Abstract
Entry of herpes simplex virus into the cell requires the interaction of gD with one of its receptors, herpesvirus entry mediator or nectin 1, and the intervention of gB, gH, or gL, required to execute fusion of the virion envelope with cell membranes. The gD ectodomain is organized in two structurally and functionally differentiated regions. The N terminus (residues 1-260) carries the receptor binding sites, and the C terminus (residues 260-310) functions as the pro-fusion domain (PFD), which is required for viral infectivity and fusion but not for receptor binding. The objective of our studies is to elucidate how gD links receptor recognition to the triggering of fusion. Here, we show that PFD is made of subdomains 1 and 2 (amino acids 260-285 and 285-310). Each one partially contributed to herpes simplex virus infectivity. By means of glutathione S-transferase (GST) fusion proteins, we show that PFD bound soluble forms of gD, truncated at residue 260 (gD260t) or downstream. Both PFD subdomains bound gD260t, highlighting multiple contact sites between the N and C termini of gD. When gD260t was in complex with either receptor, it failed to bind GST-PFD. In turn, the receptors did not bind GST-PFD, irrespective of whether they were in complex with gD. Thus, gD260t interacted with the C terminus only if unbound to the receptor. We propose that (i) before receptor binding, gD adopts a “closed” conformation in which the N and C termini interact; and (ii) on encounter with a receptor, gD modifies its conformation and the N and C termini are released from reciprocal interactions (“opened” conformation) and enabled to trigger fusion.
Entry of herpesviruses into the cell requires the concerted activity of a minimum of three glycoproteins. In particular, herpes simplex virus 1 (HSV-1) entry requires gD as the receptor-binding glycoprotein, and the trio of gH, gL, and gB is cumulatively required for fusion execution (1-6).
The gD ectodomain is organized in two structurally and functionally differentiated regions. The N terminus (amino acids 1-260) carries the receptor-binding sites, and the C terminus (amino acids 260-310) functions as the pro-fusion domain (PFD). gD interacts with two alternative receptors belonging to unrelated protein families, the herpesvirus entry mediator (HVEM) and the nectins (3-5, 7, 8). The crystal structure of a truncated form of gD was resolved up to amino acid 259 and revealed an Ig-folded central portion with extensions (9). The N-terminal extension formed a hairpin in the crystal of gD in complex with HVEM and contained all of the contact residues to HVEM (10). Critical residues for the interaction with nectin 1 are comprised in the first 250 residues of gD (11-13). Because soluble forms of gD truncated at residues 250 or 260 (gD250t and gD260t) bind soluble forms of the receptors, the receptor-binding sites cumulatively lie in the N terminus (amino acids 1-260) (6, 14) (for a schematic drawing, see Fig. 1).
Fig. 1.
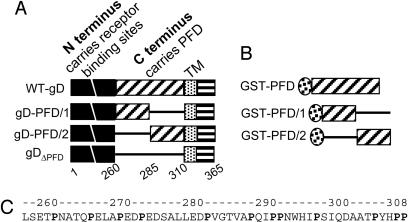
Schematic diagram of gD (A) and GST-PFD (B) constructs and PFD sequence (C). (A) Top line, linear map of WT-gD, with N terminus (amino acids 1-260) carrying receptor-binding sites (black) and C terminus (amino acids 260-310) carrying the PFD (diagonal lines), the transmembrane (TM) (dotted), and the C-tail (horizontal lines) regions. The regions are not shown to scale. The numbers indicate the region coordinates. gDΔPFD carries gD amino acids 1-260 and 50 aa from CD8, derived from the sequence upstream of TM (defined as CD8 amino acids 261-310), which replace gD amino acids 261-310. gD-PFD/1 carries gD amino acids 1-285 and CD8 amino acids 285-310. gD-PFD/2 carries gD amino acids 1-260 and 285-310 and CD8 amino acids 261-285. In all constructs, the TM and C-tail are from gD. (B) GST-PFD carries GST fused to gD amino acids 260-310. GST-PFD/1 carries GST fused to a segment composed of gD amino acids 260-285 and CD8 amino acids 285-310. GST-PFD/2 carries GST fused to a segment composed of CD8 amino acids 260-285 and gD amino acids 285-310. (C) PFD sequence and its coordinates.
The molecular events that follow receptor recognition by gD and precede the execution of fusion, referred to as triggering of fusion, are poorly understood. Two findings highlighted that the functions of gD in HSV-1 entry are twofold, i.e., to interact with its receptors, and to signal receptor recognition to the downstream glycoproteins and thus trigger fusion. The first observation is that soluble forms of gD suffice to complement the infectivity of a noninfectious gD-null HSV mutant. This finding provides compelling evidence that the gD ectodomain (the sole portion of gD required for viral infectivity) must encode two functions, i.e., the receptor binding and a signaling activity of receptor binding to downstream glycoproteins. The second is the evidence that, in addition to the receptor-binding sites, the gD ectodomain encodes the PFD, which is required for viral infectivity and fusion but not for receptor binding. PFD is located in the C terminus (amino acids 260-310) (Fig. 1 and ref. 15). Its replacement by heterologous sequences (e.g., CD8 and pseudorabies virus gD) or proline substitution reduced infectivity and fusion (15, 16). Its amino acids 260-285 subdomain was recognized as critical to induce fusion in cells cotransfected with gB, gH, or gL (16).
A key role in fusion execution is played by gH, a conserved glycoprotein in the Herpesviridae family (17-19). gH has structural features that are typical of viral fusion glycoproteins, i.e., its ectodomain carries a hydrophobic α-helix (residues 377-397) with attributes of an internal fusion peptide, and downstream of it, two heptad repeats with a propensity to form coiled coils (refs. 20 and 21 and T. Gianni and G.C.-F., unpublished work). Specifically, the hydrophobic α-helix is an essential domain in HSV gH. Its entire or partial deletion or the substitution of a few residues abolishes viral infectivity and fusion. The α-helix can be replaced with the fusion peptide from HIV gp41 or from the glycoprotein of vesicular stomatitis virus with rescue of the gH activities (20, 21). Mutational analysis of the predicted heptad repeats also shows that these sequences are critical to gH function. Synthetic peptides with the sequence of the heptad repeats inhibit virus infectivity (ref. 21 and T. Gianni and G.C.-F., unpublished work).
The objective of this study was to shed light on how gD links receptor recognition to the triggering of fusion. We defined subdomains 1 and 2 as encompassing amino acids 260-285 (PFD/1) and 285-310 (PFD/2), respectively. Each subdomain contributed to virion infectivity. To investigate the binding properties of PFD, we constructed fusion proteins made of glutathione S-transferase (GST) and the PFD (GST-PFD) or its subdomains (GST-PFD/1 and GST-PFD/2). GST-PFD physically interacted with gD260t or longer forms of gD. Both GST-PFD/1 and GST-PFD/2 bound gD260t, highlighting multiple contacts. Unexpectedly, when gD260t was in complex with soluble forms of either receptor, it failed to bind GST-PFD. The binding properties of PFD are compatible with the possibility that, before receptor binding, gD folds back on itself and the N and C termini physically interact such that gD adopts a “closed” conformation. On encounter with a receptor, the closed conformation is lost, and the N and C termini are released from reciprocal interactions and are enabled to trigger fusion.
Materials and Methods
Cells and Viruses. Cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% FCS. The receptor-negative J, J-nectin 1, and J-HVEM cells were described in refs. 5 and 15. The gD-F-gDβ virus (22) was grown in gD-expressing R6 cells (gD-/+ stock). HSV-1 facial (F) was described in ref. 23.
Constructs. First, we derived gD-PFD/2; the gD1-260CD8 construct was amplified up to amino acid 285 with primers 5′-CCCTCTAGACTCGAGCGTTCCGGTATGGGGG-3′ and 5′-CAAGTTTGGTGGGATTTGCGGCGCCACCTGCGACGCGATGGTGGGCGCCGGTGT-3′. Amplimer 2, derived by amplification of the gD gene with primers 5′-CTCTTGGAGGACCCCGTGGGGACGCCCCTGTCCCTGCGCCCAGAGGCG-3′ and 5′-GCGGTTTAAACTGAATTCTCTAGTAAAACAGGGG-3′, contained the gD amino acid 285-stop codon. Each amplimer contained overlapping sequences (contained in the primers) with the other amplimer. The amplimers were mixed and further amplified with the external primers. The product was cloned into pcDNA3.1(-) at the XhoI and EcoRI sites. To generate gDΔPFD and gD-PFD/1, we inserted an Asp718 site at amino acid 310 of gD1-260CD8. Digestion with Asp718 and EcoRI removed the transmembrane (TM) and cytoplasmic tail (C-tail) of CD8, which were replaced with the TM and C-tail gD sequences, and amplified with primers 5′-CATCCCCCGGGTACCCCGAACAACATGG-3′ and 5′-GCGGTTTAAACTGAATTCTCTAGTAAAACAGGGG-3′, thus generating gDΔPFD. To generate gD-PFD/1, the latter construct was PCR-amplified from amino acid 285 to the stop codon with primers 5′-GGTCTCTTTTGTCTCGAGCGTTCCGGTATGGGGG-3′ and 5′-CGCCTCTGGGCGCAGGGACAGGGGCGTCCCCACGGGGTCCTCCAAGAG-3′. A second amplification product, containing gD amino acids 1-285 plus the natural gD signal sequence (amino acids -25 to -1), was derived with primers 5′-CTCTTGGAGGACCCCGTGGGGACGCCCCTGTCCCTGCGCCCAGAGGCG-3′ and 5′-GCGGTTTAAACTGAATTCTCTAGTAAAACAGGGG-3′. The two amplimers contained overlapping sequences and were mixed and amplified with external primers. The final product was cloned in pcDNA3.1(-) at the XhoI and EcoRI sites.
To generate the constructs expressing GST fused to PFD or subdomains 1 and 2, first, we generated GST-PFD by amplifying amino acids 260-310 of WT-gD with primers 5′-GGAGCTGTCCGAGGAATTCAACGCCAC-3′ and 5′-CATGTTGTTCGGGGTCTCGAGGGGATGGTAAGGCG-3′ and cloning at EcoRI and XhoI sites of pGEX-4T-1. For the GST-PFD/1, the 260- to 310-aa region of gD-PFD/1, consisting of gD amino acids 260-285 plus CD8 amino acids 286-310, was amplified with the primers 5′-GGAGCTGTCCGAGGAATTCAACGCCAC-3′ and 5′-CCCATGT TGT TCGGCTCGAGGA AGTCCAGCCC-3′ and cloned into EcoRI and XhoI sites of pGEX-4T-1. For GST-PFD/2, a similar strategy was followed, except that the amplification product was generated by using gD-PFD/2 as template and the primers 5′-GCTGTCCGAGACCGAATTCCTGCCAGCGAAGCCC-3′ and 5′-CATGTTGTTCGGGGTCTCGAGGGGATGGTAAGGCG-3′.
Production and Purification of GST Fusion Proteins and Binding Experiments. GST fusion proteins were expressed as described in ref. 24 and were purified with reduced glutathione (GSH)-Sepharose beads (Amersham Pharmacia). Aliquots containing 40 ng of the various forms of gD in buffer A (PBS containing protease inhibitors, 0.1 mM Nα-(p-tosyl)-l-lysine chloromethyl ketone, 0.1 mM N-tosyl-l-phenylalanyl chloromethyl ketone, 1 mM CaCl2, and 1 mM MgCl2) were mixed with immobilized GST or GST-PFD, GST-PFD/1, or GST-PFD/2. The beads were washed four times with buffer A containing 1% Nonidet P-40 (Sigma) and 1% sodium deoxycholate and were eluted with 10 mM GSH. The eluted proteins were separated by SDS/PAGE, blotted, and reacted with mAb H170 to gD (Rumbaugh-Goodwin Institute for Cancer Research, Plantation, FL). When appropriate, GST-PFD proteins were detected by reactivity to mAb DL6 directed to gD amino acids 272-279 (25). In the assays where gD260t was premixed with soluble receptors (2 h at 4°C), the mixture was absorbed to GSH-beads. Replicate aliquots of GSH-eluted proteins were separated by SDS/PAGE and blotted, and the blots were developed with mAb H170 to gD, anti-His antibody to HVEMt, CK6 mAb to nectin 1-Fc (26), or anti-human IgG to nectin 3-Fc. Soluble forms of receptors, HVEMt, nectin 1-Fc, and nectin 3-Fc were described in refs. 27-29. To measure the binding of uninfected and HSV-1(F)-infected Vero cell proteins to GST-PFD, cells were labeled with [35S]methionine and [35S]cysteine from 5 to 19 h after infection. Lysates made in PBS containing 1% Triton X-100 were reacted with GST-PFD bound to GSH-beads and were eluted with GSH. SDS/PAGE-separated proteins were analyzed with a phosphoimager (Bio-Rad).
Cell ELISA (CELISA). CELISA was performed as described in refs. 30 and 31. COS cells in 48 wells were transfected with plasmids encoding WT-gD or mutant gD (125, 250, or 375 ng per well, corresponding to 1×, 2×, and 3× the regular amount); reacted with mAb H170, HVEMt, or nectin 1-Fc; and fixed with formaldehyde followed by anti-mouse peroxidase, anti-His6-peroxidase, or anti-human peroxidase.
Cell-Cell Fusion Assay. The luciferase-based cell-cell fusion assay was performed in triplicates as described in refs. 20 and 31. The total amount of transfected DNA was made equal by the addition Erb-2 plasmid DNA (32). The target cells were COS, J-HVEM, or J-nectin 1. The β-galactosidase-based cell-cell fusion assay was described in ref. 33.
Mutagenesis. Mutagenesis was performed in pEA102 (a plasmid carrying gD in pcDNA3.1-) (15, 34) with the following primers: gD259, 5′-GCTGCCCCCGGAGCTCTCCGCGACCCTCAACGCCACGCAG; gD266, 5′-CCCAACGCCACGCAGCTAGCACTCGCCCCGGAAGC; gD270, 5′-GCAGCCAGAACTCGCGCTAGCAGCCCCCGAGGATTC; and gD273, 5′-CTCGCCCCGGA AGCGCTAGCGGCT TCGGCCCTCTTGGAG.
Results
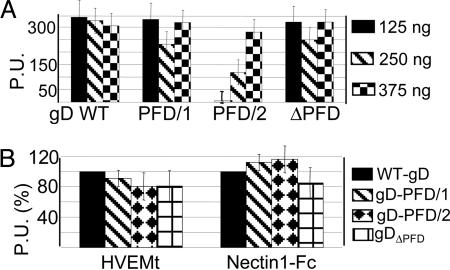
Functional Subdomains in gD-PFD. To identify functional subdomains of PFD, we generated two gD chimeric proteins that carried, downstream of residue 259, either PFD/1 (amino acids 260-285) or PFD/2 (amino acids 285-310) (Fig. 1 A). In the constructs, gD-PFD/1 and gD-PFD/2, the TM and C-tail regions were from gD, and the gD missing sequences were replaced by the corresponding sequence of CD8 (Fig. 1 A). A chimera in which the entire PFD was replaced by the corresponding sequences of CD8 (gDΔPFD) differed from gD(1-260)CD8 (15) in that the TM and C-tail regions were from gD but not from CD8 (Fig. 1 A). The chimeric forms of gD, cloned in pcDNA3.1(-), were analyzed for their ability to reach the plasma membrane, and in three functional assays, i.e., the binding to soluble forms of HVEM (HVEMt) and nectin 1 (nectin 1-Fc) (5, 27), the cell-cell fusion, and the infectivity complementation. All chimeric gDs reached the plasma membrane as measured by CELISA (Fig. 2A) and by immunofluorescence of paraformaldehyde-fixed cells (data not shown). In the case of gD-PFD/2, 3-fold higher amounts of plasmid (3×) were transfected to achieve a WT level of plasma membrane expression; gD-PFD/2 was transfected at 3× higher concentrations relative to WT-gD in all subsequent experiments, except when otherwise stated. The binding to HVEMt and nectin 1-Fc, measured by CELISA, showed an extent of binding similar to that of WT-gD (Fig. 2B).
Fig. 2.
Cell surface expression and binding to HVEMt and nectin 1-Fc of gD chimeric proteins. (A) Cell surface expression quantified by CELISA. COS cells, in 48 wells, were transfected with plasmids encoding WT or chimeric forms of gD at the indicated amounts, corresponding to 1×, 2×, or 3× the regular amount. Cells were fixed with 4% paraformaldehyde at 24 h after transfection and were reacted with mAb H170, anti-mouse IgG-peroxidase, and o-phenylenediamine. Results are expressed as peroxidase units (P.U.). Vertical bars denote SD. Columns represent the average of triplicates. Three independent experiments were run. (B) Binding of COS cells expressing the chimeric or WT-gD to HVEMt and nectin 1-Fc. Details were as in A, except that cells were reacted with the soluble receptors, anti-His6-peroxidase or anti-human peroxidase, to detect HVEMt and nectin 1-Fc, respectively, and o-phenylenediamine.
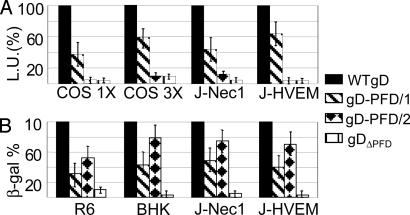
For the cell-cell fusion assay, baby hamster kidney cells were cotransfected with the gD-PFD chimera or WT-gD plus gB, gH, gL, and β-galactosidase plasmids (18, 33) and were stained with 5-bromo-4-chloro-3-indolyl β-galactopyranoside (data not shown). For the luciferase-based cell-cell fusion assay (31), each chimeric gD or WT-gD was cotransfected with gB, gH, gL, and the T7 polymerase in COS cells. The target COS, J-nectin 1, or J-HVEM cells were transfected with a T7-promoter-driven luciferase gene; COS cells were transfected at either 1× or 3× the amounts of plasmids (Fig. 3A). The two assays concordantly showed that gD-PFD/1 was partially active in the fusion assay; the activity increased when the plasmid amounts were increased 3×. By contrast, gD-PFD/2 was inactive at either concentration. gDΔPFD was inactive at either concentration, in accordance with the analogous gD(1-260)CD8 (15). The results suggest a certain degree of differentiation in fusion activity between the two PFD subdomains, and only subdomain 1 was in part sufficient for this function, in agreement with ref. 16.
Fig. 3.
Cell-cell fusion and infectivity complementation of chimeric gD proteins. (A) Luciferase-based cell-cell fusion assay. Effector COS cells were transfected with plasmids for gH, gL, gB, chimeric, or WT-gD and T7 polymerase at 125 or 375 ng of each plasmid, corresponding to 1× and 3× amounts, respectively. Target cells [COS, J-nectin 1 (J-Nec1), and J-HVEM] were transfected with T7-luciferase. The negative control lacked gD (data not shown). The luciferase activity was expressed as luciferase units (L.U.). (B) Infectivity complementation. COS cells were transfected with chimeric or WT-gD and infected 4 h later with a gD-/+ stock of FgDβ (3 plaque-forming units per cell). The negative control consisted of cells not transfected with gD. Progeny virus was titrated at 24 h in gD-expressing cells (R6) or was quantified as β-galactosidase in baby hamster kidney, J-nec1, or J-HVEM cells. Details are as in the legend to Fig. 2.
Next, we determined whether the same chimeric forms of gD were capable of complementing infectivity. Each chimera, or WT-gD, was transfected into COS cells, and 4 h later, the cells were infected with the gD-deletion virus FgDβ (22). When the virus is grown in noncomplementing cells, noninfectious gD-/- progeny are produced. When the virus is grown in cells expressing gD, gD complements the virus (gD-/+ stock) and confers infectivity. If gD is partially defective, the complemented virions exhibit a reduced infectivity. As shown in Fig. 3B, gD-PFD/1 was partially active in the infectivity complementation, consistent with the partial cell-fusion activity. The complementation activity did not change whether the gD-PFD/1-encoding plasmid was transfected at 1× or 3× amounts (data not shown). Surprisingly, gD-PFD/2, which exhibited no cell-fusion activity, partially complemented infectivity. Similar results were obtained irrespective of the cell line and of the receptor expressed in the cells where the complemented virions were titrated. The results indicate that PFD cannot be narrowed down, because subdomain 1 (amino acids 260-285) exhibits partial cell fusion and infectivity activities. The subdomain 2 (amino acids 286-310) is not sufficient for cell fusion but is sufficient for partial complementation of infectivity, in agreement with the finding that Pro residues 288, 291, 292, and 305 were critical residues for infectivity (15). The discrepancy between the results of the cell-cell fusion and infectivity complementation assays indicates that the two assays mirror each other but are not necessarily identical.
Effects of Pro and Glu Substitutions in Subdomain 1. The PFD sequence (Fig. 1C) reveals a high content of Pro residues, most of which are adjacent to Glu residues in PFD/1. Some Pro in PFD/2 (amino acids 288, 291, 292, and 305) are critical for infectivity (15). Here, the Pro residues of PFD/1 were replaced by Leu and the Glu by Ala (both are nonconservative substitutions). The mutants 1-4 carried the following substitutions, E259A-P261L, PE266-267LA, PE270-271LA, and PED273-274-275LAA, respectively. Their cell-surface expression showed no major defect. The cell-cell fusion activity of mutant 1 was increased by 25%, whereas that of mutants 2-4 was reduced by 20-25% (data not shown). In the infectivity complementation, none of the mutants exhibited a significant defect (data not shown). Thus, the Pro and Glu residues do not represent critical residues in PFD/1.
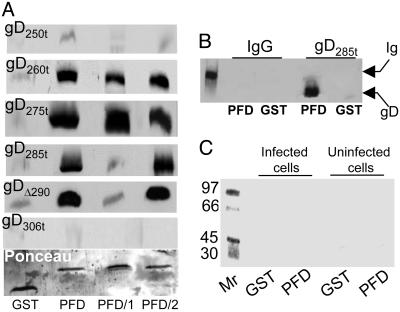
GST-PFD Fusion Proteins Bound Truncated Forms of gD. To ascertain whether the PFD can bind truncated forms of gD, we derived proteins carrying GST fused to the entire PFD, or subdomains 1 and 2, named GST-PFD, GST-PFD/1, and GST-PFD/2. These fusion proteins carried gD amino acids 260-310, 260-285, and 285-310, respectively, and were reacted with the following soluble forms of gD, gD234t, gD250t, gD260t, gD275t, gD285t, gDΔ290-299t, and gD306t, which differ in binding properties to the receptors. gD234t binds soluble HVEM but not nectin 1. All other soluble forms of gD bind both receptors at high affinity except gD306t, which exhibits a 100-fold lower affinity (14, 27). The proteins bound by the GST-PFD fusion proteins and eluted with GSH were analyzed by immunoblotting (Fig. 4A). In the eluted fractions, the amounts of GST and GST-PFD fusion proteins were ascertained to be similar by Ponceau staining. The binding to an unrelated glycosylated protein, murine IgG, served as a specificity control (Fig. 4B). The results were as follows:
Fig. 4.
Binding of GST-PFD to soluble forms of gD (A) and lack of binding to infected and uninfected cell proteins (C) and unrelated protein (B). (A) Immunoblots of soluble forms of gD bound by GST-PFD fusion proteins. Each form of gD (40 ng) was reacted with GST or the chimeric proteins GST-PFD (PFD), GST-PFD/1 (PFD/1), or GST-PFD/2 (PFD/2), previously immobilized on GSH-beads. The beads were pelleted and extensively washed. The GSH-eluted proteins were separated by SDS/PAGE, blotted, and reacted with mAb H170. Blots were stained with Ponceau (Sigma) to control that equivalent amounts of GST fusion proteins were loaded (one example shown). (B) GST or GST-PFD (PFD) immobilized on GSH-beads was incubated with murine IgG or gD285t as positive control. The GSH-eluted proteins were reacted with mAb H170 and anti-mouse Ab. The leftmost lane shows the migration position of the Ig heavy chain. (C) Autoradiographic image of a gel showing proteins from uninfected or HSV-1(F)-infected cells bound to GST or GST-PFD (PFD). Vero cells were labeled with [35S]methionine and [35S]cysteine, solubilized with 1% Triton X-100, and absorbed to GST or GST-PFD, previously immobilized on GSH-beads. The GSH-eluted proteins were analyzed by autoradiography. The leftmost lane shows the migration positions of Mr markers.
GST-PFD formed complexes with gD260t and gD275t. These complexes provide evidence for a physical interaction between the C terminus, which functions as PFD, and the N terminus. Because GST-PFD/1 and GST-PFD/2 each bound gD260t or gD275t, the results further indicate that there are at least two independent contact sites of PFD to the N terminus of gD, contributed by subdomains 1 and 2, respectively. We refer to them as the PFD/1- and PFD/2-binding site on gD N terminus.
gD250t and gD234t failed to bind GST-PFD, GST-PFD/1, or GST-PFD/2, implying that the 250- to 260-aa sequence is either part of the PFD-binding site on the gD N terminus or provides a conformation suitable to create the PFD-binding site.
The binding pattern of the GST-PFD fusion proteins to gD285t differed from that to gD260t or gD275t, in that GST-PFD and GST-PFD/2 but not GST-PFD/1 maintained the ability to form complexes. Of note, PFD/1 and gD285t share amino acids 260-285. The lack of GST-PFD/1 binding to gD285t suggests a competition between the endogenous 260- to 285-aa sequence in gD285t and PFD/1 and implies that the PFD/1-binding site on gD285t already is occupied by the endogenous PFD/1 sequence.
GST-PFD fusion proteins did not bind gD306t. Inasmuch as gD306t carries overlapping sequences with PFD, PFD/1, and PFD/2, the lack of binding suggests that the endogenous PFD sequences in gD306t occupy their binding sites and hinder the interaction with GST-PFD fusion proteins.
gDΔ290-299t is truncated at amino acid 305, and amino acids 290-299 are replaced by GKIFP. In terms of binding to GST-PFD fusion proteins, gDΔ290-299t behaved like gD285t. We interpret this result to indicate that, because of the deletion-insertion, the endogenous 285- to 305-aa sequence is modified such that it no longer binds the PFD/2-binding site on the N terminus.
Three series of experiments indicate that the bindings, when present, were specific: (a) the lack of binding of gD234t, gD250t, and gD306t; (b) the lack of binding of GST alone to any gD; and (c) the lack of binding of GST-PFD to murine IgG (Fig. 4B).
To ascertain whether GST-PFD can specifically bind any protein from HSV-1-infected or uninfected cells, cells were labeled with [35S]methionine and [35S]cysteine, and the lysates were reacted with GST-PFD or GST alone. Neither gD nor other proteins were specifically bound (Fig. 4C). The lack of binding of full-length gD to the GST-PFD fusion proteins mirrors the lack of binding of gD306t and suggests that the PFD/1 and PFD/2 binding sites on the N terminus already are occupied by the endogenous sequences, even in full-length gD.
Previously, we showed that some Pro residues in PFD/2 (amino acids 288, 291, 292, and 305) are critical for infectivity (15). The PQ288-289LA and PP291-292LA substitutions described in ref. 15 were introduced in GST-PFD. The mutants did not exhibit any defect in binding to gD260t (data not shown), suggesting that the mutations affected steps in entry and fusion, likely the triggering of fusion and not the binding of PFD to gD N terminus.
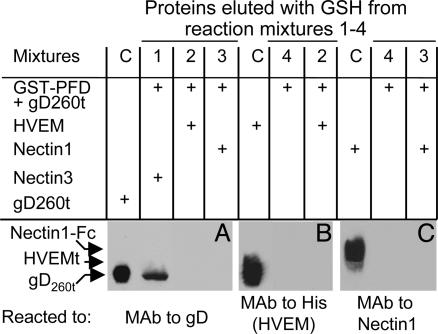
gD260t in Complex with Nectin 1-Fc or HVEMt Failed to Bind GST-PFD. To ascertain whether, when bound to its receptors, gD260t maintained the ability to physically interact with GST-PFD, replicate aliquots of gD260t were mixed with HVEMt or nectin 1-Fc. Binding to nectin 3-Fc served as negative control. The complexes were then absorbed to GSH-beads preloaded with GST-PFD. The GSH-eluted proteins were analyzed for the presence of gD (Fig. 5A), HVEMt (Fig. 5B), and nectin 1-Fc (Fig. C). The results were as follows. The fractions eluted from the mixture of gD260t plus HVEMt (reaction mixture 2) contained neither gD (Fig. 5A) nor HVEMt (Fig. 5B). Similarly, the fractions eluted from the mixture of gD260t plus nectin 1-Fc (reaction mixture 3) contained neither gD (Fig. 5A) nor nectin 1-Fc (Fig. 5C). When gD260t was premixed with an irrelevant receptor (nectin 3-Fc) (reaction mixture 1), the eluted fraction contained gD (Fig. 5A) but not nectin 3-Fc (not shown). The soluble nectin 1-Fc failed to bind GST-PFD, irrespective of whether it was premixed with gD260t in a molar ratio of 1:1 (data not shown) or in a large excess of soluble receptor (molar ratio of receptor to gD, 40:1) (experiment shown in Fig. 5). The order of addition of the reagents was irrelevant. Thus, if soluble nectin 1 was added to a preformed complex made of gD260t and GST-PFD, gD was not found in the GST-PFD-bound fraction. Altogether, these results clearly indicate that gD260t failed to interact with GST-PFD when in complex with either one of its receptors.
Fig. 5.
Immunoblots of proteins bound by GST-PFD and eluted with GSH. Aliquots of gD260t (40 ng each) alone (reaction mixture 4) or mixed with soluble HVEMt (reaction mixture 2), nectin 1-Fc (reaction mixture 3), or nectin 3-Fc (reaction mixture 1) for 2 h at 4°C were reacted with GSH-beads preloaded with GST-PFD. Replicate aliquots of the bound proteins, eluted with GSH, were separated by SDS/PAGE and blotted, and the blots were developed with mAb H170 to gD (A), anti-His antibody to HVEMt (B), CK6 mAb to nectin 1-Ig (C), or anti-human IgG to nectin 3-Ig (data not shown). Input controls (C) contained the indicated proteins (gD260t in A, HVEMt in B, and nectin 1-Fc in C) and were loaded in SDS/PAGE for control of migration position and immunoreactivity. It can be seen that, when gD260t was premixed to soluble HVEMt (reaction mixture 2) or nectin 1-Ig (reaction mixture 3), neither gD260t (A) nor the receptors (B and C) were present in the GSH-eluted proteins.
Discussion
It is convenient to discuss the results presented in this report in the light of a model that describes the interaction between the two functional regions of the HSV-1 gD ectodomain, the N terminus, which carries the receptor-binding sites, and the C terminus, which functions as PFD, and its consequences on fusion. In essence, we propose the following model: (i) When not bound to one of its receptors, gD folds back on itself and enables physical interactions between the N and C termini. We propose to name this conformation as closed. (ii) Upon gD binding to either receptor, the interaction between the N and C termini is lost, the termini are released from reciprocal restrains, and gD switches to an “opened” conformation. The opening enables receptor-dependent triggering of the fusion of the viral envelope with the cell membrane. In support of this model are the following data and conclusions:
PFD of HSV-1 gD is made of subdomains 1 and 2, comprising residues 260-285 and 285-310, respectively. Each subdomain partially contributed to virus entry, whereas only the subdomain 1 exhibited cell-cell fusion activity. The latter finding is in agreement with the report of Zago et al. (16). Mutational analysis of Pro and Glu, the most characteristic sequence pattern of subdomain 1, did not highlight any critical role of these amino acids in the infectivity complementation and resulted in a modest reduction in the cell-cell fusion assay. The lack of phenotype of these mutants is in agreement with the observation that serial 5-aa deletions in this region failed to identify a specific sequence requirement (16). The properties of the PFD/1 subdomain contrast with those of PFD/2, which carries some Pro that is critical for infectivity (15). Their mutagenesis did not alter PFD-binding to gD N terminus, implying a role at subsequent steps, likely in the triggering of fusion.
A GST-PFD fusion protein specifically bound gD260t or longer forms of gD. GST fusion proteins of subdomains 1 and 2 were capable of binding gD260t independently of each other, highlighting multiple contact sites of PFD on the N terminus. None of the GST-PFD proteins bound gD250t, possibly because the 250- to 260-aa sequence is either part of the PFD-binding site on gD N terminus or provides a conformation suitable to create the PFD-binding site. An effect due to abnormal configuration of gD250t is ruled out by the ability of gD250t to bind either receptor with high affinity and to inhibit virus entry, i.e., to compete with full-length gD (27). Remarkably, in the crystal structures of gD, the last resolved amino acid, residue 259, is at one extreme edge of the molecule on the opposite side relative to the hairpin (9). Taken together, current binding studies and the structural data suggest that at or around amino acid 260, gD folds back over the N terminus, and that the N and C termini interact.
The gD structures originally described by Carfi et al. (9) lacked the last 50 aa that constitute the PFD. The coordinates of this region were recently resolved by the same group (Herpesvirus Workshop, Reno, NV, July 25-30, 2004, and personal communication). The structure indeed shows that the C terminus of gD ectodomain wraps around the N terminus and partially occludes the HVEM-binding site. Thus, the conclusions drawn from current binding studies are in agreement with and are reinforced by the recent structural data. A further line of evidence argues that the N and C termini of the gD ectodomain are in proximity. Thus, the epitope recognized by mAb AP7 is destroyed by substitutions-deletions at the N or C termini (35-37). The current model of folding of the gD C terminus toward the N terminus accounts for the low affinity of gD306t to the receptors (14), which may result from a limited access of the receptors to their binding sites, consequent to the binding of the C terminus to its binding site on the N terminus. The lack of binding of GST-PFD, GST-PFD/1, and GST-PFD/2 to gD306t is in accordance with this view. Our results further show that full-length gD extracted from infected cell lysates also failed to bind the GST-PFD fusion proteins, i.e., its binding properties could not be differentiated from those of gD306t. Together, these considerations support the view that the folded conformation of gD reflects the conformation that the full-length glycoprotein adopts in the infected cells, and by extension, in virions before entry. We propose that the folded conformation is considered a closed conformation. Clearly, the complex of gD260t with GST-PFD mimics the closed conformation.
gD260t in complex with soluble forms of either receptor failed to bind the GST-PFD. These results highlight one of few documented modifications to gD upon receptor binding. We interpret these findings as a further indication that (i) the closed conformation reflects the conformation of the glycoprotein in the absence of, and therefore before receptor binding; and (ii) the change in conformation releases the PFD from the N terminus and leads to an opened conformation. We envision that the uncoupled PFD and N terminus are now enabled to promote the activities of the downstream fusogenic glycoproteins. It remains to be clarified whether the PFD, the N terminus in complex with its receptor, or both regions serve as the trigger of fusion. Alternatively, it was proposed that the role of PFD is to provide a flexible stalk between the domain carrying the receptor-binding sites and the membrane, which enables conformational changes in gD (16).
The steps that ensue after gD binding to its receptor are unclear at present. They may involve the formation of a complex between the gD receptor and one or more of the downstream glycoproteins or the disassembly of a preexisting complex. As mentioned in the Introduction, gH exhibits two important attributes, i.e., a candidate fusion peptide and, downstream of it, two functional heptad repeats with a propensity to form coiled coils (refs. 20 and 21 and T. Gianni and G.C.-F., unpublished work). Such structures are typical of type 1 fusion glycoproteins (38). Their presence in HSV-1 gH makes this glycoprotein the likely candidate executor of HSV fusion. Because viral fusion glycoproteins are dead ends in the fusion process and do not undergo renaturation, such attributes place gH at the end of the chain that starts with the gD-receptor complex and culminates in fusion. As a corollary, gB is likely to serve as a link in the chain. Understanding this chain requires the elucidation of the role of gB and gL in fusion and possibly the isolation of supramolecular complexes made by subassemblies of the glycoproteins.
To our knowledge, the activation of fusion through the transition from a closed to an opened conformation is unusual among viral fusion systems. However, it exists among fusion proteins of exocytic vesicles. In particular, syntaxin 1, one of the soluble N-ethylmeleimide-sensitive factor (NSF) attachment protein receptor (SNARE) proteins, and one essential protein in synaptic membrane fusion, undergoes a transition from closed to opened conformation during exocytosis (38-40). In the default closed conformation, the C terminus folds upon the N terminus, which forms a complex with munc-18-1 proteins. The closed conformation is incompatible with assembly of the SNARE core complex, a four-helix bundle required for fusion. During exocytosis, a large conformational switch mediates the transition from the syntaxin-munc-18-1 complex to the SNARE core complex (39, 40). Despite these analogies, there is a major difference between gD and syntaxin. Syntaxin is a component of the actual fusion complex, whereas gD plays the role of receptor-binding glycoprotein and of trigger of fusion but is not thought to be a component of the HSV-fusion complex. The Slyy1-Munc proteins, which cooperate with SNARE proteins, also adopt closed and opened conformations (38); they are soluble proteins and, therefore, bear a weaker analogy to gD than does syntaxin.
Taking these models and our results into consideration, we propose that the opening of gD is part of the mechanism that signals receptor recognition to the downstream glycoproteins responsible for fusion execution and that this signaling is a critical event in the triggering of fusion.
Acknowledgments
We thank Drs. Roselyn Eisenberg, Gary Cohen (University of Pennsylvania, Philadelphia), and Andrea Carfi (Istituto Ricerche di Biologia Molecolare, Rome) for invaluable reagents and for sharing with us unpublished data, and we are grateful to Elisabetta Romagnoli for help with cell cultures. This work was supported by the Fondo per gli Investimenti della Ricerca di Base Autonomous and Coordinated Projects, Cofin-Ministero dell'Istruzione, dell'Università e della Ricerca 40% 2002 and 2003, Consiglio Nazionale delle Ricerche-Functional genomics, University of Bologna 60%, and the Fondo Pallotti.
Author contributions: D.F., C.F., and G.C.-F. designed research; D.F. and C.F. performed research; D.F., C.F., and G.C.-F. analyzed data; and G.C.-F. wrote the paper.
Abbreviations: g, glycoprotein; GSH, reduced glutathione; GST, glutathione S-transferase; HSV, herpes simplex virus; HVEM, herpesvirus entry mediator; PFD, pro-fusion domain; TM, transmembrane; C-tail, cytoplasmic tail; CELISA, cell ELISA.
References
- 1.Campadelli-Fiume, G., Cocchi, F., Menotti, L. & Lopez, M. (2000) Rev. Med. Virol. 10, 305-319. [DOI] [PubMed] [Google Scholar]
- 2.Spear, P. G., Eisenberg, R. J. & Cohen, G. H. (2000) Virology 275, 1-8. [DOI] [PubMed] [Google Scholar]
- 3.Geraghty, R. J., Krummenacher, C., Cohen, G. H., Eisenberg, R. J. & Spear, P. G. (1998) Science 280, 1618-1620. [DOI] [PubMed] [Google Scholar]
- 4.Montgomery, R. I., Warner, M. S., Lum, B. J. & Spear, P. G. (1996) Cell 87, 427-436. [DOI] [PubMed] [Google Scholar]
- 5.Cocchi, F., Menotti, L., Mirandola, P., Lopez, M. & Campadelli-Fiume, G. (1998) J. Virol. 72, 9992-10002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krummenacher, C., Rux, A. H., Whitbeck, J. C., Ponce-de-Leon, M., Lou, H., Baribaud, I., Hou, W., Zou, C., Geraghty, R. J., Spear, P. G., et al. (1999) J. Virol. 73, 8127-8137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Warner, M. S., Geraghty, R. J., Martinez, W. M., Montgomery, R. I., Whitbeck, J. C., Xu, R., Eisenberg, R. J., Cohen, G. H. & Spear, P. G. (1998) Virology 246, 179-189. [DOI] [PubMed] [Google Scholar]
- 8.Lopez, M., Cocchi, F., Menotti, L., Avitabile, E., Dubreuil, P. & Campadelli-Fiume, G. (2000) J. Virol. 74, 1267-1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carfi, A., Willis, S. H., Whitbeck, J. C., Krummenacher, C., Cohen, G. H., Eisenberg, R. J. & Wiley, D. C. (2001) Mol. Cell 8, 169-179. [DOI] [PubMed] [Google Scholar]
- 10.Connolly, S. A., Landsburg, D. J., Carfi, A., Wiley, D. C., Eisenberg, R. J. & Cohen, G. H. (2002) J. Virol. 76, 10894-10904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou, G., Avitabile, E., Campadelli-Fiume, G. & Roizman, B. (2003) J. Virol. 77, 3759-3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zago, A. & Spear, P. G. (2003) J. Virol. 77, 9695-9699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Connolly, S. A., Landsburg, D. J., Carfi, A., Whitbeck, C. J., Zuo, Y., Wiley, D. C., Cohen, G. H. & Eisenberg, R. J. (2005) J. Virol. 79, 1282-1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Whitbeck, J. C., Muggeridge, M. I., Rux, A. H., Hou, W., Krummenacher, C., Lou, H., van Geelen, A., Eisenberg, R. J. & Cohen, G. H. (1999) J. Virol. 73, 9879-9890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cocchi, F., Fusco, D., Menotti, L., Gianni, T., Eisenberg, R. J., Cohen, G. H. & Campadelli-Fiume, G. (2004) Proc. Natl. Acad. Sci. USA 101, 7445-7450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zago, A., Jogger, C. R. & Spear, P. G. (2004) Proc. Natl. Acad. Sci. USA 101, 17498-17503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gompels, U. & Minson, A. (1986) Virology 153, 230-247. [DOI] [PubMed] [Google Scholar]
- 18.Turner, A., Bruun, B., Minson, T. & Browne, H. (1998) J. Virol. 72, 873-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Forrester, A., Farrell, H., Wilkinson, G., Kaye, J., Davis Poynter, N. & Minson, T. (1992) J. Virol. 66, 341-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gianni, T., Martelli, P. L., Casadio, R. & Campadelli-Fiume, G. (2005) J. Virol. 79, 2931-2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gianni, T., Menotti, L. & Campadelli-Fiume, G. (2005) J. Virol. 79, 7042-7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ligas, M. W. & Johnson, D. C. (1988) J. Virol. 62, 1486-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ejercito, P. M., Kieff, E. D. & Roizman, B. (1968) J. Gen. Virol. 2, 357-364. [DOI] [PubMed] [Google Scholar]
- 24.Kawaguchi, Y., Bruni, R. & Roizman, B. (1997) J. Virol. 71, 1019-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Isola, V. J., Eisenberg, R. J., Siebert, G. R., Heilman, C. J., Wilcox, W. C. & Cohen, G. H. (1989) J. Virol. 63, 2325-2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krummenacher, C., Baribaud, I., Ponce De Leon, M., Whitbeck, J. C., Lou, H., Cohen, G. H. & Eisenberg, R. J. (2000) J. Virol. 74, 10863-10872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krummenacher, C., Nicola, A. V., Whitbeck, J. C., Lou, H., Hou, W., Lambris, J. D., Geraghty, R. J., Spear, P. G., Cohen, G. H. & Eisenberg, R. J. (1998) J. Virol. 72, 7064-7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fabre, S., Reymond, N., Cocchi, F., Menotti, L., Dubreuil, P., Campadelli-Fiume, G. & Lopez, M. (2002) J. Biol. Chem. 277, 27006-27013. [DOI] [PubMed] [Google Scholar]
- 29.Cocchi, F., Lopez, M., Menotti, L., Aoubala, M., Dubreuil, P. & Campadelli-Fiume, G. (1998) Proc. Natl. Acad. Sci. USA 95, 15700-15705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Menotti, L., Cocchi, F. & Campadelli-Fiume, G. (2002) Virology 301, 6-12. [DOI] [PubMed] [Google Scholar]
- 31.Pertel, P. E., Fridberg, A., Parish, M. L. & Spear, P. G. (2001) Virology 279, 313-324. [DOI] [PubMed] [Google Scholar]
- 32.Rovero, S., Amici, A., Carlo, E. D., Bei, R., Nanni, P., Quaglino, E., Porcedda, P., Boggio, K., Smorlesi, A., Lollini, P. L., et al. (2000) J. Immunol. 165, 5133-5142. [DOI] [PubMed] [Google Scholar]
- 33.Avitabile, E., Lombardi, G. & Campadelli-Fiume, G. (2003) J. Virol. 77, 6836-6844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Menotti, L., Casadio, R., Bertucci, C., Lopez, M. & Campadelli-Fiume, G. (2002) J. Virol. 76, 5463-5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dean, H. J., Terhune, S. S., Shieh, M. T., Susmarski, N. & Spear, P. G. (1994) Virology 199, 67-80. [DOI] [PubMed] [Google Scholar]
- 36.Chiang, H. Y., Cohen, G. H. & Eisenberg, R. J. (1994) J. Virol. 68, 2529-2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Minson, A. C., Hodgman, T. C., Digard, P., Hancock, D. C., Bell, S. E. & Buckmaster, E. A. (1986) J. Gen. Virol. 67, 1001-1013. [DOI] [PubMed] [Google Scholar]
- 38.Jahn, R., Lang, T. & Sudhof, T. C. (2003) Cell 112, 519-533. [DOI] [PubMed] [Google Scholar]
- 39.Dulubova, I., Sugita, S., Hill, S., Hosaka, M., Fernandez, I., Sudhof, T. C. & Rizo, J. (1999) EMBO J. 18, 4372-4382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Misura, K. M., Scheller, R. H. & Weis, W. I. (2000) Nature 404, 355-362. [DOI] [PubMed] [Google Scholar]