Abstract
The peacock blenny Salaria pavo is notorious for its extreme male sexual polymorphism, with large males defending nests and younger reproductive males mimicking the appearance and behavior of females to parasitically fertilize eggs. The lack of a reference genome has, to date, limited the understanding of the genetic basis of the species phenotypic plasticity. Here, we present the first reference genome assembly of the peacock blenny using PacBio HiFi long-reads and Hi-C sequencing data. The final assembly of the S. pavo genome spanned 735.90 Mbp, with a contig N50 of 3.69 Mbp and a scaffold N50 of 31.87 Mbp. A total of 98.77% of the assembly was anchored to 24 chromosomes. In total, 24,008 protein-coding genes were annotated, and 99.0% of BUSCO genes were fully represented. Comparative analyses with closely related species showed that 86.2% of these genes were assigned to orthogroups. This high-quality genome of S. pavo will be a valuable resource for future research on this species’ reproductive plasticity and evolutionary history.
Subject terms: Genome, Data publication and archiving, Genomics
Background & Summary
Phenotypic plasticity, which allows organisms to respond to their environment without altering their underlying genotype, is widespread in nature and is considered a potential adaptation for thriving in changing environments1,2. Hence, it is hypothesized that phenotypic plasticity plays a key role in the evolution of phenotypic diversity among species2. Remarkable examples of morphological, physiological, habitat, and behavioral diversity have been described in teleost fish, representing the richest evolutionary radiation of vertebrates, with more than 30,000 species described3. The African cichlids are a classical example of this phenomenon, where adaptive radiation allowed species into a variety of ecological niches over a short evolutionary time span4,5. However, less focus has been given to the genomic mechanisms underlying plastic responses within species expressed either within (e.g. sex change in the bluehead wrasse Thalassoma bifasciatum6) or across (e.g. temperature effects on sex ratios in the zebrafish Danio rerio7) generations.
The peacock blenny Salaria pavo (Teleostei: Blenniidae; Fig. 1), a small intertidal fish found in the rocky shores of the Mediterranean and adjacent Atlantic regions8, is a remarkable example of reproductive plasticity. In this species, nest-holder males are larger than females and exhibit well-developed secondary sexual characters (SSCs), such as a head crest and a sex-pheromone-producing anal gland, which they use to attract females to their nests for spawning9,10, providing sole parental care to eggs until hatching11,12. The intensity of mating competition varies among populations and is primarily influenced by the abundance of nest sites, which impacts mate availability during the breeding season, leading to the appearance of polymorphic reproductive phenotypes. In populations inhabiting coastal lagoons, nest sites are scarce (e.g. Ria Formosa, southern Portugal) and a strong intrasexual competition for mates is present, leading to a sex role reversal in courtship behavior, with females becoming the courting sex13–15. The scarceness of nests leads smaller and younger males to adopt an alternative male reproductive tactic (ART), behaving as female-mimics to approach nests defended by nest-holder males and sneak fertilizations16. During the breeding season, males’ territory is restricted to the nest, with females competing for access by displaying elaborate courtship behaviors, consisting of stereotypic movements and a transient nuptial coloration, more often than nest-holder males13,14,17. Sneaker males switch to nest-holders after their first breeding season18, encompassing a transitional male morph which is reproductively inactive. Hence, in this species, the same male can express both male and female reproductive behavior during its lifetime. In contrast, rocky shore populations have abundant nest sites (e.g. Adriatic Sea, Gulf of Trieste), with nest-holder males establishing nests in rock crevices and aggressively defending a territory around the nest, where they display courtship behavior towards females, who usually assume a passive role in courtship responding with changes in coloration and few displays before entering the nest to spawn (i.e. ‘conventional’ sex roles)15. Interestingly, experiments under similar laboratory conditions indicate that the expression of courtship behavior in this species is plastic19, suggesting a genomic regulation of this behavior. This species thus exhibits high polymorphism in mating strategies, with plastic behavioral phenotypes in both sexes, and developmental plasticity in males (i.e. life-history traits).
Fig. 1.

Peacock blenny Salaria pavo nest-holder male, inside the nest, being courted by two females on the right and one sneaker male on the left.
In total, 402 species belonging to 59 genera have been identified within the Blenniidae family (Fishbase3). The peacock blenny is one of the two species currently recognized within the Salaria genus, alongside Salaria basilisca20. Previously, the genus included several freshwater species until a recent taxonomic revision by Vecchioni et al.21, which reclassified these species under the newly established genus Salariopsis. The phylogeographic history of the genus Salaria is complex, with the marine ancestor of S. pavo thought to have diverged from the ancestor of the freshwater lineage in the Middle Miocene22. Notably, within the Salariinae subfamily, male ARTs have been described in at least three additional species besides S. pavo: Scartella cristata23, Salaria fluviatilis24, and Parablennius sanguinolentus parvicornis25, though the expression of ARTs varies among species.
Although the ecology, behavior, and physiology of S. pavo are well characterized, with brain transcriptomic profiles linked to each male morphotype and females26, a complete genome assembly has been lacking, limiting the understanding of the genomic mechanisms associated with phenotypic plasticity in this species. Furthermore, epigenetic mechanisms have been proposed as key drivers of plasticity, allowing for rapid genome modulation in response to environmental cues through changes in epigenetic marks in gene regulatory networks that drive whole-organism performance27. Therefore, we constructed a high-quality chromosome-level genome for S. pavo using a combination of DNBSEQ short reads, HiFi long reads, and Hi-C data. We also annotated the genome for repetitive elements, protein-coding genes and non-coding RNAs (ncRNAs). This genome offers the possibility for further studies on the genomic mechanisms underlying the behavioral and developmental plasticity observed in this species, and contributes to the genome database of the combtooth blennies, allowing further research on male ARTs, and evolutionary and biogeographic analyses within this family.
Methods
Fish sampling
One nest-holder male peacock blenny, S. pavo, collected at Culatra Island (Ria Formosa Natural Park, 36°59′N, 7°51′W, Portugal; for a description of the sampling area, see13) was euthanized using an overdose of buffered MS-222 (tricaine methanesulfonate anesthetic; Sigma-Aldrich), the blood and longitudinal muscle sampled immediately and snap-frozen in liquid nitrogen following BGI tissue sampling guidelines. Additionally, seven tissues from the same individual were dissected out – gill together with branchial arch, liver, testis, muscle, kidney, heart, and whole brain together with pituitary – and snap-frozen for RNA analysis. All samples were stored at −80°C until extraction. Fish procedures were performed in accordance with accepted veterinary practice under a “Group-1” license issued by the Portuguese National Authority for Animal Health (DGAV), permit number 0421/000/000/2013.
Library construction and sequencing
Sample extraction, library preparation and sequencing were performed by BGI Tech Solutions (Hong Kong, China). For short-read sequencing, gDNA extracted from muscle tissue was randomly fragmented with an insert size of 350 bp and sequenced using the DNBSEQ-G400 platform to generate 150 bp paired-end reads. The raw short reads were filtered by SOAPnuke v2.1.728 to remove adapters and low-quality reads with the parameters: -n 0.001, -l 10, -q 0.5, --adaMR 0.25,--apolyX 50, and --aminReadLen 150. A total of 79.1 Gb of clean data resulting in 107.49-fold coverage of the S. pavo genome was obtained (Table 1).
Table 1.
Statistics of sequencing data generated by DNBSEQ and PacBio.
| Libraries | Total length (bp) | Read count | Read length (bp) | Sequence coverage (X) |
|---|---|---|---|---|
| DNBSEQ reads | 79,104,612,300 | 263,682,041 | 150 | 107.49 |
| PacBio reads | 49,319,533,384 | 2,494,162 | 19,789 | 67.02 |
| Total | 128,424,145,684 | 266,176,203 | — | 174.51 |
For HiFi (high-fidelity) sequencing, high-molecular-weight (HMW) gDNA obtained from the blood sample was sheared to approximately 15 Kb selected using Sage ELF before preparing the PacBio HiFi library. The genomic library was sequenced in CCS mode on the Pacific Biosciences Sequel II System using two cells. A total of 49.3 Gb of long clean reads were generated, with mean lengths of 19.5 Kb and 20.0 Kb, respectively, resulting in 67.02-fold coverage of the S. pavo genome (Table 1).
For Hi-C sequencing, cells obtained from muscle tissue were treated with the crosslinking agent formaldehyde to fix the DNA and its binding proteins. After cell lysis, the DNA was digested with the restriction enzyme DpnII and marked with a biotinylated residue29. The blunt ends of the crosslinked DNA fragments were then ligated using DNA ligase to form circular molecules, which were subsequently purified and sheared. The target DNA was captured through a biotin-streptavidin-mediated pull-down. Following the construction of the Hi-C library, sequencing was performed on the DNBSEQ-G400 platform as 150 bp paired-end reads. Quality control and filtering of the sequencing data were conducted using SOAPnuke v2.1.728 with the parameters: -n 0.001, -l 20, -q 0.3,–adaMR 0.25,–polyX 50, and–minReadLen 150. After filtering, 96.08 Gb of clean data were obtained, with Q20 and Q30 scores of 98.25% and 93.83%, respectively (Table 2).
Table 2.
Statistics of Hi-C clean data.
| Clean Reads | Clean Bases | Q20 (%) | Q30 (%) | GC (%) |
|---|---|---|---|---|
| 320,268,665 | 96,080,599,500 | 98.25 | 93.83 | 41.20 |
Strand-specific mRNA libraries were prepared for each tissue and sequenced on the DNBSEQ-G400 platform as 150 bp paired-end reads. Raw data with adapter sequences or low-quality sequences were filtered with SOAPnuke v2.1.728 using the following parameters: -n 0.001, -l 20, -q 0.4, --adaMR 0.25, --polyX 50, and --minReadLen 150. A total of 50.0 Gb of clean data was generated – gill together with branchial arch (7.2 Gb), liver (7.2 Gb), testis (7.2 Gb), muscle (6.8 Gb), kidney (7.2 Gb), heart (7.2 Gb), and whole brain together with pituitary (7.2 Gb). These RNA-seq datasets will be assembled into a reference transcriptome and the transcripts used for structural and gene annotation of the assembled genome.
Genome survey and de novo genome assembly
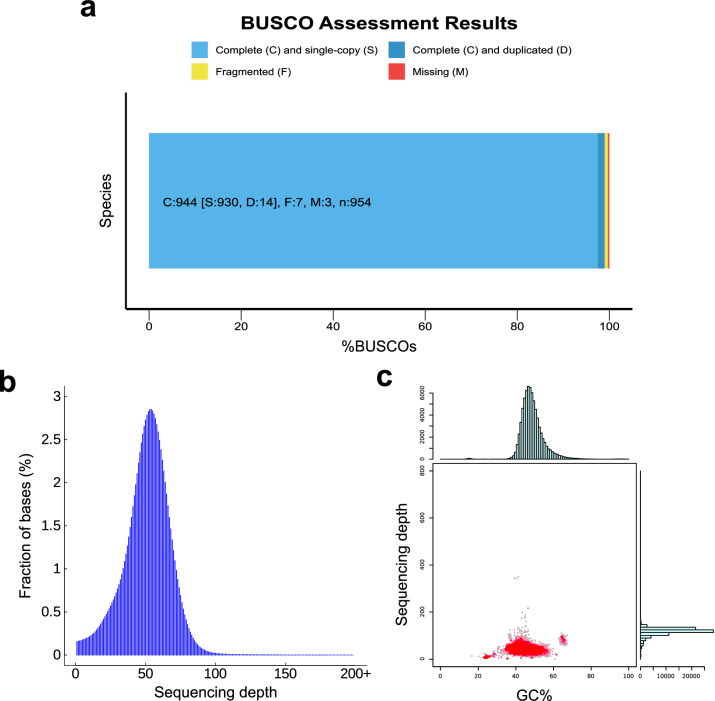
K-mer analysis was performed on 38 Gb of DNBSEQ sequencing data using Jellyfish v2.2.730 with K = 17. The genome size calculated from the K-mer number/depth was approximately 738.04 Mbp, with a corrected genome size of 717.65 Mbp, a genome heterozygosity rate of 0.55%, and a repeat sequence proportion of 32.55%. Subsequently, a preliminary assembly was conducted using SOAPdenovo2 v2.4231 with K = 41, resulting in a contig N50 of 148 bp, a total length of 1,306,005,430 bp, and a GC content of 40.72%. The 49.0 Gb of clean HiFi reads generated by Pacbio were then used in Hifiasm v0.19.532 with default parameters to obtain the de novo assembly of the S. pavo genome. The assembled genome had a total length of 735.90 Mbp, with a contig N50 of 3.69 Mbp and a scaffold N50 of 31.87 Mbp (Table 3). BUSCO v5.4.533,34 was used to evaluate the completeness of the genome assembly with the Metazoa_odb10 dataset (954 markers). The evaluation results showed that 99.0% of the sequences in the reference dataset had a complete ortholog in S. pavo genome, including 97.5% complete and single-copy genes and 1.5% complete and duplicate genes, 0.7% of the genes were reported as fragmented and 0.3% of the genes were completely missing, which indicates a high level of genome completeness (Fig. 2).
Table 3.
Genome assembly statistics.
| Type | Contig (bp) | Scaffold (bp) |
|---|---|---|
| Total Number | 709 | 61 |
| Total Length | 735,896,725 | 735,961,525 |
| Average Length | 1,037,936 | 12,064,943 |
| Max Length | 12,769,392 | 40,169,789 |
| Min Length | 13,387 | 18,985 |
| N50 Length | 3,694,337 | 31,867,360 |
| N50 Number | 56 | 11 |
| N90 Length | 524,579 | 22,483,029 |
| N90 Number | 234 | 22 |
Fig. 2.
Genome assembly assessment. (a) BUSCO evaluation results. (b) Sequence depth distribution plot. (c) GC content and depth distribution plot.
Hi-C scaffolding and chromosome anchoring
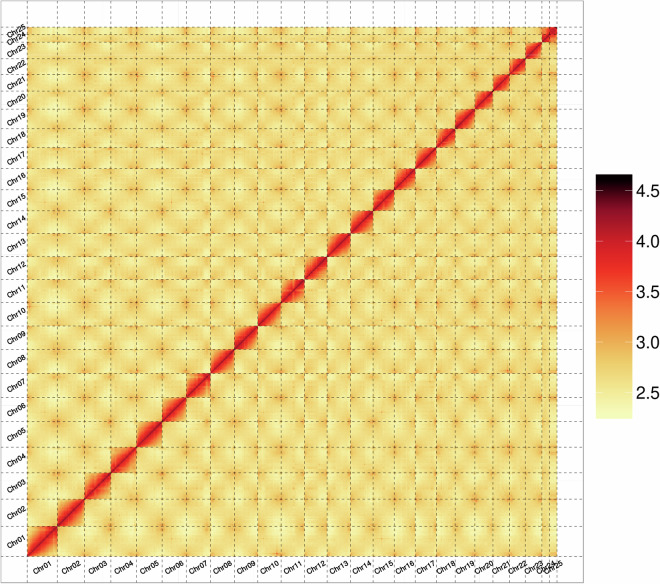
The Hi-C sequencing data was assembled using the ALLHiC v0.9.8 pipeline35 with the following parameters: --minREs 50 --maxlinkdensity 3 --NonInformativeRatio 2. Interaction signals between contigs were calculated based on the Hi-C data, and contigs were grouped according to the strength of these signals. Genetic algorithms were then used for random optimization to sort and orient the contigs within each group, thereby achieving genome anchoring and clustering. Finally, the heatmap was manually corrected using Juicebox v2.20.0036 to obtain a chromosome-level genome. The assembly consisted of 25 chromosome-level sequences and 37 unanchored sequences (Fig. 3). The genome-wide Hi-C analysis shows that the last two chromosomes are similar in length and relatively shorter than the other chromosomes and are independently distinguishable while exhibiting a strong interaction with each other. Since the sequenced individual is male, and based on the observed patterns of interaction, the last two chromosomes are inferred to be the X and Y chromosomes. Consequently, the karyotype is 2n = 48, indicating a haploid genome of 24 chromosomes in accordance with the published karyotype for this species37,38. The sequences anchored to chromosomes had a total length of 726,921,114 bp, out of a total genome length of 735,961,425 bp, resulting in 98.77% of the assembled sequences being assigned to chromosomes (Table 4).
Fig. 3.
Genome-wide Hi-C heatmap of S. pavo.
Table 4.
Assembly statistics for Hi-C.
| Class | Scaffold Number | Total Length (bp) |
|---|---|---|
| place | 25 | 726,921,114 |
| unplace | 37 | 9,040,311 |
| total | 62 | 735,961,425 |
| Coverage rate | 98.77% | |
Repeat and ncRNA annotation
First, the de novo genome-specific repeat library was generated using RepeatModeler v2.0.339. Afterwards, RepeatMasker v4.1.2-p140 was employed to identify and annotate repetitive sequences in the genome by comparing them to both the de novo repeat database and the RepBase database41. The results showed that 26.87% of the genome consisted of repeat sequences, with long interspersed nuclear elements (LINEs), short interspersed nuclear elements (SINEs), long terminal repeats (LTRs), and DNA transposons accounting for 0.09%, 4.18%, 6.85%, and 14.06% of the genome, respectively (Table 5). Based on the structural characteristics of tRNA, tRNAscan-SE v1.442 was used to identify tRNA sequences in the genome. Due to the high conservation of rRNA genes, the rRNA sequences of closely related species Blennius ocellaris43, Lipophrys pholis44, and Salarias fasciatus45 were used as reference sequences to identify rRNA in the genome through NCBI-BLAST + v2.2.2646 alignment. The miRNA and snRNA information in the genome was predicted and annotated using the covariance model of Rfam v14.147. Finally, a total of 5,148 tRNAs, 5,849 rRNAs, 2,678 miRNAs, and 536 snRNAs were annotated in the S. pavo genome (Table 6).
Table 5.
Statistics of repetitive sequences in the S. pavo genome.
| Type | Number | Length (bp) | Percentage (%) |
|---|---|---|---|
| SINE | 6,840 | 668,490 | 0.09 |
| LINE | 186,876 | 30,745,474 | 4.18 |
| LTR | 291,227 | 50,398,843 | 6.85 |
| DNA | 861,528 | 103,442,202 | 14.06 |
| Unknown | 39,800 | 4,073,104 | 0.55 |
| Total | 1,341,271 | 197,738,351 | 26.87 |
Table 6.
Statistics of non-coding RNA annotation in the S. pavo genome.
| Type | Copy (w) | Average length (bp) | Total length (bp) | % of genome | |
|---|---|---|---|---|---|
| miRNA | miRNA | 2,678 | 139.814 | 374,423 | 0.051 |
| tRNA | tRNA | 5,148 | 74.666 | 384,379 | 0.052 |
| rRNA | rRNA | 5,849 | 179.937 | 1,052,449 | 0.143 |
| 18S | 211 | 808.152 | 170,520 | 0.023 | |
| 28S | 1,734 | 266.874 | 462,760 | 0.063 | |
| 5.8S | 96 | 156.000 | 14,976 | 0.002 | |
| 5S | 3,808 | 106.143 | 404,193 | 0.055 | |
| snRNA | snRNA | 536 | 149.159 | 79,949 | 0.011 |
| CD-box | 172 | 122.023 | 20,988 | 0.003 | |
| HACA-box | 81 | 160.173 | 12,974 | 0.002 | |
| splicing | 226 | 154.186 | 34,846 | 0.005 | |
| scaRNA | 56 | 197.964 | 11,086 | 0.002 | |
| Unknown | 1 | 55.000 | 55 | 0.000 | |
Protein-coding gene prediction and annotation
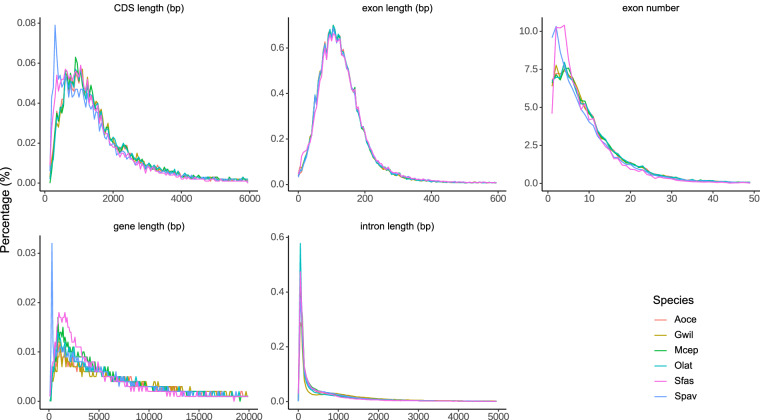
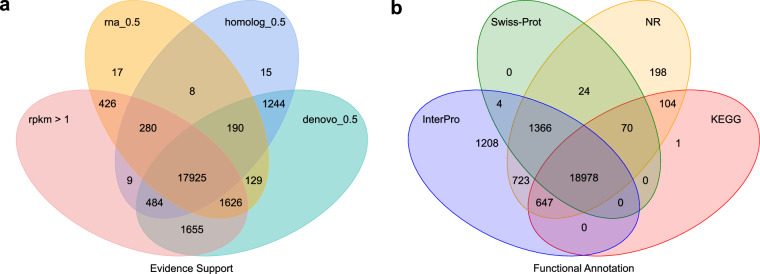
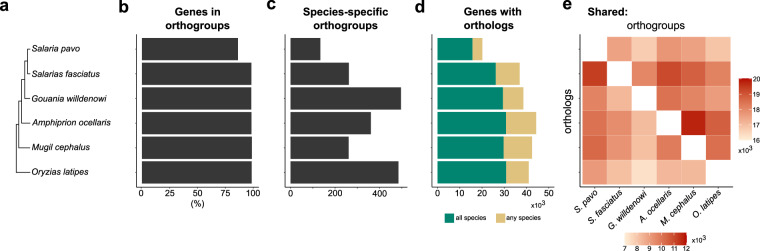
Based on the genome annotation information of closely related species S. fasciatus45 (Sfas), Gouania willdenowi48 (Gwil), Amphiprion ocellaris49 (Aoce), Mugil cephalus50 (Mcep), and the model species Oryzias latipes51 (Olat), Augustus v3.552 and SNAP v2013.11.2953 were trained for de novo gene prediction of S. pavo genome. Additionally, gene homology prediction was conducted by aligning S. pavo genome with the protein-coding sequences of the previously mentioned species using BLAST + 46 and Genewise v2.4.154. Furthermore, the RNA-seq data was assembled using Trinity v2.8.555, and the transcripts used to predict the gene structures by aligning the sequences with S. pavo genome using BLAT56. Finally, the gene sets predicted by these three methods were deduplicated using EVidenceModeler v1.1.157 and refined with PASA v2.5.358, resulting in the prediction of 24,008 genes. RNA-Seq reads were aligned to the genome using HISAT2 v2.2.159 with default parameters to obtain baseline gene expression support for the predicted genes. The average transcript length obtained from the three methods was 14,092.38 bp, while the average CDS, exon, and intron lengths were 1,641.78 bp, 173.35 bp, and 1,469.85 bp, respectively (Table 7). In comparison with S. fasciatus, G. willdenowi, A. ocellaris, M. cephalus, and O. latipes, the results showed that the number of genes, average gene length, average CDS length, average exon length, and average intron length in S. pavo are similar to those of these species, indicating the high-quality and accuracy of the S. pavo genome assembly (Fig. 4 & Table 8). The gene sets were then compared with the Swiss-Prot, NR, Pfam, KEGG, and InterPro protein databases using DIAMOND v0.8.2260/BLAST46 (E-value < 10-5), annotating the functions of 20,442, 22,110, 18,806, 19,800, and 22,926 genes, respectively (Fig. 5). After integrating and deduplicating the results, a total of 23,323 genes (97.2% of all genes) were functionally predicted. Orthology inference was performed between predicted peptides of the S. pavo genome (N = 24,008) and annotated peptides (including all alternative splice versions) in S. fasciatus45 (N = 39,222), G. willdenowi48 (N = 43,478), A. ocellaris49 (N = 47,242), M. cephalus50 (N = 44,626), and O. latipes51 (N = 44,766), using OrthoFinder v2.5.561 with the multiple sequence alignment-based tree inference (MSA)62 option (Fig. 6). The results showed that 86.2% of the predicted genes (20,700/24,008) in S. pavo could be assigned to orthogroups (Fig. 6b). Among these, only a low percentage – 2.3% (556/20,700) – were present in species-specific orthogroups (Fig. 6c), while the majority of the (15,750/20,700) had orthologs in all species (Fig. 6d). Additionally, the ratio of shared orthologs and orthogroups supports the expected phylogeny (Fig. 6e).
Table 7.
Statistics of gene structure annotation of the S. pavo genome.
| Type | Gene Set | Number | Average transcript length (bp) | Average CDS length (bp) | Average exons per gene (bp) | Average exon length (bp) | Average intron length (bp) |
|---|---|---|---|---|---|---|---|
| De novo | Augustus | 36,282 | 8,342.07 | 1,239.89 | 6.63 | 187.10 | 1,262.19 |
| SNAP | 49,111 | 20,417.64 | 1,196.63 | 7.91 | 151.23 | 2,780.65 | |
| Homolog | Aoce | 20,658 | 13,651.11 | 1,770.44 | 9.91 | 178.71 | 1,333.88 |
| Olat | 19,859 | 13,193.08 | 1,727.30 | 9.62 | 179.59 | 1,330.43 | |
| Mcep | 20,223 | 13,803.83 | 1,771.45 | 9.94 | 178.27 | 1,346.36 | |
| Sfas | 21,074 | 11,679.72 | 1,496.00 | 8.53 | 175.48 | 1,353.30 | |
| Gwil | 19,439 | 13,385.95 | 1,725.71 | 9.72 | 177.49 | 1,336.76 | |
| RNAseq | transcripts | 46,357 | 21,938.85 | 3,670.60 | 12.27 | 299.12 | 1,620.80 |
| PASA | 42,219 | 9,513.77 | 1,232.76 | 7.47 | 165.04 | 1,280.03 | |
| EVM | EVM | 32,236 | 11,024.55 | 1,367.41 | 7.64 | 179.04 | 1,454.91 |
| PASA | PASA | 31,902 | 11,508.10 | 1,392.88 | 7.76 | 179.59 | 1,497.27 |
| Final set | Final | 24,008 | 14,092.38 | 1,641.78 | 9.47 | 173.35 | 1,469.85 |
Fig. 4.
Comparisons of the genomic elements of closely related species.
Table 8.
Statistical information on gene structures in closely related species.
| Species | Number | Average gene length (bp) | Average CDS length (bp) | Average exons per gene | Average exon length (bp) | Average intron length (bp) | Genome size (Mb) |
|---|---|---|---|---|---|---|---|
| Salaria pavo | 24,008 | 14,092.38 | 1,641.78 | 9.47 | 173.35 | 1,469.85 | 735.96 |
| Salarias fasciatus | 25,108 | 12,012.46 | 1,516.02 | 8.78 | 172.74 | 1,349.79 | 797.49 |
| Gouania willdenowi | 22,774 | 19,931.28 | 1,797.42 | 10.39 | 173.04 | 1,931.73 | 937.15 |
| Amphiprion ocellaris | 23,040 | 18,584.98 | 1,846.01 | 10.56 | 174.74 | 1,750.18 | 863.47 |
| Mugil cephalus | 23,268 | 13,897.58 | 1,826.60 | 10.49 | 174.10 | 1,271.73 | 634.85 |
| Oryzias latipes | 22,094 | 16,477.55 | 1,842.13 | 10.57 | 174.26 | 1,529.15 | 734.04 |
Fig. 5.
Gene prediction and functional annotation of the S. pavo genome. (a) Venn diagram of the gene sets predicted using a gene overlap greater than 50% between two or more of the following methods: de novo, genes supported by EVM-integrated de novo predictions; homolog, genes supported by EVM-integrated homologous predictions; RNA, genes supported by EVM-integrated RNA-seq data predictions. Gene predictions had a baseline gene expression, RPKM, greater than 1. (b) Venn diagram of functional annotation based on different protein databases: InterPro, Swiss-Prot, NR, and KEGG. The numbers represent the gene count.
Fig. 6.
Summary of orthology inference of predicted proteins between S. pavo and closely related species. (a) Evolutionary relationship among S. pavo, S. fasciatus, G. willdenowi, A. ocellaris, M. cephalus, and O. latipes. (b) Number of genes per species that could be placed in an orthogroup. (c) Number of orthogroups that are specific to each species. (d) Total number of peptides with orthologs in at least one other species. (e) Heatmap of the number of orthogroups containing each species pair (top right) and one-to-one orthologs between each species (bottom left).
Data Records
The assembly and all DNA and RNA sequencing data have been deposited at NCBI under BioProject accession number PRJNA1151695. This project contains the GenBank chromosome level assembly SPavo_v1.1 (GCA_043647535.163), the transcriptomic sequencing data (SRR3040355464, SRR3040355565, SRR3040355666, SRR3040355767, SRR3040355868, SRR3040355969, SRR3040356070), DNBSEQ WGS sequencing data (SRR3041524071), the PacBio sequencing data (SRR3041669572), and the Hi-C sequencing data (SRR3041793073). The genome annotation files are available in Figshare74.
Technical Validation
Genome assembly assessment
The completeness of the genome was evaluated using BUSCO, which revealed that 99.0% of the BUSCO genes were complete in the assembled genome (Fig. 2), indicating a high genome completeness. Short-read libraries were aligned to the assembled genome using BWA v0.7.875 to determine alignment rate and coverage depth. The results showed an alignment rate of 97.36% and coverage of 99.74%, demonstrating high accuracy of the genome assembly. Genome assembly quality was further assessed using Merqury v1.376, resulting in a QV (quality value) of 40.5061, indicating genome accuracy exceeding 99.99%. In summary, the evaluation results from these software tools collectively indicate a high level of consistency, completeness, and accuracy in the assembled genome.
Hi-C assembly validation
The genome anchoring rate was 98.77%, and the Hi-C heatmap displayed a well-ordered pattern of interaction contacts (Fig. 3). These findings collectively indicate that the Hi-C-assisted assembly process yielded high-quality outcomes.
Genome annotation
Gene prediction using EVidenceModeler, integrating de novo, homologous, and RNA-seq data, resulted in the prediction of 24,008 genes in the S. pavo genome (Fig. 5a), with an average transcript length of 14,092.38 bp (Table 7). Comparative analysis with closely related species showed that S. pavo has a similar number of genes and comparable average gene, CDS, exon, and intron lengths (Table 8). Additionally, 97.2% of the predicted genes could be functionally annotated (Fig. 5b). Orthology inference with closely related species showed that 86.2% of the predicted genes in S. pavo could be assigned to orthogroups (Fig. 6). Together, these results indicate a high-quality and accuracy of the S. pavo genome assembly.
Acknowledgements
The authors thank António Roleira and Magda Teles for the technical assistance during sample collection. This work was supported by the National key R&D Program of China (2023YFE0106200), the FCT-FDCT “FISHMUC—Bioactive properties of external mucus isolated from coastal fish of Macao and Portugal”, through FCT-Fundação para a Ciência e a Tecnologia, project reference MACAU/0003/2019, and Macao Science and Technology Development Fund (FDCT), project reference 0005/2019/APJ. S.D.C. was supported by the postdoctoral fellowship FDCT0001/2021/APD.
Author contributions
D.G. conceived the research project. S.D.C. collected the samples and curated the data, C.J. and L.S. performed the analyses. S.D.C, C.J., L.S., L.Z., D.G. wrote and revised the manuscript.
Code availability
All software and pipelines used in this study were executed according to the manual and protocols of the published bioinformatic tools. The versions of the software have been given in the Methods.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Sara D. Cardoso, Chunxi Jiang.
Contributor Information
Libin Zhang, Email: zhanglibin@qdio.ac.cn.
David Gonçalves, Email: david.goncalves@usj.edu.mo.
References
- 1.Pigliucci, M. Phenotypic Plasticity: Beyond Nature and Nurture. (Johns Hopkins University Press, London, 2001).
- 2.West-Eberhard, M. J. DevelopmentalPlasticity and Evolution. (Oxford University Press, New York, 2003).
- 3.Froese, R, & Pauly, D, editors. FishBase. World Wide Web Electronic Publication (2024). Available at: https://www.fishbase.org (Version June, 2024).
- 4.Ronco, F. et al. Drivers and dynamics of a massive adaptive radiation in cichlid fishes. Nature589, 76–81 (2021). [DOI] [PubMed] [Google Scholar]
- 5.McGee, M. D. et al. The ecological and genomic basis of explosive adaptive radiation. Nature586, 75–79 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Todd, E. V. et al. Stress, novel sex genes, and epigenetic reprogramming orchestrate socially controlled sex change. Sci. Adv.5, eaaw7006 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Valdivieso, A., Ribas, L., Monleón-Getino, A., Orbán, L. & Piferrer, F. Exposure of zebrafish to elevated temperature induces sex ratio shifts and alterations in the testicular epigenome of unexposed offspring. Environ. Res.186, 109601 (2020). [DOI] [PubMed] [Google Scholar]
- 8.Zander, C. D. Blenniidae. in Fishes of the North-eastern Atlantic and the Mediterranean (eds. et al) 1096–1112 (UNESCO, Paris, 1986).
- 9.Barata, E. N. et al. Putative pheromones from the anal glands of male blennies attract females and enhance male reproductive success. Anim. Behav.75, 379–389 (2008). [Google Scholar]
- 10.Gonçalves, D., Barata, E. N., Oliveira, R. F. & Canário, A. V. M. The role of male visual and chemical cues on the activation of female courtship behaviour in the sex‐role reversed peacock blenny. J. Fish Biol.61, 96–105 (2002). [Google Scholar]
- 11.Fishelson, L. Observations on littoral fishes of Israel. I. Behaviour of Blennius pavo Risso (Teleostei: Blenniidae). Isr. J. Ecol. Evol.12, 67–80 (1963). [Google Scholar]
- 12.Patzner, R. A., Seiwald, M., Adlgasser, M. & Kaurin, G. The reproduction of Blennius pavo (Teleostei, Blenniidae) V. Reproductive behavior in natural environment. Zool. Anz.216, 338–350 (1986). [Google Scholar]
- 13.Almada, V. C., Gonçalves, E. J., Santos, A. J. & Baptista, C. Breeding ecology and nest aggregations in a population of Salaria pavo (Pisces: Blenniidae) in an area where nest sites are very scarce. J. Fish Biol.45, 819–830 (1994). [Google Scholar]
- 14.Saraiva, J. L., Barata, E. N., Canário, A. V. M. & Oliveira, R. F. The effect of nest aggregation on the reproductive behaviour of the peacock blenny Salaria pavo. J. Fish Biol.74, 754–762 (2009). [DOI] [PubMed] [Google Scholar]
- 15.Saraiva, J. L., Pignolo, G., Gonçalves, D. & Oliveira, R. F. Interpopulational variation of the mating system in the peacock blenny Salaria pavo. Acta Etholog15, 25–31 (2012). [Google Scholar]
- 16.Gonçalves, E. J., Almada, V. C., Oliveira, R. F. & Santos, A. J. Female Mimicry as a Mating Tactic in Males of the Blenniid Fish Salaria Pavo. J. Mar. Biol. Assoc. U.K.76, 529–538 (1996). [Google Scholar]
- 17.Almada, V. C., Gonçalves, E. J., Oliveira, R. F. & Santos, A. J. Courting females: ecological constraints affect sex roles in a natural population of the blenniid fish Salaria pavo. Anim. Behav.49, 1125–1127 (1995). [Google Scholar]
- 18.Fagundes, T. et al. Birth date predicts alternative life‐history pathways in a fish with sequential reproductive tactics. Funct. Ecol.29, 1533–1542 (2015). [Google Scholar]
- 19.Saraiva, J. L., Gonçalves, D. M., Simões, M. G. & Oliveira, R. F. Plasticity in reproductive behaviour in two populations of the peacock blenny. Behaviour148, 1457–1472 (2011). [Google Scholar]
- 20.Belaiba, E., Marrone, F., Vecchioni, L., Bahri-Sfar, L. & Arculeo, M. An exhaustive phylogeny of the combtooth blenny genus Salaria (Pisces, Blenniidae) shows introgressive hybridization and lack of reciprocal mtDNA monophyly between the marine species Salaria basilisca and Salaria pavo. Mol. Phylogenet. Evol.135, 210–221 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Vecchioni, L. et al. Multi-Locus Phylogenetic Analyses of the Almadablennius Clade Reveals Inconsistencies with the Present Taxonomy of Blenniid Fishes. Diversity14, 53 (2022). [Google Scholar]
- 22.Almada, V. C. et al. Phylogenetic analysis of Peri-Mediterranean blennies of the genus Salaria: Molecular insights on the colonization of freshwaters. Mol. Phylogenet. Evol.52, 424–431 (2009). [DOI] [PubMed] [Google Scholar]
- 23.Neat, F. C., Locatello, L. & Rasotto, M. B. Reproductive morphology in relation to alternative male reproductive tactics in Scartella cristata. J. Fish Biol.62, 1381–1391 (2003). [Google Scholar]
- 24.Neat, F. C., Lengkeek, W., Westerbeek, E. P., Laarhoven, B. & Videler, J. J. Behavioural and morphological differences between lake and river populations of Salaria fluviatilis. J. Fish Biol.63, 374–387 (2003). [Google Scholar]
- 25.Santos, R. S. Parentais e satélites: tácticas alternativas de acasalamento nos machos de Blennius sanguinolentus Pallas (Pisces: Blenniidae). Arquipélago, Sér. Ciênc. Nat.6, 119–146 (1985). [Google Scholar]
- 26.Cardoso, S. D., Gonçalves, D., Goesmann, A., Canário, A. V. M. & Oliveira, R. F. Temporal variation in brain transcriptome is associated with the expression of female mimicry as a sequential male alternative reproductive tactic in fish. Mol. Ecol.27, 789–803 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Duncan, E. J., Gluckman, P. D. & Dearden, P. K. Epigenetics, plasticity, and evolution: How do we link epigenetic change to phenotype? J. Exp. Zoöl. Part B: Mol. Dev. Evol.322, 208–220 (2014). [DOI] [PubMed] [Google Scholar]
- 28.Chen, Y. et al. SOAPnuke: a MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. GigaScience7, 1–6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Belaghzal, H., Dekker, J. & Gibcus, J. H. Hi-C 2.0: An optimized Hi-C procedure for high-resolution genome-wide mapping of chromosome conformation. Methods123, 56–65 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marçais, G. & Kingsford, C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics27, 764–770 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luo, R. et al. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaScience1, 1–6 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheng, H., Concepcion, G. T., Feng, X., Zhang, H. & Li, H. Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat. Methods18, 170–175 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seppey, M., Manni, M. & Zdobnov, E. M. BUSCO: Assessing Genome Assembly and Annotation Completeness. Methods Mol. Biol.1962, 227–245 (2019). [DOI] [PubMed] [Google Scholar]
- 34.Manni, M., Berkeley, M. R., Seppey, M., Simão, F. A. & Zdobnov, E. M. BUSCO Update: Novel and Streamlined Workflows along with Broader and Deeper Phylogenetic Coverage for Scoring of Eukaryotic, Prokaryotic, and Viral Genomes. Mol. Biol. Evol.38, 4647–4654 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang, X., Zhang, S., Zhao, Q., Ming, R. & Tang, H. Assembly of allele-aware, chromosomal-scale autopolyploid genomes based on Hi-C data. Nat. Plants5, 833–845 (2019). [DOI] [PubMed] [Google Scholar]
- 36.Durand, N. C. et al. Juicebox Provides a Visualization System for Hi-C Contact Maps with Unlimited Zoom. Cell Syst3, 99–101 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cataudella, S. & Civitelli, M. V. Cytotaxonomical consideration of the genus Blennius (pisces-perciformes). Experientia31, 167–169 (1975). [DOI] [PubMed] [Google Scholar]
- 38.Garcia, E., Alvarez, M. C. & Thode, G. Chromosome relationships in the genus Blennius (Blenniidae Perciformes) C-banding patterns suggest two karyoevolutional pathways. Genetica72, 27–36 (1987). [Google Scholar]
- 39.Flynn, J. M. et al. RepeatModeler2 for automated genomic discovery of transposable element families. Proc. Natl. Acad. Sci.117, 9451–9457 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tarailo‐Graovac, M. & Chen, N. Using RepeatMasker to Identify Repetitive Elements in Genomic Sequences. Curr. Protoc. Bioinform.25, Unit 4.10 (2009). [DOI] [PubMed] [Google Scholar]
- 41.Jurka, J. et al. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res.110, 462–467 (2005). [DOI] [PubMed] [Google Scholar]
- 42.Chan, P. P., Lin, B. Y., Mak, A. J. & Lowe, T. M. tRNAscan-SE 2.0: improved detection and functional classification of transfer RNA genes. Nucleic Acids Res49, 9077–9096 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.NCBI GenBankhttps://identifiers.org/ncbi/insdc.gca:GCA_963422515.1 (2023).
- 44.NCBI GenBankhttps://identifiers.org/ncbi/insdc.gca:GCA_963383615.1 (2023).
- 45.NCBI GenBankhttps://identifiers.org/ncbi/refseq.gcf:GCF_902148845.1 (2019).
- 46.Camacho, C. et al. BLAST+: architecture and applications. BMC Bioinform10, 421 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kalvari, I. et al. Rfam 14: expanded coverage of metagenomic, viral and microRNA families. Nucleic Acids Res49, D192–D200 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.NCBI GenBankhttps://identifiers.org/ncbi/refseq.gcf:GCF_900634775.1 (2019).
- 49.NCBI GenBankhttps://identifiers.org/ncbi/refseq.gcf:GCF_022539595.1 (2023).
- 50.NCBI GenBankhttps://identifiers.org/ncbi/refseq.gcf:GCF_022458985.1 (2022).
- 51.NCBI GenBankhttps://identifiers.org/ncbi/refseq.gcf:GCF_002234675.1 (2019).
- 52.Nachtweide, S. & Stanke, M. Multi-Genome Annotation with AUGUSTUS. Methods Mol. Biol.1962, 139–160 (2019). [DOI] [PubMed] [Google Scholar]
- 53.Leskovec, J. & Sosič, R. SNAP: A General Purpose Network Analysis and Graph Mining Library. ACM Trans. Intell. Syst. Technol.8, 1–20 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Birney, E. & Durbin, R. Using GeneWise in the Drosophila Annotation Experiment. Genome Res10, 547–548 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Grabherr, M. G. et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol.29, 644–652 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kent, W. J. BLAT—The BLAST-Like Alignment Tool. Genome Res12, 656–664 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Haas, B. J. et al. Automated eukaryotic gene structure annotation using EVidenceModeler and the Program to Assemble Spliced Alignments. Genome Biol9, R7 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jia, H. et al. PASA: Identifying More Credible Structural Variants of Hedou12. IEEE/ACM Trans. Comput. Biol. Bioinform.17, 1493–1503 (2019). [DOI] [PubMed] [Google Scholar]
- 59.Kim, D., Paggi, J. M., Park, C., Bennett, C. & Salzberg, S. L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol.37, 907–915 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Buchfink, B., Xie, C. & Huson, D. H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods12, 59–60 (2015). [DOI] [PubMed] [Google Scholar]
- 61.Emms, D. M. & Kelly, S. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol.20, 238 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Emms, D. M. & Kelly, S. STRIDE: Species Tree Root Inference from Gene Duplication Events. Mol. Biol. Evol.34, 3267–3278 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.NCBI GenBankhttps://identifiers.org/ncbi/insdc.gca:GCA_043647535.1 (2024).
- 64.NCBI Sequence Read Archivehttps://identifiers.org/ncbi/insdc.sra:SRR30403554 (2024).
- 65.NCBI Sequence Read Archivehttps://identifiers.org/ncbi/insdc.sra:SRR30403555 (2024).
- 66.NCBI Sequence Read Archivehttps://identifiers.org/ncbi/insdc.sra:SRR30403556 (2024).
- 67.NCBI Sequence Read Archivehttps://identifiers.org/ncbi/insdc.sra:SRR30403557 (2024).
- 68.NCBI Sequence Read Archivehttps://identifiers.org/ncbi/insdc.sra:SRR30403558 (2024).
- 69.NCBI Sequence Read Archivehttps://identifiers.org/ncbi/insdc.sra:SRR30403559 (2024).
- 70.NCBI Sequence Read Archivehttps://identifiers.org/ncbi/insdc.sra:SRR30403560 (2024).
- 71.NCBI Sequence Read Archivehttps://identifiers.org/ncbi/insdc.sra:SRR30415240 (2024).
- 72.NCBI Sequence Read Archivehttps://identifiers.org/ncbi/insdc.sra:SRR30416695 (2024).
- 73.NCBI Sequence Read Archivehttps://identifiers.org/ncbi/insdc.sra:SRR30417930 (2024).
- 74.Cardoso, S. D., Jiang, C., Sun, L., Zhang, L. & Gonçalves, D. Chromosome-level genome assembly of the highly-polymorphic peacock blenny (Salaria pavo). figshare10.6084/m9.figshare.26854312 (2024). [DOI] [PMC free article] [PubMed]
- 75.Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rhie, A., Walenz, B. P., Koren, S. & Phillippy, A. M. Merqury: reference-free quality, completeness, and phasing assessment for genome assemblies. Genome Biol21, 245 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- NCBI GenBankhttps://identifiers.org/ncbi/insdc.gca:GCA_963422515.1 (2023).
- NCBI GenBankhttps://identifiers.org/ncbi/insdc.gca:GCA_963383615.1 (2023).
- NCBI GenBankhttps://identifiers.org/ncbi/refseq.gcf:GCF_902148845.1 (2019).
- NCBI GenBankhttps://identifiers.org/ncbi/refseq.gcf:GCF_900634775.1 (2019).
- NCBI GenBankhttps://identifiers.org/ncbi/refseq.gcf:GCF_022539595.1 (2023).
- NCBI GenBankhttps://identifiers.org/ncbi/refseq.gcf:GCF_022458985.1 (2022).
- NCBI GenBankhttps://identifiers.org/ncbi/refseq.gcf:GCF_002234675.1 (2019).
- NCBI GenBankhttps://identifiers.org/ncbi/insdc.gca:GCA_043647535.1 (2024).
- NCBI Sequence Read Archivehttps://identifiers.org/ncbi/insdc.sra:SRR30403554 (2024).
- NCBI Sequence Read Archivehttps://identifiers.org/ncbi/insdc.sra:SRR30403555 (2024).
- NCBI Sequence Read Archivehttps://identifiers.org/ncbi/insdc.sra:SRR30403556 (2024).
- NCBI Sequence Read Archivehttps://identifiers.org/ncbi/insdc.sra:SRR30403557 (2024).
- NCBI Sequence Read Archivehttps://identifiers.org/ncbi/insdc.sra:SRR30403558 (2024).
- NCBI Sequence Read Archivehttps://identifiers.org/ncbi/insdc.sra:SRR30403559 (2024).
- NCBI Sequence Read Archivehttps://identifiers.org/ncbi/insdc.sra:SRR30403560 (2024).
- NCBI Sequence Read Archivehttps://identifiers.org/ncbi/insdc.sra:SRR30415240 (2024).
- NCBI Sequence Read Archivehttps://identifiers.org/ncbi/insdc.sra:SRR30416695 (2024).
- NCBI Sequence Read Archivehttps://identifiers.org/ncbi/insdc.sra:SRR30417930 (2024).
Data Availability Statement
All software and pipelines used in this study were executed according to the manual and protocols of the published bioinformatic tools. The versions of the software have been given in the Methods.