ABSTRACT
Shenxian–Shengmai (SXSM) is a Chinese patent medicine used in the treatment of sick sinus syndrome (SSS). However, its active chemical compounds and the underlying molecular mechanisms remain unclear. In this study, we researched the underlying mechanisms of SXSM in treating SSS. We conducted network analysis and molecular docking to identify the small molecules and core targets responsible for the therapeutic efficacy of SXSM on SSS. In vitro experiments were performed to verify the potential therapeutic mechanism. Network pharmacological analysis identified 17 core targets. Among these, BMP4, KCNH2, KCNMA1, and KCNQ1 were identified to be involved in various biological processes, such as the formation and regulation of the cardiac pacemaking system and potassium ion transmembrane transport. The experimental analysis revealed that SXSM could upregulate the expression of the Bmp4/Tbx3/Hcn4 pathway and the expression of Kcnh2, Kcnma1, and Kcnq1 channels, which protected and improved the pacemaking function of pacemaker cells (P cells) and increased the heart rate. These findings provide a scientific basis in the study of the mechanism of traditional Chinese medicine in the treatment of SSS.
Keywords: BMP4, molecular docking, network pharmacology, potassium ion channel, sick sinus syndrome
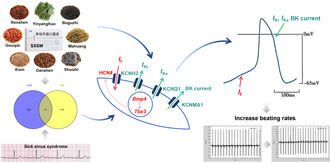
Shenxian–Shengmai (SXSM) is the first Chinese patent medicine that has been specially designed for sick sinus syndrome. It could increase the expression of the Bmp4/Tbx3/Hcn4 pathway and Kcnh2, Kcnq1, Kcnma1 potassium channels. These regulations may shorten the action potential duration of P cells and increase the heart rate.
1. Introduction
Sick sinus syndrome (SSS) refers to a variety of arrhythmias, such as sinus bradycardia, sinus arrest, and sinoatrial block, as the main manifestations caused by the disorder of the pacemaking and/or conduction function of the sinus node tissues. These disorders reduce the heart rate, which may result in major cardiovascular events, thromboembolism, inadequate heart rate response to stress or exercise (chronotropic incompetence), or any other symptom requiring pacemaker implantation (De Ponti et al. 2018). SSS is a commonly reported arrhythmia and, frequently, the cause for cardiac consultation. With the aging of the population, the number of newly diagnosed cases of SSS in the United States is projected to increase from 78,000 in 2012 to 172,000 by 2060 (Chen and Wu 2023). The clinical manifestations of SSS include lightheadedness, palpitation, dyspnea on exertion, chest tightness, and chronic fatigue. In severe cases, consciousness disturbance, syncope, and even death have been reported (Makita et al. 2014). Because SSS refers to a series of diseases, it involves several different pathophysiological mechanisms, including degenerative fibrosis, inherited primary arrhythmia syndromes caused by gene mutations, ion channel diseases, atrial myopathies such as amyloidosis and connective tissue diseases, and secondary metabolic disorders, among others. Therefore, the etiological and effective treatment of SSS is of great significance. The goal of treatment in SSS is to increase the heart rate, normalize cardiac output, and maintain the perfusion of the brain and other terminal organs to meet physiological needs. For patients with symptoms directly attributable to SSS, cardiac pacing can increase the heart rate and improve the related symptoms. However, pacemaker therapy cannot be fully popularized because of the high medical cost involved. At the same time, although pacemaker implantation is a relatively low‐risk cardiac procedure, it may be challenged by procedural complications and death directly related to the implantation; also, the implanted battery and leads have been reported to have long‐term management implications (Strathmore et al. 2017; Lee et al. 2018; Montgomery and Ellis 2018; Liu et al. 2020). In addition, several studies have estimated that about 30% of pacemakers have been implanted in other indications besides Class I and IIa. In this group of patients, the continuous need for pacing remains debatable, and how the pacing treatment can be interrupted remains unclear. In patients unwilling to undergo pacemaker implantation and in those who are not candidates for permanent pacing, oral medicine can be considered for the treatment of symptomatic SSS (Kusumoto et al. 2019). Currently, no mainstream medicine is deemed suitable for the long‐term treatment of SSS, primarily owing to the limitation of the dosage forms and the side effects (Li, Zhang, and Shuai 2014; Greene et al. 2018). Therefore, for patients receiving conservative treatment, scientifically proven safe and effective traditional Chinese medicine modalities are urgently needed.
Shenxian–Shengmai (SXSM) is the first Chinese patent medicine that has been specially designed for SSS with precise and effective formulation and quality. It is also known to exert curative effects on SSS, with minimal side effects (Hu, Chen, and Hua 2012; Hu et al. 2016; Liu et al. 2018). It is composed of 8 Chinese medicinal materials, namely Ginseng Radix et Rhizoma (Renshen), Epimedii Folium (Yinyanghuo), Psoraleae Fructus (Buguzhi), Lycii Fructus (Gouqizi), Ephedrae Herba (Mahuang), Asari Radix et Rhizoma (Xixin), Salviae Miltiorrhizae Radix et Rhizoma (Danshen) (Table 1), and Hirudo (Shuizhi). However, it involves numerous chemical compounds and a complicated metabolic process, which makes it difficult to explore its therapeutic targets and the action mechanisms against SSS.
TABLE 1.
Ingredients of SXSM used with accepted names.
| Latin name of herbal drugs | Chinese name | Latin name of medicinal plants | Family | Medication part |
|---|---|---|---|---|
| Ginseng Radix et Rhizoma | Renshen | Panax ginseng C. A. Mey. | Araliaceae | Root |
| Epimedii Folium | Yinyanghuo | Epimedium brevicornu Maxim. | Berberidaceae | Leaf |
| Psoraleae Fructus | Buguzhi | Psoralea corylifolia L. | Leguminosae | Berry |
| Lycii Fructus | Gouqizi | Lycium barbarum L. | Solanaceae | Berry |
| Ephedrae Herba | Mahuang | Ephedra intermedia Schrenk et C. A. Mey. | Ephedraceae | Stem |
| Asari Radix et Rhizoma | Xixin | Asarum sieboldii Miq. | Aristolochiaceae | Root |
| Salviae Miltiorrhizae Radix et Rhizoma | Danshen | Salvia miltiorrhiza Bge. | Lamiaceae | Root |
Traditional Chinese medicine prescription contains several natural chemical ingredients that are a valuable source of Chinese original drug research. The treatment of complex diseases needs to develop from a single target to a comprehensive and systematic network regulation, which is the advantage of traditional Chinese medicine treatment. In recent years, improvement in the understanding of the relationship between medicine targets and biological networks has assisted in the identification of several drugs with multiple targets that can interact with each other from the perspective of biological regulatory networks. Consequently, the concept of “network pharmacology” has been proposed. The present study employed the network pharmacology method as a comprehensive and systematic analysis tool and aided in the identification of active compounds present in SXSM, the exploration of the associated targets, and the analysis of the underlying mechanism involved in the treatment of SSS at the molecular level. Following this, molecular docking and in vitro experiments were employed to explain the main underlying mechanism.
2. Materials and Methods
2.1. Network Pharmacology Analysis
2.1.1. Collection of Compounds and Targets of SXSM
Six herbs of SXSM (namely, Ginseng Radix et Rhizoma, Epimedii Folium, Lycii Fructus, Ephedrae Herba, Asari Radix et Rhizoma, and Salviae Miltiorrhizae Radix et Rhizoma) were searched using the TCMSP (https://old.tcmsp‐e.com/tcmsp.php). The active compounds were screened according to oral availability (OB) ≥ 30% and drug‐like properties (DL) ≥ 0.18 (Ru et al. 2014). Data of the compounds of Psoraleae Fructus and Hirudo were collected from the literature, and the active compounds were screened by GI absorption, Lipinski, Ghose, Veber, Egan, and Muegge on the SwissADME database (http://www.swissadme.ch/). The SMILES molecular format of the compounds was uploaded to the SwissTargetPrediction database (http://www.swissadme.ch/) to investigate the targets (Gfeller et al. 2014).
2.1.2. Collection of the Targets of SSS
The keywords “sick sinus syndrome,” “sinus node dysfunction,” “bradyarrhythmia,” and “bradycardia” were searched in OMIM (https://omim.org/), DrugBank (https://go.drugbank.com/), and DisGeNET (https://www.disgenet.org/) databases. The results obtained were combined with those from the literature.
2.1.3. Protein–Protein Interaction (PPI) Network Construction
The potential targets of SXSM and SSS‐related targets were intersected to obtain the common targets with reference to the online Venn 2.1 diagram (http://bioinformatics.psb.ugent.be/webtools/Venn/). The protein name of the common targets was uploaded to the String database (https://cn.string‐db.org/), choose “homo sapiens” for “organisms”, choose “0.4” for “minimum required interaction score” and hide disconnected nodes in the network to obtain the PPI network information. The analysis results were saved in the “TSV” format. The results were input into the Cytascape 3.7.2 software, and the network analyzer function was selected under the “tools” option to analyze the topological parameters of each node in the network (Shannon et al. 2003; Liu et al. 2023). The core targets were screened according to the condition such that the degree, betweenness centrality, and closeness centrality attribute values were greater than the median values.
2.1.4. GO Enrichment and KEGG Pathway Analysis
The core targets were further analyzed with reference to the DAVID 6.8 database (https://David.ncifcrf.gov/) for gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses (Huang Da, Sherman, and Lempicki 2009a, 2009b). We selected biological process (BP), cellular component (CC), and molecular function (MF) for GO enrichment analyses and performed KEGG enrichment analyses to target the gene pathway. Rank from small to large according to the p value, and the top 20 results (p < 0.05) were added to the bar charts.
2.2. Molecular Docking
The target protein names of KCNH2, KCNQ1, and KCNMA1 were entered in the RCSB PDB database to search for and select the appropriate protein crystal structure and then saved in the pdb format. The target protein name of BMP4 was entered in AlphaFold Protein Structure Database to download the pdb file of the predicted structure. The information about target proteins is shown in Table S1. The PyMOL 2.4.2 software was used to remove water and phosphate from the protein molecules. The PubChem database was employed to download the compound structures, and Chem3D software was used to create the corresponding 3D structures. AutoDockTools 1.5.6 software was used to convert the pdb format of the compound and the target protein file into the pdbqt format, and the active pockets were searched. The Vina script was used to calculate the molecular binding energy and perform molecular docking (Trott and Olson 2010). Finally, the visual analysis was performed with PyMOL 2.4.2 and Discovery Studio 2021.
2.3. In Vitro Experiments
2.3.1. Reagents
SXSM was purchased from Buchang Pharmaceutical Co. Ltd. (Shandong, China, CFDA national drug standard no. WS3‐065 (Z‐010)‐2003 (Z); CFDA approval no. Z20080183; batch no. 1010466770). All Chinese medicinal materials of SXSM were purchased from Anhui Guangyintang Chinese Medicine Co. Ltd. and AnguoKangshenghua Medicine Ltd. (Baoding, China). The cardiomyocyte culture medium, CardioExcyte 96‐electrode plate, and the cardiomyocyte plating solution were obtained from Help Stem Cell Innovation (Nanjing, China). Dulbecco's Modified Eagle's medium (DMEM)/F12 and penicillin/streptomycin were obtained from Hyclone (USA), and fetal bovine serum was obtained from Sijiqing Biotechnology (Beijing, China). Phosphate‐buffered saline (PBS) was purchased from Solarbio Biotechnology (Beijing, China). siRNA sequences were designed and synthesized from Ribo Bio (Guangzhou, China). Lipofectamine RNAiMAX reagent and Trizol reagent were obtained from Thermo Fisher Scientific (USA). The TaKaRa PrimeScript RT Master Mix reagent and TaKaRa TB Green Premix Ex Taq II reagent were obtained from Takara Bio (Japan).
2.3.2. Cell Culture
The HL‐1 cell line was obtained from Procell Life Science & Technology Co. Ltd. (Hubei, China). These cells were seeded onto cell culture dishes and cultured in the DMEM/F12 medium supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin. The medium was refreshed every 48 h. These cells were maintained in an incubator at 37°C under a 5% CO2 atmosphere. The cells were further categorized into the control, si‐NC, model, SXSM, and si‐NC + SXSM groups. The cells in the si‐NC and si‐NC + SXSM groups were transfected with nonsense siRNA. The cells in the model and SXSM groups were transfected with the siRNA targets on Bmp4, Kcnh2, Kcnq1, and Kcnma1, respectively. At 24 h of transfection, the cells in the SXSM and si‐NC + SXSM groups were treated with 1 mL/L SXSM. The appropriate concentration used was inferred from the results of CCK‐8 cell viability determination, as suggested by Zhang et al. (2021). The hiPSC‐AMs were obtained from Help Stem Cell Innovation (Nanjing, China) and used to simulate P cells and cultured in the cardiomyocyte culture medium that was refreshed every 48 h (Mandel et al. 2012). The cells were stored in an incubator at 37°C under a 5% CO2 atmosphere. We assigned the cells to the control, si‐NC, model, and SXSM groups. The si‐NC cells were transfected with nonsense siRNA, while the cells in the model and SXSM groups were transfected with the siRNA targets of BMP4, KCNH2, KCNQ1, and KCNMA1, respectively. Finally, the cells in the SXSM group were treated with 1 mL/L SXSM after 24 h of siRNA transfection.
2.3.3. siRNA Transfection
Lipofectamine RNAiMAX reagent was used to transfect the siRNA‐carrying plasmid into HL‐1 cells and hiPSC‐AMs. We employed real‐time quantitative PCR (RT‐qPCR) to detect the interference efficiency of each siRNA sequence and obtain the siRNA sequence with better interference efficiency. The target sequences are presented in Table 2.
TABLE 2.
siRNA target sequence.
| Cell | siRNA | Target sequence |
|---|---|---|
| HL‐1 | si‐Bmp4 | CCAGACTAGTCCATCACAA |
| si‐Kcnh2 | GGACATCCTTATCAATTTC | |
| si‐Kcnq1 | CCATCATTGACCTCATCGT | |
| si‐Kcnma1 | CCTGTTAGATGGTCCCTTT | |
| hiPSC‐AMs | si‐BMP4 | CGAGACTGGTCCACCACAA |
| si‐KCNH2 | CCGTAAGTTCATCATCGCC | |
| si‐KCNQ1 | GCCAAGAAGAAATTCCAGC | |
| si‐KCNMA1 | GGACCAAGACGATGATGAT |
2.3.4. Real‐Time Quantitative PCR
The total RNA of HL‐1 cells was extracted with the Trizol reagent. Purified RNA was reverse‐transcribed by the TaKaRa PrimeScript RT Master Mix reagent under the following reaction conditions: reverse transcription reaction at 37°C for 15 min, followed by an inactivation reaction of reverse transcriptase at 85°C for 5 s. RT‐qPCR was performed using the TaKaRa TB Green Premix Ex Taq II reagent on the 7500 Real‐Time PCR System (Applied Biosystems; Thermo Fisher Scientific). The RT‐qPCR amplification procedure was as follows: stage 1: pre‐denaturation at 95°C for 30 s; Stage 2: denaturation at 95°C for 5 s; annealing at 60°C for 34 s; and repeat for 40 cycles. β‐actin was used for endogenous control, and the relative expression of mRNA was quantified using the 2−△△CT method. The primer sequences used in this study are listed in Table 3.
TABLE 3.
RT‐qPCR primer sequence.
| Target gene | Forward primer (5′–3′) | Reverse primer (5′–3′) |
|---|---|---|
| Bmp4 | ATTCCTGGTAACCGAATGCTG | CCGGTCTCAGGTATCAAACTAGC |
| Tbx3 | AGCGGGGTACAGAGATGGTC | TTGGCCTTTTTATCCAGTCCAG |
| Hcn4 | GACCTGACGATGCTGTTGCT | CGGAAGTTGAGGACCAAGTCA |
| Kcnh2 | CCTGCGATTTCCTGCATGG | AGAGGCGATCTCCACTTTGC |
| Kcnma1 | ATCCTCACCCTAATACGGACC | GGACCATCTAACAGGGCTAAC |
| Kcnq1 | CAGTGTCCTGTCCACTATTGAGC | ACCAGAGGCGGACCACATAT |
2.3.5. Measurement of Beating Rates and FPD
hiPSC‐AMs were seeded on the CardioExcyte 96‐electrode plate coated with the cardiomyocyte plating solution at the density of 5 × 104 cells/well and maintained in the cardiomyocyte culture medium. The medium was refreshed every 48 h and cultured continuously for 12 days until hiPSC‐AMs reached a stable state. At 48 h of siRNA transfection, the beating rates and FPD of hiPSC‐AMs in each group were analyzed by the CardioExcyte 96 system.
2.3.6. Data Analyses
All experiments were repeated thrice. Statistical analyses were performed with SPSS 26.0. The results obtained are presented as the mean ± SEM, and statistical comparisons were performed using one‐way analysis of variance (ANOVA). p < 0.05 was considered to indicate statistical significance.
3. Results
3.1. PPI Network for Common Targets of SXSM and SSS
The present study identified 947 potential targets of SXSM and 883 targets of SSS. A total of 55 common targets were identified (Figure 1). The PPI network is depicted in Figure 2. The layout is in accordance with the area and node color. The nodes represent the potential targets. The bigger and redder the nodes, the relatively larger are the degrees. The wider the edge between nodes, the greater the intensity of the red color and the stronger the interaction between the nodes. According to the topological parameters, 17 core targets were screened out, which included PPARG, COMT, MTOR, CACNA1C, KCNH2, BMP4, KCNQ1, KCNMA1, PTPN11, ADRB2, CACNA2D1, CACNA1H, CHRM1, ICAM1, VCAM1, CASP8, and CYP2D6.
FIGURE 1.
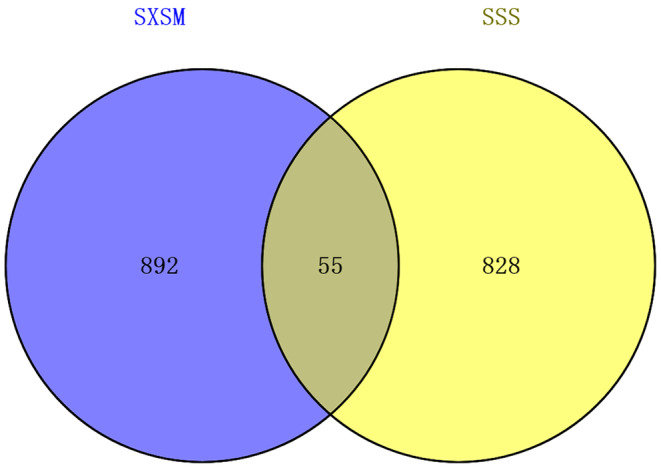
Venn diagram of the potential targets of SXSM and the targets of SSS.
FIGURE 2.
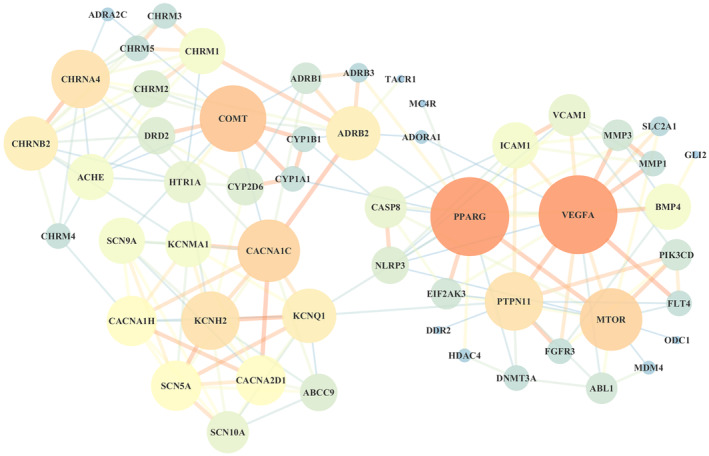
The PPI network of the potential targets of SXSM in SSS treatment.
3.2. Compound–Target Network
Cytoscape 3.7.2 was used to construct the compound‐target network of SXSM (Figure 3). The layout is in accordance with the shape and area of the nodes. The ellipse nodes represent herbs, the octagon represents active compounds, the hexagon nodes represent common active compounds of different herbs, and the diamond nodes represent the potential targets of SXSM. The larger the node, the closer is the connection with other nodes.
FIGURE 3.
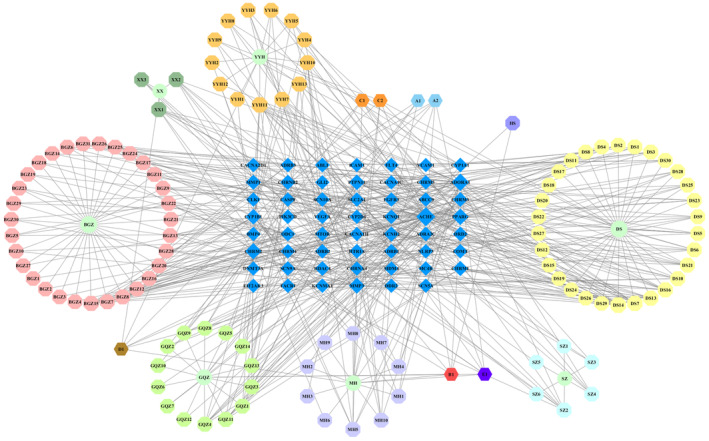
The compound‐target network of SXSM in SSS treatment.
3.3. GO Enrichment Analysis
Furthermore, 17 core targets were analyzed using the DAVID database for GO enrichment, and 81 GO entries were obtained (p < 0.05), which included 62 BP terms, 15 CC terms, and eight MF terms (Figure 4). BP related to SSS was mainly concentrated in the regulation of membrane potential, regulation of ion transmembrane transport, heart development, calcium ion import, potassium ion transmembrane transport, and aging. CC mainly involved the ion channel complex, plasma membrane protein complex, membrane raft, transmembrane transporter complex, voltage‐gated calcium channel complex, voltage‐gated potassium channel complex, synaptic membrane, sarcolemma, apical plasma membrane, and cell surface. MF mainly included scaffold protein binding, voltage‐gated calcium channel activity, drug binding, voltage‐gated potassium channel activity, alpha‐actinin binding, and protein domain‐specific binding.
FIGURE 4.
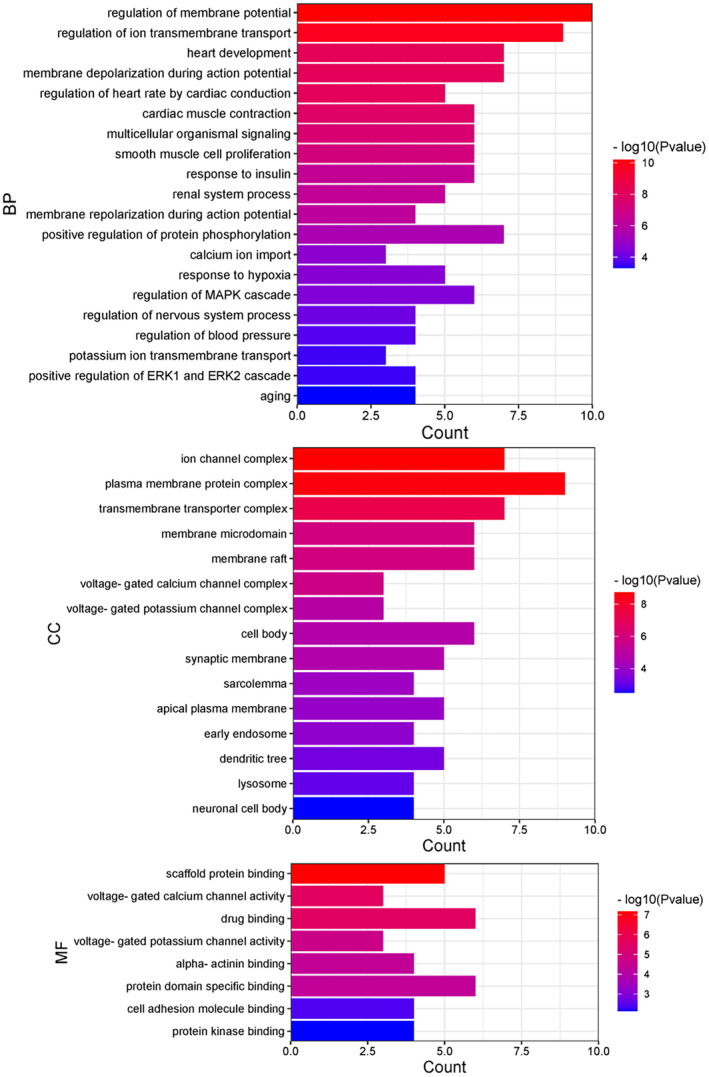
The GO enrichment analysis for 17 core targets, including BP, CC, and MF.
3.4. KEGG Pathway Analysis
The DAVID database was used to analyze 17 core targets for the KEGG pathway analysis, and 11 pathways were obtained (p < 0.05; Figure 5). Among these, the targets closely related to SSS were mainly concentrated in adrenergic signaling in cardiomyocytes, calcium‐signaling pathway, renin secretion, TNF‐signaling pathway, cGMP‐PKG‐signaling pathway, and cAMP‐signaling pathway.
FIGURE 5.
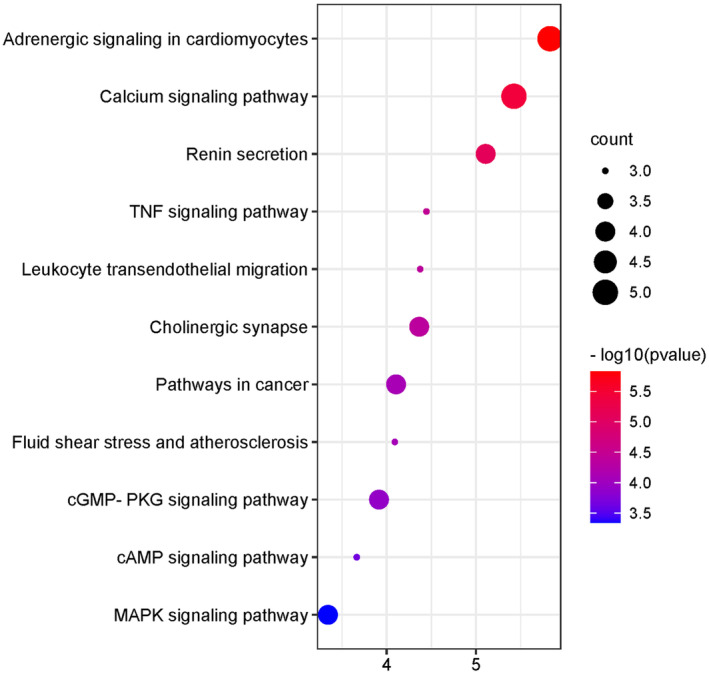
The KEGG pathway analysis of 17 core targets.
3.5. Molecular Docking
The results of compound‐target network analysis revealed that 16 compounds of SXSM were closely related to the core targets BMP4, KCNH2, KCNQ1, and KCNMA1. To simulate the binding modes of these compound molecules and target proteins, molecular docking was performed (Table 4). The binding energy is shown in Table 5, and the parameter of molecular docking is shown in Table S2. Generally, binding energy < −5.0 kcal/mol is indicative of a good binding activity (Wang et al. 2023a, 2023b). In order to further explain the binding of small molecules to protein receptors, these molecules were sorted according to the minimum binding energy. Next, the small molecules with the best binding activity toward BMP4, KCNH2, KCNQ1, and KCNMA1 were selected. Particularly, this included erythrinin A and BMP4, corylidin and KCNH2, danshenol B and KCNQ1, and sugiol and KCNMA1. The interaction between the molecules and the target protein was further subjected to visual analysis (Figures 6, 7, 8, 9).
TABLE 4.
The compounds that interact with BMP4, KCNH2, KCNQ1, and KCNMA1.
| NO. | Name | Structure | Target |
|---|---|---|---|
| BGZ15 | Bavachin |
|
BMP4 |
| BGZ28 | Erythrinin A |
|
BMP4 |
| XX1 | 4,9‐Dimethoxy‐1‐vinyl‐beta‐carboline |
|
BMP4 KCNH2 |
| HS | (E,E,E,E)‐Squalene |
|
KCNH2 |
| YYH11 | 8‐Prenyl‐flavone |
|
KCNH2 |
| BGZ5 | Corylidin |
|
KCNH2 |
| BGZ10 | 12,13‐Epoxybakuchiol |
|
KCNH2 |
| GQZ4 | Atropine |
|
KCNH2 |
| GQZ5 | 24‐Methylenelophenol |
|
KCNH2 |
| MH2 | Squalene |
|
KCNH2 |
|
MH7 DS13 |
Eriodictyol |
|
KCNH2 |
| DS21 | (1R)‐5‐hydroxy‐1,6,6‐trimethyl‐2,7,8,9‐tetrahydro‐1H‐naphtho[1,2‐g][1]benzofuran‐10,11‐dione |
|
KCNH2 |
| DS14 | Danshenol A |
|
KCNQ1 |
| DS15 | Danshenol B |
|
KCNQ1 |
| DS6 | Arucadiol |
|
KCNMA1 |
| DS28 | Sugiol |
|
KCNMA1 |
TABLE 5.
Analysis of the binding energy of 16 molecules with 4 target proteins.
| Active ingredient | Target protein | The lowest binding energy (kcal/mol) |
|---|---|---|
| Bavachin | BMP4 | −7.3 |
| Erythrinin A | BMP4 | −8 |
| 4,9‐Dimethoxy‐1‐vinyl‐beta‐carboline | BMP4 | −6 |
| 4,9‐Dimethoxy‐1‐vinyl‐beta‐carboline | KCNH2 | −5.2 |
| (E,E,E,E)‐Squalene | KCNH2 | −3.6 |
| 8‐Prenyl‐flavone | KCNH2 | −6 |
| Corylidin | KCNH2 | −6.6 |
| 12,13‐Epoxybakuchiol | KCNH2 | −4.8 |
| Atropine | KCNH2 | −5.3 |
| 24‐Methylenelophenol | KCNH2 | −5.7 |
| Squalene | KCNH2 | −3.6 |
| Eriodictyol | KCNH2 | −5.7 |
| (1R)‐5‐hydroxy‐1,6,6‐trimethyl‐2,7,8,9‐tetrahydro‐1H‐naphtho[1,2‐g][1]benzofuran‐10,11‐dione | KCNH2 | −6 |
| Danshenol A | KCNQ1 | −6.1 |
| Danshenol B | KCNQ1 | −6.2 |
| Arucadiol | KCNMA1 | −7.7 |
| Sugiol | KCNMA1 | −7.9 |
FIGURE 6.
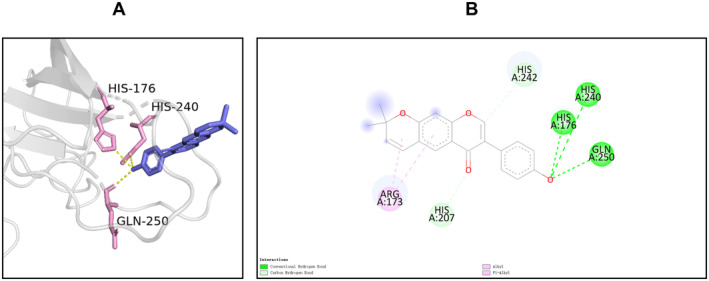
The binding mode of erythrinin A at the binding site of BMP4. (A) Schematic diagram of the 3D mode. (B) Schematic diagram of the 2D mode. BMP4 forms a hydrogen bond with erythrinin A through its His‐240, His‐176, and Gln‐250. Meanwhile, His‐242 and erythrinin A formed a weak hydrogen bond with the Pi bond as a donor; His‐207 and erythrinin A formed C–H interaction, and Arg‐173 and erythrinin A formed alkyl interaction and PI.
FIGURE 7.
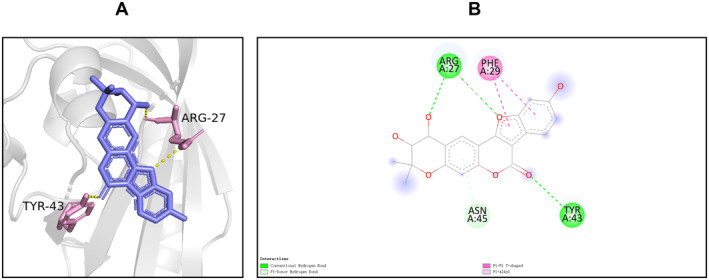
The binding mode of corylidin at the binding site of KCNH2. (A) The schematic diagram of the 3D mode. (B) Schematic diagram of the 2D mode. Through its Arg‐27 and Tyr‐43, KCNH2 formed a hydrogen bond with corylidin, while Asn‐45 and corylidin formed a weak hydrogen bond with the Pi bond as the donor; Phe‐29 and corylidin formed a T‐type Pi–Pi interaction, and Arg‐27 and corylidin formed a Pi‐alkyl interaction.
FIGURE 8.
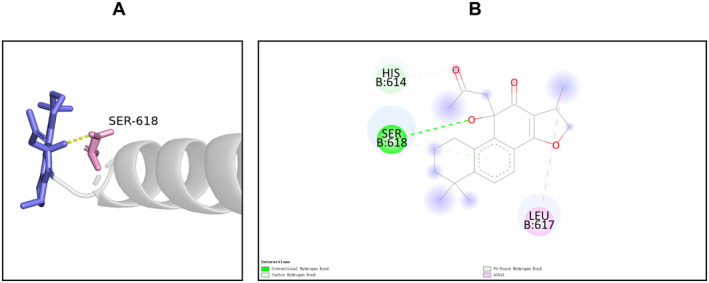
The binding mode of danshenol B at the binding site of KCNQ1. (A) The schematic diagram of the 3D mode. (B) The schematic diagram of the 2D mode. KCNQ1 formed a hydrogen bond with danshenol B through its Ser‐618. Meanwhile, Ser‐618 and danshenol B formed a weak hydrogen bond with Pi bond as the donor; His‐614 and danshenol B formed a C‐H interaction, while Leu‐627 and danshenol B formed an alkyl interaction.
FIGURE 9.
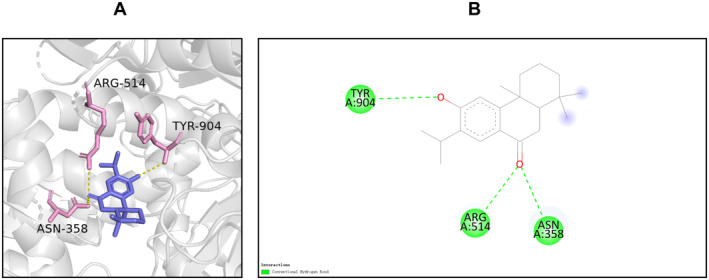
The binding mode of sugiol at the binding site of KCNMA1. (A) The schematic diagram of the 3D mode. (B) The schematic diagram of the 2D mode. KCNMA1 interacts with sugiol through its Tyr‐904, Arg‐514, and Asn‐358 to form hydrogen bonds.
3.6. Effect of SXSM on the Expression of Bmp4, Tbx3, and Hcn4 in HL‐1 Cells
To verify whether SXSM could regulate the Bmp4/Tbx3/Hcn4 pathway, we used HL‐1 cells to simulate the P cells of SAN, which knocked down the expression of Bmp4 with siRNA and treated with SXSM. The mRNA expressions of Bmp4, Tbx3, and Hcn4 in HL‐1 cells in each group were determined by RT‐qPCR (Figure 10). The results revealed that, in comparison with the si‐NC group, the expressions of Bmp4, Tbx3, and Hcn4 were reduced in the si‐Bmp4 group (p < 0.01). When compared with the model group, the expressions of Bmp4, Tbx3, and Hcn4 were increased in the SXSM group (p < 0.01).
FIGURE 10.
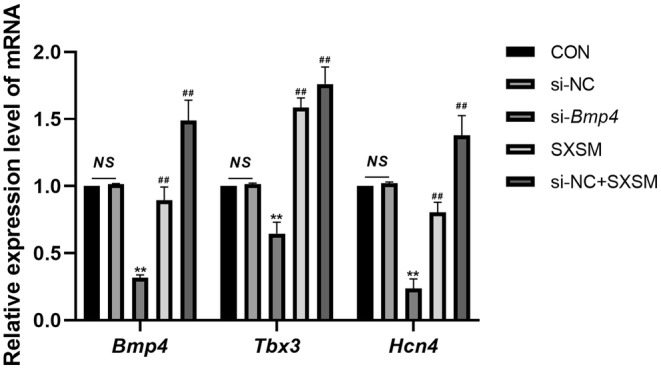
Expressions of mRNA of Bmp4, Tbx3, and Hcn4 detected by RT‐qPCR. Comparison of the RT‐qPCR results. **p < 0.01, when compared with the si‐NC group. ## p < 0.01, when compared with the si‐Bmp4 group. NS, Not statistically significant.
3.7. Effect of SXSM on the Expression of Kcnh2, Kcnq1, and Kcnma1 in HL‐1 Cells
To verify whether SXSM could regulate the expression of Kcnh2, Kcnq1, and Kcnma1, we specifically knocked down their mRNA levels in the HL‐1 cell line with siRNA and added SXSM. The expressions of Kcnh2, Kcnq1, and Kcnma1 in HL‐1 cells in each group were determined by PT‐qPCR (Figure 11). The results showed that, in comparison with the si‐NC group, the expressions of Kcnh2, Kcnq1, and Kcnma1 were reduced in the model group (p < 0.01). When compared with the model group, the expressions of Kcnh2, Kcnq1, and Kcnma1 were increased in the SXSM group (p < 0.05, p < 0.01).
FIGURE 11.
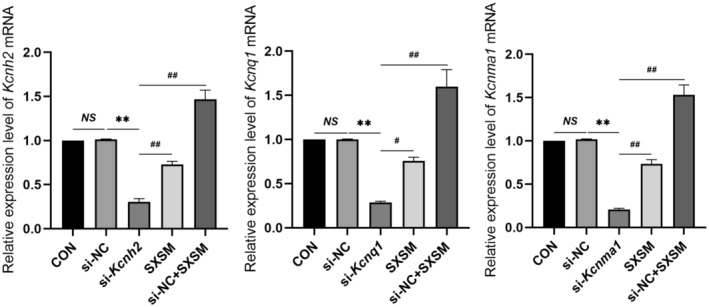
Expression of the mRNA of three genes detected by RT‐qPCR. Comparison of the RT‐qPCR results. **p < 0.01, # p < 0.05, ## p < 0.01. NS, Not statistically significant.
3.8. Effect of SXSM on the Beating Rates and FPD of hiPSC‐AMs
We used autonomously beating hiPSC‐AMs to simulate the P cells of SAN, knocked down the expressions of BMP4 by siRNA, and added SXSM. We then determined the beating rates and FPD of hiPSC‐AMs in each group. The electrophysiological assay results showed that, in comparison with the control group, the beating rates were reduced and the FPD was prolonged in the model group (Figure 12A–C; p < 0.01). After the addition of SXSM, the FPD was shortened and the beating rates were increased (Figure 12A–C; p < 0.05, p < 0.01). To verify whether SXSM could increase the beating rates of hiPSC‐AMs by regulating the expression of KCNH2, KCNQI, and KCNMA1, we specifically knocked down their mRNA levels in hiPSC‐AMs with siRNA and added SXSM. The results suggested that, in comparison with the control group, the beating rates were reduced and the FPD was prolonged in the model group (Figure 13A–C; p < 0.01). When compared with the model group, the FPD was shortened and the beating rates increased in the SXSM group (Figure 13A–C; p < 0.05, p < 0.01).
FIGURE 12.
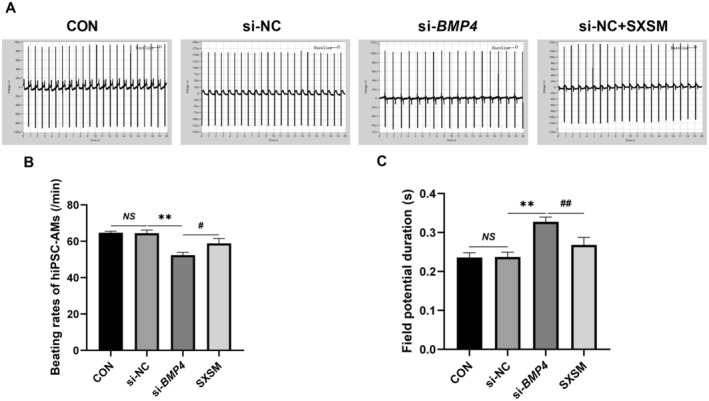
FPD and the beating rates in hiPSC‐AMs. (A) Representative field potential traces in hiPSC‐AMs. (B, C) Comparison of the beating rates and FPD in each group. **p < 0.01, # p < 0.05, ## p < 0.01. NS, Not statistically significant.
FIGURE 13.
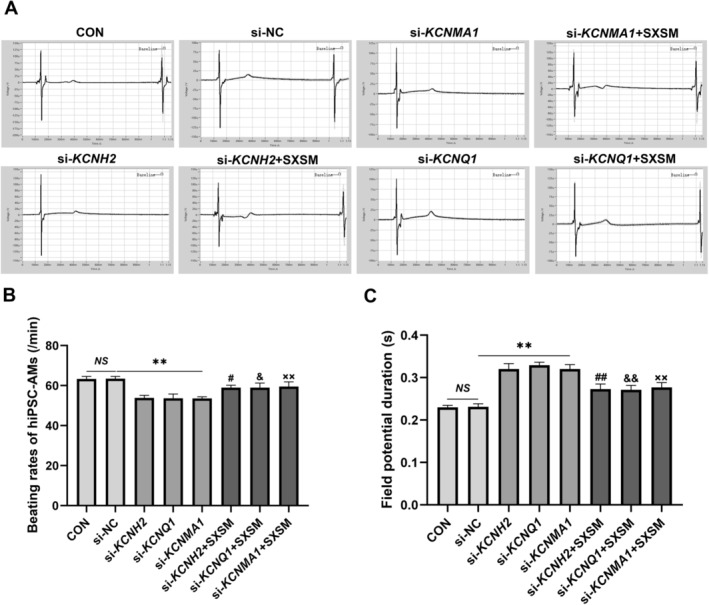
FPD and the beating rates in hiPSC‐AMs. (A) Representative field potential traces in hiPSC‐AMs. (B, C) Comparison of the beating rates and FPD in each group. **p < 0.01. # p < 0.05, ## p < 0.01, when compared with the si‐KCNH2 group. & p < 0.05, && p < 0.01, when compared with the si‐KCNQ1 group. ×× p < 0.01, when compared with the si‐KCNMA1 group. NS, Not statistically significant.
4. Discussion
The pathophysiology of SSS is varied and usually involves complex electrophysiological and structural remodeling. The sinoatrial node is comprised of a complex matrix of pacemaker cells, transitional cells, endothelial cells, fibroblasts, and extracellular scaffolding and is characterized by a unique ion channel and connexin expression profile that results in chronotropic automaticity. The underlying mechanism of SSS mostly involved the dysfunction of P cells in SAN (Liu and Hou 2023). The formation and maintenance of cardiac pacemaking functions are usually regulated by a series of specific genes. These genes regulate SAN‐specific ion channels, which play a fundamental role in regulating the action potential of P cells, thereby affecting the heart rate. The present study focused on SXSM, a Chinese patent medicine, which is known to exhibit remarkable curative effects and has a good safety profile for use in the treatment of SSS. This study discusses the underlying mechanism responsible for its therapeutic effect on SSS by using a combination of network pharmacology and in vitro experiments.
In this study, we screened 55 potential targets of SXSM for SSS through network pharmacology and analyzed the core mechanisms by GO and KEGG enrichment analyses. We observed that the targets of SXSM in the treatment of SSS were mostly concentrated in the function of the regulation of the membrane potential, while the second was the regulation of ion transmembrane transport, suggesting that these are the main mechanisms of SXSM involved in SSS treatment. The membrane potential of myocardial cells changes with the opening and closing of specific ion channels, which, in turn, generates the action potential duration (APD). The APD of P cells in SAN is categorized into the depolarization, repolarization, and spontaneous depolarization phases. When the spontaneous depolarization reaches the threshold membrane potential, it triggers a new action potential, such that rhythmic excitation is continuously generated. Therefore, the shorter the APD, the more rhythmic excitations P cells automatically generate, and the faster the heart rate. HCN4, CACNAIC, CACNA1H, CACNA2D1, KCNH2, KCNQ1, and KCNMA1 are the ion channels that play a major role in the regulation of APD (Hennis et al. 2022; Charpentier et al. 2010; Guo et al. 2011; Schmidt and Peyronnet 2018). Heart development is another important molecular mechanism, and BMP4 is the key target during the cardiomyocyte differentiation process (Wang et al. 2023a, 2023b). Therefore, we selected BMP4, KCNH2, KCNQ1, and KCNMA1 as the key targets in this study to explain the main mechanism of SXSM involved in the treatment of SSS.
During the development of the SAN, the genetic program for pacemaker cells is promoted while the genetic program that promotes chamber specification is inhibited; this process is tightly controlled by a series of transcription factors (Van Eif et al. 2018). In recent years, the role of bone morphogenetic protein (BMP) in animal embryo development and cell function has been widely explored (Chen, Zhao, and Mundy 2004). Among these, Bmp4 is known to regulate the formation of P cells in SAN, which is adequate and necessary for the induction of the Hcn4 expression (Hashem et al. 2013). BMP4 treatment promoted efficient differentiation into spontaneously beating embryoid bodies (EBs) in hiPSCs lines; these cells may develop into SAN (Kimura et al. 2019). It has been previously reported that the loss of function of Bmp4 could lead to abnormal heart structure and even embryo death (Uchimura et al. 2009). Past studies have reported that Bmp4 could reprogram cardiac myocytes into pacemaker‐like cells (Haraguchi et al. 2015). Bmp4 was also considered a new strategy for the construction of biological pacemakers (Sun et al. 2015). It has previously been shown that Bmp4 could bind to the promoter of transcription factor Tbx3 and promote its expression. Tbx3 is usually expressed in SAN, atrioventricular node (AVN), and the proximal part of His bundle. During the formation and maintenance of the cardiac pacemaking and conduction systems, it can inhibit the gene expression in myocardial working cells, thereby separating them from the cardiac chamber (Hoogaars et al. 2004; Wu et al. 2019). Further experiments revealed that Tbx3 could reprogram terminally differentiated working cardiomyocytes, thereby facilitating biological pacemaker formation (Wiese et al. 2009; Bakker et al. 2012; Jensen et al. 2013; Zhao et al. 2020). Tbx3 could activate the expression of Hcn4 and other pacemaker cell‐specific genes. Hcn4 is usually considered to be the molecular marker of the cardiac pacemaking system and serves as an important research target for biological pacemaking (Saito et al. 2015; Li et al. 2020). It has been previously shown that the Hcn4 mutation could lead to SSS (Milanesi et al. 2006; Laish‐Farkash et al. 2010). Several clinical studies have reported that the loss of function of Hcn4 could result in bradycardia (Hoogaars et al. 2007; Verkerk and Wilders 2015). Saito et al., for instance, transplanted mouse embryonic stem cell‐derived cardiomyocytes overexpressing Hcn4 into the rat model for bradycardia and reported an increase in the heart rate (Saito et al. 2018). The HCN4 channel regulates the mixed Na+/K+ current, I f, which is referred to as the “pacemaker current,” and plays an important role in regulating neuronal excitability and cardiac autonomic rhythm (DiFrancesco 2010). The HCN4 channel is activated during the spontaneous depolarization phase of APD. When the HCN4 channel opens, the enhancement of the inward current leads to membrane depolarization. When the spontaneous depolarization reaches the potential threshold (−50 mv), the calcium channel on the cell membrane gets activated and a new APD is generated. Therefore, the more the Hcn4 channel is expressed, the more I f current passes through the membrane, and higher is the autorhythmicity of P cells.
In order to verify whether SXSM could improve the SAN function by regulating the Bmp4/Tbx3/Hcn4 pathway, HL‐1 cells were used to simulate P cells in vitro. The results of RT‐qPCR indicated that SXSM could increase the expression of Bmp4, transcription factor Tbx3, and ion channel Hcn4 (p < 0.01). Because hiPSC‐AMs have a spontaneous beating rhythm, we used hiPSC‐AMs to simulate P cells of SAN to observe whether the promotion of SXSM on the expression of the Bmp4/Tbx3/Hcn4 pathway affects the spontaneous beating rate. The knockdown of BMP4 resulted in a reduction of the beating rates of hiPSC‐AMs, while the FPD was prolonged. After the addition of SXSM, the FPD was shortened and the beating rates were increased. We, therefore, speculated that SXSM may increase the expression of the downstream Hcn4 channel by promoting the expression of the Bmp4/Tbx3/Hcn4 pathway, thus increasing the I f current density, shortening the APD, and increasing the heart rate.
The effect of the repolarization phase on the heart rate in the APD of P cells is also extremely important. The potassium channels play an important role in promoting P‐cell membrane repolarization during APD. KCNH2, KCNQ1, and KCNMA1 are the main ion channels involved in this process. The HERG channel encoded by KCNH2 and the KvLQT1 channel encoded by KCNQ1 are the main members of the α‐subunit of voltage‐gated potassium channels. Particularly, KCNH2 is involved in the regulation of rapidly activating delayed rectifier potassium current I Kr (Corponi et al. 2019). In comparison, KCNQ1 regulates the slowly activating delayed rectifier potassium channel current, I Ks. In the repolarization phase of the action potential phase 3, the potassium channels are activated and opened, leading to potassium ion outflow. When the membrane potential reaches about −50 mV, the depolarization is re‐activated to complete an APD. Therefore, the more the potassium channels are expressed, the more potassium ion outflow in the repolarization phase, and the faster the spontaneous beating rates of P cells. Potassium channel blocker E‐4031 can decrease the pacemaking rate of SAN by reducing the maximum repolarization potential, which, in turn, affects the full activation of I f. It has previously been reported that mutations in KCNH2 can cause SSS and long QT syndrome (LQTS) (Yang et al. 2022). There are two types of mutations in KCNQ1: Haploinsufficient KCNQ1 mutations and dominant‐negative mutations. In these two mutations, the I Ks reduction may range from mild (> 50%) to significant (< 93.75%) (Schwartz et al. 2021). Clinical studies have reported that the KCNQ1 mutation can cause sinus bradycardia (Henrion et al. 2012; Ki et al. 2014). Furthermore, several studies have confirmed that KCNQ1 dysfunction can cause SSS and other heart diseases, such as long QT interval syndrome and abnormal internode conduction (Demolombe et al. 2001; Hong et al. 2005; Zhou et al. 2019). KCNMA1 encodes a large conductance voltage potassium channel that is calcium‐sensitive, which regulates the export of potassium ions and contributes to the repolarization of the membrane potential (Basrai et al. 2002). Past clinical studies have reported that the loss of function of this channel can slow down the heart rate, which further contributes to the development of SSS (Imlach et al. 2010; Lai et al. 2014; Pineda et al. 2021).
To verify whether SXSM could increase the heart rate by regulating these potassium ion channels, in the present study, the expression of Kcnh2, Kcnq1, and Kcnma1 was knocked down in HL‐1 cells. Following this, SXSM was added, which increased the expression of Kcnh2, Kcnq1, and Kcnma1 (p < 0.05, p < 0.01). Meanwhile, the expression of KCNH2, KCNQ1, and KCNMA1 was knocked down in hiPSC‐AMs; the results indicated that a decrease in KCNH2, KCNQ1, and KCNMA1 expression could prolong the FPD and lower the beating rates of hiPSC‐AMs, suggesting that decreasing the expression of KCNH2, KCNQ1, and KCNMA1 channels may slow down the heart rate by prolonging APD. After the addition of SXSM, the FPD was shortened and the beating rates were increased. All these results indicated that SXSM may shorten the APD of P cells by increasing the expression of these pacemaking‐related potassium channels and increasing the heart rate (Figure 14).
FIGURE 14.
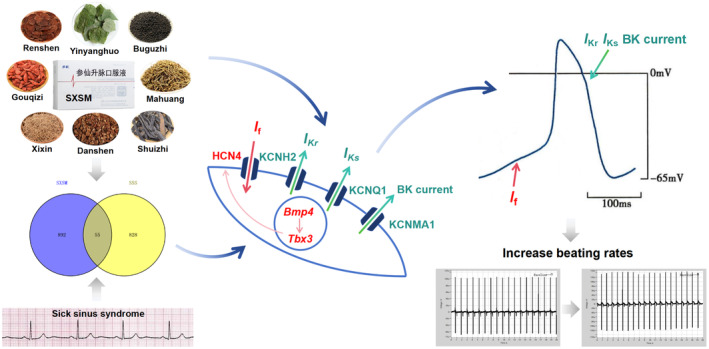
Summary of the main mechanism of SXSM in the treatment of SSS.
Molecular docking results further provided a visual analysis of the interaction between 16 active compounds of SXSM and BMP4, KCNH2, KCNMA1, and KCNQ1. Most of these compounds have been reported in references (Xiang et al. 2018; Zong et al. 2024). The present results suggested that most compound molecules exhibited a good binding effect toward these four targets. In the future, we plan to isolate and purify these compounds to explore their therapeutic mechanisms for SSS toward increasing the efficiency and reducing the toxicity of SXSM.
5. Conclusion
SSS is a set of diseases with abnormal cardiac pacing; with the progression of this disease, it may lead to end‐organ hypoperfusion. In patients who are not candidates for pacemaker therapy, SXSM is the first Chinese patent medicine specially designed for SSS that has curative effects and low side effects. Our study analyzed the potential mechanisms of SXSM involved in the treatment of SSS based on network pharmacology prediction. We found that SXSM may increase the heart rate by regulating P cell gene programming and the transmembrane transport of potassium ions. Specifically, SXSM could regulate the Bmp4/Tbx3/Hcn4 pathway and increase the expression of the Hcn4 channel. Meanwhile, it increased the expression of three potassium channels, namely Kcnh2, Kcnq1, and Kcnma1. These regulations may shorten the APD of P cells and increase the heart rate.
Author Contributions
Ping Hou: conceptualization (supporting), data curation (supporting), formal analysis (supporting), funding acquisition (lead), methodology (supporting), project administration (supporting), supervision (supporting), validation (supporting), writing – original draft (supporting), writing – review and editing (supporting). Heng Zhang: data curation (supporting), formal analysis (supporting), methodology (supporting), software (supporting). Dong‐Yu Min: formal analysis (supporting), methodology (supporting), software (supporting). Jie Wu: data curation (supporting), formal analysis (supporting). Chen Chen: data curation (supporting), formal analysis (supporting). Jie Wang: data curation (supporting), formal analysis (supporting). Yong‐Ping Lu: data curation (supporting), software (supporting). Ying‐Jia Yao: data curation (supporting), software (supporting). Ling‐Kang Li: data curation (supporting). Yue Liu: conceptualization (lead), data curation (lead), formal analysis (lead), funding acquisition (supporting), methodology (lead), project administration (lead), software (lead), supervision (lead), writing – original draft (lead), writing – review and editing (lead).
Conflicts of Interest
The authors declare no conflicts of interest.
Supporting information
Table S1. Information of target proteins for molecular docking.
Table S2. The parameter and result of molecular docking.
Funding: This work was supported by the National Natural Science Foundation of China (81874403).
Data Availability Statement
The data that support the findings of this study are available on request from the corresponding author.
References
- Bakker, M. L. , Boink G. J., Boukens B. J., et al. 2012. “T‐Box Transcription Factor Tbx3 Reprogrammes Mature Cardiac Myocytes Into Pacemaker‐Like Cells.” Cardiovascular Research 94: 439–449. [DOI] [PubMed] [Google Scholar]
- Basrai, D. , Kraft R., Bollensdorff C., Liebmann L., Benndorf K., and Patt S.. 2002. “BK Channel Blockers Inhibit Potassium‐Induced Proliferation of Human Astrocytoma Cells.” Neuroreport 13: 403–407. [DOI] [PubMed] [Google Scholar]
- Charpentier, F. , Mérot J., Loussouarn G., and Baró I.. 2010. “Delayed Rectifier K(+) Currents and Cardiac Repolarization.” Journal of Molecular and Cellular Cardiology 48: 37–44. [DOI] [PubMed] [Google Scholar]
- Chen, D. , Zhao M., and Mundy G. R.. 2004. “Bone Morphogenetic Proteins.” Growth Factors 22: 233–241. [DOI] [PubMed] [Google Scholar]
- Chen, M. , and Wu Q.. 2023. “Roles and Mechanisms of Natural Drugs on Sinus Node Dysfunction.” Biomedicine & Pharmacotherapy 164: 114777. [DOI] [PubMed] [Google Scholar]
- Corponi, F. , Fabbri C., Boriani G., et al. 2019. “Corrected Qt Interval Prolongation in Psychopharmacological Treatment and Its Modulation by Genetic Variation.” Neuropsychobiology 77: 67–72. [DOI] [PubMed] [Google Scholar]
- De Ponti, R. , Marazzato J., Bagliani G., Leonelli F. M., and Padeletti L.. 2018. “Sick Sinus Syndrome.” Cardiac Electrophysiology Clinics 10: 183–195. [DOI] [PubMed] [Google Scholar]
- Demolombe, S. , Lande G., Charpentier F., et al. 2001. “Transgenic Mice Overexpressing Human KvLQT1 Dominant‐Negative Isoform. Part 1: Phenotypic Characterisation.” Cardiovascular Research 50: 314–327. [DOI] [PubMed] [Google Scholar]
- DiFrancesco, D. 2010. “The Role of the Funny Current in Pacemaker Activity.” Circulation Research 106: 434–446. [DOI] [PubMed] [Google Scholar]
- Gfeller, D. , Grosdidier A., Wirth M., Daina A., Michielin O., and Zoete V.. 2014. “Swisstargetprediction: A Web Server for Target Prediction of Bioactive Small Molecules.” Nucleic Acids Research 42: W32–W38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene, S. C. , Halmer T., Carey J. M., Rissmiller B. J., and Musick M. A.. 2018. “Theophylline Toxicity: An Old Poisoning for a New Generation of Physicians.” Turkish Journal of Emergency Medicine 18: 37–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, J. , Wang T., Yang T., et al. 2011. “Interaction Between the Cardiac Rapidly (I Kr) and Slowly (I Ks) Activating Delayed Rectifier Potassium Channels Revealed by Low K+‐Induced hERG Endocytic Degradation.” Journal of Biological Chemistry 286: 34664–34674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraguchi, Y. , Matsuura K., Shimizu T., Yamato M., and Okano T.. 2015. “Simple Suspension Culture System of Human Ips Cells Maintaining Their Pluripotency for Cardiac Cell Sheet Engineering.” Journal of Tissue Engineering and Regenerative Medicine 9: 1363–1375. [DOI] [PubMed] [Google Scholar]
- Hashem, S. I. , Lam M. L., Mihardja S. S., White S. M., Lee R. J., and Claycomb W. C.. 2013. “SHOX2 Regulates the Pacemaker Gene Program in Embryoid Bodies.” Stem Cells and Development 22: 2915–2926. [DOI] [PubMed] [Google Scholar]
- Hennis, K. , Biel M., Fenske S., and Wahl‐Schott C.. 2022. “Paradigm Shift: New Concepts for HCN4 Function in Cardiac Pacemaking.” Pflügers Archiv 474: 649–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrion, U. , Zumhagen S., Steinke K., et al. 2012. “Overlapping Cardiac Phenotype Associated With a Familial Mutation in the Voltage Sensor of the KCNQ1 Channel.” Cellular Physiology and Biochemistry 29: 809–818. [DOI] [PubMed] [Google Scholar]
- Hong, K. , Piper D. R., Diaz‐Valdecantos A., et al. 2005. “De Novo KCNQ1 Mutation Responsible for Atrial Fibrillation and Short QT Syndrome In Utero.” Cardiovascular Research 68: 433–440. [DOI] [PubMed] [Google Scholar]
- Hoogaars, W. M. , Engel A., Brons J. F., et al. 2007. “Tbx3 Controls the Sinoatrial Node Gene Program and Imposes Pacemaker Function on the Atria.” Genes & Development 21: 1098–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogaars, W. M. , Tessari A., Moorman A. F., et al. 2004. “The Transcriptional Repressor Tbx3 Delineates the Developing Central Conduction System of the Heart.” Cardiovascular Research 62: 489–499. [DOI] [PubMed] [Google Scholar]
- Hu, J. , Chen S., and Hua X.. 2012. “Clinical Research of Shenxian‐Shengmai Oral Liquid on Sinus Bradycardiac Patients With Sick Sinus Syndrome (In Chinese).” Chinese Traditional Patent Medicine 35: 79. [Google Scholar]
- Hu, Y. , Mao J., Hou Y., Du Y., Wang Y., and Zhu M.. 2016. “Systematic Evaluation and Meta‐Analysis of the Efficacy and Safety of Shenxian‐Shengmai Oral Liquid in the Treatment of Bradyarrhythmia (In Chinese).” China Journal of Traditional Chinese Medicine and Pharmacy 31: 1059–1063. [Google Scholar]
- Huang Da, W. , Sherman B. T., and Lempicki R. A.. 2009a. “Bioinformatics Enrichment Tools: Paths Toward the Comprehensive Functional Analysis of Large Gene Lists.” Nucleic Acids Research 37: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Da, W. , Sherman B. T., and Lempicki R. A.. 2009b. “Systematic and Integrative Analysis of Large Gene Lists Using DAVID Bioinformatics Resources.” Nature Protocols 4: 44–57. [DOI] [PubMed] [Google Scholar]
- Imlach, W. L. , Finch S. C., Miller J. H., Meredith A. L., and Dalziel J. E.. 2010. “A Role for BK Channels in Heart Rate Regulation in Rodents.” PLoS One 5: e8698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen, B. , Wang T., Christoffels V. M., and Moorman A. F.. 2013. “Evolution and Development of the Building Plan of the Vertebrate Heart.” Biochimica et Biophysica Acta 1833: 783–794. [DOI] [PubMed] [Google Scholar]
- Ki, C. S. , Jung C. L., Kim H. J., et al. 2014. “A Kcnq1 Mutation Causes Age‐Dependant Bradycardia and Persistent Atrial Fibrillation.” Pflügers Archiv 466: 529–540. [DOI] [PubMed] [Google Scholar]
- Kimura, M. , Furukawa H., Shoji M., and Shinozawa T.. 2019. “Increased Mesodermal and Mesendodermal Populations by BMP4 Treatment Facilitates Human iPSC Line Differentiation Into a Cardiac Lineage.” Journal of Stem Cells and Regenerative Medicine 15: 45–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusumoto, F. M. , Schoenfeld M. H., Barrett C., et al. 2019. “2018 ACC/AHA/HRS Guideline on the Evaluation and Management of Patients With Bradycardia and Cardiac Conduction Delay: Executive Summary: A Report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines, and the Heart Rhythm Society.” Journal of the American College of Cardiology 74: 932–987. [DOI] [PubMed] [Google Scholar]
- Lai, M. H. , Wu Y., Gao Z., Anderson M. E., Dalziel J. E., and Meredith A. L.. 2014. “BK Channels Regulate Sinoatrial Node Firing Rate and Cardiac Pacing In Vivo.” American Journal of Physiology. Heart and Circulatory Physiology 307: H1327–H1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laish‐Farkash, A. , Glikson M., Brass D., et al. 2010. “A Novel Mutation in the hcn4 Gene Causes Symptomatic Sinus Bradycardia in Moroccan Jews.” Journal of Cardiovascular Electrophysiology 21: 1365–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J. Z. , Ling J., Diehl N. N., et al. 2018. “Mortality and Cerebrovascular Events After Heart Rhythm Disorder Management Procedures.” Circulation 137: 24–33. [DOI] [PubMed] [Google Scholar]
- Li, X. , Zhang J. Q., and Shuai J. W.. 2014. “Isoprenaline: A Potential Contributor in Sick Sinus Syndrome‐Insights From a Mathematical Model of the Rabbit Sinoatrial Node.” Scientific World Journal 2014: 540496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. , Wang K., Li Q., and Zhang H.. 2020. “Biological Pacemaker: From Biological Experiments to Computational Simulation.” Journal of Zhejiang University Science B 21: 524–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J. , Wen L., Yao S., Zheng P., Zhao S., and Yang J.. 2020. “Adverse Clinical Events Caused by Pacemaker Battery Depletion: Two Case Reports.” BMC Cardiovascular Disorders 20: 344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, S. , Tian G., Chen J., et al. 2018. “Traditional Chinese Medicine for Bradyarrhythmia: Evidence and Potential Mechanisms.” Frontiers in Pharmacology 9: 324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, W. , Dai J., Chen X., Du N., and Hu J.. 2023. “Integrated Network Pharmacology and In‐Silico Approaches to Decipher the Pharmacological Mechanism of Dioscorea Septemloba Thunb in Treating Gout and Its Complications.” Combinatorial Chemistry & High Throughput Screening. [DOI] [PubMed] [Google Scholar]
- Liu, Y. , and Hou P.. 2023. “Research Progress on the Mechanism of Bradyarrhythmia and the Treatment of Traditional Chinese Medicine (In Chinese).” Chinese Archieves of Traditional Chinese Medicine 41: 157–163. [Google Scholar]
- Makita, S. , Onoda T., Ohsawa M., et al. 2014. “Bradycardia Is Associated With Future Cardiovascular Diseases and Death in Men From the General Population.” Atherosclerosis 236: 116–120. [DOI] [PubMed] [Google Scholar]
- Mandel, Y. , Weissman A., Schick R., et al. 2012. “Human Embryonic and Induced Pluripotent Stem Cell‐Derived Cardiomyocytes Exhibit Beat Rate Variability and Power‐Law Behavior.” Circulation 125: 883–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milanesi, R. , Baruscotti M., Gnecchi‐Ruscone T., and DiFrancesco D.. 2006. “Familial Sinus Bradycardia Associated With a Mutation in the Cardiac Pacemaker Channel.” New England Journal of Medicine 354: 151–157. [DOI] [PubMed] [Google Scholar]
- Montgomery, J. A. , and Ellis C. R.. 2018. “Longevity of Cardiovascular Implantable Electronic Devices.” Cardiac Electrophysiology Clinics 10: 1–9. [DOI] [PubMed] [Google Scholar]
- Pineda, S. , Nikolova‐Krstevski V., Leimena C., et al. 2021. “Conserved Role of the Large Conductance Calcium‐Activated Potassium Channel, K(Ca)1.1, in Sinus Node Function and Arrhythmia Risk.” Circulation: Genomic and Precision Medicine 14: e003144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ru, J. , Li P., Wang J., et al. 2014. “Tcmsp: A Database of Systems Pharmacology for Drug Discovery From Herbal Medicines.” Journal of Cheminformatics 6: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito, Y. , Nakamura K., Yoshida M., et al. 2015. “Enhancement of Spontaneous Activity by hcn4 Overexpression in Mouse Embryonic Stem Cell‐Derived Cardiomyocytes – A Possible Biological Pacemaker.” PLoS One 10: e0138193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito, Y. , Nakamura K., Yoshida M., et al. 2018. “Hcn4‐Overexpressing Mouse Embryonic Stem Cell‐Derived Cardiomyocytes Generate a New Rapid Rhythm in Rats With Bradycardia.” International Heart Journal 59: 601–606. [DOI] [PubMed] [Google Scholar]
- Schmidt, C. , and Peyronnet R.. 2018. “Voltage‐Gated and Stretch‐Activated Potassium Channels in the Human Heart: Pathophysiological and Clinical Significance.” Herzschrittmachertherapie & Elektrophysiologie 29: 36–42. [DOI] [PubMed] [Google Scholar]
- Schwartz, P. J. , Moreno C., Kotta M. C., et al. 2021. “Mutation Location and I Ks Regulation in the Arrhythmic Risk of Long QT Syndrome Type 1: The Importance of the KCNQ1 S6 Region.” European Heart Journal 42: 4743–4755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon, P. , Markiel A., Ozier O., et al. 2003. “Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks.” Genome Research 13: 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strathmore, N. F. , Swerdlow C. D., Tompkins C., and Wazni O.. 2017. “2017 HRS Expert Consensus Statement on Cardiovascular Implantable Electronic Device Lead Management and Extraction.” Heart Rhythm 14: e503–e551. [DOI] [PubMed] [Google Scholar]
- Sun, C. , Yu D., Ye W., et al. 2015. “The Short Stature Homeobox 2 (SHOX2)‐bone Morphogenetic Protein (Bmp) Pathway Regulates Dorsal Mesenchymal Protrusion Development and Its Temporary Function as a Pacemaker During Cardiogenesis.” Journal of Biological Chemistry 290: 2007–2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott, O. , and Olson A. J.. 2010. “Autodock Vina: Improving the Speed and Accuracy of Docking With a New Scoring Function, Efficient Optimization, and Multithreading.” Journal of Computational Chemistry 31: 455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchimura, T. , Komatsu Y., Tanaka M., McCann K. L., and Mishina Y.. 2009. “Bmp2 and Bmp4 Genetically Interact to Support Multiple Aspects of Mouse Development Including Functional Heart Development.” Genesis 47: 374–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Eif, V. W. W. , Devalla H. D., Boink G. J. J., and Christoffels V. M.. 2018. “Transcriptional Regulation of the Cardiac Conduction System.” Nature Reviews. Cardiology 15: 617–630. [DOI] [PubMed] [Google Scholar]
- Verkerk, A. O. , and Wilders R.. 2015. “Pacemaker Activity of the Human Sinoatrial Node: An Update on the Effects of Mutations in HCN4 on the Hyperpolarization‐Activated Current.” International Journal of Molecular Sciences 16: 3071–3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, F. , Yin L., Zhang W., Tang Y., Wang X., and Huang C.. 2023a. “The Method of Sinus Node‐Like Pacemaker Cells From Human Induced Pluripotent Stem Cells by BMP and Wnt Signaling.” Cell Biology and Toxicology 39: 2725–2741. [DOI] [PubMed] [Google Scholar]
- Wang, X. , Zhang X., Li J., et al. 2023b. “Network Pharmacology and LC‐MS Approachs to Explore the Active Compounds and Mechanisms of Yuanjiang Decoction for Treating Bradyarrhythmia.” Computers in Biology and Medicine 152: 106435. [DOI] [PubMed] [Google Scholar]
- Wiese, C. , Grieskamp T., Airik R., et al. 2009. “Formation of the Sinus Node Head and Differentiation of Sinus Node Myocardium Are Independently Regulated by tbx18 and Tbx3.” Circulation Research 104: 388–397. [DOI] [PubMed] [Google Scholar]
- Wu, L. , Du J., Jing X., et al. 2019. “Bone Morphogenetic Protein 4 Promotes the Differentiation of Tbx18‐Positive Epicardial Progenitor Cells to Pacemaker‐Like Cells.” Experimental and Therapeutic Medicine 17: 2648–2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang, W. , Suo T. C., Yu H., et al. 2018. “A New Strategy for Choosing “Q‐Markers” via Network Pharmacology, Application to the Quality Control of a Chinese Medical Preparation.” Journal of Food and Drug Analysis 26: 858–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Z. , Ma Y., Huang J., et al. 2022. “Digenic Heterozygous Mutations of KCNH2 and SCN5A Induced Young and Early‐Onset Long QT Syndrome and Sinoatrial Node Dysfunction.” Annals of Noninvasive Electrocardiology 27: e12889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H. , Hao M., Li L., et al. 2021. “Shenxian‐Shengmai Oral Liquid Improves Sinoatrial Node Dysfunction Through the Pkc/Nox‐2 Signaling Pathway.” Evidence‐based Complementary and Alternative Medicine: 5572140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, H. , Wang F., Tang Y., et al. 2020. “Hcn2 and tbx3 Reprogram Human‐Induced Pluripotent Stem Cells‐Derived Cardiomyocytes Into Pacemaker‐Like Cells.” DNA and Cell Biology 39: 289–298. [DOI] [PubMed] [Google Scholar]
- Zhou, X. , Bueno‐Orovio A., Schilling R. J., et al. 2019. “Investigating the Complex Arrhythmic Phenotype Caused by the Gain‐Of‐Function Mutation KCNQ1‐G229D.” Frontiers in Physiology 10: 259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong, S. Y. , Di Z. B., Liu Q. Q., et al. 2024. “Rapid Analysis of Chemical Constituents in Shenxianshengmai Oral Liquid by UHPLC‐Q Exactive Focus MS/MS (In Chinese).” World Science and Technology‐Modernization of Traditional Chinese Medicine and Materia Medica 26: 218–228. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Information of target proteins for molecular docking.
Table S2. The parameter and result of molecular docking.
Data Availability Statement
The data that support the findings of this study are available on request from the corresponding author.


