Abstract
Filoviruses, including Ebola, Marburg, Sudan, and Taï Forest viruses, are zoonotic pathogens that can cause severe viral hemorrhagic fever and death. Developing vaccines that provide durable, broad immunity against multiple filoviruses is a high global health priority. In this Phase 1 trial, we enrolled 60 healthy U.S. adults and evaluated the safety, reactogenicity and immunogenicity of homologous and heterologous MVA-BN®-Filo and Ad26.ZEBOV prime-boost schedules followed in select arms by MVA-BN®-Filo boost at 1 year (NCT02891980). We found that all vaccine regimens had acceptable safety and reactogenicity. The heterologous prime-boost strategy elicited superior Ebola binding and neutralizing antibody, antibody-dependent cellular cytotoxicity (ADCC), and cellular responses compared to homologous prime-boost. The MVA-BN®-Filo boost administered at 1 year resulted in robust humoral and cellular responses that persisted through 6-month follow-up. Overall, our data demonstrated that a heterologous Ad26.ZEBOV/MVA-BN®-Filo prime-boost was safe and immunogenic and established immunologic memory primed to respond after re-exposure. Clinicaltrials.gov, NCT02891980, registered September 1, 2016.
Subject terms: Viral infection, Vaccines
Introduction
The filovirus family contains multiple zoonotic viruses (e.g., Ebola virus (EBOV), Sudan virus (SUDV), Marburg virus (MARV), Taï Forest virus (TAFV), Bundibugyo virus, Ravn virus) that have been associated with sporadic human cases or outbreaks of severe hemorrhagic fever with high case fatality rates1. Outbreaks of Ebola Virus Disease (EVD) have occurred almost yearly since 1994. The largest outbreak between December 2013 and June 20162 involved predominantly Guinea, Sierra Leone, and Liberia with over 28,652 cases and 11,325 deaths3,4. Subsequent filovirus cases have included a large outbreak of EVD in 2018-2020, and smaller sporadic outbreaks of SUDV1 and MARV5. Most recently, an outbreak of MARV has been reported in Rwanda with 66 illnesses and 15 deaths as of November 20246.
The development of vaccines that elicit durable immune responses across the breadth of multiple filoviruses is a high global public health priority. The recombinant vesicular stomatitis virus–Zaire Ebola virus (rVSV-EBOV) vaccine (ERVEBO®) is the only Ebola vaccine approved in the U.S. based on clinical efficacy, but its ability to cross-protect against other filoviruses (e.g., SUDV, MARV) remains doubtful7,8. One of the leading vaccination strategies attempting to generate a broader immunological response to the filovirus family utilizes a heterologous two-dose vaccination strategy involving an adenovirus 26-vectored vaccine expressing Ebola glycoprotein (GP) (Ad26.ZEBOV) followed 8 weeks later by a modified vaccinia Ankara-vectored vaccine expressing GPs from EBOV, SUDV, and MARV (Supplementary Fig. 1), and the nucleoprotein from TAFV (Supplementary Fig. 2) (MVA-BN-Filo). The GPs are predominant immunogens capable of inducing filovirus-specific antibody responses9, while the nucleoprotein harbors highly conserved filovirus T-cell epitopes and is a key driver of T-cell responses following filovirus natural infection10. This vaccination regimen is approved by the European Medicines Agency for the prevention of EBOV and has the potential to elicit humoral responses against the glycoproteins of three major filoviruses and provide cross-protective cellular immunity11. This approach has been well tolerated and immunogenic in Phase 1 and 2 clinical trials, including in HIV-infected patients, children, and adolescents12–17. It offers multiple possible advantages, including filovirus multivalency, replication deficiency (eliminating vaccine shedding concerns) and multiple storage options. Common adverse effects of MVA-BN-Filo and Ad26.ZEBOV observed in clinical trials included fatigue, headache, myalgias, arthralgias, and injection site reactions18,19.
The objectives of this study were to evaluate the safety, reactogenicity, and immunogenicity of a homologous or heterologous prime-boost series of Ad26.ZEBOV and MVA-BN-Filo given 4 weeks apart, followed in selected arms by a boost at 1 year with MVA-BN-Filo. Since the correlates of protection against filoviruses are incompletely understood, we sought to evaluate multiple aspects of the immune response including binding, neutralizing and functional antibody-dependent cellular cytotoxicity (ADCC) responses, plasmablast responses, and T cell responses after Ad26.ZEBOV and MVA-BN-Filo vaccination. In addition, we sought to utilize systems vaccinology20,21 approaches to identify gene expression biomarkers that may predict vaccine immunogenicity.
Results
Study participants
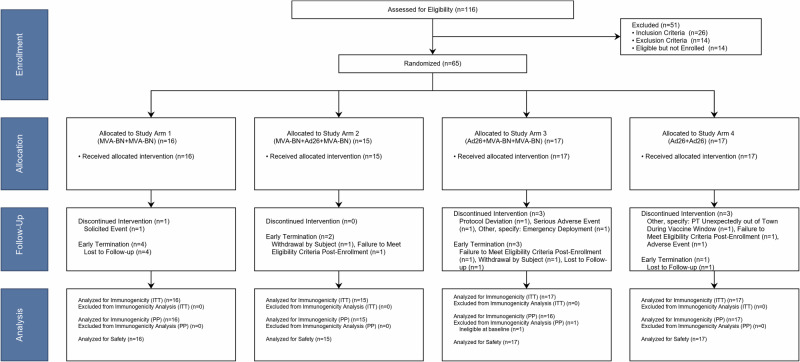
From April 11, 2017 to September 25, 2017, 116 individuals were screened for the study at a single U.S. site (Emory University, Atlanta, GA) and 65 were enrolled into one of four Study Arms (Fig. 1). Some minor imbalances were noted in the allocations for attributes that were not included in enrollment stratification (e.g., predominance of females in Arm 1). The participants had a mean age of 27.8 years [standard deviation (SD) 5.7] and were of predominantly white race (80.0%) and non-Hispanic ethnicity (88.0%) (Table 1). Seven individuals discontinued vaccinations during the study: one from Study Arm 1, three in Study Arm 3, and three in Study Arm 4. Ten individuals terminated the study early, of whom six were due to lost follow-up (Fig. 1). The trial completed on March 21, 2019.
Fig. 1. CONSORT flow diagram.
Planned enrollment was 60 with potential replacement of participants that did not receive the second dose. Final enrollment was 65 to replace 5 participants who did not receive a second dose.
Table 1.
Demographic Characteristics of Patients by Randomization Status
| Variable | Statistic | Study Arm 1 (MVA-BN + MVA-BN) (N = 16) | Study Arm 2 (MVA-BN + Ad26 + MVA-BN) (N = 15) | Study Arm 3 (Ad26 + MVA-BN + MVA-BN) (N = 17) | Study Arm 4 (Ad26 + Ad26) (N = 17) | All Participants (N = 65) |
|---|---|---|---|---|---|---|
| Age (Years) | Mean | 25.3 | 27.8 | 30.3 | 27.5 | 27.8 |
| Standard Deviation | 5.1 | 6.0 | 5.5 | 5.6 | 5.7 | |
| Median | 25 | 26 | 30 | 27 | 27 | |
| IQR | 21.5–27 | 23–32 | 26–34 | 23–29 | 23–31 | |
| BMI (kg/m2) | Mean | 23.6 | 24.6 | 25.6 | 24.9 | 24.7 |
| Standard Deviation | 3.5 | 4.6 | 4.6 | 4.2 | 4.2 | |
| Median | 23.3 | 24.1 | 24.4 | 24.3 | 24.1 | |
| IQR | 20.4–25.9 | 20.8–26.4 | 23.4–28.6 | 20.8–28.8 | 20.8–27.1 | |
| Variable | Characteristic | Study Arm 1 (MVA-BN + MVA-BN) (N = 16) n (%) | Study Arm 2 (MVA-BN + Ad26 + MVA-BN) (N = 15) n (%) | Study Arm 3 (Ad26 + MVA-BN + MVA-BN) (N = 17) n (%) | Study Arm 4 (Ad26 + Ad26) (N = 17) n (%) | All Participants (N = 65) n (%) |
| Sex | Male | 5 (31.3) | 9 (60.0) | 10 (58.8) | 7 (41.2) | 31 (47.7) |
| Female | 11 (68.8) | 6 (40.0) | 7 (41.2) | 10 (58.8) | 34 (52.3) | |
| Ethnicity | Not Hispanic or Latino | 13 (81.3) | 14 (93.3) | 15 (88.2) | 15 (88.2) | 57 (87.7) |
| Hispanic or Latino | 3 (18.8) | 1 (6.7) | 2 (11.8) | 2 (11.8) | 8 (12.3) | |
| Race | Asian | - | 4 (26.7) | 1 (5.9) | 2 (11.8) | 7 (10.8) |
| Black or African American | - | - | - | 2 (11.8) | 2 (3.1) | |
| White | 14 (87.5) | 11 (73.3) | 15 (88.2) | 12 (70.6) | 52 (80.0) | |
| Multi-Racial | 2 (12.5) | - | 1 (5.9) | 1 (5.9) | 4 (6.2) |
N = Number of Participants in the Safety Population.
Vaccine reactogenicity & safety
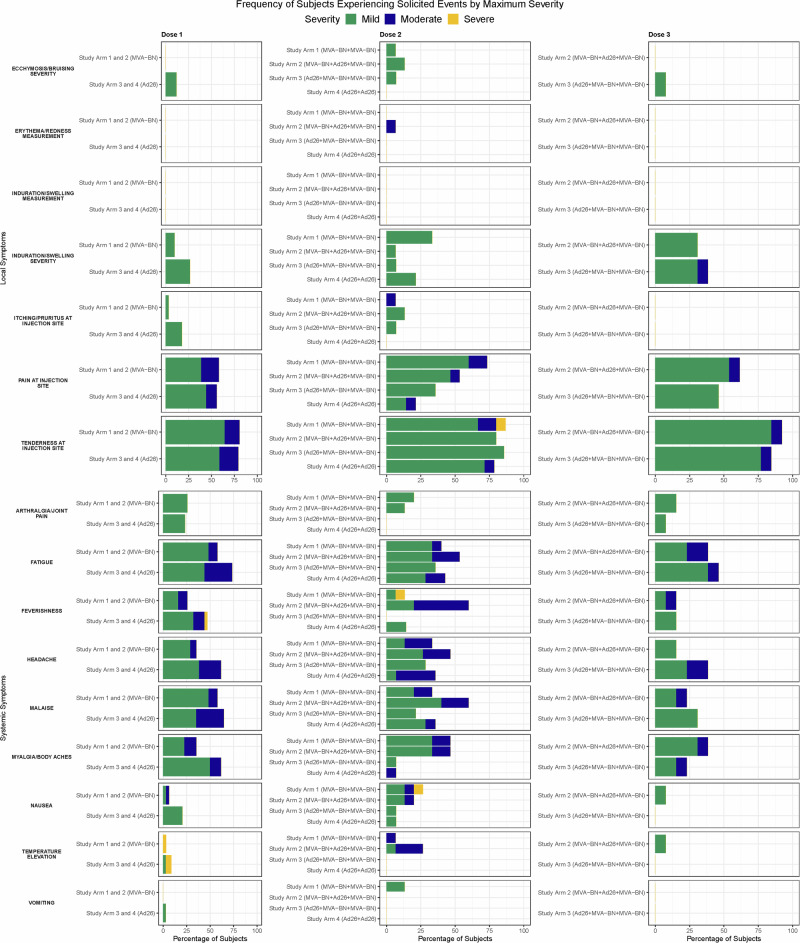
Local solicited adverse events (AE) occurred in nearly all participants (95.0%), and in all participants who received at least one dose of MVA-BN-Filo (Supplementary Table 1, Fig. 2). Systemic solicited AEs occurred in 87.7% of participants ranging from 82.4% to 93.3% across Study Arms. The most frequent local and systemic solicited AEs across all treatment arms were injection site pain (95.4%), tenderness (90.8%), malaise (76.9%), fatigue (75.4%), myalgias (64.6%), and headache (61.5%) (Supplementary Table 2, Fig. 2). Most events were mild to moderate in severity and occurred within the first 1–3 days after vaccination; however, severe reactions were observed in three participants (19.0%) in Study Arm 1 (elevated oral temperature post-dose 1, feverishness, nausea, tenderness at injection site post-dose 2), one participant (6.0%) in Study Arm 3 (elevated oral temperature post-dose 1), and two participants (12%) in Study Arm 4 (elevated oral temperature, feverishness post-dose 1) (Fig. 2). No severe reactogenicity was observed after the third dose in any of the Study Arms (Fig. 2).
Fig. 2. Frequency of subjects experiencing solicited adverse events by maximum severity, by vaccine dose and Study Arm.
Local and Systemic Reactions. Green, mild severity; Blue, moderate; Yellow, severity. Study Arm 1, MVA-BN + MVA-BN; Study Arm 2, MVA-BN + Ad26 + MVA-BN; Study Arm 3, Ad26 + MVA-BN + MVA-BN; Study Arm 4, Ad26 + Ad26.
Unsolicited AEs after any dose that were considered related to the vaccine occurred in nine (13.8%) participants. These occurred most frequently in Study Arm 1 (25%) and Study Arm 4 (17.6%) (Supplementary Table 3, Supplementary Fig. 3). Most were mild to moderate in severity. A single severe unsolicited AE of positional lightheadedness occurred 2 days after the second dose in a Study Arm 1 participant; the event was considered related to vaccination and lasted for 2 days. Other safety evaluations, including vital signs (Supplementary Table 4) and laboratory analyses (Supplementary Tables 5–7), are detailed in the Supplementary Material.
Four participants experienced an AE that resulted in treatment withdrawal or study discontinuation prior to the second study vaccination. These included one unrelated serious adverse event (SAE) of abdominal pain due to an ovarian cyst; one unrelated moderate viral upper respiratory tract infection; one related severe lymphopenia on Day 15 after prime dosing with MVA-BN-Filo in Study Arm 1; and one related moderate diastolic hypertension on Day 28 after prime dosing with Ad26.ZEBOV in Study Arm 4 (Fig. 1). No AEs of special interest were observed in the study. A single SAE of abdominal pain was reported on Day 29 in a female participant in Study Arm 3 which was considered unrelated to vaccine and instead due to a large ovarian cyst that required surgery. No pregnancies and no deaths occurred during the study.
Immunogenicity outcomes
EBOV glycoprotein (GP) antibody responses: enzyme-linked immunosorbent assay (ELISA) and virus neutralizing assay (VNA)
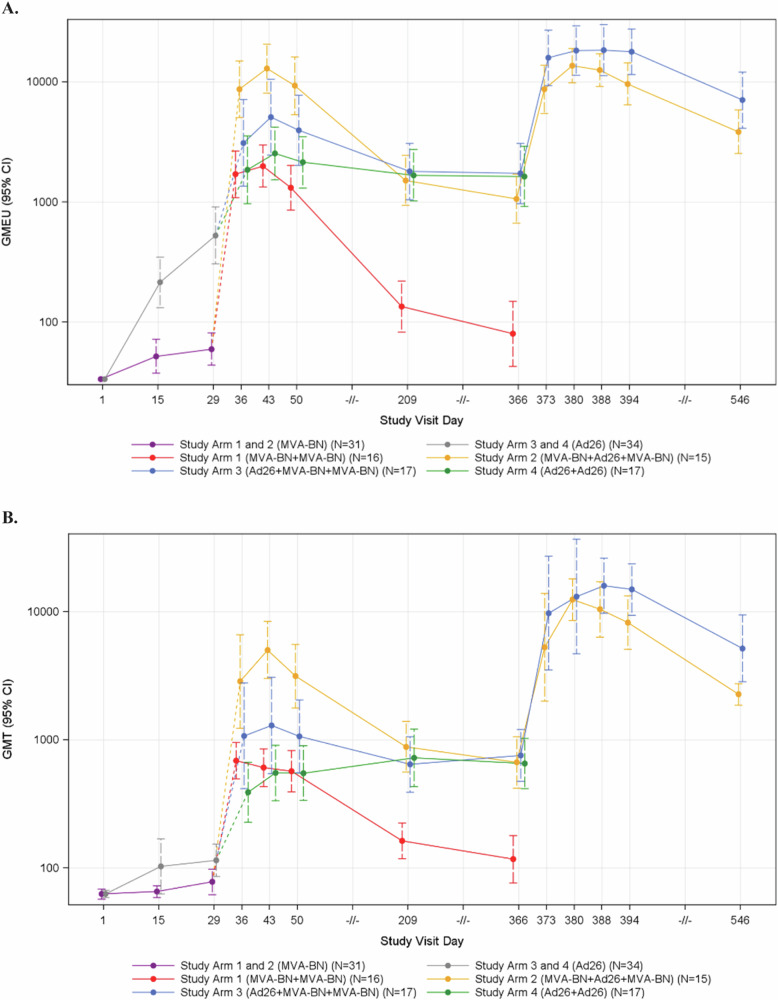
At baseline, EBOV-GP binding and pseudovirus neutralizing antibodies were not detectable. In general, the peak EBOV ELISA and VNA geometric mean (GM) antibody responses were highly correlated with one another with Spearman correlation coefficients of 0.58, 0.91, and 0.84 for peak post-first vaccination, peak post-second vaccination, and peak post-third vaccination response, respectively (Supplementary Figs. 4–6). After a single dose of vaccine, participants who received Ad26.ZEBOV (Study Arms 3 and 4) achieved higher peak EBOV-GP ELISA and VNA antibody responses than those who first received MVA-BN-Filo (Study Arms 1 and 2) (ELISA GMT 507.5 vs. 62.3, p < 0.001; VNA GMT 164.4 vs. 83.2, p = 0.016) (Fig. 3, Supplementary Table 8). After the second dose of vaccine, heterologous vaccine recipients (Study Arms 2 and 3) had higher peak EBOV-GP ELISA and VNA antibody responses than homologous recipients (Study Arms 1 and 4), with the greatest responses observed in Study Arm 2 (MVA-BN-Filo + Ad26.ZEBOV) (Supplementary Table 8). The EBOV antibody responses in the MVA-BN-Filo homologous Study Arm 1 declined back to baseline by 6 months after the second dose while the Ad26.ZEBOV homologous Study Arm 4 remained stable from 1 month after the second dose through 12 months after the second dose. The EBOV-GP ELISA and VNA antibody responses for Study Arm 4 were similar to those observed in the heterologous study arms (Study Arms 2 and 3) at Days 209 and 365 (Fig. 3, Supplementary Tables 9, 10).
Fig. 3. Binding and neutralizing antibody responses against EBOV GP by Study Day and Treatment Arm; Intent-to-Treat Analysis Population.
A Enzyme-linked immunosorbent assay (ELISA) Antibody Results (ELISA Units/mL) Against EBOV GP by Study Visit Day and Treatment Arm. B Virus Neutralization Antibody (VNA) Titer against EBOV GP by Study Day and Treatment Arm.
After receiving the third dose (i.e., an MVA-BN-Filo booster dose at 1 year), Study Arm 3 (Ad26.ZEBOV + MVA-BN-Filo + MVA-BN-Filo) showed further increase in peak EBOV-GP ELISA responses compared to peak post-dose 2 (p < 0.001), while Study Arm 2 (MVA-BN-Filo + Ad26.ZEBOV + MVA-BN-Filo) did not (p = 0.894) (Fig. 3A, Supplementary Table 8). However, peak EBOV VNA geometric mean titers (GMT) were greater for both arms 3 and 2 after the third dose than after the second dose (p = 0.010 and p < 0.001, respectively), suggesting additional maturation of the antibody responses with this late boost (Fig. 3B, Supplementary Table 8). The peak EBOV VNA GMTs after the third vaccination did not statistically differ between Study Arm 2 and Study Arm 3 (VNA GMTs 14,892.2 and 22,103.8, respectively, p = 0.120; Supplementary Table 8). EBOV-GP ELISA and VNA antibody responses remained durable through 6 months after the third dose at levels that exceeded the Day 209 levels (6 months after the second dose) (Fig. 3).
Antibody-dependent cellular cytotoxicity
An ADCC assay was developed (Supplementary Methods, Supplementary Figs. 7, 8) to measure titers against EBOV, MARV, and SUDV GP. Detectable EBOV-GP ADCC responses (i.e., exceeding baseline levels) were observed in over 75% of participants at Day 43 for the heterologous vaccine arms (Study Arms 2 and 3), which persisted through 12-month follow-up (Supplementary Table 11, Supplementary Fig. 9). Interestingly, in addition to the heterologous vaccine arms, detectable EBOV-GP ADCC antibody titers were also observed in about 40% of participants at Day 43 (Day 15 post-second vaccination) and 70% of participants at Day 209 (6 months after the second dose) in both homologous study arms (Study Arms 1 and 4) with detectable antibodies persisting through 12-month follow-up. EBOV-GP ADCC antibodies persisted in over two-thirds of participants at Day 365. In Study Arms 2 and 3, all participants developed an EBOV-GP ADCC response by 2 weeks after the third dose and these ADCC antibodies remained detectable through 6 months after the third dose in nearly all participants.
ADCC responses were detected in only a few participants against MARV GP beginning at Day 209. Even after the third dose, only 31% of participants in Study Arms 2 and 3 retained detectable responses at 6 months (Supplementary Table 11, Supplementary Fig. 9). Unlike the responses to MARV, ADCC responses to SUDV-GP were detectable in some participants by 2 weeks after the first dose, including in 17.6% of participants in Study Arms 3 and 4 (received prime dose of Ad26.ZEBOV) (Supplementary Table 11, Supplementary Fig. 9). Increases in those with detectable SUDV-GP ADCC response were observed after the third dose in Study Arms 2 and 3, and were maintained through 6 months follow-up in 23% and 31% of participants respectively.
Plasmablast responses
Frequencies of IgA and IgG secreting plasmablasts against EBOV-GP, SUDV-GP, and MARV-GP were similar to baseline levels 7 days after the first and third doses of vaccine in all Study Arms (Supplementary Table 12). After the second dose of vaccine, detectable plasmablast responses producing IgA and IgG against EBOV GP were observed at Day 36 (Day 8 post-second vaccination) only in Study Arm 2 participants (MVA-BN-Filo+Ad26.ZEBOV). These EBOV-GP IgA and IgG plasmablast responses weakly correlated with the peak anti-EBOV-GP ELISA antibody responses after the second dose (Spearman correlation IgA 0.581, p = 0.023; IgG 0.568, p = 0.047) and had a slightly weaker correlation with peak anti-EBOV-GP VNA titers (IgA 0.472, p = 0.076; IgG 0.490, p = 0.064). Additionally, MARV or SUDV-GP plasmablast responses occurred in only a few participants.
T cell responses
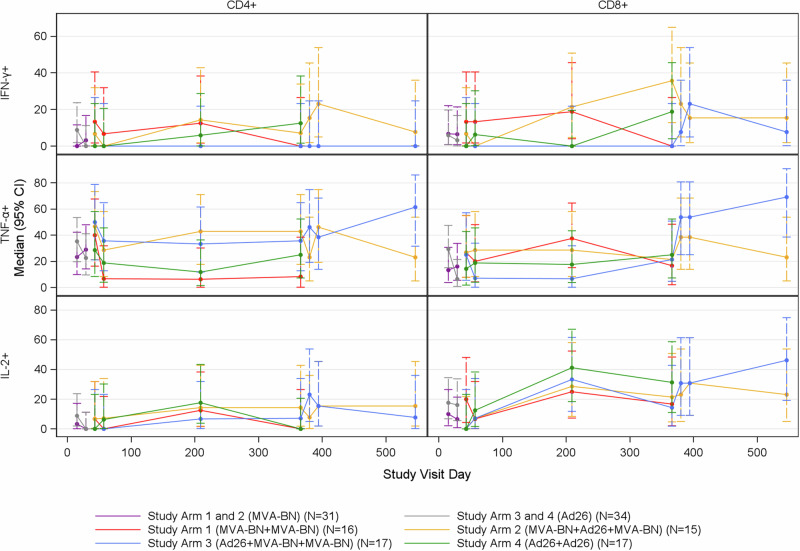
The percentage of activated T cells (CD4+ and CD8+) expressing select cytokines (IFN-γ, TNF-α, IL-2) following stimulation with EBOV peptide pools was measured using intracellular cytokine staining (ICS) (Supplementary Fig. 10). Normalized percent activation values were compared to a cutoff to classify participants as positive or negative responders at each post-vaccination day (Supplemental Materials). The maximum response across post-vaccination days following each dose was used to classify participants as positive or negative responders for each vaccination (Fig. 4, Supplementary Figs. 11–13). In general, the percent positive responders for IFN-γ and IL-2 were greater for CD8+ cells than for CD4+ T cells while the percent positive responders for TNF-α responses for CD8+ and CD4+ T cells were similar against EBOV GP antigen (Supplementary Fig. 14). IFN-γ CD4+ and CD8+ percent activated cells were similar to baseline in all of the study arms, although a few individuals in some of the study arms had detectable responses. IL-2 CD8+ percent activated cells were present at Day 209 for all study arms. Both heterologous boost vaccine Study Arms 2 and 3 achieved higher than baseline IL-2 CD8+ responses at Day 380 (Day 15 post-third dose) that remained present through the 6 months after the third dose.
Fig. 4. CD4+ and CD8 + EBOV T-cell responses induced by MVA-BN-Filo and Ad26.ZEBOV vaccines.
Figure shows percent positive responders based on percent activated T-cells compared to pre-vaccination by Study Day, Cell Type, Cytokine, and Treatment Group. INF-γ (top), TNF-α (middle), and L-2 (bottom) in CD4 + (left) and CD8 + (right) cells following EBOV stimulation; Intent-to-Treat Analysis Population. The error bars represent 95% confidence intervals.
The greatest T cell responses, in terms of percent positive responders based on percent activated T-cells and peak difference in percent activated T-cells against EBOV GP antigen, were TNF-α CD4+ and CD8+ responses observed in the heterologous vaccine arms (Study Arms 2 and 3) following the second and third vaccinations (Supplementary Tables 13, 14). The TNF-α CD4+ peak percent activated cells for Study Arm 2 were 0.035 post-dose 2 and 0.025 post-dose 3, while TNF-α CD8+ peak percent activated cells were 0.095 post-dose 2 and 0.068 post-dose 3. Study Arm 3 did not differ significantly compared to Study Arm 2 (p > 0.05 for all comparisons), with TNF-α CD4+ peak percent activated cells of 0.028 post-dose 2 and 0.024 post-dose 3; and TNF-α CD8+ peak percent activated cells of 0.043 post-dose 2 and 0.243 post-dose 3. At Day 366, all study arms except Study Arm 1 (MVA-BN-Filo+MVA-BN-Filo) had higher than pre-vaccination TNF-α CD4+ T cell activation levels. Responses were observed by 2 weeks after the third dose. At Day 546 (Day 181 post-third dose), TNF-α CD4+ and CD8+ percent activated cells after EBOV peptide pool stimulation in both heterologous study arms exceeded baseline levels and had greatest durability in Study Arm 3.
The percent of activated T cells (CD4+ and CD8+) expressing certain cytokines (IFN-γ, TNF-α, IL-2) were also assessed following stimulation with MARV, SUDV, and TAFV peptide pools (Supplementary Tables 15–17, Supplementary Figs. 11–14). Detectable responses were most frequently observed for TNF-α and IFN-γ. Following SUDV stimulation, the IFN-γ CD8+ peak response was higher after dose 2 in Study Arm 2 (MVA-BN-Filo+Ad26.ZEBOV) compared to Study Arm 1 (MVA-BN-Filo+MVA-BN-Filo) (p = 0.009) and the IFN-γ CD4+ and CD8+, and the TNF-α and IL-2 CD4+ peak responses were higher after dose 2 in Study Arm 2 (MVA-BN-Filo+Ad26.ZEBOV) compared to Study Arm 4 (Ad26.ZEBOV+Ad26.ZEBOV) (p < 0.05 for all comparisons). Responses were comparable between Study Arms 2 and 3 after the third dose. Following MARV stimulation, no differences were observed between the Study Arms at any timepoint. After TAFV stimulation, the IFN-γ CD8+ peak responses after the second dose of vaccine were greater for Study Arm 1 than Study Arm 3 (p = 0.012). No statistical differences were observed between the Study Arms at any other timepoint across the cytokines tested.
Gene expression biomarkers predicting post-second vaccination peak VNA antibody titer
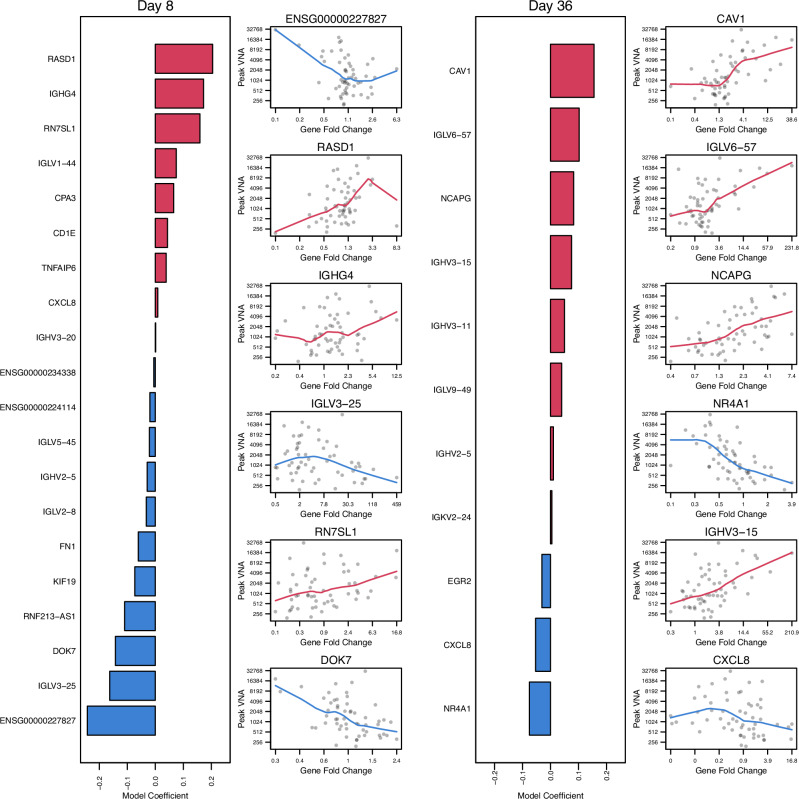
To identify genes expressed in PBMCs that predicted the magnitude of the log2 peak EBOV-GP VNA antibody titer after the second vaccination, regularized logistic linear regression models were fit separately for gene expression log2 fold changes (LFC) observed at Days 2, 4, 8, 15, and 29 after the first vaccination and for Days 2, 4, 8, 15 after the second vaccination. The models using gene expression LFC at Days 8 post-first and post-second vaccination had the best predictive performance. Both models selected 20 and 11 genes, respectively, and selected multiple immunoglobulin-related genes (Supplementary Tables 18, 19, Fig. 5). Aside from the immunoglobulin signature, the top genes selected by each model (in terms of the magnitude of the estimated regression coefficient) included ENSG00000227827 (PKD1 pseudogene, negatively correlated), RASD1 (positively correlated), RN7SL1 (positively correlated), DOK7 (negatively correlated) at Day 8 post-first vaccination and CAV1 (positively correlated), NCAPG (positively correlated), NR4A1 (negatively correlated), and CXCL8 (negatively correlated) at Day 8 post-second vaccination (Fig. 5). The models using Day 2 post-first vaccination and Days 2 and 4 post-second vaccination (Days 30 and 32) also selected predictive genes, but these fitted models had lower predictive performance compared to the Day 8 models. However, IFI27 encoding for Interferon alpha-inducible protein 27 was selected in all 3 of these earlier response models. This gene is known to be activated by type-1 interferon signaling, has antiviral properties, and plays a role in apoptosis. IFI27’s association with peak EBOV-GP VNA antibody titer was mixed. It was negatively associated at Day 2 post-first vaccination and positively associated at Day 2 and Day 4 post-second vaccination (Supplementary Tables 18, 19, Fig. 5).
Fig. 5. Multivariate gene log2 fold change (LFC) responses at Day 8 and Day 36 that best predicted the magnitude of peak log2 virus neutralization assay (VNA) antibody titer post-second vaccination.
The bar plots show the estimated linear regression coefficients for genes selected by the best model. Scatterplots that summarize the marginal association between gene expression fold change and peak VNA response, including locally estimated scatterplot smoothing (loess) trend lines, are shown for the top six genes selected by each model.
Discussion
This Phase 1 randomized study compared homologous and heterologous Ad26.ZEBOV and MVA-BN-Filo prime-boost strategies with an MVA-BN-Filo booster dose at 1 year in heterologous study arms12,13,15–17,22. Overall, the homologous and heterologous strategies were safe, with mostly mild-to-moderate reactogenicity. The most common adverse events were previously reported18,19, and included pain and tenderness at the injection site, fatigue, malaise, and myalgias. The resultant immune responses were dependent upon the vaccination regimen. Peak EBOV-GP antibody responses after the second dose were much greater with a heterologous boost approach in comparison to a homologous approach13. The advantage of a heterologous approach has been observed with other vaccine antigens and platforms23, and may be attributable to stimulating different innate and adaptive immune pathways24. Antibody responses (as measured by ELISA, VNA, and ADCC for EBOV) were durable through 1 year after the first vaccination, as has been observed by others for the heterologous study arms12,14, and interestingly also for the homologous Ad26-ZEBOV arm. In contrast, antibody responses to the homologous MVA-BN-Filo group rapidly waned, which has similarly been observed with mPox antibody responses to the MVA-BN (Jynneos) two-dose vaccine series25. At 12 months, the MVA-BN-Filo booster dose resulted in robust EBOV GP ELISA responses in both study arms. These antibodies remained at much higher titers through 6 months after the third dose than those observed at 6 months after the second dose. Although correlates of protection for filoviruses are incompletely understood, EBOV GP binding antibodies have been shown to correlate with vaccine-induced protection against EBOV in animal models26–28. Our study suggests that the heterologous two-dose series results in establishment of robust immunological memory that is primed to respond after re-exposure with filovirus antigens or the MVA-BN-Filo vaccine.
Assessment of other components of the immunological response (e.g., ADCC, plasmablast, cell-mediated) provided additional novel insights. Although others have published data using flow cytometry to assess cytokine markers of ADCC responses29,30, we developed novel functional ADCC assays for this study31 that we have also developed for other pathogens (e.g., SARS-CoV-2, influenza, Zika)32–35. ADCC responses to Ebola were observed in >75% of participants at Day 43 in the heterologous arms and these generally persisted up to 1 year. After the late MVA-BN-Filo boost, ADCC responses were observed in all participants within 2 weeks after boost that persisted. ADCC responses to Sudan and Marburg glycoproteins were present in a smaller percentage of participants, including in Study Arms 3 (Ad26.ZEBOV + MVA-BN-Filo + MVA-BN-Filo) and 4 (Ad26.ZEBOV+Ad26.ZEBOV), which may have been attributable to cross-reactive antibodies. Although ADCC has correlated with protection against a number of other pathogens (e.g., HIV, influenza)36–40, the role of these Fc-effector antibodies in protection against filoviruses remains uncertain. Although others have observed robust antibody-secreting cell (ASC) responses with Ebola infection or after VSVΔG-ZEBOV vaccination41,42, we observed minimal ASC responses at 1 week after both vaccinations except after the second vaccination in Study Arm 2 (MVA-BN-Filo+Ad26.ZEBOV) which predicted the peak EBOV-GP ELISA result. Since the correlation was modest and only present in a single arm, plasmablasts are likely not a high-yield biomarker to predict ultimate EBOV serologic responses to the vaccines tested in this study.
In terms of cellular immunity, the greatest T cell responses were TNF-α CD4+ and CD8+ responses to EBOV observed in the heterologous vaccine arms following the second and third vaccinations, corroborating previous findings of T cell immunity following heterologous Ad26.ZEBOV + MVA-BN-filo extending for at least 1 year following vaccination14. On Day 181 following the third dose, TNF-α CD4+ and CD8+ responses were most frequent in Arm 3 (Ad26.ZEBOV + MVA-BN-Filo + MVA-BN-Filo) across all antigens. Thus, this regimen elicited superior breadth and durability of T cell responses among the study arms. Although animal data have supported the importance of antibody-mediated protection against Ebola in studies with deoxyribonucleic acid (DNA) prime-adenovirus boost, Ebola virus-like particles, vesicular stomatitis virus (VSV)-vectored, and parainfluenza virus-vectored vaccines27, a recombinant adenovirus serotype 5 (Ad5) EBOV vaccine protected macaques from infection primarily through the activity of CD8+ T cell responses43. Acute protection from EBOV in macaques was also elicited by a chimpanzee Ad3 vector EBOV vaccine, and protection correlated with antibody responses, while long-term protection in this study required functional CD8+ T cell responses and was enhanced with a modified vaccinia Ankara (MVA) boost44. Therefore, there is currently evidence for antibody-mediated protection and for cell-mediated protection against Ebola virus. Notably, in the patients cared for at Emory University in 2014, the resolution of viremia correlated with the presence of activated CD8+ T cells, which preceded detectable immunoglobulin (Ig) G antibody responses42.
Regarding transcriptomic responses, after the second vaccination, changes in gene expression in three genes (CXCL10, IDO1, CCL2) and one gene (IFI27) at 1 and 3 days post vaccination, respectively, which are known to be involved in cytokine production, interferon signaling, and T-cell regulation, were identified as early gene expression signatures that predicted the magnitude of the log2 peak VNA titer after the second vaccination. CCL2 encodes the monocyte chemoattractant protein 1, which is known to recruit dendritic cells, monocytes, and memory B-cells to the site of inflammation45. CXCL10 encodes the Interferon gamma-induced protein 10 (IP-10) cytokine, which is involved in chemoattraction of monocytes, dendritic cells, NK-cells, and T-cells46. IDO1 has been shown to induce T regulatory cell development in vitro and negatively regulate T cell activation47. IFI27 which was predictive at Day 4 is known to be activated by type-1 interferon signaling, has antiviral properties and plays a role in apoptosis. It has been linked with severity of COVID-19, indicating a key role in early infection48. Thus, our analysis showed that important transcriptome biomarkers were identified as early as 24 h after vaccination and that these biomarkers may play important roles in regulating innate and adaptive immune signaling/processes.
Important limitations to these data include that the study was conducted in healthy adults which may limit generalizability, although others have now assessed this approach in children, adolescents, and HIV-infected patients15,16. The study was relatively small which limits precision and the ability to assess for rare side effects (e.g., thrombosis with thrombocytopenia syndrome observed after receipt of Ad26-vectored COVID-19 vaccination, neurological events observed in the Phase 2, myocarditis which can occur with MVA), although no similar events were observed12,49. Immunogenicity was assessed using established EBOV-GP ELISA and VNA assays, but standardized ELISA and VNA antibody assays were not available against other filoviruses (e.g., MARV, SUDV, TAFV) at the time the study was conducted, limiting the ability to assess for the breadth of cross-reactive antibodies elicited to filovirus GPs. ADCC and plasmablast assays were based on the GP antigen and were not measured for TAFV. CMI assessments utilized ICS and flow cytometry rather than ELISpot assessments of IFN-γ which other groups have utilized13,14, and memory T and B cell responses were not measured. No correlate of protection has been definitively established for filoviruses, and this study did not evaluate vaccine efficacy. Finally, a late boost was only performed with MVA-BN-Filo and late boosting of vaccine recipients of homologous strains was not performed.
In conclusion, this study of Ad26.ZEBOV and MVA-BN-Filo vaccines demonstrated that the heterologous vaccination strategy with delayed MVA-BN-Filo boost was safe with acceptable reactogenicity and resulted in robust EBOV GP ELISA and VNA antibody responses. Additionally, ADCC and cellular immune responses were observed, particularly in heterologous vaccine recipients. The detection of transcriptomic activation of cellular pathways that correlated with the peak VNA antibody titers after the second vaccination provides potential novel insights for understanding the gene activation associated with subsequent immunological responses. Such immunological and gene activation data could help identify new correlates of protection and disease. The heterologous vaccination strategy could be particularly effective in countries in which emergence and reemergence of any viruses from within the filovirus family could occur, as it could prime the immune system for a robust response to an MVA-BN-Filo booster administered in the time of need.
Online methods
Trial design and participants
This study was reviewed and approved by the Emory University Institutional Review Board (IRB00087894) and all participants provided written informed consent. The NCT number was NCT02891980, “A Safety Trial to Test MVA-BN(R)-Filo and Ad26.ZEBOV Vaccines in Healthy Volunteers,” registered September 1, 2016. This was a Phase 1, double-blinded randomized trial to evaluate the safety and immunogenicity of two heterologous and two homologous prime-boost regimens using MVA-BN®-Filo and Ad26.ZEBOV administered in different sequences at Days 1 and 29 in healthy adult participants aged 18–45 years. The two heterologous prime-boost arms (Study Arms 2 and 3) also received a late MVA-BN®-Filo boost at Day 366. Participants and study staff were blinded to a participant’s study vaccine assignment within study vaccination schedule (e.g., enrollment into Study Arm 1 or 4 versus 2 or 3 was known). Participants randomized to Study Arms 1 and 4 (2-dose schedule) were followed through ~12 months after the first study vaccination, while those randomized to Groups 2 and 3 (3-dose schedule) continued follow-up through ~18 months after the first study vaccination (6 months after the third study vaccination). This study planned for 60 participants to be randomized 1:1:1:1 to one of four Study Arms (i.e., 15 per arm), but participants who did not receive the second study product dose were replaced.
A thorough medical history, physical examination, an electrocardiogram (ECG), and laboratory tests were obtained at screening. Enrollment criteria included healthy adult participants aged ≥18 to ≤45 years without a history of Ebola disease, exposure to Ebola or prior vaccination with an Ebola vaccine, and receipt of any Ad26-based vaccine or vaccinia (smallpox)-based vaccine. See Supplementary Methods for additional information. Participants were followed as per their randomization assignment.
Study product and administration
MVA-BN®-Filo dose was administered at 1 × 108 international units (IU) per dose as a 0.5 mL intramuscular (IM) injection in the deltoid, and the Ad26.ZEBOV dose was administered at 5 × 1010 virus particles (VP) per dose as a 0.5 mL IM injection in the deltoid. The Adenovirus 26-vectored vaccine (Ad26.ZEBOV) expresses the EBOV glycoprotein, while the modified vaccinia Ankara-vectored vaccine expresses glycoproteins from EBOV, SUDV, and MARV, and the nucleoprotein from Taï Forest virus (MVA-BN-Filo).
Data collection
The occurrence of solicited injection site and systemic reactogenicity events was measured from the time of study vaccination through Day 8 after each vaccination. Unsolicited AEs were collected from vaccination through Day 29 after each vaccination. SAEs, AEs of special interest (AESIs), and AEs related to blood draws were collected from enrollment through the end of the study.
Assessment of responses
The study included phlebotomy for transcriptomics prior to each dose of vaccine (Day 1), and Days 2, 4, 8, 15, and 29 after each dose of vaccine. Plasmablast responses were obtained prior to each vaccination (Day 1) and Day 8 after each dose of vaccine. Serology and cellular responses were obtained prior to each dose of vaccine (Day 1), and at Days 8, 15, and 29 after each vaccination. Durability data was obtained at 6 and 11 months after the second dose in all participants and at 6 months after the third dose in Study Arms 2 and 3.
Binding and neutralizing antibody responses
Antibody responses were assessed using anti-EBOV-GP (Kikwit) IgG enzyme-linked immunosorbent assay (ELISA) (measured in ELISA Units (EU) per mL) at Battelle, Columbus, Ohio as previously described50. The EBOV GP (Makona) pseudovirus viral neutralization assay (VNA) (measured in IC50 titers) was performed at Monogram Biosciences, Inc, San Francisco, CA as previously described28. Briefly, EBOV GP pseudoviruses were generated using a replication defective retroviral vector containing a luciferase reporter gene. Pseudoviruses were incubated with serial dilutions of participant sera and used to infect humab embryonic kidney (HEK) 293 cell cultures. Luciferase activity was measured at ~72 h postviral inoculation to determine serum neutralizing antibody titers. These were summarized on a per-visit, fold change (relative to pre-vaccination), and peak response level following first, second, and third vaccination (as applicable).
Antibody-dependent cell-mediated cytotoxicity (ADCC) responses
To measure ADCC responses, dual-reporter target cell lines with tetracycline-inducible expression of green fluorescent protein, luciferase, and individual filovirus GPs (EBOV, SUDV, MARV, TAFV) were generated as previously described for other pathogens (Supplementary Methods)32. The effector cells for the assay were CD16-176V-NK-92 cells (obtained from Fox Chase Cancer Center, Philadelphia, PA) which have high surface expression of CD16. These cells were maintained in Mylocult media supplemented with 200 IU/mL of recombinant human IL-2 (R&D Systems, Minneapolis, MN).
To perform ADCC assays, serum samples were serially diluted 1:3 with a starting dilution of 1:20. Serum was mixed with target cells and NK effector cells with an effector:target (E:T) ratio of 2:1 and incubated for 4 h at 37 °C. Following incubation, Britelite Plus luciferase reagent (PerkinElmer) was added and Relative luminescence units (RLUs) were read on a luminometer (TopCount NXT Luminescence Counter). To calculate the percent ADCC, we determined the percent lysis of target cells using the following formula: ADCC (%) = [RLU × (no antibody) − RLU (with antibody)]/RLU (no antibody)] × 100. The endpoint ADCC titer was defined as the highest serum dilution at which >24% ADCC was observed for a given sample.
Plasmablast responses
To measure total and Ebola-specific (EBOV GP, SUDV GP, MARV GP) plasmablast responses, an ELISpot assay was implemented as previously described8,51. Briefly, 96-well ELISpot plates (EMD Millipore, Billerica, MA) were coated with 100 μl of either polyclonal coating antibodies (goat anti-human IgA + IgG + IgM (H + L), 10 μg/ml, Jackson Immuno Research, West Grove, PA), or with 3.0 μg/ml of recombinant EBOV GP, SUDV GP, or MARV GP (IBT Bioservices, Gaithersburg, MD) and incubated at 4 °C overnight. Plates were blocked with RPMI media (Cellgro, Manassas, VA) supplemented with 10% heat inactivated fetal bovine serum (Sigma-Aldrich, St. Louis, MO) for 2 h at 37 °C. Following washing, freshly isolated PBMCs (5 × 105 cells per well), were added to the wells and plates incubated overnight at 37 °C in 5% CO2. Following incubation, plates were washed four times with phosphate buffered saline (PBS) and four times with PBS supplemented with 0.05% Tween 20 (PBS-T). Subsequently, 100 μl of 1:1000 diluted biotin-conjugated donkey anti-human IgG and goat anti-human IgA and IgM (Jackson) were added to each well and plates incubated at room temperature for 2 h. Following another washing with PBS-T/PBS, plates were incubated with 100 μl of 1:1000 diluted horseradish peroxidase conjugated avidin D (Vector Laboratories, Burlingame, CA) for 1 h at RT. Plates were thoroughly washed and the reaction was developed with the addition of 100 μl/well of AEC Substrate (BD Biosciences, San Diego, CA). Reactions were allowed to develop at RT and stopped, when spots became visible in the wells with highest density of the cells (generally 5–8 min), by discarding the reaction mix and washing the plates under running cold water. Plates were allowed to dry and were read in an ELISpot reader (CTL, Shaker Height, OH). Final data was calculated as antibody secreting cells (ASC) per million PBMC.
Cellular responses by Intracellular cytokine staining (ICS)
ICS of PBMCs collected at various time points (pre-vaccination and post-vaccination) were assessed for the production of interferon-γ (IFN-γ), tumor necrosis factor α (TNF-α), and interleukin-2 (IL-2) from T cells stimulated with antigen-specific peptide pools. Customized peptide pools of Ebola virus Glycoprotein (EBOV GP, Zaire Mayinga strain, GenBank # AF086833.2), Sudan virus Glycoprotein (SUDV GP, Sudan Gulu strain, Genbank # AY729654), Marburgvirus Glycoprotein (MARV GP, Marburgvirus Musoke strain, GenBank # DQ217792.1) and Taï Forest virus Nucleoprotein (TAF NP, Taï Forest ebolavirus, GenBank # KU182910.1), were synthesized by Genescript. Each pool had 40 peptides, which are each 15 amino acids with overlapping by 11 amino acids.
Detailed methods are described in Supplementary Methods. Briefly, PBMCs were thawed and incubated with peptide pools for 6 h in the presence of Brefeldin A (BD Bioscience, Cat # 555028). The final peptide concentration was 2 μg/mL for each peptide. Stimulated cells with Staphylococcal Enterotoxin B (SEB) or DMSO (peptide diluent) was used as a positive or negative control, respectively. Then, cells were washed and stained with viability dye (Zombie aqua, BioLegend, Cat # 423102). Next, cells were surface stained with BV605-CD3 (HIT3a, BD Bioscience, Cat # 5647120), eFluor 450-CD4 (OKT-4, eBioscience, Cat# 48-0048-42), APC-Cy7-CD8 (RPA-T8, BD Bioscience, Cat # 557760), PE-CF594-CCR7 (150503, BD Bioscience, Cat # 562381), and PE-Cy7-CD45RA (HI100, BD Bioscience, Cat# 560675) for 30 minites at 4 °C, followed by permeabilization using Cytofix/Cytoperm (BD Bioscience, Cat # 555028). To quantify cytokines, PE-IFN-γ (4S.B3, BioLegend, Cat # 502509), FITC-IL-2 (MQ1-17H12, BioLegend, Cat # 500304), and Alexa 647-TNF-α (MAb11, BioLegend, Cat # 502916) were used. Fluorescence intensity was measured using an LSRII cytometer (BD Biosciences) and a Symphony A5 (BD Biosciences). Data analysis was performed using FlowJo software (Tree Star). Samples were classified as positive or negative for a given cytokine based on pre-defined cut-off values established by the laboratory.
Transcriptomics
For transcriptomics, a single aliquot per time point and subject was analyzed. PBMCs were lysed in Buffer RLT (Qiagen) containing 1% b-mercaptoethanol and stored at −70 °C for later extraction. RNA was isolated from each sample using the RNeasy Mini kit (Qiagen) with on-column DNase digestion at the Emory Transcriptomics Core. RNA quality was assessed using an Agilent Bioanalyzer and 500 ng of total RNA was used as input for library preparation using the TruSeq mRNA Stranded Kit (Illumina) according to the manufacturer’s instructions. As an internal control, an ERCC ExFold RNA Spike-In Mix (ThermoFisher) was also added to each sample well according to manufacturer instructions. Libraries were validated by capillary electrophoresis on an Agilent 4200 TapeStation, pooled at equimolar concentrations, and sequenced on an Illumina HiSeq3000 at 100SR, yielding 25–30 million reads per sample. All reads were extracted using standard Illumina sequencing demultiplexing (bcl2fastq) with no processing, filtering, or cleaning.
Statistical analyses
Sample size
The sample size for this study was selected to obtain preliminary estimates of vaccine safety and immunogenicity in a time sensitive manner. The study was not designed to test a specific null hypothesis.
ELISA and neutralizing antibody analyses
For each study arm and analysis population, titers or ELISA units/mL results and fold changes for applicable study visits were summarized by tabulating the number of observations, geometric mean, and 95% CI of the geometric mean (based on Student’s t-distribution), geometric standard deviation, median, first and third quartile, minimum, and maximum. In addition, peak titers were compared in a pairwise fashion between the four study arms using a two-sided Welch’s t-test.
The estimate of the population mean difference in log2 peak value following each vaccination and the associated two-sided 95% confidence intervals (CI) for each comparison were calculated using the Welch–Satterthwaite method. The difference was also presented on the original scale representing the ratio of the geometric means and associated 95% CI. To assess if the third vaccine dose for Study Arms 2 and 3 resulted in statistically significantly increased or decreased peak values following third vaccination, a two-sided paired t-test was used to compare mean log2 peak values post third and post second vaccination within Study Arms 2 and 3.
Antibody-dependent cellular cytotoxicity analyses
Prior to analysis, ADCC titers that were negative (titer < 1:20) were assigned a titer of one half the lowest dilution (i.e., 1:10). Post-vaccination samples for participants that were negative, i.e., had a titer below 1:20 at baseline were classified as positive if the respective post-vaccination day sample was positive, i.e., had a titer greater than or equal to 1:20. Post-vaccination samples for participants that were positive at baseline were classified as positive if the respective post-vaccination showed ≥4-fold increase compared to pre-first vaccination (Day 1). For each analysis population (ITT and PP), ADCC titer fold change compared to pre-vaccination was summarized using descriptive statistics (Minimum, Q1, Median, 95% CI of the median, Q3, and Maximum) by study arm and post-vaccination time point. The non-parametric bootstrap method with 1,000 bootstrap replicates was used to obtain 95% CIs of the median for each treatment arm and study visit combination. In addition, positive response on a per-visit basis was summarized using n/NNM (%) and associated Clopper-Pearson 95% CIs where n represented the number of participants who had a positive response at the visit and NNM represented the number of participants for that particular study visit and treatment combination with non-missing laboratory results.
For each participant, peak ADCC fold change response following first vaccination was defined as the maximum post-first vaccination fold change (for Days 15 and 29 relative to post-first vaccination) compared to pre-first vaccination (Day 1). For each participant, peak ADCC fold change response following second vaccination was defined as the maximum fold change post-second vaccination (for Days 15, 29, and 181 post-second vaccination and Day 366 post-first vaccination) compared to pre-first vaccination (Day 1). For each participant in Study Arms 2 and 3, peak ADCC fold change response following third vaccination was defined as the maximum fold change post-third vaccination (for Days 15, 29, and 181 post-third vaccination) compared to pre-first vaccination (Day 1). A two-sided Wilcoxon Rank-Sum test (normal approximation) was applied to assess difference in peak ADCC titer fold change compared to pre-vaccination between treatment arms for each analysis population and dose. Results were summarized using the number of participants in each arm, medians of the peak fold change response for each arm, the Wilcoxon Rank-Sum test statistic and p value. In addition, peak positive response (the number of participants who had a positive response at any point in the postvaccination period) was compared in a pairwise fashion between study arms using a two-sided Fisher’s exact test. Results were summarized using n/NNM (%), Odds Ratio, and P value where n represented the number of participants who had a positive response and NNM represented the number of participants for that particular peak response and treatment comparison with non-missing laboratory results.
Cell-mediated immunity treatment arm comparisons
Additional summaries of this data for the exploratory endpoint included the comparison of the peak difference in the percentage of activated T cells post-first, post-second, and post-third vaccination. A two-sided Wilcoxon Rank-Sum test (normal approximation) was used to assess the statistical significance of applicable pairwise study arm comparisons. Results were summarized using the number of participants in each arm, medians of the peak response for each arm, the Wilcoxon Rank-Sum test statistic and p value. In addition, peak positive response (the number of participants who had a positive response at any point in the post-vaccination period) was compared in a pairwise fashion between study arms using a two-sided Fisher’s exact test.
Results were summarized using n/NNM (%), Odds Ratio, and p value where NNM represented the number of participants for that particular peak response and treatment comparison with non-missing laboratory results.
Plasmablast analysis
Prior to analysis, technical replicates were aggregated using the mean. The lower limit of detection (LOD) was specified as 7 cells per million. The upper limit of quantification (ULOQ) was specified as 85 cells per million. Results for cell populations that were below the LOD were imputed using 0.5 x LOD = 3.5 cells per million. Cell populations too numerous to count were imputed using the 2 × ULOQ = 170 cells per million. Analysis was performed for the exploratory plasmablast analysis population. The number of secreting plasmablast cells per million and fold change relative to pre-vaccination were summarized for each antibody type and Ebolavirus GP antigen, applicable timepoint, and study arm using the minimum, 25th percentile (Q1), median, 95% CI of the median, 75th percentile (Q3), and maximum. The non parametric bootstrap method with 1000 bootstrap replicates was used to obtain 95% CIs of the median. For each dose, antibody type, and Ebolavirus GP antigen, a two-sided Wilcoxon Rank-Sum test was performed to assess the difference in peak fold change in secreting plasmablast cells per million compared to pre-vaccination between treatment arms. Correlations between the change in the number of secreting plasmablasts and peak humoral and cellular assay results were assessed using Spearman correlation.
Gene expression biomarkers predicting peak VNA titer
Regularized linear regression models were fit to determine gene expression fold-change responses that best predict peak virus neutralization assay (EBOV GP VNA) antibody titer post-second vaccination using the glmnet R package (Version 3.0.2) [17]. To avoid overfitting (n « # genes and collinearity among genes) and to facilitate variable selection, an elastic net regularization step (combination of L1 Lasso and L2 ridge penalization, a = 0.5) was applied. The analysis was carried out across treatment arms. Six-fold stratified cross validation (sets of 10 participants) was used to determine the optimum regularization parameter that minimizes the model mean squared error and peak EBOV GP VNA titer was utilized for stratifying participants by antibody response using deciles. In both cases, the input gene set was based on log2 fold change in LCPM for DE genes that were identified using binomial models as implemented in edgeR for any timepoint and comparison (within and between study arm FDR < 0.05, FC ≥ 1.5). Regularized linear regression models were fit for Days 2, 4, 8, 15, 29 (relative change compare to first vaccination) and Days 30, 32, 36 (relative change compared to second vaccination).
Supplementary information
Acknowledgements
We thank the participants who consented to participate in this study. We particularly acknowledge the Children’s Healthcare of Atlanta and Emory University Pediatric Biostatistics Core and Flow Cytometry Core for assistance. We thank the teams of Emory Children’s Center-Vaccine Research Clinic (ECC-VRC) and Hope Clinic of the Emory Vaccine Center for their efforts on behalf of this study. We also thank the Division of Microbiology and Infectious Diseases (DMID) and The Emmes Company, LLC for their support of this study. This study was funded by contracts with the National Institute of Allergy and Infectious Diseases at the National Institutes of Health to the Emory Vaccine and Treatment Evaluation Units [VTEU]: HHSN272201300018I (Rouphael). Additional support was provided by the Georgia Research Alliance (GRA), the Emory University School of Medicine, and Childrens Healthcare of Atlanta (CHOA). The content is solely the responsibility of the authors and does not necessarily reflect the official views of the National Institutes of Health or the Biomedical Advanced Research and Development Authority (BARDA).
Author contributions
C.A.R.: Investigation, Writing—Original Draft, Writing—Review & Editing. I.Y.: Investigation, Writing—Original Draft, Writing—Review & Editing. C.K.: Investigation, Writing—Original Draft, Writing—Review & Editing. J.Y.: Investigation, Writing—Original Draft, Writing—Review & Editing. S.K.: Investigation, Writing—Review & Editing. E.P.: Investigation, Project Administration, Writing—Review & Editing. K.S.: Investigation, Project Administration, Writing—Review & Editing. T.G.: Investigation, Writing—Review & Editing. H.H.: Investigation, Writing—Review & Editing. K.S.: Investigation, Writing—Review & Editing. P.S.: Conceptualization, Methodology, Resources, Supervision, Writing—Review & Editing. C.M.: Formal analysis, Writing—Review & Editing. V.A.: Project Administration, Writing—Review & Editing. K.M.T.: Conceptualization, Methodology, Writing—Review & Editing. J.B.G.: Formal Analysis, Data Curation, Visualization, Writing—Original Draft, Writing—Review & Editing. C.E.G.: Formal Analysis, Data Curation, Visualization, Writing—Original Draft, Writing—Review & Editing. R.A.J.: Conceptualization, Methodology, Writing—Review & Editing. S.L.: Investigation, Formal Analysis, Data Curation, Writing—Review & Editing. K.M.: Formal Analysis, Writing—Review & Editing. S.B.: Formal Analysis, Writing—Review & Editing. E.A.O.: Formal Analysis, Writing—Review & Editing. X.C.: Investigation, Formal Analysis, Writing—Review & Editing. L.J.A.: Supervision, Investigation, Writing—Review & Editing. J.W.: Supervision, Investigation, Writing—Review & Editing. M.S.: Supervision, Investigation, Writing—Review & Editing. N.R.: Supervision, Investigation, Writing—Original Draft, Writing—Review & Editing. E.J.A.: Conceptualization, Methodology, Formal Analysis, Investigtion, Resources, Writing—Original Draft, Writing—Review & Editing, Supervision, Funding acquisition. All authors reviewed the manuscript.
Data availability
The deidentified datasets used and/or analyzed during the current study, the study protocol, and statistical analysis plan will be made available from the corresponding author upon reasonable request.
Competing interests
E.J.A has consulted for Pfizer, Sanofi Pasteur, GSK, Janssen, Moderna, and Medscape, and his institution receives funds to conduct clinical research unrelated to this manuscript from MedImmune, Regeneron, PaxVax, Pfizer, GSK, Merck, Sanofi-Pasteur, Janssen, and Micron. He also serves on a safety monitoring board for Kentucky BioProcessing, Inc. and Sanofi Pasteur. He serves on a data adjudication board for WCG and ACI Clinical. His institution has also received funding from NIH to conduct clinical trials of COVID-19 vaccines. He is now employed by Moderna and has stock/stock options. N.R. has consulted for EMMES, Moderna, Sanofi, Seqirus, GSK, and ICON and her institution receives funds to conduct clinical research unrelated to this manuscript from Pfizer, Sanofi, Quidel, Lilly, Merck, and Vaccine Company as well as NIH to conduct translational clinical studies and interventional clinical trials. C.A.R.’s institution has received funds to conduct clinical research unrelated to this manuscript from BioFire Inc, GSK, MedImmune, Micron, Janssen, Merck, Moderna, Novavax, PaxVax, Pfizer, Regeneron, Sanofi-Pasteur. Her institution has received funding from NIH to conduct clinical trials of Moderna and Janssen COVID-19 vaccines. She is co-inventor on patented RSV vaccine technology, which has been licensed to Meissa Vaccines, Inc. with royalties received. I.Y. reported research funding from the Centers for Disease Control and Prevention, National Institutes of Health, and Gates Foundation; funding to her institution to conduct clinical research from Merck, Moderna, Pfizer outside the submitted work; honorarium for advisory board from Merck and Sanofi Pasteur. All other authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41541-024-01042-4.
References
- 1.Sprecher, A. Understanding the key to outbreak control—Sudan virus disease in Uganda. N. Engl. J. Med. 10.1056/NEJMp2213975 (2022). [DOI] [PubMed]
- 2.Baize, S. et al. Emergence of Zaire Ebola virus disease in Guinea. N. Engl. J. Med.371, 1418–1425 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Rugarabamu, S. et al. Forty-two years of responding to Ebola virus outbreaks in sub-Saharan Africa: a review. BMJ Glob. Health5, e001955 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. 2014-2016 Ebola Outbreak in West Africa, http://www.cdc.gov/vhf/ebola/history/2014-2016-outbreak/index.html.
- 5.Sohan, M., Shahriar, M., Bhuiyan, M. A. & Islam, M. R. Recent outbreak of Marburg virus disease: could it be a threat for global public health? Health Sci. Rep.6, e971 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. Marburg Outbreak in Rwanda Situation Summary, https://www.cdc.gov/marburg/situation-summary/index.html (2024).
- 7.Jones, S. M. et al. Live attenuated recombinant vaccine protects nonhuman primates against Ebola and Marburg viruses. Nat. Med.11, 786–790 (2005). [DOI] [PubMed] [Google Scholar]
- 8.Singh, K. et al. A bivalent, spherical virus-like particle vaccine enhances breadth of immune responses against pathogenic Ebola viruses in rhesus macaques. J. Virol.94. 10.1128/JVI.01884-19 (2020). [DOI] [PMC free article] [PubMed]
- 9.Gunn, B. M. et al. Survivors of Ebola virus disease develop polyfunctional antibody responses. J. Infect. Dis.221, 156–161 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sakabe, S. et al. Analysis of CD8+ T cell response during the 2013–2016 Ebola epidemic in West Africa. Proc. Natl. Acad. Sci. USA115, E7578–E7586 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zahn, R. et al. Ad35 and ad26 vaccine vectors induce potent and cross-reactive antibody and T-cell responses to multiple filovirus species. PLoS ONE7, e44115 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pollard, A. J. et al. Safety and immunogenicity of a two-dose heterologous Ad26.ZEBOV and MVA-BN-Filo Ebola vaccine regimen in adults in Europe (EBOVAC2): a randomised, observer-blind, participant-blind, placebo-controlled, phase 2 trial. Lancet Infect. Dis.21, 493–506 (2021). [DOI] [PubMed] [Google Scholar]
- 13.Milligan, I. D. et al. Safety and immunogenicity of novel adenovirus type 26- and modified vaccinia ankara-vectored Ebola vaccines: a randomized clinical trial. JAMA315, 1610–1623 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Winslow, R. L. et al. Immune responses to novel adenovirus type 26 and modified vaccinia virus Ankara-vectored Ebola vaccines at 1 year. JAMA317, 1075–1077 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Anywaine, Z. et al. Safety and immunogenicity of 2-dose heterologous Ad26.ZEBOV, MVA-BN-Filo Ebola vaccination in children and adolescents in Africa: a randomised, placebo-controlled, multicentre Phase II clinical trial. PLoS Med.19, e1003865 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barry, H. et al. Safety and immunogenicity of 2-dose heterologous Ad26.ZEBOV, MVA-BN-Filo Ebola vaccination in healthy and HIV-infected adults: A randomised, placebo-controlled Phase II clinical trial in Africa. PLoS Med.18, e1003813 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anywaine, Z. et al. Safety and immunogenicity of a 2-dose heterologous vaccination regimen with Ad26.ZEBOV and MVA-BN-Filo Ebola vaccines: 12-month data from a phase 1 randomized clinical trial in Uganda and Tanzania. J. Infect. Dis.220, 46–56 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.European Medicines Association. Mvabea: EPAR- Medicine Overview. https://www.ema.europa.eu/en/documents/overview/mvabea-epar-medicine-overview_en.pdf (2020).
- 19.European Medicines Association. Zabdeno (Ad26.ZEBOV-GP, recombinant). https://www.ema.europa.eu/en/documents/overview/zabdeno-epar-medicine-overview_en.pdf (2020).
- 20.Bernstein, A., Pulendran, B. & Rappuoli, R. Systems vaccinomics: the road ahead for vaccinology. OMICS15, 529–531 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pulendran, B. Systems vaccinology: probing humanity’s diverse immune systems with vaccines. Proc. Natl. Acad. Sci. USA111, 12300–12306 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mutua, G. et al. Safety and immunogenicity of a 2-dose heterologous vaccine regimen with Ad26.ZEBOV and MVA-BN-Filo Ebola vaccines: 12-month data from a phase 1 randomized clinical trial in Nairobi, Kenya. J. Infect. Dis.220, 57–67 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nguyen, T. T. et al. Reactogenicity and immunogenicity of heterologous prime-boost immunization with COVID-19 vaccine. Biomed. Pharmacother.147, 112650 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Orlova, O. V., Glazkova, D. V., Bogoslovskaya, E. V., Shipulin, G. A. & Yudin, S. M. Development of modified vaccinia virus Ankara-based vaccines: advantages and applications. Vaccines10. 10.3390/vaccines10091516 (2022). [DOI] [PMC free article] [PubMed]
- 25.Collier, A.-R. Y. et al. Decline of Mpox Antibody responses after modified vaccinia Ankara–Bavarian Nordic vaccination. JAMA. 10.1001/jama.2024.20951 (2024). [DOI] [PMC free article] [PubMed]
- 26.Wong, G. et al. Immune parameters correlate with protection against Ebola virus infection in rodents and nonhuman primates. Sci. Transl. Med.4, 158ra146 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sullivan, N. J., Martin, J. E., Graham, B. S. & Nabel, G. J. Correlates of protective immunity for Ebola vaccines: implications for regulatory approval by the animal rule. Nat. Rev. Microbiol7, 393–400 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roozendaal, R. et al. Nonhuman primate to human immunobridging to infer the protective effect of an Ebola virus vaccine candidate. npj Vaccines5, 112 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wagstaffe, H. R. et al. Antibody-dependent natural killer cell activation after Ebola vaccination. J. Infect. Dis.223, 1171–1182 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wagstaffe, H. R. et al. Ebola virus glycoprotein stimulates IL-18-dependent natural killer cell responses. J. Clin. Investig.130, 3936–3946 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Singh, K., Marasini, B., Chen, X. & Spearman, P. A novel Ebola virus antibody-dependent cell-mediated cytotoxicity (Ebola ADCC) assay. J. Immunol. Methods460, 10–16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen, X. et al. Development and optimization of a Zika virus antibody-dependent cell-mediated cytotoxicity (ADCC) assay. J. Immunol. Methods488, 112900 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rostad, C. A. et al. Functional antibody responses to SARS-CoV-2 variants in children with COVID-19, MIS-C, and after two doses of BNT162b2 vaccination. J. Infect. Dis. 10.1093/infdis/jiac215 (2022). [DOI] [PMC free article] [PubMed]
- 34.Chen, X. et al. The development and kinetics of functional antibody-dependent cell-mediated cytotoxicity (ADCC) to SARS-CoV-2 spike protein. Virology559, 1–9 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen, X. et al. Functional antibody-dependent cell mediated cytotoxicity (ADCC) responses to vaccine and circulating influenza strains following vaccination. Virology569, 44–55 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Asokan, M. et al. Fc-mediated effector function contributes to the in vivo antiviral effect of an HIV neutralizing antibody. Proc. Natl. Acad. Sci. USA117, 18754–18763 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haynes, B. F. et al. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N. Engl. J. Med.366, 1275–1286 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Su, B. et al. Update on Fc-mediated antibody functions against HIV-1 beyond neutralization. Front. Immunol.10, 2968 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zheng, Z. et al. Contribution of Fc-dependent cell-mediated activity of a vestigial esterase-targeting antibody against H5N6 virus infection. Emerg. Microbes Infect.9, 95–110 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gao, R., Sheng, Z., Sreenivasan, C. C., Wang, D. & Li, F. Influenza A virus antibodies with antibody-dependent cellular cytotoxicity function. Viruses12. 10.3390/v12030276 (2020). [DOI] [PMC free article] [PubMed]
- 41.Lai, L. et al. Emergency postexposure vaccination with vesicular stomatitis virus-vectored Ebola vaccine after needlestick. JAMA313, 1249–1255 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McElroy, A. K. et al. Human Ebola virus infection results in substantial immune activation. Proc. Natl. Acad. Sci. USA112, 4719–4724 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sullivan, N. J. et al. CD8+ cellular immunity mediates rAd5 vaccine protection against Ebola virus infection of nonhuman primates. Nat. Med.17, 1128–1131 (2011). [DOI] [PubMed] [Google Scholar]
- 44.Stanley, D. A. et al. Chimpanzee adenovirus vaccine generates acute and durable protective immunity against ebolavirus challenge. Nat. Med.20, 1126–1129 (2014). [DOI] [PubMed] [Google Scholar]
- 45.Singh, S., Anshita, D. & Ravichandiran, V. MCP-1: Function, regulation, and involvement in disease. Int. Immunopharmacol.101, 107598 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu, M. et al. CXCL10/IP-10 in infectious diseases pathogenesis and potential therapeutic implications. Cytokine Growth Factor Rev.22, 121–130 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Merlo, L. M. F. et al. Differential roles of IDO1 and IDO2 in T and B cell inflammatory immune responses. Front. Immunol.11, 1861 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shojaei, M. et al. IFI27 transcription is an early predictor for COVID-19 outcomes, a multi-cohort observational study. Front. Immunol.13, 1060438 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shay, D. K. et al. Safety monitoring of the Janssen (Johnson & Johnson) COVID-19 vaccine—United States, March-April 2021. Mmwr70, 680–684 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rudge, T. L. Jr. et al. Development, qualification, and validation of the Filovirus Animal Nonclinical Group anti-Ebola virus glycoprotein immunoglobulin G enzyme-linked immunosorbent assay for human serum samples. PLoS ONE14, e0215457 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wrammert, J. et al. Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature453, 667–671 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The deidentified datasets used and/or analyzed during the current study, the study protocol, and statistical analysis plan will be made available from the corresponding author upon reasonable request.