Abstract
Predicting elemental cycles and maintaining water quality under increasing anthropogenic influence requires knowledge of the spatial drivers of river microbiomes. However, understanding of the core microbial processes governing river biogeochemistry is hindered by a lack of genome-resolved functional insights and sampling across multiple rivers. Here we used a community science effort to accelerate the sampling, sequencing and genome-resolved analyses of river microbiomes to create the Genome Resolved Open Watersheds database (GROWdb). GROWdb profiles the identity, distribution, function and expression of microbial genomes across river surface waters covering 90% of United States watersheds. Specifically, GROWdb encompasses microbial lineages from 27 phyla, including novel members from 10 families and 128 genera, and defines the core river microbiome at the genome level. GROWdb analyses coupled to extensive geospatial information reveals local and regional drivers of microbial community structuring, while also presenting foundational hypotheses about ecosystem function. Building on the previously conceived River Continuum Concept1, we layer on microbial functional trait expression, which suggests that the structure and function of river microbiomes is predictable. We make GROWdb available through various collaborative cyberinfrastructures2,3, so that it can be widely accessed across disciplines for watershed predictive modelling and microbiome-based management practices.
Subject terms: Microbiome, Water microbiology, Carbon cycle, Metagenomics
GROWdb defines US river microbiomes at the genome level.
Main
Earth’s surface is dominated by water, much of it the oceans, that is known to buffer against anthropogenic climate change through microorganisms dictating the fate of ocean-absorbed carbon4,5. Although the oceans and their microorganisms have been extensively studied globally by large scientific consortia (such as the Tara Oceans Consortium6), other elements of Earth’s water system, such as rivers, are relatively understudied. This is problematic, as rivers (1) offer an important nexus of nutrient transport across terrestrial and aquatic interfaces7; (2) are hotspots for biogeochemical processes that contribute substantially to global terrestrial carbon and nitrogen budgets, ultimately influencing global greenhouse gas emissions, eutrophication and acidification7–9; and (3) have immediate societal impacts on sustainable energy, agriculture, environmental health and human health10,11. Microbial metabolisms dictate river ecosystem functioning with major influence on carbon (C) respiration and sequestration, nitrogen (N) cycling and uptake, food webs and pollutants12–14. Given these important contributions, there is a growing need to better resolve the ecology and biogeochemical contributions of microorganisms across diverse river systems.
Despite being critical modulators of biogeochemistry, river microbiomes remain undersampled15. For example, a majority of river microbiome studies relies on 16S rRNA gene analysis (Supplementary Data 1). Although these single-gene studies have advanced understanding of riverine microbial community diversity and membership16–18, they lack information on poorly characterized lineages and are limited in their ability to functionally link microorganisms to biogeochemical processes. There are several studies with metagenomics (n = 49) that provide functional attributes of river microbiomes, but these rarely recover metagenome-assembled genomes (MAGs), masking the contributions of novel members of the microbiome. Three studies used genome-resolved expression methods, hindering the ability to estimate the metabolic processes active in river systems (Supplementary Data 1). Finally, in terms of sampling, most studies focus on a single site or stream network, leaving the applicability of microbiome findings across river systems uncertain. To establish a transferable functional understanding of river microbiomes, there is a need to genomically resolve the taxonomy, metabolic potential and expression of river microbiomes at scale.
To meet this need, we developed a crowd-sourced, distributed sampling effort to increase and standardize river surface water microbiome sampling. We then compiled these sequencing results, along with their paired geospatial data, into the large-scale GROWdb. An emphasis of GROWdb is a publicly available and ever-expanding microbial genome database. GROWdb represents, to our knowledge, the first microbial, river-focused resource parsed at various scales from genes, to MAGs, to the community level, including genome and expression-based measurements. GROWdb is based on a crowd-sourced, network-of-networks approach to move beyond a small collection of well-studied rivers, towards a spatially distributed, global network of systematic observations.
Construction of GROWdb
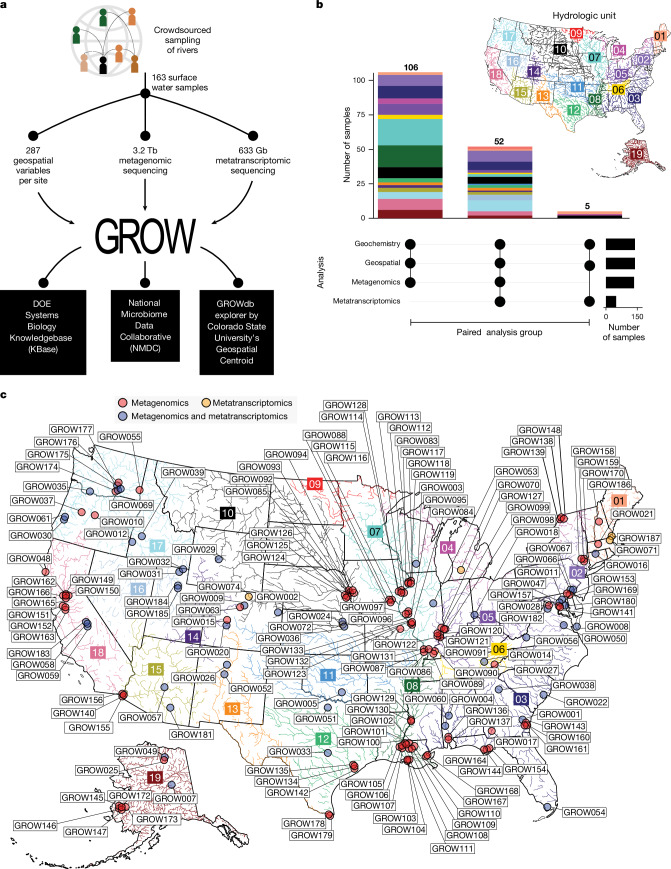
To establish the GROWdb, more than 100 teams were crowdsourced to collect 163 samples at 106 sites across US rivers, with teams chosen on the basis of field site locations (Methods). This approach led to around 3.8 terabases (Tb) of metagenomic and metatranscriptomic sequencing data to go with extensive (up to 287) geochemical and geospatial measurements at each site (Fig. 1a,b and Supplementary Data 1). Geospatial parameters were obtained using latitude and longitude for sampling locations as queries and included land use and other watershed characteristics (for example, stream order, watershed size), while geochemical information was collected at the same time as sampling (Methods). Through this process, we aimed to capture community-level, genome-resolved microbiome variations in taxonomy, function and gene expression in the context of geographical and environmental gradients across the United States. The effort resulted in surface water sampling that covered 90% of US watersheds (n = 21 as determined by hydrologic unit 2) (Fig. 1c) and spanned diverse ecoregions, stream orders and watershed sizes (Extended Data Fig. 1). In summary, GROWdb integrates genomics, biogeochemistry and a range of contextual environmental variables to enable a predictive framework of microbiomes and their biogeochemical contributions.
Fig. 1. Distributed sampling and sequencing of rivers enabled the construction of the GROWdb.
a, The workflow, denoting the number of samples and the resulting datasets made up of geospatial and microbiome (metagenomics, metatranscriptomics) data. GROWdb data are accessible through KBase, NMDC and the GROWdb Explorer. b, The number of samples with paired data types (denoted as filled black circles below) coloured by hydrologic unit, and the number of samples per analysis. c, GROW sampling across the United States. The points mark the sampling location. Colour coding represents the microbiome analysis performed (metagenomics, red; metatranscriptomics, yellow; paired metagenomics and metatranscriptomics, blue). The boxed numbers and the corresponding river colours indicate hydrologic unit (HUC-2).
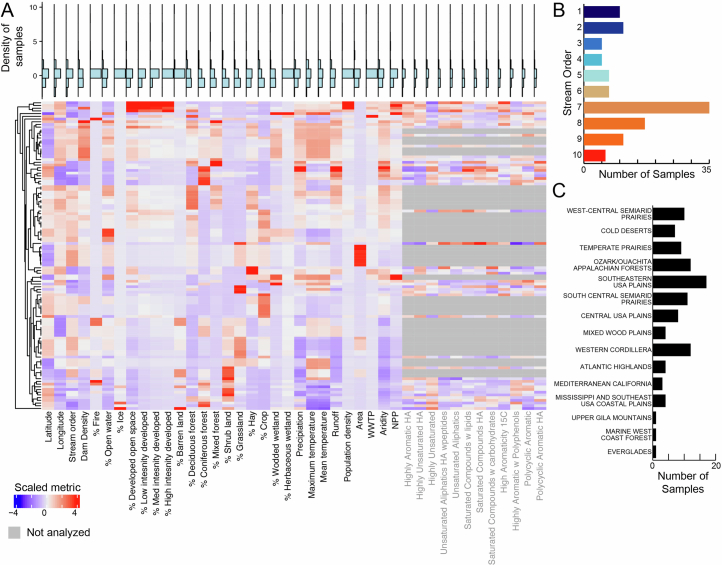
Extended Data Fig. 1. Distribution of river characteristics sampled in GROWdb.
A) Heatmap of geospatial and chemical parameters sampled in GROWdb, where columns are environmental and variables and rows are corresponding samples within GROWdb. Each variable has been scaled by subtracting the vector mean for each variable and dividing by its standard deviation. Variables in grey text were determined by FTICR. Blue histogram plots above highlight the distribution of samples for each variable, with high values at the top of the plot. Histogram plots of key variables used throughout the main text including stream order (B) and ecoregion (C) are also shown.
To ensure data accessibility, we provide four access points for user engagement with GROWdb (Fig. 1a). First, all reads and MAGs are publicly hosted at the National Center for Biotechnology (NCBI), enabling transferability to resources that pull and incorporate this content. Datasets underlying GROWdb are freely available and searchable through the National Microbiome Data Collaborative (NMDC)2 data portal, linking to other data types (for example, metabolome) to allow for broader synthesis where available. GROWdb MAGs are available as an annotated genomic collection in the freely accessible KBase3 cyberinfrastructure. Here users can access sample information and gene- and MAG-level annotations, profile functional summaries and genome-scale models in a point-and-click interface. Last, to help with data exploration, we distilled the taxonomic and functional insights from GROWdb into a web-accessible format called GROWdb Explorer, enabling the rapid profiling of taxonomic and functional distributions across the dataset. GROWdb version 1 can be accessed across platforms (Fig. 1a), making this microbiome content available in an expanding repository to incorporate and unify global river multi-omic data for the future.
Over 3,000 surface-water MAGs recovered
To identify the key microbial players and functions in surface water river microbiomes, we constructed a genome database composed of MAGs. Our sequencing represents, on average, threefold more sequencing per sample compared with published riverine metagenome studies, thereby increasing the sensitivity for detecting the breadth of microbial functions encoded in these systems (Extended Data Fig. 2). From these sequencing data, we assembled and reconstructed 3,825 medium- and high-quality MAGs, which were dereplicated into 2,093 MAGs at 99% identity (Extended Data Fig. 3 and Supplementary Data 2). On the basis of read mapping, the majority (mean, 52%) of metagenomic reads mapped back to this surface-water-derived MAG database, signifying that the underlying sequencing reads were well represented by the genomic database.
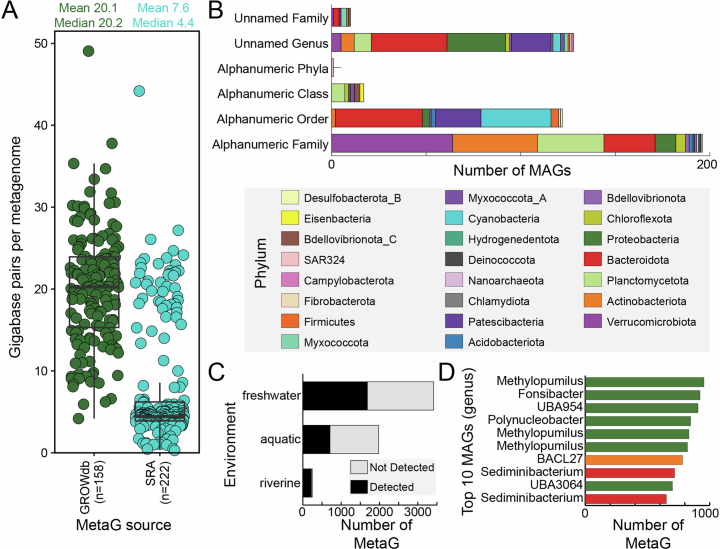
Extended Data Fig. 2. GROWdb comparison to other metagenomics data sources.
A) Sequencing depth comparison of GROWdb metagenomes (n = 158) to SRA metagenomes classified as riverine (n = 222) shows 3x increase in average sequencing depth for GROWdb. Each point represents a single metagenome, with mean and median values listed at the top of the graph. For the boxplot, upper and lower box edges extend from the first to third quartile and the line in the middle represents the median. The whiskers are 1.5 times the interquartile range and every point outside this range represents an outlier. B) Stacked bar chart shows novelty of GROWdb MAGs when compared to GTDB (r207). Each MAG was placed at the highest level of novelty, with no assignment within a taxonomic level (e.g., unnamed family or genus) being highest level of novelty and alpha numeric identifiers being the second highest (e.g., UBA lineages). Bars are coloured by Phylum. C) Stacked bar chart shows the proportion of SRA metagenomes that a GROWdb lineage (95% identity) was detected (black) or not detected (grey) within an SRA environment category. D) The top ten MAGs most frequently detected across river surface water related SRA metagenomes are displayed at the genus level on the bar chart, with colours denoting phyla (key above).
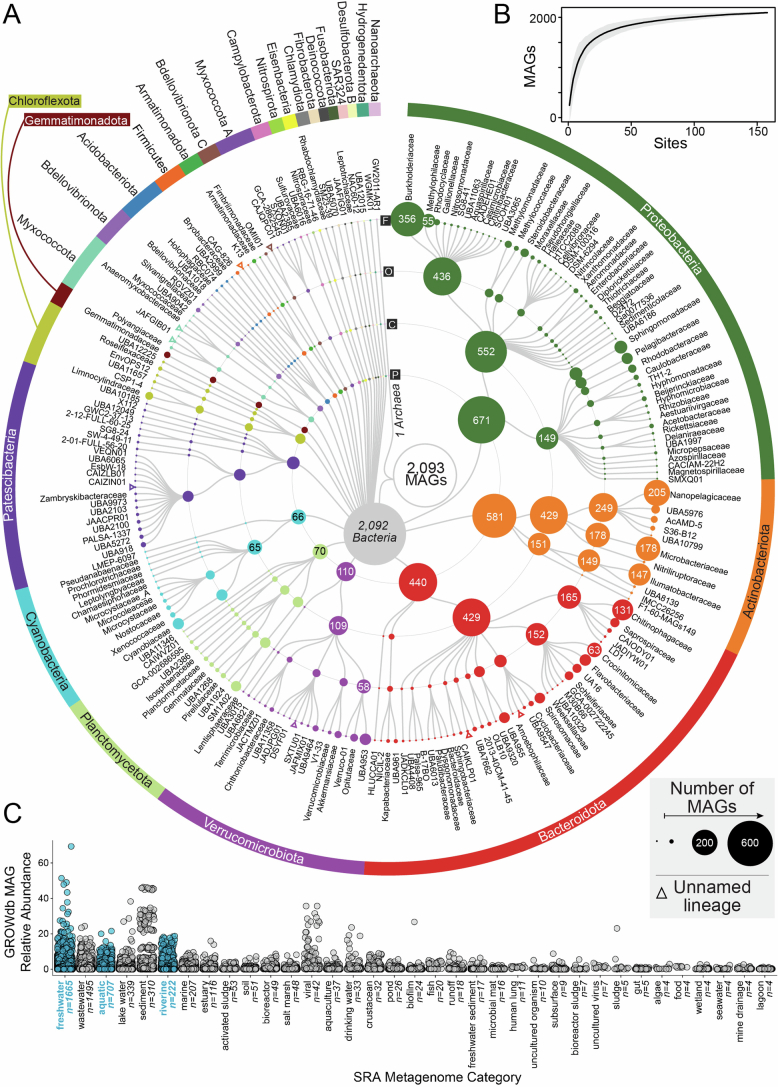
Extended Data Fig. 3. Taxonomic diversity of 2,093 unique surface water metagenome assembled genomes (MAGs) in GROWdb.
A) Cladogram shows GROWdb MAGs taxonomy with each sequential ring noting taxonomy level (Phylum, P; Class, C; Order, O; Family, F). Circle size indicates the number of genomes within a given taxonomy level and is further noted by MAG number inside the circle when sampling at that taxonomic position exceeds 50 MAGs sampled. Colours highlight phylum level taxonomy denoted on the outermost ring. Open triangles represent unnamed lineages within a particular level of taxonomy. B) MAG accumulation curve where the black line represents the mean richness of 100 random permutations and grey shading represents standard deviation. C) One dimensional boxplot displays the environments GROWdb MAGs were detected in across 266,764 metagenomes in the Sequence Read Archive with each point representing a single MAG. Upper and lower box edges extend from the first to third quartile and the line in the middle represents the median. The whiskers are 1.5 times the interquartile range and every point outside this range represents an outlier. Environments are ordered by number of metagenomes GROWdb MAGs were detected in from left to right, with the number of metagenomes also noted along the x-axis. Freshwater related environments are highlighted in blue.
The dereplicated MAG database (n =2,093) contained genomes from 27 phyla, many of which represent the most abundant and cosmopolitan lineages in rivers19–21. Beyond providing genomic resources for these ecologically known taxa, the GROWdb MAGs provide genomic resources for many less-well-known taxa. A subset of our genomes represented novel lineages, including 10 families and 128 genera across 16 phyla (Extended Data Figs. 2 and 3). Moreover, a large proportion of MAGs belonged to lineages defined only by alphanumeric names (for example, uncultured bacterial and archaeal genomes, UBA22) at the phylum (n = 1), class (n = 17), order (n = 121) and family (n = 196) levels (Extended Data Fig. 2). Notably, a MAG accumulation analysis suggests comprehensive sampling of river surface water microbial communities (Extended Data Fig. 3). To compare GROWdb MAGs in this study derived from US watersheds, we have compiled MAGs from other biogeography studies with freshwater MAGs23–25, as well as 23 GROW metagenomes from sites outside the United States (Supplementary Note 1). This meta-analysis revealed vast differences in genomic membership between lakes and rivers, and the relative undersampling of rivers compared to lakes (Extended Data Fig. 4). Together these findings underscore the importance of analysing river metagenomes across varied geographical and environmental gradients to recover the breadth of river bacterial diversity.
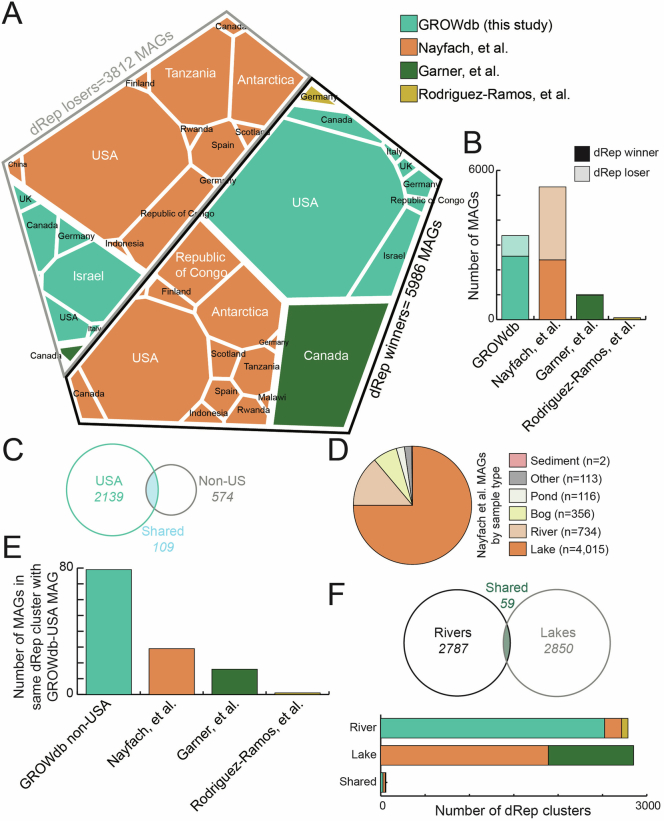
Extended Data Fig. 4. GROWdb comparison to global freshwater MAGs.
In order to compare GROWdb MAGs in this study derived from United States watersheds, we have compiled MAGs from other biogeography studies with freshwater MAGs24–26, as well as included 1,286 additional MAGs derived from 23 metagenomes released in this study, including 6 countries beyond the United States (UK, Canada, Italy, Germany, Israel, Republic of the Congo). A) Area plot shows the dereplication results of 9,798 MAGs from freshwater sources (lakes and rivers) at 99% identity. This dereplication status, with winner defined as the best MAG representative of the cluster, is reported by outline colour with black outline denoting winner and grey outline denoting loser. The area plot within these sections has area size proportional to MAGs recovered, divided first by study (colour in legend), then by country (noted on area plot). B) Stacked bar chart summarizes area plot in A by study, with GROW contributing the most representative MAGs (dRep winners). C) Venn diagram shows the number of MAG representatives (dRep winners) derived from rivers only (does not include lakes) by location, with USA being compared to Non-US sites. Circle area is sized by number of MAGs. D) Nayfach, et al. is a comprehensive catalogue of Earth’s microbiomes that includes 52,215 MAGs, of which we retain 5,336 MAGs from aquatic freshwater environments for our global comparison, excluding soil, sediment, and wastewater related habitats within the aquatic freshwater ecosystem to more directly compare to surface water GROWdb. Pie chart shows the breakdown of sample types for this subset of MAGs, highlighting that a majority are derived from lake systems. E) Bar chart shows clustering of MAGs from other studies with GROWdb non-US studies, with bars coloured by study. F) Venn diagram compares the number of MAG representatives (dRep winners) derived from rivers and lakes across studies, with circle area sized by number of MAGs. Stacked bar chart below summarizes these results by study. All data for this comparison is reported in Supplementary Data 2 (tab 5), with MAG files available at Zenodo.
To highlight the relevance of GROWdb, we analysed 266,764 public metagenome datasets in the Sequence Read Archive (SRA) to reveal that GROWdb MAGs were detected in 90% of metagenomes classified as riverine and 46% of metagenomes classified as freshwater, aquatic or riverine. We verified that the most prevalent phyla and genera in GROWdb had parallel representation in publicly available metagenomes (Extended Data Fig. 2). Moreover, GROWdb members were detected from other environments including wastewater, lake water, sediment, marine, estuary, activated sludges and soil, supporting the notion that rivers contain diverse communities across habitats acting as integrators across landscapes (Extended Data Fig. 3). Likewise, consistent with other studies25, GROWdb MAGs showed minimal overlap with sediment metagenomes, with 16% of MAGs being detected in this interconnected yet distinct river compartment. This affirms the growing distinction between surface water and sediment microbial communities, further articulating how suspended surface water microorganisms probably originate from diverse, non-native sources. The comparison to publicly available data also underscored the need for this river-based microbiome study, as there were only half and one-third as many freshwater-related metagenomes in comparison to their soil and ocean counterparts, respectively, in the SRA. Moreover, this analysis highlighted the importance of standardized metadata practices for data reuse, as more than 10% of metagenomes in the publicly available set had vague classifications such as metagenome or bacterium, making the data unusable. GROWdb ascribes to standardized protocols and metadata practices26,27, making interoperability a hallmark of this resource and permitting meta-analysis with other studies, which is of utmost importance as our ability to scale multi-omics methods rapidly increases.
Core river microbiome features
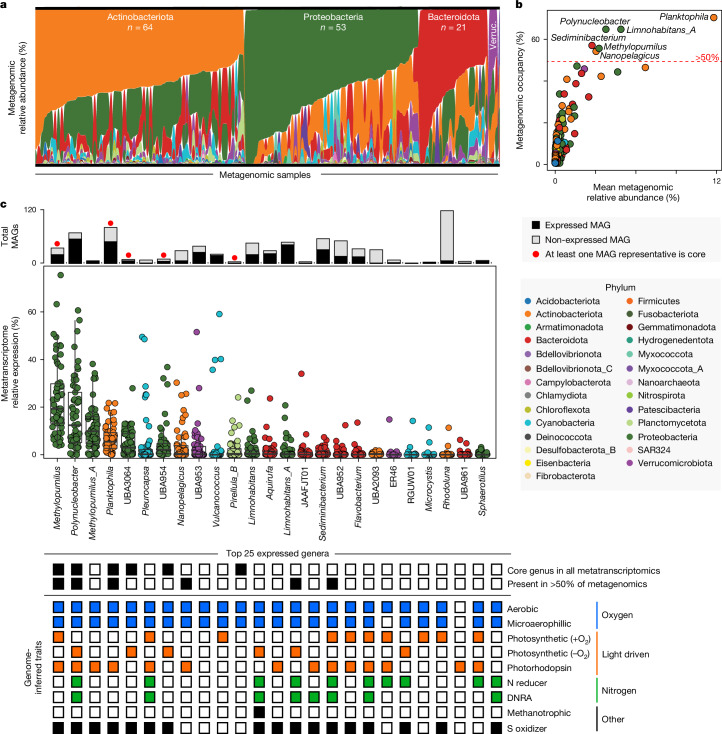
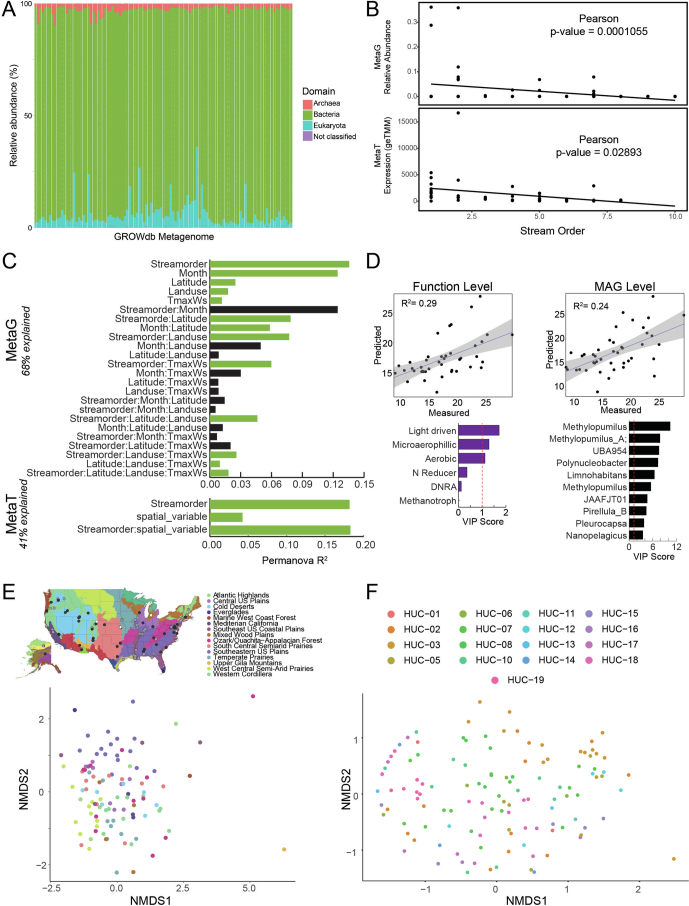
We identified core and dominant features of metagenomes and metatranscriptomes across rivers. In terms of relative abundance across microbiomes, members of the Actinobacteria, Proteobacteria, Bacteroidota and Verrucomicrobiota dominated all samples as determined by metagenomics (Fig. 2a). Within these phyla, genera that were the most cosmopolitan (high occupancy) across samples were also the most abundant members of these communities (Fig. 2b). This was especially true for MAGs affiliated with the genus Planktophilia, a well-known freshwater microorganism28, which were present in 70% of the GROW metagenomes and had the highest mean relative abundance across samples at 12%. Five other genera, including Limnohabitans_A, Polynucleobacter, Methylopumilus, Nanopelagicus and Sediminibacterium, were also present in more than 50% of metagenomes.
Fig. 2. Core lineages and functions across river microbiomes.
a, Phyla metagenomic relative abundance across samples, with each sample organized by the most dominant phyla from top to bottom along the y axis. The samples are grouped by the dominant phyla along the x axis. The Actinobacteriota, Proteobacteria, Bacteroidota and Verrucomicrobiota (Verruc.) phyla are the most dominant across samples. b, The metagenomic relative abundance versus metagenomic occupancy (the percentage of metagenomes that a genus was present in); the points represent each genus in GROWdb and are coloured by phylum. Genera detected in more than 50% of samples (red dashed line) are named. c, The top 25 most transcribed (highest metatranscriptomic expression) genera are shown by box plots, with each point representing a single metatranscriptome (n = 57 metatranscriptomes). The upper and lower box edges extend from the first to third quartile, the centre line represents the median and the whiskers are 1.5× the interquartile range; points outside this range represent outliers. The stacked bar chart above box plots indicates the number of MAGs in GROWdb within each genus and is coloured by detection in metatranscriptomes (black, expressed; grey, non-expressed). A red circle above the bar indicates that one of the genomes was core across metatranscriptomes, as defined as having gene expression in every sample. For each of the top 25 expressed genera, the black boxes represent those that were detected in 100% of metatranscriptomes (core genera) and in more than 50% of metagenomes. The inferred genomic potential of each genus is indicated below, including aerobic respiration (blue), light-driven energy metabolism (orange), nitrogen metabolism (green) and other metabolisms (methanotrophy and sulfur oxidation, black). DNRA, dissimilatory nitrate reduction to ammonium.
For the subset of samples with paired metatranscriptomes, we evaluated the microorganisms that were most transcriptionally active. To focus on the most relevant lineages, we limited our analyses to MAGs that were expressing genes in at least 10% of the samples. These resulted in a quarter of the 2,093 MAGs being considered active, including at least one representative from 19 out of the 27 phyla in GROWdb. The six most pertinent genera identified by metagenomics (Fig. 2b) also belonged to the top 25 genera with the highest mean gene expression (Fig. 2c), indicating that prevalence, dominance and activity were in agreement. Furthermore, three of these pertinent lineages (Methylopumilus, Polynucleobacter, Planktophilia), as well as members of Pirellula B, and two alphanumeric genera of Burkholderiaceae (UBA3064, UBA954) were transcriptionally active in every metatranscriptome, here denoted as the core, active genera. Notably, this was not an aggregate genus-level effect, because each of these genera apart from Polynucleobacter had a single MAG representative that was expressed in every metatranscriptome, indicating that some microbial strains have widespread metabolic activity across rivers. Here we show how analyses of GROWdb enable us to constrain the thousands of microbial genomes to a set of six genera with genes detected in every transcriptome, revealing lineages and metabolic pathways that could represent diagnostic or metabolism targets needing accurate representation in biogeochemical models moving forward.
To understand the effects of these core, transcriptionally active genera in modulating river biogeochemistry, we used genomic content to assign metabolic traits to each MAG, inventorying the capacity to use oxygen, light, nitrogen, sulfur and other key energy generation systems (Extended Data Fig. 5 and Supplementary Data 3). We found that the core and most expressed genera had the capacity for aerobic respiration and the use of light as an energy source, capturing energy through high-yield oxygenic or anoxygenic photosystems or simple, low-yield photorhodopsins. In fact, of the top 25 most active genera, more than 90% were capable of aerobic respiration or light-driven metabolism, with many encoding multiple light-harvesting mechanisms (Fig. 2c and Extended Data Fig. 6). In addition to heterotrophy and autotrophy, many of these core active lineages had the capacity to aerobically oxidize inorganic electron donors such as sulfur and possibly methane, the latter through a divergent particulate methane monooxygenase (Methods). Last, half of these most active genera had the capacity for nitrogen reduction through respiration or by dissimilatory nitrate reduction to ammonium (Methods). Together, the encoding of both aerobic and anaerobic energy systems, and light-driven metabolisms among the many core, active taxa highlight the metabolic redundancy contained in river surface waters.
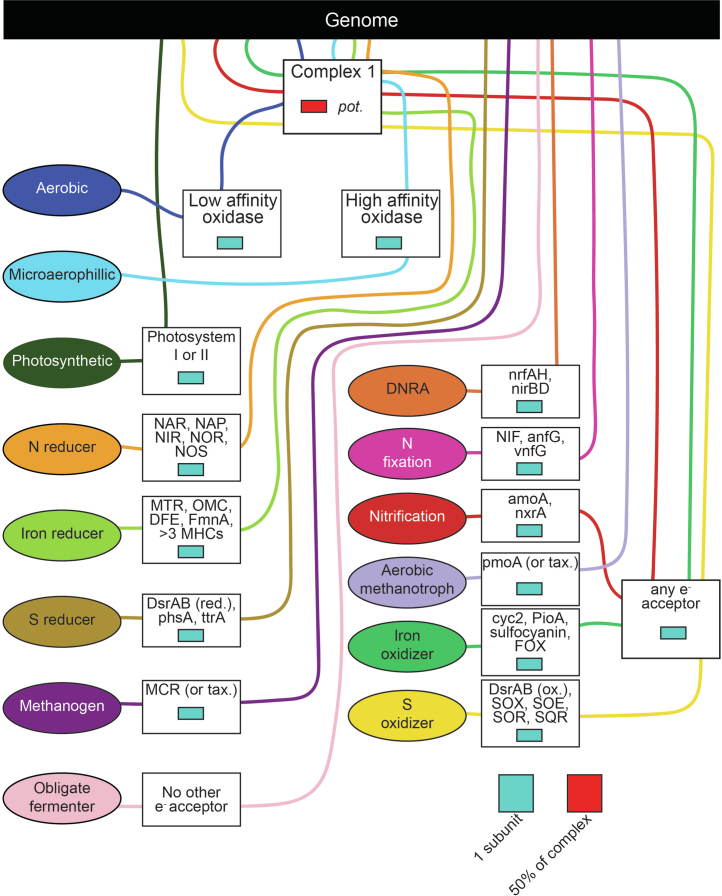
Extended Data Fig. 5. Metabolic trait assignment ruleset.
Each trait is defined by a set of genes and the percent of genes required for that function. Lines flow from the genome (top black box) to traits (ovals), passing through boxes of gene requirements to be consider TRUE for that particular trait.
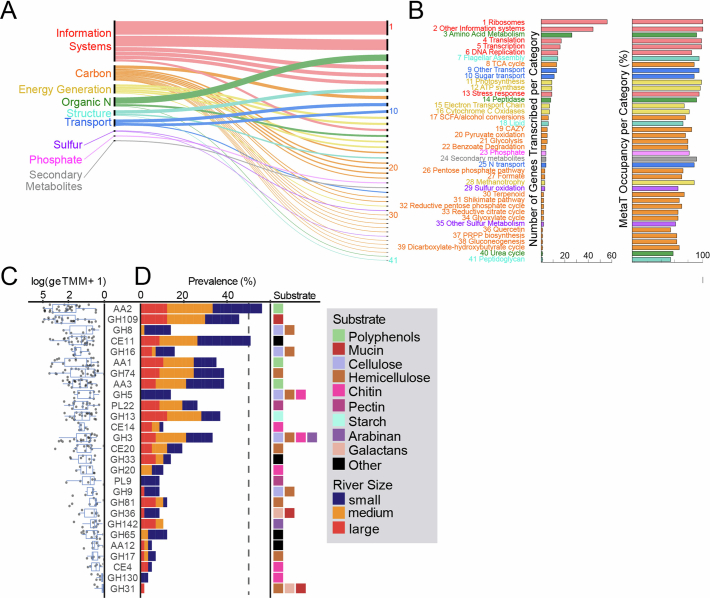
Extended Data Fig. 6. Gene level expression across rivers.
Genes detected in more than 50% of metatranscriptomes, with gene functions (n = 365) grouped by broad categories (n = 9, A) and refined to subcategories (n = 41, B). Thickness of lines and line order in A show the number of functions within a particular category (right) and subcategory (left). A and B are linked by subcategory number (1–41). For each of the 41 subcategories, the number of genes and occupancy defined as the percentage of samples detected across metatranscriptomes is shown by bar charts. Hypothetical and genes with unknown annotations are not shown, albeit 21 genes with these annotations were considered core or expressed in all metatranscriptomes. C) Focusing on carbon, carbohydrate-active enzyme (CAZyme) family gene expression is shown across river metatranscriptomes (n = 57) as log-transformed expression (geTMM). In the box plot, upper and lower box edges extend from the first to third quartile and the line in the middle represents the median. The whiskers are 1.5 times the interquartile range and every point outside this range represents an outlier. D) The prevalence of each CAZyme family across the metatranscriptomes is shown by stacked bar plots, which represent the fraction of river metatranscriptomes with expression for each family, with bar colour corresponding to river size as denoted in the legend. The dotted line marks 50% of metatranscriptome samples. At right, the substrate type for each CAZyme family is given based on the DRAM metabolism summary; see Shaffer and Borton et al for substrate logic91. If more than one box is present, the CAZyme family can act upon multiple substrate types.
Some critical river biogeochemical processes such as nitrification were represented by GROWdb MAGs but were not sampled in the top 25 most active genera. In surface waters, nitrification appeared to be catalysed by bacteria, a finding consistent with taxonomy profiles from our unassembled reads in which archaea made up less than 3% of the relative abundance across samples (Extended Data Fig. 7). We identified one MAG within the bacterial Nitrosomonas genus that encoded genes for ammonia oxidation (the first step in nitrification). We note this genome also included genes to produce the greenhouse gas nitrous oxide (N2O), a finding consistent with other ammonia oxidizing bacteria29.
Extended Data Fig. 7. GROWdb membership and structure across geospatial parameters.
A) Stacked bar chart of the singleM profiles of GROWdb metagenomic reads, with bars coloured by domain. By domain, the most reads are assigned to the Bacteria (mean=91.1%), followed by Eukaryota (mean=6.1%), Archaea (mean=2.6%), and Unknown (mean=0.2%). B) Correlations of Patescibacteria relative abundance (metagenomics, top) and expression (metatranscriptomics, bottom) with stream order. Correlation significance was tested in R using cor.test (two-sided), with p-values shown. C) Permutational analysis of variance (PERMANOVA) results for metagenomes (metaG) and metatranscriptomes (metaT) indicate that drivers of community structure and expression, respectively. These drivers and their interactions explain 68% of the metagenome and 41% of the metatranscriptome variance. Bar height represents the R2, with green bars denoting significant drivers (p < 0.05), while black bars are not significant drivers. D) Sparse Partial Least Squares (sPLS) regressions show significant function (top) and MAG level (bottom) expression predictions of watershed maximum temperature, with key variables (Variable Importance Projection >1) denoted in bar graphs below. Fitted regression line is shown with grey shading representing 95% confidence interval. E) Non-metric multidimensional scaling of genome resolved metagenomic Bray-Curtis distances shows clustering of microbial communities by ecoregion (classified by Omernik II), with sampling location depicted on map above (mrpp, p < 0.001). Abbreviations: NPOC, Non-Purgable Organic Carbon; DNRA, Dissimilatory Nitrite Reduction to Ammonia; WWTP Density, Waste Water Treatment Plant Density; NPP, Net Primary Production. F) Non-metric multidimensional scaling of genome resolved metagenomic Bray-Curtis distances shows clustering of microbial communities by hydrological unit (HUC-2), with sampling location depicted on map on Fig. 1c.
Two other GROWdb MAGs contained genes for nitrite oxidation (the second step in nitrification) with taxonomy assignments to the Nitrospira_D genus and an unassigned species within the Palsa1315 genus of the Nitrospiraceae family (Supplementary Note 2). With these genomes being up to 95% complete, we infer that comamomox30 is unlikely, as these MAGs contained genes for nitrite oxidation but lacked genes for ammonia oxidation. These two nitrite oxidizers were detected in 14–88% of the metatranscriptome samples, including detection of transcripts for the key protein in nitrite oxidation. Each of the three nitrifier MAGs contained genes for combating reactive oxygen species (superoxide dismutase, catalase and/or peroxidase) and a photolyase gene involved in the repair of damage caused by exposure to ultraviolet light, all adaptations that are probably important in surface waters31. Overall, our findings uncover nitrifier metabolic potential and expression in rivers, which are under-represented in genomic databases compared with nitrifiers from soil and marine habitats.
Although not core members, we also detected 17 Patescibacterial MAGs that were transcriptionally active from the 48 total MAGs sampled in this phylum. These genomes all lacked the capacity for aerobic or anaerobic respiration and were inferred to be anoxic, obligate fermenters, consistent with previous genomic reports32 from this phylum that to date lacks any pure-culture, characterized representatives. Given that surface waters are oxic, we verified that the abundance patterns reported here were consistent with other river metagenome and amplicon-based studies33,34, in which these lineages accounted for up to 7% of the relative abundance in river surface water communities. It is possible that these obligately anaerobic members exist as symbionts, or thrive in lower-oxygen niches associated with biofilms on suspended particles, or hyporheic environments in which oxygen can be depleted during dissolved organic matter decomposition35,36. In support of the latter, we observed that relative abundance and expression of Patescibacteria significantly decreased with river size (Extended Data Fig. 7), suggesting that these obligate fermenters were more active in shallow waters when there is greater exchange between water and the stream bed37.
Emerging contaminants
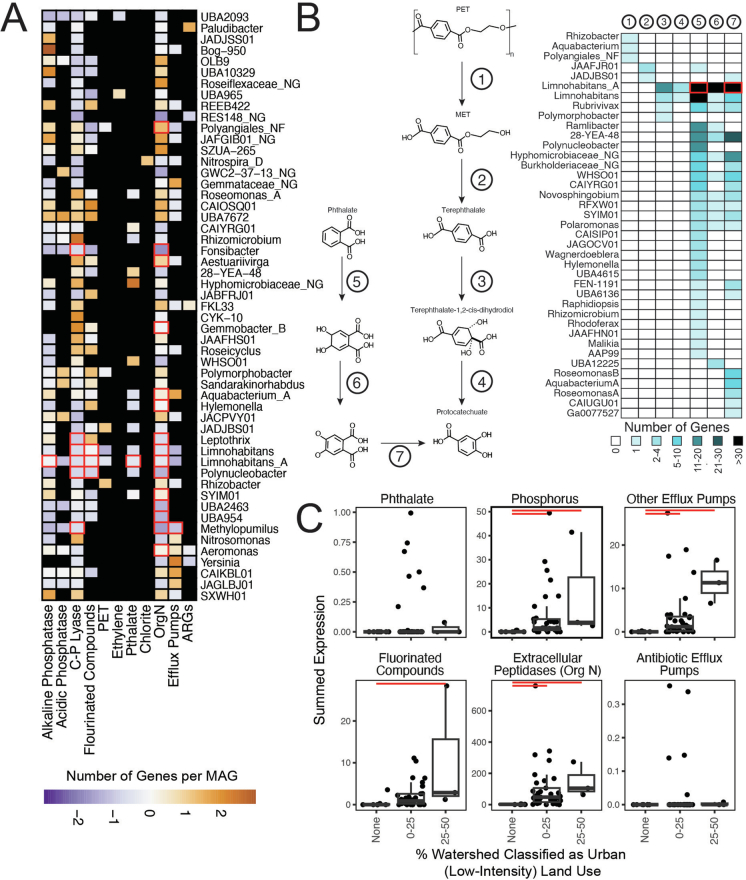
Given the threat of emerging contaminants (for example, pharmaceuticals, pesticides and plastics) to the environment and human heath, we hypothesized that GROWdb MAGs would encode and express genes related to transformations of these compounds to which river microorganisms are continuously exposed. Specifically, we identified microbial genes related to antibiotics, disinfection by-products, fluorinated compounds, fertilizers and microplastics based on their relevance to river systems38–41. In total, we classified 261 gene types related to emerging contaminants from GROWdb MAGs into 11 categories (Extended Data Fig. 8 and Supplementary Data 4). This resulted in gene recovery related to antibiotic resistance (n = 1,587), terephthalate and phthalate metabolism (n = 405) and fluorinated compounds (n = 1,194), while genes for phosphorus (n = 10,717) and organic nitrogen (n = 149,676) metabolism served as an indicator for fertilizer transformations. This provides extensive evidence for the ability of river microorganisms to interact with emerging contaminants across river ecosystems, as they are ultimately responsible for the depuration and nutrient removal in rivers.
Extended Data Fig. 8. GROWdb inventory of Emerging Contaminant Genes.
A) Heatmap shows the genomic potential for emerging contaminant transformation categories by genus, with number of genes normalized to the number of genomes within a genus and scaled by column. Black boxes indicate no detection of a related gene, while red box outlines indicate expression of a gene within at least six metatranscriptomes. B) Several genera encoded the potential for Terephthalate and Phthalate microplastic related metabolisms, with the entire pathway from polyethylene terephthalate (PET) and Phthalate shown. Heatmap corresponds to the pathway with steps 1–7, where box colour indicates the number of genes encoded per genus. Red outlines indicate expression of a gene within at least six metatranscriptomes. C) Emerging contaminant gene expression categories were related to land use, with significant relationships detected among the percent of the watershed classified as low-intensity, urban impacted shown by horizontal red bars (p-value < 0.05). Each point represents a single metatranscriptome (n = 43). Boxplot upper and lower box edges extend from the first to third quartile and the line in the middle represents the median. The whiskers are 1.5 times the interquartile range and every point outside this range represents an outlier. A similar trend was shown with high-impact urban land use, but lacked power based on number of samples. Significance (p-value < 0.05) is noted by red bar.
As rivers flow with heavy antibiotic burdens, antibiotic resistance develops rapidly and disseminates into various environmental compartments42. Antibiotic production is also part of natural competition in these complex communities. We catalogued 1,587 antibiotic-resistance genes (ARGs) recovered from 1,135 (54.3%) MAGs in GROWdb, representing 25 different Phyla (Supplementary Data 4). As our analysis was MAG focused, these numbers may represent a floor on ARG prevalence in rivers, as they do not include plasmid-encoded ARGs. These candidate ARGs represent 25 broad antimicrobial-resistance gene families as defined by the Comprehensive Antibiotic Resistance Database (CARD)43. Individual MAGs sometimes coded ARGs from multiple gene families and targeting multiple drug classes. Most (n = 1,219) candidate ARGs were homologues of proteins coded in glycopeptide resistance (van) gene clusters, which occurred in 955 distinct MAGs. However, the vast majority of these genes did not occur in canonical van gene clusters, and did not occur in close proximity to obvious biosynthetic gene clusters, as is the case in known Gram-positive actinomycete producers44. Although single van genes have been shown to be sufficient for conferring resistance to glycopeptide antibiotics44, the function of this large new pool of candidate van homologues remains to be determined.
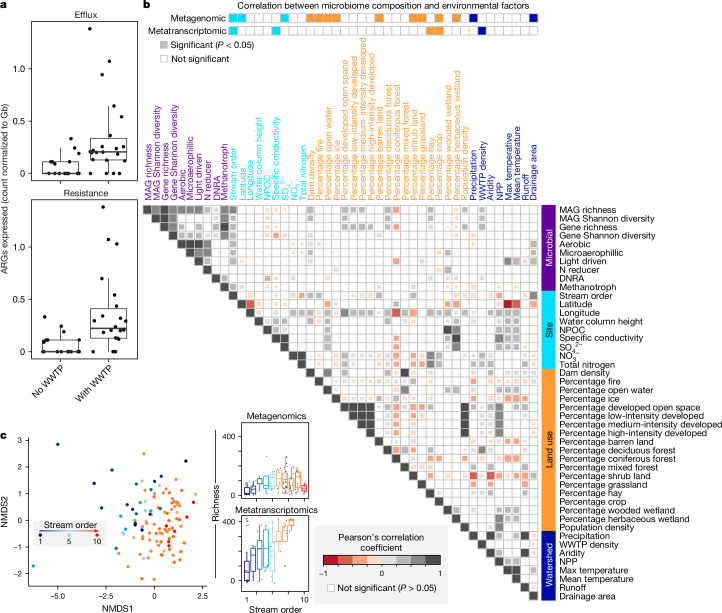
Thirty per cent of the ARGs had evidence of expression in metatranscriptomes of one or more samples, with antibiotic target alteration and antibiotic efflux pumps being the most widely expressed. Expression of ARGs was variable across samples, with 11 samples having at least 20 ARGs with evidence of expression. Given that wastewater treatment plants (WWTPs) have been shown to be an accumulation point for antimicrobial resistance38,45, we hypothesized that the presence and expression of ARGs would be related to the density of WWTPs in the watershed. Our findings show that the presence of WWTPs within a watershed resulted in more expression of ARGs, and this correlation also held for efflux pumps specifically (Fig. 3a and Extended Data Fig. 8).
Fig. 3. Patterns and drivers of river microbiome composition and function.
a, The number of efflux pumps (top) and ARGs (bottom) expressed at sites without or with impact from WWTPs, normalized to Gb of metatranscriptomic sequencing per sample (n = 43 metatranscriptomes). b, Metagenomic and metatranscriptomic composition, function and diversity were related to 36 selected site, land-use or watershed variables using Mantel tests (top two rows). This was followed with pairwise comparisons using Pearson’s correlation (heat map in b), with only significant values shown, as determined using the two-sided cor.test in R. Variables are coloured by category, including microbial (purple), site or local (light blue), land-use (orange) and watershed metrics (dark blue). For pairwise comparisons of microbial data, metatranscriptomic metrics were used for diversity and function abundance calculations. c, Microbial community diversity was significantly associated with stream order as depicted by non-metric multidimensional scaling of genome resolved metagenomic Bray–Curtis distances (left, beta-diversity) and Pearson correlations of richness to stream order (right, alpha-diversity) with points (n = 105) coloured by stream order. For the box plots, the upper and lower box edges extend from the first to third quartile, the centre line represents the median and the whiskers are 1.5× the interquartile range; points outside this range represent outliers.
Beyond antibiotics, river microbiomes encoded the capacity for the transformation of other emerging contaminants including those derived from fertilizers (phosphorus and organic N), microplastics (ethylene, poly(ethylene) terephthalate and terephthalate), disinfection by-products (chlorite) and fire retardants (fluorinated compounds)38. Extracellular peptidases for organic nitrogen transformations and C-P lyases for freeing phosphorus were the most widely encoded and expressed (Extended Data Fig. 8). This omnipresence across river organisms is probably due to the necessity of nitrogen and phosphorus compounds for microbial life in general. We also saw genes associated with transformation of other emerging contaminants including fluorinated compounds, as well as ethylene and phthalate metabolisms. Genes for defluorination (dehalogenases) were encoded across many river microorganisms and expressed in members of the Limnohabitans and Limnohabitans_A genera, and in a core member Polynucleobacter (Extended Data Fig. 8). Notably, the full pathway for polyethylene terephthalate degradation to protocatechuate was collectively encoded across multiple organisms, with lower parts of the pathway expressed in Limnohabitans_A. As these emerging contaminants are derived from anthropogenic influences, we suspected that expression of these genes might be correlated to land use, finding urban influences to be driving the expression of these genes in river microbiomes (Extended Data Fig. 8). River surface water microbiomes exhibit a vast capability to transform a wide array of emerging contaminants, with urban influences driving the expression of these genes, unveiling an intriguing intersection of microbial ecology and environmental pollution.
Continental-scale patterns
One of the strengths of our sampling design was the spatial, chemical and physical variables that accompanied our microbiome sampling, enabling us to contextualize the factors driving microbial biogeography at the continental scale. Previous studies have done this using taxonomy alone16,18,46 but, to our knowledge, these analyses have not incorporated functional gene-trait information. We hypothesized that river microbial communities exhibit spatial patterns at the continental scale of inquiry, and that these patterns would be predictable from hydrobiogeochemical, geographical and land-management factors. Every sample had a paired suite of more than 250 physical, chemical and spatial variables (for example, stream size, latitude, total nitrogen), which we used to identify the potential drivers of microbiome structure and expressed function (Supplementary Data 1).
Of all of the river site variables examined (Fig. 3b), stream order—a numerical ranking of the relative river size that spans small headwater streams (low order 1–3) to larger rivers such as sections of Mississippi river (high order 8–12)—was the most important controller of microbiome composition. River size was more important than latitudinal position or total carbon, which are often cited as controllers of microbiomes across other habitats47,48. Both metagenomes and metatranscriptomes were structured by stream order (Fig. 3b,c), providing evidence in favour of the river continuum concept (RCC)1, described below. After stream order, expressed microbial functional profiles were also influenced by watershed air temperature (both mean and maximum derived from geospatial data not taken at the time of sampling), area and total runoff (Fig. 3).
Given this relationship with air temperature, we sought to understand which functional traits and microorganisms most contributed to these community-level observations. Regression-based modelling showed that light-driven metabolisms, followed by aerobic processes, were the most important variables, predictive of mean and maximum watershed air temperature (Extended Data Fig. 7). The most important organismal predictors of maximum watershed temperature were the core active lineages like Methylopumilis, UBA954, Polynucleobacter and Limnohabitans that were actively transcribing genes for light-harvesting metabolisms (Fig. 2c and Extended Data Fig. 7). Our findings show that light-harvesting metabolisms are critical to energy generation in rivers and suggest that climate influences on water temperature may have a defining role in the niches of these microorganisms. However, the impact of light, which often varies with temperature in river systems and influences microbial resource availability, cannot be discounted. These findings are consistent with reports from marine systems49, hinting at an emerging rule set shared across aquatic microbiomes.
Beyond environmental factors, we also observed that geographical position had a role in structuring river microbiomes. For example, microbial community genomic membership was structured across ecoregions defined by Omernik level II ecoregions50, a classification system used to delineate distinct ecological regions based on similar environmental characteristics, providing a standardized framework for understanding ecological patterns and processes across landscapes. Notably, drier-climate, mixed-grass river microbiomes shared similar microbial communities that were distinct from those derived from wet to subtropical regions (Extended Data Fig. 7). Similarly, hydrologic unit code (HUC), a classification system for watersheds in the United States shown in Fig. 1c, recognized distinct microbial communities from continental subregions (Extended Data Fig. 7). These findings support earlier work showing that river microbial communities are inoculated from the landscape, and this terrestrial influence has an important role in downstream community assembly processes17. Note that the spatial structuring was not observed at the expressed functional level, indicating that microbial changes are compensated by functional equivalence at this continental scale. This finding suggests that taxonomic information may not be best suited for translation of microbiome content into management indicators, unless incorporated into an eco-regional framework as has been suggested for soil health indicators51.
To use microbiota information as sentinels for monitoring human and environmental health in river systems, a greater understanding of bacterial community structure, function and variability in lotic systems is required52. Although each of these land-use and watershed variables independently exhibited significant relationships with surface water microbial community composition and expression (Fig. 3b), our focus extended beyond their individual impacts. We aimed to understand the combined contributions of the most influential factors identified in explaining variation in both microbial community structure and expression. Moreover, based on factors like temperature acting as a significant driver of microbial community function (Extended Data Fig. 7), we hypothesized that time of year (season and month) may have a role. We found that stream order category, month, latitude, land use and maximum watershed temperature and their interactions explained a significant proportion of the variation in the microbial community composition at the metagenome level (R2 = 0.69; Extended Data Fig. 7c). Notably, stream order and month explained the most variation relative to other variables and all interactions. Metatranscriptome composition when tested with the same variable set did not show the same result, as only stream order and spatial location (taking into account latitude and longitude) were significant drivers (R2 = 0.41). Overall, the results suggest that multiple environmental factors, including geographical and land use variables, have important roles in shaping microbial community composition and expression. Analyses using GROWdb provide a framework for the environmental factors and determinant mechanisms that shape riverine communities.
River continuum concept
The RCC provides a framework for integrating predictable and observable biological features of flowing water systems, and further characterizing how biodiversity changes along a river system1. Specifically, the RCC postulates that, as rivers increase in size, the influences of terrestrial inputs will decrease. It also assumes that biological richness will initially increase with stream order complexity due to maximum interface with the landscape, but then decrease along with river width and discharge. Support for the applicability of the RCC to microbial communities has been observed as decreased microbial 16S rRNA gene richness occurring across stream order gradients in the Thames19, Mississippi52 and Amazon53 rivers. Given the expansion of our dataset from individual rivers, and the addition of functional resolved processes, we hypothesized that the RCC would extend to functional potential and expression patterns across continental scales.
First, we were interested in how microbial richness at the metagenome and metatranscriptome level changed across the stream-order gradient and whether these followed rules like 16S rRNA richness-based studies from single rivers. At the metagenome level, overall genome richness peaked at stream order 6 (Fig. 3c). At the metatranscriptome level, richness increased with stream order and peaked at stream order 8, the highest stream order profiled by metatranscriptomics (Fig. 3c). Metagenome results were consistent with previous reports of the RCC in which stream order peaks in mid-sized streams52. To our knowledge, this is the first report of genome-resolved metatranscriptomics across rivers and suggests that genome-inferred transcriptional richness may be governed by a different set of environmental controls than gene presence at the continental scale.
One major control on biological diversity described by the RCC is variability in sunlight exposure. Lower-order streams are often characterized by thick shore vegetation or overhanging trees that limit sunlight penetration and restrict phytoplankton and benthic microalgae primary production1,54. Consistent with this idea, we observed a statistically significant increase in light-driven microbial metabolisms when moving from lower-order streams to higher-order rivers (Fig. 3b). Moreover, the RCC proposes that the ratio of photosynthesis to respiration (P/R) increases in medium-sized rivers but is decreased in the smallest and largest rivers due to light limitations from riparian vegetation occlusion and turbidity, respectively. Using microbial gene expression coupled to genome-resolved lifestyle information, we estimated P/R ratios, revealing the highest P/R ratio in rivers with stream orders of 6–8, providing tentative support for this concept. However, the robustness of this P/R indicator would need further evaluation in larger-order rivers (such as 9–12), which are undersampled in this metatranscriptome dataset.
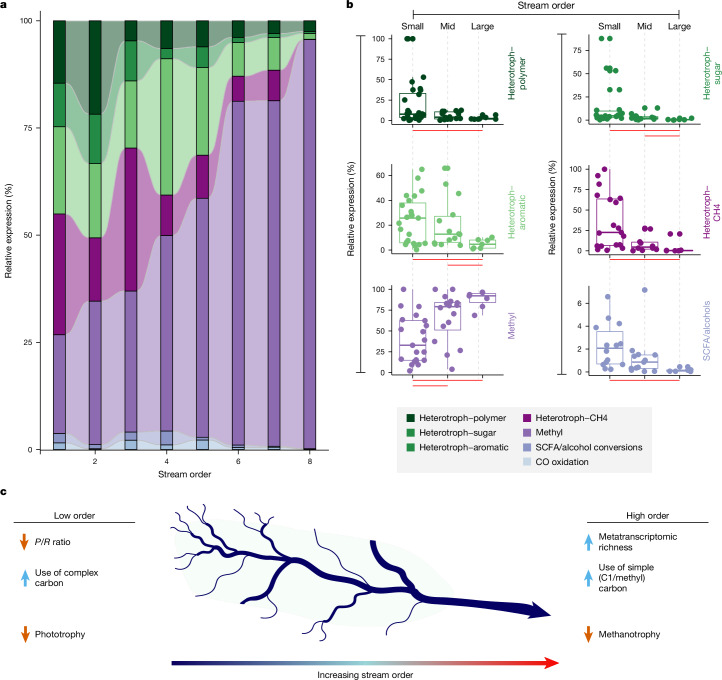
Another ecological control described by the RCC is a downstream decrease in the importance of terrestrial carbon inputs. We hypothesized that gene expression would show that microbial carbon usage reflects decreasing impacts of terrestrial inputs with river size. To resolve changes in microbial metabolism across a stream-order gradient, we defined carbon-usage patterns based on microbial gene expression in GROWdb MAGs. Our findings show significant differences in expressed microbial carbon usage following the stream-order gradient (Fig. 4a and Extended Data Fig. 6). Specifically, transcripts of genes targeting polymers, aromatics and sugars are upregulated in low-order streams, while methylotrophy gene transcripts, primarily from methanol oxidation, are increased in higher-order rivers (Fig. 4b and Supplementary Data 4). Methanol is probably autochthonous in these systems, derived from river phytoplankton biomass55 or microbial metabolism of aromatic allochthonous plant litter56,57. Our findings show that the inferred microbial metabolisms related to carbon usage follow the expected decrease in impact of terrestrial inputs proposed by the RCC, but we acknowledge that more research is needed to validate these insights, especially from higher-order rivers.
Fig. 4. Microbial lifestyle and carbon use are structured along a stream-order gradient.
a, The relative expression of microbial lifestyles (defined in the Methods) across stream-order gradient. b, One-dimensional box plots correspond to data in a, with each point (n = 53) representing a single sample and streams grouped by small (1–3), mid (4–6) and large (7–8) orders. For the box plots, the upper and lower box edges extend from the first to third quartile, the centre line represents the median and the whiskers are 1.5× the interquartile range; points outside this range represent outliers. Significant differences in expression between small-, mid- and large-order streams were determined using Kruskal–Wallis tests and are denoted by horizontal red bars below each plot (P < 0.01). Exact P values are reported in Supplementary Data 4. c, The stream-order model highlights changes in microbial expression from small-order (left) to large-order (right) streams.
In summary, river systems were once thought of as passive pipes, transporting water from terrestrial to marine systems. As a result, it was regarded that rivers were viewed as mere conduits, lacking substantial biogeochemical activity and offering little predictive capability58. Instead, we show that river microbiomes and encoded functionalities are not haphazardly distributed but are instead structured by river size, ecological region and land management regimes. This study also supports the application of the RCC to microbial communities and provides evidence that landscape patterns in river microbiomes are grounded in mechanistic changes in genomic function. We show that microbial richness both in terms of genome potential and expression, as well as expressed functional attributes, follow RCC tenets and are moulded by the physical–geomorphic environment (Fig. 4c). This application of GROWdb to the RCC adds a view of how microbial metabolism changes across rivers.
Conclusion
Changing climate impacts rivers through altered precipitation intensity, surface runoff, flooding, fires, sea level rise and droughts, and all of these have direct impacts on human health, agriculture, energy production and ecosystem resiliency59. Moreover, two-thirds of drinking water in the United States comes from surface river waters. Consequently, river management is expected to be one of the most politically charged topics in decades to come60. Microorganisms are master orchestrators of nutrient and energy flows that will probably dictate water quality under current and future water scenarios.
GROWdb is an effort to comprehensively understand river microbiomes, integrating genomics, biogeochemistry and environmental variables. Through the generation of over 3.8Tb of sequencing data, GROWdb provides insights into the taxonomic and functional diversity of microbial communities in river surface waters. The database includes over 2,000 microbial genomes, revealing both known and novel taxa and their metabolic abilities. Importantly, GROWdb demonstrates the prevalence of aerobic and light-driven energy metabolisms across river microbiomes, identifying the core microbial players and their contributions to biogeochemical processes. Moreover, the project identifies river microbiomes as reservoirs for genes related to emerging contaminants, highlighting their relationship with land use. By analysing biogeographical patterns at the continental scale, GROWdb underscores the influence of stream order, geographical location and environmental factors on microbial community structure and function. This study not only confirms the applicability of the RCC to microbial communities but also reveals mechanistic insights into how microbial metabolism changes along river gradients. Overall, GROWdb provides a valuable resource for understanding and managing river ecosystems in the face of environmental change.
To rapidly construct a large-scale river microbiome catalogue, we crowdsourced the data acquisition using standardized sampling, processing, sequencing and analysis to enable cross-site comparisons and modular augmentation. This product and its many data access and synthesis sites reduces the computational barriers for expediting the translation of reads to functional content. GROWdb offers a genome-centric window into river microbiota and a FAIR-use cyberinfrastructure-powered platform for future researchers. We envision that this genomic infrastructure will pave the way for future developments in water quality monitoring and identifying biomarkers indicative of land use or water quality changes. Collectively, GROWdb fills a major knowledge gap in the current understanding of microbial diversity and function in river ecosystems—observations that can be integrated into predictive watershed scale models.
Methods
Sample collection through crowdsourcing and standardization of workflows
To build GROWdb, we used two approaches to obtain samples from across US rivers. One was a network-of-networks61 approach based on sampling efforts of the Worldwide Hydrobiogeochemistry Observation Network for Dynamic River Systems (WHONDRS) consortium62, which is designed to facilitate the development of transferable scientific understanding and mutual benefit across stakeholders26,27. The WHONDRS sampling itself was based on sending free sampling kits, along with standardized protocols, to interested researchers globally. These researchers volunteered their time to collect samples and sent the samples back for processing using consistent methods to enable cross-site comparisons, interoperable data and transferable understanding. Samples from the WHONDRS consortium contributed 44% of the metagenomes and all the metatranscriptomes in GROWdb. Moreover, WHONDRS data included Fourier transform ion cyclotron resonance mass spectrometry data and were collected and analysed as described previously63, with data analysis specific to this paper reported online (https://data.ess-dive.lbl.gov/datasets/doi:10.15485/2439202). We note that all WHONDRS samples were collected over a period of 6 weeks in the summer of 2019, meaning that all the metatranscriptomes reported in this Article were collected during this sampling period.
Samples collected under the WHONDRS 2019 sampling campaign are described (Supplementary Data 1) and were reported previously63. In brief, we recruited collaborators based on geographical sampling priorities, and these sample collectors selected sampling sites within 100 m of a gauge station that measured river discharge, height or pressure. Geochemical data collected under the WHONDRS 2019 sampling campaign are available at ESS-DIVE, and the methods were described previously64. For microbiome analyses, at each site, approximately 1 l of surface water was sampled using a 60 ml syringe and was filtered through a 0.22 μm sterivex filter (EMD Millipore). The filters were capped, filled with 3 ml of RNAlater and shipped to the Pacific Northwest National Laboratory on blue ice within 24 h of collection. Surface water samples and filters were immediately frozen at −20 °C after receiving for nucleic acid extraction, respectively.
To build GROWdb, beyond WHONDRS, the second sampling approach was through a collaboration with the US Geological Survey (USGS) National Water Quality Network (NWQN)65. This long-term water-quality monitoring program characterized consistent information on streamflow and water-quality conditions. Data were collected to assess the status and trends of water-quality conditions at large inland and coastal river sites, as well as in small streams indicative of urban, agricultural and reference conditions65. The methods of sample collection used by the NWQN conform to the USGS National Field Manual for the Collection of Water-Quality Data66, and DNA was collected using the 0.22 μm Sterivex-GP filter (EMD Millipore). Here we provided kits integrated with USGS protocols for river sample processing with samples preserved as described previously67. All of the samples were stored on ice and stored at −20 °C until nucleic acid extraction.
A key component of this analysis was the standardization that occurred in data processing and analyses. For WHONDRS samples, DNA and RNA were co-extracted at single facility at Colorado State University. DNA and RNA were coextracted from filters at Colorado State University using the ZymoBIOMICS DNA/RNA Miniprep kit (Zymo Research, R2002) coupled with the RNA Clean & Concentrator-5 kit (Zymo Research, R1013). The samples were eluted in 40 μl and stored at −20 °C until sequencing (Supplementary Note 4). For NWQN samples, DNA was extracted using a standard phenol–chloroform extraction protocol68. The Community Sequencing Project provided by the Joint Genome Institute (JGI) ensured that sequencing protocols and methodologies were consistent across the project. Owing to the extensive geographical distribution of data collection for most sites, replicate sequencing experiments were not conducted at the same sites. All of the metagenomes and 23% of the metatranscriptomes were provided by JGI, with the balance of metatranscriptomes processed at University of Colorado Anschutz using the same kits and methods as specified by the JGI. Lastly, sequence data processing for each sample was performed using identical methods, using the GROWdb standard operating procedures documented on GitHub69. Collectively, the use of crowdsourced approaches, JGI support and standardized methodologies resulted in GROWdb, a compendium of river microbiome data, an endeavour that would not have been possible to execute in this time frame by a single laboratory alone.
Acquisition of geospatial data
The watershed statistics for each sample were primarily obtained from the Environmental Protection Agency’s StreamCat database70 and the National Hydrography Plus Version 2 (NHDPlus V2) Dataset using the nhdplusTools package71 in R. StreamCat provides over 600 consistently computed watershed metrics for all waterbodies identified in the USGS NHDPlus V2 geospatial framework, making it a suitable data source for the broad spectrum of sample locations in this study. For watershed metrics that were not included in StreamCat (that is, dominant Omernik ecoregion, mean net primary production and mean aridity index), we first delineated each sample’s watershed using nhdplusTools, then used the terra package72 to aggregate the additional datasets across each site’s watershed accordingly. This approach is consistent with SteamCat’s geospatial methodology.
Last, we collected streamflow data for sites that had a nearby stream gauge. For locations without an identified co-located stream gauge (WHONDRS typically co-located their sample sites with a stream gauge), we identified USGS stream gauges within 10 km upstream or downstream of our sampling locations using the dataRetrieval and nhdplusTools packages. All stream gauges were then manually verified for their applicability to each sampling site (for example, verifying that there were no dams between the site and the stream gauge, a major confluence). A complete list of datasets included in our analysis is provided in Supplementary Data 1. The complete R workflow for this geospatial analysis is available at GitHub73.
Metagenomic assembly, binning and annotation
At the JGI, genomic DNA was prepared for metagenomic sequencing using plate-based DNA library preparation on the PerkinElmer Sciclone NGS robotic liquid handling system. In brief, 1 ng of DNA was fragmented and adapter ligated using the Nextera XT kit (Illumina) and unique 8 bp dual-index adapters (IDT, custom design). The ligated DNA fragments were enriched with 12 cycles of PCR and purified using Coastal Genomics Ranger high-throughput agarose gel electrophoresis size selection to 450–600 bp. The prepared libraries were sequenced using Illumina NovaSeq sequencer according to a 2 × 150 nucleotide indexed run program.
Our metagenome workflow is described and visualized (Extended Data Fig. 9 and Supplementary Note 3). In brief, the resulting fastq files were assembled and binned using the accessible GROWdb pipelines released on GitHub69. To maximize genome recovery, three assemblies were performed on each set of fastq files and binned separately: (1) read trimming with sickle (v.1.33)74, assembly with MEGAHIT (v.1.2.9)75 and binning with metabat276 (v.2.12.1); (2) read trimming with sickle (v.1.33), random filtering to 25% of reads, assembly with IDBA-UD77 (v.1.1.0) and binning with metabat276 (v.2.12.1); (3) bins derived from the JGI-IMG pipeline78 (that used metaSPAdes79 and metabat276) were downloaded. All of the resulting bins were assessed for quality using checkM80 (v.1.1.2) and medium and high-quality MAGs with >50% completion and <10% contamination were retained. The resulting 3,284 MAGs across all samples and assemblies were dereplicated at 99% identity using dRep81 (v.2.6.2) to obtain the dereplicated first version of the GROW database (n = 2,093 MAGs). MAG taxonomy was assigned using GTDB-tk82 (v.2.1.1, r207) and annotated using DRAM (v.1.4.4)83.
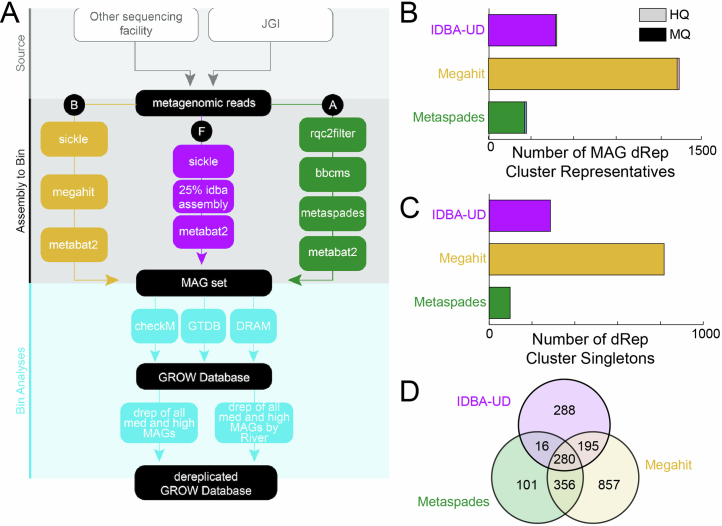
Extended Data Fig. 9. GROWdb metagenomic analysis pipeline and results.
A) Metagenomic pipeline for GROWdb that resulted in three assemblies per sample (A, B, and F), with all parameters and version used outlined in methods and on GitHub (10.5281/zenodo.11041178). B) Stacked bar graph shows the number of medium (MQ) and high (HQ) quality dereplicated MAG representatives recovered from each assembly type. C) Bar graph shows the number of singleton dereplicated MAG representatives from each assembly type. D) Venn diagram compares the number of dereplicated MAG cluster representatives (dRep winners) recovered from each assembly type with overlaps indicating MAGs within the same cluster were recovered from multiple assembly types.
To quantify MAG relative abundance across samples, trimmed metagenomic reads were mapped to the dereplicated MAG set using Bowtie284 and output as SAM files, which were then converted to sorted BAM files using samtools. Sorted BAM files were then filtered to paired reads only with a 95% identity match using reformat.sh. To obtain the mean coverage for each MAG, we used CoverM85 (-m trimmed_mean). The mean coverage table was then filtered to MAGs that had at least 60% coverage across a MAG with at least 3× coverage within a sample, using additional CoverM85 outputs (-m relative_abundance --min-covered-fraction 0.6 and -m reads_per_base, respectively). CoverM outputs were merged in R; the script is available on the GROWdb GitHub69.
Metatranscriptomic mapping and analysis
RNA was prepared for metatranscriptome sequencing according to JGI established protocols. In brief, rRNA was removed from 10 ng of total RNA using Qiagen FastSelect probe sets for bacterial, yeast and plant rRNA depletion (Qiagen) with RNA blocking oligo technology. The fragmented and rRNA-depleted RNA was reverse transcribed to create first-strand cDNA using the Illumina TruSeq Stranded mRNA Library prep kit (Illumina) followed by second-strand cDNA synthesis, which incorporates dUTP to quench the second strand during amplification. The double-stranded cDNA fragments were then A-tailed and ligated to JGI dual-indexed Y-adapters, followed by an enrichment of the library through 13 cycles of PCR. The prepared libraries were quantified using the KAPA Biosystems’ next-generation sequencing library qPCR kit and run on the Roche LightCycler 480 real-time PCR instrument. Sequencing of the flowcell was performed on the Illumina NovaSeq sequencer following a 2 × 150 nucleotide indexed run program.
The resulting fastq files were mapped using Bowtie284 (-D 10 -R 2 -N 1 -L 22 -i S,0,2.50) to the dereplicated GROWdb. SAM files were transformed to BAM files using samtools, filtered to 97% ID using reformat.sh and name sorted using samtools. Transcripts were counted for each gene using feature-counts86. Counts were transformed to geTMM (gene length corrected trimmed mean of M-values) in R using edgeR package87. Genes were considered if they were expressed in 10% of samples. Core calculations in Fig. 2 had an additional requirement to express at least 20 genes per genome.
Microbial metabolism trait and carbon usage classification
To classify microbial genes and genomes based on their carbon metabolism, we curated the metabolism assignments made by DRAM83 using rulesets to assign genomes to functional guilds (Extended Data Fig. 5). For example, genomes were classified by respiratory capacity based on the presence of >50% of the subunits required for complex 1 of the electron-transport chain and the presence at least one gene for an electron acceptor. As such, for a genome to be classified as a microaerophile, we required the genome to have more than 50% of complex 1 subunit and at least one subunit of a low-affinity cytochrome oxidase. Likewise, if a genome did not have more than 50% of the subunits required for complex 1 of the electron-transport chain or the potential for any electron acceptor, it was classified as an obligate fermenter (Extended Data Fig. 5). All calls made by the defined rule set were checked manually to account for misbins, low bit scores and genome incompleteness.
From the DRAM output, we further assigned genomes as capable of carbon fixation if they encoded >70% of one of six seven carbon fixation pathways. We then assigned each MAG in each river metatranscriptome as a photoautotroph, photoheterotroph, chemolithoautotrophy, heterotroph or mixotroph by assessing the gene expression in that system. We then focused in on genes required for using different carbon substrates in the genomes identified for heterotrophy. We assigned carbon gene expression into the following categories: polymer, sugar, aromatic compound, methanotrophy, methylotrophy, short chain fatty acid utilization and carbon monoxide utilization using DRAM assigned rules. Carbon usage curation scripts are available on the GROWdb GitHub69. P/R ratios were defined by the ratio of expression of light-driven energy metabolisms (aerobic photosynthesis, anaerobic photosynthesis and photorhodopsins) divided by aerobic respiration metabolisms (aerobic respiration and microaerophilic respiration).
Phylogenetic analyses were performed to refine the annotation of nitrogen related metabolism including genes annotated as respiratory nitrate reductase (nar), nitrite oxidoreductase (nxr), ammonia monooxygenase (amo) or methane monooxygenase (pmo) to improve the assignment the nitrogen cycling capabilities of GROW MAGs. Specifically, Nxr/Nar and PmoA/AmoA amino acid reference sequences were downloaded30,88,89 and this set of reference sequences was combined with amino acid sequences of homologues from the GROWdb, aligned separately using MUSCLE (v.3.8.31) and run through a Python script for generating phylogenetic trees (ProtPipeliner; https://github.com/WrightonLabCSU/Protpipeliner/tree/main)90,91. ProtPipeliner runs as follows: (1) alignments are curated with minimal editing by GBLOCKS92; (2) model selection is conducted via ProtTest93; and (3) maximum-likelihood phylogeny for alignments are conducted using RAxML94 v.8.3.1 with 100 bootstrap replicates. This resulted in two phylogenies, one for Nxr/Nar and one for Pmo/Amo, that were visualized using iTOL95 (https://itol.embl.de/shared/wrighton_lab) and were used to refine the homology-based gene annotations in the MAG database. Raw tree files are also available as newick files available at Zenodo (10.5281/zenodo.8173286).
For in silico predictions of ARGs, GROWdb-predicted proteins were searched for homology to proteins in the Comprehensive Antibiotic Resistance Database (CARD; v.3.2.7, downloaded June 2023) using the Resistance Gene Identifier (RGI; v.6.0.2)43. RGI was run locally in protein input mode with distributed input and default parameters and with the ‘include loose’ option. However, the final list of candidate ARGs analysed here includes only proteins identified by RGI as ‘perfect’ or ‘strict’ hits, and includes only protein homologue models (that is, no protein variant models were included in the analysis). Other contaminant annotations were derived from DRAM annotations with the list of targeted genes included (Supplementary Data 4).
SRA analysis
To analyse the distribution of microbial lineages recovered by GROW across public datasets, the Sandpiper96 database (https://sandpiper.qut.edu.au) was used as a basis96. At the time of analysis, it contained metagenomes that were publicly available on 15 December 2021. Reanalysis of these datasets was performed with SingleM 1.0.0beta796. The ‘supplement’ subcommand was first used to add 95% ANI dereplicated GROW MAGs to the SingleM96 reference metapackage built with GTDB RS07-207 (10.5281/zenodo.7582579). The ‘renew’ subcommand was then used to reanalyse all metagenomes present in the Sandpiper database, outputting a taxonomic profile, detailing the microbial lineages and unclassified lineages in each metagenome, together with their relative abundance.
To search for public metagenomes in which GROW MAGs were present, taxonomic profiles of metagenomes containing microbial lineages that had an associated GROW MAG (either novel or already represented in GTDB) were further analysed. To reduce the incidence of false identification, we required at least two microbial lineages represented by a GROW MAG to be present and the combined relative abundance to be >1%. Metadata of metagenomes containing GROW MAGs were gathered using Kingfisher ‘annotate’ (https://github.com/wwood/kingfisher-download).
Statistical analysis
Geospatial variables were categorized into site or local, land-use or watershed characteristic groups and combined with microbial data to generate the biogeography dataset (Fig. 3b). Biogeographical patterns were assessed in three ways: (1) a pairwise Pearson correlation matrix was calculated for all variables using cor.test to test for significance, with all correlations with P > 0.05 removed; (2) for each variable non-microbial variable, a distance matrix was calculated using the Euclidean distance metric and then individual mantel tests were conducted to assess the correlation between the variable distance matrix and a Bray–Curtis distance matrix of metatranscriptome or metagenome MAG abundance; (3) PERMANOVA was conducted using the adonis2 function with 999 permutations to assess the influence of various environmental predictors on microbial community expression. For (3), spatial distance metrics were calculated and assessed against microbial communities as either latitude, longitude or through a primary spatial variable calculated as the first principal component of latitude and longitude. Likewise, a collective land use variable was calculated as the as the first principal component of land-use metrics in Fig. 3b. Several models were run, with the two reported in the text as model 1: effects of stream order, month, land use and maximum temperature on microbial community composition; and model 2: effects of stream order and spatial variable on microbial community composition. Note that spatial variables often covary with abiotic and biotic factors; thus, correlations make it challenging to disentangle whether shifts in the relative abundances of specific microbial taxa are directly influenced by temperature or by concurrent changes in other factors that also affect river microbial communities. Here we provide multiple levels of testing, to evaluate those variables in a pairwise manner, as well as collectively.
Metagenomic and metatranscriptomic composition, function and diversity were related to 36 selected site, land-use or watershed variables using Mantel tests (top two rows). This was followed with pairwise comparisons using Pearson’s correlation (heat map Fig. 3b). Variables are coloured by category including microbial (purple), site or local (light blue), land-use (orange) and watershed metrics (dark blue). For pairwise comparisons of microbial data, metatranscriptomic metrics were used for diversity and function abundance calculations.
All data analysis and visualization was done in R (v4.2.1) with the following packages: stats (v.4.1.1), vegan (v.2.6), ggplot2 (v.3.3.6), ComplexUpset (v.2.8.0), tidyr (v.1.2.0), dplyr (v.1.0.9), corrplot (v.0.92), pheatmap (v.1.0.12), RColorBrewer (v.1.1-3), pls (v.2.8), edgeR (v.3.16). Scripts for figure generation and data analysis are available on GitHub69. Map data were derived from publicly available data sources: (1) Fig. 1b,c and Extended Data Fig. 7 were generated using the state boundaries developed using the tigris (https://github.com/walkerke/tigris); (2) Fig. 1b,c was generated using the flowlines from National Hydrography Plus Version 271; and (3) Extended Data Fig. 7 was generated using the ecoregions50 provided from https://www.epa.gov/eco-research/ecoregions.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Online content
Any methods, additional references, Nature Portfolio reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at 10.1038/s41586-024-08240-z.
Supplementary information
Tab 1: table summary of river microbiome studies detailing the title, authors and microbiome methods used. Highlighted studies are genome-resolved multi-omics like GROWdb, with only eight rivers surveyed. Tab 2: metagenome and metatranscriptome information. Tab 3: geospatial data (n = 287 variables) for each sample. Tab 4: list of datasets included in geospatial analysis. Tab 5: Chloroplast contamination analysis.
Tab 1: table summary of GROWdb MAGs including taxonomy and quality information. Tab 2: table summary of GROWdb novelty assigned by GTDB-Tk. Tab 3: GROWdb MAG relative abundance across metagenomes. Tab 4: core analysis of GROWdb genera including metagenomic and metatranscriptomic occupancy and mean abundance/expression. Tab 5: global MAG analysis of 9,798 MAGs derived from GROWdb efforts, along with other biogeography studies.
Tab 1: annotation summary of GROWdb MAGs from DRAM. Tab 2: GROWdb whole-genome trait calls. Tab 3: functional genes used to make whole-genome trait calls. Tab 4: output of resistance gene identifier containing list of predicted ARGs. Tab 5: metatranscriptomic expression of identified ARGs by category.
Tab 1: list of genes profiled for emerging contaminants, with ARGs listed in tabs 4–5. Tab 2: annotations of emerging contaminant genes for GROWdb MAGs. Tab 3: table of emerging contaminant genes by genus normalized to the number of genomes. Tab 4: output of resistance gene identifier containing list of predicted ARGs. Tab 5: metatranscriptomic expression of identified ARGs by category. Tab 6: carbon usage analysis in Fig. 4. Tab 7: exact P values for Kruskal–Wallis statistical tests in Fig. 4.
Acknowledgements
Samples were sequenced and processed as a part of the Genome Resolved Open Watersheds database (GROWdb) effort to sequence global watersheds. A portion of the samples and data used in this manuscript were generated as part of the USGS NWQN program in collaboration with B.C.C. We thank M. Riskin and USGS scientists for sample collection; and L. Fine, C. Kellogg, J. Payet, D. URycki and a team of undergraduate interns at Oregon State University for DNA sample extraction. A portion of samples and data used in this manuscript were generated as a part of the WHONDRS global crowdsourced Summer 2019 Sampling (S19S) and we thank those who participated in the design and implementation of that effort. We thank T. Claffey and R. Wolfe for server management and Z. Crockett for generation of the sample set Digital Object Identifier. This work was partially supported by awards from US Department of Energy (DOE) Office of Science, Office of Biological and Environmental Research (BER) grants DE-SC0023084 (M.A.B., B.B.M., M.J.W., K.C.W.), DE-SC0021350 (M.A.B., D.M.S., C.S.H., C.S.M., K.C.W.), and DE-AC02-05CH11231 (M.A.B., E.W.C). B.C.C., T.B. and S.P.G. were partially supported by US National Science Foundation awards DEB1840243, EAR1836768 and DEB1457794. Funding support also was provided by start-up funding to K.C.W. from Colorado State University. A portion of this work was also performed by M.A.B. under a subcontract to K.C.W. from the River Corridor Science Focus Area (RC-SFA) at Pacific Northwest National Laboratory (PNNL) and funded by the DOE BER Environmental System Science (ESS) Program. PNNL is operated by Battelle Memorial Institute for the DOE under contract no. DE-AC05-76RL01830. WHONDRS efforts described in this Article along with J.C.S. A.E.G., and R.E.D. were also funded under the RC-SFA at PNNL by DOE BER ESS. Metagenomic and metatranscriptomic sequencing was performed at the JGI under a Community Science Program and the University of Colorado Anschutz’s Genomics Shared Resource. The work (proposal 10.46936/10.25585/60001289) conducted by the US DOE JGI (https://ror.org/04xm1d337), a DOE Office of Science User Facility, is supported by the Office of Science of the US DOE operated under contract no. DE-AC02-05CH11231. Work conducted at the Genomics Shared Resource at the University of Colorado was supported by the Cancer Center Support Grant (P30CA046934). The work conducted by the National Microbiome Data Collaborative (https://ror.org/05cwx3318) is supported by the Genomic Science Program in the US DOE, Office of Science, Office of Biological and Environmental Research (BER) under contract numbers DE-AC02-05CH11231 (LBNL), 89233218CNA000001 (LANL) and DE-AC05-76RL01830 (PNNL). The work conducted by the DOE Systems Biology Knowledgebase (KBase) is funded by the US DOE, Office of Science, Office of Biological and Environmental Research under award numbers DE-AC02-05CH11231, DE-AC02-06CH11357, DE-AC05-00OR22725 and DE-AC02-98CH10886.
Extended data figures and tables
Author contributions
M.A.B., J.C.S. and K.C.W. conceptualized, designed and supervised the study. M.A.B., K.R.W., F.L., J.N.E., J.P.F., T.B., R.A.D., A.E.G., M.J.W., E.K.H., C.P., S.R., E.A.E.-F., S.P.G., E.M.W.-C., C.S.H., M.R.V.R., B.C.C., J.C.S. and K.C.W. performed and supervised experimental work to generate data. M.A.B., B.B.M., K.R.W., B.J.W., A.C.M., D.M.S., I.L., R.D. and C.S.M. analysed and visualized the data. M.A.B., K.R.W., A.P., F.L., A.F., J.N.E., J.P.F., R.D., A.E.G. and E.M.W.-C. curated the data. M.A.B. and K.C.W. drafted the manuscript, with contributions from B.B.M., A.C.M., M.B.S., C.S.M. and J.C.S. All of the authors read, commented on and edited the manuscript, as well as approved its final form.
Peer review
Peer review information
Nature thanks Tom Battin, Jack Gilbert and Serina Robinson for their contribution to the peer review of this work.
Data availability
The data underlying GROWdb are accessible across multiple platforms to ensure many levels of data use and structure are widely available. First, all reads and MAGs are publicly hosted at the National Center for Biotechnology (NCBI) under BioProject PRJNA946291. Second, all data presented in this Article, including MAG annotations, phylogenetic tree files, antibiotic-resistance gene database files and expression data tables are available at Zenodo97 (10.5281/zenodo.8173286). Data visualized as maps were derived from publicly available data sources: (1) state boundaries developed using the tigris R package (https://github.com/walkerke/tigris); (2) flowlines from National Hydrography Plus Version 271; (3) ecoregions50 provided from https://www.epa.gov/eco-research/ecoregions. Beyond the content listed above, our aim for GROWdb was to maximize data use by making the data available in searchable and interactive platforms including the National Microbiome Data Collaborative (NMDC) data portal, the Department of Energy’s Systems Biology Knowledgebase (KBase)3 and a GROW-specific user interface released here, GROWdb Explorer. Each platform provides different ways to interact with data in the GROWdb. GROWdb was a flagship project for the newly formed NMDC. Specifically, individual GROWdb datasets (metagenomes, metatranscriptomes and so on) are easily accessible and searchable through the NMDC data portal98 (https://data.microbiomedata.org/), where they are systematically connected to each other and to a rich suite of sample information, other data collected on the same samples and standard analysis results, following findable, accessible, interoperable and reusable data practices26. GROWdb is also a publicly available collection (https://narrative.kbase.us/collections/GROW) within KBase3, with samples, MAGs and corresponding genome-scale metabolic models found in the KBase narrative structure (10.25982/109073.30/1895615). Access within KBase allows for immediate access and reuse of data, including comparison to private data analyses using KBase’s 500+ analysis tools, in a point and click format. GROWdb Explorer is a graphical user interface built through the Colorado State University Geospatial Centroid (https://geocentroid.shinyapps.io/GROWdatabase/), enabling users to search and graph microbial and spatial data simultaneously. Here the microbial data, metabolite and geospatial data are included. The microbial data were distilled into functional gene information, so that biogeochemical contributions and the microorganisms catalysing them can be assessed and visualized rapidly across the dataset. In summary, GROWdb represents to our knowledge the first publicly available genome collection from rivers and offers data that can be leveraged across microbiome studies. GROWdb is an expanding repository to incorporate and unify global river multi-omic data for the future.
Code availability
All scripts involved with microbial data generation, processing, curation and visualization are available at GitHub and Zenodo99 (https://github.com/jmikayla1991/Genome-Resolved-Open-Watersheds-database-GROWdb/tree/main, 10.5281/zenodo.11041178). Code for geospatial analysis and GROWdb Explorer are available at GitHub (https://github.com/rossyndicate/GROWdb). Code for figures and data analysis are available in Zenodo100 (10.5281/zenodo.11188634).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Mikayla A. Borton, Email: mborton@colostate.edu
Kelly C. Wrighton, Email: wrighton@colostate.edu
Extended data
is available for this paper at 10.1038/s41586-024-08240-z.
Supplementary information
The online version contains supplementary material available at 10.1038/s41586-024-08240-z.
References
- 1.Vannote, R. L., Minshall, G. W., Cummins, K. W., Sedell, J. R. & Cushing, C. E. The river continuum concept. Can. J. Fish. Aquat. Sci.37, 130–137 (1980). [Google Scholar]
- 2.Wood-Charlson, E. M. et al. The National Microbiome Data Collaborative: enabling microbiome science. Nat. Rev. Microbiol.18, 313–314 (2020). [DOI] [PubMed] [Google Scholar]
- 3.Arkin, A. P. et al. KBase: the United States Department of Energy Systems Biology Knowledgebase. Nat. Biotechnol.36, 566–569 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cavicchioli, R. et al. Scientists’ warning to humanity: microorganisms and climate change. Nat. Rev. Microbiol.17, 569–586 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hutchins, D. A. & Fu, F. Microorganisms and ocean global change. Nat. Microbiol.2, 17058 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Sunagawa, S. et al. Tara Oceans: towards global ocean ecosystems biology. Nat. Rev. Microbiol.18, 428–445 (2020). [DOI] [PubMed] [Google Scholar]
- 7.Battin, T. J. et al. River ecosystem metabolism and carbon biogeochemistry in a changing world. Nature613, 449–459 (2023). [DOI] [PubMed] [Google Scholar]
- 8.Kroeze, C., Dumont, E. & Seitzinger, S. P. New estimates of global emissions of N2O from rivers and estuaries. Environ. Sci.2, 159–165 (2005). [Google Scholar]
- 9.Butman, D. & Raymond, P. A. Significant efflux of carbon dioxide from streams and rivers in the United States. Nat. Geosci.4, 839–842 (2011). [Google Scholar]
- 10.Anderson, E. P. et al. Understanding rivers and their social relations: a critical step to advance environmental water management. WIREs Water6, e1381 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mishra, A., Alnahit, A. & Campbell, B. Impact of land uses, drought, flood, wildfire, and cascading events on water quality and microbial communities: a review and analysis. J. Hydrol.596, 125707 (2021). [Google Scholar]
- 12.Rodríguez-Ramos, J. A. et al. Genome-resolved metaproteomics decodes the microbial and viral contributions to coupled carbon and nitrogen cycling in river sediments. mSystems7, e00516-22 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ghosh, D., Ghosh, A. & Bhadury, P. Arsenic through aquatic trophic levels: effects, transformations and biomagnification—a concise review. Geosci. Lett.9, 20 (2022). [Google Scholar]
- 14.Boddicker, A. M. & Mosier, A. C. Genomic profiling of four cultivated Candidatus Nitrotoga spp. predicts broad metabolic potential and environmental distribution. ISME J.12, 2864–2882 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chu, H., Gao, G.-F., Ma, Y., Fan, K. & Delgado-Baquerizo, M. Soil microbial biogeography in a changing world: recent advances and future perspectives. mSystems5, e00803-19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stadler, M. & del Giorgio, P. A. Terrestrial connectivity, upstream aquatic history and seasonality shape bacterial community assembly within a large boreal aquatic network. ISME J.16, 937–947 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crump, B. C., Amaral-Zettler, L. A. & Kling, G. W. Microbial diversity in arctic freshwaters is structured by inoculation of microbes from soils. ISME J.6, 1629–1639 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ruiz-González, C., Niño-García, J. P. & del Giorgio, P. A. Terrestrial origin of bacterial communities in complex boreal freshwater networks. Ecol. Lett.18, 1198–1206 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Read, D. S. et al. Catchment-scale biogeography of riverine bacterioplankton. ISME J.9, 516–526 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Savio, D. et al. Bacterial diversity along a 2600 km river continuum. Environ. Microbiol.17, 4994–5007 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Payne, J. T., Millar, J. J., Jackson, C. R. & Ochs, C. A. Patterns of variation in diversity of the Mississippi river microbiome over 1,300 kilometers. PLoS ONE12, e0174890 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parks, D. H. et al. Recovery of nearly 8,000 metagenome-assembled genomes substantially expands the tree of life. Nat. Microbiol.2, 1533–1542 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Garner, R. E. et al. A genome catalogue of lake bacterial diversity and its drivers at continental scale. Nat. Microbiol.8, 1920–1934 (2023). [DOI] [PubMed] [Google Scholar]
- 24.Nayfach, S. et al. A genomic catalog of Earth’s microbiomes. Nat. Biotechnol.39, 499–509 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rodríguez-Ramos, J. et al. Spatial and temporal metagenomics of river compartments reveals viral community dynamics in an urban impacted stream. Front. Microbiomes2, 1199766 (2023).
- 26.Wilkinson, M. D. et al. The FAIR guiding principles for scientific data management and stewardship. Sci. Data3, 160018 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goldman, A. E., Emani, S. R., Pérez-Angel, L. C., Rodríguez-Ramos, J. A. & Stegen, J. C. Integrated, coordinated, open, and networked (ICON) science to advance the geosciences: introduction and synthesis of a special collection of commentary articles. Earth Space Sci.9, e2021EA002099 (2022). [Google Scholar]
- 28.Jezbera, J., Sharma, A. K., Brandt, U., Doolittle, W. F. & Hahn, M. W. ‘Candidatus Planktophila limnetica’, an actinobacterium representing one of the most numerically important taxa in freshwater bacterioplankton. Int. J. Syst. Evol. Microbiol.59, 2864–2869 (2009). [DOI] [PubMed] [Google Scholar]
- 29.Stein, L. Y. Insights into the physiology of ammonia-oxidizing microorganisms. Curr. Opin. Chem. Biol.49, 9–15 (2019). [DOI] [PubMed] [Google Scholar]
- 30.Daims, H. et al. Complete nitrification by Nitrospira bacteria. Nature528, 504–509 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu, S. et al. Comammox Nitrospira within the Yangtze River continuum: community, biogeography, and ecological drivers. ISME J.14, 2488–2504 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wrighton, K. C. et al. Fermentation, hydrogen, and sulfur metabolism in multiple uncultivated bacterial phyla. Science337, 1661–1665 (2012). [DOI] [PubMed] [Google Scholar]
- 33.Lian, Y., Zhen, L., Chen, X., Li, Y. & Li, X. Microbial biomarkers as indication of dynamic and heterogeneous urban water environments. Environ. Sci. Pollut. Res.10.1007/s11356-022-24539-8 (2022). [DOI] [PubMed]
- 34.Regina, A. L. A. et al. A watershed impacted by anthropogenic activities: microbial community alterations and reservoir of antimicrobial resistance genes. Sci. Total Environ.793, 148552 (2021). [DOI] [PubMed] [Google Scholar]
- 35.Ploug, H., Kühl, M. & Buchholzcleven, B. Anoxic aggregates—an ephemeral phenomenon in the pelagic environment? Aquat. Microb. Ecol.13, 285–294 (1997). [Google Scholar]
- 36.Böckelmann, U., Manz, W., Neu, T. R. & Szewzyk, U. Characterization of the microbial community of lotic organic aggregates (‘river snow’) in the Elbe River of Germany by cultivation and molecular methods. FEMS Microbiol. Ecol.33, 157–170 (2000). [DOI] [PubMed] [Google Scholar]
- 37.Battin, T. J. et al. Biophysical controls on organic carbon fluxes in fluvial networks. Nat. Geosci.1, 95–100 (2008). [Google Scholar]
- 38.Gomes, I. B., Maillard, J.-Y., Simões, L. C. & Simões, M. Emerging contaminants affect the microbiome of water systems—strategies for their mitigation. Npj Clean Water3, 39 (2020). [Google Scholar]
- 39.Li, J., Liu, H. & Paul Chen, J. Microplastics in freshwater systems: a review on occurrence, environmental effects, and methods for microplastics detection. Water Res.137, 362–374 (2018). [DOI] [PubMed] [Google Scholar]
- 40.Mdee, A. et al. The top 100 global water questions: results of a scoping exercise. One Earth5, 563–573 (2022). [Google Scholar]
- 41.Zrimec, J., Kokina, M., Jonasson, S., Zorrilla, F. & Zelezniak, A. Plastic-degrading potential across the global microbiome correlates with recent pollution trends. mBio12, e0215521 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jia, S. et al. Fate of antibiotic resistance genes and their associations with bacterial community in livestock breeding wastewater and its receiving river water. Water Res.124, 259–268 (2017). [DOI] [PubMed] [Google Scholar]
- 43.Alcock, B. P. et al. CARD 2023: expanded curation, support for machine learning, and resistome prediction at the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res.51, D690–D699 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yushchuk, O., Binda, E. & Marinelli, F. Glycopeptide antibiotic resistance genes: distribution and function in the producer Actinomycetes. Front. Microbiol.11, 1173 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pal, A., He, Y., Jekel, M., Reinhard, M. & Gin, K. Y.-H. Emerging contaminants of public health significance as water quality indicator compounds in the urban water cycle. Environ. Int.71, 46–62 (2014). [DOI] [PubMed] [Google Scholar]
- 46.Lear, G. et al. The biogeography of stream bacteria. Glob. Ecol. Biogeogr.22, 544–554 (2013). [Google Scholar]
- 47.Dickey, J. R. et al. The utility of macroecological rules for microbial biogeography. Front. Ecol. Evol.9, 633155 (2021).
- 48.Smith, L. C. et al. Large-scale drivers of relationships between soil microbial properties and organic carbon across Europe. Glob. Ecol. Biogeogr.30, 2070–2083 (2021). [Google Scholar]
- 49.DeLong, E. F. Microbial community genomics in the ocean. Nat. Rev. Microbiol.3, 459–469 (2005). [DOI] [PubMed] [Google Scholar]
- 50.Omernik, J. M. Ecoregions of the conterminous United States. Ann. Assoc. Am. Geogr.77, 118–125 (1987). [Google Scholar]
- 51.Fine, A. K., van Es, H. M. & Schindelbeck, R. R. Statistics, scoring functions, and regional analysis of a comprehensive soil health database. Soil Sci. Soc. Am. J.81, 589–601 (2017). [Google Scholar]
- 52.Henson, M. W. et al. Nutrient dynamics and stream order influence microbial community patterns along a 2914 kilometer transect of the Mississippi River. Limnol. Oceanogr.63, 1837–1855 (2018). [Google Scholar]
- 53.Satinsky, B. M. et al. Metagenomic and metatranscriptomic inventories of the lower Amazon River, May 2011. Microbiome3, 39 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Maiolini, B. & Bruno, M. C. The River Continuum Concept revisited: Lessons from the Alps (Innsbruck Univ. Press, 2023).
- 55.Mincer, T. J. & Aicher, A. C. Methanol production by a broad phylogenetic array of marine phytoplankton. PLoS ONE11, e0150820 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McInerney, M. J. et al. Physiology, ecology, phylogeny, and genomics of microorganisms capable of syntrophic metabolism. Ann. N. Y. Acad. Sci.1125, 58–72 (2008). [DOI] [PubMed] [Google Scholar]
- 57.Schink, B. & Zeikus, J. G. Microbial methanol formation: a major end product of pectin metabolism. Curr. Microbiol.4, 387–389 (1980). [Google Scholar]
- 58.Cole, J. J. et al. Plumbing the global carbon cycle: integrating inland waters into the terrestrial carbon budget. Ecosystems10, 172–185 (2007). [Google Scholar]
- 59.Gudmundsson, L. et al. Globally observed trends in mean and extreme river flow attributed to climate change. Science371, 1159–1162 (2021). [DOI] [PubMed] [Google Scholar]
- 60.Hundley N. Jr Water and the West: The Colorado River Compact and the Politics of Water in the American West (Univ. California Press, 2009).
- 61.Arora, B. et al. Building cross-site and cross-network collaborations in critical zone science. J. Hydrol.618, 129248 (2023). [Google Scholar]
- 62.Stegen, J. C. & Goldman, A. E. WHONDRS: a community resource for studying dynamic river corridors. mSystems3, e00151-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Garayburu-Caruso, V. A. et al. Using community science to reveal the global chemogeography of river metabolomes. Metabolites10, 518 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Toyoda, J. et al. WHONDRS Summer 2019 Sampling Campaign: Global River Corridor Surface Water FTICR-MS, NPOC, and Stable Isotopes10.15485/1603775 (2020).
- 65.US Geological Survey. In Book 9: Techniques for Water-Resources Investigations Ch. A4 pubs.er.usgs.gov/publication/twri09A4 (2006).
- 66.Lee, C. J. & Henderson, R. J. Tracking Water Quality in U. S. Streams and Rivershttps://pubs.usgs.gov/publication/fs20213019 (USGS, 2020).
- 67.Crump, B. C., Kling, G. W., Bahr, M. & Hobbie, J. E. Bacterioplankton community shifts in an Arctic lake correlate with seasonal changes in organic matter source. Appl. Environ. Microbiol.69, 2253–2268 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kellogg, C. T. E., McClelland, J. W., Dunton, K. H. & Crump, B. C. Strong seasonality in Arctic estuarine microbial food webs. Front. Microbiol.10, 2628 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Borton, M. A. Genome Resolved Open Watersheds database (GROWdb). Githubhttps://github.com/jmikayla1991/Genome-Resolved-Open-Watersheds-database-GROWdb (2023).
- 70.Hill, R. A., Weber, M. H., Leibowitz, S. G., Olsen, A. R. & Thornbrugh, D. J. The Stream-Catchment (StreamCat) Dataset: a database of watershed metrics for the conterminous United States. JAWRA52, 120–128 (2016). [Google Scholar]
- 71.Blodgett, D., Johnson, J. M. & Bock, A. Generating a reference flow network with improved connectivity to support durable data integration and reproducibility in the coterminous US. Environ. Model. Softw.165, 105726 (2023). [Google Scholar]
- 72.Hijmans, R. J. et al. Package ‘terra’ (2022).
- 73.Willi, K. R., Matthew R. V. & ROSS. Genome Resolved Open Watersheds Database (GROWdb) Geospatial data puller. Githubhttps://github.com/rossyndicate/GROWdb (2023).
- 74.Joshi, N. A. & Fass, J. N. Sickle: a windowed adaptive trimming tool for FASTQ files using quality. Githubhttps://github.com/najoshi/sickle (2011).
- 75.Li, D., Liu, C.-M., Luo, R., Sadakane, K. & Lam, T.-W. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics31, 1674–1676 (2015). [DOI] [PubMed] [Google Scholar]
- 76.Kang, D. D. et al. MetaBAT 2: an adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ7, e7359 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Peng, Y., Leung, H. C. M., Yiu, S. M. & Chin, F. Y. L. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics28, 1420–1428 (2012). [DOI] [PubMed]
- 78.Clum, A. et al. DOE JGI metagenome workflow. mSystems6, e00804-20 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nurk, S., Meleshko, D., Korobeynikov, A. & Pevzner, P. A. metaSPAdes: a new versatile metagenomic assembler. Genome Res.27, 824–834 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Parks, D. H., Imelfort, M., Skennerton, C. T., Hugenholtz, P. & Tyson, G. W. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res.25, 1043–1055 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Olm, M. R., Brown, C. T., Brooks, B. & Banfield, J. F. dRep: a tool for fast and accurate genomic comparisons that enables improved genome recovery from metagenomes through de-replication. ISME J.11, 2864–2868 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chaumeil, P.-A., Mussig, A. J., Hugenholtz, P. & Parks, D. H. GTDB-Tk: a toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics36, 1925–1927 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shaffer, M. et al. DRAM for distilling microbial metabolism to automate the curation of microbiome function. Nucleic Acids Res.48, 8883–8900 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Woodcroft, B. J. CoverM. Githubhttps://github.com/wwood/CoverM (2023).
- 86.Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 87.Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tavormina, P. L., Orphan, V. J., Kalyuzhnaya, M. G., Jetten, M. S. M. & Klotz, M. G. A novel family of functional operons encoding methane/ammonia monooxygenase-related proteins in gammaproteobacterial methanotrophs. Environ. Microbiol. Rep.3, 91–100 (2011). [DOI] [PubMed] [Google Scholar]
- 89.Rochman, F. F. et al. Novel copper-containing membrane monooxygenases (CuMMOs) encoded by alkane-utilizing Betaproteobacteria. ISME J.14, 714–726 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Borton, M. A. et al. Coupled laboratory and field investigations resolve microbial interactions that underpin persistence in hydraulically fractured shales. Proc. Natl Acad. Sci. USA115, E6585–E6594 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Solden, L. M. et al. New roles in hemicellulosic sugar fermentation for the uncultivated Bacteroidetes family BS11. ISME J.11, 691–703 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Castresana, J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol.17, 540–552 (2000). [DOI] [PubMed] [Google Scholar]
- 93.Abascal, F., Zardoya, R. & Posada, D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics21, 2104–2105 (2005). [DOI] [PubMed] [Google Scholar]
- 94.Stamatakis, A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics30, 1312–1313 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Letunic, I. & Bork, P. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res.49, W293–W296 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Woodcroft, B. J. et al. SingleM and Sandpiper: robust microbial taxonomic profiles from metagenomic data. Preprint at bioRxiv10.1101/2024.01.30.578060 (2024).
- 97.Borton, M. A. et al. Data for ‘A functional microbiome catalogue crowdsourced from North American rivers’. Zenodo10.5281/zenodo.8173286 (2024). [DOI] [PMC free article] [PubMed]
- 98.Eloe-Fadrosh, E. A. et al. The National Microbiome Data Collaborative Data Portal: an integrated multi-omics microbiome data resource. Nucleic Acids Res.50, D828–D836 (2022). [Google Scholar]
- 99.Borton, M. A. et al. Data generation scripts for ‘A functional microbiome catalogue crowdsourced from North American rivers’. Zenodo10.5281/zenodo.11041178 (2024).
- 100.Borton, M. A. et al. Figure generation code for ‘A functional microbiome catalogue crowdsourced from North American rivers’. Zenodo10.5281/zenodo.11188634 (2024).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tab 1: table summary of river microbiome studies detailing the title, authors and microbiome methods used. Highlighted studies are genome-resolved multi-omics like GROWdb, with only eight rivers surveyed. Tab 2: metagenome and metatranscriptome information. Tab 3: geospatial data (n = 287 variables) for each sample. Tab 4: list of datasets included in geospatial analysis. Tab 5: Chloroplast contamination analysis.
Tab 1: table summary of GROWdb MAGs including taxonomy and quality information. Tab 2: table summary of GROWdb novelty assigned by GTDB-Tk. Tab 3: GROWdb MAG relative abundance across metagenomes. Tab 4: core analysis of GROWdb genera including metagenomic and metatranscriptomic occupancy and mean abundance/expression. Tab 5: global MAG analysis of 9,798 MAGs derived from GROWdb efforts, along with other biogeography studies.
Tab 1: annotation summary of GROWdb MAGs from DRAM. Tab 2: GROWdb whole-genome trait calls. Tab 3: functional genes used to make whole-genome trait calls. Tab 4: output of resistance gene identifier containing list of predicted ARGs. Tab 5: metatranscriptomic expression of identified ARGs by category.
Tab 1: list of genes profiled for emerging contaminants, with ARGs listed in tabs 4–5. Tab 2: annotations of emerging contaminant genes for GROWdb MAGs. Tab 3: table of emerging contaminant genes by genus normalized to the number of genomes. Tab 4: output of resistance gene identifier containing list of predicted ARGs. Tab 5: metatranscriptomic expression of identified ARGs by category. Tab 6: carbon usage analysis in Fig. 4. Tab 7: exact P values for Kruskal–Wallis statistical tests in Fig. 4.
Data Availability Statement
The data underlying GROWdb are accessible across multiple platforms to ensure many levels of data use and structure are widely available. First, all reads and MAGs are publicly hosted at the National Center for Biotechnology (NCBI) under BioProject PRJNA946291. Second, all data presented in this Article, including MAG annotations, phylogenetic tree files, antibiotic-resistance gene database files and expression data tables are available at Zenodo97 (10.5281/zenodo.8173286). Data visualized as maps were derived from publicly available data sources: (1) state boundaries developed using the tigris R package (https://github.com/walkerke/tigris); (2) flowlines from National Hydrography Plus Version 271; (3) ecoregions50 provided from https://www.epa.gov/eco-research/ecoregions. Beyond the content listed above, our aim for GROWdb was to maximize data use by making the data available in searchable and interactive platforms including the National Microbiome Data Collaborative (NMDC) data portal, the Department of Energy’s Systems Biology Knowledgebase (KBase)3 and a GROW-specific user interface released here, GROWdb Explorer. Each platform provides different ways to interact with data in the GROWdb. GROWdb was a flagship project for the newly formed NMDC. Specifically, individual GROWdb datasets (metagenomes, metatranscriptomes and so on) are easily accessible and searchable through the NMDC data portal98 (https://data.microbiomedata.org/), where they are systematically connected to each other and to a rich suite of sample information, other data collected on the same samples and standard analysis results, following findable, accessible, interoperable and reusable data practices26. GROWdb is also a publicly available collection (https://narrative.kbase.us/collections/GROW) within KBase3, with samples, MAGs and corresponding genome-scale metabolic models found in the KBase narrative structure (10.25982/109073.30/1895615). Access within KBase allows for immediate access and reuse of data, including comparison to private data analyses using KBase’s 500+ analysis tools, in a point and click format. GROWdb Explorer is a graphical user interface built through the Colorado State University Geospatial Centroid (https://geocentroid.shinyapps.io/GROWdatabase/), enabling users to search and graph microbial and spatial data simultaneously. Here the microbial data, metabolite and geospatial data are included. The microbial data were distilled into functional gene information, so that biogeochemical contributions and the microorganisms catalysing them can be assessed and visualized rapidly across the dataset. In summary, GROWdb represents to our knowledge the first publicly available genome collection from rivers and offers data that can be leveraged across microbiome studies. GROWdb is an expanding repository to incorporate and unify global river multi-omic data for the future.
All scripts involved with microbial data generation, processing, curation and visualization are available at GitHub and Zenodo99 (https://github.com/jmikayla1991/Genome-Resolved-Open-Watersheds-database-GROWdb/tree/main, 10.5281/zenodo.11041178). Code for geospatial analysis and GROWdb Explorer are available at GitHub (https://github.com/rossyndicate/GROWdb). Code for figures and data analysis are available in Zenodo100 (10.5281/zenodo.11188634).