Abstract
Biologically produced protein drugs are generally susceptible to degradation by proteases and often exhibit immunogenicity. To address this issue, mirror-image peptide/protein binders consisting of D-amino acids have been developed so far through the mirror-image phage display technique. Here, we develop a mirror-image protein binder derived from a monobody, one of the promising protein scaffolds, utilizing two notable technologies: chemical protein synthesis and TRAP display, an improved version of mRNA display. A sequential workflow of initial screening followed by affinity maturation, facilitated by TRAP display, generates an L-monobody with high affinity (KD = 1.3 nM) against monocyte chemoattractant protein-1 (MCP-1) D-enantiomer. The chemically synthesized D-monobody demonstrates strong and specific binding to L-MCP-1 and exhibits pharmaceutically favorable properties such as proteolytic resistance, minimal immune response, and a potent inhibitory effect on MCP-1-induced cell migration. This study elevates the value of mirror-image peptide/protein binders as an alternative modality in drug discovery.
Subject terms: Antibody fragment therapy, Synthetic biology
In this work, a mirror-image protein binder derived from a monobody targeting MCP-1 is generated via in vitro display technology and chemical synthesis. It exhibits pharmaceutically promising properties, including protease resistance, negligible immunogenicity, and a potent inhibitory effect on cell migration.
Introduction
Mirror-image proteins have emerged as attractive molecular materials in the fields of structural and synthetic biology. These proteins consist of D-amino acids with an achiral glycine and were prepared exclusively by synthetic peptide chemistry1,2. Racemic or quasi-racemic protein crystallography utilizes these mirror-image proteins to aid in the crystallization of desired proteins through the formation of centrosymmetric crystals, thereby facilitating structural determination3,4. By virtue of their chiral catalytic features, mirror-image enzymes have been synthesized to evaluate and utilize their unique catalytic activities5–8. Recently, intensive studies have been conducted aiming at the creation of a mirror-image central dogma through the synthesis of mirror-image key components such as polymerases9,10 and ribosomal proteins11.
Since mirror-image peptides and proteins are more resistant to proteolytic degradation and exhibit lower immunogenicity than their native counterparts12,13, these D-configured polypeptides are also considered promising drug candidates in the field of pharmaceutical sciences. While the development of L-configured peptide/protein binders against desired molecules has become relatively accessible, mainly thanks to reliable screening technologies like phage display14, discovering D-peptide/protein binders remains challenging.
The first milestone study to obtain a mirror-image polypeptide binder was established by Kim and coworkers in 1996 as a technique named mirror-image phage display (MIPD)15. In MIPD, a randomized L-peptide library displayed on phage surfaces is screened against chemically synthesized D-configured target proteins. Given that the interaction between the identified L-peptide and the target D-protein mirrors that of their enantiomeric counterparts according to the law of symmetry, a D-peptide binder can subsequently be generated. After this first report of MIPD to isolate a D-peptide binder against the Src homology 3 domain of c-Src, this strategy has been employed to obtain D-peptide binders against various mirror-image protein targets, such as amyloid peptide Aβ16, MDM217, programmed cell death ligand 1 (PD-L1)18 and others19–24. However, the affinities of these D-peptide binders against the targets are generally moderate (i.e., KD ranging from sub-µM to double-digit µM on average, as low as double-digit nM), although additional efforts such as multimerization can make these D-peptide binders more potent25,26. One promising solution to increase target affinity is the use of protein scaffolds27,28, which usually have larger and more rigid interaction surfaces than linear peptide-based binders. Indeed, Kent and coworkers employed a 56-residue protein scaffold, the B1 domain of streptococcal protein G (GB1), in MIPD targeting the angiogenic protein vascular endothelial growth factor (VEGF-A), to obtain a mirror-image protein binder with a KD of 85 nM29. Although this initial binder was not stable at physiological temperatures, extensions of both the N- and C-termini improved the thermal stability and also the binding affinity (KD = 6 nM)30. More recently, two different three-helix bundle scaffolds, the protein G-derived GA domain (53-residue) and the protein A-derived Z domain (58-residue), were screened by MIPD against D-VEGF-A. The individually obtained binders were chemically crosslinked to form a D-configured heterodimer, which exhibited a KD of 0.08 nM due to the multivalency effect and also showed inhibitory activity on tumor growth31.
Another key factor in obtaining high-affinity peptide/protein binders frequently is the library diversity used in the display selection. Generally, there is a correlation between the initial diversity of the library and the probability of acquiring low KD binders32. In this respect, mRNA display33,34 can generate an even larger diversity (~ 1013) than phage display (~ 1011), thereby increasing the probability of identifying high-affinity peptide/protein binders. However, conventional mRNA display requires complicated operations including stepwise transcription, puromycin linker ligation, and translation in separate tubes, leading to time-consuming experimental procedures. To address this issue, we developed an improved version of mRNA display, called TRAP (transcription−translation coupled with association of puromycin linker) display, in which a polypeptide library conjugated with each mRNA sequence is automatically produced via tandem reactions and used for the isolation of macrocyclic peptide binders with double-digit nM affinities35. More recently, a fibronectin type III domain-derived protein scaffold, named monobody36, was employed in further improved TRAP display selections to generate binders with strong affinities ranging from sub-nM to single-digit nM against multiple targets including SARS-CoV-2 spike protein, epidermal growth factor receptor 1 (EGFR1), human epidermal growth factor receptor 2 (HER2)37 and optineurin38.
In this study, we establish a mirror-image TRAP display to create D-configured monobody-based binders powered by chemical protein synthesis, in which peptide segments prepared by solid-phase peptide synthesis (SPPS) are assembled by chemoselective peptide ligation reactions39–41. We choose monocyte chemoattractant protein-1 (MCP-1) as a target protein, which is known to be related to the pathogenesis of many disease conditions such as cancers, infectious diseases, diabetes, cardiovascular diseases, and more42. We isolate high-affinity L-monobody clones with single-digit nM KD against D-configured MCP-1 via TRAP display selection and subsequent affinity maturation experiments. Chemical synthesis using a double Cys substitution approach enables the preparation of a mirror-image monobody with high purity and minimal effort. Comparative evaluation of the mirror-image monobody with the L-monobody reveals properties such as high proteolytic resistance, undetectable immunogenicity, and potent inhibitory activity against the interaction between MCP-1 and its receptor C-C chemokine receptor type 2 (CCR2) in a cultured cell environment. Furthermore, the D-configured monobody inhibits chemotactic migration of monocytes as potently as previously developed anti-MCP-1 antibody, carlumab, which has been tested in clinical trials43. Together, these results showcase a substantial advance in the development of mirror-image peptide/protein binders. We also refer to a parallel study by Schmidt et al.44 that developed D-monobodies targeting the SH2 domain of the leukemic Bcr-Abl tyrosine kinase by utilizing MIPD. Our two studies employ distinct yet complementary approaches to achieve a common goal: the straightforward development of a functional mirror-image monobody that targets a therapeutically significant molecule. The convergence of these approaches not only underscores the robustness of the platforms for developing mirror-image binding proteins but also collectively demonstrates the viability of mirror-image binders.
Results
Chemical synthesis of mirror-image MCP-1
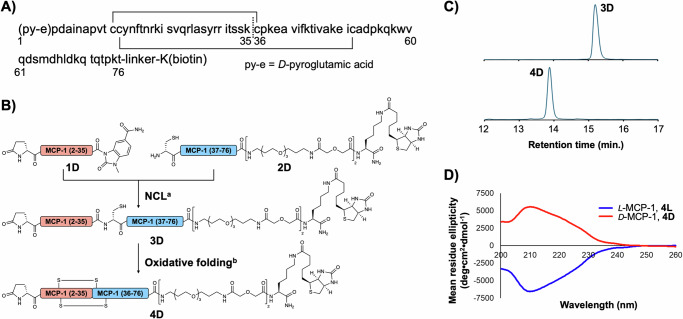
Mature MCP-1 is produced after cleavage of the N-terminal 23-residue putative signal peptide45 and identified as a 76-residue protein with N-terminal pyroglutamic acid46 (Fig. 1A), which is an occasionally observed post-translational modification47. For the synthesis of D-configured MCP-1, the amino-acid sequence was divided into two peptide segments at the K35/C36 junction according to the previous report48, which employed native chemical ligation (NCL)49 for peptide assembly (Fig. 1B). First, we prepared the N-terminal segment 1D and the C-terminal segment 2D via Fmoc-SPPS (Supplementary Figs. 1, 2). While 1D includes a C-terminal N-acyl-N’-methylacylurea (MeNbz)50 as a thioester precursor, 2D contains an N-terminal Cys residue for NCL and C-terminal biotinylated Lys residue for the streptavidin pull-down used in binder selection. Segments 1D and 2D were ligated under conventional NCL conditions including mercaptophenylacetic acid (MPAA) as the thiol catalyst51, tris(carboxyethyl)phosphine (TCEP) as the reducing agent, and guanidine hydrochloride salt (Gn·HCl) as the denaturing agent. We obtained 3D in 39% isolated yield (Supplementary fig. 3), which was then folded under air oxidation conditions to yield the desired D-MCP-1, 4D in 56% isolated yield after HPLC purification (Supplementary Fig. 4). The folding was confirmed by a change in the HPLC retention time (Fig. 1C). Following the same synthetic procedure, we also prepared L-configured MCP-1 4L and 4L' with and without a C-terminal biotin linker, respectively (Supplementary Figs. 5–12). Circular dichroism (CD) spectroscopic analysis of the biotinylated D- and L-MCP-1 revealed inverted CD spectra, indicating mutually mirror-imaged conformations (Fig. 1D).
Fig. 1. Synthesis and evaluation of biotinylated D-MCP-1.
A Amino-acid sequence of MCP-1 with N-terminal pyroglutamic acid, pE, and C-terminal biotinylated lysine. Two disulfide bonds (solid lines) and a ligation site (a dashed line) are shown. B Synthetic scheme of biotinylated D-MCP-1 via NCL followed by air oxidation. Reaction conditions: (a) 200 mM MPAA, 50 mM TCEP, 6 M Gn·HCl, 0.2 M phosphate (pH 6.5), 37 °C; (b) AcOH/H2O, then NH3 aq. C Comparison of HPLC retention time between purified 3D and 4D. HPLC peaks were monitored at 220 nm in the linear gradient with water/acetonitrile containing 0.1% TFA. The gradient of HPLC: acetonitrile 20–40% for 20 min. D CD spectra of synthetic MCP-1. 4D and 4 L (10 µM) dissolved in PBS (pH7.4) were measured. Source data are provided as a Source Data file.
Monobody selection against D-MCP-1 by TRAP display
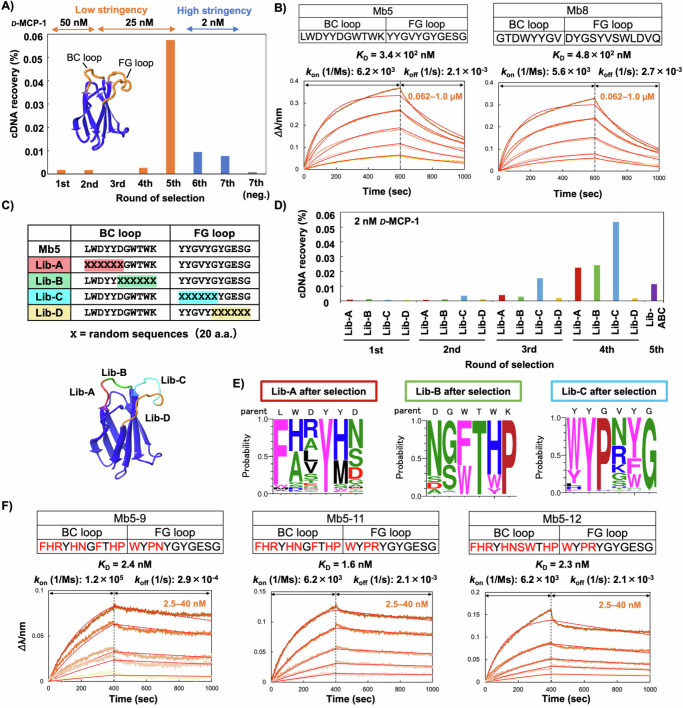
Previously, we obtained high-affinity monobody clones (sub-nM to nM KD) against several protein targets via TRAP display37,38. For the first-round selection against the biotinylated D-MCP-1 4D, we employed the same monobody mRNA library as in the previous study37, where Gly-, Ser-, Trp-, and Tyr-rich random residues were introduced into the BC (8 or 10 residues) and FG (10 or 12 residues) loops of monobody to increase the probability of obtaining high-affinity clones52. Notably, in vitro transcription (i.e., mRNA library construction) was performed separately from the in vitro translation to maximize the diversity of the monobody library. The purified library mRNAs were translated and conjugated into monobody-mRNA complexes in a reconstituted in vitro translation system53 coupled with in situ puromycin-mediated crosslinking (Supplementary Table 1). After reverse transcription to generate monobody-cDNA/mRNA conjugates, non-specific binders such as bead binders and streptavidin binders were removed from the monobody-cDNA/mRNA library by treatment with streptavidin-coated magnetic beads (negative selection). The library was then incubated with 50 nM of the biotinylated D-MCP-1, and the binding clones were pulled down with streptavidin beads to recover D-MCP-1 binders. cDNA of the recovered monobody conjugates was PCR-amplified and applied to the next round. From the second-round selection, consecutive transcription-translation reactions coupled with puromycin conjugation35,37 were utilized to accelerate the selection procedure. We repeated the selection procedures and observed an enrichment of the cDNA recovery rate after the fifth-round selection (Fig. 2A). To obtain higher affinity clones, two additional rounds of selection were conducted with increased selection stringency (2 nM biotinylated D-MCP-1). The seventh-round selection was conducted both with and without D-MCP-1, and the recovered cDNAs were sequenced by an NGS technique.
Fig. 2. Selection of monobodies against D-MCP-1 using TRAP display.
A Progress of the TRAP display selection. After each round of selection, the recovered cDNA was quantified by real-time PCR. The recovery of cDNA (%) was calculated by dividing the amount of cDNA recovered after the pull-down with D-MCP-1 by the amount of PuL in the translation mixture. From the sixth round, the selection pressure was increased by decreasing the target concentration from 25 nM to 2 nM. B Determination of kinetic parameters of Mb5 and Mb8 by BLI. D-MCP-1 was immobilized on a streptavidin-sensor chip, and Nus-Tag fused Mb5 or Mb8 (0.062, 0.125, 0.25, 0.50, 1.0 µM) were used in the kinetic analysis. The data were fitted to a 1:1 binding model. C Sequences and spatial arrangement of saturation mutagenesis libraries for affinity maturation of Mb5. Saturation mutagenesis (X) was introduced using NNK codons (N = A, C, G, T; K = G or T; 32 codons/20 aa) at six consecutive residues in the BC and FG loops. D Progress of TRAP display selection at each library in the affinity maturation. In the fifth round selection, selection stringency was increased by extending washing time was applied against mixed library derived from libraries A, B, and C. E The probability of amino acids at each position in the loops of the selected clones shown by WebLogo. F Determination of kinetic parameters of three Mb5 mutants by BLI. D-MCP-1 was immobilized on a streptavidin-sensor chip, and Mb5-9, 5-11, 5-12 (2.5, 5.0, 10, 20, 40 nM) were used in the kinetic analysis. The data are fitted to a 1:1 binding model. Abbreviations: BLI, Bio-layer interferometry; Lib, library; TRAP, transcription–translation coupled with association of puromycin linker. Source data are provided as a Source Data file.
Amino acid sequences of the BC and FG loops of the enriched monobodies were aligned based on the order of the relative abundance after the seventh round of selection with D-MCP-1 (Supplementary Table 2). Among the top 8 clones (Mb1 ~ 8), 7 clones were chosen for further studies of recombinant expression and binding tests, while Mb6 was excluded due to its low P/N value, which refers to a ratio obtained by dividing a relative abundance in positive selection by that in negative selection and could indicate a tendency of non-specific binding. The 7 clones were successfully expressed and isolated with good purity (Supplementary Fig. 13A). We screened the recombinant monobodies using biolayer interferometry (BLI) analysis and found that Mb5 and Mb8 showed higher intensities than the others against D-MCP-1 immobilized on the BLI sensors (Supplementary Fig. 13B). Notably, the high relative abundance values do not necessarily promise the high affinity probably because monobody molecules in TRAP display selection is surrounded by a different environment from those after E. coli expression. To determine the kinetic parameters and dissociation constants, BLI sensorgrams with different concentrations of Mb5 and Mb8 were obtained and evaluated by a global fitting algorithm. As a result, sub-µM KD values of 0.34 and 0.48 µM were calculated for Mb5 and Mb8, respectively (Fig. 2B). This result was unexpected for us, as high-affinity binders with nM or sub-nM KD were reproducibly selected via TRAP display in our previous studies37,38. This discrepancy might be attributed to the less favorable binding patterns between D- and L-proteins compared to those between L-proteins. To validate this hypothesis, we also conducted TRAP display selection against L-MCP-1 and obtained single-digit nM clones, which have more than 100-fold affinities than those obtained in D-MCP-1 selection (Supplementary Fig. 14).
To obtain higher affinity clones against D-MCP-1, we conducted an in vitro affinity maturation of Mb5, following our previous study, which selected multiple high-affinity clones with sub-nM KD against variants of the SARS-CoV-2 spike protein54. We prepared four kinds of mRNA libraries, each incorporating saturation mutagenesis at 6 consecutive amino acid residues in the regions of the BC or FG loop (Fig. 2C). Notably, the required diversity of 6 random amino acids (i.e., 6.4 × 107) is readily covered by our library construction. We used a low concentration of biotinylated D-MCP-1 (2 nM) from the first-round selection to exclude low-affinity binders. After the fourth round of selection, increases in cDNA recovery rate were observed from libraries A, B, and C, whereas no enrichment was detected from library D (Fig. 2D). As for library D, the lack of critical mutations that could improve the affinity in the last six residues of the FG loop, suggests that the original sequence of Mb5 was already near optimal. The cDNAs from libraries A, B, and C after the fourth round were equivalently mixed, and then the fifth-round selection was conducted with an extended washing time to enrich binders with slow dissociation kinetics (i.e., low koff value). The recovered cDNA was sequenced and analyzed by Weblogo55 to display the appearance ratio of 20 amino acids at each position (Fig. 2E). The results not only clarified essential residues for target recognition and/or conformational stability of the monobody but also identified the mutation-tolerant sites in each loop sequence. Specifically, we observed dominant mutations at the first residue of the BC loop (Leu to Phe), the last residue of the BC loop (Lys to Pro), and the third residue of the FG loop (Gly to Pro), which could improve the affinity by stabilizing the loop structures. By selecting amino acids with a high appearance ratio from each library and combining them, we designed Mb5-9. We also designed Mb5-10, which has a Gly-to-Ser mutation at the 7th position and a Phe-to-Trp mutation at the 8th position in the BC loop, as these mutations appeared in one of the top 10 sequences from library B. Mb5-11, which includes an Arg substitution at the 4th position, was designed based on its appearance ratio as the second most abundant sequence in library C. In addition, we combined these mutations to construct Mb5-12. Mb5-9, -10, -11, and -12 exhibited significantly stronger binding signals and slower dissociation rates compared to those with fewer mutations (Mb5-1 to Mb5-8, Supplementary Fig. 15), indicating that the combination of these mutations was crucial for increasing affinity. Kinetic parameters, determined by BLI with different concentrations of Mb5-9, Mb5-11, and Mb5-12, followed by fitting analysis, revealed that Mb5-11 had the best KD value of 1.6 nM (Fig. 2F). These results clearly demonstrate the efficacy of affinity maturation in obtaining optimized, high-affinity binders.
Synthesis of mirror-image monobody
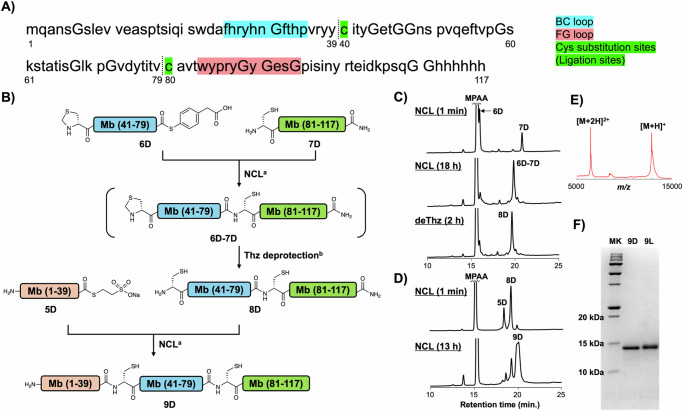
With the high-affinity clones against D-MCP-1 in hand, we then undertook the synthesis of the D-monobody that is supposed to bind to L-MCP-1 by symmetry. The full-length monobody sequence was divided into four peptide segments at junctions including Ala residues (Supplementary Fig. 16A), which can be converted from Cys via a free radical-based desulfurization reaction56 after assembly. In our initial attempt, we conducted a C-to-N one-pot ligation employing NCL-compatible allyloxycarbonyl (Alloc) protecting groups as the N-terminal Cys protecting groups57,58 combined with allyl-protected Asp, which can be deprotected by the same organometal complex as Alloc deprotection59. However, in the first ligation between the C-terminal 2 segments, we failed to produce a detectable product peak in analytical HPLC, likely due to an invisible aggregation even in the presence of 6 M Gn·HCl. Consequently, we revised the order of peptide ligation as shown in Supplementary Fig. 16B, reaching the final product via seven-step reactions including four HPLC purifications. The total yield of the final product 8D’ was less than 1%, likely due to the multiple purification steps, and the purity of 8D’ was compromised by inseparable byproducts in the final HPLC purification step (Supplementary Fig. 16C).
Recently, we developed a chemically synthesized anti-GFP monobody variant containing two Cys substitutions, which simplify the synthesis of the monobody from three peptide segments and eliminate the need for a desulfurization step after peptide assembly60. Importantly, this synthetic L-configured monobody variant maintained its affinity for the target protein, GFP. Consequently, we employed this Cys substitution strategy to synthesize the mirror-image form of Mb5-11 (Fig. 3). The N-terminal segment, 5D, which has a C-terminal alkyl thioester moiety, was synthesized via NaNO2-mediated hydrazide activation61,62. The middle segment, 6D, containing N-terminal thiazolidine (Thz) moiety as a protected Cys residue in addition to a C-terminal thioester, was synthesized through hydrazide activation and subsequent thioesterification63. The acetylacetone-mediated thioesterification was chosen because Thz moiety is incompatible with the standard NaNO2-mediated thioesterification conditions64. For the synthesis of the C-terminal segment, 7D, which contains an N-terminal cysteine and a C-terminal amide, we attached 6 × His sequence at the C-terminal region via a Gly linker for potential solubilizing and/or affinity tag. These peptide segments were synthesized through standard Fmoc SPPS, and the purified segments were analyzed by HPLC and MALDI-TOF mass spectrometry (Supplementary Figs. S17–19). The resulting peptides were assembled from the C-terminus to the N-terminus by NCL (Fig. 3B). The first NCL was conducted between 6D and 7D under standard NCL conditions and a new peak corresponding to the ligation product was observed in analytical HPLC after 18 h (Fig. 3C). Then, the pH of the reaction mixture was adjusted to 4, and methoxyamine was added to the reaction mixture to convert the Thz into Cys in a one-pot reaction. After 2 h of methoxyamine treatment, the obtained product 8D was purified and isolated in 29% yield (Supplementary Fig. 20). A second NCL between 5D and 8D was then conducted to afford 10.4 mg of full-length Cys mutant monobody, 9D in 14% isolated yield (4.0% total yield) (Fig. 3D, E). Through the identical synthetic scheme, we also synthesized an L-configured monobody 9 L, an enantiomer of 9D (Supplementary Figs. 21, 22), and obtained 10.5 mg of the product in 7.2% total yield. To our delight, SDS-PAGE analysis revealed that the purity of 9D and 9 L was significantly improved (Fig. 3F), compared to the previous synthetic scheme shown in Supplementary Fig. 16C.
Fig. 3. Chemical synthesis of D-monobody.
A Target sequence derived from Mb5−11. BC- and FG-loop and Cys substituted sites (ligation sites) are highlighted in blue, pink, and green, respectively. B Synthetic scheme of D-monobody, 9D via C-to-N 3 segment ligation. Reaction conditions: (a) 100 mM MPAA, 50 mM TCEP, 6 M Gn·HCl, 0.2 M phosphate (pH 6.8), 37 °C; (b) 200 mM methoxyamine (pH 4.0) in addition to the NCL condition. C HPLC monitoring of the one-pot reaction of the 1st NCL and subsequent thiazolidine deprotection. HPLC peaks were monitored at 220 nm in the linear gradient with water/acetonitrile containing 0.1% TFA. The gradient of HPLC: acetonitrile 10–60% for 30 min. D HPLC monitoring of the 2nd NCL. HPLC peaks were monitored at 220 nm in a linear gradient with water/acetonitrile containing 0.1% TFA. The gradient of HPLC: acetonitrile 10–60% for 30 min. E MALDI-TOF/MS spectrum of peptide 9D after purification. MS(MALDI-TOF) m/z: [M + H]+ Calcd for C576H849N161O175S3 12926.1; Found 12927.3. F SDS-PAGE analysis of 9D and 9 L, whose synthetic data are shown in Supplementary Fig. 21 and 22. MK: molecular weight ladder marker. Source data are provided as a Source Data file.
Evaluation of mirror-image monobody
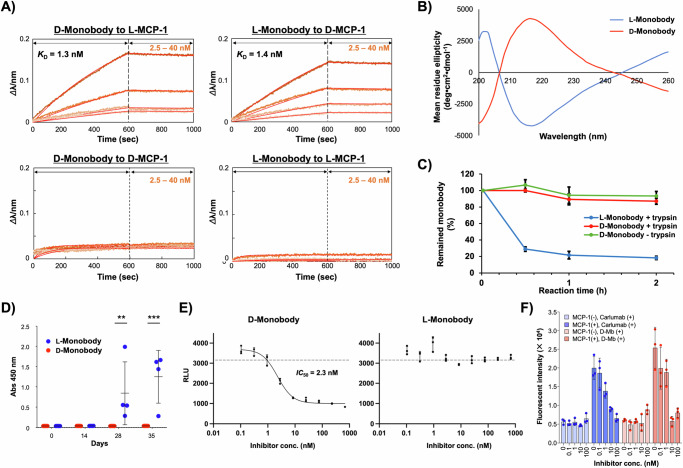
First, we evaluated the binding affinity and specificity of the synthetic monobodies by BLI. Biotinylated L- or D-MCP-1, synthesized as described above, was immobilized on the sensor and different concentrations of D- or L-configured monobodies were analyzed (Fig. 4A). As a result, D-monobody 9D exhibited a KD value of 1.3 nM against L-MCP-1, which is almost identical to the affinity of recombinant Mb5-11 to D-MCP-1 as described in Fig. 2F. Likewise, 9 L also bound to D-MCP-1 with a KD value of 1.4 nM. On the other hand, 9D and 9 L did not show any binding to D- and L-MCP-1, respectively. These results suggest that the synthetic monobodies recognize their target proteins in enantioselective manners. Furthermore, 9D did not show significant binding against pharmaceutically important human proteins of interleukin-6 (IL-6), leukemia inhibitory factor (LIF), IL-6 receptor (IL-6R), cluster of differentiation 266 (CD266), cytotoxic T-lymphocyte associated protein 4 (CTLA4) and programmed cell death protein 1 (PD-1) (Supplementary Fig. 23), indicating that the recognition of 9D against L-MCP-1 is target-selective. Notably, no folding procedures were undertaken on these synthetic monobody samples before BLI analysis. When we evaluated the binding affinity after a dialysis-based folding procedure, the calculated KD value was 1.2 nM, indicating an ignorable difference from the sample without the folding procedure (Supplementary Fig. 24). Therefore, we assume that the monobody established in this study could automatically fold into the proper conformation. CD spectrometry of these synthetic monobodies dissolved in PBS without a folding procedure indicated the existence of monobody-like β-sheet structure as observed in the previous study60 (Fig. 4B). Importantly, 9D and 9 L showed mutually inverted spectra, suggesting that 9D possesses the mirror-image conformation of 9 L.
Fig. 4. Evaluation of mirror-image monobody.
A Binding affinity and specificity analyzed by BLI. L-MCP-1 or D-MCP-1 was immobilized on a streptavidin-sensor chip, and synthetic monobody 9D or 9 L (2.5, 5.0, 10, 20 nM) was used in the kinetic analysis. B CD spectra of synthetic monobodies. 9D and 9 L (5 µM) dissolved in PBS (pH7.4) were measured. C Tryptic digestion of synthetic monobodies. 9D and 9 L (5 µM) dissolved in PBS (pH 7.4) containing 1% PEG were incubated with trypsin (300 nM) for 0.5, 1, and 2 h. The data are presented as mean ± SD, n = 3 biological replicates. D Evaluation of the immunogenicity of 9D and 9L . A 1:1,000 dilution of immunized plasma from each mouse collected on days 0, 14, 28, and 35 was added to the 9D or 9L-coated plates. Generation of antibodies against 9D or 9 L was detected using HRP-anti-mouse IgG (H + L). Bars represent mean ± SD calculated from independent experiments (4 mice for 9L: 5 mice for 9D). Statistical analysis was performed by two-way ANOVA followed by Sidak’s multiple comparisons test. **, p = 0.0047; ***, p < 0.0001. E MCP-1/CCR2 inhibition assay with cultured cells using the PathHunter® β-Arrestin eXpress GPCR Assay kit (DiscoverX). The cultured cells were incubated at 37 °C for 90 min with 9D or 9 L (ranging from 0.10 to 680 nM in a 3-fold serial dilution) and 7 nM L-MCP-1. Chemiluminescence was detected in a plate reader. The experiments were performed in 96-well plates with three wells for each condition (n = 3, biological replicates) and the bars represent mean ± SD. The dotted horizontal line denotes basal RLU as shown in Supplementary Fig. S26. F Cell migration assay was performed with and without MCP-1, and with or without the inhibitor (i.e., carlumab or D-Monobody, 9D), using three independent wells for each condition to obtain biological replicates (n = 3). Data are expressed as mean ± SD. Inhibitor concentrations were varied at 0, 0.1, 1, 10, and 100 nM. Cells that migrated to the empty chamber were quantified by Calcein-AM staining. Source data are provided as a Source Data file.
To investigate the proteolytic stability, we conducted a protease degradation assay of the synthetic monobodies using trypsin as a typical protease. After incubation at 37 °C for 0.5, 1, and 2 h, the tryptic digestion solution was analyzed by SDS-PAGE (Supplementary Fig. 25), and the acquired band intensities were quantified by image processing. As a result, about 90% of full-length 9 L was degraded within 2 h, whereas 9D remained almost intact after 2 h incubation (Fig. 4C). This clearly demonstrated the higher protease resistance of the D-monobody compared to that of L-monobody. These results support previous studies showing that other mirror-image peptides and proteins are less degradable than L-configured native ones.
Next, the immunogenicity of 9D was assessed in comparison with 9 L according to the previously reported procedure60. Intraperitoneal immunization of BALB/c mice was performed with administration of 9D or 9 L emulsified in Freund’s complete adjuvant at day 0, and incomplete adjuvant at days 14 and 28. After plasma samples were collected at days 0, 14, 28, and 35, the level of anti-9D or anti-9L antibody was measured by ELISA (Fig. 4D). Generation of anti-monobody IgG antibody was observed at day 28 from the plasma samples of 9L-immunized mice and the average antibody levels further increased at day 35. On the other hand, the plasma samples from 9D-immunized mice did not show any ELISA signals even at day 35. These results are totally consistent with the previous study showing less immunogenicity of D-monobody than that of L-monobody60. Given that antibody production begins with the degradation of protein antigens into peptides in antigen-presenting cells, the low immunogenicity of mirror-image protein could be attributed to the high proteolytic stability.
In order to represent the biological relevance of D-monobody, the inhibitory activity of 9D on the interaction between MCP-1 and CCR2 was evaluated using PathHunter® β-Arrestin eXpress GPCR Assay kit, which includes CHO-K1 cells expressing CCR2 on their cell membrane. In this assay, upon MCP-1 binding to CCR2, which is fused with a peptide fragment of β-galactosidase, β-arrestin fused with the opposite β-galactosidase fragment is recruited to the intracellular region of CCR2, followed by fragment complementation of β-galactosidase, leading to the emission of chemiluminescence. Therefore, if the monobody binding to MCP-1 competes with CCR2 binding, the luminescence signal should decrease. Initially, we investigated the synthetic L-MCP-1-mediated CCR2 activation, which led to the luminescence emission. A significant increase in luminescence signal was observed only in the presence of 7 nM L-MCP-1, indicating proper signal transduction triggered by this synthetic MCP-1 (Supplementary Fig. 26). Then, we performed the assay by titrating chemically synthesized 9D and 9 L with nine different concentrations in the presence of synthetic L-MCP-1 (Fig. 4E). A clear decrease in luminescence signals was observed depending on the concentration of 9D, whereas nearly constant signals were detected at all concentrations of 9 L. The estimated IC50 value of 9D was 2.3 nM, suggesting the structural stability and MCP-1-specific inhibitory activity of the mirror-image monobody 9D against living cells in culture media.
Finally, we evaluated the inhibitory effect of 9D on MCP-1-induced chemotaxis of cultured monocytes. Carlumab, an anti-MCP-1 antibody that has been tested in clinical trial43, was used as a well-developed inhibitor. THP-1 monocyte cell line was stimulated by recombinant MCP-1 in the presence or absence of the inhibitors and migrated cells were quantified by Calcein-AM staining. While, in the absence of MCP-1, neither 9D or carlumab did not affect the migration of THP-1 cells, both effectively inhibited MCP-1-induced migration in a concentration-dependent manner (Fig. 4F). Therefore, we reasoned that the inhibitory effect of the synthetic D-monobody 9D is comparable to the IgG antibody.
Discussion
By virtue of chemical protein synthesis and TRAP display, we discovered a pharmaceutically promising anti-MCP-1 mirror-image monobody with high affinity, proteolytic resistance, and undetectable immunogenicity. Notably, potent inhibitory activities were observed in biologically relevant environments as comparable to carlumab, which has been used in clinical studies.
During the clone selection process, although the affinity maturation process successfully decreased the dissociation constants to single-digit nM, the initial TRAP display selection by using a library with a diversity of over 1013 generated low-affinity monobody clones against D-MCP-1 (as low as 340 nM for KD). This result was unexpected to us because several sub-nM affinity monobody clones have been obtained in the previous TRAP display selection without affinity maturation37,38. We considered two possibilities to explain the result: (1) unique character of MCP-1 protein or (2) limited patterns of interaction between D- and L-configured polypeptides. Generally speaking, the binding affinities of enriched clones from the same library would vary depending on the target characteristics such as molecular size, shape, and surface charge. Particularly, when the target protein is small, isolation of strong binders would become more challenging due to the limited binding sites available on the surface of the target protein32. Therefore, it seems reasonable that high-affinity clones were not obtained against the 76-residue MCP-1, which is relatively small compared to previously reported targets consisting of over 200 residues such as SARS-CoV-2 spike protein, EGFR1, and HER237. However, this first possibility was rejected because high-affinity clones with single-digit nM KD were obtained by the same procedure of selection using L-MCP-1 as the target (Supplementary Fig. 14). Another possibility is that the interactions between D- and L-configured polypeptides could be less favorable than those found in L-configured polypeptides. Although the second hypothesis would be novel and interesting, there is currently a limited amount of data available regarding the interactions between D- and L-polypeptides. In the future, the selection of more mirror-image binders and the determination of their structures will provide insights into enantioselective binding, as demonstrated in the parallel study44.
Consistent production of mirror-image peptide/protein binders is hindered by limited accessibility to reliable setups and techniques for in vitro selection and chemical protein synthesis. Given that higher library diversity tends to yield high-affinity binders against the same target, efforts to increase library diversity, such as optimizing mRNA sequences65, would be the one of promising approaches. Continuous development of methodologies for chemical protein synthesis is also necessary to efficiently prepare D-configured target and binder proteins. The results reported here demonstrate the successful generation of a mirror-image protein binder with high affinity against a pharmaceutically important target protein through chemical protein synthesis and TRAP display. While a previous study highlighted mirror-image mRNA display that produced a moderate D-peptide binder with a dissociation constant of 261 nM66, the current study, by leveraging a protein scaffold and a highly diverse library, demonstrates the production of a mirror-image binder with high affinity (KD = 1.3 nM), significant inhibitory activity on the interaction between MCP-1 and CCR2 (IC50 = 2.3 nM), and potent inhibitory effect on cell migration comparable to carlumab. We anticipate that the combinatorial use of matured screening and synthesis methodologies will lead to the revitalization of mirror-image binder development in the drug discovery field.
Methods
Materials for TRAP display
Oligonucleotides were purchased from Fasmac Co. and Ltd., Nippon Bio Service. The sequences of the primers and synthetic DNAs are listed in Supplementary Data 1. The preparation of the E. coli reconstituted cell-free translation system, Pfu-S DNA polymerase, and Moloney murine leukemia virus reverse transcriptase (MMLV) have been described in previous reports37,53,67,68. The composition of the translation system is listed in Supplementary Table 1.
In vitro selection of monobody clones against D-MCP-1 by TRAP display
For the first-round selection, the puromycin linker (PuL)/mRNA complex was prepared by annealing HEX-mPuL (4 µM) with the FN3 mRNA library (4.8 µM)37 in annealing buffer (25 mM HEPES-K pH 7.8, 200 mM potassium acetate) by heating the solution (200 µL) to 95 °C for 3 min and cooling to 25 °C. The HEX-mPuL /mRNA complex (126 µL) was added to the E. coli reconstituted cell-free translation system, resulting in a final reaction volume of 500 µL. The mixture was incubated at 37 °C for 30 min. Subsequently, 41.7 µL of 200 mM EDTA (pH 8.0) was added to the translation mixture. Reverse transcription buffer (41.1 µL; 0.78 M Tris-HCl pH 8.4, 1.16 M KCl, 0.37 M MgCl2 and 0.08 M DTT), 5 mM dNTPs (66.7 µL), 100 μM FN3S.R29 (10 µL), and 28.7 μM HMLV (27.5 µL) were added to the translation mixture, and the resulting solution was incubated at 42 °C for 15 min. The buffer was exchanged to HBST buffer [50 mM HEPES-K pH 7.5, 300 mM NaCl, 0.05% (v/v) Tween 20] using Zeba™ Spin Desalting Columns. In order to remove the bead binders, the resulting solution was mixed with beads from a 100 µL suspension of Dynabeads M-280/M-270 streptavidin (1:1) (Thermo Fisher Scientific) at 25 °C for 10 min. The supernatant was mixed with 1.75 µL of 20 µM D-MCP-1-biotin (f.c. 50 nM) and then incubated at 25 °C for 10 min. After recovering the target proteins with beads from 200 µL of Dynabeads M-270 streptavidin, the resulting beads were washed with 1 mL of HBST buffer thrice, and PCR premix [10 mM Tris-HCl pH 8.4, 50 mM KCl, 0.1% (v/v) Triton X-100, 2 mM MgCl2, 0.25 mM each dNTP] (690 µL) was added, and heated at 95 °C for 2 min. The amount of the eluted cDNAs was quantified by SYBR green-based quantitative PCR using T7SD8M2.F44 and FN3Lip.R20 as primers. The eluted cDNAs were PCR-amplified using T7SD8M2.F44, G5S-4Gan21-3.R42, and Pfu-S DNA polymerase and was purified by phenol/chloroform extraction and isopropanol precipitation. The DNA was dissolved in 10 mM HEPES-K pH 7.8.
For the second-round selection, the resulting DNA (25 nM, 4.0 µL) was added to the TRAP system, and the reaction mixture (20 µL) was incubated at 37 °C for 30 min. After the reaction, 4 µL of 100 mM EDTA (pH 8.0) was added to the translation mixture. A reverse transcription mixture (12 µL; 150 mM Tris-HCl pH 8.4, 225 mM KCl, 75 mM MgCl2 and 16 mM DTT, 1.5 mM dNTPs, 7.5 μM FN3S.R29, and 3.4 μM HMLV) was then added to the translation mixture, and the resulting solution was incubated at 42 °C for 15 min. The buffer was exchanged for HBST buffer using Zeba™ Spin Desalting Columns. To remove bead binders, the resulting solution was mixed thrice with beads from 10 µL of Dynabeads M-280/M-270 streptavidin (1:1) at 25 °C for 20 min. The half volume of the supernatant (16 µL) was mixed with 0.4 µL of 1 µM D-MCP-1-biotin (f.c. 25 nM). After incubating at 25 °C for 10 min, the solution was mixed with beads from 0.2 µL of Dynabeads M-270 streptavidin at 25 °C for 1 min. The beads were washed with 10 µL of HBST buffer thrice, and the PCR premix was added to the beads. Quantitation of cDNA and amplification and purification of DNA were performed using the same procedure as described for the first-round selection.
For the third to fifth-round selections, the procedure was performed in a manner similar to that of the second round, except for the volume of the reaction mixtures, which was 5 µL. For the sixth to seventh-round selections, the solution for the selection was diluted 16 times with HBST to reduce the concentration of D-MCP-1-biotin to 2 nM. In the seventh round, the pulldown step was performed both with and without D-MCP-1-biotin, and the recovered cDNAs were subsequently sequenced using an Ion Torrent instrument (Thermo Fisher Scientific) (Supplementary Data 2).
Selection against L-MCP-1-biotin was conducted with the same procedure as that of D-MCP-1-biotin.
Affinity maturation of Mb5
For the first round of selection, 1 µM mRNA/HEX-mPuL was added to a reconstituted translation system, and the reaction mixture (5 µL) was incubated at 37 °C for 30 min. After the reaction, 1 µL of 100 mM EDTA (pH 8.0) was added to the translation mixture. A reverse transcription mixture (3 µL; 150 mM Tris-HCl pH 8.4, 225 mM KCl, 75 mM MgCl2, and 16 mM DTT, 1.5 mM each dNTP, 7.5 μM FN3S.R29 primer, and 3.4 μM MMLV) was added to the translation mixture, and the resulting solution was incubated at 42 °C for 15 min. The buffer was changed to HBST buffer using Zeba™ Spin Desalting Columns. To remove the bead binders, the resulting solution was mixed with beads from 10 µL of Dynabeads M-280/M-270 streptavidin (1:1) (Thermo Fisher Scientific) at 25 °C for 20 min. This step was repeated another two times. The supernatant (9 µL) was diluted with the HBST buffer (81 µL) and mixed with 0.9 µL of 200 nM D-MCP-1-biotin (f.c. 2 nM); the resulting solution was incubated at 25 °C for 3 min. The target proteins were collected by mixing with beads from 0.5 µL of Dynabeads M-270 streptavidin for 1 min. The collected beads were washed with 10 µL of the HBST buffer for 1 min two times, and 100 µL of PCR premix was added. The beads were heated at 95 °C for 5 min, and the amount of eluted cDNA was quantified by SYBR green-based quantitative PCR using T7SD8M2.F44 and FN3Lip.R20 as primers. The eluted cDNA was PCR-amplified using T7SD8M2.F44, G5S-4Gan21-3.R42, and Pfu-S DNA polymerase and purified by phenol/chloroform extraction and isopropanol precipitation. From the following selection, the resulting DNA (about 5 nM final concentration) was added to the TRAP system, and the reaction mixture (5 µL) was incubated at 37 °C for 30 min. The subsequent procedures were similar to those described above. In the final round of selection, after washing twice with 10 µL of HBST, the beads were extensively washed by incubating them in 200 µL of HBST at 25 °C for 20 min The sequences of the recovered DNA were determined using an Ion Torrent instrument (Thermo Fisher Scientific).
Affinity measurement of monobody clones against D-MCP-1 and L-MCP-1 by BLI
Affinity measurement was performed on biotinylated D-MCP-1, or L-MCP-1 immobilized on a streptavidin biosensor (ForteBio) using the Octet system (ForteBio) as described in the manufacturer’s instructions. The analyte monobody was dissolved in water to prepare a 10 μM of monobody solution, and the buffer was changed to buffer D (50 mM HEPES (pH 7.5), 300 mM NaCl, 0.05% [vol/vol] tween 20, 0.1% [wt/vol] PEG) using Zeba™ Spin Desalting Columns. The protein concentration was measured at A280, according to the molar extinction coefficient estimated from the amino acid composition. The monobody solution was stored at − 80 °C and was used for the following binding assay after dilution with buffer D. The binding assay was performed at 25 °C in buffer A. Each step in the binding assay was as follows: equilibration for 60 s, association for 400 s or 600 s, and dissociation for 600 s.
General protocol of SPPS for D-monobody segments
Automated solid-phase peptide synthesis was performed by using Initiator + Alstra (Biotage). Fmoc-protected D- and L-amino acids containing standard side-chain protecting groups were used in Fmoc SPPS. For Fmoc group deprotection, 20% piperidine, 0.1 M HOBt in DMF was treated for 5 min twice. For coupling, HBTU as an activator and DIEA as a base were used. For capping, 5% anhydrous acetic acid in DMF was used. Coupling of amino acids other than His, Arg, and Cys derivatives: Fmoc-protected amino acids (4 equiv) were activated with HBTU (3.9 equiv) and DIEA (8 equiv) in DMF and transferred to the resin (coupling time: 5 min at 75 °C, single coupling). Coupling of His, Arg, and Cys derivatives: Fmoc-protected amino acids (4 equiv) were activated with HBTU (3.9 equiv) and DIEA (8 equiv) in DMF and transferred to the resin (coupling time: 60 min at room temperature, double coupling). The isolated yields of each peptide were estimated by using the molecular weights of TFA salt at the N-terminal amine and the sidechains of Arg, Lys, and His.
As amino-acid building blocks, Fmoc-protected monomers were used as shown below: Fmoc-D-Ala-OH, Fmoc-D-Arg(Pbf)-OH, Fmoc-D-Asn(Trt)-OH, Fmoc-D-Asp(Ot-Bu)-OH, Fmoc-D-Cys(Trt)-OH, Fmoc-D-Gln(Trt)-OH, Fmoc-D-Glu(Ot-Bu)-OH, Fmoc-Gly-OH, Fmoc-D-His(Trt)-OH, Fmoc-D-Ile-OH, Fmoc-D-Leu-OH, Fmoc-D-Lys(Boc)-OH, Fmoc-D-Met-OH, Fmoc-D-Phe-OH, Fmoc-D-Pro-OH, Fmoc-D-Ser(t-Bu)-OH, Fmoc-D-Thr(t-Bu)-OH, Fmoc-D-Trp(Boc)-OH, Fmoc-D-Tyr(t-Bu)-OH, Fmoc-D-Val-OH.
Synthesis of D-monobody1–39 (5D) by Fmoc-SPPS
2-Chlorotrityl chloride resin was swelled with DMF for 30 min. Fmoc-NHNH2 (1.1 equiv) and DIEA (dissolved in DMF, 2.2 equiv) were added to the resin and reacted at room temperature for 12 h. Then, MeOH was added and reacted for 10 min. The resin was subsequently washed with DMF and DCM for each 3 times. After Fmoc quantification, the target peptide sequence was elongated by Initiator + Alstra. For peptide recovery and global deprotection, the obtained resin was treated with TFA/TIS/H2O (95/2.5/2.5) for 2 h at room temperature. Then, the TFA solution was obtained by filtration, and 10 times the volume of cold diethyl ether was added. The tube was vortexed well and centrifuged 10,000 × g at 3 °C for 5 min. Ether was removed by decantation. This precipitation was washed with diethyl ether twice, and the precipitation was dried in vacuo. The solid crude peptide was dissolved in 6 M Gn·HCl and 0.2 M NaH2PO4 at pH 3.0 (peptide concentration: ~ 2.5 mM calculated from resin loading). The solution was cooled to − 17 °C and NaNO2 was added (10 equiv against peptide). The mixture was stirred at − 17 °C for 15 min, and then 125 mM MESNa aq. (50 equiv against each peptide) was added to the reaction mixture. The pH was adjusted to 6.5–6.8 with 6 M NaOH aq. and the solution was stirred at room temperature for 30 min. Then, the peptide solution was purified by preparative HPLC (Nacalai Cosmosil 5C18-AR-II column) to provide 5D (5.7 mg, 0.7% isolated yield from resin). MALDI-TOF/MS spectrum of 5D after purification. MS(MALDI-TOF) m/z: [M + H]+ Calcd for C204H296N56O62S3 4622.1; Found 4621.7.
Synthesis of D-monobody40–79 (6D) by Fmoc-SPPS
The same SPPS procedure described above was employed for resin preparation and peptide elongation. Manual synthesis was performed by the following protocol: For Fmoc group deprotection, the resin was treated with 20% piperidine, 0.1 M HOBt in DMF for 5 min twice. For coupling of Thz, Boc-Thz-OH (5 equiv) were activated with HATU (4.9 equiv) and DIEA (10 equiv) in DMF and transferred to the resin (coupling time; 2 h at room temperature). For peptide recovery and global deprotection, the obtained resin was treated with TFA/thioanisole/EDT/anisole (90/5/3/2) for 2 h at room temperature. Then, the TFA solution was obtained by filtration, and 10 times the volume of cold diethyl ether was added. The tube was vortexed well and centrifuged 10,000 × g at 3 °C for 5 min. Ether was removed by decantation. This precipitation was washed with diethyl ether twice, and the precipitation was dried in vacuo. The peptide precipitation was dissolved in 6 M Gn·HCl, 100 mM MPAA, and 0.2 M NaH2PO4 at pH 3.0 (peptide concentration: ~ 1 mM calculated from resin loading). Then, acetylacetone was added (2.5 equiv against peptide), and the solution was stirred at 37 °C. For analysis of each reaction, an aliquot (1.0 ~ 2.0 μL) of each reaction mixture was diluted by water containing 0.1% TFA and injected into analytical HPLC. After the thioesterification was completed, the peptide solution was purified by preparative HPLC (Phenomenex Jupiter C4 300 column) to provide 6D (20.2 mg, 6.2% isolated yield from resin). MS(MALDI-TOF) m/z: [M + H]+ Calcd for C188H290N44O63S2 4239.7; Found 4239.7.
Synthesis of D-monobody80–117 (7D) by Fmoc-SPPS
Rink amide-PEG resin (0.24 mmol/g, 0.20 mmol, Watanabe Chemical) was swelled with DMF for 30 min. The resin was subsequently washed with DMF and DCM for each 3 times. The target peptide sequence was elongated by Initiator + Alstra. The resin was then treated with TFA/TIS/H2O/DMB (90/2.5/2.5/5) for 2 h at room temperature. Then, the TFA solution was obtained by filtration, and 10 times the volume of cold diethyl ether was added. The tube was vortexed well and centrifuged 10,000 × g at 3 °C for 5 min. Ether was removed by decantation. This precipitation was washed with diethyl ether twice, and the precipitation was dried in vacuo. Then, the peptide solution dissolved in water/acetonitrile mixture containing 0.1% TFA was purified by preparative HPLC (Nacalai Cosmosil Protein-R column) to provide 7D (64.7 mg, 5.9% isolated yield from resin loading). MS(MALDI-TOF) m/z: [M + H]+ Calcd for C195H277N61O55S1 4388.7; Found 4389.5.
Synthesis of D-monobody40–117 (8D) by peptide ligation
1 mM peptides 6D (17.0 mg, 3.72 μmol) and 1 mM 7D (20.5 mg, 3.72 μmol) were dissolved in MPAA ligation buffer [100 mM MPAA, 50 mM TCEP, 6 M Gn·HCl, 200 mM phosphate (pH 6.5)], and the reaction mixture was incubated for 18 h at 37 °C. 1 M HCl was added to the reaction mixture to adjust the pH into 4. Then, methoxyamine·HCl was added (200 equiv against peptide) and the solution was stirred for 2 h at 37 °C. For analysis of each reaction, an aliquot (1.0 μL) of each reaction mixture was injected into analytical HPLC. The mixture was purified by preparative HPLC (Phenomenex Jupiter C4 300 column) to provide 8D (10.4 mg, 1.06 μmol, 29% isolated yield). MS(MALDI-TOF) m/z: [M + H]+ Calcd for C374H559N105O116S2 8447.2; Found 8446.9.
Synthesis of D-monobody1–117 (9D) by peptide ligation
1 mM peptides 5D (5.6 mg, 1.06 μmol) and 1 mM 8D (10.4 mg, 1.06 μmol) were dissolved in MPAA ligation buffer [100 mM MPAA, 50 mM TCEP, 6 M Gn·HCl, 200 mM phosphate (pH 6.5)], and the reaction mixture was incubated for 13 h at 37 °C. For analysis of each reaction, an aliquot (1.0 μL) of each reaction mixture was diluted by TCEP (4.0 μL), DTT (5.0 μL) and injected into analytical HPLC. The mixture was purified by preparative HPLC (Phenomenex C4 300 column) to provide 9D (2.2 mg, 0.146 μmol, 14% isolated yield). MS(MALDI-TOF) m/z: [M + H]+ Calcd for C576H849N161O175S3 12926.1; Found 12927.3.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
This study was financially supported by the Japan Society for the Promotion of Science (JSPS) KAKENHI JP20H04704, JP21H00278, JP22KK0128, JP24H01893 (G.H.); JP20K21252, JP22H02747, JP22K19376, JP24K01628 (S.O. and M.N.); JP22KJ1842 (N.I.); and JP23H05456 (H.M.). This study was also supported by JST PRESTO JPMJPR19K6 (G.H.), AMED JP22ama121009 (G.H.), JP20ak0101144 (S.O. and M.N.), JP21zf0127004 (H.M.), the Tokyo Biochemical Research Foundation (S.O.), Astellas Foundation for Research on Metabolic Disorders (S.O.), Takeda Science Foundation (S.O.), and TERUMO Life Science Foundation (S.O.). We acknowledge H.M. for their personal financial contribution to the Nagoya University Foundation, which is associated with Nagoya University, the author’s affiliated institution. This was a one-time donation and not from grant funds. N.I. is grateful for JSPS Research Fellowships for Young Scientists.
Author contributions
G.H., M.N., S.O., and H.M. proposed the idea and designed the experiments, analyzed all the results, and wrote the manuscript. T.N., N.I., S.S., and Y.U. carried out chemical synthesis and evaluation of D- and L-proteins. S.M. performed TRAP display selection and affinity maturation. M.B.-S. and K.H. performed a cell migration assay. M.N. carried out an immunogenicity assay. All authors read and discussed the manuscript.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
All data generated in this study are provided in the Supplementary Information/Source Data file. Supplementary Information is provided in this paper. Source Data are provided as a Source Data file. Source data are provided in this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Gosuke Hayashi, Email: hayashi@chembio.nagoya-u.ac.jp.
Motohiro Nonaka, Email: nonaka.motohiro.4r@kyoto-u.ac.jp.
Shinya Oishi, Email: soishi@mb.kyoto-phu.ac.jp.
Hiroshi Murakami, Email: murah@chembio.nagoya-u.ac.jp.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-54902-x.
References
- 1.Harrison, K., Mackay, A. S., Kambanis, L., Maxwell, J. W. C. & Payne, R. J. Synthesis and applications of mirror-image proteins. Nat. Rev. Chem.7, 383–404 (2023). [DOI] [PubMed] [Google Scholar]
- 2.Lander, A. J., Jin, Y. & Luk, L. Y. P. D-peptide and D-protein technology: Recent advances, challenges, and opportunities. ChemBioChem24, e202200537 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yeates, T. O. & Kent, S. B. H. Racemic protein crystallography. Annu. Rev. Biophys.41, 41–61 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Kent, S. B. H. Racemic & quasi-racemic protein crystallography enabled by chemical protein synthesis. Curr. Opin. Chem. Biol.46, 1–9 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Milton, R. C. D., Milton, S. C. F. & Kent, S. B. H. Total chemical synthesis of a D-enzyme: The enantiomers of HIV-1 protease show reciprocal chiral substrate specificity. Science256, 1445–1448 (1992). [DOI] [PubMed] [Google Scholar]
- 6.Weinstock, M. T., Jacobsen, M. T. & Kay, M. S. Synthesis and folding of a mirror-image enzyme reveals ambidextrous chaperone activity. Proc. Natl. Acad. Sci. USA111, 11679–11684 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ding, R. C., Shi, W. W. & Zheng, J. S. Chemically synthetic d-sortase enables enzymatic ligation of d-peptides. Org. Lett.25, 4857–4861 (2023). [DOI] [PubMed] [Google Scholar]
- 8.Zhang, G. W. & Zhu, T. F. Mirror-image trypsin digestion and sequencing of D-proteins. Nat. Chem.16, 592–598 (2024). [DOI] [PubMed] [Google Scholar]
- 9.Wang, Z. M., Xu, W. L., Liu, L. & Zhu, T. F. A synthetic molecular system capable of mirror-image genetic replication and transcription. Nat. Chem.8, 698–704 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Xu, Y. & Zhu, T. F. Mirror-image T7 transcription of chirally inverted ribosomal and functional RNAs. Science378, 405–411 (2022). [DOI] [PubMed] [Google Scholar]
- 11.Ling, J. J. et al. Mirror-image 5S ribonucleoprotein complexes. Angew. Chem. Int. Ed.59, 3724–3731 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poduslo, J. F., Curran, G. L., Kumar, A., Frangione, B. & Soto, C. -sheet breaker peptide inhibitor of Alzheimer’s amyloidogenesis with increased blood-brain barrier permeability and resistance to proteolytic degradation in plasma. J. Neurobiol.39, 371–382 (1999). [PubMed] [Google Scholar]
- 13.Dintzis, H. M., Symer, D. E., Dintzis, R. Z., Zawadzke, L. E. & Berg, J. M. A comparison of the immunogenicity of a pair of enantiomeric proteins. Proteins16, 306–308 (1993). [DOI] [PubMed] [Google Scholar]
- 14.Scott, J. K. & Smith, G. P. Searcjomg for peptide ligands with an epitope library. Science249, 386–390 (1990). [DOI] [PubMed] [Google Scholar]
- 15.Schumacher, T. N. M. et al. Identification of D-peptide ligands through mirror-image phage display. Science271, 1854–1857 (1996). [DOI] [PubMed] [Google Scholar]
- 16.Wiesehan, K. et al. Selection of D-Amino-Acid peptides that bind to Alzheimer’s disease amyloid peptide Aβ1-42 by mirror image phage display. ChemBioChem4, 748–753 (2003). [DOI] [PubMed] [Google Scholar]
- 17.Liu, M. et al. A left-handed solution to peptide inhibition of the p53-MDM2 interaction. Angew. Chem. Int. Ed.49, 3649–3652 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang, H. N. et al. Blocking of the PD-1/PD-L1 interaction by a D-peptide antagonist for cancer immunotherapy. Angew. Chem. Int. Ed.54, 11760–11764 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Eckert, D. M., Malashkevich, V. N., Hong, L. H., Carr, P. A. & Kim, P. S. Inhibiting HIV-1 entry: Discovery of D-peptide inhibitors that target the gp41 coiled-coil pocket. Cell99, 103–115 (1999). [DOI] [PubMed] [Google Scholar]
- 20.Díaz-Perlas, C. et al. Protein chemical synthesis combined with mirror-image phage display yields d-peptide EGF ligands that block the EGF-EGFR interaction. ChemBioChem20, 2079–2084 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Zhou, X. M. et al. A novel d-peptide identified by mirror-image phage display blocks TIGIT/PVR for cancer immunotherapy. Angew. Chem. Int. Ed.59, 15114–15118 (2020). [DOI] [PubMed] [Google Scholar]
- 22.Li, Z. X. et al. Novel strategy utilizing extracellular cysteine-rich domain of membrane receptor for constructing D-peptide mediated targeted drug delivery systems: A case study on Fn14. Bioconjug. Chem.28, 2167–2179 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Malhis, M. et al. Potent Tau aggregation inhibitor D-peptides selected against Tau-repeat 2 using mirror image phage display. ChemBioChem22, 3049–3059 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Callahan, A. J. et al. Mirror-image ligand discovery enabled by single-shot fast-flow synthesis of D-proteins. Nat. Commun.15, 1813 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Welch, B. D., VanDemark, A. P., Heroux, A., Hill, C. P. & Kay, M. S. Potent D-peptide inhibitors of HIV-1 entry. Proc. Natl. Acad. Sci. USA104, 16828–16833 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Welch, B. D. et al. Design of a potent D-peptide HIV-1 entry inhibitor with a strong barrier to resistance. J. Virol.84, 11235–11244 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vazquez-Lombardi, R. et al. Challenges and opportunities for non-antibody scaffold drugs. Drug Discov. Today20, 1271–1283 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Gebauer, M. & Skerra, A. Engineered protein scaffolds as next-generation therapeutics. Annu. Rev. Pharmacol. Toxicol.60, 391–415 (2020). [DOI] [PubMed] [Google Scholar]
- 29.Mandal, K. et al. Chemical synthesis and X-ray structure of a heterochiral {D-protein antagonist plus vascular endothelial growth factor} protein complex by racemic crystallography. Proc. Natl. Acad. Sci. USA109, 14779–14784 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Uppalapati, M. et al. A potent D-protein antagonist of VEGF-A is nonimmunogenic, metabolically stable, and longer-circulating in vivo. ACS Chem. Biol.11, 1058–1065 (2016). [DOI] [PubMed] [Google Scholar]
- 31.Marinec, P. S. et al. A non-immunogenic bivalent D-protein potently inhibits retinal vascularization and tumor growth. ACS Chem. Biol.16, 548–556 (2021). [DOI] [PubMed] [Google Scholar]
- 32.Kamalinia, G., Grindel, B. J., Takahashi, T. T., Millward, S. W. & Roberts, R. W. Directing evolution of novel ligands by mRNA display. Chem. Soc. Rev.50, 9055–9103 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roberts, R. W. & Szostak, J. W. RNA-peptide fusions for the in vitro selection of peptides and proteins. Proc. Natl. Acad. Sci. USA94, 12297–12302 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nemoto, N., MiyamotoSato, E., Husimi, Y. & Yanagawa, H. In vitro virus: Bonding of mRNA bearing puromycin at the 3’-terminal end to the C-terminal end of its encoded protein on the ribosome in vitro. FEBS Lett.414, 405–408 (1997). [DOI] [PubMed]
- 35.Ishizawa, T., Kawakami, T., Reid, P. C. & Murakami, H. TRAP Display: A high-speed selection method for the generation of functional polypeptides. J. Am. Chem. Soc.135, 5433–5440 (2013). [DOI] [PubMed] [Google Scholar]
- 36.Koide, A., Bailey, C. W., Huang, X. L. & Koide, S. The fibronectin type III domain as a scaffold for novel binding proteins. J. Mol. Biol.284, 1141–1151 (1998). [DOI] [PubMed] [Google Scholar]
- 37.Kondo, T. et al. Antibody-like proteins that capture and neutralize SARS-CoV-2. Sci. Adv.6, eabd3916 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamano, K. et al. Optineurin provides a mitophagy contact site for TBK1 activation. EMBO J.43, 754–779 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dawson, P. E. & Kent, S. B. Synthesis of native proteins by chemical ligation. Annu. Rev. Biochem.69, 923–960 (2000). [DOI] [PubMed] [Google Scholar]
- 40.Bondalapati, S., Jbara, M. & Brik, A. Expanding the chemical toolbox for the synthesis of large and uniquely modified proteins. Nat. Chem.8, 407–418 (2016). [DOI] [PubMed] [Google Scholar]
- 41.Kulkarni, S. S., Sayers, J., Premdjee, B. & Payne, R. J. Rapid and efficient protein synthesis through expansion of the native chemical ligation concept. Nat. Rev. Chem.2, 0122 (2018). [Google Scholar]
- 42.Singh, S. & Anshita, D. Ravichandiran. MCP-1: Function, regulation, and involvement in disease. Int. Immunopharmacol.101, 107598 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li, H., Wu, M. & Zhao, X. Role of chemokine systems in cancer and inflammatory diseases. MedComm3, e147 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmidt, N. et al. Development of mirror-image monobodies targeting the oncogenic BCR::ABL1 kinase. Nat. Commun.10.1038/s41467-024-54901-y (2024). [DOI] [PMC free article] [PubMed]
- 45.Furutani, Y. et al. Cloning and sequencing of the cDNA for human monocyte chemotactic and activating factor (MCAF). Biochem. Biophys. Res. Commun.159, 249–255 (1989). [DOI] [PubMed] [Google Scholar]
- 46.Robinson, E. A. et al. Complete amino acid sequence of a human monocyte chemoattractant, a putative mediator of cellular immune reactions. Proc. Natl. Acad. Sci. USA86, 1850–1854 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lai, Z. W., Petrera, A. & Schilling, O. Protein amino-terminal modifications and proteomic approaches for N-terminal profiling. Curr. Opin. Chem. Biol.24, 71–79 (2015). [DOI] [PubMed] [Google Scholar]
- 48.Grygiel, T. L. R. et al. Synthesis by native chemical ligation and crystal structure of human CCL2. Biopolymers94, 350–359 (2010). [DOI] [PubMed] [Google Scholar]
- 49.Dawson, P. E., Muir, T. W., Clarklewis, I. & Kent, S. B. H. Synthesis of proteins by native chemical ligation. Science266, 776–779 (1994). [DOI] [PubMed] [Google Scholar]
- 50.Blanco-Canosa, J. B., Nardone, B., Albericio, F. & Dawson, P. E. Chemical protein synthesis using a second-generation N-acylurea linker for the preparation of peptide-thioester precursors. J. Am. Chem. Soc.137, 7197–7209 (2015). [DOI] [PubMed] [Google Scholar]
- 51.Johnson, E. C. B. & Kent, S. B. H. Insights into the mechanism and catalysis of the native chemical ligation reaction. J. Am. Chem. Soc.128, 6640–6646 (2006). [DOI] [PubMed] [Google Scholar]
- 52.Wojcik, J. et al. A potent and highly specific FN3 monobody inhibitor of the Abl SH2 domain. Nat. Struct. Mol. Biol.17, 519–527 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shimizu, Y. et al. Cell-free translation reconstituted with purified components. Nat. Biotechnol.19, 751–755 (2001). [DOI] [PubMed] [Google Scholar]
- 54.Kondo, T. et al. Monobodies with potent neutralizing activity against SARS-CoV-2 Delta and other variants of concern. Life Sci. Alliance5, e202101322 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Crooks, G. E., Hon, G., Chandonia, J. M. & Brenner, S. E. WebLogo: A sequence logo generator. Genome Res.14, 1188–1190 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wan, Q. & Danishefsky, S. J. Free-radical-based, specific desulfurization of cysteine: a powerful advance in the synthesis of polypeptides and glycopolypeptides. Angew. Chem. Int. Ed.46, 9248–9252 (2007). [DOI] [PubMed] [Google Scholar]
- 57.Kamo, N., Hayashi, G. & Okamoto, A. Triple function of 4-mercaptophenylacetic acid promotes one-pot multiple peptide ligation. Angew. Chem. Int. Ed.57, 16533–16537 (2018). [DOI] [PubMed] [Google Scholar]
- 58.Kamo, N. et al. Organoruthenium-catalyzed chemical protein synthesis to elucidate the functions of epigenetic modifications on heterochromatin factors. Chem. Sci.12, 5926–5937 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kamo, N., Hayashi, G. & Okamoto, A. Efficient peptide ligation between allyl-protected Asp and Cys followed by palladium-mediated deprotection. Chem. Commun.54, 4337–4340 (2018). [DOI] [PubMed] [Google Scholar]
- 60.Iwamoto, N. et al. Design and synthesis of monobody variants with low immunogenicity. ACS Med. Chem. Lett.14, 1596–1601 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fang, G. M. et al. Protein chemical synthesis by ligation of peptide hydrazides. Angew. Chem. Int. Ed.50, 7645–7649 (2011). [DOI] [PubMed] [Google Scholar]
- 62.Zheng, J. S., Tang, S., Qi, Y. K., Wang, Z. P. & Liu, L. Chemical synthesis of proteins using peptide hydrazides as thioester surrogates. Nat. Protoc.8, 2483–2495 (2013). [DOI] [PubMed] [Google Scholar]
- 63.Flood, D. T. et al. Leveraging the Knorr pyrazole synthesis for the facile generation of thioester surrogates for use in native chemical ligation. Angew. Chem. Int. Ed.57, 11634–11639 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sato, K. et al. Direct synthesis of N-terminal thiazolidine-containing peptide thioesters from peptide hydrazides. Chem. Commun.54, 9127–9130 (2018). [DOI] [PubMed] [Google Scholar]
- 65.Umemoto, S., Kondo, T., Fujino, T., Hayashi, G. & Murakami, H. Large-scale analysis of mRNA sequences localized near the start and amber codons and their impact on the diversity of mRNA display libraries. Nucleic Acids Res.51, 7465–7479 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Huang, L. Y. et al. Highly selective targeting of hepatic stellate cells for liver fibrosis treatment using a D-enantiomeric peptide ligand of Fn14 identified by mirror-image mRNA display. Mol. Pharm.14, 1742–1753 (2017). [DOI] [PubMed] [Google Scholar]
- 67.Ohashi, H., Shimizu, Y., Ying, B. W. & Ueda, T. Efficient protein selection based on ribosome display system with purified components. Biochem. Biophys. Res. Commun.352, 270–276 (2007). [DOI] [PubMed] [Google Scholar]
- 68.Reid, P. C., Goto, Y., Katoh, T. & Suga, H. Charging of tRNAs using ribozymes and selection of cyclic peptides containing thioethers. Methods Mol. Biol.805, 335–348 (2012). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
All data generated in this study are provided in the Supplementary Information/Source Data file. Supplementary Information is provided in this paper. Source Data are provided as a Source Data file. Source data are provided in this paper.