Abstract
Soil salinization poses a substantial threat to global food security, particularly under the influence of climate change, and is recognized as one of the most urgent challenges in land degradation. This study aims to elucidate the challenges associated with managing arid and semi-arid saline-alkali lands in China’s Ningxia province and propose feasible solutions. To assess moss crust colonization, we measured changes in organic matter and chlorophyll levels. Additionally, we investigated the impact of an interlayer composed of Goji berry root bark using liquid chromatography-mass spectrometry analysis, biological enzyme activity analysis, and metagenomic sequencing. A total of 45 endophytes were isolated from the moss crust. The most significant colonization of moss crusts was observed when the Goji berry root bark was used as the interlayer, resulting in a significant increase in chlorophyll content. Several responses were identified as pivotal factors facilitating moss crust growth when the Goji berry root bark was used as the interlayer. In saline-alkali soil, the Goji berry root bark interlayer increased the activities of sucrase, urease, and alkaline phosphatase. Metagenomic data analysis revealed variations in the relative abundance of microorganisms at the phylum level, although these differences were not statistically significant. Evaluation of the impact of physical isolation and moss crust transplantation on the ecological restoration of saline-alkali soil using liquid chromatography-tandem mass spectrometry and metagenomic sequencing indicated that the Goji berry root bark as a physical isolation method promotes moss crust colonization in saline-alkali soil and increases soil organic matter and nutrient elements, offering valuable insights for the ecological management of saline-alkali land and serving as a reference for future research in this field.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00248-024-02473-1.
Keywords: Biological moss crusts, Microbial composition, Macrogenome, Physical and chemical properties of saline-alkali soil, Saline-alkali land treatment, Root bark of Goji berries (the Goji berry root bark)
Introduction
The physical, chemical, and biological properties of soil are influenced by its type and its interactions with environmental factors, including salinization [1, 2]. Soil salinization is caused by specific natural or human-induced conditions, leading to the concentration of soluble salts on the soil surface and an increase in soil conductivity. Natural leaching processes are often insufficient to remove the excess salt from the soil profile. Essentially, rising groundwater levels, driven by evaporation, redistribute highly soluble salts both horizontally and vertically on the surface. Additionally, inadequate drainage and high evaporation rates contribute to salt accumulation on the soil surface exceeding normal levels [3–6]. Excessive salt in saline-alkali soil can adversely affect soil structure, leading to a decline in several soil properties, including soil compaction, particle dispersion, reduced water permeability, decreased microbial activity, inhibition of organic matter accumulation, and subsequent effects on enzyme activity. These alterations lead to a reduction in carbon, nitrogen, and phosphorus content in the soil, significantly impacting soil fertility and creating an unfavorable environment for crop growth. Thus, soil salinization poses a major threat to the sustainable development of agriculture [7]. The accumulation of excessive salt in the soil disrupts ion distribution in plant roots, causing physiological dehydration and oxidative stress. Salt stress adversely affects plants through osmotic stress, ionic toxicity, and nutrient deficiency, resulting in inhibited growth and reduced crop yields [8–13].
Improving and utilizing saline-alkali land offers several benefits, including increased crop yields, mitigating food shortages, and enhancing the agricultural ecological landscape, ultimately improving the quality of life for individuals [14–21]. Considerable research on the physical and chemical characteristics, as well as the structure of saline-alkali soil, highlighted the potential of tailored strategies to address varying levels of salinity and alkalinity [9–18]. The technology for improving saline-alkali land can be broadly categorized into three primary approaches: physical improvement, chemical improvement, and ecological improvement. In recent years, numerous studies have investigated the ecological functions of biological soil crusts (BSCs), which were found to have a significant impact in various bioclimatic regions, affecting sandy landscapes, soil ecology, soil hydrology, and soil biogeochemical cycles. The formation of BSCs plays a crucial role in improving soil physical and chemical properties, enhancing soil stability, and fostering soil fertility; thus, hold substantial ecological importance in the realms of environmental preservation and the revitalization of terrestrial ecosystems [22]. BSCs significantly influence ecological processes occurring on the soil surface, such as soil surface stabilization, enhancement of soil physical and chemical properties, regulation of soil hydrological processes, promotion of seed germination, establishment and growth of vascular plants, and enhancement of soil biodiversity [23]. BSCs secrete organic acids that facilitate soil formation. Through photosynthesis, BSCs capture atmospheric carbon (C) and nitrogen (N) to effectively improve soil fertility. Moreover, BSCs alter the roughness, porosity, water retention, and aggregation of the soil surface, profoundly impacting water utilization and redistribution. The secreted polysaccharides facilitate soil particle cohesion, thereby enhancing soil aggregation and stability. Additionally, BSCs mitigate soil erosion caused by wind and water, reduce nutrient loss, provide a favorable environment for colonizing other plant communities, promote vegetation succession, and contribute to ecosystem stability [24].
The Yinchuan Plain, situated in Ningxia province, China, falls within the semi-arid and arid region of the middle temperate zone. This area experiences low precipitation and high evaporation rates, accounting for only 0.4% of the nation’s total water resources. Both natural factors and human activities have contributed to the degradation of the ecological environment in the Yinchuan Plain over time. Notably, soil salinization represents the most significant constraint on local agricultural production. Consequently, addressing challenges such as population growth, diminishing cultivable land, and food shortages requires immediate measures to expand cultivable land resources, optimize the utilization of saline-alkali land, and promote ecological agriculture in China [13]. Hence, this study investigates the microbial composition of soil moss crusts collected from the periphery of saline-alkali land in the Ningxia province of China to analyze their colonization on saline-alkali soil, elucidate the underlying reasons for the challenges in managing arid and semi-arid saline-alkali lands, and propose practical solutions.
Materials and Methods
Collection of Moss Crust Samples, Saline-Alkali Soil, and the Isolation Layer
The moss crusts used in this experiment were collected from three different areas: Shapotou Hongwei Experimental Area (N37° 45′, E104° 79′), Helan Mountain (N38° 60′, E106° 03′), and Xixia District of Ningxia province (N38° 29′, E106° 5′). These areas represent arid and semi-arid saline-alkali lands situated at the periphery of saline-alkali soil. Sections of moss crusts, approximately 1.0 cm thick, were collected using a sterile shovel. Afterward, the samples were dried in the shade, and any extraneous debris was manually removed. Subsequently, the moss crusts were combined with the soil to create the seed source.
Saline-alkali soil samples were procured from two distinct locations: N38° 24′, E106° 25′ and N38° 24′, E106° 25′.
The isolation layer: The interlayers used in this experiment include common soil, the root bark of Goji berries, green grasses, leaves, grape peels, and locust flowers, which were supplied by the Key Laboratory of Chemical Engineering and Technology of the State Ethnic Affairs Commission. The treatment procedures applied to these interlayers were identical to those employed for the saline-alkali soil samples.
Preparation of the Test Medium
Fungal culture medium (Potato Dextrose Agar, PDA) [25]: 200 g of potatoes was diced into small pieces and boiled for 30 min, then filtered through six layers of gauze and adjusted to a final volume of 1000 mL. Add 20 g glucose, 3 g peptone, 1 g magnesium sulfate, 0.5 g dipotassium hydrogen phosphate, and 1 g yeast extract, stir well, then add 18 g AGAR powder, and sterilize the filtrate at 115 °C for 30 min.
Actinomycete culture medium [26]: The modified Gao’s No. 1 medium of 37.5 g was taken and diluted to 1000 mL. After mixing, it was divided into conical bottles and sterilized at 121 °C for 20 min.
Preparation of salt-tolerant culture medium: Sodium chloride was added into both the PDA culture medium and the actinomycete culture medium. Then, the sodium chloride concentration was adjusted to the following levels: 0%, 3%, 6%, 9%, 12%, 15%, 18%, and 21%.
Experimental Instruments
In this study, various equipment and instruments were utilized, including an Agilent 6530 liquid chromatography-mass spectrometer, a Mettler LE438 pH meter, an ultrasonic cleaner SB-5200DTS from Ningbo Xinzhi Biotechnology Co., Ltd, a conductivity meter DDS-307 from Shanghai Yixin Scientific Instrument Co., Ltd, and a super pure water machine Smart-s15 from Shanghai Hetai Instrument Co., Ltd. Additionally, a multifunctional full-wavelength microplate spectrometer (Spectromax M2/M2e) from Meigu Molecular Instruments (Shanghai) Co., Ltd, a microscope SN-1200W from Shenzhen Sannuo Xinu Technology Co., Ltd, a rotary evaporator R1002-VN from Agilent Technology Co., Ltd, and a magnetic stirrer JB-1A from Shanghai Precision Scientific Instrument Co., Ltd, were employed. Furthermore, the study utilized an ultraviolet spectrophotometer UV180G from Beijing Aode Instrument Equipment Co., Ltd, a polarizing microscope BM-500 from Sanuo Instrument Co., Ltd, a full automatic Kjeldahl nitrogen analyzer NKD6260 from Shanghai Wanghai Environmental Technology Co., Ltd, and a flame atomic absorption spectrophotometer ZA3300 from Hitachi Scientific Instruments (Beijing) Co., Ltd.
Experimental Methods
Isolation of Endophytic Bacteria from Moss Crusts
Moss Crust Treatment and Inoculation
After drying the moss crust, soil and other impurities were removed, leaving only the moss, which was subsequently cleaned with distilled water. Then, 1 g of the moss was placed within a sterilized ultra-clean workbench. The moss was rinsed thrice with sterile water, followed by a 1-min immersion in 75% alcohol. After another round of rinsing with sterile water, the surface was dried using sterile filter paper. The moss was then finely ground using a mortar and sequentially diluted into five gradients (10−1, 10−2, 10−3, 10−4, and 10−5). Using an Eppendorf pipette, 0.5 mL of the dilution was added onto both PDA plates and Gauze’s No. 1 plates containing 1% potassium dichromate, supplemented with 120 µL/mL of penicillin and streptomycin. Coating rods were used to ensure even distribution of the solution, and each gradient was prepared in triplicate.
Microbial Culture
The sealed plates were placed into an incubator set at a temperature of 28 °C, initially for upright cultivation for 1 day, after which they were inverted. The PDA medium was cultured for 15–20 days, while the Gauze’s No. 1 medium was cultivated for 5–7 days. Throughout the cultivation period, the growth of the colonies was observed. Details regarding the screening of the interlayer cultivation method are shown in the Supporting Information, and that of the control group and experimental groups are shown in Table S1.
Strain Separation
Actinomycetes were cultured using Gauze’s No. 1 medium, while fungi were cultured using PDA medium. This procedure was repeated thrice to ensure the colonies remained uncontaminated by foreign bacteria. Microscopic examination was conducted to validate the purity of the colonies.
Strain Preservation
The identified strain from the single colony was inoculated onto a slant culture medium using an inoculation ring, allowed to grow under normal conditions during the incubation period, and subsequently stored in a refrigerator at 4 °C. For spore-producing actinomycetes, the spores were rinsed with 20% glycerol and preserved in a low-temperature refrigerator. For sporogenous fungi, the spores were washed with 50% glycerol and similarly stored in a low-temperature refrigerator.
Optical Microscope
On an ultra-clean table, sterilized cover glasses were inserted into each culture medium at a 45° angle, with 3–5 pieces placed into each medium. Then, the culture dishes were incubated in a constant temperature incubator for 1–2 days. Afterward, the cover glasses were carefully removed based on the growth of the strain and microscopically observed. Clear images of the mycelium were captured and recorded for subsequent analysis.
Salt Resistance Test
The isolated strains were inoculated into media containing different salt concentrations. After incubating for 3–5 days at 30 °C, the growth of the strains was observed and recorded. The experiment was performed in triplicate.
Diversity Analysis and Identification of Endophytic Bacteria
The isolated bacteria underwent sequencing for the 16S rDNA region, while the fungi were identified using ITS sequencing, both conducted by Shenzhen Micro Science Alliance Technology Group Co., Ltd. The sequencing process included the following specific steps:
Extraction of Genomic Deoxyribonucleic Acid (DNA), Genome Polymerase Chain Reaction (PCR) Amplification, PCR Amplification Reaction System, and Sequence Analysis
Extraction of Genomic DNA
Extraction of bacterial genomic DNA: First, 1.0 × 109 bacterial cultures (1 mL bacterial solution OD600 was 1–1.5) were collected with 2-mL centrifuge tubes, centrifuged with 12,000 rotational speed (rpm) for 30 s, and the supernatant was discarded. Secondly, 150 µL Buffer S containing RNase A was used for suspension precipitation. Then, 20 µL of lysozyme storage solution was added, mixed evenly, and allowed to stand at room temperature for 5 min. Then, add 30 µL 0.25 M EDTA (pH 8.0), mix well, and ice bath for 5 min. After adding 450 µL Buffer G-A, vortex oscillation for 15 s, water bath at 65 °C for 10 min. Then, 400 µL Buffer G-B and 1 mL Buffer DV (pre-cooled at 4 °C) were added, mixed, and centrifuged for 2 min. After that, the upper phase is discarded as much as possible, and the interphase precipitation and the lower phase are retained. Subsequently, 1 mL of 4 °C pre-cooled Buffer DV was added and centrifuged for 2 min. Discard the upper phase, transfer the lower phase to the filter (the filter is placed in a 2-mL centrifuge tube), and centrifuge it at 12,000 rpm for 1 min. After that, the filter was discarded, and 400 µL Buffer BV was added to the filtrate to make it mixed evenly. It was transferred into a preparation tube and centrifuged at 12,000 rpm for 1 min. The filtrate was discarded, and the preparation tube was placed back into the original 2-mL centrifuge tube; 500 µL Buffer W1 was added and centrifuged for 1 min. Then, the filtrate was discarded, the preparation tube was placed back into the original 2-mL centrifuge tube, and 700 µL Buffer W2 was added and centrifuged for 1 min. Wash once with 700 µL of Buffer W2 in the same way. The filtrate was discarded, and the prepared tube was placed back into the original 2-mL centrifuge tube, centrifuged at 12,000 rpm for 1 min. Finally, the preparation tube was placed in another clean 1.5-mL centrifuge tube, and 100–200 µL of Eluent or deionized water was added to the center of the silica membrane. After standing at room temperature for 1 min, the DNA was eluted by centrifugation at 12,000 rpm for 1 min.
Extraction of fungal genomic DNA: Firstly, a certain sample was scraped from the plate and placed in a 2-mL centrifuge tube, and 200 µL buffer GA was added to oscillate until the bacteria were completely suspended. Secondly, 2 µL Proteinase K solution was added to the tube and mixed well. Then, 220 µL Buffer GB was added, oscillated for 15 s, and placed at 70 °C for 10 min. After the solution was strained and clear, it was briefly centrifuged to remove the water droplets on the inner wall of the tube cover. After removing the supernatant, add 220 μL of anhydrous ethanol to the test tube, fully oscillate and mix 15 s, at this time may appear flocculent precipitate, briefly centrifuge to remove water beads on the inner wall of the tube cap. Subsequently, the solution and flocculent precipitate obtained in the previous step were added to an adsorption column CB3. After centrifugation at 12,000 rpm for 30 s, the waste liquid was discarded, and the adsorption column CB3 was placed in a collection tube. Then, 500 µL Buffer GD was added to the adsorption column CB3, centrifuged at 12,000 rpm for 30 s, the waste liquid was poured away, and the adsorption column CB3 was put into the collection tube. At the same time, 600 µL rinse solution PW was added to the adsorption column CB3, centrifuged at 12,000 rpm for 30 s, and the waste liquid was discarded. The adsorption column CB3 was placed in a collection tube. After repeating this procedure, the adsorption column CB3 was placed back into the collection tube, centrifuged at 12,000 rpm for 2 min, and the waste liquid was poured away. After that, the adsorption column CB3 was placed at room temperature for several minutes to completely dry the residual rinse solution in the adsorption material. Finally, the adsorption column CB3 was transferred into a clean centrifuge tube, and 50 ~ 200 µL elution buffer TE was added to the middle of the adsorption membrane in the air, and the solution was collected into the centrifuge tube after being placed at room temperature for 2–5 min and centrifuged at 12,000 rpm for 2 min.
Genome PCR Amplification
The extracted DNA was used as an amplification template for PCR amplification. The bacterial primers were 27F: 5-AGAGTTTGATCCTGGCTCAG-3 and 1492R: 5-GGTTACCTTGTTACGACTT-3. The fungal primers were ITS1: TCCGTAGGTGAACCTGCGG and ITS4: TCCTCCGCTTATTGATATGC.
PCR Amplification Reaction System
1.0 µL genomic DNA (20 ng/µL), 5.0 µL 10 × Buffer (containing 2.5 mM Mg2+), 1.0 µL Taq polymerase (5 µ/µL), 1.0 µL dNTP (10 mM), 1.5 µL 27F primer (10 µM) and 1.5 µL 1492R primer (10 µM) (1.5 µL ITS1 primer (10 µM) and 1.5 µL ITS4 primer (10 µM)), and 39.0 µL ddH2O were added to the 0.2-mL centrifuge tube, and the mixture was gently mixed. The droplets on the tube wall were collected by instantaneous centrifugation to the bottom of the tube, and the PCR reaction was carried out on the PCR amplification instrument. After the reaction was completed, 3 µL of the PCR product was detected by 1% agarose gel electrophoresis to confirm the PCR amplified fragment.
Sequence Analysis
The NCBI BLAST program was used to compare the spliced sequence file with the data in the NCBI nucleic acid database, and the species information with the greatest similarity to the sequence of the species to be tested was obtained, which was the identification result.
Enlarged Cultivation of Optimal Combination
Expanded cultivation: Trays with diameters of 6, 9, and 12 cm were prepared for the extensive culture of the optimal experimental group and its control group, as well as the direct saline-alkali soil group and its control group. The culture period for all groups was 60 days. The amount of soil sample, isolation zone, moss crust fragments added, and the amount of water applied was increased proportionally according to the cultivation area, and the cultivation method was the same as above. For other information, please view the supporting information.
Chemical Analysis
High-Performance Liquid Chromatography Quadrupole Time-of-Flight Tandem Mass Spectrometry (HPLC-Q-TOF MS) Analysis
To extract the soil sample, 20 g of soil was combined with 50 mL of ultrapure water and subjected to ultrasonic extraction at 60 °C for 30 min, which was repeated three times. Then, the mixture was centrifuged and filtered. From the resulting supernatant, 10 mL was extracted for further analysis through micro solid-phase extraction. The compounds extracted were eluted sequentially using 2 mL of water, followed by 50% methanol–water and 95% methanol–water. The eluents were then subjected to liquid quality analysis to determine their composition.
HPLC Condition, Mass Spectrum Condition, and Molecular Network Analysis Methods
HPLC conditions: The liquid chromatography is performed using an Agilent 1200 (USA) automatic injection system. The method of two-phase gradient elution uses chromatographic acetonitrile (A) + pure water solution (B) as the mobile phase. Within 0–40 min, phase A gradually increases from 5% (V/V) to 95% (V/V) and is eluted with 95% acetonitrile water for 5 min. After passing through the chromatographic column, the eluent was divided into three equal parts using a diode array detector (DAD) and a mass detector (Q-TOF). The column temperature chamber is set to 20 °C, the injection volume is 5 µL, and the flow rate is 1.0 mL/min.
Mass spectrometry conditions: High-purity nitrogen (liquid nitrogen) is used as the atomization and drying gas, and high-purity helium is used as the collision gas. The collection method is to separate the detection of positive and negative ions. The capillary voltage is 4.5 kV, the spray pressure is 2.0 bar, the flow rate of dry gas (N2) is 4.0 L/min, the dry gas temperature is 180 °C, the ion energy of the fourth pole is 3.0 eV, the ion transmission time is 80 µs, the pre-pulse storage time is 5 µs, the collision gas is argon, and the collision energy is 30 eV. The scanning range of primary mass spectrometry is m/z 300–2000, and the scanning range of secondary mass spectrometry is m/z 20–2000. Using automatic secondary mode, select ions with an abundance greater than 104 for secondary mass spectrometry collection.
Molecular network analysis methods: The raw secondary mass spectrometry data file obtained by the LC–MS/MS method for sample collection was processed using ProteoWizard software (http://proteowizard.sourceforge.net/). After converting the data file into.mzXML format, import it into GNPS (https://gnps.ucsd.edu/ProteoSAFe/static/gnps-splash.jsp) and perform a search for similar components and molecular clustering, with network parameters set as follows: (1) MS/MS ion error of 0.5 Da; (2) mother ion error 1.0 Da; (3) the lowest ion pair score is 0.6; (4) the minimum number of matching ions is 4; (5) the minimum number of matching ions is 10 for networked Top K. Visualize and analyze the results using Cytoscape software (http://www.cytoscape.org/), then use the GNPS data platform to analyze the molecular network and obtain known compounds. Based on the correlation between nodes in the molecular network, compare the similarities and differences in mass spectrometry data between related nodes, and obtain analogues or other unknown compounds of known compounds.
Enzyme Activity Evaluation
Before conducting the soil enzyme activity test, the dried and sieved soil samples were prepared following the kit’s instructions, including a pre-test to ensure the precision and reliability of the testing procedure.
Sucrase (S-SC) is an enzyme that catalyzes the hydrolysis of complex carbohydrates into monosaccharides, which react with 3,5-dinitrosalicylic acid to produce brownish-red amino compounds, exhibiting light absorption characteristics at 540 nm. The enzymatic products of S-SC are closely linked to the organic matter content in the soil. To determine S-SC activity, the soil sample was initially air-dried and sieved through a 30–50 mesh sieve, following which 0.1 g of the sample (performed in triplicate) was carefully measured. 3,5-Dinitrosalicylic acid was added to the samples, followed by thorough shaking and placement in a 37 °C water bath for 15 min. After adding a required reagent, the mixture was incubated at a constant temperature for 24 h. Subsequently, the samples were centrifuged at 10,000 rpm and 4 °C for 5 min. The resulting supernatant was diluted tenfold, and the reagent was added and mixed thoroughly, followed by boiling for 5 min and subsequent cooling. The enzymatic activity was then measured using a microplate reader.
Soil urease (S-UE) is an enzyme responsible for the hydrolysis of urea, yielding ammonia and carbonic acid. The activity of S-UE serves as an indicator of soil nitrogen status. In this method, urea was used as the substrate, and the produced NH3-N was quantified using indophenol blue colorimetry, resulting in the formation of a blue indophenol complex. To conduct the test, 0.25 g of the soil sample (performed in triplicate) was measured and mixed with 125 µL of toluene. The mixture was thoroughly shaken and allowed to stand at room temperature for 15 min. Subsequent steps were carried out following the provided protocol. Dilutions were prepared as needed, and measurements were taken at 630 nm.
Soil alkaline phosphatase (S-AKP) is an enzyme that facilitates the mineralization of organic phosphorus compounds in soil, influencing the decomposition and transformation of organic phosphorus. It serves as an indicator for assessing the direction and intensity of soil phosphorus biological transformation. Under alkaline conditions, S-AKP catalyzes the hydrolysis of disodium phenyl phosphate, producing phenol and disodium hydrogen phosphate. The enzyme’s activity can be determined by quantifying the amount of phenol generated. To determine S-AKP activity, air-dried soil was first passed through a 30–50 mesh sieve. Next, 0.1 g of soil (in triplicate) was weighed and combined with 0.05 mL of toluene, followed by the subsequent steps outlined in the provided instructions and guidelines.
Stress Resistance Test on Moss Crust
The study assessed the stress resistance of moss crusts under saline and alkaline conditions by quantifying the levels of protein, polysaccharide, and malondialdehyde.
The bicinchoninic acid (BCA) method was used, and Cu2+ was reduced to Cu+. The BCA reagent, when combined with Cu+, formed a blue-purple complex with its maximum absorption at 562 nm. Accumulation of soluble proteins in plants under stress plays a crucial role in maintaining cellular water and safeguarding essential cell components and biofilm. To perform the determination step, approximately 0.1 g of the sample was weighed and mixed with 1 mL of distilled water. The mixture was homogenized in an ice bath, centrifuged at 12,000 rpm for 10 min at 4 °C, and the resulting supernatant was collected for the test. The subsequent steps were conducted according to the instructions provided in the kit, and the absorbance was measured at 562 nm.
Polysaccharides were quantified by hydrolyzing sugars into monosaccharides using concentrated sulfuric acid, rapidly dehydrating them to form sugar aldehyde derivatives, and then condensing them with phenol to yield yellow compounds. Accumulation of soluble sugars (SS) enhances plant resistance to harsh environments and improves water retention by increasing cell osmotic potential, thereby maintaining membrane system integrity and stability. To determine polysaccharide content, approximately 3 mg of the sieved sample was mixed with 2 mL of distilled water. The mixture was heated in a boiling water bath (95–100 °C) for 2 h, followed by cooling at room temperature. After centrifugation at 8000 rpm for 5 min, the sediment was retained. Subsequently, 1 mL of 80% ethanol was added to the sediment, mixed thoroughly, and centrifuged again at 8000 rpm for 5 min. The supernatant was discarded, and 2 mL of distilled water was added to the sediment. The mixture was heated in a boiling water bath until dissolution, preparing the polysaccharide solution for testing. The subsequent steps adhered to the kit’s instructions, and absorbance was measured at 488 nm.
Malondialdehyde (MDA) is produced as a result of peroxidation in plant tissues during injury or adverse conditions, providing an indicator of cell membrane damage. To perform this assessment, approximately 0.1 g of the sample was precisely weighed and combined with 1 mL of an extract. The mixture was homogenized within an ice bath and subsequently centrifuged at 4 °C and 12,000 rpm for 10 min. The resulting supernatant was collected and maintained on ice for testing. The subsequent steps were conducted in accordance with the kit’s instructions, with absorbance measurements recorded at 532 nm and 600 nm, respectively.
Data Processing
The growth status of the crusts and the morphology of endophytic bacteria were documented using Sannuo image analysis software. Data obtained were processed using Microsoft Excel 2010. Statistical analyses, including t-tests and single-factor variance, were conducted using SPSS 17.0 software to assess the significance of soil physical and chemical properties, crust growth status, and enzyme activity (P < 0.05, P < 0.01). Histograms were generated using Origin 2019 software. For DNA sequencing of the isolated endophytic bacteria, the ABI3730-XL sequencer was employed, and the phylogenetic tree was constructed utilizing the neighbor-joining method in the MEGA software. Qualitative analysis of soil extract liquid data was performed using Qualitative Navigator B.08.00 software, while molecular network analysis was conducted using the GNPS data platform. High-throughput sequencing of soil sample microorganisms was executed on the Illumina PE 150 platform. Data quality control was implemented using KneadData software, and data quality assessment was performed using FastQC.
Results
Microbial Composition Analysis of Biological Crust
Endophytic Bacteria from Moss Crusts
A total of 19 endophytic bacterial strains were isolated from moss crusts and designated as A1–A19 (Fig. S1 and Table S2). The analysis revealed that the 19 bacterial strains belonged to four different genera, indicating a moderate level of diversity. However, the species diversity was relatively limited, possibly influenced by the physical and chemical properties, as well as the composition of the crusts, which may have affected the nutrient availability required for the growth and reproduction of endophytic bacteria. Among the isolated strains, 15 strains were classified under the Bacillus genus, two strains under Agrobacterium, one strain under Priestia, and one strain under Metabacillus. According to the salt tolerance experiment and the classification standard of halophilic bacteria, strains A6, A7, and A13 were identified as weakly halophilic bacteria, strains A8 and A11 as moderately halophilic bacteria, and strain A18 as an extremely halophilic bacterium.
Endophytic Fungi from Moss Crusts
Twenty-six endophytic fungi were isolated and labeled as A20–A45 (Fig. S2 and Table S3). The resulting PCR products revealed that these fungi belonged to 14 different genera, showcasing notable diversity. Among these genera, Aspergillus was the most abundant, accounting for 34.6% of the total. Fusarium was represented by three strains, accounting for 11.5%. Pyrenochaetopsis and Curvularia each accounted for two strains, contributing to 7.7% of the total for each genus. The remaining genera (Listeria, Striaticonidium, Rhizomucor, Chaetomium, Thielavia, Neoascochyta, Phaeosphaeria, Humicola, Paraphoma, and Didymella) were represented by one strain each, accounting for 3.8% of the total. According to the salt tolerance test and the classification standard of halophilic bacteria, strains A26, A37, and A42 were categorized as weakly halophilic bacteria, while strains A22, A24, A36, and A44 were classified as moderately halophilic bacteria. Lastly, strain A40 was identified as an extremely halophilic bacterium.
Physical and Chemical Properties of Saline-Alkali Soil
The basic characteristics of Helan saline-alkali soil are outlined in the Supporting Information (Table S4). Compared to typical farmland [4, 7], the organic matter content was found to be lower. Organic matter comprises a range of nutrients necessary for crop growth, which can directly or indirectly supply nitrogen, boron, potassium, calcium, magnesium, sulfur, and other trace elements essential for crop development. A reduced organic matter content indicates that even though the total substance content in the soil is relatively normal, there is limited decomposition and utilization.
The total nitrogen content in the saline-alkali soil was assessed using the Kjeldahl method, revealing a nitrogen compound content within the normal range. The determination of total phosphorus content, conducted through molybdenum antimony anti-colorimetry, exhibited a relatively high phosphorus content. Conversely, the determination of total potassium content, using flame atomic spectrophotometry, displayed a lower potassium content than what is considered normal [4, 7]. These findings collectively indicated that the saline-alkali soil in Helan County was unsuitable for conventional plant and crop cultivation due to its elevated salt levels, strong alkalinity, diminished organic matter content, and limited nutrient availability, rendering it agriculturally unproductive.
Potential Application of Moss Crust to Saline-Alkali Soil Treatment
Isolation Zone with the Growth State of Moss Crust as an Indicator
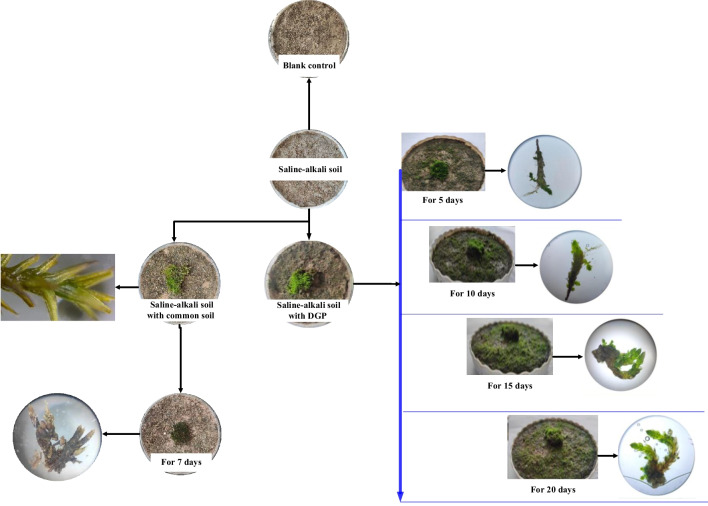
In the presence of saline-alkali soil and the addition of various isolation zones, only the moss crust fragments within saline-alkali soil, saline-alkali soil mixed with common soil, and saline-alkali soil with the Goji berry root bark demonstrated outward growth from the mature moss crust as the central point, suggesting that mature moss crusts release signaling factors or compounds that facilitate the development of these crust fragments (Fig. 1). Microscopic examination revealed the emergence of new buds from the crusts during the planting process, indicating a distinctive mode of reproduction. The mature moss crusts displayed robust growth with a green coloration. Additionally, other plant seedlings were observed sprouting on the saline-alkali soil, confirming the role of biological crusts in promoting seed germination. Conversely, crusts in the other isolation zones initially appeared green but gradually turned yellow and dried out over time, indicating a state of decay. The mature moss crusts in these zones also underwent a transition from dark green to withered.
Fig. 1.
Effects of isolation zones on the growth state of moss crusts. The blank control represents the parallel processing for 7 days, the saline-alkali soil with grape peels is the implicit control, and the saline-alkali soil with the Goji berry root bark promotes moss crust growth
Impact of Different Isolation Zones on Moss Crust Coverage
The moss crust coverage varied significantly among the different treatments. Particularly, the conditions of saline-alkali soil, saline-alkali soil with common soil, and saline-alkali soil with the Goji berry root bark led to the successful colonization of moss crusts, yielding coverage percentages of 3.05%, 5.12%, and 9.10%, respectively (P < 0.01). Among the isolation zones, the Goji berry root bark treatment displayed the highest moss crust coverage, indicating that the Goji berry root bark isolation zone provided the most favorable conditions for moss crust colonization (Fig. 1).
Effect of Different Isolation Zones on Chlorophyll Content of Moss Crusts
Figure S3 illustrates the changes in chlorophyll content in moss crusts subjected to different belt treatments. The results reveal varying chlorophyll content levels across different treatments. In general, the chlorophyll content of moss crusts increased under all conditions. Specifically, the increases in chlorophyll content were 24.43%, 19.75%, 2.29%, 10.36%, 8.02%, 13.83%, and 7.36%, respectively, for treatments without and with different isolation zones. Notably, the treatment without an isolation zone exhibited the highest increase in chlorophyll content, reaching 24.43%.
Significant differences (P < 0.05) were observed between all experimental groups and the control groups, with saline-alkali soil (group 1 in Table S1), saline-alkali soil with green grasses (group 3 in Table S1), saline-alkali soil with the Goji berry root bark (group 5 in Table S1), and saline-alkali soil with grape peels (group 6 in Table S1) showing highly significant differences (P < 0.01). The increase in chlorophyll content in moss crusts suggests that although some fragments in the isolation zone did not successfully colonize and exhibited signs of yellowing and decline, they were not completely lifeless but rather in a state of suspended animation. This adaptive response of moss crusts to salt stress allows the cells to regain their physiological functions under suitable conditions, indicating that salt stress does not entirely strip mosses of their physiological capabilities and they may have the potential for recovery.
Effects of Moss Crust Addition on Soil Physical and Chemical Properties
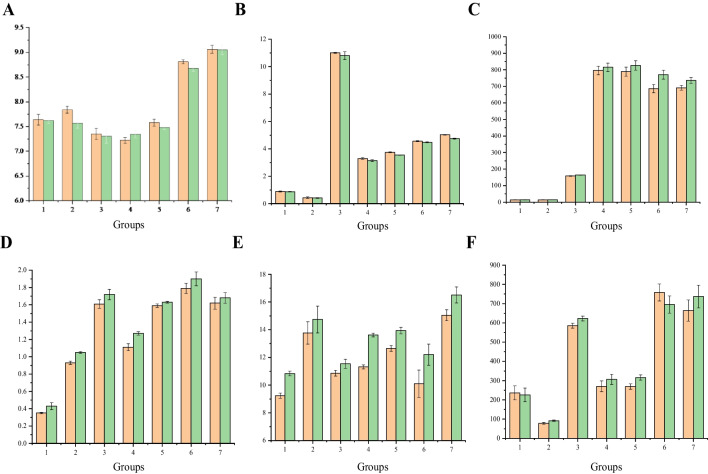
Effects of Moss Crusts in Different Isolation Zones on Soil pH
The pH changes of saline-alkali soil under different isolation zones are shown in Fig. 2A, indicating that different isolation zones exert divergent influences on saline-alkali soil pH. Incorporating green grasses, leaves, and the Goji berry root bark isolation belts contributes to pH reduction in saline-alkali soil treatment. In contrast, the addition of common soil, grape peels, and locust flowers isolation belts leads to pH elevation in saline-alkali soil treatment, with a notable increase associated with the grape peels and locust flowers isolation belts, which may explain why moss crusts could not be planted.
Fig. 2.
Effects of moss crusts in different isolation zones on soil properties: A soil pH; B soil conductivity, sm/cm; C organic matter content, g/Kg; D total phosphorus content, g/Kg; E total nitrogen content, g/Kg; and F soluble nitrogen content, mg/Kg. Groups 1 to 7 correspond to Fig. S1. Yellow columns represent control groups, while green columns represent test groups
The addition of moss crusts has a notable impact on the pH of saline-alkali soil. When moss crusts were added under the saline-alkali soil treatment and different isolation zones, the pH of the saline-alkali soil decreased by 0.26%, 3.44%, 0.54%, 1.32%, 1.48%, and 0.11%, respectively, in all cases except for the lefts isolation zone. Significantly, the saline-alkali soil treated with common soil and the saline-alkali soil treated with the Goji berry root bark showed significant differences compared to the control group (P < 0.05). Overall, the addition of moss crusts consistently reduced the pH of saline-alkali soil in the Mingjiao alpine permafrost region, supporting the general understanding of the impact of biological crusts on soil physical and chemical properties in this region.
Effect of Moss Crusts in Different Isolation Zones on Soil Conductivity
Changes in soil conductivity under different isolation zones are presented in Fig. 2B. The conductivity of saline-alkali soil exhibits significant variation when subjected to different isolation zones. The addition of the common soil isolation decreased the soil’s conductivity, while the incorporation of other isolation belts increased soil conductivity. Remarkably, the inclusion of a green grass isolation belt can substantially elevate the soil’s conductivity by several orders of magnitude, potentially explaining the challenges in establishing moss crusts in such conditions. Conversely, despite the notably high conductivity observed in the Goji berry root bark isolation zone (reaching 3.75 ms/cm), moss crusts can successfully colonize this area, underscoring their capacity to thrive in high-salinity environments and emphasizing their resilience and adaptability.
The addition of moss crusts significantly impacts the electrical conductivity of saline-alkali soil. Moss crusts introduced beneath saline-alkali soil in various isolation zones lead to a notable reduction in soil conductivity, with percentage decreases of 0.62%, 4.55%, 1.82%, 4.55%, 5.33%, 1.75%, and 5.57%, respectively. Among these diverse isolation zones, the addition of moss crusts under locust flowers isolation zones results in the most substantial reduction in soil conductivity. The saline-alkali soil with lefts and saline-alkali soil with the Goji berry root bark experimental groups exhibit significant differences from the control group (P < 0.05). Overall, these findings underscore the effective role of moss crusts in lowering the electrical conductivity of saline-alkali soil, consistent with prior observations in reference [27] regarding the influence of biological crusts on soil properties across various soil and vegetation environments.
Effect of Moss Crusts in Different Isolation Zones on Organic Matter
Figure 2C illustrates the changes in organic matter content in saline-alkali soil after adding moss crusts in different isolation zones, showing that the organic matter content varied significantly in different isolation zones. Except for the common soil and green grass isolation belts, the addition of other belts leads to a significant increase in organic matter content, ranging from 13.51 to 788.76 g/kg. Furthermore, the addition of moss crusts also impacted the organic matter content of saline-alkali soil. Adding moss crusts under the saline-alkali soil and different isolation zones led to increased organic matter content of 8.88%, 7.29%, 3.54%, 2.43%, 4.79%, 12.29%, and 6.40%, respectively. The most significant increase in organic matter content was observed under the grape peel isolation zone. All experimental groups, except for the saline-alkali soil with green grass, displayed significant differences compared to the control group (P < 0.05), with the saline-alkali soil under lefts conditions showing highly significant differences (P < 0.01). These findings highlight the efficacy of moss crusts in substantially increasing the organic matter content of saline-alkali soil, consistent with the impact of biological crusts on organic carbon content in sandy soil under varying rainfall conditions.
Effect of Moss Crusts in Different Isolation Zones on Total Phosphorus
Figure 2D illustrates the changes in total phosphorus content in saline-alkali soil after adding moss crusts in different isolation zones. The total phosphorus content significantly varied among the isolation zones, doubling under different conditions, with the highest increment of 1.79 g/kg observed in the grape peel isolation zone. The introduction of moss crusts also influenced the total phosphorus content of saline-alkali soil, resulting in increments of 22.86%, 12.90%, 6.83%, 14.41%, 2.52%, 6.15%, and 3.70% under different isolation zones. The saline-alkali soil with moss crusts displayed the most significant increase in total phosphorus content, and the experimental group under the common soil isolation zone exhibited a significant difference compared to the control group (P < 0.05). These findings highlight the efficacy of moss crusts in enhancing the total phosphorus content of saline-alkali soil, aligning with previous research on moss crusts’ impact on the physical and chemical properties of topsoil during sand-fixing vegetation succession [28].
Effect of Moss Crusts in Different Isolation Zones on Total Nitrogen
The change in total nitrogen content under different isolation belt conditions is shown in Fig. 2E. There were significant variations in total nitrogen content among the different isolation zones. The addition of various isolation belts contributed to an increase in total nitrogen content, with the most substantial increment (up to approximately 5 times) observed in the locust flowers isolation zone. The presence of moss crusts also influenced the total nitrogen content of saline-alkali soil. The addition of moss crusts under saline-alkali soil and different isolation zones increased total nitrogen content to 17.32%, 7.04%, 6.16%, 20.32%, 10.11%, 20.79%, and 9.70%, respectively. Moss crusts added under the saline-alkali soil with grape peel conditions induced the most significant increase in total nitrogen content in saline-alkali soil, with the experimental groups of saline-alkali soil, saline-alkali soil with lefts, saline-alkali soil with the Goji berry root bark, and saline-alkali soil with grape peels exhibiting a highly significant difference from the control (P < 0.01). Overall, these findings demonstrate that the incorporation of moss crusts can enhance the total nitrogen content of saline-alkali soil, aligning with the findings of a previous study regarding the impact of biological soil crusts on vegetation and the physical and chemical properties of artificial grassland in the source area of the Yellow River [29].
Effect of Moss Crusts in Different Isolation Zones on Hydrolyzed Nitrogen
The content change of hydrolyzed nitrogen added to moss crusts under different isolation zones is presented in Fig. 2F. The results indicate significant variations in hydrolyzed nitrogen content among the different isolation zones. The introduction of different isolation zones led to an increase in hydrolyzed nitrogen content in saline-alkali soil, with the exception of the common soil isolation zone. The most substantial increase in hydrolyzed nitrogen content was observed when the grape peel isolation zone was added, which suggests that although the total nitrogen content increases with the addition of the common soil isolation zone, the availability of organic nitrogen for absorption and utilization remains limited. The presence of moss crusts also affected the hydrolyzed nitrogen content in saline-alkali soil under different isolation zone conditions, resulting in both increases and decreases. The most significant increase in hydrolyzed nitrogen content in saline-alkali soil occurred under the saline-alkali soil with the Goji berry root bark conditions, with a rise of 17.39%. Moreover, significant differences (P < 0.05) were observed between the experimental groups and the control under all conditions. These findings imply that the addition of moss crusts has minimal effects on the hydrolyzed nitrogen content in saline-alkali soil.
Analysis of the Reasons Why the Goji Berry Root Bark Acts as the Isolation Belt to Promote the Moss Crust to Settle on Saline-Alkali Soil
Collectively, our analysis highlights the efficacy of the Goji berry root bark isolation zone in facilitating moss crust colonization on saline-alkali soil. Consequently, we examined the chemical composition, active enzyme levels, and stress resistance of the saline-alkali soil control group, the saline-alkali soil experimental group, the saline-alkali soil with the Goji berry root bark control group, and the saline-alkali soil with the Goji berry root bark experimental group to gain preliminary insights into the mechanisms promoting moss crust colonization.
Analysis Results of HPLC-q-TOF MS-Assisted Molecular Networking Method
Figure S4 displays the total ion diagrams under the negative ion mode for the saline-alkali soil with the Goji berry root bark group. The diagrams represent samples washed with 50% methanol–water (Fig. S4A) and samples washed with 95% methanol–water (Fig. S4B). The figures present the total ion diagrams for the saline-alkali soil control group, the saline-alkali soil experimental group, the saline-alkali soil with the Goji berry root bark control group, and the saline-alkali soil with the Goji berry root bark experimental group, from top to bottom. Under the 50% methanol–water washing mode to remove negative ions from the samples, both the saline-alkali soil experimental group and the control group exhibited strong peaks in the collection time range of 37–49 min. However, there were differences in the mass spectrometry data spectra between the two groups. The peak areas of the experimental group were lower than those of the control group, and there were distinct responses at 39.833 min and 44.565 min. In the saline-alkali soil with the Goji berry root bark experimental group and the control group, the primary peak emergence time was mainly between 39 and 49 min. The ion peaks were similar, and the primary peaks were the same. However, the experimental group displayed a unique ion peak at 35.209 min, although its responsiveness was not high. These observations suggest minimal differences in the chemical composition of the 50% methanol eluted fractions between the two groups under the negative ion mode, with relatively low ion abundance. Additionally, strong peaks were observed in the saline-alkali soil experimental group and the control group at 40–45 min collection time under the 95% methanol elution fraction. The collected mass spectrometry data spectra differed between the two groups. The peak areas of the experimental group were lower than those of the control group, and there were distinct specific responses at 49.255 min and 44.565 min. Similarly, in the saline-alkali soil with the Goji berry root bark experimental group and the control group, the primary peak emergence time was mainly between 40 and 50 min. The ion peaks were similar, and the primary peaks were the same. Compared to the control group, the experimental group had specific ion peaks at 27.922 min and 40.997 min, although their responsiveness was not high.
In addition, compared to both the saline-alkali soil and the saline-alkali soil with the Goji berry root bark, the saline-alkali soil with the Goji berry root bark treated with 50% methanol–water washing mode displayed a pronounced and strong response at 40.632 min, suggesting a potential role of the saline-alkali soil with the Goji berry root bark in facilitating moss crust colonization.
Figure S5 displays the total ion diagram under the positive ion mode, demonstrating the samples washed with 50% methanol–water (Fig. S5A) and 95% methanol–water (Fig. S5B) and showing the total ion diagram of the saline-alkali soil control group, the saline-alkali soil experimental group, the saline-alkali soil with the Goji berry root bark control group, and the saline-alkali soil with the Goji berry root bark experimental group from top to bottom. Under the positive ion mode, all four experimental groups exhibited higher ion abundance in the distillate ions washed with 95% methanol–water. In the total ion diagrams of the 50% methanol–water elution samples, both the saline-alkali soil experimental group and the control group displayed strong peaks in the collection time range of 32–47 min, with distinct differences in their collected ion spectra. The peak area of the experimental group was lower than that of the control group, but it exhibited a strong response at 39.194 min and 44.565 min. In the saline-alkali soil with the Goji berry root bark experimental group and the control group, the peak time was primarily between 39 and 49 min, and the ion peaks exhibited certain differences. Compared to the control group, the experimental group showed unique ion peaks at 39.722 min and 43.254 min, with a strong response. These observations indicate noticeable distinctions between the spectra of the saline-alkali soil and the saline-alkali soil with the Goji berry root bark, suggesting differences in the chemical composition of the 50% methanol-eluted fractions under the negative ion mode. In the 50% methanol elution fraction, there was generally a distinct separation between the two treatment chromatograms. In the saline-alkali soil experimental group and the control group, there was a strong and distinct peak at the collection time of 32–47 min, with their collected ion chromatograms being similar. The peak area of the experimental group was lower than that of the control group, but there was a strong and specific response at 41.886 min. In the saline-alkali soil with the Goji berry root bark experimental group and the control group, the peak time was mainly between 28 and 47 min, and the ion peaks were similar, with the main peak being the same. Compared to the control group, the experimental group exhibited special ion peaks at 14.213 min and 24.158 min, although their intensities were not high. When comparing the saline-alkali soil and the saline-alkali soil with the Goji berry root bark as a whole, the saline-alkali soil with the Goji berry root bark displayed a distinct and strong response at 28.687 min and 39.728 min in the 95% methanol–water elution sample, which may be related to its promotion of moss crust colonization.
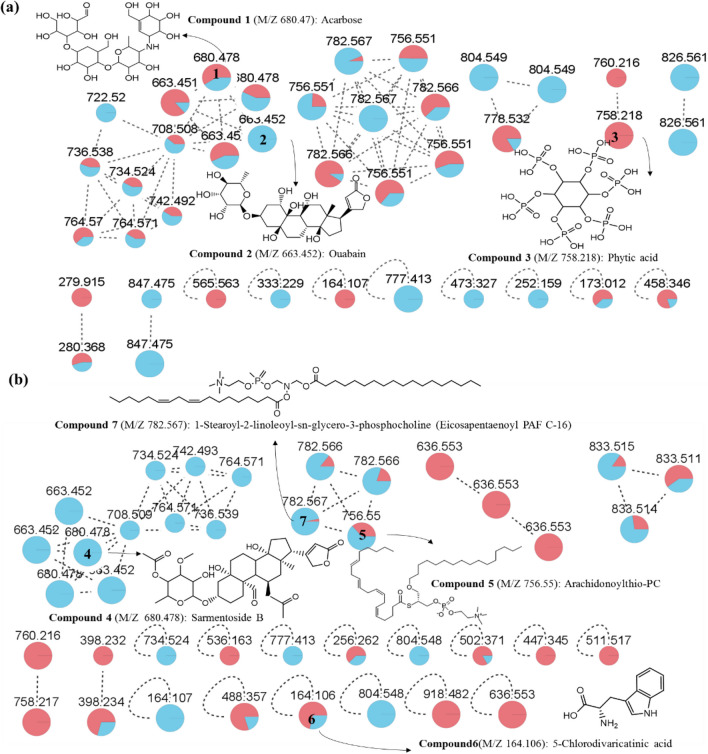
To analyze the impact of adding exotica on the composition of soil crust compounds, we utilized HPLC-q-TOF MS data from the saline-alkali soil control group and the saline-alkali soil with the Goji berry root bark control group to construct a molecular network diagram (Fig. 3), resulting in the prediction of ten compounds (Fig. S6): acarbose (1); ouabain (2); phytic acid (3); sarmentose B (4); arachidonic acid thio-PC (5); 5-chlorovanillic acid (6); 1-stearoyl-2-linoleoyl-sn-glycero-3-phosphate (7); 3,7-epoxycaryophyllan-6-one (8); [6-[5,7-dihydroxy-2-(4-hydroxyphenyl)−4-oxochromen-3-yl] oxy-3,4-dihydroxy-5-(3,4,5-trihydroxybenzoyl) oxyxan-2-yl] methyl 3,4,5-trihydroxybenzoate (9); and parfumine (10). Among these compounds, phytic acid plays a significant role in promoting soil quality and improving saline-alkali soil conditions. It gradually enhances the soil’s remediation ability and creates favorable conditions for moss crust formation and planting. Additionally, phytic acid’s acidic nature helps neutralize saline-alkali land and maintains the soil’s acid–alkali balance. On the other hand, arachidonic acid thio-PC, as a fatty acid, facilitates energy storage for moss crusts and promotes their biological nitrogen fixation.
Fig. 3.
a, b Molecular network analysis of HPLC-q-TOF MS data conducted on the GNPS platform. Red nodes represent HPLC-q-TOF MS data from the saline-alkali soil control group, while blue nodes represent the saline-alkali soil with the Goji berry root bark group
Effect of Moss Crust on Soil Enzyme Activity
Figure S7 illustrates the changes in soil enzyme activity before and after adding moss crusts under saline-alkali soil and saline-alkali soil with the Goji berry root bark conditions. The addition of the Goji berry root bark isolation zone led to an increase in soil enzyme S-SC activity from 39.477 to 126.521 U/g, S-UE activity from 1417.549 to 1818.190 U/g, and S-AKP activity from 1710.007 to 3098.511 U/g. These changes corresponded to the increase in soil organic matter, total nitrogen, and total phosphorus content observed following the addition of the isolation zone, as shown above.
The addition of moss crusts also had an impact on soil enzyme activity. Under the conditions of saline-alkali soil and saline-alkali soil with the Goji berry root bark, the activity of soil enzyme S-SC increased by 31.72% and 6.84%, respectively, with a significant difference observed between the experimental group and the control group (P < 0.05). Conversely, the activity of soil enzyme S-UE showed a downward trend, with a decline rate of 60.13% and 14.57%, respectively. It is important to note that enzymes related to nitrogen transformation in soil include not only S-UE but also proteases and other invertases. Thus, reducing S-UE activity may increase the activity of proteases and other enzymes upon adding moss crusts. Furthermore, the addition of moss crusts to saline-alkali soil led to a decrease in the activity of soil enzyme S-AKP, but under the Goji berry root bark isolation zone condition, S-AKP activity increased by 27.99%, which was significantly different from the control group (P < 0.01).
Stress Resistance Comparison of Moss Crust Under Saline-Alkali Conditions
As shown in Fig. S8, the content of malondialdehyde, protein, and polysaccharides in the moss crusts exhibit different patterns. The malondialdehyde content was 14.335, 21.167, and 17.959 nmol/g; the protein content was 1.893, 2.291, and 3.067 mg/g; and the polysaccharide content was 71.983, 73.058, and 72.677 mg/g, respectively. Each group represents different conditions: normal conditions, the saline-alkali soil moss crusts, and the saline-alkali soil with the Goji berry root bark moss crusts under stress. The differences in malondialdehyde content among the groups were highly significant (P < 0.01). Malondialdehyde is one of the lipid peroxides of the cell membrane, which is an important indicator of the degree of system damage. The increased malondialdehyde content in the saline-alkali soil and the saline-alkali soil with the Goji berry root bark moss crusts indicates a certain degree of damage to the cell membrane’s physiological function. However, the increase in malondialdehyde content was lower in the saline-alkali soil with the Goji berry root bark group, suggesting that moss crusts under this condition have a stronger ability to scavenge free radicals and exhibit antioxidative properties.
The protein and polysaccharide contents in the moss crusts display significant differences among the three conditions (P < 0.05). In both the saline-alkali soil and saline-alkali soil with the Goji berry root bark conditions, there was an increase in protein and polysaccharide contents compared to the normal condition, suggesting that moss crusts can maintain osmotic pressure and water balance within their structures by accumulating substances that aid in osmoregulation, enabling them to maintain regular metabolic functions within the saline-alkali environment. Under the saline-alkali soil with the Goji berry root bark condition, the moss crust showed a lower increase in malondialdehyde content while displaying a notable increase in protein and polysaccharide content, particularly in terms of protein, which may contribute to the improved colonization of moss crusts in this specific condition.
Metagenomic Data Analysis
Horizontal Community Composition of Soil Microflora
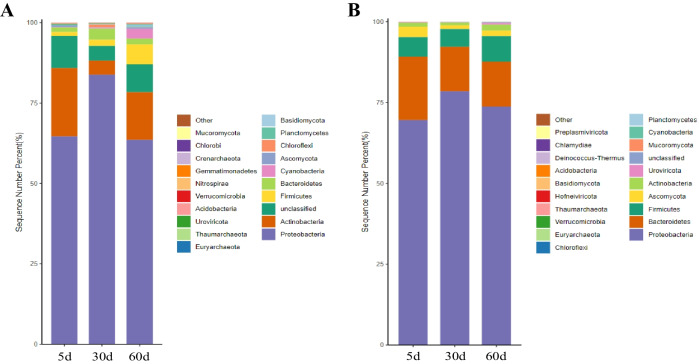
Compared with the saline-alkali soil and the Goji berry root bark in the three time periods (Fig. 4), the addition of the Goji berry root bark isolation zone increased the abundance of Proteobacteria, Bacteroides, Firmicutes, Ascomycetes, and Urophage. Among them, the significant increase in Proteobacteria abundance suggests its high sensitivity to environmental changes. The negative correlation between Proteobacteria and soil pH indicates that adding the Goji berry root bark isolation zone and establishing moss crusts may reduce soil pH, enabling the survival of moss crusts in high-salinity environments. It further indicates that the Goji berry root bark isolation zone may promote the colonization of moss crusts by enhancing the abundance of salt-tolerant bacteria, such as Proteobacteria. Bacteroides can degrade cellulose, and their increase may promote the degradation of the Goji berry root bark, providing nutrients needed for moss crust colonization. Firmicutes can produce spores and resist dehydration and extreme environments. Therefore, the addition of the Goji berry root bark isolation zone may promote the successful establishment of moss crusts by enhancing their stress tolerance through the increased abundance of Firmicutes.
Fig. 4.
Temporal shifts in soil microflora horizontal community composition at 5, 30, and 60 days. A, B The negative control and the saline-alkali soil with the Goji berry root bark group, respectively
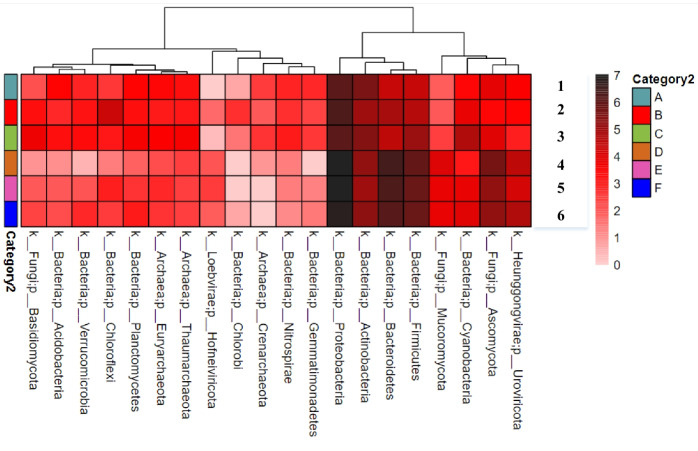
The heatmap clustering results for the six groups of data over the three periods in both the saline-alkali soil and the Goji berry root bark conditions are shown in Fig. 5. These results reveal no significant differences between the groups, indicating that although the relative abundance of soil microorganisms at the phylum level may vary across different time periods and with or without the presence of the Goji berry root bark isolation belt, these observed differences may not reach statistical significance.
Fig. 5.
Heatmap clustering results at the gate classification level for six groups. Groups 1–3 represent samples from soil at 5, 30, and 60 days, respectively, and groups 4–6 represent samples from crust at 5, 30, and 60 days, respectively
Diversity of Soil Microbial Community
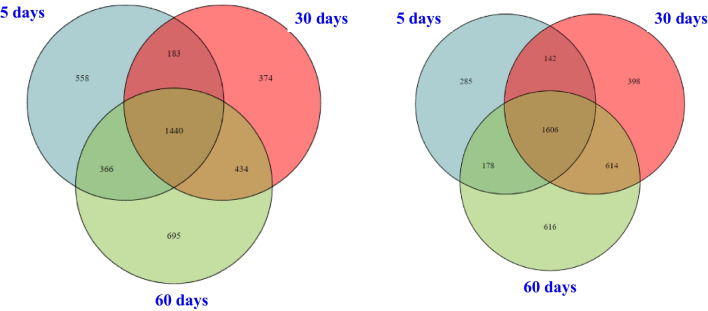
The Venn diagram in Fig. 6 provides an overview of the shared and unique soil microorganisms at the species level between the saline-alkali soil and the Goji berry root bark. In the three periods of the saline-alkali soil, a total of 4050 OTUs were identified, with 1440 OTUs (35.56% of the total) shared among them. Additionally, we observed 258 specific OTUs in the Goji berry root bark01 (6.77% of the total), 398 specific OTUs in the Goji berry root bark02 (10.44%), and 616 specific OTUs in the Goji berry root bark03 (16.16%). Notably, the dominant OTUs across all three time periods include Stenotrophomonas maltophilia, Enterobacter hormaechei, Sphingobacterium multivorum, Chryseobacterium balustinum, and Acrobat sp.−77.
Fig. 6.
Venn plots of the species level at 5, 30, and 60 days. The left figure corresponds to the saline-alkali soil control group, while the right one corresponds to the saline-alkali soil with the Goji berry root bark group
Each of the three Goji berry root bark periods exhibited its own unique set of bacteria. Specifically, in the Goji berry root bark01, we identified specific bacteria such as Vagococcus fluvialis, Klebsiella sp. P1CD1, and Bacillus sp. 275, while Bacillus sp-CR71, Acinetobacter sp.-Ac-14, and Enterobacter cloacae complex sp.-ECL78 were unique to the Goji berry root bark02. Meanwhile, the Goji berry root bark03 had its distinct bacteria, including Rhodococcus sp. WB1, Fusarium cf. Solani PUF001, and Fusarium cf. solani PUF007. In the saline-alkali soil group, unique OTUs were present in each of the three periods: 558 unique OTUs in saline-alkali soil01 (13.78% of the total), 374 unique OTUs in saline-alkali soil02 (9.23%), and 695 unique OTUs in saline-alkali soil03 (17.16%). For instance, unique bacteria in saline-alkali soil01 included Pseudomonas stutzeri, uncultured bacteria, Hydrogenophaga sp., Stenotrophomonas sp. PAMC25021, Rhodobium sp. S41, and Qipengyuania vulgaris. In the saline-alkali soil02, Acrobat sp., Ochrobactrum sp., and Paracoccus sp. were unique, while uncultured Clostridium sp., Clostridium distributium, and Bradyrhizobium sp. CCBAU 517 were specific to the saline-alkali soil03. Overall, the total number of OTUs across the three time periods of the Goji berry root bark amounted to 3812, with 1606 OTUs (42.13% of the total) shared among them, indicating that the diversity of the soil microbial community varies across different time periods.
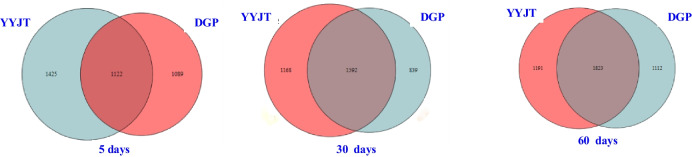
The Venn diagram in Fig. 7 illustrates the overlap and distinctiveness of soil microorganisms at the species level between the saline-alkali soil and the Goji berry root bark in different periods. Notably, the total number of OTUs in the saline-alkali soil was higher than that in the Goji berry root bark, and this number increased over time for both environments. Specifically, the saline-alkali soil exhibited total OTUs numbering 3636, 3599, and 4126 across the three time periods, demonstrating an increase in diversity with time. In contrast, the Goji berry root bark had a total of 1122, 1592, and 1823 OTUs, also displaying an increasing trend in diversity with time.
Fig. 7.
Venn plots comparing the species at 5, 30, and 60 days. “YYJT” corresponds to samples from the saline-alkali soil control group, and “the Goji berry root bark” corresponds to samples from the saline-alkali soil with the Goji berry root bark group
Among the strains common to the three stages of the Goji berry root bark, Sphingobacterium multivorum stood out as it produced acid and maintained intestinal microecological balance, as well as competitively inhibited pathogenic microorganisms, generated beneficial metabolites, and activated acid protease activity. Furthermore, it contributes to lowering the pH of saline-alkali soil, thereby promoting the successful colonization of moss crusts.
Functional Diversity of Soil Microorganisms
Figure 8 shows the annotation of six groups of soil microorganisms in the KEGG primary metabolic pathway across three time periods for both treatments. Among these groups, metabolism was the most abundant, accounting for 70.94%, 71.03%, 72.23%, 69.99%, 69.68%, and 70.37%, respectively, indicating that the primary function of soil microorganisms was related to metabolism, reflecting their role in the activation process. The abundance of metabolism in the saline-alkali soil was higher than in the Goji berry root bark in each time period, and the abundance of the two treatments increased gradually with time. In terms of human diseases, the abundance in the saline-alkali soil was lower than in the Goji berry root bark across all periods. The abundance of human diseases in the saline-alkali soil decreased gradually over time, while in the Goji berry root bark, it initially increased and then decreased. Cellular processes showed an initial increase and then a decrease in both treatments over time, with the abundance in the Goji berry root bark being higher than in the saline-alkali soil in each period. Regarding environmental information processing, the abundance was 2.43%, 2.60%, 2.38%, 3.48%, 3.48%, and 3.20%, respectively. The abundance in the Goji berry root bark was higher than in the saline-alkali soil in each period. The genetic information processing of the abundance of the saline-alkali soil in each period was higher than that of the Goji berry root bark, while the genetic information processing of the abundance of the two processes in each period did not significantly change. In organic systems, both treatments showed an initial decrease, then an increase over time, with the abundance in the saline-alkali soil greater than in the Goji berry root bark at each time point.
Fig. 8.
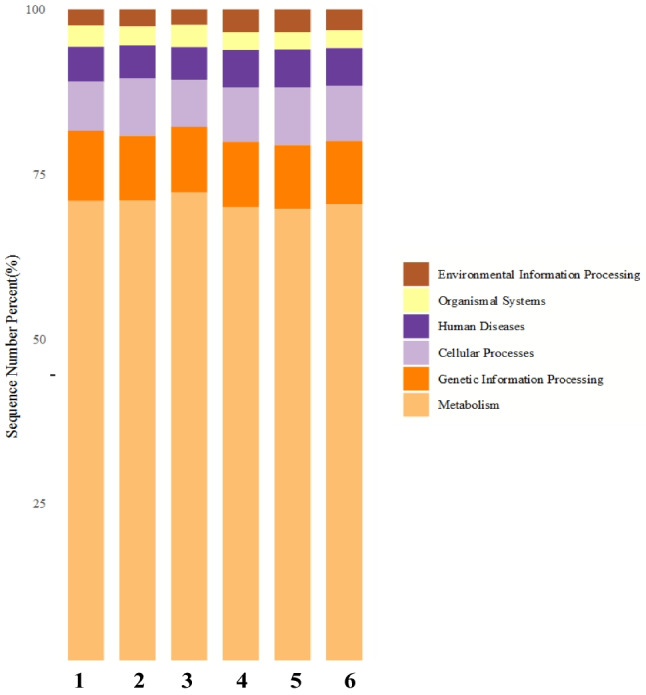
Bar graph depicting the level 1 hierarchical abundance of the KEGG metabolic pathway. Groups 1–3 represent samples from the soil at 5, 30, and 60 days, while groups 4–6 represent samples from the crust at 5, 30, and 60 days of Fig. 5, respectively
Figure 9 presents the gene ontology (GO) category proportions for six samples: the Goji berry root bark01, the Goji berry root bark02, the Goji berry root bark03, saline-alkali soil01, saline-alkali soil02, and saline-alkali soil03, which were analyzed for gene ontology (GO) categories. The overall composition of genes in the top 10 abundances for GO were as follows: 0016021 [CC] membrane, GO: 0005737 [CC] cytoplasmic gene, GO: 0005524 [MF] ATP binding gene, GO: 0003677 [MF] DNA binding gene, GO: 005886 [CC] plasma membrane gene, GO: 006351 [BP] DNA template gene, GO: 003700 [MF] transcription factor activity, sequence-specific DNA binding gene, GO: 0046872 [MF] metal ion binding gene, GO: 0006412 [BP] transformation gene, and GO: 0003735 [MF] ribosome structure composition gene were analyzed. These categories represent molecular function (MF), biological process (BP), and cell components (CC).
Fig. 9.
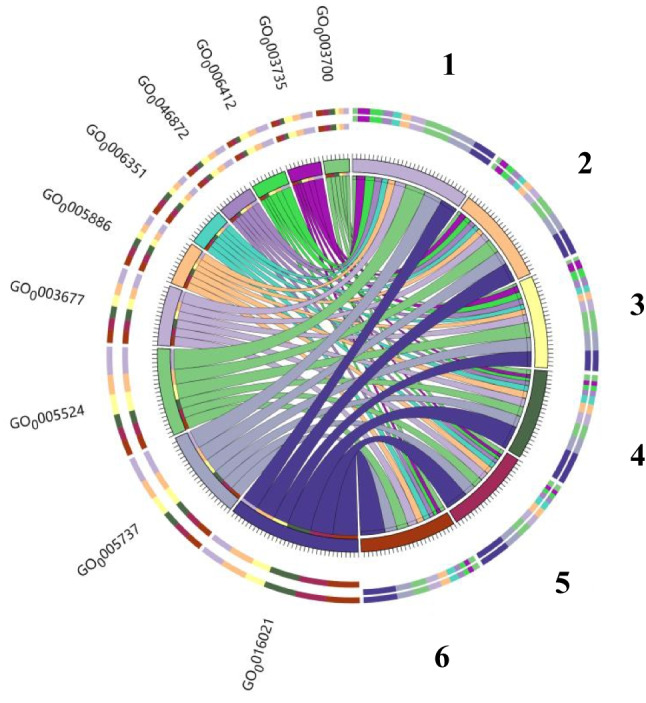
Circos distribution map of six samples in gene ontology, depicting the top 10 abundant categories. Groups 1–3 represent samples from the soil at 5, 30, and 60 days, while groups 4–6 represent samples from the crust at 5, 30, and 60 days of Fig. 5, respectively
Taken together, the addition of the Goji berry root bark isolation band resulted in a higher gene abundance related to GO: 0016021 (integral component of the membrane), GO: 0003700 (transcription factor activity), and GO: 006351 (DNA template gene), suggesting that the Goji berry root bark isolation band may enhance crust colonization by promoting the synthesis of beneficial microbial biofilms and increasing microbial genetic expression.
Discussion
Soil salinization is a pressing global environmental concern that leads to the degradation of valuable soil resources. In China, saline-alkali land is prevalent and extensive, with its expansion driven by both natural processes and human activities. This makes it an area with substantial potential for development and utilization, attracting growing attention from researchers in the field [2, 30, 31]. In the past, physical and chemical methods have been predominantly used to control saline-alkali land, which achieved certain control effects. However, these measures often focus on engineering solutions that address the symptoms rather than the root causes, as they tend to provide short-term effects and can be easily reversible, failing to bring about fundamental improvements to the physical and chemical properties of the soil [32, 33]. In recent years, biological treatments have emerged as a promising alternative, including approaches such as phytoremediation and microbial remediation, with a particular emphasis on phytoremediation [34, 35]. While biological crusts have been traditionally used in the reclamation of arid desert areas, moss crusts, due to their remarkable stress resistance, offer the potential to significantly enhance soil properties and thrive in extreme environments [36–39]. This study takes advantage of the unique characteristics of moss crusts and explores their application in the management of saline-alkali land to provide a solid scientific foundation for the future development and utilization of saline-alkali land.
Bacterial communities in BSCs have been investigated in many studies worldwide. Among them, owing to the significant colonization status, the 16S rRNA gene amplicon sequencing method was used to explore the bacterial diversity and community composition of BSCs [40–43]. However, there has been no consensus on the trends in bacterial diversity variation during BSC development. In this study, we employed the endophytic bacteria isolation method to isolate and purify endophytic bacteria from moss crusts. A total of 45 strains were successfully isolated, comprising 19 endophytic bacteria and 26 endophytic fungi. These isolated strains belonged to various genera, including Bacillus, Agrobacterium, Pristella, Aspergillus, Fusarium, Acanthoderma, Curvularia, Listeria, Perilacia, and Rhizomucor. To assess their salt and alkaline tolerance, we conducted thorough analyses of the isolated strains. Our findings revealed that most of these strains exhibited relatively weak halophilic characteristics, while a smaller subset, such as strains A23 and A62, demonstrated extreme halophilic properties. The presence of these salt-tolerant microorganisms within moss crusts suggests their potential for colonization in saline-alkali land, providing a valuable basis for future experiments and investigations in this domain.
This present study fills a significant research gap by investigating the colonization of moss crusts under various isolation zones, including the absence of an isolation zone, which had not been explored previously. The findings reveal that the development of moss crusts has a significant impact on the physical and chemical properties of saline-alkali soil. Notably, moss crusts reduce the pH of the soil, with the most substantial pH decrease occurring under the common soil isolation zone. Moreover, the electrical conductivity of the saline-alkali soil was significantly decreased in the presence of moss crusts, particularly under the locust flowers isolation zone. Additionally, adding moss crusts increased organic matter contents, especially when combined with the grape peel isolation zone. Total phosphorus contents in saline-alkali soil also significantly increased, particularly under saline-alkali soil conditions. Furthermore, moss crust colonization contributed to higher total nitrogen content in saline-alkali soil, particularly when grape peels were involved, although it had little effect on hydrolyzed nitrogen content. Overall, the presence of moss crusts significantly enhances soil properties, improving soil fertility and suitability for crop growth. Among the isolation conditions tested, the Lycium barbarum root bark (the Goji berry root bark) isolation zone stands out for its effectiveness in promoting moss crust colonization, as indicated by a significant increase in chlorophyll content and maximum moss crust coverage.
Several significant findings were observed following a comparison of the experimental and control groups within the saline-alkali soil and the saline-alkali soil with the Goji berry root bark. Notably, the HPLC-q-TOF MS-assisted molecular network analysis revealed distinct and strong responses at specific retention times, such as 40.632 min in the negative ion plot for the saline-alkali soil with the Goji berry root bark subjected to 50% methanol–water washing. In the positive ion plot, the saline-alkali soil with the Goji berry root bark treated with 95% methanol–water exhibited specific and intense responses at 28.687 min and 39.728 min, hinting at potential compounds associated with the promotion of moss crust colonization. Furthermore, the introduction of the Goji berry root bark isolation zone led to increased soil enzyme activities, particularly S-SC, S-UE, and S-AKP, thereby facilitating moss crust colonization by promoting enzymatic processes. Interestingly, moss crust colonization was found to enhance S-SC activity while reducing S-UE activity. Additionally, the Goji berry root bark isolation zone contributed to a decline in malondialdehyde content, an increase in protein and polysaccharide content, and enhanced salt stress resistance in moss crusts.
Metagenomic data from three time periods under saline-alkali soil and saline-alkali soil with the Goji berry root bark moss crust addition were thoroughly analyzed. The incorporation of the Goji berry root bark isolation belt notably increased the abundance of the bacterial community within the microbial composition at the phylum level. While variations in the relative abundance of soil microorganisms were observed across the six groups, these differences were not significant. Functional analysis of soil microbial activities, focusing on the primary metabolic pathways within the KEGG framework, indicates that the Goji berry root bark isolation zone likely enhances moss crust colonization by stimulating the production of beneficial microbial biofilms and increasing microbial genetic expression. Moreover, in GO analysis, it was evident that the presence of the Goji berry root bark isolation band correlated with a higher abundance of genes associated with integral membrane components (GO: 0016021), transcription factor activity (GO: 0003700), and DNA template genes (GO: 006351).
Conclusion
The effects of physical isolation and moss crust transplantation on the ecological rehabilitation of saline-alkali soil were assessed through LC–MS and metagenomic sequencing techniques. The utilization of the Goji berry root bark root bark as a physical isolation method was found to enhance moss crust colonization in saline-alkali soil while also elevating soil organic matter and nutrient levels. These findings offer valuable insights into the ecological management of saline-alkali land and provide a reference for future research in this field.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Home for Researchers editorial team (www.home-for-researchers.com) for language editing service.
Author Contributions
Zhi-bo Jiang and Huan-huan Guo performed the data analysis; Hui Zhang and Jing-jing Tian performed the formal analysis; Zhi-bo Jiang and Hui Zhang performed the validation; Zhi-bo Jiang and Xiao-li Ma wrote the manuscript. All authors reviewed the manuscript.
Funding
This work was co-funded by the National Natural Science Foundation of China (No. 82160672), the Outstanding Youth Program of Ningxia Natural Science Foundation (No. 2022AAC05041 and 2023AAC05048), the Key R&D Projects in Ningxia (Talent Introduction Special Project, No. 2021BEB04019), and the Ningxia Natural Science Foundation (No. 2021AAC03210 and 2019AAC03113).
Data Availability
No datasets were generated or analyzed during the current study.
Declarations
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Tugel AJ, Herrick JE, Brown JR, Mausbach MJ, Puckett W, Hipple K (2005) Soil change, soil survey, and natural resources decision making: a blueprint for action. Soil Sci Soc Am J 69(3):738–747 [Google Scholar]
- 2.Zhang K, Chang L, Li G, Li Y (2023) Advances and future research in ecological stoichiometry under saline-alkali stress. Environ Sci Pollut Res Int 30(3):5475–5486 [DOI] [PubMed] [Google Scholar]
- 3.Zhang B, Wang N (2021) Study on the harm of saline-alkali land and its improvement technology in China IOP Conference Series: Earth and Environmental Science. IOP Conf Ser: Earth Environ Sci 692(4):042053 [Google Scholar]
- 4.Jaiswal B, Singh S, Agrawal SB, Lokupitiya E, Agrawal M (2022) Improvements in soil physical, chemical and biological properties at natural saline and non-saline sites under different management practices. Environ Manage 69(5):1005–1019 [DOI] [PubMed] [Google Scholar]
- 5.Gao Y, Zou H, Wang B, Yuan F (2022) Progress and applications of plant growth-promoting bacteria in salt tolerance of crops. Int J Mol Sci 23(13):7036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qin H, Li Y, Huang R (2020) Advances and challenges in the breeding of salt-tolerant rice. Int J Mol Sci 21(21):8385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jaiswal B, Singh S, Agrawal SB, Agrawal M (2021) Assessment of physiological, biochemical and yield responses of wheat plants under natural saline and non-saline field conditions. Physiol Mol Biol Plants 27(10):2315–2331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Y, He W, He J, Shen Z (2016) Present situation and the prospect of improvement and utilization of saline-alkali land in Northwest. Ningxia Eng Technol 15(4):404–408 (in Chinese) [Google Scholar]
- 9.Negacz K, Malek Ž, Vos A, Vellinga P (2022) Saline soils worldwide: identifying the most promising areas for saline agriculture. J Arid Environ 203:104775 [Google Scholar]
- 10.Liu G, Zhang X, Wang X, Shao H, Yang J, Wang X (2017) Soil enzymes as indicators of saline soil fertility under various soil amendments. Agr Ecosyst Environ 237:274–279 [Google Scholar]
- 11.Simpson RM, Mason K, Robertson K, Müller K (2019) Relationship between soil properties and enzyme activities with soil water repellency. Soil Res 57(6):689–702 [Google Scholar]
- 12.Sanchez-Hernandez JC, Notario Del Pino J, Capowiez Y, Mazzia C, Rault M (2018) Soil enzyme dynamics in chlorpyrifos-treated soils under the influence of earthworms. Sci Total Environ 612:1407–1416 [DOI] [PubMed] [Google Scholar]
- 13.Li F, Guo Y, Wang X, Xing X, Yang J (2011) Effect of different soil improvement measures on microbial diversity and alfalfa biomass in saline-alkali soil in Ningxia. J Soil Water Conserv 25(5):107–111 (In Chinese) [Google Scholar]
- 14.Teo HM, Aziz A, Wahizatul AA, Bhubalan K, SitiNordahliawate MS, Muhamad SCI, Lee CN (2022) Setting a plausible route for saline soil-based crop cultivations by application of beneficial halophyte-associated bacteria: a review. Microorganisms 10(3):657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y, Tian S, Zuo Z (2021) Research of desalting effect of improved underground drainage pipe in saline-alkali soil of Ningxia. J Ningxia Univ (Nat Sci Ed) 42(4):456–462 (In Chinese) [Google Scholar]
- 16.Dong S, Wan S, Kang Y, Miao J, Li X (2021) Different mulching materials influence the reclamation of saline soil and growth of the Lycium barbarum L. under drip irrigation in a saline wasteland in northwest China. Agr Water Manage 247:106730 [Google Scholar]
- 17.Sha Y, Huang Z, Wei Z (2022) Impact of microbial agent broadcast application on the microbial community structure of saline-alkali soil in Shizuishan of Ningxia. Chin Agr Sci Bull 38(34):82–90 (In Chinese) [Google Scholar]
- 18.Rezapour S, Asadzadeh F, Barin M, Nouri A (2022) Organic amendments improved the chemical-nutritional quality of saline-sodic soils. Int J Environ Sci Technol 19:4659–4672 [Google Scholar]
- 19.Borowik A, Wyszkowska J, Kucharski J (2022) Bacteria and soil enzymes supporting the valorization of forested soils. Materials 15(9):3287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang C, Zhang Q, Wang F, Liang W (2017) Toxicological effects of dimethomorph on soil enzymatic activity and soil earthworm (Eisenia fetida). Chemosphere 169:316–323 [DOI] [PubMed] [Google Scholar]
- 21.Du J, Hou F, Zhou Q (2021) Response of soil enzyme activity and soil bacterial community to PCB dissipation across different soils. Chemosphere 283:131229 [DOI] [PubMed] [Google Scholar]
- 22.Teng J, Jian RL, Zhao Y (2017) Impact of sand burial on bacterial community structure and diversity within biocrusts dominated by Bryum argenteum. Acta Ecol Sin 37(7):2179–2187 [Google Scholar]
- 23.Liu Y, Yang H, Jia R, Li Y (2019) Effects of human trampling biocrusts on soil enzyme activities. J Desert Res 34(4):54–63 (In Chinese) [Google Scholar]
- 24.Dong J, Li Y, Li X, Li B, Guo Q, Bu C (2019) Effect of vegetation type on biocrusts and the underlying soil nutrients in Muus sandland. J Soil Water Conserv 26(2):112–117 (In Chinese) [Google Scholar]
- 25.Luiza de Farias V, Ximenes Monteiro K, Rodrigues S, André NarcisoFernandes F, Adolfo Saavedra Pinto G (2010) Comparison of Aspergillus niger spore production on Potato Dextrose Agar (PDA) and crushed corncob medium. J Gen Appl Microbiol 56(5):399–402 [DOI] [PubMed] [Google Scholar]
- 26.Ma PY, Geng WL, Ji HY, Yue BW, Liu C, Wang S, Jiang ZB, Chen J, Wu XL (2022) Native endophytes of Tripterygium wilfordii-mediated biotransformation reduces toxicity of celastrol. Front Microbiol 13:810565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang J, Xu M, Zou X, Xu M (2019) Effects of biological crufts on soil properties under different soil and vegetation habitats. J Soil Water Conserv 33(5):323–328 (In Chinese) [Google Scholar]
- 28.Li YY, Ma XJ, Li XJ (2020) Changes in physicochemical properties of moss crusts and the underlying soil during the succession of sand-binding-vegetation. J Lanzhou Univ: Nat-Sci 56(8):463–470 (In Chinese) [Google Scholar]
- 29.Sun HF, Li XL, Jin LQ, Li CL, Zhan GJ (2020) Effects of biological soil crusts on the physical and chemical properties of soil and vegetation of artificial grassland in the Yellow River source zone. Acta Agrestia Sin 28(2):509–520 (In Chinese) [Google Scholar]
- 30.Duan Q, Zhu Z, Wang B, Chen M (2022) Recent progress on the salt tolerance mechanisms and application of Tamarisk. Int J Mol Sci 23(6):3325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu L, Wang B (2021) Protection of halophytes and their uses for cultivation of saline-alkali soil in China. Biology (Basel) 10(5):353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu M, Sun Q, Liu Q, He G, Wang C, He K (2023) Biochar decreases fertilizer leaching and promotes miscanthus growth in saline-alkaline soil. Plants (Basel) 12(20):3649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu R, Liang B, Zhao H, Zhao Y (2023) Impacts of various amendments on the microbial communities and soil organic carbon of coastal saline-alkali soil in the Yellow River Delta. Front Microbiol 14:1239855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu X, Guo L, Wang S, Wang X, Ren M, Zhao P, Huang Z, Jia H, Wang J, Lin A (2023) Effective strategies for reclamation of saline-alkali soil and response mechanisms of the soil-plant system. Sci Total Environ 905:167179 [DOI] [PubMed] [Google Scholar]
- 35.Ma J, Xie Y, Sun J, Zou P, Ma S, Yuan Y, Ahmad S, Yang X, Jing C, Li Y (2023) Co-application of chitooligosaccharides and arbuscular mycorrhiza fungi reduced greenhouse gas fluxes in saline soil by improving the rhizosphere microecology of soybean. J Environ Manage 345:118836 [DOI] [PubMed] [Google Scholar]
- 36.Weber B, Belnap J, Büdel B, Antoninka AJ, Barger NN, Chaudhary VB, Darrouzet-Nardi A, Eldridge DJ, Faist AM, Ferrenberg S, Havrilla CA, Huber-Sannwald E, Malam Issa O, Maestre FT, Reed SC, Rodriguez-Caballero E, Tucker C, Young KE, Zhang Y, Zhao Y, Zhou X, Bowker MA (2022) What is a biocrust? A refined, contemporary definition for a broadening research community. Biol Rev Camb Philos Soc 97(5):1768–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jia R, Gao Y, Zhao L, Zhang T, Guo H, You W, Duan Y (2022) Differential influences of wind-blown sand burial on bacterial and fungal communities inhabiting biological soil crusts in a temperate desert. China Microorganisms 10(10):2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen N, Yu K, Jia R, Teng J, Zhao C (2020) Biocrust as one of multiple stable states in global drylands. Sci Adv 6(39):eaay3763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen N, Liu X, Zheng K, Zhang C, Liu Y, Lu K, Jia R, Zhao C (2019) Ecohydrological effects of biocrust type on restoration dynamics in drylands. Sci Total Environ 687:527–534 [DOI] [PubMed] [Google Scholar]
- 40.Zhou H, Li L, Liu Y (2023) Biological soil crust development affects bacterial communities in the Caragana microphylla community in alpine sandy areas. Front Microbiol 14:1106739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Garcia-Pichel F (2023) The microbiology of biological soil crusts. Annu Rev Microbiol 77:149–171 [DOI] [PubMed] [Google Scholar]
- 42.Barrera A, Acuña-Rodríguez IS, Ballesteros GI, Atala C, Molina-Montenegro MA (2022) Biological soil crusts as ecosystem engineers in antarctic ecosystem. Front Microbiol 13:755014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ghiloufi W, Seo J, Kim J, Chaieb M, Kang H (2019) Effects of biological soil crusts on enzyme activities and microbial community in soils of an arid ecosystem. Microb Ecol 77(1):201–216 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No datasets were generated or analyzed during the current study.