Abstract
Multiple myeloma (MM) remains incurable despite novel therapeutics. A major contributor to the development of relapsed/refractory and resistant MM is extraosseous extramedullary disease (EMD), whose molecular biology is still not fully understood. We analyzed 528 MM patients who presented to our institution between 2014 and 2021 and who had undergone molecular testing. We defined EMD as organ plasmacytoma distinct from bones and evaluated patients for the development of EMD with the goal of defining their molecular characteristics. Here, we show that RAS/BRAF mutations are likely essential for the development of EMD. Our results also indicate that the underlying reason for the negative outcomes in patients with poor prognostic factors such as duplication 1q and deletion 17p is largely due to the development of EMD. However, the presence of TP53 mutation remains a poor prognostic factor regardless of EMD development. Furthermore, mutation sites of TP53 were different between EMD versus non-EMD patients, with gain-of-function mutations enriched in patients with EMD. Our data highlights distinct molecular abnormalities in patients with EMD and provides potential mechanistic insights for novel therapeutic targets for the future.
Subject terms: Cancer genomics, Myeloma, Myeloma, Genetics research
Introduction
Multiple myeloma (MM) is a bone marrow (BM)-based, multifocal neoplastic proliferation of plasma cells and Extramedullary disease (EMD) is defined by plasma cell neoplasms that arise in tissues distinct from bone [1]. Treatment of MM continues to evolve, resulting in dramatically improved patient outcomes in recent years. Despite these advances, cure remains elusive [2], with EMD playing a major role in the development of relapsed/refractory MM (RRMM) [3, 4] resistant to novel therapies. Typically, EMD is seen in advanced MM, but rarely patients can present with EMD at diagnosis. The prevalence of EMD is approximately 10–15% in all RRMM patients, with only 0.5–6.4 of cases presenting at diagnosis [5–9]. EMD is associated with poor prognosis and is clinically distinct from MM without EMD [3, 10, 11]. Previous reports suggest that EMD can arise in any organ, with the site of origin varying from patient to patient [11, 12]. Although EMD has been defined by plasmacytomas distinct from bone, there are studies and trials which include para-skeletal plasmacytomas as EMD, while others do not [12–15]. The mechanisms involved in the pathogenesis of EMD remain unknown and unclear diagnostic criteria may hinder the understanding of it. Previous studies have reported risk factors for EMD, with some suggesting the potential role of RAS/BRAF mutations in the intramedullary to extramedullary transition in a limited number of patients [16, 17]. Patients with RAS/BRAF mutations showed a higher likelihood of developing EMD compared to patients without this mutation [16–19]. Prevalence of RAS/BRAF mutations increases in RRMM patients [20]. Mutations in TP53, deletion 17p (del (17p)), and duplication 1q (dup (1q)) are well-known poor prognostic factors, and previous reports have suggested that these risk factors are enriched in EMD patients [16, 21–23]. However, the consequent impact of these abnormalities on EMD remains unknown [20]. Thus, defining the molecular characteristics of EMD is critical to a better understanding of EMD biology and will help with advancing treatment of these patients. In the present retrospective study, we report the landscape of molecular features and clinical outcomes of MM patients with EMD from a cohort of 528 MM patients who had undergone molecular testing at a single center.
Methods
Patient cohort
Five hundred twenty-eight MM patients who presented to Massachusetts General Hospital between 2014 and 2021 were evaluated if they had molecular profiling completed on at least one sample (BM and/or EMD and/or bone plasmacytoma). All patients voluntarily provided informed consent approved by the Institutional Review Board for molecular testing. Clinical data was retrospectively collected from electronic medical records. Histological analysis, fluorescence in situ hybridization (FISH), and mutational analysis were conducted on BM aspirates, BM biopsies, and EMD tumor specimens when available.
Diagnosis of EMD
We defined EMD by the presence of plasmacytomas in organs and soft tissue distinct from bones, based on imaging data. The diagnosis of EMD was determined by imaging (CT, PET CT, or MRI) as part of clinical care (Fig. 1A). In some cases, EMD was confirmed by biopsy. Plasma cell leukemia was not included.
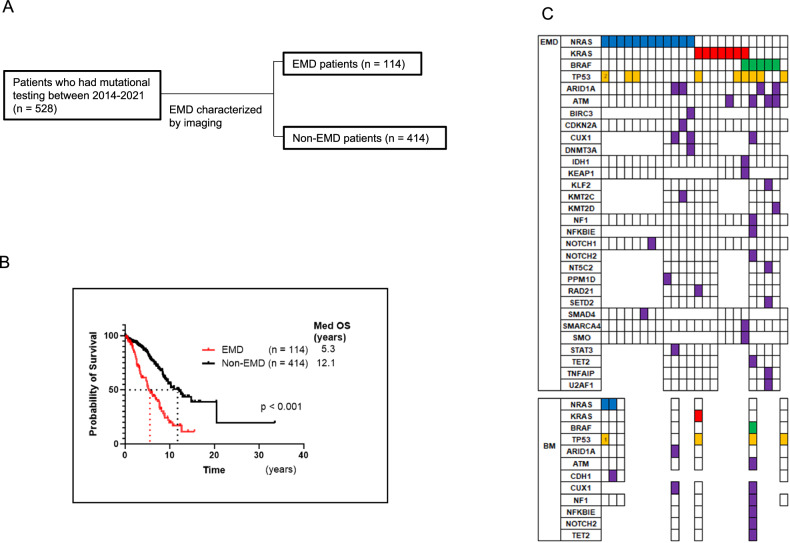
Fig. 1. RAS/BRAF mutations are enriched in extramedullary disease (EMD).
A Study overview of myeloma patients with mutational testing and EMD development characterized by imaging. B Overall survival (OS) of EMD patients (n = 114) versus non-EMD patients (n = 414) demonstrated a median OS of 5.3 years vs 12.1 years (p < 0.001). C Mutational profile of 24 EMD and 7 paired bone marrow (BM) samples. Each column represents a patient sample, and the different rows highlight the mutated genes: 11 patients harbored an NRAS mutation, 7 had a KRAS mutation and 5 patients had a BRAF mutation with 1 patient harboring both BRAF and KRAS mutations.
Mutational analysis
Nucleic acid extraction was performed using Promega Maxwell columns from BM and/or EMD samples obtained from MM patients. Heme panel which is a Multiplexed mutational analysis was conducted with primers designed to cover 91 (until 2018) or 111 (after 2019) genes (Supplementary Table 1), using Anchored Multiplex PCR for single nucleotide variant and insertion/deletion in genomic DNA using ArcherDx platform and Illumina NextSeq. The threshold of tumor cell frequency in BM aspirates and biopsies was determined as 9% based on previous laboratory cutoffs. If samples had a mutation at any time, we defined them as positive for that mutation.
Fluorescent in situ hybridization
Fluorescent in situ hybridization (FISH) was performed at Mayo Clinic laboratories. The probes are listed in Supplementary Table 2. We followed Mayo Clinic laboratories’ interpretation regarding the positivity.
Statistical analysis
Median survival rates for the whole group, and by mutation status, were calculated using the Kaplan-Meier method, while the log-rank test was used to compare survival. In the descriptive analysis, p-values were calculated using Fisher’s exact test. The Kolmogorov-Smirnov test was used to compare the distributions of mutations. All analyses were conducted using Prism software, and a p-value ≤ 0.05 was considered statistically significant.
Results
Mutational testing reveals RAS/BRAF mutations are likely essential for the development of EMD
Five hundred and twenty-eight patients who underwent mutation analysis at our institution were included in this study. All patients were included in the OS study. Data from 528 patients, with a median follow-up of 3.8 years is presented here (Table 1). Overall, 114 patients (21.6%) were classified as having EMD as plasmacytomas distinct from bones by imaging, while 414 patients (78.4%) did not show any evidence of EMD (Fig.1B). Patients with EMD had poorer prognosis compared to those without EMD; median OS was 5.3 years for 114 EMD patients vs 12.1 years for 414 non-EMD patients (p < 0.001) (Fig. 1B). The median time of developing EMD was 3.2 years, and once patients developed EMD, median survival was only 10.1 months (Data not shown). No significant differences were found between EMD and non-EMD patients regarding MM type, gender, and International Staging System (ISS) stage (Table 1). However, age at diagnosis was significant with younger patients developing EMD (mean age = 59.6) compared to non-EMD patients (mean age = 65.2), (p < 0.001). Age stratification showed that the EMD incidence rate decreases with age at MM diagnosis, especially after the age of 50 years (Supplementary Fig. 1A). To compensate for the influence of the duration of follow-up, we analyzed the data by cumulative incidence curve. The data revealed that MM diagnosis at the age of 50 and under was at a significantly higher risk of EMD development (p = 0.027) (Supplementary Fig. 1B). Treatment details are now included in supplementary information (Supplementary Table 3) as no firm conclusions could be drawn due to patient heterogeneity.
Table 1.
Patient Demographics.
| EMD patients | Non-EMD patients | ||
|---|---|---|---|
| Number | 114 | 414 | |
| Follow-up (years) | |||
| Range | 0–15.5 | 0–33.5 | |
| (Median) | 3.5 | 3.9 | |
| Age at Diagnosis | |||
| Range | 28–93 | 29–86 | p < 0.001 |
| (Median) | 60 | 65 | |
| Sex | |||
| Male | 72 (63.2%) | 230 (55.6%) | P = 0.17 |
| Female | 42 (36.8%) | 184 (44.4%) | |
| Myeloma type | |||
| IgG | 53 (46.5%) | 244 (58.9%) | |
| IgA | 26 (22.8%) | 77 (18.6%) | |
| IgD | 1 (0.9%) | 4 (1.0%) | |
| Light Chain | 30 (26.3%) | 84 (20.3%) | |
| Non-secretory | 3 (2.6%) | 5 (1.2%) | |
| ISS | |||
| 1 | 38 (33.3%) | 149 (36.0%) | |
| 2 | 23 (20.2%) | 102 (24.6%) | |
| 3 | 34 (29.8%) | 115 (27.8%) | |
| Unknown | 19 (16.7%) | 48 (11.6%) |
Among 114 EMD patients, 64 had biopsies of EMD tissue, of whom 24 underwent mutational testing. Twenty-three of 24 samples (95.8%) harbored at least one of the RAS/BRAF mutations (NRAS, KRAS, and BRAF); 12 had NRAS mutations, 6 had KRAS mutations, and 4 had BRAF mutations, and 1 had both KRAS and BRAF mutations (Fig. 1C). For BM, samples with less than 9% tumor cells were excluded from mutation analysis evaluation as mentioned. As a result, there were 7 patients with paired BM samples. Interestingly, two EMD-NRAS-positive patients did not have NRAS mutations in their BM (Fig. 1C). The tumor cell ratios in BM of those two patients were 70% and 75%, and the sequencing coverages were 267x and 183x, respectively, which is sufficient to eliminate false negatives. For both patients, EMD samples were collected prior to BM samples. One patient received chemotherapy between those two-time points, and the other patient had focal radiation therapy at the EMD site. Additionally, one of the patients had two TP53 mutations in the EMD site but only had one in the BM. These mutational discrepancies may suggest the heterogeneity of tumor cells throughout the body and indicate that RAS/BRAF-mutated cells predispose to the development of EMD.
The prognosis of patients with BM-RAS/BRAF is largely defined by EMD development
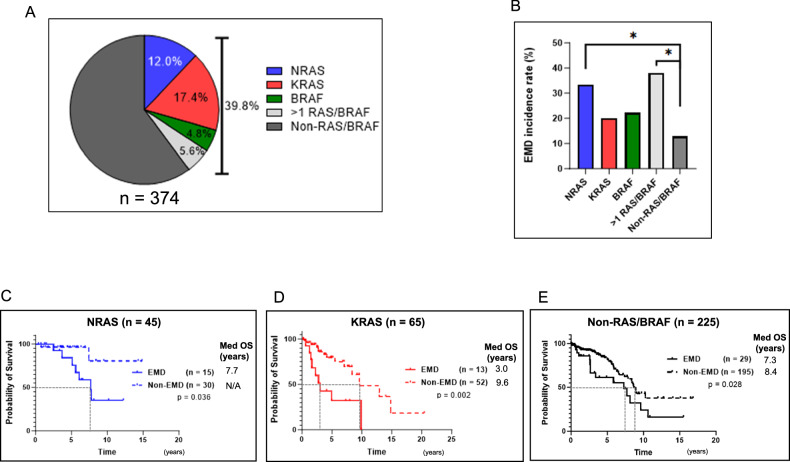
Since RAS/BRAF mutations were enriched in EMD samples, we compared the incidence of EMD and OS of BM-NRAS, KRAS, BRAF single-positive patients and non-RAS/BRAF patients. There were 374 patients whose BM samples had more than 9% of tumor cells (198 patients had samples taken at diagnosis and 45 patients had samples taken at multiple time points). One hundred and forty-nine of 374 patients (39.8%) harbored BM-RAS/BRAF mutations; 45 single-NRAS, 65 single-KRAS, 18 single-BRAF, and 21 had more than 1 mutation present throughout their clinical course (Fig. 2A). The incidence rates of EMD were significantly higher in patients with BM-NRAS and with more than 1 mutation than non-RAS/BRAF patients (Fig. 2B). Generally, the incidence of EMD increases with recurrences, and the majority (54/114, 47.3%) of patients developed EMD after three or more recurrences in our cohort (Data not shown). Therefore, we examined the percentage of BM-RAS/BRAF mutations in relation to disease progression. Similarly, the percentage of patients with BM-RAS/BRAF mutation gradually increased with each relapse, and the highest rate was observed at the time of 3 or more recurrences in EMD patients (61.1%) (Supplementary Fig. 2). Even considering the percentage of RAS/BRAF mutations is associated with relapses, the percentage of RAS/BRAF mutations in EMD samples (95.1%) was significantly higher than that in BM of EMD patients with three or more recurrences (61.1%) (p = 0.004). This may suggest the existence of clonal heterogeneity and the evolutional processes between BM and EMD. We also analyzed the impact of EMD on OS by BM mutation status. Among 45 BM-NRAS, 65 BM-KRAS patients, and 225 non-BM-RAS/BRAF patients, EMD patients had poorer OS than non-EMD patients (Fig. 2C–E). No statistically significant difference was found in 18 patients with BM-BRAF patients and 21 patients with more than 1 mutation by the development of EMD, but this may be as a consequence of limited patient numbers (Data not shown). Among non-EMD patients, neither BM-KRAS nor NRAS had a negative impact on prognosis. These results indicate that the development of EMD has a greater impact on patient’s prognosis than the BM mutation status itself.
Fig. 2. Impact of BM-RAS/BRAF mutations on the development of EMD and overall survival (OS).
A Bone marrow mutational status of 374 in patients who met the criteria for plasma cell cut-off of 9%: single-NRAS (12.0%), single-KRAS (17.4%), single-BRAF (4.8%), and >1 mutation (5.6%)). B EMD incidence rate by BM mutational status (n = 374: single-NRAS (33.3%), single-KRAS (20.0%), single-BRAF (22.2%), >1 mutation (38.1%) and non-RAS/BRAF (12.9%)) (p < 0.05). C OS of 45 BM-NRAS patients with EMD (Median OS: 7.7 years vs Not reached) (p = 0.036). D OS of 65 BM-KRAS patients with EMD (Median OS: 3.0 years vs 9.6 years) (p = 0.002). E OS of 225 Non-BM-RAS/BRAF patients with EMD (Median OS: 7.3 years vs 8.7 years) (p = 0.028).
Poor prognosis of patients with dup (1q) and del (17p) is conferred by the development of EMD
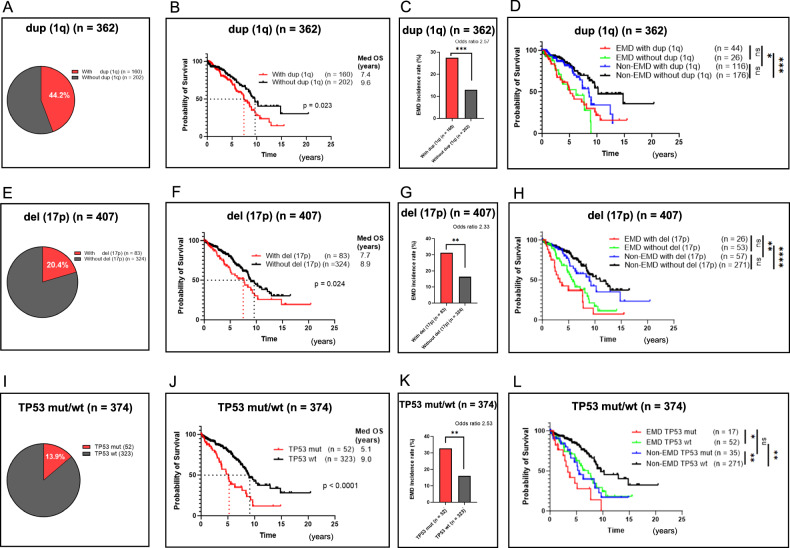
Since the presence of EMD impacts patients’ prognosis, we analyzed molecular abnormalities other than RAS/BRAF mutations in EMD and non-EMD patients. We had three EMD samples analyzed for FISH with the only common abnormality being dup (1q), suggesting a possible underlying contribution of this molecular abnormality to EMD development. In addition to dup (1q), we analyzed del (17p), and TP53 mutation status in bone marrow samples. Among the 362 patients whose BM was analyzed for dup (1q), 160 patients (44.2%) were positive (Fig. 3A). As previously reported, patients with dup (1q) had a poorer prognosis than those without it (Median OS 7.4 years vs 9.6 years, p = 0.023) (Fig. 3B). Patients with dup (1q) also had a higher incidence of EMD compared to those without it (27.5% vs 12.9%, p < 0.001, Odds ratio 2.57) (Fig. 3C). Next, we classified patients into four groups: EMD with and without dup (1q), and non-EMD with and without dup 1q, in order to examine the association between high-risk features, EMD development, and OS. Surprisingly, the presence of dup (1q) did not differentially affect OS in either EMD or non-EMD groups (Fig. 3D). TP53 is on chromosome 17 and both del (17p) and TP53 mutation are reported as risk factors on MM. Regarding del (17p), 83 patients (20.4%) were positive among the 407 patients whose BM was analyzed (Fig. 3E). Similarly, patients with del (17p) also had poorer prognosis compared with those without it (Median OS 7.7 years vs 8.9 years, p = 0.024) (Fig. 3F) and had a higher incidence of EMD (31.3% vs 16.4%, p = 0.003, Odds ratio 2.33) (Fig. 3G). Interestingly, the presence of del (17p) did not differentially affect OS in EMD versus non-EMD patients (Fig. 3H). This suggests that poor prognosis of del (17p) is largely conferred by the development of EMD. With respect to TP53 mutation, it was present in 52 out of 374 patients (13.9%) (Fig.3I). Similar to the dup (1q) and del (17p) data, patients had a poorer prognosis compared to patients with TP53 wild-type (wt) (Median OS 5.1 years vs 9.0 years, p < 0.0001) (Fig. 3J) and these patients had a higher incidence of EMD as well (32.7% vs 16.1%, p = 0.003, Odds ratio 2.33) (Fig. 3K). However, unlike the other poor prognostic factors, there was a statistical difference between TP53 mut and wt even after stratification by EMD development (Fig. 3L). Additionally, there was no statistically significant difference in OS between EMD (Median OS 3.4 years) and non-EMD (Median OS 5.5 years) if the patients had TP53 mutation (p = 0.07). Although the number is limited, these data may indicate that the reason why dup (1q) and del (17p) present a poor prognosis may be largely due to their propensity to develop EMD. On the other hand, TP53 mutations have a negative impact on OS regardless of EMD development.
Fig. 3. Poor outcomes are noted in EMD patients with or without duplication 1q (dup (1q)) and deletion 17p (del (17p)).
TP53 mutation, however, portends a poor outcome regardless of the development of EMD. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001; ns, not significant. A Percentages of patients with and without dup (1q) (n = 362, 44.2% vs 55.8%). B OS of patients with and without dup (1q) (Median OS 7.4 years vs 9.6 years, p = 0.023). C EMD incidence rates of patients with and without dup (1q) (27.5% vs 12.9%, p < 0.001). D OS of EMD and non-EMD patients with and without dup (1q) (Median OS 5.1 years (red), 6.2 years (green), 8.5 years (blue), 10.2 years (black)). E Percentages of patients with and without del (17p) (n = 407, 20.4% vs 79.6%). F OS of patients with and without del (17p) (Median OS 7.7 years vs 8.9 years, p = 0.024). G EMD incidence rates of patients with and without del (17p) (31.3% vs 16.4%, p = 0.003). H OS of EMD and non-EMD patients with and without del (17p) (Median OS 3.4 years (red), 5.8 years (green), 9.0 years (blue), 11.0 years (black)). I Percentages of patients with TP53 mutation (mut) and TP53 wild-type (wt) (n = 374, 13.9% vs 86.1%). J OS of patients with TP53 mut and TP53 wt (Median OS 5.1 years vs 9.0 years, p < 0.0001). K EMD incidence rates of patients with TP53 mut and wt (32.7% vs 16.1%, p = 0.007). L OS of EMD and non-EMD patients with TP53 mut and TP53 wt (Median OS 3.4 years (red), 5.5 years (green), 7.3 years (blue), 9.6 years (black)).
The site of TP53 mutation, but not RAS/BRAF, likely affects EMD development
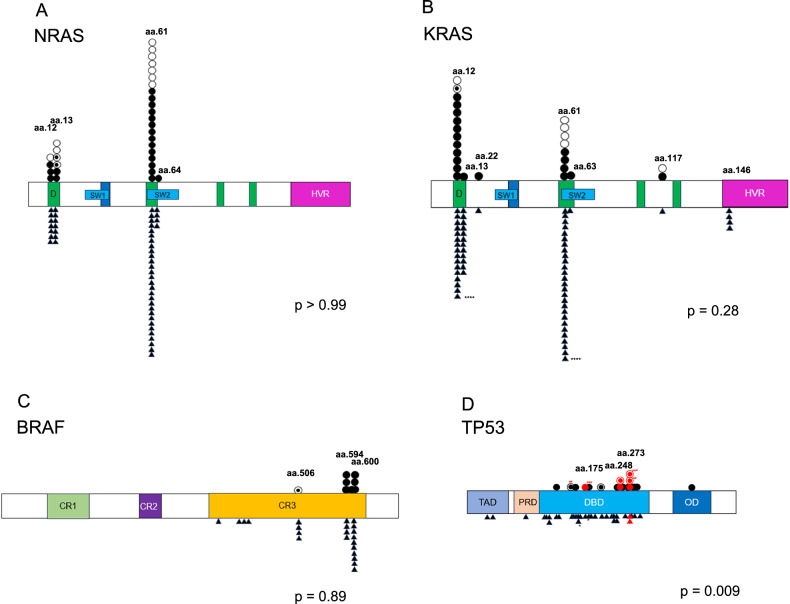
We next sought to understand the influence of RAS/BRAF and TP53 mutation sites on EMD occurrence. All mutations from BM and EMD samples were plotted (Fig. 4A–D). Our results showed that most of the mutations were seen in recurrent mutation sites and no difference was found between EMD and non-EMD patients among RAS/BRAF mutation sites (Fig. 4A–C). In contrast, there were differences in TP53 mutation sites between EMD and non-EMD patients. Non-EMD patients showed mutations distributed throughout the gene, while EMD patients had more mutations in the C-terminus of the TP53 gene (Fig. 4D) (p = 0.009). One patient showed two TP53 mutations in both C- and N-terminus in the EMD sample but only an N-terminal TP53 mutation in a subsequent BM sample. Notably, six of 17 EMD patients had TP53 mutations accumulated in the sites of aa175, 248, and 273 which are reported as gain-of-function mutation sites in other cancers, while only 2 were found in 35 non-EMD patients. (35.3%: 5.7%, p = 0.011). This suggests that TP53 mutations have different molecular effects depending on their sites and gain-of-function may contribute to the development of EMD.
Fig. 4. Mutation sites differ with respect to TP53, but not RAS/BRAF, in EMD versus non-EMD patients.
Individual data points of mutations from EMD patients (above the gene bar) and non-EMD patients (below the gene bar) are represented(〇EMD tissue of EMD patients, ●BM of EMD patients, ◉Both BM and EMD tissue of EMD patients, ▲BM of non-EMD patients). The bar represents D (DNA binding lesion); SW (switch region); HVR, (hypervariable region); CR, (conserved region); TAD, (transactivation domain); PRD, (proline rich domain); DBD, (DNA binding domain); OD, (oligomerization domain). One non-EMD patient (*) and three EMD patients had 2 mutations in their samples (**, ***, and ****, respectively). Red symbols represent gain-of-function mutations. A Plots demonstrating NRAS mutation sites (p > 0.99). B Plots demonstrating KRAS mutation sites (p = 0.28). C Plots demonstrating BRAF mutation sites (p = 0.89). D Plots demonstrating TP53 mutation sites (p = 0.009).
Discussion
Several studies have conducted mutational analyses in MM patient samples and demonstrated significant heterogeneity [24–27]. The clinical course of MM patients also varies widely across patients, which underscores the use of an individualized treatment approach. In recent years, there has been an unmet need to elucidate the molecular mechanisms underlying the development of EMD, one of the most refractory and intractable conditions of MM even with the availability of novel T cell redirected therapies. Myeloma typically occurs in older people and its incidence in patients below the age of 50 is low. In the past, there has been a multi-center retrospective study which showed that patients younger than 50 showed favorable factors with regard to ISS stage, as well as a fair prognosis [28]. However, according to our data, there was no clear correlation between ISS staging and EMD development. Our cumulative incidence data revealed that patients who were diagnosed with MM under the age of 50 were at a higher risk for EMD development even in the era of new drugs. Going forward, larger prospective studies will be needed to confirm our preliminary findings.
Numerous past studies have reported clinical data on EMD; however, sample sizes were often limited and the definition of EMD has not been clear or consistent [5–8]. With the definition of EMD as organ plasmacytoma distinct from bones, our data shows a strong relationship between EMD and RAS/BRAF mutations observed in MM patients. It is known that RAS/BRAF mutations are usually observed in advanced-stage MM rather than MGUS, SMM, or newly diagnosed MM, and previous data indicate that RAS/BRAF mutations are acquired throughout the clinical course with disease progression as a consequence of clonal evolution [29]. In solid cancers, commonly occurring missense mutations in three members of RAS family genes result in their constitutive activation, and importantly, these mutations serve as predictors of poor patient survival [30, 31]. This is largely due to the association of RAS/BRAF mutations with the presence of metastases at diagnosis and with inherent or acquired resistance to treatments [32–34]. Our data indicates that EMD incidence is higher in BM-RAS/BRAF patients than non-BM-RAS/BRAF patients and the development of EMD is a poor prognostic factor regardless of the BM mutational status. Having a RAS/BRAF mutation alone does not result in a uniform clinical phenotype, nor does it directly determine prognosis. In our cohort, EMD patients with BM-KRAS had the worst outcome, with an OS of 3.0 years. Several studies have reported that the prognosis of patients who had KRAS mutations is worse than that of non-RAS/BRAF patients [35–37]. The poorer OS among EMD patients may be a contributing factor to the unfavorable prognosis of BM-KRAS patients in previous studies.
Whole genome sequencing studies have revealed that the genetic landscape of metastasis in solid cancers is distinct from the original site and is thought to be an evolutionary process [38, 39]. It has been reported that multiple myeloma patients have a mixture of clones in their BM and the dominance of these clones changes throughout the clinical course, leading to refractory disease [40]. Interestingly, in our cohort, two EMD-NRAS-positive patients did not have NRAS mutations in their BM. Also, one of those EMD samples had two TP53 mutations, one of which was not detectable in BM. Of note, both BM samples were acquired after EMD samples, and one patient only had radiotherapy without systemic therapy between the two biopsies. Previous studies have shown similar results, but the authors concluded that detection limitations may have occurred owing to the low tumor burden in the BM [16]. However, BM samples collected in our study met the 9% tumor cell frequency threshold, which is sufficient to detect mutational burden. Although RAS/BRAF mutations are likely essential to the development of EMD and incidence of EMD is enriched in BM-RAS/BRAF patients, there was no difference in the site of RAS/BRAF mutation between EMD and non-EMD patients. It is also possible that patients who were negative for RAS/BRAF mutations in BM may have had mutated cells at an undetectable level which was not a dominant clone. These data may suggest a temporal and spatial heterogeneity of MM cells in patients.
Although RAS/BRAF mutations are likely essential for the development of EMD, not all BM-RAS/BRAF patients develop EMD suggesting that the mutation alone is not sufficient to develop EMD. It is well known that dup (1q), del (17p), and TP53 mutations are poor prognostic factors, and these are enriched in EMD patients which we confirmed in our cohort. Surprisingly, our detailed analyses revealed that EMD is a poor prognostic factor even in patients with dup (1q) and del (17p). In addition, there was no statistical difference in OS with or without the presence of these risk factors in both EMD and non-EMD patient groups. This indicates that the manner in which these are poor prognostic factors is largely due to the propensity to the development of EMD. TP53 is one of the most significant tumor suppressor genes, and its mutation is often observed in a variety of cancers [41]. It consists of four domains, with most mutations contained in the DNA binding domain [42]. Some studies have shown the distinct functions of TP53 N-terminus and C-terminus, with the C-terminus controlling site-specific DNA binding and structural changes within the central DNA binding domain. It is reported that the TP53 gene has not only loss-of-function but also gain-of-function mutations [43]. Those gain-of-function mutations are known to promote cancer progression and metastasis in xenograft models and are associated with poor clinical outcomes in patients [44–47]. In the context of myeloma, it has been previously reported that mutations of TP53 are associated with poor clinical outcomes, as they are linked with more aggressive and advanced forms of the disease, but the exact molecular role of TP53 mutations in MM patients is unclear, including the mechanism of loss-of-function and gain-of-function mutations [47]. Interestingly, we found significant differences in the site of TP53 mutations between EMD and non-EMD patients; non-EMD patients had mutations distributed throughout the gene, while EMD patients showed more mutations near the C-terminus, and accumulated in the sites of aa175, 248, and 273. These specific mutations are known as gain-of-function mutations. Currently, the function of TP53 mutations in MM is poorly understood and no risk classification based on the site of TP53 mutation exists. Our data indicates that the site of TP53 mutations has a molecularly important impact on the development of EMD and that specific mutations may help to elucidate the underlying mechanisms of EMD. It also suggests that not all TP53 mutations should be considered equal and that depending on mutation site the prognostic significance may vary. To our knowledge, this is the first report suggesting the relevance between EMD and TP53 gain-of-function mutations. This analysis encompasses both newly diagnosed and RRMM; therefore, a more extensive and homogeneous prospective study in the future would be desirable. This was also why no specific conclusions could be drawn on impact of treatment on our current cohort.
In summary, EMD sites may have different mutational landscapes from the BM in MM which suggests that RAS/BRAF mutations may play a critical role in the development of EMD, coupled with other associated molecular abnormalities, especially dup (1q) and del (17p), and site-specific TP53 mutations. By integrating clinical outcomes and molecular features, we have revealed the clinical impact of EMD on MM patients and its association with the influences of known risk factors. These results will enable us to provide more accurate EMD risk stratification and provide insights for novel approaches in the treatment of MM.
Supplementary information
Acknowledgements
RN-M is supported by the Japan Society for the Promotion of Science. NSR is supported by the Paula and Rodger Riney Foundation.
Author contributions
RN-M and NSR were responsible for the conceptualization and acquisition of clinical information. VN performed the mutational analysis. RN-M and NH analyzed the data. TF, RS, FR, KO, CP, KF, FR, and DST provided helpful discussion. ARB, DC, AJY, and NSR contributed to the sample collection and patient treatments. RN-M, R-HS, and NSR wrote the manuscript and all the authors reviewed the manuscript.
Data availability
The data that support the findings of this study are available upon request from the corresponding author. The data is not publicly available due to privacy and ethical restrictions.
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
This study was approved by the Institutional Review Board of Massachusetts General Hospital (15-190). All methods were performed in accordance with the Declaration of Helsinki, the relevant guidelines, and regulations. Informed consent was obtained from all individual participants in this analysis.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41408-024-01190-9.
References
- 1.E.Campo NLH, Jaffe ES, Pileri SA, Stein H, Thiele J. WHO classification of tumors of haematopoietic and lymphoid tissues. (IARC Press; 2016).
- 2.O’Donnell EK, Shapiro YN, Yee AJ, Nadeem O, Hu BY, Laubach JP, et al. Quality of life, psychological distress, and prognostic perceptions in patients with multiple myeloma. Cancer 2022;128:1996–2004. [DOI] [PubMed] [Google Scholar]
- 3.Bhutani M, Foureau DM, Atrash S, Voorhees PM, Usmani SZ. Extramedullary multiple myeloma. Leukemia 2020;34:1–20. [DOI] [PubMed] [Google Scholar]
- 4.Dima D, Ullah F, Mazzoni S, Williams L, Faiman B, Kurkowski A, et al. Management of relapsed-refractory multiple myeloma in the era of advanced therapies: evidence-based recommendations for routine clinical practice. Cancers. 2023;15:2160. [DOI] [PMC free article] [PubMed]
- 5.Pintoffl JP, Weisel K, Schulze M, Maksimovic O, Claussen CD, Kramer U, et al. Role of dynamic contrast-enhanced sonography for characterization and monitoring of extramedullary myeloma: comparison with serologic data. J Ultrasound Med 2013;32:1777–88. [DOI] [PubMed] [Google Scholar]
- 6.Cavo M, Terpos E, Nanni C, Moreau P, Lentzsch S, Zweegman S, et al. Role of (18)F-FDG PET/CT in the diagnosis and management of multiple myeloma and other plasma cell disorders: a consensus statement by the International Myeloma Working Group. Lancet Oncol 2017;18:e206–e17. [DOI] [PubMed] [Google Scholar]
- 7.Montefusco V, Gay F, Spada S, De Paoli L, Di Raimondo F, Ribolla R, et al. Outcome of paraosseous extra-medullary disease in newly diagnosed multiple myeloma patients treated with new drugs. Haematologica 2020;105:193–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blade J, Beksac M, Caers J, Jurczyszyn A, von Lilienfeld-Toal M, Moreau P, et al. Extramedullary disease in multiple myeloma: a systematic literature review. Blood Cancer J 2022;12:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosinol L, Beksac M, Zamagni E, Van de Donk N, Anderson KC, Badros A, et al. Expert review on soft-tissue plasmacytomas in multiple myeloma: definition, disease assessment and treatment considerations. Br J Haematol 2021;194:496–507. [DOI] [PubMed] [Google Scholar]
- 10.Terpos E, Rezvani K, Basu S, Milne AE, Rose PE, Scott GL, et al. Plasmacytoma relapses in the absence of systemic progression post-high-dose therapy for multiple myeloma. Eur J Haematol 2005;75:376–83. [DOI] [PubMed] [Google Scholar]
- 11.Usmani SZ, Heuck C, Mitchell A, Szymonifka J, Nair B, Hoering A, et al. Extramedullary disease portends poor prognosis in multiple myeloma and is over-represented in high-risk disease even in the era of novel agents. Haematologica 2012;97:1761–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Short KD, Rajkumar SV, Larson D, Buadi F, Hayman S, Dispenzieri A, et al. Incidence of extramedullary disease in patients with multiple myeloma in the era of novel therapy, and the activity of pomalidomide on extramedullary myeloma. Leukemia 2011;25:906–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu P, Davies FE, Boyd K, Thomas K, Dines S, Saso RM, et al. The impact of extramedullary disease at presentation on the outcome of myeloma. Leuk Lymphoma 2009;50:230–5. [DOI] [PubMed] [Google Scholar]
- 14.Varettoni M, Corso A, Pica G, Mangiacavalli S, Pascutto C, Lazzarino M. Incidence, presenting features and outcome of extramedullary disease in multiple myeloma: a longitudinal study on 1003 consecutive patients. Ann Oncol 2010;21:325–30. [DOI] [PubMed] [Google Scholar]
- 15.Deng H, Liu M, Yuan T, Zhang H, Cui R, Li J, et al. Efficacy of humanized Anti-BCMA CAR T cell therapy in relapsed/refractory multiple myeloma patients with and without extramedullary disease. Front Immunol 2021;12:720571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rasmussen T, Kuehl M, Lodahl M, Johnsen HE, Dahl IM. Possible roles for activating RAS mutations in the MGUS to MM transition and in the intramedullary to extramedullary transition in some plasma cell tumors. Blood 2005;105:317–23. [DOI] [PubMed] [Google Scholar]
- 17.de Haart SJ, Willems SM, Mutis T, Koudijs MJ, van Blokland MT, Lokhorst HM, et al. Comparison of intramedullary myeloma and corresponding extramedullary soft tissue plasmacytomas using genetic mutational panel analyses. Blood Cancer J 2016;6:e426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andrulis M, Lehners N, Capper D, Penzel R, Heining C, Huellein J, et al. Targeting the BRAF V600E mutation in multiple myeloma. Cancer Discov 2013;3:862–9. [DOI] [PubMed] [Google Scholar]
- 19.Liu Y, Jelloul F, Zhang Y, Bhavsar T, Ho C, Rao M, et al. Genetic basis of extramedullary plasmablastic transformation of multiple myeloma. Am J Surg Pathol 2020;44:838–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kortum KM, Mai EK, Hanafiah NH, Shi CX, Zhu YX, Bruins L, et al. Targeted sequencing of refractory myeloma reveals a high incidence of mutations in CRBN and Ras pathway genes. Blood 2016;128:1226–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang H, Sloan S, Li D, Keith Stewart A. Multiple myeloma involving central nervous system: high frequency of chromosome 17p13.1 (p53) deletions. Br J Haematol 2004;127:280–4. [DOI] [PubMed] [Google Scholar]
- 22.Qu X, Chen L, Qiu H, Lu H, Wu H, Qiu H, et al. Extramedullary manifestation in multiple myeloma bears high incidence of poor cytogenetic aberration and novel agents resistance. Biomed Res Int 2015;2015:787809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Billecke L, Murga Penas EM, May AM, Engelhardt M, Nagler A, Leiba M, et al. Cytogenetics of extramedullary manifestations in multiple myeloma. Br J Haematol 2013;161:87–94. [DOI] [PubMed] [Google Scholar]
- 24.Maura F, Bolli N, Angelopoulos N, Dawson KJ, Leongamornlert D, Martincorena I, et al. Genomic landscape and chronological reconstruction of driver events in multiple myeloma. Nat Commun 2019;10:3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chapman MA, Lawrence MS, Keats JJ, Cibulskis K, Sougnez C, Schinzel AC, et al. Initial genome sequencing and analysis of multiple myeloma. Nature 2011;471:467–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O’Donnell E, Mahindra A, Yee AJ, Nardi V, Birrer N, Horick N, et al. Clinical grade “SNaPshot” genetic mutation profiling in multiple myeloma. EBioMedicine 2015;2:71–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walker BA, Boyle EM, Wardell CP, Murison A, Begum DB, Dahir NM, et al. Mutational spectrum, copy number changes, and outcome: results of a sequencing study of patients with newly diagnosed myeloma. J Clin Oncol 2015;33:3911–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ludwig H, Durie BG, Bolejack V, Turesson I, Kyle RA, Blade J, et al. Myeloma in patients younger than age 50 years presents with more favorable features and shows better survival: an analysis of 10 549 patients from the International Myeloma Working Group. Blood 2008;111:4039–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Egan JB, Shi CX, Tembe W, Christoforides A, Kurdoglu A, Sinari S, et al. Whole-genome sequencing of multiple myeloma from diagnosis to plasma cell leukemia reveals genomic initiating events, evolution, and clonal tides. Blood 2012;120:1060–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu J, Pfarr N, Endris V, Mai EK, Md Hanafiah NH, Lehners N, et al. Molecular signaling in multiple myeloma: association of RAS/RAF mutations and MEK/ERK pathway activation. Oncogenesis 2017;6:e337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Frank AJ, Dagogo-Jack I, Dobre IA, Tait S, Schumacher L, Fintelmann FJ, et al. Management of lung cancer in the patient with interstitial lung disease. Oncologist 2023;28:12–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stephen AG, Esposito D, Bagni RK, McCormick F. Dragging Ras back in the ring. Cancer Cell 2014;25:272–81. [DOI] [PubMed] [Google Scholar]
- 33.Renaud S, Seitlinger J, Falcoz PE, Schaeffer M, Voegeli AC, Legrain M, et al. Specific KRAS amino acid substitutions and EGFR mutations predict site-specific recurrence and metastasis following non-small-cell lung cancer surgery. Br J Cancer 2016;115:346–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nazarian R, Shi H, Wang Q, Kong X, Koya RC, Lee H, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature 2010;468:973–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu P, Leong T, Quam L, Billadeau D, Kay NE, Greipp P, et al. Activating mutations of N- and K-ras in multiple myeloma show different clinical associations: analysis of the Eastern Cooperative Oncology Group Phase III Trial. Blood 1996;88:2699–706. [PubMed] [Google Scholar]
- 36.Smith D, Armenteros E, Percy L, Kumar M, Lach A, Herledan G, et al. RAS mutation status and bortezomib therapy for relapsed multiple myeloma. Br J Haematol 2015;169:905–8. [DOI] [PubMed] [Google Scholar]
- 37.Chng WJ, Gonzalez-Paz N, Price-Troska T, Jacobus S, Rajkumar SV, Oken MM, et al. Clinical and biological significance of RAS mutations in multiple myeloma. Leukemia 2008;22:2280–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rabbie R, Ferguson P, Wong K, Couturier DL, Moran U, Turner C, et al. The mutational landscape of melanoma brain metastases presenting as the first visceral site of recurrence. Br J Cancer 2021;124:156–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paul MR, Pan TC, Pant DK, Shih NN, Chen Y, Harvey KL, et al. Genomic landscape of metastatic breast cancer identifies preferentially dysregulated pathways and targets. J Clin Invest 2020;130:4252–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Keats JJ, Chesi M, Egan JB, Garbitt VM, Palmer SE, Braggio E, et al. Clonal competition with alternating dominance in multiple myeloma. Blood 2012;120:1067–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shajani-Yi Z, de Abreu FB, Peterson JD, Tsongalis GJ. Frequency of somatic TP53 mutations in combination with known pathogenic mutations in colon adenocarcinoma, non-small cell lung carcinoma, and gliomas as identified by next-generation sequencing. Neoplasia 2018;20:256–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kato S, Han SY, Liu W, Otsuka K, Shibata H, Kanamaru R, et al. Understanding the function-structure and function-mutation relationships of p53 tumor suppressor protein by high-resolution missense mutation analysis. Proc Natl Acad Sci Usa 2003;100:8424–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dittmer D, Pati S, Zambetti G, Chu S, Teresky AK, Moore M, et al. Gain of function mutations in p53. Nat Genet 1993;4:42–6. [DOI] [PubMed] [Google Scholar]
- 44.Lang GA, Iwakuma T, Suh YA, Liu G, Rao VA, Parant JM, et al. Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell 2004;119:861–72. [DOI] [PubMed] [Google Scholar]
- 45.Schulz-Heddergott R, Stark N, Edmunds SJ, Li J, Conradi LC, Bohnenberger H, et al. Therapeutic ablation of gain-of-function mutant p53 in colorectal cancer inhibits Stat3-mediated tumor growth and invasion. Cancer Cell 2018;34:298–314.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cooks T, Pateras IS, Jenkins LM, Patel KM, Robles AI, Morris J, et al. Mutant p53 cancers reprogram macrophages to tumor supporting macrophages via exosomal miR-1246. Nat Commun 2018;9:771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Flynt E, Bisht K, Sridharan V, Ortiz M, Towfic F, Thakurta A. Prognosis, biology, and targeting of TP53 dysregulation in multiple myeloma. Cells. 2020;9:287. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available upon request from the corresponding author. The data is not publicly available due to privacy and ethical restrictions.