Abstract
Papillomaviruses are small DNA viruses that are associated with benign and malignant epithelial lesions, including >95% of cervical cancers and ≈20% of head and neck cancers. Because papillomavirus replication and virion production are tied to epithelial cell differentiation, infectious papillomavirus virion production has been limited to cumbersome organotypic cultures and mouse xenografts. Consequent difficulties in obtaining useful amounts of wild-type or mutant human papillomavirus (HPV) virions have greatly limited studies on many aspects of papillomavirus biology. To overcome these limitations, we developed a system to encapsidate the full-length papillomaviral genome into infectious virions, independently of viral DNA replication and epithelial differentiation. This transient-transfection-based system produces >1,000 times more infectious virus per cell culture dish than the much more labor-intensive organotypic culture. Furthermore, we show that this method allows the facile generation of infectious particles containing wild-type, mutant, or chimeric papillomaviral genomes, overcoming barriers to studying many facets of replication, host interactions, and vaccine and drug development, which has been limited by the insufficient availability of infectious virions.
Keywords: infection, virions, virus packaging, capsid, DNA encapsidation
Papillomaviruses are nonenveloped, double-stranded DNA viruses with ≈8-kb, circular genomes, 55-nm spherical capsids, wide distribution in higher vertebrates, and tight species specificity (1). Human papillomaviruses (HPVs), of which there are >100 genotypes, infect and replicate in cutaneous or mucosal epithelia, inducing benign lesions, including warts that are self-limiting and normally regress over time (2, 3). A subset of mucosotropic HPVs, the high-risk genotypes, such as HPV16, HPV18, and HPV31, are causally associated with anogenital cancers, including nearly all, if not all, cervical carcinomas, a leading cause of cancer death among women worldwide (1, 4, 5). Also, high-risk HPVs (in particular, HPV16) are associated with 20-30% of head and neck cancers (6, 7), the sixth most common cancer in the United States, of which the survival rate is <50% and has not improved for decades (8).
The HPV life cycle is tightly linked to epithelial differentiation (9, 10). HPVs initially infect cells of the poorly differentiated, proliferative, basal compartment of stratified epithelia. Here, the viral genome sets up residence as a low-copy nuclear plasmid, a subset of viral genes (the early genes) are expressed at low levels, and no progeny virus is made. However, as infected basal cells divide and daughter cells migrate into the suprabasal compartment for terminal differentiation, the productive stage of the viral life cycle is initiated (10, 11). Here, the virus reprograms suprabasal cells to support high-copy-number amplification of the viral genome, the viral structural genes encoding the major L1 and minor L2 capsid proteins are expressed, and progeny virions are assembled and then released into the environment from the most superficial layers of the epithelia. The requirement for terminal differentiation of epithelial cells to support the productive stage of the viral life cycle precludes obtaining infectious virus particles from conventional cell culture. Consequently, the only prior methods capable of producing infectious papillomavirus virions were organotypic raft culture (12-15), by which small quantities of artificial skin can be produced in cell culture, or the use of xenografts in mouse skin (16-18). However, these methods are technically demanding, time-consuming, and variable, and they produce relatively low virus yields and require access to epithelial cell populations or human tissue in which the viral genotype of interest persists as a nuclear plasmid. These limitations have severely restricted the availability of infectious HPV for basic and clinical research.
Recently, Buck et al. reported an approach for producing papilloma virus-like particles (VLPs) in which reporter plasmids were encapsidated into bovine papillomavirus (BPV) capsid proteins expressed in transiently transfected mammalian cells (19). Here, high-level expression of papillomavirus L1 and L2 capsid proteins was achieved by using codon-optimized synthetic genes in 293TT human embryonic kidney cells, which stably express SV40 large T antigen to enhance replication of SV40 origin-containing plasmids. When cotransfected into 293TT cells with L1- and L2-expression plasmids, target plasmids of <6-7 kb were efficiently encapsidated into the resulting VLPs. This method allowed the differentiation-independent encapsidation of reporter genes. However, Buck et al. concluded that intracellular packaging of target plasmids into VLPs by this approach was limited by a strong size discrimination to target plasmids of ≤6-7 kb in size (19), which is much less than the natural viral genome size of ≈8 kb.
In this article, we describe extension of the transient-transfection method to achieve the successful and efficient packaging of the full-length HPV genomes into HPV16 capsids to generate virus particles. We demonstrate that the resulting HPV virions are highly infectious in their natural, host epithelial cells. Importantly, a single 10-cm dish of 293T cells, after DNA preparation and a simple 2-day transient transfection, yielded >1,000 times more infectious virus than a much more labor-intensive and time-consuming organotypic raft culture. By providing a ready, genetically manipulatable source of infectious papillomavirus virions, this approach makes it possible to study HPV replication and vaccine and drug development, which has been limited or blocked by the difficulty of producing infectious HPV virions.
Materials and Methods
Plasmids. pEF399, containing the complete W12E HPV16 genome (20), and pHPV31b, containing the HPV31b genome (21), have been described. pcDNA-HPV16L1 and pcDNA-HPV16L2, expressing codon-optimized HPV16 L1 and L2 (22), respectively, were generously provided by Martin Muller (Forschungsschwerpunkt fur Angewandte Tumorvirologie, Heidelberg, Germany). pXULL, coexpressing both codon-optimized HPV16 L1 and L2, was generously provided by John Schiller (National Institutes of Health, Bethesda). pSEAP (pSEAP-control) for expression of secreted alkaline phosphatase (SEAP) was purchased from Clontech. We cloned 5.0-, 5.9-, and 6.8-kb PCR-generated subgenomic fragments of the HPV16 W12E genome into XhoI- and SacI-cleaved pRL-null (Promega). These HPV subgenomic fragments, deleted for nucleotides 4,296-7,120, 4,296-6,300, and 4,296-5,400 of the HPV genome, respectively, were generated by PCR using pfu DNA polymerase along with the following primers: TTATAAAGTTGGGTAGCCGATGCACG (sense) and TCTACAACTGCTAAACGCAAAAAA (5.0 kb), CTGGATATTTGTACATCTATTTGC (5.9 kb), or TCTTTATCAGGTTATATTCC (6.8 kb) (anti-sense). PCR was performed for 32 cycles of the following: 1 min of denaturation at 94°C, 1 min of annealing at 58°C, and 14 min of polymerization at 72°C, followed by 20 min of extension at 72°C. For transfection into 293TT and 293T cells, the full-length or subgenomic HPV DNAs were released from the bacterial vector by restriction-enzyme digestion (BamHI for full-length HPV16 and HPV31b genomes, and XhoI for subgenomic HPV16 clones) and recircularized by ligation under conditions that favor monomeric ligation events (23).
Cell Lines. Human embryonic kidney cell line 293T (American Type Culture Collection) and its enhanced SV40 T antigen-expressing daughter cell line 293TT (19) (obtained from John Schiller) were maintained in DMEM supplemented with 10% FBS (Invitrogen). Immortalized human keratinocyte cell line HaCaT (24) was maintained in F-media (three parts F-12 and one part DMEM; Invitrogen), supplemented with 10% FBS. W12E cells, clone 20850 (25, 26), harboring extrachromosomal HPV16 genomes, were maintained on mitomycin C (4 μg/ml; Sigma)-treated 3T3 mouse fibroblasts in F-media supplemented with 0.4 μg/ml hydrocortisone (Calbiochem), 0.1 nM cholera toxin (ICN), 5 μg/ml insulin (Sigma), 25 μg/ml adenine (Sigma), 10 ng/ml epidermal growth factor (R & D Systems), and 5% FBS, as described (25).
Virus Packaging and Purification. We cotransfected 293T or 293TT cells, which were plated in a 10-cm dish 1 day before, with 10 μg of each HPV16 capsid protein expression plasmid as indicated, as well as one of the target DNAs for encapsidation, such as the full-length HPV16 genome (from 12 μg of starting plasmid) or pSEAP (10 μg) by using Lipofectamine 2000 (Invitrogen). After 48 h at 37°C, cells were harvested and resuspended in PBS with 9.5 mM MgCl2. Cells were lysed by adding Brij58 to 0.25%, followed by 0.3% benzonase (Sigma) and 2 units per 100 μl of exonuclease V (Epicenter Technologies, Madison, WI) and incubating at 37°C for 24 h to remove unpackaged cellular and viral DNA and to allow capsid maturation (48). The lysate was incubated on ice for 10 min with 0.17 volumes of 5 M NaCl and centrifuged at 2,000 × g for 10 min at 4°C. The resulting cleared supernatant was loaded on top of a 27-33-39% Optiprep/PBS/0.8 M NaCl step gradient and centrifuged in an SW50.1 rotor at 215,000 × g for 3.5 h at 16°C. Fractions (200 μl each) were collected by puncturing the bottom of the tube. Aliquots of 10 μl from each fraction were analyzed by Western blotting. Briefly, proteins fractionated by SDS/PAGE were transferred to poly-vinylidene difluoride (PVDF) paper (Amersham Pharmacia). HPV16 L1 protein bands were visualized by chemiluminescence using mouse anti-HPV16 L1 antibody CAMVIR-1 (Abcam, Cambridge, MA) and goat anti-mouse IgG conjugated with horseradish peroxidase (Jackson ImmunoResearch). To assay for DNA encapsidation, DNA was extracted from purified virions by using a QIAquick PCR purification kit (Qiagen, Valencia, CA) or phenol/chloroform/proteinase K (27). DNA extracts from each fraction were run on 0.8% agarose gel and visualized by SYBR green I (Sigma) staining. The copy number of encapsidated DNA was quantified with serially diluted standard DNA. Capsid protein levels were measured by SDS/PAGE and silver staining using defined dilutions of BSA as concentration standards.
Infectivity Assay. Infectivity of packaged HPV16 and HPV31b virions was examined by RT-PCR by amplifying the splice junction region in a class of HPV early mRNAs, hereafter designated E1 E4, that share a common splice junction between E1 and E4 ORFs. HaCaT cells were infected with dilutions of packaged virus overnight at 37°C, washed twice with PBS, and incubated for 48 h at 37°C. Total RNA was isolated by using the RNeasy total RNA purification kit (Qiagen), treated with RQ DNase I (Promega) to remove possible DNA contaminants, purified again on RNeasy columns to remove DNase I, and quantified by spectrophotometer. Double-stranded cDNA was synthesized from 20 μg of total RNA with oligo(dT) by using a SuperScript cDNA synthesis kit (Invitrogen), and PCR was performed with TaqDNA polymerase (Promega). Oligonucleotide primers (Table 1, which is published as supporting information on the PNAS web site) were designed by using the primer3 primer-design program (28), synthesized by MWG Biotech (High Point, NC), and used at 0.5 μM for PCR amplification for 36 cycles of the following steps: 1 min of denaturation at 94°C, 1 min of annealing at 55°C, and 2 min of polymerization at 72°C, followed by a 10-min extension at 72°C. For the following nested PCR, 2% of the first-round PCR products were used in 30 cycles of amplification. Alternatively, the HPV signal was amplified for one round of 42 cycles of 30 s of denaturation at 94°C, 1 min of annealing at 55°C, and 1 min of polymerization at 72°C, followed by 7 min of extension at 72°C. The final PCR products were analyzed by 1% agarose gel electrophoresis and ethidium bromide staining.
The activity of pSEAP-control-encapsidating pseudovirions was measured by using a Phospha-Light chemiluminescent alkaline phosphatase assay (Applied Biosystems). After adding pSEAP-control pseudovirions, 293T cells were incubated for 48 h. We collected 50 μl of culture supernatant (processed according to the manufacturer's protocol), and luminescence was measured for 1 s in a luminometer.
Capsid Disruption and Virus Neutralization. HPV capsids were disrupted by treating with 200 mM NaHCO3 (pH 9.6) at 4°C for 16 h (29), followed by dialysis with PBS for 24 h with a Slide-A-Lyzer dialysis cassette (Pierce). In parallel, an equal amount of virions was processed in PBS (pH 7.0) as a control. HPV16-neutralizing antibodies H16.7E, H16.E70, and H16.V5 were generously provided by Neil Christensen (Pennsylvania State University, Hershey). Diluted virus preparations were incubated with one of the neutralizing antibodies at 1:100 dilutions for 1 h at 4°C with rotating before infecting human keratinocytes. Mouse anti-HPV16 L1 H16.J4 (30) and anti-HIV-1 Gag (31, 32) IgG antibodies were used as isotype controls.
Results
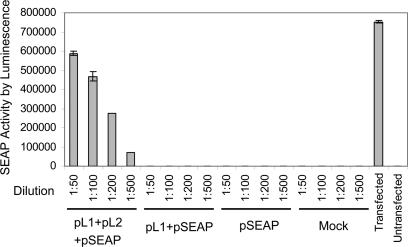
Gene Delivery and Expression Efficiency of HPV Capsids. To establish that we could efficiently assemble functional capsids by expressing the L1 and L2 capsid protein genes of the oncogenic HPV genotype HPV16, we measured the efficiency with which we could package and deliver by infection a 5.1-kb reporter plasmid, pSEAP, expressing SEAP. pSEAP was cotransfected with two codon-optimized HPV16 L1 and L2 expression plasmids, pcDNA3.1(+)-HPV16L1 and pcDNA3.1(+)-HPV16L2 (22), into 293T cells. At 2 days after transfection, pseudovirions were extracted, treated with benzonase endonuclease and exonuclease V to degrade unencapsidated DNA, and fractionated on Optiprep gradients. Comparable levels of L1 capsid protein expression were found by Western blotting after transfection of L1+L2 or L1 alone (data not shown). We then assayed for effective delivery of packaged pSEAP by monitoring SEAP activity after exposure of 293T cells to pseudovirion preparations. SEAP activity was nearly linear with dilution of the infecting pseudovirus stock over the tested range, retaining substantial activity ≈700-fold above background after 500-fold dilution (Fig. 1). Although major capsid protein L1 assembles capsids by itself, this delivery relied on coexpression of minor capsid protein L2 as well (Fig. 1), consistent with prior findings (19, 33). Significantly, SEAP activity after infection with ≈1.5 × 108 copies of L1/L2 encapsidated pSEAP DNA was nearly as high as that in 293T cells after Lipofectamine 2000 transfection with ≈3.4 × 1011 copies of unencapsidated pSEAP-control DNA. Thus, delivery of SEAP plasmid in the context of these HPV16 L1/L2 pseudovirions was ≥2,000-fold more efficient, on a per plasmid copy basis, than an efficient transfection method.
Fig. 1.
Gene transfer and expression by HPV16 pseudovirions. The histogram shows SEAP activity levels in cell culture supernatants 48 h after treating 293T cells with the stated dilutions of virus particles purified by Optiprep gradient fractionation from cells mock-transfected or transfected with the indicated combinations of reporter plasmid pSEAP and independent expression plasmids for HPV16 capsid proteins L1 and L2. For the direct transfection positive control (second bar from the right), SEAP activity was measured directly on 293T cells transfected with 1 μg of pSEAP DNA.
HPV16 Genome Can Be Packaged Efficiently to Generate High Yields of Papillomavirus. Buck et al. reported (19) that the size of the target DNA packaged by BPV capsid proteins L1 and L2 in their transient-transfection system was limited to <6-7 kb. Therefore, it was predicted that full-length, ≈8-kb HPV genomes would not be encapsidated efficiently in this system. To determine the size limit of HPV16 viral DNA encapsidation, we generated three progressively truncated derivatives of the HPV16 genome, with nested deletions in the L1 and L2 ORFs (Fig. 2A). In the smallest 5.0-kb derivative, the entire L1 and L2 ORFs were deleted, leaving intact the long control region and early genes. The 5.9-kb HPV16 subgenomic clone retains the 3′ portion of the L1 gene. The 6.8-kb subgenomic clone has an intact L1 ORF but is deleted for most of the L2 ORF. These HPV16 genome derivatives, as well as the full-length HPV16 genome, were amplified in Escherichia coli plasmids, excised by XhoI digestion, and recircularized by ligation. Each HPV genome derivative then was transfected into 293TT cells together with HPV16 L1 and L2 expression plasmids. Equivalent results were obtained by using separate expression plasmids pcDNA3.1(+)-HPV16L1 and pcDNA3.1(+)-HPV16L2 for L1 and L2 (22), or a combined L1+L2 expression plasmid, pXULL (generously provided by John Schiller). After a 48-h incubation, the cells were lysed, treated with benzonase endonuclease and exonuclease V to degrade unencapsidated viral and cellular DNA, and fractionated on Optiprep gradients.
Fig. 2.
Encapsidation of the HPV16 genome and its derivatives by HPV16 L1 and L2 capsid proteins. (A) Map of HPV16 deletion derivatives generated by PCR-mediated deletions (gaps) extending to the indicated HPV16 genome position. Each target DNA was cotransfected into 293TT cells with HPV16 L1 and L2 expressing plasmids. After Optiprep gradient purification of the virus particles, the encapsidated DNA was analyzed by 0.8% agarose gel electrophoresis and SYBR green staining (B and C), and HPV16 L1 capsid proteins were visualized by Western blotting (C). For the gradient of C and similar gradients, virus particles from the peak fraction of encapsidated, full-length HPV16 DNA (arrow) were isolated for infectivity assays.
The results (Fig. 2 B and C) showed that HPV16 L1 and L2 expression protected not only all three truncated derivatives but also full-length HPV genomic DNA from nuclease, producing DNA bands that were visible by direct staining with the intercalating fluorescent stain, SYBR green I, and that cosedimented in gradients with authentic HPV virions. No such DNA recovery and protection was observed if L1 and L2 expression was omitted, consistent with the DNase-resistant viral genomes having been encapsidated into virus particles, or if L1 was expressed without L2 (19, 33). Western blotting (Fig. 2C) showed that L1 was distributed over a wider range of the gradient (1.12-1.20 g/ml) than was the DNA (1.17 g/ml), with a major peak immediately above the sedimenting DNA (i.e., at a lower density, 1.14 g/ml), as expected for empty HPV capsids and as found in CsCl gradients. Optiprep gradient fractionation of BPV encapsidation products showed a similar density range (≤1.2 g/ml) for DNA-containing particles, although in that case, empty capsids were unexpectedly found at higher density than DNA-containing capsids (19). Surprisingly, the efficiency with which the full-length and the various truncated HPV16 genomes were encapsidated into the virus particles was equal. Thus, in contrast to prior observations made with reporter plasmids and BPV L1 and L2 (19), HPV16 L1 and L2 encapsidated HPV16 genomic DNAs ranging 5-8 kb in size, with comparable efficiency in multiple repeated experiments. HPV16 virus packaging also was performed with transfection of 293T or 293TT cells to compare efficiency of target DNA encapsidation in the presence of lower and higher levels, respectively, of SV40 T antigen (19). However, even with L1- and L2-expression plasmid pXULL, containing an SV40 replication origin, there was no significant difference between 293T and 293TT cells in HPV16 genome encapsidation (data not shown).
To measure the copy number of encapsidated HPV genomes, DNA extracted from the virus particles was analyzed by agarose gel electrophoresis, in comparison with HPV16 genomic DNA standards of known concentrations. Transfecting a single 10-cm dish of 293T or 293TT cells yielded an average of 2 × 109 copies of nuclease-resistant HPV16 genomic DNA encapsidated into HPV16 L1/L2 capsid proteins, with a variation of ≤2-fold over many repeated transfections. To calculate the efficiency of HPV DNA encapsidation per capsid formed, the gradient-purified, packaged virus stock containing DNase-resistant DNA was analyzed by SDS/PAGE and the levels of HPV L1 and L2 proteins present were compared by silver staining to serially diluted BSA protein standards. These results showed that ≈8 × 1010 assembled HPV particles were collected from one 10-cm dish of ≈7 million cells, and that ≈3% of these virus particles contained HPV16 genomic DNA. This encapsidation efficiency is similar to that obtained when HPV particles are produced in organotypic cultures, in which 5-6% of the capsids contain HPV genomic DNA (33). However, each such organotypic culture yields in the range of only 107 particles in total. Thus, our results show that 2 days after transfection of a single 10-cm dish of cells, 1,000-fold more HPV DNA-containing virions were produced than from a much more labor-intensive, 2-week organotypic raft culture.
To test whether different HPV genotypes could be packaged by the same method, we cotransfected full-length, recircularized HPV31b genomic DNA along with pXULL, and found that these HPV31 genomes also were packaged efficiently into HPV16 L1/L2 capsids (data not shown). Thus, the results are not HPV16-specific, and the same approaches can be applied to produce virions encapsidating other HPV genotypes.
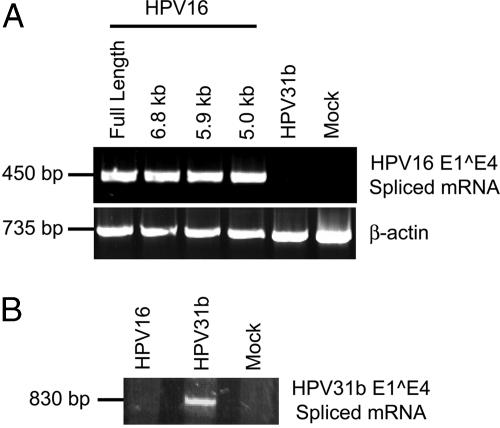
Infectivity of Packaged, Full-Length HPV16 Genomes. The infectivity of the HPV16 virions that were produced in 293TT cells was tested in HaCaT cells, an immortalized human keratinocyte cell line. At 48 h after inoculation, cells were harvested, total RNA was purified, and RT-PCR was performed to detect one class of spliced viral mRNAs, an established measure for HPV infectivity (34-36). The expected PCR product from the E1 E4 spliced mRNAs was seen (Fig. 3), showing that the virus particles generated by using the transient-transfection-based method were infectious. Infection with 100- to 6,250-fold serial dilutions of the starting virus stock, corresponding to 100 to 1.6 HPV16 viral genome equivalents (vge) per cell revealed that <10 vge per cell of virion stock from 293TT cell transfection still clearly showed detectable infection by single-round RT-PCR (Fig. 3A). This result further confirms that the yield of virus, as measured above by the quantity of DNase resistant viral genomes present in Optiprep-fractionated virus stock, was >1,000-fold greater than the amount of virus typically obtained from rafts, whereas the specific infectivity is comparable (37, 38).
Fig. 3.
Infectivity of encapsidated, full-length HPV16 DNA. Virions encapsidating the HPV16 W12 genome were produced as in Fig. 2C and inoculated onto HaCaT cells. At 48 h after inoculation, cells were harvested, total RNA was isolated, and a single round of RT-PCR (A) or RT-PCR, followed by a second round of nested PCR (B and C), was performed with PCR primers (see Table 1) to detect E1 E4 spliced mRNAs. We amplified β-actin mRNA simultaneously as an internal standard. (A) Cells were inoculated with 100 to 1.6 vge per cell as indicated and assayed for infection by single round RT-PCR. (B) As indicated, HaCaT cells were mock-inoculated or inoculated with 100 vge per cell of virions that were untreated or treated with high-pH carbonate buffer for 24 h at 4°C or with unencapsidated, full-length HPV16 DNA. Total RNA extracted from HPV16-positive W12E cells was used as a positive control. (C) Before inoculating at 100 vge per cell, virions were incubated for 1 h at 4°C with 1:100 dilutions of HPV16 L1-neutralizing antibodies H16.7E or H16.V5. A nonneutralizing, anti-HPV16L1 antibody and an anti-HIV Gag antibody were used as isotypic negative controls.
Above, we showed that the infectivity of the HPV16 virion preparations was nuclease-resistant. To confirm further that viral gene expression detected in the infection assay did not arise wholly or partially from direct transfection of naked DNA not packaged by HPV16 capsid proteins, we treated virus stocks with 200 mM NaHCO3 (pH 9.6), which efficiently and irreversibly disrupts HPV capsids (29). After treatment, the preparations were dialyzed against PBS and added to HaCaT cells. This pretreatment of virus stocks at high pH abolished viral mRNA expression in inoculated HaCaT cells, whereas untreated, but dialyzed, control HPV16 retained a strong E1 E4 mRNA expression signal (Fig. 3B). In parallel, we inoculated HaCaT cells with 0.1 μg of unencapsidated HPV16 genomic DNA, a 200-fold excess over the level of HPV16 DNA in the gradient-purified, DNase-treated virion preparations that successfully initiate infection. As expected, even this great excess of unencapsidated HPV16 genomic DNA did not lead to any PCR-detectable expression of E1 E4 mRNAs (Fig. 3B), showing that successful infection and viral gene expression under these conditions require HPV16 DNA encapsidation.
To validate further that we were producing infectious HPV particles, we monitored the effect of pretreating the HPV16 virion preparations with L1-specific HPV16 neutralizing antibodies, H16.7E, H16.E70, and H16.V5 (15, 30, 39), before inoculating HaCaT cells. Each of these neutralizing antibodies efficiently blocked the ability of the HPV preparations to induce E1 E4 mRNA expression, whereas virus incubated with non-specific isotype control antibodies such as nonneutralizing anti-HPV16L1 and HIV-1 p24 antibodies, showed unimpaired infectivity (Fig. 3C). Thus, the susceptibility of our infectious preparations from HPV DNA-transfected 293TT cells to chemical disruption and highly specific neutralizing antibodies paralleled that of natural HPV virions.
To examine the minimum HPV16 genome sequence required for early gene expression, we infected HaCaT cells with virion particles containing the 5.0-, 5.9-, and 6.8-kb HPV16 genome derivatives. All three derivatives showed expression of E1 E4 spliced mRNA in HaCaT cells (Fig. 4A), whereas HPV31b- and mock-infected cells did not, suggesting that the HPV16 L1 and L2 ORF sequences are not necessary for early viral gene transcription.
Fig. 4.
Induction of early gene expression by encapsidated HPV16 genome derivatives and infectivity of encapsidated, full-length HPV31b genomic DNA. Virions were prepared as in Fig. 2 by cotransfecting HPV16 L1- and L2-expression plasmids and either the HPV16 genome deletion derivatives of Fig. 1 A (A) or the HPV31b full-length genome (B), and they were inoculated onto HaCaT cells at 100 vge per cell. After a 48-h incubation, cells were harvested, total RNA was isolated, and nested RT-PCR was performed with PCR primers to detect E1 E4 spliced mRNAs of HPV16 (A) or HPV31b (B) (see Table 1).
Infectivity of Packaged HPV31b. As noted above, nuclease treatment and gradient fractionation showed that HPV16 L1 and L2 capsid proteins successfully encapsidated the full-length HPV31b genomic DNA. To test the infectivity of HPV31b genome packaged in HPV16 virus particles, we added these chimeric virions to HaCaT cells and performed RT-PCR with HPV31b-specific primers. The expected signal for the E1 E4 spliced mRNAs of HPV31b was revealed from extracted total RNA by one round of amplification, whereas HaCaT cells infected with virions made by cotransfecting L1 and L2 expression plasmids with full-length HPV16 genomes or no HPV genomic DNA did not express HPV31b RNA transcripts (Fig. 4B). These results indicate that mammalian cell transfection for HPV packaging is not restricted to one genotype but could be applied to other high- and low-risk HPV genotypes.
Discussion
HPVs are of major clinical importance because they are the most common sexually transmitted pathogen and cause prevalent human cancers. The dependence of HPV replication on epithelial cell differentiation has greatly limited the ability to produce infectious HPV for both basic and clinical research. Here, we have described a facile method for efficiently producing large quantities of HPV particles that display normal infectious properties. This method offers significant advantages over prior methods such as organotypic cultures by providing an >1,000-fold increased yield of virus in much less time. In addition to 2 days for transfection and cell growth, this method requires some additional time to isolate HPV expression plasmids and genomic DNA for transfection, but sufficient DNA for many transfections can be prepared at once. Virus of any desired genotype can be generated, and furthermore, because there is no reliance on replication of the viral genome in the cells in which packaging occurs, one can encapsidate HPV DNAs bearing essentially any desired mutation.
Contrary to the original transient-transfection-based study, which found that the efficiency with which BPV-1 capsids encapsidated target DNAs fell off rapidly for DNAs of >6-7 kb (19), we found that full-length, 7.9-kb HPV16 and HPV31 genomes were incorporated into HPV16 L1 and L2 capsid proteins as efficiently as 5- to 7-kb deletion derivatives (Fig. 2B) or smaller reporter plasmids. This result suggests either that the HPV16 based capsids produced here have different packaging constraints than the BPV1 capsids produced in the earlier study, or that differences in the DNA being packaged contribute to differences in packaging efficiency. The latter possibility could reflect the presence of packaging signals in the HPV genome that permit efficient packaging of full-length or nearly full-length genomes. A region of the BPV1 genome has been reported to contribute to DNA encapsidation (40). However, this region conferred only a modest increase in packaging efficiency in the BPV1 L1/L2-based, transient-transfection assay (19). Whether this and/or other regions of HPV genomes confer efficient encapsidation of full-length viral genomes in the HPV16 L1/L2-based transient-transfection assays described here is worthy of further investigation. The packaging efficiencies of full-length versus truncated versions of the HPV16 genome suggests that any such packaging signal(s) is not located within the L1 and L2 ORFs. An alternative hypothesis is that non-HPV-derived sequences can inhibit packaging efficiency. Regardless of the reason, whereas prior results indicated that in vivo encapsidation by papillomavirus capsid proteins was promiscuous with respect to target DNA sequence and driven primarily by a size discrimination mechanism predicted to greatly limit encapsidation of full-length viral genomes (19), we find that size discrimination does not prevent efficient encapsidation of full-length HPV genomes.
Although HPV16 is the most prevalent genotype in HPV pathogenesis and oncogenesis, other HPVs including high- and low-risk mucosotropic virus such as HPV6, 11, 18, and 31 are of clinical importance because they cause sexually transmitted disease, and for the high-risk genotypes, cancer (1, 5). Thus, it would also be valuable if a single approach could produce infectious virions for multiple HPV genotypes. Toward this end we demonstrated the ability to generate infectious HPV16 L1/L2 virus particles harboring either HPV16 or HPV31 genomes. Furthermore, the transient-transfection-based system described here should produce virtually any papillomavirus of interest in high yield, because the L1 and L2 proteins of all other tested papillomaviruses can self-assemble into capsids, and because highly efficient expression of papillomaviral proteins in mammalian cells can now be routinely achieved by using codon optimized versions of the cognate genes (22, 41, 42). In addition to providing infectious virus for basic research studies on other papillomavirus genotypes, such infectious HPV stocks would be highly useful for evaluating the successful generation of neutralizing antibodies to other genotypes in preclinical and clinical vaccine studies.
The abilities presented here to rapidly produce infectious HPV virions in large amounts and, moreover, to incorporate essentially any desired mutation in the HPV DNA so packaged (e.g., Fig. 2 A and B and data not shown), make possible previously unapproachable studies of virus-host interactions in early phases of the HPV life cycle. For example, although some potential cellular receptors for HPV entry have been identified (43, 44), their relevance to establishment of clinical HPV infections remains controversial. The procedures described here would allow biochemical testing of virus attachment to various candidate cell-surface receptors by using infectious particles, as well as mutagenesis, to map and characterize cell binding motifs in HPV capsid proteins. Also, wild-type HPV or HPV derivatives prepared with the methods presented here could be used in experiments to identify and characterize host factors critical for virus entry and other steps crucial for initiating infection and gene expression by the full HPV genome, such as uncoating, virion protein-mediated HPV DNA trafficking to nucleus, initial transcription, etc. Understanding these mechanisms should identify therapeutic targets for antiviral drug development.
Another important advantage is that, because the procedures described here encapsidate HPV genomes directly isolated from bacteria, such DNAs could contain bacterially directed Dam and Dcm methylation. Because mammalian host cells lack such methylation, this approach allows discrimination by methylation-specific restriction enzymes between the infecting HPV founder genomes and subsequently replicated copies (45). This approach would facilitate studies of early steps in viral DNA replication after infection, about which little is known.
Current HPV vaccine strategies are directed to raising neutralizing antibodies to prevent initial HPV infection. The approaches described here open the possibility of producing live, attenuated HPV vaccines to more effectively trigger both humoral and cell-mediated immune responses. Such cell-mediated immune responses, including cytotoxic T cell activation, may provide more effective prophylaxis against papillomaviral infections and also offer therapeutic potential for the large numbers of patients who are already infected with HPV.
The ability to package HPV genomes with engineered changes into infectious HPV virions also should facilitate the study of other important aspects of immune responses to HPV. Some HPV16 early genes (such as E5, E6, and E7) have been reported to have immunosuppressive effects (46, 47), implying that host immune responses against intact infectious HPV might differ radically from those against empty HPV capsids. Such differences can now be evaluated.
Supplementary Material
Acknowledgments
We thank Chris Buck and John Schiller for helpful comments and pXULL plasmid and 293TT cells, Martin Muller for the pcDNA3.1(+)-HPV16L1 and pcDNA3.1(+)-HPV16L2 plasmids, Neil Christensen for antibodies against HPV16 L1, and Michelle Ozbun and members of our laboratory for comments on the manuscript. This work was supported by National Cancer Institute Grant CA22443. P.A. is an Investigator of the Howard Hughes Medical Institute.
Author contributions: D.P., P.F.L., and P.A. designed research; D.P. performed research; D.P., P.F.L., and P.A. analyzed data; and D.P., P.F.L., and P.A. wrote the paper.
Abbreviations: HPV, human papillomavirus; BPV, bovine papillomavirus; SEAP, secreted alkaline phosphatase; vge, viral genome equivalents.
References
- 1.Burd, E. M. (2003) Clin. Microbiol. Rev. 16, 1-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Longworth, M. S. & Laimins, L. A. (2004) Microbiol. Mol. Biol. Rev. 68, 362-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McMurray, H. R., Nguyen, D., Westbrook, T. F. & McAnce, D. J. (2001) Int. J. Exp. Pathol. 82, 15-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.zur Hausen, H. (1996) Biochim. Biophys. Acta 1288, F55-F78. [DOI] [PubMed] [Google Scholar]
- 5.zur Hausen, H. (1999) Eur. J. Cancer 35, 1878-1885. [DOI] [PubMed] [Google Scholar]
- 6.Gillison, M. L. & Shah, K. V. (2001) Curr. Opin. Oncol. 13, 183-188. [DOI] [PubMed] [Google Scholar]
- 7.Gillison, M. L. & Lowy, D. R. (2004) Lancet 363, 1488-1489. [DOI] [PubMed] [Google Scholar]
- 8.Hunter, K. D., Parkinson, E. K. & Harrison, P. R. (2005) Nat. Rev. Cancer 5, 127-135. [DOI] [PubMed] [Google Scholar]
- 9.Stubenrauch, F. & Laimins, L. A. (1999) Semin. Cancer Biol. 9, 379-386. [DOI] [PubMed] [Google Scholar]
- 10.zur Hausen, H. (2002) Nat. Rev. Cancer 2, 342-350. [DOI] [PubMed] [Google Scholar]
- 11.Lowy, D. R. & Howley, P. M. (2001) in Fields Virology, eds. Knipe, D. M. & Howley, P. M. (Lippincott-Raven, Philadephia), Vol. 2, pp. 2231-2264. [Google Scholar]
- 12.Dollard, S. C., Wilson, J. L., Demeter, L. M., Bonnez, W., Reichman, R. C., Broker, T. R. & Chow, L. T. (1992) Genes Dev. 6, 1131-1142. [DOI] [PubMed] [Google Scholar]
- 13.Meyers, C., Frattini, M. G., Hudson, J. B. & Laimins, L. A. (1992) Science 257, 971-993. [DOI] [PubMed] [Google Scholar]
- 14.McLaughlin-Drubin, M. E., Wilson, S., Mullikin, B., Suzich, J. & Meyers, C. (2003) Virology 312, 1-7. [DOI] [PubMed] [Google Scholar]
- 15.McLaughlin-Drubin, M. E., Christensen, N. D. & Meyers, C. (2004) Virology 322, 213-219. [DOI] [PubMed] [Google Scholar]
- 16.Bonnez, W., DaRin, C., Borkhuis, C., de Mesy Jensen, K., Reichman, R. C. & Rose, R. C. (1998) J. Virol. 72, 5256-5261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sexton, C. J., Williams, A. T., Topley, P., Shaw, R. J., Lovegrove, C., Leigh, I. & Stables, J. N. (1995) J. Gen. Virol. 76, 3107-3112. [DOI] [PubMed] [Google Scholar]
- 18.McBride, A. A., Dlugosz, A. & Baker, C. C. (2000) Proc. Natl. Acad. Sci. USA 97, 5534-5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buck, C. B., Pastrana, D. V., Lowy, D. R. & Schiller, J. T. (2004) J. Virol. 78, 751-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flores, E. R., Allen-Hoffmann, B. L., Lee, D., Sattler, C. A. & Lambert, P. F. (1999) Virology 262, 344-354. [DOI] [PubMed] [Google Scholar]
- 21.Hummel, M., Hudson, J. B. & Laimins, L. A. (1992) J. Virol. 66, 6070-6080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leder, C., Kleinschmidt, J. A., Wiethe, C. & Muller, M. (2001) J. Virol. 75, 9201-9209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Genther, S. M., Sterling, S., Duensing, S., Munger, K., Sattler, C. & Lambert, P. F. (2003) J. Virol. 77, 2832-2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boukamp, P., Petrussevska, R. T., Breitkreutz, D., Hornung, J., Markham, A. & Fusenig, N. E. (1988) J. Cell Biol. 106, 761-771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jeon, S., Allen-Hoffmann, B. L. & Lambert, P. F. (1995) J. Virol. 69, 2989-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doorbar, J., Parton, A., Hartley, K., Banks, L., Crook, T., Stanley, M. & Crawford, L. (1990) Virology 178, 254-262. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook, J. & Russell, D. W. (2001) in Molecular Cloning, eds. Sambrook, J. & Russell, D. W. (Cold Spring Harbor Lab. Press, Woodbury, NY), Vol. 1, pp. 6.23-6.27. [Google Scholar]
- 28.Rozen, S. & Skaletsky, H. J. (2000) in Bioinformatics Methods and Protocols: Methods Mol. Biol., eds. Krawetz, S. & Misener, S. (Humana, Totowa, NJ), pp. 365-386.
- 29.McCarthy, M. P., White, W. I., Palmer-Hill, F., Koenig, S. & Suzich, J. A. (1998) J. Virol. 72, 32-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Christensen, N. D., Dillner, J., Eklund, C., Carter, J. J., Wipf, G. C., Reed, C. A., Cladel, N. M. & Galloway, D. A. (1996) Virology 223, 174-184. [DOI] [PubMed] [Google Scholar]
- 31.Chesebro, B., Wehrly, K., Nishio, J. & Perryman, S. (1992) J. Virol. 66, 6547-6554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Toohey, K., Wehrly, K., Nishio, J., Perryman, S. & Chesebro, B. (1995) Virology 213, 70-79. [DOI] [PubMed] [Google Scholar]
- 33.Holmgren, S. C., Patterson, N. A., Ozbun, M. A. & Lambert, P. F. (2005) J. Virol. 79, 3938-3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.White, W. I., Wilson, S. D., Bonnez, W., Rose, R. C., Koenig, S. & Suzich, J. A. (1998) J. Virol. 72, 959-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ozbun, M. A. (2002) J. Virol. 76, 11291-11300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ozbun, M. A. (2002) J. Gen. Virol. 83, 2753-2763. [DOI] [PubMed] [Google Scholar]
- 37.Lee, J. H., Yi, S. M. P., Anderson, M. E., Berger, K. L., Welsh, M. J., Klingerhutz A. J. & Ozbun, M. A. (2004) Proc. Natl. Acad. Sci. USA 101, 2094-2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Patterson, N. A., Smith, J. L. & Ozbun, M. A. (2005) J. Virol. 79, 6838-6847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.White, W. I., Wilson, S. D., Palmer-Hill, F. J., Woods, R. M., Ghim, S. J., Hewitt, L. A., Goldman, D. M., Burke, S. J., Jenson, A. B., Koenig, S. & Suzich, J. A. (1999) J. Virol. 73, 4882-4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou, J., Stenzel, D. J., Sun, X. Y. & Frazer, I. H. (1993) J. Gen. Virol. 74, 763-768. [DOI] [PubMed] [Google Scholar]
- 41.Zhou, J., Liu, W. J., Peng, S. W., Sun, X. Y. & Frazer, I. (1999) J. Virol. 73, 4972-4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mossadegh, N., Gissmann, L., Muller, M., Zentgraf, H., Alonso, A. & Tomakidi, P. (2004) Virology 326, 57-66. [DOI] [PubMed] [Google Scholar]
- 43.Shafti-Keramat, S., Handisurya, A., Kriehuber, E., Meneguzzi, G., Slupetzky, K. & Kirnbauer, R. (2003) J. Virol. 77, 13125-13135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Selinka, H. C., Giroglou, T., Nowak, T., Christensen, N. D. & Sapp, M. (2003) J. Virol. 77, 12961-12967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brune, W. & Durst, M. (1995) J. Invest. Dermatol. 104, 277-281. [DOI] [PubMed] [Google Scholar]
- 46.Barnard, P. & McMillan, N. A. (1999) Virology 259, 305-313. [DOI] [PubMed] [Google Scholar]
- 47.Borchers, A., Braspenning, J., Meijer, J., Osen, W., Gissmann, L. & Jochmus, I. (1999) Arch. Virol. 144, 1539-1556. [DOI] [PubMed] [Google Scholar]
- 48.Buck, C. B., Thompson, C. D., Pang, Y. S., Lowy, D. R. & Schiller, J. T. (2005) J. Virol. 79, 2839-2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.