Abstract
Purpose
The treatment landscape for chronic myeloid leukemia (CML) has been revolutionized by the introduction of imatinib, a tyrosine kinase inhibitor, which has transformed the disease from a fatal condition into a manageable chronic illness for a substantial number of patients. Despite this, some individuals do not respond adequately to the treatment, and others may experience disease progression even with continued therapy. This study examined how CYP2C8*3 (G416A; rs11572080) and ABCG2 C421A (rs2231142) single nucleotide polymorphisms (SNPs) affect the plasma trough concentration and therapeutic response of imatinib in Egyptian CML patients.
Methods
The study included fifty patients with chronic-phase CML, who were categorized into two groups: responders (n = 26) and non-responders (n = 24), according to their BCR-ABL1 transcription levels after 12 months of imatinib treatment. Genotyping of the CYP2C8*3 and ABCG2 C421A polymorphisms was performed using polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP), while plasma trough concentrations were determined through high-performance liquid chromatography with ultraviolet-diode array detection (HPLC-UV/DAD).
Results
Patients with the CA genotype of ABCG2 C421A showed significantly higher mean plasma trough concentrations of imatinib (CA: 1731 ± 424.7 ng/mL; CC: 1294 ± 381.3 ng/mL; p = 0.0132) and demonstrated a better molecular response compared to those with the CC genotype (p = 0.0395).
Conclusion
The ABCG2 C421A polymorphism significantly influenced imatinib plasma trough concentrations and molecular responses in Egyptian chronic-phase CML patients. Genotyping of this polymorphism in these patients could assist in optimizing imatinib therapy, predicting more favorable treatment outcomes, and enabling the development of more personalized treatment plans.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00280-024-04723-y.
Keywords: CML, Imatinib, CYP2C8*3, ABCG2
Introduction
Chronic myeloid leukemia (CML) is a malignant tumor that results from the abnormal proliferation of hematopoietic stem cells within the bone marrow. It constitutes approximately 15% of leukemia cases in adults [1].
A hallmark of this disease is the “Philadelphia (Ph) chromosome”, which is produced by a balanced reciprocal translocation between the long arms of chromosomes 9 and 22 [t (9; 22) (q34; q11)] [2].
This genetic translocation transposes the Abelson murine leukemia 1 (ABL1) gene from chromosome 9 to the breakpoint cluster region (BCR) gene on chromosome 22, resulting in the generation of the BCR-ABL1 fusion oncogene that encodes the BCR-ABL1 oncoprotein [3].
The treatment of CML was revolutionized in 2001 with the introduction of imatinib, a tyrosine kinase inhibitor (TKI). This advancement transformed CML from a life-threatening illness into a manageable condition for most patients [4, 5].
Despite this significant improvement in CML outcomes achieved with imatinib, some patients either experience disease progression during therapy or fail to respond adequately to the treatment [6].
Treatment failure with imatinib can be attributed to several mechanisms, including the upregulation of BCR-ABL1, the appearance of additional cytogenetic abnormalities, and mutations within the kinase domain of BCR-ABL1 [7]. Furthermore, variability in imatinib pharmacokinetics could influence its efficacy, as a correlation between imatinib exposure and clinical outcomes has been demonstrated [8–10].
Imatinib exhibits significant inter-patient variability in trough concentrations [11]. This variability may be attributed to differences in drug-metabolizing enzyme activity, the function of influx/efflux transporters [12], interaction with other drugs [13], and patient non-adherence [14].
It is predominantly metabolized into N-desmethyl imatinib (NDMI), by cytochrome P450 (CYP) 3A4 isozyme [15]. Furthermore, CYP2C8 is integral to imatinib metabolism, especially when CYP3A4 experiences auto-inhibition at imatinib’s steady state [16, 17].
Other isozymes, including CYP1A2, CYP2D6, CYP2C9 and CYP2C19, also contribute to imatinib metabolism, but their roles are relatively minor [18].
Active efflux of imatinib from cells is carried out by adenosine triphosphate-binding cassette (ABC) transporters, notably ABCB1 and ABCG2. Conversely, the uptake of imatinib into cells is facilitated by the human organic cation transporter-1 (OCT1), also known as solute carrier family 22 member 1 (SLC22A1) [19].
Thus, genetic polymorphisms in the CYP2C8 and ABCG2 genes are likely to affect intracellular or systemic concentrations of imatinib, potentially altering its therapeutic efficacy.
A notable gap in research existed regarding these polymorphisms in Egyptian CML patients. Therefore, this study aims to evaluate the influence of CYP2C8*3 (G416A; rs11572080) and ABCG2 C421A (rs2231142) single nucleotide polymorphisms (SNPs) on plasma trough concentration and therapeutic response to imatinib in Egyptian patients with CML.
Materials and methods
Study design
This observational, cross-sectional study was conducted at the Medical Oncology Department of the South Egypt Cancer Institute (SECI), Assiut, Egypt.
The study protocol was reviewed and approved by the Ethics Committee of the Faculty of Medicine at Assuit University, Egypt (Institutional Review Board number: 17200117) and was registered in the ClinicalTrials.gov database (Identification Number: NCT03262974).
Chemicals and kits
GeneJet Whole Blood Genomic DNA Purification Mini Kit (Thermo Scientific, Lithuania), COSMO PCR Master Mix (Willowfort, England), Forward and reverse primers (Macrogen, South Korea), Restriction enzymes: BseRI and HpyCH4III (New England Biolabs, USA), Gel loading dye purple (6X) (New England Biolabs, USA), DNA ladder (GeneDireX, Taiwan) were purchased. Agarose powder (Bioline, USA), Tris base, Boric acid, ethylenediaminetetraacetic acid (EDTA) and ethidium bromide (Sigma-Aldrich, USA).
Imatinib mesylate (CAS No. 220127-57-1) and propranolol hydrochloride (CAS No. 318-98-9, the internal standard) were purchased from AK Scientific, Inc., USA. Acetonitrile and methanol (Sigma-Aldrich, Germany) were of HPLC grade. Water was purified by a Milli-Q Gradient A10 water purification system (Merck Millipore, USA).
Patients
Patients were enrolled from the outpatient clinic of the Medical Oncology Department at the SECI, Egypt. The included patients had a diagnosis of Ph chromosome-positive CML, were aged over 18 years, and had been treated with imatinib for at least 12 months with good compliance to treatment. Exclusion criteria encompassed patients in accelerated or blastic phases, those taking medications that induce or inhibit liver microsomal enzymes (e.g., Ketoconazole, Phenytoin, and Valproic acid), individuals with poor compliance, and pregnant women. All patients provided informed consent before participation in the study.
Patient data, including age, sex, age at CML diagnosis, and BCR-ABL1 transcript levels at 12 months post-start of imatinib treatment, were collected during their regular follow-up visits.
The molecular response to imatinib was assessed using the International Scale, which quantifies the ratio of BCR-ABL1 transcripts to ABL1 transcripts, expressed as a percentage of BCR-ABL1 on a logarithmic scale. This scale corresponds to reductions of 2, 3, 4, 4.5, and 5 (equating to 1%, 0.1%, 0.01%, 0.0032%, and 0.001%, respectively) relative to the standardized baseline established in the International Randomized Study of Interferon versus STI571 (IRIS) [20].
In accordance with the European LeukemiaNet recommendations for CML treatment, treatment failure is defined as BCR-ABL1 levels exceeding 1% after 12 months of imatinib therapy [21].
Accordingly, patients in this study were classified into two groups based on their BCR-ABL1 transcript levels 12 months after initiating imatinib treatment: responders, with transcript levels of 1% or below, who continued imatinib therapy, and non-responders, with transcript levels above 1%, who were transitioned to second-generation TKIs such as nilotinib or dasatinib.
Blood sampling
Peripheral blood samples were collected from patients into EDTA tubes during their routine follow-up visits at the outpatient clinic, concurrent with their regular laboratory tests.
Two milliliters were collected from all patients for DNA extraction and subsequent genotyping. An additional two milliliters were collected from patients receiving imatinib for at least 12 months, approximately 24 ± 3 h after their last dose but before the next dose (trough sample), following 28 consecutive days of administration [9].
Samples were then centrifuged at 2500 RPM (1000 ×g) for 15 min [22] using a benchtop centrifuge (Rotofix 32 A, Hettich, Germany). Plasma samples obtained were stored at -80 °C until subsequent measurement of trough concentrations.
All blood samples were collected within the same timeframe; however, there was a lapse between the molecular response evaluation and the blood sample collection for each patient, which depended on the duration of their imatinib treatment.
DNA extraction and genotyping
Whole blood DNA was extracted using a commercial extraction kit following the manufacturer’s instructions. The concentration and purity of the extracted DNA were assessed using a microplate spectrophotometer (Epoch, BioTek Instruments Inc., USA). Ratios of A260/A280 ranging from 1.7 to 1.9 indicate pure template DNA, optimal for PCR [23]. Extracted DNA samples were stored at -80 °C until genotyping analysis.
The CYP2C8*3 (G416A; rs11572080) [24] and ABCG2 C421A (rs2231142) [25] polymorphisms were genotyped using the polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) method, as previously described. Detailed primer sequences, PCR thermal conditions, and restriction enzymes used are provided in the Online Resource 1.
The PCR was performed using a thermal cycler (Veriti 96 Well Thermal Cycler, Applied Biosystems, USA). The resulting PCR products were then digested with appropriate restriction enzymes according to the manufacturer’s protocol.
The digested products were separated by electrophoresis on 3% agarose gel using a horizontal electrophoresis unit (Biometra Compact M, Analytik Jena, Germany) and visualized under ultraviolet (UV) light at 312 nm wavelength using a UV transilluminator (UVstar, Analytik Jena, Germany). Gel images were captured and documented using a gel documentation system (BioDocAnalyze, Analytik Jena, Germany). DNA extraction and PCR-RFLP analyses were carried out at the Medical Research Center, Assiut University, Assiut, Egypt.
Measurement of plasma trough concentration of imatinib
Plasma trough concentrations were measured using the high-performance liquid chromatography with ultraviolet-diode array detection (HPLC-UV/DAD) method described previously [26]. The analysis was performed using a Waters 2695 Alliance Separations Module equipped with a 996 PDA detector (Waters Corporation, USA), with the detector wavelength set at 265 nm. Chromatographic separation was achieved on a C18 reversed-phase analytical column (Inertsil ODS 4 analytical column, 250 mm × 4.6 mm internal diameter, five µm particle size; GL Sciences, Japan) using isocratic elution. Chromatogram processing, data generation, and concentration calculations were conducted using Empower 3 chromatography data software (Waters Corporation, USA). These measurements were carried out at Nawah Scientific Inc., Cairo, Egypt.
Statistical analysis
Possible deviation from Hardy–Weinberg equilibrium (HWE) of various genotypes was assessed using Chi-square test.
The Shapiro–Wilk test was employed to evaluate the normality of variable distributions. Continuous variables that followed a normal distribution were reported as mean ± standard deviation (SD), whereas those deviating from normality were presented as median and range. Categorical variables were reported in terms of frequency and percentage.
Fisher’s exact test was utilized to compare the frequencies of various genotypes between imatinib responders and non-responders. Additionally, the unpaired Student’s t-test was applied to assess differences in plasma trough concentrations of imatinib across patients with different genotypes.
GraphPad Prism software version 9.5.1 (GraphPad Software Inc., USA) was used to perform statistical analyses and create graphical representations. Statistically significant differences were indicated by a p-value of less than 0.05.
Results
Patients’ characteristics
Table 1 presents the demographic and clinical characteristics of the patients. A total of 50 Egyptian patients with Ph chromosome-positive CML in the chronic phase were included in the study. Of whom, 30 (60%) were females. The mean age of the patients at enrollment was 43.84 ± 13.30 years. At the time of CML diagnosis, the mean age was 38.48 ± 13.25 years. Twenty-six patients (52%) were responders to imatinib, with a mean treatment duration of 5.58 ± 2.16 years. The remaining 24 patients (48%) were non-responders.
Table 1.
Demographic and clinical characteristics of Egyptian patients with CML (n = 50)
| Sex: | |
|
- Male, n (%) - Female, n (%) |
20 (40%) 30 (60%) |
| Age (years), mean ± SD | 43.84 ± 13.30 |
| Age at diagnosis (years), mean ± SD | 38.48 ± 13.25 |
| Molecular response to imatinib at 12 months: | |
|
- Responders (BCR-ABL1 transcript level ≤ 1%), n (%) - Non-responders (BCR-ABL1 transcript level > 1%), n (%) |
26 (52%) 24 (48%) |
| Duration of treatment with imatinib (years), mean ± SD | 5.58 ± 2.16 |
| BCR-ABL1transcript level at 12 month (%): | |
|
- Responders, median (range) - Non-responders, median (range) |
0.2 (0-0.9) 30 (2-100) |
Genotype and allele frequency
Table 2 presents the frequency of different genotypes and alleles of CYP2C8*3 G416A and ABCG2 C421A polymorphisms in our patients. For the CYP2C8*3 G416A polymorphism, the frequencies of the GG (CYP2C8*1/*1, homozygous wild type) and GA (CYP2C8*1/*3, heterozygous type) genotypes were 76% and 24%, respectively. The frequency of the variant allele (A allele) was 12%. For the ABCG2 C421A polymorphism, the frequencies of the CC (homozygous wild) and CA (heterozygous) genotypes was 78% and 22%, respectively. The frequency of the variant allele (A allele) was 11%. In the present study, we did not detect the AA (homozygous variant) genotype for either CYP2C8*3 G416A or ABCG2 C421A polymorphisms. All reported frequencies did not significantly deviate from the HWE (p > 0.05).
Table 2.
Genotype and allele frequencies of CYP2C8*3 and ABCG2 C421A polymorphisms among Egyptian patients with CML
| SNP | Genotype | Frequency (%) | Allele | Frequency (%) | HWE p-value |
|---|---|---|---|---|---|
| CYP2C8*3 (G416A; rs11572080) | GG | 76 | G | 88 | 0.3349 |
| GA | 24 | A | 12 | ||
| ABCG2 C421A (rs2231142) | CC | 78 | C | 89 | 0.3821 |
| CA | 22 | A | 11 |
Influence of different genotypes on imatinib trough concentrations
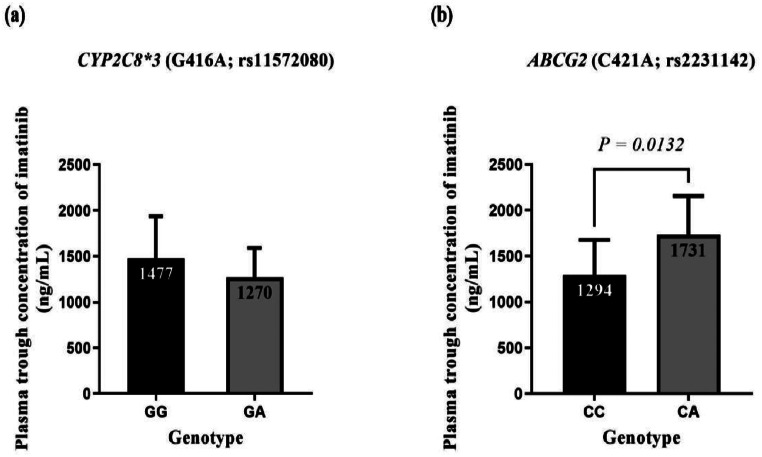
It was observed that there was no statistically significant difference in the mean plasma trough concentration of imatinib in patients carrying different genotypes of the CYP2C8*3 polymorphism (GG: 1477 ± 459.8 vs. GA: 1270 ± 321.5; p = 0.3995) (Fig. 1a).
Fig. 1.
Plasma trough concentrations of imatinib in different genotypes of CYP2C8*3 (a) and ABCG2 C421A (b) polymorphisms in CML patients
On the other hand, patients carrying the CA genotype of the ABCG2 C421A polymorphism had a statistically significant higher mean plasma trough concentration of imatinib compared to those carrying the CC genotype (CA: 1731 ± 424.7 ng/mL vs. CC: 1294 ± 381.3 ng/mL; p = 0.0132) (Fig. 1b).
Data are presented as mean ± SD. No statistically significant difference was found in the mean plasma trough concentration of imatinib between the different genotypes of CYP2C8*3 polymorphism (p = 0.3995). A statistically significant difference was observed in the mean plasma trough concentration of imatinib between the CC and CA genotypes of ABCG2 C421A polymorphism (p = 0.0132). Results were compared using the unpaired Student’s t-test.
Influence of different genotypes on molecular response of imatinib
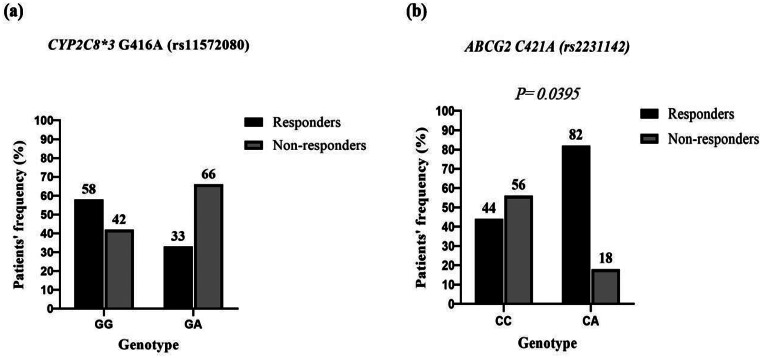
Regarding the genotypes of the CYP2C8*3 polymorphism, no statistically significant difference was observed in their distribution between responder and non-responder patients (p = 0.1902) (Fig. 2a).
Fig. 2.
Frequency of different genotypes of CYP2C8*3 (a) and ABCG2 C421A (b) polymorphisms in imatinib responders and non-responders
However, a statistically significant difference was noted between the two patient groups regarding the distribution of different genotypes of the ABCG2 C421A polymorphism with predominance of the CA genotype in responder patients (p = 0.0395) (Fig. 2b).
Assessment of molecular response to imatinib based on the BCR-ABL1 transcript level at 12 months. Responders (n = 26); Non-responders (n = 24). No statistically significant difference was found between the frequency of GG and GA genotypes of CYP2C8*3 in the two groups (p = 0.1902). However, There was a statistically significant difference between the frequency of CC and CA genotypes of ABCG2 C421A in the two groups (p = 0.0395). Results were compared using Fisher’s exact test.
Discussion
Despite the transformative impact of imatinib on the treatment of CML, a substantial proportion of CML patients (30–35%) exhibit inadequate response, either as suboptimal responders or as resistant to the treatment [27]. Various factors contribute to these outcomes [28], including genetic polymorphisms that impact drug metabolism and transport [29].
In this study, we focused on two key SNPs, CYP2C8*3 (G416A; rs11572080) and ABCG2 C421A (rs2231142), which may influence imatinib pharmacokinetics. These genetic variations can alter plasma trough concentrations of imatinib and impact treatment outcomes in Egyptian patients with chronic-phase CML.
Over the past three decades, many methods have been developed for SNP genotyping, with PCR-RFLP being a widely used and cost-effective option [30]. In this study, we utilized PCR-RFLP to genotype target SNPs in all 50 patients. However, this technique has limitations, including its dependence on specific restriction enzymes to distinguish alleles, which can restrict its effectiveness when suitable enzymes are unavailable or show non-specific binding. Additionally, the multi-step process is time-intensive, increasing the risk of contamination and error, and is less efficient than high-throughput methods like real-time PCR and next-generation sequencing [31].
Many authors recommend HPLC with UV detection for measuring imatinib plasma concentrations, as it is cost-effective, straightforward, and suitable for routine laboratory use. Studies have also shown that HPLC-UV provides results comparable to those obtained with LC-MS/MS [32, 33]. Additionally, incorporating a diode-array detector (DAD) into HPLC-UV systems improves specificity by allowing spectral comparisons and peak purity verification, making HPLC-UV/DAD sufficiently sensitive and specific for imatinib analysis [34].
The CYP2C8 enzyme, comprising 7% of the liver’s CYP content, is crucial for metabolizing about 20% of commonly prescribed medications. It is encoded by the CYP2C8 gene, situated on chromosome 10q24 [35].
This isozyme shows significant genetic diversity, with various SNPs identified, including CYP2C8*2, *3, and *4, which account for most of the non-synonymous variability of CYP2C8 in humans [36, 37].
Among these SNPs, CYP2C8*3 stands out as the most extensively studied functional polymorphism. It encompasses two non-synonymous variants, G416A (Arg139Lys; rs11572080) and A1196G (Lys399Arg; rs10509681), often found in complete or near-complete linkage disequilibrium [38].
In vitro studies suggest that CYP2C8*3 is linked to enhanced enzyme activity, which may expedite the conversion of imatinib to NDMI [11, 39].
However, in vivo studies indicate substrate-specific effects, including enhanced metabolism of drugs like pioglitazone and repaglinide, and reduced metabolism of ibuprofen [40].
In the context of our study on Egyptian CML patients, we did not observe a significant influence of the CYP2C8*3 polymorphism on imatinib’s plasma trough concentration or molecular response.
Nonetheless, we noted a trend towards lower imatinib concentrations in patients carrying the GA (CYP2C*1/*3) genotype, particularly prevalent among non-responders. These findings suggest a potential detrimental impact of this genotype on imatinib pharmacokinetics and clinical efficacy.
To our knowledge, this study is the first to investigate the impact of CYP2C8*3 (G416A; rs11572080) on imatinib therapy outcomes in Egyptian CML patients, highlighting the need for further validation through larger studies.
Results of the present study align with those of Verboom et al. [41], who similarly found no significant effect of the CYP2C8*3 on imatinib pharmacokinetics in Dutch patients with GIST or CML.
Conversely, studies by Barratt et al. [42] and Dalle Fratte et al. [43] reported increased metabolic ratios of NDMI/imatinib in CML and GIST patients carrying the CYP2C8*3 allele, contrasting with our findings. Further research in diverse populations and larger cohorts is crucial to elucidate the full spectrum of CYP2C8*3’s impact on imatinib metabolism and treatment response.
The ABCG2, also known as the breast cancer resistance protein, belongs to the subfamily G of the ABC efflux transporter superfamily and mediates ATP-dependent efflux of diverse molecules across cell membranes [44]. This protein is encoded by the ABCG2 gene, which is situated on chromosome 4q22.1 [45].
This transporter is extensively distributed in the human body. It operates in the apical membrane of enterocytes, where it restricts intestinal absorption; in the sinusoidal membrane of hepatocytes, where it facilitates hepatobiliary excretion; and in the apical membrane of proximal tubular cells in the kidney, where it contributes to uric acid elimination [46]. Furthermore, it significantly affects the pharmacokinetics of diverse compounds including anticancer drugs, antibiotics, antivirals and analgesics [47].
Numerous SNPs have been identified in the ABCG2 gene, with C421A (rs2231142) in exon 5 being extensively studied. This SNP involves a substitution of glutamine with lysine at codon 141 (Q141K) [48].
In both in vitro and in vivo studies, this polymorphism was reported to generally reduce ABCG2 protein expression. Additionally, some studies indicate that it may also diminish ATPase activity, leading to compromised transport function [49].
Given its location on the apical membrane of hepatocytes, ABCG2 is likely essential for the excretion of imatinib. Consequently, genetic variations in the ABCG2 gene may affect the pharmacokinetics and clinical response to imatinib [18]. However, the precise effect of the ABCG2 C421A SNP on imatinib’s plasma trough concentration and therapeutic response remains a subject of debate.
In this study, we found that patients with the CA genotype of the ABCG2 C421A polymorphism showed significantly higher plasma trough concentrations of imatinib compared to those with the CC genotype. Additionally, the CA genotype was more prevalent among responder patients, suggesting a potential role of this polymorphism in enhancing the therapeutic efficacy of imatinib.
Results of the present study align with those of Takahashi et al. [50], who reported higher imatinib trough concentrations in Japanese CML patients with the CA or AA genotypes compared to those with the CC genotype. However, they did not find a significant association between this SNP and treatment response.
Seong and his colleagues [18] identified a potential association between the ABCG2 C421A variant and an increased rate of major molecular response in Korean CML patients. However, they did not observe a significant effect on imatinib trough concentrations.
Jiang et al. [51] conducted a meta-analysis that included seven studies with nearly 2,200 patients, which further reinforced the association between the ABCG2 C421A variant allele and a higher clinical response rate.
Similarly, Alves et al. [52] observed that the CC genotype was associated with imatinib resistance, while the CA genotype was linked to a favorable response in Portuguese CML patients.
Findings of the current study are comparable with those of Cheng et al. [53], who examined the influence of the ABCG2 C421A polymorphism on imatinib plasma concentration and response in 190 Chinese CML patients. They found that individuals with the CA or AA genotype exhibited higher imatinib concentrations and more favorable cytogenetic and molecular responses compared to those with the CC genotype. This finding supports our observation that the CA genotype is linked to higher plasma trough concentrations of imatinib and an improved therapeutic response.
In contrast, several studies have found no significant association between the ABCG2 C421A SNP and imatinib response. For instance, Francis et al. [54] reported no significant impact of this SNP on imatinib trough concentration in Indian CML patients.
Similarly, Omran et al. [55] and Rajamani et al. [5] reported no significant influence of the ABCG2 C421A SNP on imatinib response in Egyptian and Indian CML patients, respectively. Additionally, Sabri et al. [56] found no significant association between ABCG2 gene expression and response to imatinib in their study on Egyptian CML patients.
Recent studies by Nouri et al. [57] and Mohammadi et al. [58] also concluded that the ABCG2 C421A SNP had no impact on imatinib response in Iranian CML patients.
The present study faced several limitations. The primary constraint was the small sample size, which might have hindered the identification of some genotypes within the patient population. Thus, large and multicenter studies are necessary. Furthermore, financial restrictions limited our investigation to a selected number of polymorphisms.
Conclusion
The current study revealed that the ABCG2 C421A (rs2231142) polymorphism significantly impacted both the plasma trough concentration and molecular response to imatinib in Egyptian patients with chronic-phase CML. Patients carrying the CA genotype showed higher plasma imatinib concentrations and more favorable treatment outcomes compared to those with the CC genotype. In contrast, the CYP2C8*3 (G416A; rs11572080) polymorphism did not significantly affect imatinib pharmacokinetics or clinical outcomes in the study population. These findings suggest that genotyping for the ABCG2 C421A SNP could be a valuable tool in optimizing imatinib therapy for CML patients, allowing for more personalized treatment strategies.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Author contributions
S.A.M., M.H.A., H.A.E., and A.M.R.H. conceptualized the study and supervised the work. H.A.E. and M.S.M. collected data and recruited patients. M.S.M. conducted genetic and HPLC analyses, performed statistical analysis, and wrote the original draft. All authors reviewed and approved the final manuscript and agreed to be accountable for all aspects of the work, ensuring its accuracy and integrity.
Funding
Open access funding provided by The Science, Technology & Innovation Funding Authority (STDF) in cooperation with The Egyptian Knowledge Bank (EKB). Financial support for this study was partially provided by the Grant Office of the Faculty of Medicine at Assiut University, Egypt, under Grant Number 2017-08-30-024-R2.
Data availability
No datasets were generated or analysed during the current study.
Declarations
Declaration of generative AI and AI-assisted technologies in the writing process
In the preparation of this manuscript, generative AI, specifically OpenAI’s ChatGPT, was utilized to support the writing process. ChatGPT was employed to assist with language refinement, editing, and the restructuring of certain passages to ensure clarity and adherence to scientific writing standards. The use of AI was guided by the authors, who provided all the intellectual content and made final decisions on the presentation and interpretation of the research findings. All AI-generated text was carefully reviewed and edited by the authors to align with the scientific rigor and standards expected of the manuscript. The final content of the manuscript is the result of the authors’ original work and expertise.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Cortes J, Pavlovsky C, Saußele S (2021) Chronic myeloid leukaemia. Lancet 398(10314):1914–1926. 10.1016/S0140-6736(21)01891-8 [DOI] [PubMed] [Google Scholar]
- 2.Kang ZJ, Liu YF, Xu LZ, Long ZJ, Huang D, Yang Y, Liu B, Feng JX, Pan YJ, Yan JS, Liu Q (2016) The Philadelphia chromosome in leukemogenesis. Chin J Cancer 35:48. 10.1186/s40880-016-0108-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kumar V, Singh P, Gupta SK, Ali V, Verma M (2022) Transport and metabolism of tyrosine kinase inhibitors associated with chronic myeloid leukemia therapy: a review. Mol Cell Biochem 477(4):1261–1279. 10.1007/s11010-022-04474-0 [DOI] [PubMed] [Google Scholar]
- 4.Harivenkatesh N, Kumar L, Bakhshi S, Sharma A, Kabra M, Velpandian T, Gogia A, Shastri SS, Biswas NR, Gupta YK (2017) Influence of MDR1 and CYP3A5 genetic polymorphisms on trough levels and therapeutic response of imatinib in newly diagnosed patients with chronic myeloid leukemia. Pharmacol Res 120:138–145. 10.1016/j.phrs.2017.03.003 [DOI] [PubMed] [Google Scholar]
- 5.Rajamani BM, Benjamin ES, Abraham A, Ganesan S, Lakshmi KM, Anandan S, Karathedath S, Varatharajan S, Mohanan E, Janet NB, Srivastava VM (2020) Plasma imatinib levels and ABCB1 polymorphism influences early molecular response and failure-free survival in newly diagnosed chronic phase CML patients. Sci Rep 10(1):20640. 10.1038/s41598-020-77691-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kavanagh S, Nee A, Lipton JH (2018) Emerging alternatives to tyrosine kinase inhibitors for treating chronic myeloid leukemia. Expert Opin Emerg Drugs 23(1):51–62. 10.1080/14728214.2018.1400461 [DOI] [PubMed] [Google Scholar]
- 7.Soverini S, Mancini M, Bavaro L, Cavo M, Martinelli G (2018) Chronic myeloid leukemia: the paradigm of targeting oncogenic tyrosine kinase signaling and counteracting resistance for successful cancer therapy. Mol Cancer 17:49. 10.1186/s12943-018-0808-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Larson RA, Druker BJ, Guilhot F, O’Brien SG, Riviere GJ, Krahnke T, Gathmann I, Wang Y (2008) Imatinib pharmacokinetics and its correlation with response and safety in chronic-phase chronic myeloid leukemia: a subanalysis of the IRIS study. Blood 111(8):4022–4028. 10.1182/blood-2007-10-116475 [DOI] [PubMed] [Google Scholar]
- 9.Guilhot F, Hughes TP, Cortes J, Druker BJ, Baccarani M, Gathmann I, Hayes M, Granvil C, Wang Y (2012) Plasma exposure of imatinib and its correlation with clinical response in the Tyrosine Kinase Inhibitor Optimization and selectivity trial. Haematologica 97(5):731–738. 10.3324/haematol.2011.056200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Verheijen RB, Yu H, Schellens JH, Beijnen JH, Steeghs N, Huitema AD (2017) Practical recommendations for therapeutic drug monitoring of kinase inhibitors in oncology. Clin Pharmacol Ther 102(5):765–776. 10.1002/cpt.693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farag S, Verheijen RB, Martijn Kerst J, Cats A, Huitema AD, Steeghs N (2017) Imatinib pharmacokinetics in a large observational cohort of gastrointestinal stromal tumour patients. Clin Pharmacokinet 56:287–292. 10.1007/s40262-016-0442-2 [DOI] [PubMed] [Google Scholar]
- 12.Barratt DT, Somogyi AA (2017) Role of pharmacogenetics in personalised imatinib dosing. Transl Cancer Res 6(Suppl 10). 10.21037/tcr.2017.06.01. S1541-S1557
- 13.Shao J, Markowitz JS, Bei D, An G (2014) Enzyme-transporter-mediated drug interactions with small molecule tyrosine kinase inhibitors. J Pharm Sci 103(12):3810–3833. 10.1002/jps.24126 [DOI] [PubMed] [Google Scholar]
- 14.Lankheet NA, Desar IM, Mulder SF, Burger DM, Kweekel DM, van Herpen CM, van der Graaf WT, van Erp NP (2017) Optimizing the dose in cancer patients treated with imatinib, sunitinib and pazopanib. Br J Clin Pharmacol 83(10):2195–2204. 10.1111/bcp.13328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Filppula AM, Laitila J, Neuvonen PJ, Backman JT (2012) Potent mechanism-based inhibition of CYP3A4 by Imatinib explains its liability to interact with CYP3A4 substrates. Br J Pharmacol 165(8):2787–2798. 10.1111/j.1476-5381.2011.01717.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nebot N, Crettol S, d’Esposito F, Tattam B, Hibbs DE, Murray M (2010) Participation of CYP2C8 and CYP3A4 in the N-demethylation of imatinib in human hepatic microsomes. Br J Pharmacol 161(5):1059–1069. 10.1111/j.1476-5381.2010.00924.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Filppula AM, Neuvonen M, Laitila J, Neuvonen PJ, Backman JT (2013) Autoinhibition of CYP3A4 leads to important role of CYP2C8 in imatinib metabolism: variability in CYP2C8 activity may alter plasma concentrations and response. Drug Metab Dispos 41(1):50–59. 10.1124/dmd.112.046094 [DOI] [PubMed] [Google Scholar]
- 18.Seong SJ, Lim M, Sohn SK, Moon JH, Oh SJ, Kim BS, Ryoo HM, Chung JS, Joo YD, Bang SM, Jung CW (2013) Influence of enzyme and transporter polymorphisms on trough imatinib concentration and clinical response in chronic myeloid leukemia patients. Ann Oncol 24(3):756–760. 10.1093/annonc/mds501 [DOI] [PubMed] [Google Scholar]
- 19.Peng B, Lloyd P, Schran H (2005) Clinical pharmacokinetics of imatinib. Clin Pharmacokinet 44:879–894. 10.2165/00003088-200544080-00002 [DOI] [PubMed] [Google Scholar]
- 20.Cross NC, White HE, Müller MC, Saglio G, Hochhaus A (2012) Standardized definitions of molecular response in chronic myeloid leukemia. Leukemia 26(10):2172–2175. 10.1038/leu.2012.104 [DOI] [PubMed] [Google Scholar]
- 21.Hochhaus A, Baccarani M, Silver RT, Schiffer C, Apperley JF, Cervantes F, Clark RE, Cortes JE, Deininger MW, Guilhot F, Hjorth-Hansen H (2020) European LeukemiaNet 2020 recommendations for treating chronic myeloid leukemia. Leukemia 34(4):966–984. 10.1038/s41375-020-0776-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malhotra H, Sharma P, Bhargava S, Rathore OS, Malhotra B, Kumar M (2014) Correlation of plasma trough levels of imatinib with molecular response in patients with chronic myeloid leukemia. Leuk Lymphoma 55(11):2614–2619. 10.3109/10428194.2014.898748 [DOI] [PubMed] [Google Scholar]
- 23.Bruijns B, Hoekema T, Oomens L, Tiggelaar R, Gardeniers H (2022) Performance of spectrophotometric and fluorometric DNA quantification methods. Analytica 3(3):371–384. 10.3390/analytica3030031 [Google Scholar]
- 24.Pechandova K, Buzkova H, Matouskova O, Perlik F, Slanar O (2012) Genetic polymorphisms of CYP2C8 in the Czech Republic. Genet Test Mol Biomarkers 16(7):812–816. 10.1089/gtmb.2011.0332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.El Mesallamy HO, Rashed WM, Hamdy NM, Hamdy N (2014) High-dose methotrexate in Egyptian pediatric acute lymphoblastic leukemia: the impact of ABCG2 C421A genetic polymorphism on plasma levels, what is next? J Cancer Res Clin Oncol 140:1359–1365. 10.1007/s00432-014-1695-1 [DOI] [PubMed] [Google Scholar]
- 26.Kaza M, Piorkowska E, Filist M, Rudzki PJ (2016) HPLC-UV assay of imatinib in human plasma optimized for bioequivalence studies. Acta Pol Pharm 73(6):1495–1503 [PubMed] [Google Scholar]
- 27.Ankathil R, Azlan H, Dzarr AA, Baba AA (2018) Pharmacogenetics and the treatment of chronic myeloid leukemia: how relevant clinically? An update. Pharmacogenomics 19(5):475–493. 10.2217/pgs-2018-0017 [DOI] [PubMed] [Google Scholar]
- 28.Volpe G, Panuzzo C, Ulisciani S, Cilloni D (2009) Imatinib resistance in CML. Cancer Lett 274(1):1–9. 10.1016/j.canlet.2008.08.002 [DOI] [PubMed] [Google Scholar]
- 29.Angelini S, Soverini S, Ravegnini G, Barnett M, Turrini E, Thornquist M, Pane F, Hughes TP, White DL, Radich J, Kim DW (2013) Association between Imatinib transporters and metabolizing enzymes genotype and response in newly diagnosed chronic myeloid leukemia patients receiving imatinib therapy. Haematologica 98(2):193–200. 10.3324/haematol.2012.068676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kratochwil CF, Kautt AF, Rometsch SJ, Meyer A (2022) Benefits and limitations of a new genome-based PCR‐RFLP genotyping assay (GB‐RFLP): a SNP‐based detection method for identification of species in extremely young adaptive radiations. Ecol Evol 12(3):e8751. 10.1002/ece3.8751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jordan D, Mills D (2021) Past, present, and future of DNA typing for analyzing human and non-human forensic samples. Front Ecol Evol 9:646130. 10.3389/fevo.2021.646130 [Google Scholar]
- 32.Caterino M, Casadei GM, Arvonio R, De Francia S, Pirro E, Piccione FM, Pane F, Ruoppolo M (2013) Quantification of imatinib plasma levels in patients with chronic myeloid leukemia: comparison between HPLC–UV and LC–MS/MS. Int J Pept Res Ther 19:109–116. 10.1007/s10989-012-9321-0 [Google Scholar]
- 33.Miura M, Takahashi N (2016) Routine therapeutic drug monitoring of tyrosine kinase inhibitors by HPLC–UV or LC–MS/MS methods. Drug Metab Pharmacokinet 31(1):12–20. 10.1016/j.dmpk.2015.09.002 [DOI] [PubMed] [Google Scholar]
- 34.Roth O, Spreux-Varoquaux O, Bouchet S, Rousselot P, Castaigne S, Rigaudeau S, Raggueneau V, Therond P, Devillier P, Molimard M, Maneglier B (2010) Imatinib assay by HPLC with photodiode-array UV detection in plasma from patients with chronic myeloid leukemia: comparison with LC-MS/MS. Clin Chim Acta 411(3–4):140–146. 10.1016/j.cca.2009.10.007 [DOI] [PubMed] [Google Scholar]
- 35.Aquilante CL, Niemi M, Gong L, Altman RB, Klein TE (2013) PharmGKB summary: very important pharmacogene information for cytochrome P450, family 2, subfamily C, polypeptide 8. Pharmacogenet Genomics 23(12):721–728. 10.1097/FPC.0000000000000004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou Y, Ingelman-Sundberg M, Lauschke VM (2017) Worldwide distribution of cytochrome P450 alleles: a meta-analysis of population-scale sequencing projects. Clin Pharmacol Ther 102(4):688–700. 10.1002/cpt.685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abudahab S, Hakooz N, Tobeh N, Gogazeh E, Gharaibeh M, Al-Eitan L, Zihlif M, Dajani R (2022) Variability of CYP2C8 polymorphisms in three Jordanian populations: circassians, chechens, and Jordanian-Arabs. J Immigr Minor Health 24(5):1167–1176. 10.1007/s10903-021-01275-2 [DOI] [PubMed] [Google Scholar]
- 38.Tornio A, Backman JT (2018) Cytochrome P450 in pharmacogenetics: an update. Adv Pharmacol 83:3–32. 10.1016/bs.apha.2018.03.001 [DOI] [PubMed] [Google Scholar]
- 39.Khan MS, Barratt DT, Somogyi AA (2016) Impact of CYP2C8*3 polymorphism on in vitro metabolism of imatinib to N-desmethyl imatinib. Xenobiotica 46(3):278–287. 10.3109/00498254.2015.1060367 [DOI] [PubMed] [Google Scholar]
- 40.Aquilante CL, Kosmiski LA, Bourne DW, Bushman LR, Daily EB, Hammond KP, Hopley CW, Kadam RS, Kanack AT, Kompella UB, Le M (2013) Impact of the CYP2C8*3 polymorphism on the drug–drug interaction between gemfibrozil and pioglitazone. Br J Clin Pharmacol 75(1):217–226. 10.1111/j.1365-2125.2012.04328.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Verboom MC, Visser L, Kouwen S, Swen JJ, Diepstraten J, Posthuma WF, Gelderblom H, Van Lammeren D, Wilms EB (2017) Influence of CYP2C8 polymorphisms on imatinib steady-state trough level in chronic myeloid leukemia and gastrointestinal stromal tumor patients. Pharmacogenet Genomics 27(6):223–226. 10.1097/FPC.0000000000000282 [DOI] [PubMed] [Google Scholar]
- 42.Barratt DT, Cox HK, Menelaou A, Yeung DT, White DL, Hughes TP, Somogyi AA (2017) CYP2C8 genotype significantly alters imatinib metabolism in chronic myeloid leukemia patients. Clin Pharmacokinet 56:977–985. 10.1007/s40262-016-0505-y [DOI] [PubMed] [Google Scholar]
- 43.Dalle Fratte C, Gagno S, Roncato R, Polesel J, Zanchetta M, Buzzo M, Posocco B, De Mattia E, Borsatti R, Puglisi F, Foltran L (2023) CYP2D6 and CYP2C8 pharmacogenetics and pharmacological interactions to predict imatinib plasmatic exposure in GIST patients. Br J Clin Pharmacol 89(3):1089–1098. 10.1111/bcp.15572 [DOI] [PubMed] [Google Scholar]
- 44.Kukal S, Guin D, Rawat C, Bora S, Mishra MK, Sharma P, Paul PR, Kanojia N, Grewal GK, Kukreti S, Saso L (2021) Multidrug efflux transporter ABCG2: expression and regulation. Cell Mol Life Sci 78:6887–6939. 10.1007/s00018-021-03910-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bruckmueller H, Cascorbi I (2021) ABCB1, ABCG2, ABCC1, ABCC2, and ABCC3 drug transporter polymorphisms and their impact on drug bioavailability: what is our current understanding? Expert Opin Drug Metab Toxicol 17(4):369–396. 10.1080/17425255.2021.1896652 [DOI] [PubMed] [Google Scholar]
- 46.Woodward OM, Köttgen A, Coresh J, Boerwinkle E, Guggino WB, Köttgen M (2009) Identification of a urate transporter, ABCG2, with a common functional polymorphism causing gout. Proc Natl Acad Sci U S A 106(25):10338–11042. 10.1073/pnas.0902308106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Safar Z, Kis E, Erdo F, Zolnerciks JK, Krajcsi P (2019) ABCG2/BCRP: variants, transporter interaction profile of substrates and inhibitors. Expert Opin Drug Metab Toxicol 15(4):313–328. 10.1080/17425255.2019.1574854 [DOI] [PubMed] [Google Scholar]
- 48.Fohner AE, Brackman DJ, Giacomini KM, Altman RB, Klein TE (2017) PharmGKB summary: very important pharmacogene information for: ABCG2. Pharmacogenet Genomics 27(11):420–427. 10.1097/FPC.0000000000000348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Woodward OM, Tukaye DN, Cui J, Greenwell P, Constantoulakis LM, Parker BS, Rao A, Köttgen M, Maloney PC, Guggino WB (2013) Gout-causing Q141K mutation in ABCG2 leads to instability of the nucleotide-binding domain and can be corrected with small molecules. Proc Natl Acad Sci U S A 110(13):5223–5228. 10.1073/pnas.1219808110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takahashi N, Miura M, Scott SA, Kagaya H, Kameoka Y, Tagawa H, Saitoh H, Fujishima N, Yoshioka T, Hirokawa M, Sawada K (2010) Influence of CYP3A5 and drug transporter polymorphisms on imatinib trough concentration and clinical response among patients with chronic phase chronic myeloid leukemia. J Hum Genet 55(11):731–737. 10.1038/jhg.2010.108 [DOI] [PubMed] [Google Scholar]
- 51.Jiang ZP, Zhao XL, Takahashi N, Angelini S, Dubashi B, Sun L, Xu P (2017) Trough concentration and ABCG2 polymorphism are better to predict imatinib response in chronic myeloid leukemia: a meta-analysis. Pharmacogenomics 18(1):35–56. 10.2217/pgs-2016-0047 [DOI] [PubMed] [Google Scholar]
- 52.Alves R, Gonçalves AC, Jorge J, Marques G, Ribeiro AB, Tenreiro R, Coucelo M, Diamond J, Oliveiros B, Pereira A, Freitas-Tavares P (2022) Genetic variants of ABC and SLC transporter genes and chronic myeloid leukemia: impact on susceptibility and prognosis. Int J Mol Sci 23(17):9815. 10.3390/ijms23179815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cheng F, Cui Z, Li Q, Chen S, Li W, Zhang Y (2024) Influence of genetic polymorphisms on imatinib concentration and therapeutic response in patients with chronic-phase chronic myeloid leukemia. Int Immunopharmacol 133:112090. 10.1016/j.intimp.2024.112090 [DOI] [PubMed] [Google Scholar]
- 54.Francis J, Dubashi B, Sundaram R, Pradhan SC, Chandrasekaran A (2015) A study to explore the correlation of ABCB1, ABCG2, OCT1 genetic polymorphisms and trough level concentration with imatinib mesylate-induced thrombocytopenia in chronic myeloid leukemia patients. Cancer Chemother Pharmacol 76:1185–1189. 10.1007/s00280-015-2871-8 [DOI] [PubMed] [Google Scholar]
- 55.Omran MM, Abdelfattah R, Moussa HS, Alieldin N, Shouman SA (2020) Association of the trough, peak/trough ratio of imatinib, pyridine–N-oxide imatinib and ABCG2 SNPs 34 G > A and SLCO1B3 334 T > G with imatinib response in Egyptian chronic myeloid leukemia patients. Front Oncol 10:1348. 10.3389/fonc.2020.01348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sabri A, Omran MM, Azim SA, Abdelfattah R, Allam RM, Shouman SA (2023) Role of IDH1 (R132) mutation on imatinib toxicity and effect of ABCG2/OCT1 expression on N-desmethyl imatinib plasma level in Egyptian chronic myeloid leukemia patients. Drug Res (Stuttg) 73(3):146–155. 10.1055/a-1924-7746 [DOI] [PubMed] [Google Scholar]
- 57.Nouri N, Mehrzad V, Khalaj Z, Zaker E, Zare F, Abbasi E, Khosravi M, Kalantar SM, Salehi M (2023) Effects of ABCG2 C421A and ABCG2 G34A genetic polymorphisms on clinical outcome and response to imatinib mesylate, in Iranian chronic myeloid leukemia patients. Egypt J Med Hum Genet 24:1. 10.1186/s43042-023-00467-0 [Google Scholar]
- 58.Mohammadi F, Rostami G, Hamid M, Shafiei M, Azizi M, Bahmani H (2023) Association of ABCB1, ABCG2 drug transporter polymorphisms and smoking with disease risk and cytogenetic response to imatinib in chronic myeloid leukemia patients. Leuk Res 126:107021. 10.1016/j.leukres.2023.107021 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No datasets were generated or analysed during the current study.