias is not related to habitat type or geographic proximity of populations.
| % variation
|
|||
|---|---|---|---|
| AMOVA “group” model* | Groups† | Populations within groups | Individuals within populations |
| Lake vs. lagoon | −3.61‡ | 84.06§ | 19.55§ |
| Meromictic vs. stratified vs. holomictic | −16.29¶ | 95.91§ | 20.38§ |
| Coral vs. no coral | 1.34∥ | 79.56§ | 19.11§ |
| By island of occurrence | 7.58** | 73.39§ | 19.02§ |
Populations were grouped according to habitat type or geographic location (Table 1) to test four alternative hypotheses concerning the geographic distribution of genetic variation by using Analysis of Molecular Variance (AMOVA) in arlequin 2.0 (25).
Negative values indicate some medusae are genetically less similar to medusae within their own group than to medusae in the other group(s).
P = 0.62.
P < 0.00001; significance test based on 1,023 permutations of the datamatrix.
P = 0.38.
P = 0.87.
P = 0.31.
Fig. 2.
Relationships between geographic isolation, habitat area, and genetics of Mastigias populations. (A) Genetic distances between marine lake and lagoon populations of Mastigias medusae are positively correlated with the shortest geographic distances from the lakes to the lagoon. Regression of mean pairwise sequence difference corrected for intrapopulation variation (y) against log10-transformed shortest geographic distance (x) of lake populations from the lagoon gives y = 2.332 log10[x] -9.436 (SE on m ± 0.4752, c ± 2.380), R2 = 0.858, F1,4 = 24.072, P = 0.008. Linear regression by using untransformed geographic distances produced a weaker but still significant fit, y = 0.015x -0.177 (SE on m ± 0.004, c ± 0.642), R2 = 0.797, F1,4 = 15.741, P = 0.017. (B) Genetic diversity (#/n) of Mastigias populations is positively correlated with habitat area (Spearman's Rank correlation r = 0.975, P = 0.005, excluding OTM, which was decimated by the 1998 La Niña; ref. 28). Including OTM, r = 0.812, P = 0.050. Diversity in the lagoon (mean ± SE, n = 3) is shown for comparison but was not used to calculate correlation coefficients. Linear regression of genetic diversity on habitat area (m2) for the five lake populations excluding OTM gives y = (2.944 × 10-7)x + 0.119 (SE on m ± 0.000, c ± 0.037), R2 = 0.482, F1,3 = 2.791, P = 0.193. Linear regression of log10[diversity] on log10[area] for the five lake populations excluding OTM gives y = 0.352x -2.513 (SE on m ± 0.145, c ± 0.679), R2 = 0.662, F1,3 = 5.880, P = 0.094. (Insets) Expectations given genetic and phylogenetic consequences of island biogeography. (B Inset) Plotted on continuous linear axes (26).
Morphological data allow these hypotheses to be tested from another perspective, assuming morphology provides a proxy for underlying nuclear genetic variation and molecular evolution (29-30). In contrast to molecular diversity, morphological diversity was unrelated to habitat area (linear regression: R2 = 0.0454, F1,3 = 0.1426, P = 0.731; linear regression on log-transformed data: R2 = 0.118, F1,3 = 0.403, P = 0.571) and morphological divergence was not correlated with shortest overland distance from the lagoon (R2 = 0.0274, F1,3 = 0.0845, P = 0.7902). Morphological divergence was influenced by time since isolation, although an hypothesized linear relationship between morphological divergence and lake depth, a proxy for time, was marginally nonsignificant (y = 0.043x + 1.119 [SE on m ± 0.015, c ± 0.389], R2 = 0.739, F1,3 = 8.473, P = 0.062), suggesting time since isolation is not the only factor that has influenced the degree of morphological evolution. Indeed, the evidently rapid, obviously parallel, evolution of morphology is strong evidence of directional selection and adaptive radiation, in this case characterized by vestigialization (Fig. 3), attributable to the marine lake environment.
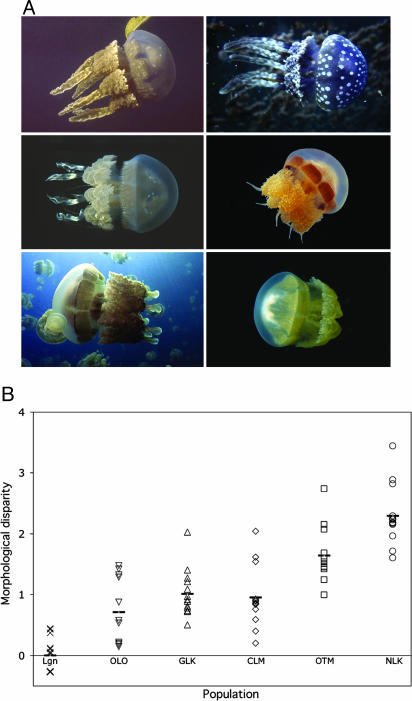
Fig. 3.
Rapid parallel morphological evolution of Mastigias medusae in marine lakes during the Holocene. (A) Typical morphologies of the ancestral lagoonal form in Ngermeuangel Cove (NCK) (Top Left) and derived marine lake morphotypes in Uet era Ongael (OLO) (Top Right), in Goby Lake (GLK) (Middle Left), Clearwater Lake (CLM) (Middle Right), Ongeim'l Tketau (OTM) (Bottom Left), and Uet era Ngermeuangel (NLK) (Bottom Right). Medusae shown are ≈15 cm bell diameter except NCK (≈20 cm) and OLO and NLK (≈10 cm). (B) Morphological disparity of medusae from the average lagoonal morphotype (y = 0) has changed unidirectionally, generally reflecting vestigialization, in marine lake populations. Horizontal bars show mean for each population.
Behavior also shows evolutionary radiation in response to novel selection pressures in marine lakes. The ancestral diel horizontal migration still evident in lagoon populations and GLK maximizes exposure of photosymbionts to sunlight. In contrast, the migration by medusae in OTM does not maximize incident sunlight, but minimizes exposure to otherwise intense predation by endemic benthic medusivorous anemones, Entacmaea medusivora (22). The diffuse migration by Mastigias in NLK (22) possibly reflects reduced dependence on autotrophic nutrition and physiological divergence of lake from lagoon medusae (31).
These behavioral, genetic, morphologic, and physiological changes in Mastigias evince incipient speciation (32). Concomitantly, previously uncharacterized endemic species of tunicate (Styela complexa), sea cucumbers (Synaptula spinifera and Holothuria cavans), crab (Orcovita saltatrix), and fishes (Cristatogobius sp., Parioglossus sp., and Antherinomorus sp.) have been found in a marine lake in Berau, Indonesia (33). Although some may represent simply the first record of new species yet to be found elsewhere, Styela complexa is thought to have speciated in situ from the widely distributed S. canopus (34). Such patterns of endemism and evolution, at subspecies (32) and species (34) levels, in marine lake taxa are highly reminiscent of patterns of endemism and evolution in species isolated on terrestrial islands and in freshwater lakes (35). The rapidity of evolution rivals that seen in limnetic fishes such as the three-spine stickleback, whose marine ancestors colonized freshwater lakes formed after the last glacial maximum (36), and the east African rift lake cichlids (ref. 37, but see ref. 38).
Thus, even though, compared with their terrestrial counterparts, marine species generally are relatively few (4) and widespread (1, 2), small peripheral populations such as those in marine lakes have considerable evolutionary potential. This potential is not a unique property of marine lake populations; conditions favorable to founder speciation are common in marine systems (39) and archipelagic-level endemism is well documented in fishes (40). Relatively short-term (millennialscale; ref. 41) geographic isolation over very short distances (23 km; ref. 42) can be an important source of marine biodiversity, diversity that subsequently may be maintained by physical and biological (3, 14, 43) factors inhibiting gene flow. Thus, marine lakes may be valuable modern analogues, or natural laboratories (27), for studying the early stages of evolution in bathymetrically complex regions, such as the Indo-West Pacific and Caribbean centers and sources of marine biodiversity (11, 44). Although this analogy implies that it is the increased isolation of ocean basins from each other, rather than their reduction in size, during low stands that is most important, it does not mean that separation should be equated with “dumbbell” vicariance because shoaling seas could conceivably divide species ranges unevenly. It also does not mean that each ocean basin should be viewed as an homogeneous unit, because even widespread species with high dispersal ability may experience differential selection across their range (45), and coastal organisms in particular inhabit only a small and highly heterogenous fraction of any ocean basin. Marine lake taxa may also have modern counterparts in extralimital populations, introduced populations, disjunct species, species with very small ranges (e.g., island endemics, edge-effect species), and the biotas of inland seas.
Geographic isolation, however, may result in a wide range of rates of evolution and speciation (46). The phenotypic divergence of Mastigias subspecies in marine lakes during the Holocene contrasts starkly with the relative morphological conservatism of allopatric subspecies of Catostylus mosaicus inhabiting coastal estuaries and bays north or west of Cape Howe, Australia, despite genetic divergence ≈1.4 million years ago (16). Of 15 morphological features measured in both species, eight differed significantly between subspecies of Mastigias [lagoon (n = 14) cf. NLK (n = 10), t test P ≤ 0.014; ref. 23], but only four differed significantly between subspecies of C. mosaicus (P ≤ 0.01, 5 ≤ n1 ≤ 7, 13 ≤ n2 ≤ 16; ref. 47) despite a 100-fold difference in divergence times. In this context, the creeping pace of morphological evolution in Catostylus subspecies in similar coastal habitats, and the predominant homoplasious pattern of parallel morphological evolution in Mastigias subspecies that independently invaded novel marine lakes, provide two different examples of why traditional morphological approaches have often failed to distinguish marine taxa and why molecular genetic analyses of marine invertebrates, including gelatinous zooplankton, are now increasingly revealing very large numbers of cryptic species (48-50) and even cryptic higher taxa (51). Evolution in the seas is clearly complex, but selection for particular phenotypes may be very strong; “rampant homoplasy” (52) suggests there may be relatively few successful solutions to life in an environment as simple and transparent as the sea (53).
Summary of Major Conclusions
Marine populations are, relative to terrestrial and freshwater populations, typically well connected (1, 2), but cases of low connectivity are not uncommon (13, 14, 39, 42). Occasional distant dispersal, for example, provides opportunities for divergence in peripheral marine environments (39, 54-56). Glacially lowered sea levels predominated throughout the Pleistocene with multiple episodes ≈100+ m below present that greatly increased geographic isolation of marine taxa for ≈10,000-15,000 years (18, 57), the time scale of evolution in Mastigias reported here, contributing to pulses of intraspecific differentiation (12-14, 16) and speciation (15, 17). However, isolating mechanisms and the early stages of speciation in marine taxa remain generally unclear, so well documented examples of any mode of evolution are valuable. Populations isolated in marine lakes provide an extraordinary opportunity to study peripatric evolution in marine taxa, including soft-bodied organisms (e.g., algae, jellyfish, and tunicates) that lack a good fossil record, and taxa that typically leave a good fossil record (e.g., gastropods). The existence of multiple independently derived populations in marine lakes and of ancestral populations in the adjacent lagoon in Palau provides an unprecedented opportunity to study genetic and phenotypic evolution (e.g., 29, 30) in representatives of most major marine phyla.
Acknowledgments
E. Basilius, L. Bell, P. Colin, W. Maech, M. Mesubed, L. Martin, and F. Toribiong assisted in the field. E. Wong assisted in the lab. L. Sharron photographed the Ongael Mastigias. Permits were issued by the Division of Marine Resources and the Koror State Government. G. Bernardi, B. Bowen, B. Grant, R. Grosberg, L. Martin, R. Paine, I. Suthers, and several anonymous reviewers gave thoughtful critiques of various versions of the manuscript. Funding was provided by the University of California, Los Angeles; International Women's Fishing Association; American Museum of Natural History (Lerner-Gray Award); British Schools and Universities Foundation; and the University of New South Wales.
Author contributions: M.N.D. and W.M.H. designed research; M.N.D. performed research; and M.N.D. and W.M.H. wrote the paper.
Abbreviations: COI, cytochrome c oxidase subunit I; GLK, Goby Lake; NLK, Uet era Ngermeuangel; OLO, Ongael Lake; OTM, Ongeim'l Tketau; TLM, Tketau Lake.
Data deposition: The DNA sequence data reported in this paper has been deposited in the GenBank database (accession nos. AY902925-AY903047).
References
- 1.Angel, M. V. (1994) Conserv. Biol. 7, 760-772. [Google Scholar]
- 2.Palumbi, S. R. (1994) Annu. Rev. Ecol. Syst. 25, 547-572. [Google Scholar]
- 3.Palumbi, S. R. & Warner, R. R. (2003) Science 299, 51-52. [DOI] [PubMed] [Google Scholar]
- 4.Pimm, S. L. & Raven, P. R. (2000) Nature 403, 843-845. [DOI] [PubMed] [Google Scholar]
- 5.Rapoport, E. H. (1994) Philos. Trans. R. Soc. London B 343, 71-78. [Google Scholar]
- 6.Jablonski, D. & Roy, K. (2003) Proc. R. Soc. London B 270, 401-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.May, R. M. (1994) Philos. Trans. R. Soc. London B 343, 105-111. [Google Scholar]
- 8.Longhurst, A. (1998) Ecological Geography of the Sea (Academic, San Diego).
- 9.Jones, G. P., Milicich, M. J., Emslie, M. J. & Lunow, C. (1999) Nature 402, 802-804. [Google Scholar]
- 10.Grosberg, R. K. & Cunningham, C. W. (2001) in Marine Community Ecology, eds. Bertness, M. D., Gaines, S. D. & Hay, M. E. (Sinauer, Sunderland, MA), pp. 61-84.
- 11.Briggs, J. C. (2004) in Frontiers of Biogeography: New Directions in the Geography of Nature, eds. Lomolino, M. V. & Heaney, L. R. (Sinauer, Sunderland, MA), pp. 239-254.
- 12.Williams, S. T., Jara, J., Gomez, E. & Knowlton, N. (2002) Integr. Comp. Biol. 42, 941-952. [DOI] [PubMed] [Google Scholar]
- 13.Barber, P. H., Palumbi, S. R., Erdmann, M. V. & Moosa, M. K. (2000) Nature 406, 692-693. [DOI] [PubMed] [Google Scholar]
- 14.Barber, P. H., Palumbi, S. R., Erdmann, M. V. & Moosa, M. K. (2002) Mol. Ecol. 11, 659-674. [DOI] [PubMed] [Google Scholar]
- 15.O'Hara, T. D. & Poore, G. C. B. (2000) J. Biogeogr. 27, 1321-1335. [Google Scholar]
- 16.Dawson, M. N. (2005) J. Biogeogr. 32, 515-533. [Google Scholar]
- 17.Jackson, J. B. C. (1995) in Extinction Rates, eds. Lawton, J. H. & May, R. M. (Oxford Univ. Press, Oxford), pp. 45-54.
- 18.Haq, B. U., Hardenbol, J. & Vail, P. R. (1987) Science 235, 1156-1167. [DOI] [PubMed] [Google Scholar]
- 19.Ocean Drilling Program (2000) Leg 189 Preliminary Report (Shipboard Scientific Party, Ocean Drilling Program, Texas A & M University, College Station, TX).
- 20.Hamner, W. M. & Hamner, P. P. (1998) Phys. Geogr. 19, 175-220. [Google Scholar]
- 21.Hamner, W. M. & Hauri, I. R. (1981) Limnol. Oceanogr. 26, 414-423. [Google Scholar]
- 22.Dawson, M. N. & Hamner, W. M. (2003) Mar. Biol. 143, 1161-1174. [Google Scholar]
- 23.Dawson, M. N. (2005) Hydrobiologia 537, 185-206. [Google Scholar]
- 24.Swofford, D. L. (2002) PAUP* 4.0b10 (Sinauer Associates, Sunderland, MA).
- 25.Schneider, S., Roessli, D. & Excoffier, L. (2000) arlequin 2.0 (Genetics and Biometry Laboratory, University of Geneva, Geneva).
- 26.Johnson, K. P., Adler, F. R. & Cherry, J. L. (2000) Evolution (Lawrence, Kans.) 54, 387-396. [DOI] [PubMed] [Google Scholar]
- 27.Hamner, W. M. (1982) Natl. Geogr. 161, 264-282. [Google Scholar]
- 28.Dawson, M. N., Martin, L. E. & Penland, L. K. (2001) Hydrobiologia 451, 131-144. [Google Scholar]
- 29.Fondon, J. W., III, & Garner, H. R. (2004) Proc. Natl. Acad. Sci. USA 101, 18058-18063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shapiro, M. D., Marks, M. E., Peichel, C. L., Blackman, B. K., Nereng, K. S. Jónsson, B., Schluter, D. & Kingsley, D. M. (2004) Nature 428, 717-723. [DOI] [PubMed] [Google Scholar]
- 31.McCloskey, L. R., Muscatine, L. & Wilkerson, F. P. (1994) Mar. Biol. 119, 13-22. [Google Scholar]
- 32.Dawson, M. N, (2005) J. Mar. Biol. Assoc. U.K. 85, 679-694. [Google Scholar]
- 33.Tomascik, T., Mah, A. J., Nontji, A. & Moosa, M. K. (1997) The Ecology of the Indonesian Seas (Periplus Editions, Hong Kong).
- 34.Kott, P. (1995) Raffles Bull. Zool. 43, 469-474. [Google Scholar]
- 35.Grant, P. R. (1998) Evolution on Islands (Oxford Univ. Press, Oxford).
- 36.Schluter, D. (1996) Philos. Trans. R. Soc. Lond., B, Biol. Sci. 351, 807-814. [Google Scholar]
- 37.Johnson, T. C., Scholz, C. A., Talbot, M. R., Kelts, K., Ricketts, R. D., Ngobi, G., Beuning, K., Ssemmanda, I. & McGill, J. W. (1996) Science 273, 1091-1093. [DOI] [PubMed] [Google Scholar]
- 38.Fryer, G. (2001) Proc. R. Soc. London Ser. B 268, 1147-1152. [Google Scholar]
- 39.Paulay, G. & Meyer, C. (2002) Integr. Comp. Biol. 42, 922-934. [DOI] [PubMed] [Google Scholar]
- 40.Randall, J. E. (1998) Zool. Stud. 37, 227-268. [Google Scholar]
- 41.Jackson, J. B. C. & Cheetham, A. H. (1999) Trends Ecol. Evol. 14, 72-77. [DOI] [PubMed] [Google Scholar]
- 42.Taylor, M. S. & Hellberg, M. E. (2003) Science 299, 107-109. [DOI] [PubMed] [Google Scholar]
- 43.Mora, C. & Sale, P. F. (2002) Trends Ecol. Evol. 17, 422-428. [Google Scholar]
- 44.Briggs, J. C. (2000) J. Biogeogr. 27, 1183-1188. [Google Scholar]
- 45.Pogson, G. H., Taggart, C. T., Mesa, K. A. & Boutilier, R. G. (2001) Evolution (Lawrence, Kans.) 55, 131-146. [DOI] [PubMed] [Google Scholar]
- 46.Near, T. J. & Benard, M. F. (2004) Evolution (Lawrence, Kans.) 58, 2798-2808. [DOI] [PubMed] [Google Scholar]
- 47.Dawson, M. N. (2005) J. Mar. Biol. Assoc. U.K., 85, 723-732. [Google Scholar]
- 48.Knowlton, N. (1993) Annu. Rev. Ecol. Syst. 24, 189-216. [Google Scholar]
- 49.Knowlton, N. (2000) Hydrobiologia 420, 73-90. [Google Scholar]
- 50.Dawson, M. N. (2004) Hydrobiologia 530-531, 249-260. [Google Scholar]
- 51.Fukami, H., Budd, A. F., Paulay, G., Solé-Cava, A., Chen, C. A., Iwao, K. & Knowlton, N. (2004) Nature 427, 832-835. [DOI] [PubMed] [Google Scholar]
- 52.Gosliner, T. M. & Ghiselin, M. T. (1984) Syst. Zool. 33, 255-274. [Google Scholar]
- 53.Hamner, W. M. (1995) Mar. Fresh. Behav. Physiol. 26, 71-89. [Google Scholar]
- 54.Palumbi, S. R. (1997) Coral Reefs 16, S47-S52. [Google Scholar]
- 55.Waters, J. M. & Roy, M. S. (2004) Syst. Biol. 53, 18-24. [DOI] [PubMed] [Google Scholar]
- 56.Meyer, C. P., Geller, J. B. & Paulay, G. (2005) Evolution (Lawrence, Kans.) 59, 113-125. [PubMed] [Google Scholar]
- 57.Shackleton N. J. (2000) Science 289, 1897-1902. [DOI] [PubMed] [Google Scholar]