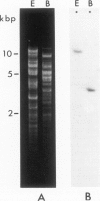
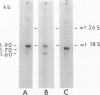
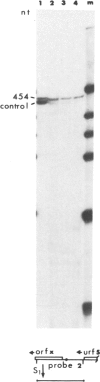
Abstract
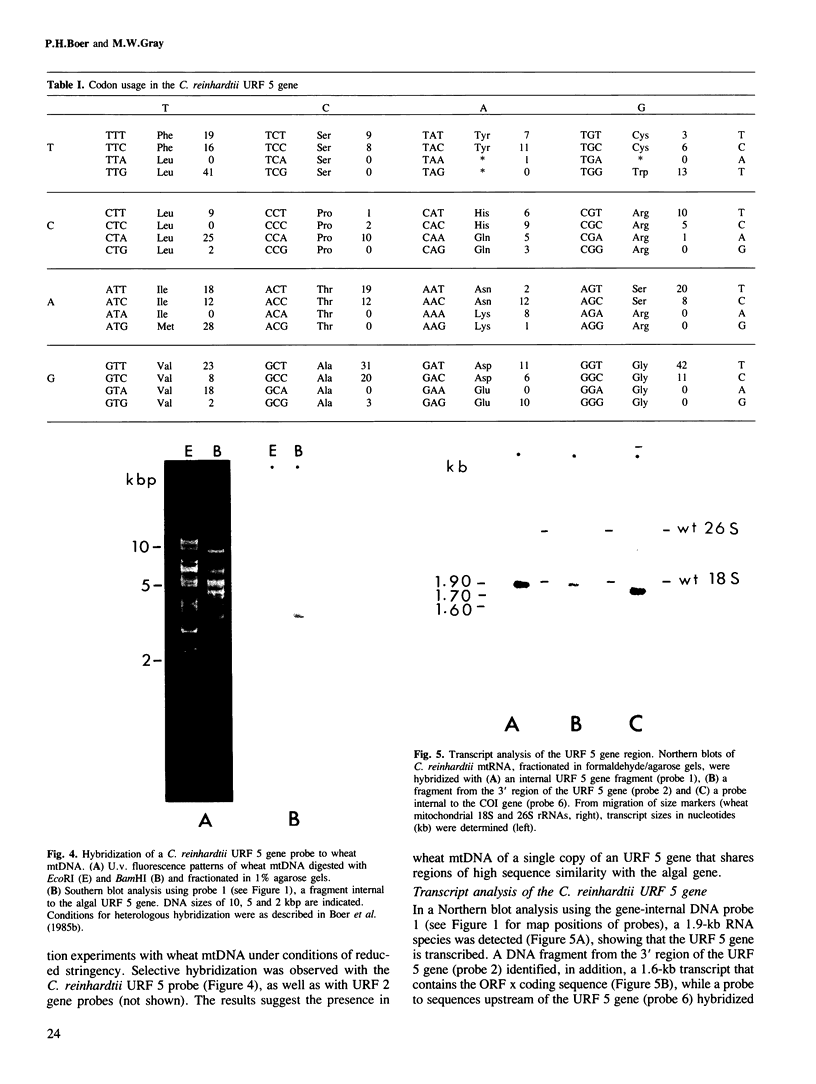
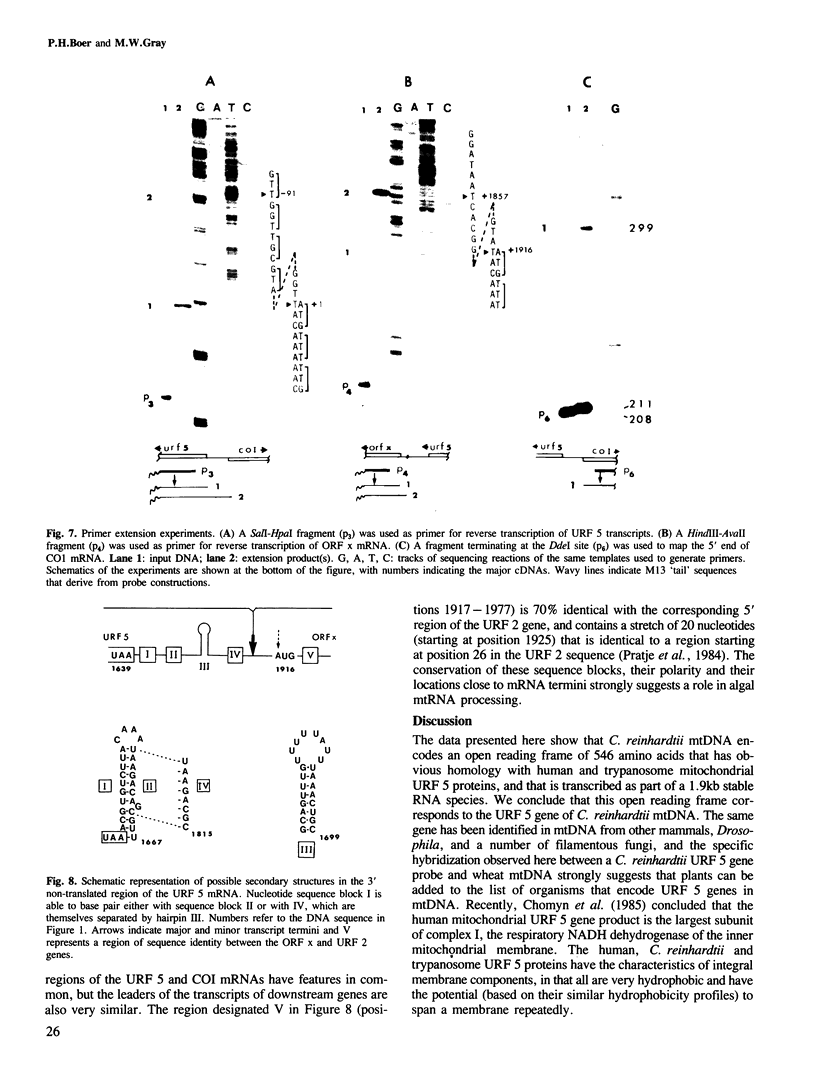
A gene homologous to unassigned reading frame (URF) 5 of the mammalian mitochondrial genome has been identified in the mitochondrial DNA of the unicellular green alga, Chlamydomonas reinhardtii. The algal URF 5 gene is closely flanked by the gene for subunit I of cytochrome oxidase (COI) and by an unidentified gene (ORF x). The URF 5 and ORF x genes are transcribed in the same direction, but opposite to that of the COI gene. Transcript analysis reveals a 1.9-kb mRNA whose major 5' terminus maps to the putative URF 5 initiation codon and whose 3' end abuts the 5' end of the ORF x transcript. Characterization of other C. reinhardtii mitochondrial RNAs suggests a general pattern of abutting transcripts and mature mRNAs having little or no 5' leader sequence. While this is reminiscent of post-transcriptional processing in animal mitochondria, different mechanisms must be employed in the two systems, since tRNA sequences (which appear to function as transcript processing signals in animal mitochondria) do not generally flank protein coding sequences in the C. reinhardtii mitochondrial genome. Nevertheless, characteristic secondary structure motifs do occur within the 3'-terminal regions of C. reinhardtii mitochondrial mRNAs, and their location close to mRNA termini suggests that such motifs may play a role in directing the precise endonucleolytic cleavage of long primary transcripts.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson S., Bankier A. T., Barrell B. G., de Bruijn M. H., Coulson A. R., Drouin J., Eperon I. C., Nierlich D. P., Roe B. A., Sanger F. Sequence and organization of the human mitochondrial genome. Nature. 1981 Apr 9;290(5806):457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- Anderson S., de Bruijn M. H., Coulson A. R., Eperon I. C., Sanger F., Young I. G. Complete sequence of bovine mitochondrial DNA. Conserved features of the mammalian mitochondrial genome. J Mol Biol. 1982 Apr 25;156(4):683–717. doi: 10.1016/0022-2836(82)90137-1. [DOI] [PubMed] [Google Scholar]
- Bibb M. J., Van Etten R. A., Wright C. T., Walberg M. W., Clayton D. A. Sequence and gene organization of mouse mitochondrial DNA. Cell. 1981 Oct;26(2 Pt 2):167–180. doi: 10.1016/0092-8674(81)90300-7. [DOI] [PubMed] [Google Scholar]
- Boer P. H., Bonen L., Lee R. W., Gray M. W. Genes for respiratory chain proteins and ribosomal RNAs are present on a 16-kilobase-pair DNA species from Chlamydomonas reinhardtii mitochondria. Proc Natl Acad Sci U S A. 1985 May;82(10):3340–3344. doi: 10.1073/pnas.82.10.3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boer P. H., McIntosh J. E., Gray M. W., Bonen L. The wheat mitochondrial gene for apocytochrome b: absence of a prokaryotic ribosome binding site. Nucleic Acids Res. 1985 Apr 11;13(7):2281–2292. doi: 10.1093/nar/13.7.2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonen L., Boer P. H., Gray M. W. The wheat cytochrome oxidase subunit II gene has an intron insert and three radical amino acid changes relative to maize. EMBO J. 1984 Nov;3(11):2531–2536. doi: 10.1002/j.1460-2075.1984.tb02168.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitenberger C. A., Browning K. S., Alzner-DeWeerd B., RajBhandary U. L. RNA processing in Neurospora crassa mitochondria: use of transfer RNA sequences as signals. EMBO J. 1985 Jan;4(1):185–195. doi: 10.1002/j.1460-2075.1985.tb02335.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger G., Helmer Citterich M., Nelson M. A., Werner S., Macino G. RNA processing in Neurospora crassa mitochondria: transfer RNAs punctuate a large precursor transcript. EMBO J. 1985 Jan;4(1):197–204. doi: 10.1002/j.1460-2075.1985.tb02336.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capaldi R. A., Vanderkooi G. The low polarity of many membrane proteins. Proc Natl Acad Sci U S A. 1972 Apr;69(4):930–932. doi: 10.1073/pnas.69.4.930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomyn A., Mariottini P., Cleeter M. W., Ragan C. I., Matsuno-Yagi A., Hatefi Y., Doolittle R. F., Attardi G. Six unidentified reading frames of human mitochondrial DNA encode components of the respiratory-chain NADH dehydrogenase. Nature. 1985 Apr 18;314(6012):592–597. doi: 10.1038/314592a0. [DOI] [PubMed] [Google Scholar]
- Chomyn A., Mariottini P., Gonzalez-Cadavid N., Attardi G., Strong D. D., Trovato D., Riley M., Doolittle R. F. Identification of the polypeptides encoded in the ATPase 6 gene and in the unassigned reading frames 1 and 3 of human mtDNA. Proc Natl Acad Sci U S A. 1983 Sep;80(18):5535–5539. doi: 10.1073/pnas.80.18.5535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark-Walker G. D., McArthur C. R., Sriprakash K. S. Location of transcriptional control signals and transfer RNA sequences in Torulopsis glabrata mitochondrial DNA. EMBO J. 1985 Feb;4(2):465–473. doi: 10.1002/j.1460-2075.1985.tb03652.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clary D. O., Wahleithner J. A., Wolstenholme D. R. Sequence and arrangement of the genes for cytochrome b, URF1, URF4L, URF4, URF5, URF6 and five tRNAs in Drosophila mitochondrial DNA. Nucleic Acids Res. 1984 May 11;12(9):3747–3762. doi: 10.1093/nar/12.9.3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clary D. O., Wolstenholme D. R. Genes for cytochrome c oxidase subunit I, URF2, and three tRNAs in Drosophila mitochondrial DNA. Nucleic Acids Res. 1983 Oct 11;11(19):6859–6872. doi: 10.1093/nar/11.19.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton D. A. Transcription of the mammalian mitochondrial genome. Annu Rev Biochem. 1984;53:573–594. doi: 10.1146/annurev.bi.53.070184.003041. [DOI] [PubMed] [Google Scholar]
- Dawson A. J., Jones V. P., Leaver C. J. The apocytochrome b gene in maize mitochondria does not contain introns and is preceded by a potential ribosome binding site. EMBO J. 1984 Sep;3(9):2107–2113. doi: 10.1002/j.1460-2075.1984.tb02098.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewey R. E., Schuster A. M., Levings C. S., Timothy D. H. Nucleotide sequence of F(0)-ATPase proteolipid (subunit 9) gene of maize mitochondria. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1015–1019. doi: 10.1073/pnas.82.4.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox T. D., Leaver C. J. The Zea mays mitochondrial gene coding cytochrome oxidase subunit II has an intervening sequence and does not contain TGA codons. Cell. 1981 Nov;26(3 Pt 1):315–323. doi: 10.1016/0092-8674(81)90200-2. [DOI] [PubMed] [Google Scholar]
- Grant D., Chiang K. S. Physical mapping and characterization of Chlamydomonas mitochondrial DNA molecules: their unique ends, sequence homogeneity, and conservation. Plasmid. 1980 Jul;4(1):82–96. doi: 10.1016/0147-619x(80)90085-2. [DOI] [PubMed] [Google Scholar]
- Gray M. W. Mitochondrial genome diversity and the evolution of mitochondrial DNA. Can J Biochem. 1982 Mar;60(3):157–171. doi: 10.1139/o82-022. [DOI] [PubMed] [Google Scholar]
- Gray M. W., Sankoff D., Cedergren R. J. On the evolutionary descent of organisms and organelles: a global phylogeny based on a highly conserved structural core in small subunit ribosomal RNA. Nucleic Acids Res. 1984 Jul 25;12(14):5837–5852. doi: 10.1093/nar/12.14.5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensgens L. A., Brakenhoff J., De Vries B. F., Sloof P., Tromp M. C., Van Boom J. H., Benne R. The sequence of the gene for cytochrome c oxidase subunit I, a frameshift containing gene for cytochrome c oxidase subunit II and seven unassigned reading frames in Trypanosoma brucei mitochondrial maxi-circle DNA. Nucleic Acids Res. 1984 Oct 11;12(19):7327–7344. doi: 10.1093/nar/12.19.7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiesel R., Brennicke A. Cytochrome oxidase subunit II gene in mitochondria of Oenothera has no intron. EMBO J. 1983;2(12):2173–2178. doi: 10.1002/j.1460-2075.1983.tb01719.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaac P. G., Jones V. P., Leaver C. J. The maize cytochrome c oxidase subunit I gene: sequence, expression and rearrangement in cytoplasmic male sterile plants. EMBO J. 1985 Jul;4(7):1617–1623. doi: 10.1002/j.1460-2075.1985.tb03828.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ise W., Haiker H., Weiss H. Mitochondrial translation of subunits of the rotenone-sensitive NADH:ubiquinone reductase in Neurospora crassa. EMBO J. 1985 Aug;4(8):2075–2080. doi: 10.1002/j.1460-2075.1985.tb03894.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao T., Moon E., Wu R. Cytochrome oxidase subunit II gene of rice has an insertion sequence within the intron. Nucleic Acids Res. 1984 Oct 11;12(19):7305–7315. doi: 10.1093/nar/12.19.7305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumazaki T., Hori H., Osawa S. Phylogeny of protozoa deduced from 5S rRNA sequences. J Mol Evol. 1983;19(6):411–419. doi: 10.1007/BF02102316. [DOI] [PubMed] [Google Scholar]
- Mariottini P., Chomyn A., Attardi G., Trovato D., Strong D. D., Doolittle R. F. Antibodies against synthetic peptides reveal that the unidentified reading frame A6L, overlapping the ATPase 6 gene, is expressed in human mitochondria. Cell. 1983 Apr;32(4):1269–1277. doi: 10.1016/0092-8674(83)90308-2. [DOI] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Michael N. L., Rothbard J. B., Shiurba R. A., Linke H. K., Schoolnik G. K., Clayton D. A. All eight unassigned reading frames of mouse mitochondrial DNA are expressed. EMBO J. 1984 Dec 20;3(13):3165–3175. doi: 10.1002/j.1460-2075.1984.tb02275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon E., Kao T. H., Wu R. Pea cytochrome oxidase subunit II gene has no intron and generates two mRNA transcripts with different 5'-termini. Nucleic Acids Res. 1985 May 10;13(9):3195–3212. doi: 10.1093/nar/13.9.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojala D., Montoya J., Attardi G. tRNA punctuation model of RNA processing in human mitochondria. Nature. 1981 Apr 9;290(5806):470–474. doi: 10.1038/290470a0. [DOI] [PubMed] [Google Scholar]
- Osinga K. A., De Vries E., Van der Horst G., Tabak H. F. Processing of yeast mitochondrial messenger RNAs at a conserved dodecamer sequence. EMBO J. 1984 Apr;3(4):829–834. doi: 10.1002/j.1460-2075.1984.tb01892.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queen C., Korn L. J. A comprehensive sequence analysis program for the IBM personal computer. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):581–599. doi: 10.1093/nar/12.1part2.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan R., Grant D., Chiang K. S., Swift H. Isolation and characterization of mitochondrial DNA from Chlamydomonas reinhardtii. Proc Natl Acad Sci U S A. 1978 Jul;75(7):3268–3272. doi: 10.1073/pnas.75.7.3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sederoff R. R. Structural variation in mitochondrial DNA. Adv Genet. 1984;22:1–108. doi: 10.1016/s0065-2660(08)60038-3. [DOI] [PubMed] [Google Scholar]
- Tabak H. F., Grivell L. A., Borst P. Transcription of mitochondrial DNA. CRC Crit Rev Biochem. 1983;14(4):297–317. doi: 10.3109/10409238309102797. [DOI] [PubMed] [Google Scholar]
- Van Ommen G. J., Groot G. S., Grivell L. A. Transcription maps of mtDNAs of two strains of saccharomyces: transcription of strain-specific insertions; Complex RNA maturation and splicing. Cell. 1979 Oct;18(2):511–523. doi: 10.1016/0092-8674(79)90068-0. [DOI] [PubMed] [Google Scholar]
- Weaver R. F., Weissmann C. Mapping of RNA by a modification of the Berk-Sharp procedure: the 5' termini of 15 S beta-globin mRNA precursor and mature 10 s beta-globin mRNA have identical map coordinates. Nucleic Acids Res. 1979 Nov 10;7(5):1175–1193. doi: 10.1093/nar/7.5.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngblom J., Schloss J. A., Silflow C. D. The two beta-tubulin genes of Chlamydomonas reinhardtii code for identical proteins. Mol Cell Biol. 1984 Dec;4(12):2686–2696. doi: 10.1128/mcb.4.12.2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruijn M. H. Drosophila melanogaster mitochondrial DNA, a novel organization and genetic code. Nature. 1983 Jul 21;304(5923):234–241. doi: 10.1038/304234a0. [DOI] [PubMed] [Google Scholar]
- de la Cruz V. F., Neckelmann N., Simpson L. Sequences of six genes and several open reading frames in the kinetoplast maxicircle DNA of Leishmania tarentolae. J Biol Chem. 1984 Dec 25;259(24):15136–15147. [PubMed] [Google Scholar]